Abstract
Circular RNAs (circRNAs) are a subclass of non-coding RNAs that lack free 3′ and 5′ ends and, thus, exist as continuous loop RNAs. Such circular transcripts have been identified for thousands of genes, are regulated in developmental stages and pathophysiological conditions, and are often expressed in a tissue- or cell-type-specific manner. For a long time, circular transcripts were considered as aberrant splicing by-products. However, high-throughput transcriptome sequencing and focused molecular characterization of individual circRNAs uncovered their ubiquity. Evidence emerges suggesting circRNAs are functional molecules. In this review, we illustrate the current knowledge of circRNA formation and circRNA detection methods. We summarize different molecular mechanisms of action and highlight circRNAs with specific roles in cardiovascular disease. Finally, we describe a number of tools for circRNA manipulation, which may be exploited for circRNA-based therapeutic interventions in the future.
Keywords: circular RNA, non-coding RNA, heart, cardiovascular disease, biomarker, therapeutic targets, RNA editing, backsplicing
Main Text
In the past several years, new sequencing technologies have demonstrated that the major part of the human genome is transcribed into RNAs. However, only 1%–2% of the genome encodes for transcripts that are further translated into proteins.1 Hence, RNA transcripts without any protein-coding potential, so called non-coding RNAs (ncRNAs), display the majority of RNA molecules, but they were considered as transcriptional junk for a long time. Within the last few years, broad transcriptome sequencing led to the discovery of a new class of non-coding RNAs: circular RNAs (circRNAs). These covalently closed single-stranded RNAs exist for thousands of genes, and they are produced by a process called backsplicing of linear precursor RNAs.2, 3, 4 The term circRNA was already introduced by Sanger et al.5 in 1976, when they characterized infectious single-stranded covalently closed RNA molecules in plant viroids. Later, circRNAs were found in all kingdoms of life.5, 6, 7 In 1991, Nigro et al.8 detected the first circular transcripts in mammals. They observed RNA transcripts from the DCC tumor suppressor gene with “scrambled exons,” which were joined at the ends. However, these single and untypical RNA molecules were initially considered as aberrant splicing by-products.8, 9, 10, 11
The extensive discovery of circRNAs in mammals, including humans, started in 2012, when advances in high-throughput sequencing and bioinformatics pipelines identified thousands of circular transcripts.2, 3, 4 Circular transcripts gained even more attention when two groups simultaneously published a function of the circRNA CDR1as (according to Memszak et al.3)/ciRS-7 (according to Hansen et al.12). This circRNA acts as a competing RNA molecule by sponging microRNAs (miRNAs). It contains more than 70 conserved binding sites for miR-7 and can, thereby, efficiently suppress miR-7. However, most circRNAs do not contain multiple miRNA-binding sites,13 and, consequently, other molecular mechanisms were elucidated for a number of circRNAs (discussed later in this review).
Today more than 32,000 human exonic circRNAs have been annotated;14 they are abundant, evolutionarily conserved, and can be specific for certain cell types as well as for developmental and disease stages,2, 3, 4, 13 and they are, therefore, believed to constitute functional molecules. Indeed, regulatory involvement of circRNAs was already observed in a variety of biological processes. Nevertheless, an almost complete sequence overlap with the linear mRNA counterparts makes the precise evaluation of circRNA expression and function more challenging in comparison to other forms of non-coding RNAs.
In this review, we discuss circRNA biogenesis and detection, circRNA functions in disease, and molecular tools to manipulate circRNA expression for the development of circRNA-based therapeutics. Although we mainly focus on cardiovascular diseases, general aspects in terms of biogenesis and function are transferable to any other field in biology.
Circle Formation and Detection
Biogenesis
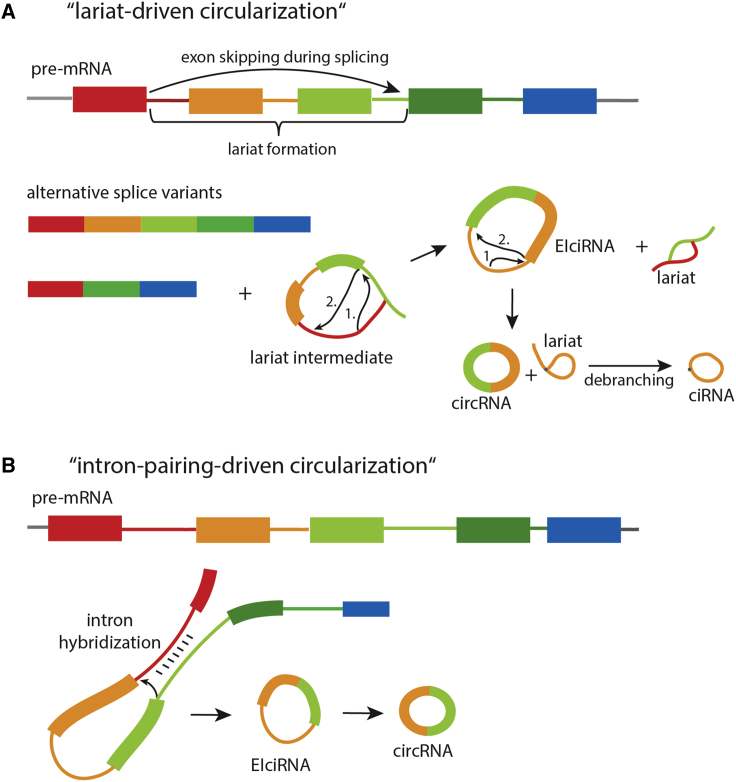
Large-scale RNA profiling has indicated that approximately 75% of the human genome is transcribed into RNA and gives rise to millions of RNA transcripts.15, 16 Retention or skipping of single exons can generate multiple distinct mRNAs from one pre-mRNA, known as alternative splicing. The formation of covalently closed exon circRNAs occurs when a 3′ end of an exon (splice donor site) is joined to a 5′ end of the same (single-exon circRNA) or an upstream exon (multiple-exon circRNA), forming a closed RNA loop. In 2013, Jeck et al.4 proposed two models of circRNA formation: “lariat-driven circularization” and “intron-pairing-driven circulation.” In lariat-driven circulation (Figure 1A), circRNAs are formed from lariat intermediates, which are produced during splicing. Further internal splicing reactions or debranching of the lariat can lead to several distinct circular transcripts: circRNAs from exons only (most often referred to as circRNAs), exon-intron circRNAs (EIciRNAs),17 and intronic circRNAs (ciRNAs)18 (Figure 1A).
Figure 1.
circRNA Formation
(A) Lariat-driven circulation, proposed by Jeck et al.,4 gives rise to circRNAs from lariat intermediate structures, which are produced during exon skipping. (B) Intron-pairing-driven circularization is a more direct circRNA formation and involves hybridization of flanking introns, which brings splice sites in close proximity.
Intron-pairing-driven circulation is based on hybridization of flanking introns, which brings both splice sites in close proximity (Figure 1B). In many cases, complementary sequences (e.g., ALU repeat elements) within the flanking regions facilitate intron hybridization and, thus, mediate exon circulation4, 19, 20 (Figure 1B). Besides the presence of ALU elements, circRNA flanking introns are often longer than the average intron length.2, 4, 21 It has been found that one gene locus can produce multiple circRNAs by using different splice donor and acceptor sites during backsplicing. Around 50% of circRNA-expressing host genes produce a single circRNA isoform, whereas other genes produce more. The Ttn gene is well known to produce at least 38 different circRNAs.22 In general, longer linear genes correlate with a higher number of circRNA isoforms.23
Interestingly, exon 2 of the gene locus is particularly often present in circRNAs. Many circRNAs contain two to six exons, however, others can harbor even around 100 exons within the backsplice site, and their sizes range from 100 bp to several kilobases.24, 25 Most circRNAs are derived from constitutive exons and are, thereby, generated at the expense of their linear counterpart.22 circRNAs also often contain the canonical splice site motif GT/AG, suggesting the involvement of the canonical spliceosome.23, 26 However, Liang et al.27 could show that circRNAs become the preferred RNA transcript when core spliceosome components (e.g., SF3b or SF3a) or transcription termination factors are inhibited. Read-through transcripts can be further extended into downstream genes and are subjected to backsplicing.27 How circularization and linear alternative splicing are mechanistically regulated remains largely unknown.
For some cirRNAs, it was shown that also RNA-binding proteins (RBPs) are involved in their formation. The RBP Quaking (QKI) seems to be involved in the formation of several different circRNAs.28, 29 Conn et al.28 found that QKI can bind to intronic QKI-binding motifs in introns flanking the circRNA-forming exons of several genes. By dimerization of two QKI molecules, QKI facilitates the hybridization of those flanking introns. The RBP Muscleblind (MBL) is involved in the formation of a circRNA from the same gene locus, circMbl. MBL acts as a splicing factor and modulates the generation of Mbl mRNA and circMbl. The protein MBL can bind to both introns flanking the circMbl as well as to several binding sites on the circMbl. Ashwal-Fluss and colleagues30 have proposed a balanced adjustment of circMbl biogenesis and MBL protein generation by the MBL protein itself.
Furthermore, the RNA-editing enzyme ADAR (adenosine deaminase acting on RNA), which mediates A-to-I substitution in RNA processing, is also linked to circRNA biogenesis. ADAR expression level negatively correlates with several circRNAs.30, 31 More recently, it was described that the RNA helicase DHX9 (DExH-Box Helicase 9), which binds double-stranded RNA (dsRNA), is involved in the formation of many circRNAs. Aktaş et al.32 observed that DHX9 specifically binds dsRNA formed by base-pairing Alu elements. Moreover, DHX9 was shown to interact with ADAR (p150 isoform). DHX9 depletion leads to an increased number of circRNA-producing genes and to an increased global circRNA expression. Overexpression of a helicase-dead DHX9 mutant in DHX9-depleted cells indicated that the helicase activity resolves the Alu-mediated secondary structure of RNA.32 Thus, DHX9 binding to circRNA-flanking Alu elements inhibits backsplicing by direct unwinding of the flanking intronic regions or recruiting ADAR enzymes that convert adenosines to inosines, or both.
Whether circRNA are generated co-transcriptionally or post-transcriptionally is still a matter of debate. Ashwal-Fluss et al.33 analyzed datasets of chromatin-bound (nascent) RNA from fly heads, and they found hundreds of head-to-tail junction reads in these datasets, suggesting that circRNAs are generated co-transcriptionally. However, using metabolic tagging of newly transcribed RNAs with 4-thiouridine (4sU), Zhang et al.34 showed that the majority of circRNAs are formed after transcription of their parent genes has completed. Consequently, according to Zhang and colleagues, only a few circRNA are produced co-transcriptionally.34 They also reported that backsplicing is extremely inefficient and far less favorable than linear splicing. They hypothesize that ligation of a downstream 5′ splice site with an upstream 3′ splice site by the spliceosome might be sterically unfavorable.
Characterization
circRNAs are generally expressed at low levels compared to mRNAs, however, some circRNAs are more abundant than their linear transcripts, and their expression can be independent of the related parental mRNA.2, 4, 24, 30, 34, 35 The lack of free ends, which are prone to exonucleolytic degradation, gives them an extraordinary stability compared to linear transcripts, and, hence, it makes them resistant to the RNA exonuclease RNase R.36 Several studies have revealed that circRNAs are most abundantly expressed in neural tissue.30, 37 This may be explained by the high resistance of circRNAs against RNA degradation and slow division rates of cell types like neurons, leading to an accumulation of circular transcripts even if the backsplicing process is less favorable than linear splicing.34
Within the cell, exonic circRNAs are predominantly found in the cytoplasm,38 whereas ciRNAs and EIciRNAs are enriched in the nucleus.17, 18 How circRNAs are released from the nucleus to the cytoplasm is not well understood yet. In 2018, Huang and colleagues39 published the first study on nuclear export of circRNAs. Using an RNAi-based approach in Drosophila DL1 cells, they screened 26 candidate proteins with known function in RNA nuclear export. The DExH/D-box helicase Hel25E was shown to be necessary and sufficient for the nuclear export of long (>811 nt) circRNAs. Hel25E depletion caused nuclear retention of long (>811 nt), but not short (>701 nt), circRNAs. In human HeLa cells, the Hel25E homologs UAP56 (DDX39B) and URH49 (DDX39A) were shown to be involved in the nuclear export of 14 circRNA candidates. UAP56 functions in the nuclear export of long circRNAs (>1,298 nt), whereas URH49 functions in the nuclear export of short circRNAs (<356 nt). A four-amino acid motif that is divergent among the Hel25E human homologs UAP56 and UAP49 (KSLN/RSFS in UAP56 and UAP49, respectively) controls the circRNA export length preference.39 How exactly the circRNA size is recognized by the four-amino acid motif is still unknown. It was reported previously that UAP56 and UAP49 have distinct protein-binding partners, and it is possible that these unique binding partners may act as size sensors in the circRNA export mechanism.39, 40 Understanding subcellular trafficking of circRNAs is just at the beginning, however, Huang et al.39 reported the first evidence that nuclear export must occur by an active process rather than diffusion.
Within the whole organism, mouse or human, circRNAs are often cell type-specifically, tissue-specifically, and/or developmental stage-specifically expressed.24 By performing RNA sequencing (RNA-seq) on ribosomal-depleted RNA from human hearts and mouse hearts, Tan et al.23 observed that circRNA-expressing genes that do not express their linear transcript in a certain tissue usually do not express the circular transcript in the same tissue. However, they reported some exceptions for the non-cardiac-expressed protein-coding genes RBL1, PIK3R2, and RP1-27O5.3, which exclusively expressed the respective circRNAs.23
Detection
Due to the lack of polyadenylated (poly(A)) tails and their non-linear conformation, circRNAs were overlooked in next-generation RNA-seq profiling, which in the past was usually done including a poly(A)+ enrichment step. However, RNA-seq in rRNA-depleted RNA led to the discovery of thousands of circRNAs, including in the heart. Tan et al.23 detected 15,318 and 3,017 cardiac circRNAs in the human and mouse hearts, respectively, using the de novo circRNA identification algorithm CIRI. The bioinformatic identification of circRNAs by aligning the sequencing data to the reference genomes is based on the presence of backsplice junction-spanning reads. Several bioinformatic algorithms were developed that allow sequence alignment of reads covering the backsplice site to the reference genome and, thereby, detecting non-linear splice events. Widely used examples include CIRCexplorer,19 CIRI,41, 42 find_circ,3 circRNA_finder,21 MapSplice,43 DCC,44 acfs,35 KNIFE,45 miARma-Seq,46 PTESFinder,47 Segemehl,48 NCLScan,49 and UROBORUS.50
While classic alignment tools reject all reads that do not arise from linear splicing, these circRNA detection tools filter for linear not-mappable reads that connect one exon with an upstream exon. circRNA detection algorithms typically use either a fragmentation- or a pseudo-reference-based method for the alignment. Fragmentation (or segmented read)-based methods split not-mappable reads into smaller fragments and align these separately. A reconstruction of the alignment pattern allows the identification of non-linear spliced transcripts. Preudo-reference-based methods rely on already existing annotation, generate putative splicing events, and try to align reads on these putative transcripts. A comprehensive overview of circRNA detection and sequence alignment tools is reviewed by Zeng et al.51 and Carrara et al.52
The increasing circRNA RNA-seq profiling in different organisms, tissues, and cell types as well as ongoing de novo detection of circRNAs generated the need for a comprehensive circRNA database that summarizes all the detected and validated circRNAs. The most widely used database called circBase was generated in 2014 by Glažar et al.53 The recent update (from 2017) includes nine RNA-seq studies in different organisms, including human and mouse. Other commonly used databases are CircNet54 and CircInteractome.55 The number of circRNA databases is still growing, with other examples including the following: circRNADb14 (circRNAs with protein-coding annotation), CSCD56 (cancer-specific circRNAs), exoRBase57 (non-coding RNAs expressed in exosomes), PlantcircBase58 (plant circRNAs), and CircFunBase59 (functional circRNAs). However, there is still a need for one comprehensive database that compiles all the information.
The individual circRNA abundance generally correlates with the abundance of its linear host gene transcript RNA, but they may also exist at much higher levels than their linear counterparts. The most abundantly expressed circRNAs within the heart are derived from key cardiac genes, including TTN (Titin), RYR2, and DMD. The TTN gene alone gives rise to up to 415 different circRNA isoforms, of which 83% originate from the I-band domain. Among those, the longest cardiac circRNA includes 153 exons. The most abundantly expressed circRNA within the heart is a cytoplasmically localized single-exon circRNA, circSLC8A1-1.23, 38 This circular transcript of exon 2 of the SLC8A1 gene, which codes for the sodium-calcium exchanger (NCX1), is one of a few circRNAs that was already discovered before the advent of huge circRNA-profiling studies. It was already discovered in 1991 that this circularized exon 2 encodes a small protein.60
Validation of circRNAs and their expression levels can be assessed by using divergent (outward-facing) primers. These primer pairs are amplifying away from each other regarding the genomic orientation, and, thereby, they only amplify a circRNA transcript without amplifying linear transcripts from that same genomic region.26 However, since detection of exons in a shuffled order is not an exclusive characteristic for circRNAs,26 circRNA validation should always rely on the combination of multiple methods to confirm the existence of backsplice junction (by PCR and sequencing), circularity (by RNase R), and expected size (northern blot). Of note, a few circRNAs were found to be degradable by RNase R, and some linear transcripts were found to be resistant to RNase R;26 therefore, caution is warranted. Additionally, circRNAs and linear transcripts can be distinguished by northern blot. A circRNA will migrate much faster than a linear RNA of the same length. If the RNA sample undergoes targeted RNase H digestion prior to gel electrophoresis, a linear RNA molecule will be cut into two parts and shows two bands, whereas a circRNA will be nicked and shows only one band on the gel. Other methods strengthening circularity are 2D gel electrophoresis and gel trap electrophoresis.26
circRNA Mode of Action
The ubiquitous presence of circRNAs revealed by large-scale RNA-seq studies in 20122 initiated the systematic search for circRNA functions. Just 1 year later, simultaneously published by two groups,3, 12 circRNA CDR1as was shown to act as a competing RNA molecule by sponging miRNAs. Shortly afterward, several other circRNAs were reported and shown to act as miRNA sponges also,61, 62, 63, 64, 65, 66, 67 strengthening the general miRNA sponge function of circRNAs. However, most circRNAs do not contain multiple miRNA-binding sites,13 thus miRNA sponging is just one of multiple potential functions. Further molecular mechansims were elucidated for a number of circRNAs, including sponging of proteins, acting as scaffolds for protein complexes, modulation of transcription and splicing, and translation to produce small peptides (Figure 2). In this section, we give a comprehensive overview of circRNA functions that are known thus far.
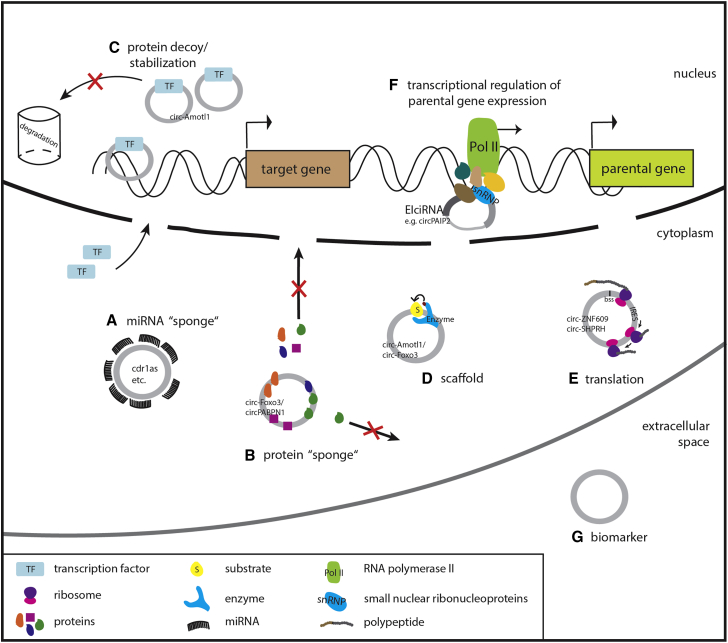
Figure 2.
Overview of circRNA Functions Discovered Thus Far
circRNAs can act as (A) miRNA sponge, (B) protein sponge, (C) protein decoy, and (D) scaffold for the formation of protein complexes. (E) A few circRNAs were shown to encode small peptides. (F) Candidates from subclasses of circular transcripts, ciRNAs and EIciRNAs, were shown to regulate parental gene expression. (G) By a thus far unknown mechanism, circRNAs are released from cells and can be detected in serum, plasma, and exosomes.
miRNA Sponging
miRNAs are a class of small RNAs with a size of ∼20 nt. Precursor miRNA (pri-miRNA) transcripts, which contain a hairpin structure covering the mature miRNA sequence, are cleaved in a two-step process, leading to an RNA duplex of ∼22 nt. The RNA duplex associates with Ago (Argonaute) proteins in the cytosol, assembling the miRNA-induced silencing complex (miRISC). The mature miRNA (guide strand) remains in the miRISC, while the passenger strand of the RNA duplex (miRNA*) is degraded. The miRISC regulates gene expression on a post-transcriptional level by interacting with miRNA-binding sites mostly in the 3′ UTR of mRNAs, leading to mRNA deadenylation, decreased mRNA stability, and reduced translation.25
The most frequently described function of circRNAs is the miRNA sponge activity. Within their nucleotide sequence, some circRNAs contain multiple miRNA-binding sites. Thus, they are able to bind miRNAs, preventing them from binding to their canonical mRNA target genes (sponge effect) (Figure 2A). The best known example, circRNA CDR1as, contains more than 70 conserved binding sites for miRNA-7 (miR-7), therefore strongly inhibiting miR-7 activity.3, 12 However, since the miR-7-binding sites are only partially complementary to miR-7, miR-7 is densely bound, but not sliced, by Ago proteins in a miR-7-dependent manner.3 Another almost complementary miRNA-binding site for miR-671, however, enables miR-671 to cleave CDR1as in an Ago-dependent manner and, thereby, directly regulate CDR1as.68
Subsequent to the cdr1as publications in 2013, multiple other circRNAs were shown to act as miRNA sponges.61, 62, 63, 64, 65, 66, 67 Using bioinformatics tools, miRNA-binding sites can be predicted within the circRNA sequence (e.g., by using CircInteractome55). Therefore, circRNAs that hold conspicuous many miRNA-binding sites for the same miRNA can be relatively easily uncovered as miRNA sponges. However, other circRNAs contain only a few binding sites for one or multiple miRNAs. circHIPK3 (circularized exon 2 of the HIPK3 gene) is significantly upregulated in liver cancer, and silencing of circHIPK3 inhibits human cell proliferation.69 Mechanistically, circHIPK3 was shown to sponge at least 9 different miRNAs with a total of 18 potential miRNA-binding sites (1–3 binding sites per miRNA). These miRNAs were identified by cloning the circHIPK3 sequence downstream of a luciferase reporter construct and, subsequently, using this reporter construct for a miRNA library screening. All nine miRNAs have been reported previously to act as growth-suppressive miRNAs. Transfection of miRNA mimics revealed that four of these nine miRNAs (miR-124, miR-193, miR-379, and miR-654) could indeed inhibit HEK293T cell proliferation.69 Another example from the cancer field, circ-ITCH was shown to share the same miRNA-binding sites (miR-7, miR-17, and miR-214) with the 3′ UTR of the ITCH mRNA and, thereby, sponge miRNAs, which otherwise suppress protein translation of the linear mRNA from the same gene locus.70
Protein Sponging
Most circRNAs do not contain multiple miRNA-binding sites.13 Not surprisingly, shortly after designating circRNAs as miRNA sponges, other molecular functions were elucidated. Already in 2014, Ashwal-Fluss et al.33 had shown that circ-Mbl contains several binding sites for the MBL protein and that circ-Mbl and MBL protein biogenesis is tightly regulated by the transcriptional products as well as MBL protein from the mbl locus itself.
Du et al.71, 72, 73 intensively studied the circRNA circ-Foxo3, a well-studied protein-interacting circRNA. Cytoplasmically distributed circ-Foxo3 is able to interact with multiple proteins in the cytoplasm.71 Anti-stress protein ID-1, anti-stress protein FAK, HIF-1α, and transcription factor E2F1 are retained in the cytoplasm (sponged by the circRNA; Figure 2B), and, hence, they cannot fulfill their anti-senescence and anti-stress functions in the nucleus, leading to increased cellular senescence. The same group focused on the association between circ-Foxo3 expression and cell cycle suppression.72 circ-Foxo3 expression levels negatively correlate with cell proliferation. Silencing circ-Foxo3 promoted cell proliferation whereas overexpression repressed cell cycle progression. Mechanistically, high circ-Foxo3 levels lead to an enhanced interaction between p21 and Cdk2 in the cytoplasm, thereby preventing Cdk2 interaction with cyclines A and E and a subsequent block of cell cycle progression.72
Another example of RBP-interacting circRNAs revealed that circRNAs are also able to regulate translation. circPABPN1 was identified as a binding target of the well-studied RBP HuR.74 Overexpression of circPABPN1 led decreased PABPN1 mRNA in HuR immunoprecipitation experiments, showing that high levels of circPABPN1 block HuR binding to the PABPN1 mRNA. HuR RNA immunoprecipitation (RIP) and HuR knockdown showed indeed that HuR regulates PABPN1 protein levels. Consequently, this study showed that circPABPN1 suppressed HuR binding to its host gene mRNA and lowered the protein level from the parental gene.74
Protein Decoy and Stabilization
While circ-Foxo3 negatively correlates with cell proliferation and acts as a protein-binding partner in the cytosol, circ-Amotl1 acts as a tumorigenic circRNA and exhibits its function in the nucleus. circ-Amotl1, highly expressed in patient tumor samples and human cancer cell lines, triggers tumorigenicity through an interaction with the oncogenic transcription factor c-Myc. circ-Amotl1 is able to bind and retain c-Myc in the nucleus, stabilize c-Myc, and upregulate c-Myc target genes, leading to increased cell proliferation, reduced apoptosis, and a highly tumorigenic phenotype.75 This study gives an example of how a circRNA can decoy single proteins to a specific cellular compartment and stabilize them (Figure 2C).
Scaffold for Protein Complexes
circRNAs that have binding sites for different RBPs can also function as scaffolds facilitating contact between two or more proteins. In a third study by Du et al.73 in 2017, circ-Foxo3 was shown to regulate apoptosis by acting as a scaffold for protein interaction. By binding p53 and the E3 ubiquitin-protein ligase Mdm2, circ-Foxo3 promotes MDM2-induced ubiquitination and the subsequent degradation of p53. With high levels of circ-Foxo3, Foxo3 protein can thereby escape MDM2 ubiquitination and proteasome degradation. High Foxo3 protein levels result in the upregulation of Foxo3 downstream target PUMA and lead to PUMA and BAX-mediated apoptosis.73
Another example is the circRNA circ-Amotl1. Besides its function in tumorigenicity by inducing c-Myc nuclear translocation,75 the same group presented circ-Amotl1 as a scaffold for the protein interaction between AKT and PDK1. circ-Amotl1 brings AKT and PDK1 in close proximity, which facilitates by phosphorylation of AKT by PDK1 and nuclear translocation of pAKT76 (Figure 2D). However, the group stated that circ-Amotl1 is exclusively expressed in human cells. Although the human Amotl1 gene is highly conserved to mouse and rat Amotl1 gene, the sequence at the backsplice junction area of circ-Amotl1 differs in humans and rodents. Of note, functional in vitro studies of cardiac repair in this report were performed in primary mouse cardiomyocytes and mouse cardiac fibroblasts, and in vivo studies were performed in a doxorubicin-induced cardiomyopathy mouse model. Therefore, the proposed functions of circ-Amotl1 to reduce apoptosis and enhance cardiac repair await validation in a human context, for instance, in human induced pluripotent stem cells (iPSC)-derived cardiomyocytes.
Translation
Investigating the possibility of circRNA translation, Jeck et al.4 performed polysome profiling by sucrose gradient centrifugation, and they detected circRNAs merely in the unbound fractions, but not in any bound fractions, suggesting that circRNAs are usually not translated.
Nevertheless, although circRNAs belong to the class of non-coding RNAs, some circRNAs can encode small peptides (Figure 2E). Translation of circRNAs was already described in the 1990s,60, 77 and three recent studies confirmed that individual circRNAs are translated.78, 79, 80 Investigating differentially expressed circRNAs in myoblast differentiation, circZNF609 was identified to control skeletal myoblast proliferation.78 circZNF609, a single-exon circRNA of exon 2 from the ZNF609 gene locus, contains two open reading frames (ORFs) spanning from two AUG start codons to a stop codon located 3 nt after the splice junction. In regard to linear exon 2, the stop codon is located upstream of the start codons, thus the open reading frame is only generated by circularization. Polysome profiling revealed that circZNF609 (hsa_circ_0000615) is bound to heavy polysome fractions. Using an overexpression construct containing a FLAG-tag immediately upstream of the stop codon showed that circZNF609 is translated, as demonstrated in anti-FLAG western blot. Since circRNAs do not contain Cap sequences, translation occurs in a Cap-independent manner.78 It has also been shown that the nucleotide modification N6-methyladenosine (m6A) can promote circRNA translation.80 Of note, two other studies reported that circZNF609 can also act as a miRNA sponge for miR-615-581 and for miR-150p.82
More recently, another group published translation of a 17-kDa peptide from the circRNA circ-SHPRH.83 circ-SHPRH (hsa_circ_0001649), previously reported as a novel biomarker and downregulated in hepatocellular carcinoma,84 was shown to encode a 146-amino acid peptide (SHPRH-146aa). Both SHPRH-146aa and full-length SHPRH protein interact with the E3 ubiquitin ligase DTL. SHPRH-146aa was shown to act as a decoy and serve as a tumor suppressor by protecting the full-length SHPRH protein from DTL-induced ubiquitination and subsequent degradation. Stabilized SHPRH, in turn, sequentially ubiquitinates PCNA (proliferating cell nuclear antigen) as an E3 ubiquitin ligase, which leads to reduced cell proliferation and tumorigenicity.83
Other Functions
In comparison to exonic circRNAs, intronic circRNAs (ciRNAs and EIciRNAs) are mainly located in the nucleus. Zhang et al.18 discovered a new function of intronic circular transcript by showing that these circRNAs can regulate host gene transcription (Figure 2F). Also some EIciRNAs (e.g., circPAIP2) can interact with U1 small nuclear ribonucleoproteins (snRNPs) and, thereby, enhance RNA polymerase II (Pol II) activity, leading to an increased transcription of the parental gene.17 However, the functional characterization of ciRNAs is still in its infancy.
In addition to defined functions in the cell cytoplasm or nucleus, circRNAs are also released from the cell. Several studies discovered circRNAs as potential new biomarkers in body fluids, such as serum, plasma, or urine samples of patients85, 86, 87 (Figure 2G). circRNAs were also shown to be enriched and stable in exosomes. qRT-PCR analysis of three circRNAs in serum samples of three healthy donors revealed that these circRNAs were predominantly located in serum-derived exosomes, but not in exosome-depleted serum,88 therefore suggesting rather active cellular release mechanisms in contrast to just passive shedding of dying cells.
circRNAs in Cardiovascular Disease
Cardiovascular diseases belong to the leading causes of death in the industrialized countries. Major pathological changes in the failing heart are cardiac hypertrophy and fibrosis.89 Several transcriptome profiling studies generated large datasets of cardiac expressed circRNAs.23, 38, 90 For instance, Werfel et al.38 performed RNA-seq analysis in rRNA-depleted libraries from human hearts (heart failure [HF] versus non-failing), mouse hearts (transverse aortic constriction [TAC] versus sham), and rat hearts (adults versus neonatal), and they detected several differentially expressed circRNAs in cardiac disease and postnatal development. Overall, the circRNA expression was slightly increased in disease condition and increased in the heart of neonates compared to adult rats.38 In addition, hundreds of differentially expressed circRNAs were detected in human thoracic aortic dissection (TAD) patients91 as well as in a myocardial infarction (MI) model of HF in mice.92 In addition to other types of non-coding RNAs, such as miRNAs and long non-coding RNAs (lncRNAs), which are already known to regulate cardiac physiology and pathophysiology,93, 94 several recent studies indicated that circRNAs are present in all cardiac cell types and also play a role in cardiovascular physiology and disease.
circRNAs with Functions in the Cardiac Muscle
circRNA CDR1as, initially mainly studied in the brain, was shown to act as a competing endogenous RNA by sponging miRNAs.3, 12 In 2016, Geng et al.66 investigated the function of the CDR1as/miR-7 pathway in a murine MI model. CDR1as expression is significantly upregulated in mice after MI as well as in hypoxia-treated primary cardiomyocytes in vitro. CDR1as overexpression was shown to increase myocyte apoptosis and infarct size after MI via reducing miR-7a activity and enhancing the expression of miR-7a target genes PARP and SP1.66
In the same year, Wang et al.95 published the first study on the role of circRNAs in cardiac hypertrophy and heart failure. They showed that the heart-related circRNA (HRCR) can bind to miR-223 and acts as a miRNA sponge to inhibit miR-223 activity, thus leading to an increased expression of the apoptosis repressor with CARD domain (ARC) protein. Adeno-associated virus (AAV) vector-mediated overexpression of HRCR attenuates isoproterenol-induced cardiac hypertrophy in vivo. Using very similar experimental settings, the same group 1 year later reported on the circRNA MFCAR (Mitochondrial Fission and Apoptosis Circular RNA),96 which acts as a miRNA sponge for miR-652. miR-652 negatively regulates the mitochondrial membrane-associated protein MTP18, whereas MTP18 knockdown reduces mitochondrial fission and suppresses cardiomyocyte apoptosis and ischemia-reperfusion (I/R) injury. Consequently, intracoronary delivery of MFCAR-small interfering RNA (siRNA) adenoviral vectors prior to I/R operation showed a beneficial effect on heart function. Since then, an increasing number of reports about circRNAs as miRNA sponges has been published. circRNA_000203 enhances the expression of fibrosis-associated genes by inhibiting the targets of miR-26b-5p, Col1a2, and CTGF in cardiac fibroblasts.97 There are more studies on circRNAs as miRNA sponges in cardiovascular disease.98, 99
As already mentioned above, circ-Foxo3 promotes cardiac senescence by interacting with anti-stress protein ID-1, anti-stress protein FAK, HIF-1α, and transcription factor E2F1 and preventing nuclear translocation of these proteins. Silencing of circ-Foxo3 inhibited senescence whereas ectopic overexpression of circ-Foxo3 induced senescence. In vivo modulation of circ-Foxo3 in doxorubicin-induced cardiomyopathy in mice showed that overexpression of circ-Foxo3 deteriorates cardiac function whereas siRNA-mediated silencing attenuated doxorubicin-induced cardiotoxicity.71
circ-Amotl1 was shown to act as a scaffold for the protein interaction between AKT and PDK1, followed by phosphorylation of AKT by PDK1 and nuclear translocation of pAKT.76 AKT activation was already shown to have a cardioprotective role in cardiovascular diseases.100, 101 Overexpression of the human circ-Amotl1 increased nuclear pAKT in vitro and in vivo. Intraperitonal injection of nanoparticles carrying the human circ-Amotl1 overexpression construct was found to be protective against doxorubicin-induced cardiomyopathy in mice, as shown by reduced left ventricle dilatation, reduced fibrosis, and reduced apoptosis.76
A recent study by Gupta et al.29 revealed that the RBP QKI inhibits doxorubicin-mediated cardiotoxicity through the regulation of several cardiac circRNAs. QKI is strongly downregulated in response to doxorubicin in vitro and in vivo. QKI knockdown increases apoptosis in primary cardiomyocytes, whereas QKI overexpression attenuates doxorubicin-induced cardiotoxicity. AAV9-mediated overexpression of QKI could prevent cardiac apoptosis and cardiac atrophy induced by doxorubicin and improved cardiac function. Modulation of QKI protein levels identified several circRNAs, including circRNAs from the Ttn, Fhod3, and Strn3 gene locus, to be expressed in a QKI-dependent manner. The Titin circRNA (circTtn-exon105-111) was further shown to act as a downstream effector of QKI. Lentivirally mediated overexpression of circRNA Ttn exons 105–111 resulted in a reduced susceptibility to doxorubicin, similar to QKI overexpression, whereas siRNA-mediated knockdown of circRNA Ttn exons 105–111 increased the susceptibility of cardiomyocytes to doxorubicin.29
The RNA Binding Motif protein 20 (RBM20) was recently shown to regulate circRNA production from the Titin gene.102 RBM20 is a known regulator of splicing of many cardiac genes. Mutations in the RBM20 gene are associated with dilated cardiomyopathy. RMB20 is required for the production of circRNAs from the I-band region of Titin, as demonstrated in RBM20-knockout mice. In addition, in a cardiac sample from a dilated cardiomyopathy (DCM) patient with a heterozygous mutation in RBM20, circRNA production from the Titin I-band region was severely altered. These data suggest a strong connection between alternative splicing and circRNA production. Mechanistically, RMB20 may exclude specific exons from the pre-mRNA and, thereby, produce a precursor substrate for circRNA production (RBM20-dependent Ttn circRNAs).102
circRNAs in the Vasculature
Just as in cardiomyocytes and fibroblasts (as described above), circRNAs can also act as miRNA sponges in endothelial cells and vascular smooth muscle cells (VSMCs).103, 104, 105 For example, hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186 and HIF-1α axis.105 Another example, circ_Lrp6 was shown to be enriched in VSMCs and act as a miRNA sponge for miR-145. As miR-145 is known to regulate VSMC physiology and homeostasis, and it was previously shown to be involved in the development of atherosclerosis, circ_Lrp6 is able to modulate VSMC migration, proliferation, and differentiation through the inhibition of miR-145. circ_Lrp6 silencing induced a pro-differentation phenotype, coherent with upregulated miR-145 levels and a reduced migration and proliferation. A small hairpin RNA (shRNA)-mediated knockdown of circ_Lrp6 was also shown to be beneficial in a mouse carotid artery stenosis model.63
Moreover, Boeckel et al.106 investigated hypoxia-regulated circRNAs, and they identified the hypoxia-induced circRNA cZNF292. siRNA-mediated knockdown of cZNF292 reduced tube formation and spheroid sprouting in vitro, suggesting a pro-angiogenic role of cZNF292. However, little is known about the mechanistic function of cZNF292 so far.106, 107
Some non-coding genes also give rise to circular non-coding transcripts, among them circTTN-As1-1, circFLG-AS1-1, circMKLN1-AS1-1, circMEF2C-AS1-1,23 and circANRIL.108, 109 circANRIL is a circular antisense transcript from the INK4A/ARF locus. The predominant circular isoform from that locus consists of exons 5–7. SNPs near the INK4A/ARF locus on chromosome 9p21.3 are linked to susceptibility of atherosclerotic vascular disease (ASVD). Patients with the coronary artery disease (CAD)-protective haplotype at 9p21 showed significantly increased expression of circANRIL in peripheral blood mononuclear cells (PBMCs) and whole blood. Interestingly, this effect was opposite to the published correlation for lncANRIL, which is downregulated in patients carrying the 9p21 protective genotype. Functional studies revealed that circANRIL overexpression increased cell apoptosis and reduced proliferation in an lncANRIL-independent manner. Mechanistically, circANRIL was shown to bind and block PES1, an essential 60S-pre-ribosomal assembly factor, thereby impairing pre-rRNA processing and ribosome biogenesis in VSMCs and macrophages. Thus, a high circANRIL level induces nucleolar stress and p53 activation, leading to the induction of apoptosis and reduced proliferation. circANRIL may act as a protective factor against atherosclerosis by controlling pre-rRNA processing and nucleolar stress in atherosclerotic plaques.
circRNAs as Novel Biomarkers in Cardiovascular Disease
Due their high stability, also in the extracellular space, circRNAs represent ideal candidates for diagnostic biomarkers. Vausort et al.85 identified the MI-associated circRNA (MIRCA) in peripheral blood of acute MI patients and age- and gender-matched healthy controls. circRNA MICRA also originates from the ZNF609 gene locus, however, compared to circZNF609,78, 81, 82, 106 MICRA includes a different exon. (MICRA is a 874-nt-long circRNA formed mainly from exon 1 of the ZNF609 gene.85) Circulating levels of MICRA in MI patients were significantly lower compared to healthy controls. Moreover, MICRA was shown to be a strong predictor of left ventricle dysfunction (4-month ejection fraction [EF] ≤40%).85 In a more recent study, patients with low MICRA levels in peripheral blood at the time of reperfusion had a higher risk of left ventricular dysfunction.86 The molecular function of MICRA is currently unknown. The authors speculate that MICRA may sponge miR-150, a miRNA known to be associated with the development of HF after acute MI.110 Interestingly, exon 2 circRNA of the ZNF609 gene locus, circZNF609, was reported to sponge miR-150-5p,82 and circulating levels of cZNF609 were downregulated in the patients with CAD and hypertension.81 Using circRNA microarray, Zhao et al.111 screened for dysregulated circRNAs in the peripheral blood of CAD patients, and they identified hsa_circ_0124644 from the ROBO2 gene as a putative new biomarker for CAD.
Manipulation Tools for circRNA-Based Therapeutic Strategies
Due to their postulated ability to regulate gene expression, circRNAs emerge as potential therapeutic tools. Consequently, molecular tools for their manipulation are currently under investigation. In addition, functional characterization of circRNAs also requires modulation of the circRNA expression in order to understand the impact of the altered circRNA expression level. This is usually assessed by gain- and loss-of-function experiments through circRNA overexpression or knockdown, respectively (Figure 3). The easiest method to inhibit circRNA expression is RNAi, which, however, still contains some challenges.71, 95, 108 siRNAs have to be complement to the backsplice site in order to be circRNA specific without affecting the host gene expression (Figure 3A). Thus, siRNA-mediated knockdown is limited to the backsplice junction, and negative effects on the parental protein-coding gene expression should always be tested for and ruled out. Naked single-stranded RNA strands are prone to degradation. Thus, especially for in vivo experiments, chemically modified antisense oligonucleotides such as GapmeRs exert a higher stability112 (Figure 3B). To our knowledge, GapmeRs haven’t been used so far to target circRNAs in vivo. However, as they are commonly used to target lncRNAs, it is conceivable that they may additonally downregulate circRNAs.25
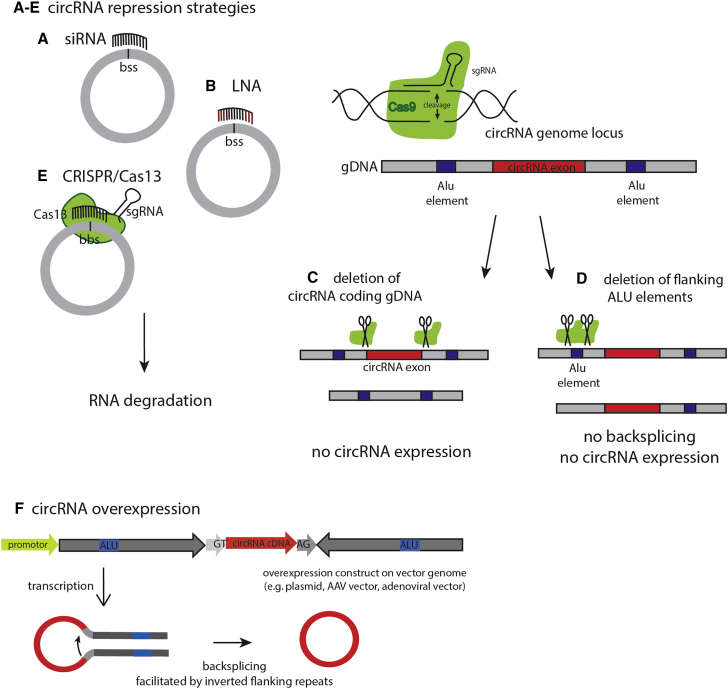
Figure 3.
Manipulation Tools for circRNA Expression
(A–E) Overview of circRNA repression strategies and (F) circRNA overexpression. circRNA knockdown is typically achieved by directing antisense oligonucleotides, such as (A) siRNAs or (B) locked-nucleic acid (LNA), to the circRNA backsplice site (bss). (C) CRISPR/Cas9-mediated deletion of the circRNA-coding genome locus prevents circRNA expression. (D) CRISPR/Cas9-mediated disruption of circRNA-flanking repeat elements prevents backsplicing and, thus, suppresses circRNA expression. (E) A circRNA-specific single-guide RNA (sgRNA) targeted against the circRNA backsplice site directs the RNase Cas13 to the circRNA, with subsequent circRNA degradation. (F) Vector constructs containing long inverted flanking repeats are typically used to enable backsplicing and produce circular transcripts from an exogenous vector.
The complete circRNA knockout by genome editing is (in most cases) more difficult, since the deletion of circRNA-involved exons would also disrupt the host gene expression. Piwecka et al.113 used the CRISPR/Cas9 genome-editing tool to remove the CDR1as locus (Figure 3C), and they generated CDR1as loss-of-function mutant mice. However, this was only possible because the CDR1as precursor RNA transcript is so efficiently backspliced that no linear transcript can be detected. Another approach of CRISPR/Cas9 genome editing to suppress circRNA expression was recently described by Zhang et al.34 By excising the intronic complement sequence (ICS) of the circGCN1L1-flanking introns (Figure 3D), circGCN1L1 expression was abolished. Removal of one ICS was sufficient to completely remove circGCN1L1 expression without effecting the linear mRNA transcription from the GCN1L1 gene locus.34
CRISPR-Cas13 was recently reported as a new tool for RNA modulation.114 The RNA-guided RNA-targeting CRISPR-Cas effector Cas13a is able to bind and degrade RNA in mammalian cells. With comparable knockdown efficiency and improved specificity compared to RNAi, CRISPR-Cas13 is a promising tool for RNA-knockdown studies, including circRNA silencing by targeting the CRISPR-Cas13 guide RNA to the circRNA backsplice site (Figure 3E). Moreover, mutations in the catalytic domain created a catalytically dead variant (dCas13), which was only binding respective RNA molecules and could be fused to any other protein domain (e.g., fluorophores for intracellular localization or RNA-editing domains like ADAR).114, 115 Even though CRISPR-Cas13 is an interesting new tool to study circRNA functional mechanisms, CRISPR-Cas13-mediated silencing is not applicable in a therapeutic context, since the Cas13 protein and guide RNA would have to be delivered into the organism with unknown side effects.
circRNA overexpression is usually achieved by vector constructs containing the circRNA sequence flanked by introns containing inverted repeats for hybridization as well as splicing signals (Figure 3F). However, this method can lead to concatemers if the RNA polymerase fails to recognize the transcription terminator site (TTS) and continues transcription in a rolling-cycle manner.116 Thus, checking the overexpression product size by northern blot is necessary to rule out concatemer expression. Engineered circRNAs containing an internal ribosomal entry site (IRES) can also drive translation of an open reading frame and, therefore, be used for protein translation.77 The overexpression construct can be delivered by any vector, e.g., plasmid transfection12 or viral vector systems like adenovirus vectors95 or AAV vectors.117
Since AAV vectors generally do not integrate into the genome, they count as relatively safe and are already used in clinical trials.118, 119 However, new delivery strategies avoiding viral vectors are arising in the field of somatic gene transfer. The advantages of non-viral systems include a faster vector production and lower biosafety risks. For example, delivery of naked RNA does not necessarily require nuclear localization (compared to DNA-based virus systems), however, linear RNA is very prone to nucleolytic degradation.120 Wesselhoeft et al.121 developed an approach to produce exogenous circRNAs in vitro for potent and stable protein expression in eukaryotic cells. The circularization system is based on self-splicing introns that can generate circRNAs up to 5 kb in length (Figure 4A). After in vitro transcription (IVT), circular products can be purified from by-products of the IVT reaction by high-performance liquid chromatography (HPLC). IVT-produced circRNA of protein-coding genes transfected in HEK293 cells can produce the respective protein. Due to their high stability against RNA degradation and/or increased translation, engineered circRNAs transfected into cell lines produced more protein than the same amount of transfected unmodified or modified linear mRNA.121 However, for future in vivo approaches, suitable delivery strategies are needed, and nucleoside modifications may further improve the stability and suppress undesired immune responses.121
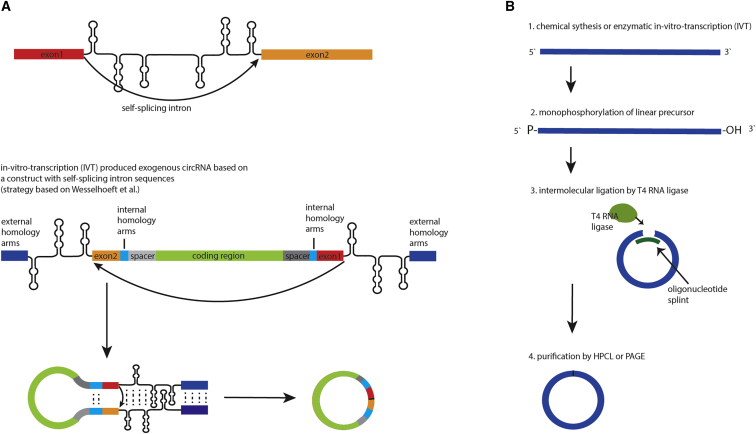
Figure 4.
circRNA IVT
(A) Wesselhoeft et al.121 generated IVT-produced circRNA based on a vector containing self-splicing intron sequences. The two halves of a self-splicing intron enable backsplicing of the interjacent sequence. (B) IVT, mono-phosphorylation of the linear precursor RNA, and intermolecular ligation by T4 RNA ligase can generate circular RNA transcripts of up to 500 nt in length.
Another in vitro method for the generation of circRNAs is the ligation of a 5′ phosphorylated RNA by using T4 RNA ligase77, 122, 123 (Figure 4B). First, linear RNA can be prepared by either chemical synthesis or IVT. Chemical synthesis allows one to introduce any backbone or base modifications, while IVT allows transcription of RNAs up to several kilobases. After generation of the linear RNA, 5′ monophospohorylation is required prior to enzymatic ligation of the RNA ends by T4 RNA ligase.123 Notably, intermolecular ligation can give rise to linear by-products as well as circular oligomers. To avoid intermolecular reactions, a low concentration of the linear precursor to favor intramolecular ligation (circulation) should be used. Moreover, adding a short oligonucleotide splint (complementary to the circRNA backsplice site) can help to increase intramolecular circularization.122 Finally, the desired circular product can be purified from site products by PAGE or HPLC.121 However, in vitro circRNA production is still in its infancy and needs further advancements, especially for the production of long circRNAs (≥500 nt). Overall, circRNA manipulation tools are a great gain in discovering circRNA function in diseases, and they have good prospects for being developed into circRNA-based therapeutic strategies in the future.
Conclusions and Future Perspectives
circRNAs, once considered to be an aberrant by-product of mRNA splicing, have become a major new class of non-coding RNAs. They are currently under intense investigation in a large number of research studies in almost all biological and biomedical fields. In contrast to their initially proposed function as miRNA sponges, solid evidence emerges that circRNAs exhibit a wide variety of molecular functions, including regulation of transcription and translation, regulation of cellular localization of interacting proteins, and even translation into small peptides. However, the knowledge about circRNA expression patterns, their mechanistic role in physiological and pathological conditions, their potential as diagnostic biomarkers, and the potential use of circRNAs as therapeutic targets is still in an early stage. A few circRNAs were shown to be critically involved in cardiac function. Whether more circRNAs play a role in different cell types of the heart remains to be investigated.
Conflicts of Interest
T.T. filed and licensed patents in the field of non-coding RNAs. T.T. is a founder and shareholder of Cardior Pharmaceuticals.
Acknowledgments
The authors acknowledge funding by the ERC Consolidator Grant Longheart, the H2020 project CardioRegenix, and the Deutsche Forschungsgemeinschaft (SFB TRR267).
Contributor Information
Christian Bär, Email: baer.christian@mh-hannover.de.
Thomas Thum, Email: thum.thomas@mh-hannover.de.
References
- 1.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 4.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kos A., Dijkema R., Arnberg A.C., van der Meide P.H., Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323:558–560. doi: 10.1038/323558a0. [DOI] [PubMed] [Google Scholar]
- 7.Arnberg A.C., Van Ommen G.J.B., Grivell L.A., Van Bruggen E.F.J., Borst P. Some yeast mitochondrial RNAs are circular. Cell. 1980;19:313–319. doi: 10.1016/0092-8674(80)90505-x. [DOI] [PubMed] [Google Scholar]
- 8.Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 9.Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 10.Cocquerelle C., Daubersies P., Majérus M.A., Kerckaert J.P., Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992;11:1095–1098. doi: 10.1002/j.1460-2075.1992.tb05148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasman Z., Been M.D., Garcia-Blanco M.A. Exon circularization in mammalian nuclear extracts. RNA. 1996;2:603–610. [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 13.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X., Han P., Zhou T., Guo X., Song X., Li Y. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep. 2016;6:34985. doi: 10.1038/srep34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hangauer M.J., Vaughn I.W., McManus M.T. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X.O., Wang H.B., Zhang Y., Lu X., Chen L.L., Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159:134–147. doi: 10.1016/j.cell.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Liang D., Wilusz J.E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 2014;28:2233–2247. doi: 10.1101/gad.251926.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westholm J.O., Miura P., Olson S., Shenker S., Joseph B., Sanfilippo P., Celniker S.E., Graveley B.R., Lai E.C. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aufiero S., van den Hoogenhof M.M.G., Reckman Y.J., Beqqali A., van der Made I., Kluin J., Khan M.A.F., Pinto Y.M., Creemers E.E. Cardiac circRNAs arise mainly from constitutive exons rather than alternatively spliced exons. RNA. 2018;24:815–827. doi: 10.1261/rna.064394.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan W.L.W., Lim B.T.S., Anene-Nzelu C.G.O., Ackers-Johnson M., Dashi A., See K., Tiang Z., Lee D.P., Chua W.W., Luu T.D. A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 2017;113:298–309. doi: 10.1093/cvr/cvw250. [DOI] [PubMed] [Google Scholar]
- 24.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beermann J., Piccoli M.-T., Viereck J., Thum T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 26.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liang D., Tatomer D.C., Luo Z., Wu H., Yang L., Chen L.-L., Cherry S., Wilusz J.E. The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting. Mol. Cell. 2017;68:940–954.e3. doi: 10.1016/j.molcel.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 29.Gupta S.K., Garg A., Bär C., Chatterjee S., Foinquinos A., Milting H., Streckfuß-Bömeke K., Fiedler J., Thum T. Quaking Inhibits Doxorubicin-Mediated Cardiotoxicity Through Regulation of Cardiac Circular RNA Expression. Circ. Res. 2018;122:246–254. doi: 10.1161/CIRCRESAHA.117.311335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rybak-Wolf A., Stottmeister C., Glažar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 31.Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R., Piechotta M., Levanon E.Y., Landthaler M., Dieterich C., Rajewsky N. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 32.Aktaş T., Avşar Ilık İ., Maticzka D., Bhardwaj V., Pessoa Rodrigues C., Mittler G., Manke T., Backofen R., Akhtar A. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–119. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 33.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y., Xue W., Li X., Zhang J., Chen S., Zhang J.L., Yang L., Chen L.L. The Biogenesis of Nascent Circular RNAs. Cell Rep. 2016;15:611–624. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 35.You X., Vlatkovic I., Babic A., Will T., Epstein I., Tushev G., Akbalik G., Wang M., Glock C., Quedenau C. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Suzuki H., Zuo Y., Wang J., Zhang M.Q., Malhotra A., Mayeda A. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 2006;34:e63. doi: 10.1093/nar/gkl151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gruner H., Cortés-López M., Cooper D.A., Bauer M., Miura P. CircRNA accumulation in the aging mouse brain. Sci. Rep. 2016;6:38907. doi: 10.1038/srep38907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Werfel S., Nothjunge S., Schwarzmayr T., Strom T.-M., Meitinger T., Engelhardt S. Characterization of circular RNAs in human, mouse and rat hearts. J. Mol. Cell. Cardiol. 2016;98:103–107. doi: 10.1016/j.yjmcc.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 39.Huang C., Liang D., Tatomer D.C., Wilusz J.E. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32:639–644. doi: 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamazaki T., Fujiwara N., Yukinaga H., Ebisuya M., Shiki T., Kurihara T., Kioka N., Kambe T., Nagao M., Nishida E., Masuda S. The closely related RNA helicases, UAP56 and URH49, preferentially form distinct mRNA export machineries and coordinately regulate mitotic progression. Mol. Biol. Cell. 2010;21:2953–2965. doi: 10.1091/mbc.E09-10-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gao Y., Zhang J., Zhao F. Circular RNA identification based on multiple seed matching. Brief. Bioinform. 2018;19:803–810. doi: 10.1093/bib/bbx014. [DOI] [PubMed] [Google Scholar]
- 42.Gao Y., Wang J., Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang K., Singh D., Zeng Z., Coleman S.J., Huang Y., Savich G.L., He X., Mieczkowski P., Grimm S.A., Perou C.M. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkq622. e178–e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng J., Metge F., Dieterich C. Specific identification and quantification of circular RNAs from sequencing data. Bioinformatics. 2016;32:1094–1096. doi: 10.1093/bioinformatics/btv656. [DOI] [PubMed] [Google Scholar]
- 45.Szabo L., Morey R., Palpant N.J., Wang P.L., Afari N., Jiang C., Parast M.M., Murry C.E., Laurent L.C., Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015;16:126. doi: 10.1186/s13059-015-0690-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andrés-León E., Núñez-Torres R., Rojas A.M. miARma-Seq: a comprehensive tool for miRNA, mRNA and circRNA analysis. Sci. Rep. 2016;6:25749. doi: 10.1038/srep25749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Izuogu O.G., Alhasan A.A., Alafghani H.M., Santibanez-Koref M., Elliott D.J., Jackson M.S. PTESFinder: a computational method to identify post-transcriptional exon shuffling (PTES) events. BMC Bioinformatics. 2016;17:31. doi: 10.1186/s12859-016-0881-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffmann S., Otto C., Doose G., Tanzer A., Langenberger D., Christ S., Kunz M., Holdt L.M., Teupser D., Hackermüller J., Stadler P.F. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol. 2014;15:R34. doi: 10.1186/gb-2014-15-2-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chuang T.-J., Wu C.-S., Chen C.-Y., Hung L.-Y., Chiang T.-W., Yang M.-Y. NCLscan: accurate identification of non-co-linear transcripts (fusion, trans-splicing and circular RNA) with a good balance between sensitivity and precision. Nucleic Acids Res. 2016;44:e29. doi: 10.1093/nar/gkv1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song X., Zhang N., Han P., Moon B.-S., Lai R.K., Wang K., Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44:e87. doi: 10.1093/nar/gkw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng X., Lin W., Guo M., Zou Q. A comprehensive overview and evaluation of circular RNA detection tools. PLoS Comput. Biol. 2017;13:e1005420. doi: 10.1371/journal.pcbi.1005420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carrara M., Fuschi P., Ivan C., Martelli F. Circular RNAs: Methodological challenges and perspectives in cardiovascular diseases. J. Cell. Mol. Med. 2018;22:5176–5187. doi: 10.1111/jcmm.13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glažar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y.-C., Li J.-R., Sun C.-H., Andrews E., Chao R.-F., Lin F.-M., Weng S.L., Hsu S.D., Huang C.C., Cheng C. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44(D1):D209–D215. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dudekula D.B., Panda A.C., Grammatikakis I., De S., Abdelmohsen K., Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xia S., Feng J., Chen K., Ma Y., Gong J., Cai F., Jin Y., Gao Y., Xia L., Chang H. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46(D1):D925–D929. doi: 10.1093/nar/gkx863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li S., Li Y., Chen B., Zhao J., Yu S., Tang Y., Zheng Q., Li Y., Wang P., He X., Huang S. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46(D1):D106–D112. doi: 10.1093/nar/gkx891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chu Q., Zhang X., Zhu X., Liu C., Mao L., Ye C., Zhu Q.H., Fan L. PlantcircBase: A Database for Plant Circular RNAs. Mol. Plant. 2017;10:1126–1128. doi: 10.1016/j.molp.2017.03.003. [DOI] [PubMed] [Google Scholar]
- 59.Meng X., Hu D., Zhang P., Chen Q., Chen M. CircFunBase: a database for functional circular RNAs. Database. 2019;2019:baz003. doi: 10.1093/database/baz003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li X.F., Lytton J. A circularized sodium-calcium exchanger exon 2 transcript. J. Biol. Chem. 1999;274:8153–8160. doi: 10.1074/jbc.274.12.8153. [DOI] [PubMed] [Google Scholar]
- 61.Rong D., Sun H., Li Z., Liu S., Dong C., Fu K., Tang W., Cao H. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271–73281. doi: 10.18632/oncotarget.19154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun M., Zhao W., Chen Z., Li M., Li S., Wu B., Bu R. Circ_0058063 regulates CDK6 to promote bladder cancer progression by sponging miR-145-5p. J. Cell. Physiol. 2019;234:4812–4824. doi: 10.1002/jcp.27280. [DOI] [PubMed] [Google Scholar]
- 63.Hall I.F., Climent M., Quintavalle M., Farina F.M., Schorn T., Zani S., Carullo P., Kunderfranco P., Civilini E., Condorelli G., Elia L. Circ_Lrp6, a Circular RNA Enriched in Vascular Smooth Muscle Cells, Acts as a Sponge Regulating miRNA-145 Function. Circ. Res. 2019;124:498–510. doi: 10.1161/CIRCRESAHA.118.314240. [DOI] [PubMed] [Google Scholar]
- 64.Zhang X., Wang S., Wang H., Cao J., Huang X., Chen Z., Xu P., Sun G., Xu J., Lv J., Xu Z. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer. 2019;18:20. doi: 10.1186/s12943-018-0935-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li M., Ding W., Tariq M.A., Chang W., Zhang X., Xu W., Hou L., Wang Y., Wang J. A circular transcript of ncx1 gene mediates ischemic myocardial injury by targeting miR-133a-3p. Theranostics. 2018;8:5855–5869. doi: 10.7150/thno.27285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Geng H.H., Li R., Su Y.M., Xiao J., Pan M., Cai X.X., Ji X.P. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on Its Target Genes Expression. PLoS ONE. 2016;11:e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun Y., Yang Z., Zheng B., Zhang X.H., Zhang M.-L., Zhao X.S., Zhao H.Y., Suzuki T., Wen J.K. A Novel Regulatory Mechanism of Smooth Muscle α-Actin Expression by NRG-1/circACTA2/miR-548f-5p Axis. Circ. Res. 2017;121:628–635. doi: 10.1161/CIRCRESAHA.117.311441. [DOI] [PubMed] [Google Scholar]
- 68.Hansen T.B., Wiklund E.D., Bramsen J.B., Villadsen S.B., Statham A.L., Clark S.J., Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011;30:4414–4422. doi: 10.1038/emboj.2011.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li F., Zhang L., Li W., Deng J., Zheng J., An M., Lu J., Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Du W.W., Yang W., Chen Y., Wu Z.-K., Foster F.S., Yang Z., Li X., Yang B.B. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur. Heart J. 2017;38:1402–1412. doi: 10.1093/eurheartj/ehw001. [DOI] [PubMed] [Google Scholar]
- 72.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Du W.W., Fang L., Yang W., Wu N., Awan F.M., Yang Z., Yang B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abdelmohsen K., Panda A.C., Munk R., Grammatikakis I., Dudekula D.B., De S., Kim J., Noh J.H., Kim K.M., Martindale J.L., Gorospe M. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang Q., Du W.W., Wu N., Yang W., Awan F.M., Fang L., Ma J., Li X., Zeng Y., Yang Z. A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ. 2017;24:1609–1620. doi: 10.1038/cdd.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zeng Y., Du W.W., Wu Y., Yang Z., Awan F.M., Li X., Yang W., Zhang C., Yang Q., Yee A. A Circular RNA Binds To and Activates AKT Phosphorylation and Nuclear Localization Reducing Apoptosis and Enhancing Cardiac Repair. Theranostics. 2017;7:3842–3855. doi: 10.7150/thno.19764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen C.Y., Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science. 1995;268:415–417. doi: 10.1126/science.7536344. [DOI] [PubMed] [Google Scholar]
- 78.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M. Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E. Translation of CircRNAs. Mol. Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu C., Yao M.-D., Li C.-P., Shan K., Yang H., Wang J.-J., Liu B., Li X.M., Yao J., Jiang Q., Yan B. Silencing Of Circular RNA-ZNF609 Ameliorates Vascular Endothelial Dysfunction. Theranostics. 2017;7:2863–2877. doi: 10.7150/thno.19353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peng L., Chen G., Zhu Z., Shen Z., Du C., Zang R., Su Y., Xie H., Li H., Xu X. Circular RNA ZNF609 functions as a competitive endogenous RNA to regulate AKT3 expression by sponging miR-150-5p in Hirschsprung’s disease. Oncotarget. 2017;8:808–818. doi: 10.18632/oncotarget.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang M., Huang N., Yang X., Luo J., Yan S., Xiao F., Chen W., Gao X., Zhao K., Zhou H. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 84.Qin M., Liu G., Huo X., Tao X., Sun X., Ge Z., Yang J., Fan J., Liu L., Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 85.Vausort M., Salgado-Somoza A., Zhang L., Leszek P., Scholz M., Teren A., Burkhardt R., Thiery J., Wagner D.R., Devaux Y. Myocardial Infarction-Associated Circular RNA Predicting Left Ventricular Dysfunction. J. Am. Coll. Cardiol. 2016;68:1247–1248. doi: 10.1016/j.jacc.2016.06.040. [DOI] [PubMed] [Google Scholar]
- 86.Salgado-Somoza A., Zhang L., Vausort M., Devaux Y. The circular RNA MICRA for risk stratification after myocardial infarction. Int. J. Cardiol. Heart Vasc. 2017;17:33–36. doi: 10.1016/j.ijcha.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vo J.N., Cieslik M., Zhang Y., Shukla S., Xiao L., Zhang Y., Wu Y.M., Dhanasekaran S.M., Engelke C.G., Cao X. The Landscape of Circular RNA in Cancer. Cell. 2019;176:869–881.e13. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jessup M., Brozena S. Heart failure. N. Engl. J. Med. 2003;348:2007–2018. doi: 10.1056/NEJMra021498. [DOI] [PubMed] [Google Scholar]
- 90.Jakobi T., Czaja-Hasse L.F., Reinhardt R., Dieterich C. Profiling and Validation of the Circular RNA Repertoire in Adult Murine Hearts. Genomics Proteomics Bioinformatics. 2016;14:216–223. doi: 10.1016/j.gpb.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zou M., Huang C., Li X., He X., Chen Y., Liao W., Liao Y., Sun J., Liu Z., Zhong L., Bin J. Circular RNA expression profile and potential function of hsa_circRNA_101238 in human thoracic aortic dissection. Oncotarget. 2017;8:81825–81837. doi: 10.18632/oncotarget.18998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu H.-J., Zhang C.-Y., Zhang S., Chang M., Wang H.-Y. Microarray Expression Profile of Circular RNAs in Heart Tissue of Mice with Myocardial Infarction-Induced Heart Failure. Cell. Physiol. Biochem. 2016;39:205–216. doi: 10.1159/000445617. [DOI] [PubMed] [Google Scholar]
- 93.Thum T., Condorelli G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ. Res. 2015;116:751–762. doi: 10.1161/CIRCRESAHA.116.303549. [DOI] [PubMed] [Google Scholar]
- 94.Bär C., Chatterjee S., Thum T. Long Noncoding RNAs in Cardiovascular Pathology, Diagnosis, and Therapy. Circulation. 2016;134:1484–1499. doi: 10.1161/CIRCULATIONAHA.116.023686. [DOI] [PubMed] [Google Scholar]
- 95.Wang K., Long B., Liu F., Wang J.-X., Liu C.-Y., Zhao B., Zhou L.Y., Sun T., Wang M., Yu T. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 96.Wang K., Gan T.-Y., Li N., Liu C.-Y., Zhou L.-Y., Gao J.-N., Chen C., Yan K.W., Ponnusamy M., Zhang Y.H., Li P.F. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111–1120. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tang C.-M., Zhang M., Huang L., Hu Z.Q., Zhu J.-N., Xiao Z., Zhang Z., Lin Q.X., Zheng X.L., -Yang M. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci. Rep. 2017;7:40342. doi: 10.1038/srep40342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhou B., Yu J.-W. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-β1. Biochem. Biophys. Res. Commun. 2017;487:769–775. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 99.Deng Y.-Y., Zhang W., She J., Zhang L., Chen T., Zhou J., Yuan Z. GW27-e1167 Circular RNA Related to PPARγ Function as ceRNA of microRNA in Human Acute Myocardial Infarction. J. Am. Coll. Cardiol. 2016;68(16, Suppl S):C51–C52. [Google Scholar]
- 100.Fujio Y., Nguyen T., Wencker D., Kitsis R.N., Walsh K. Akt promotes survival of cardiomyocytes in vitro and protects against ischemia-reperfusion injury in mouse heart. Circulation. 2000;101:660–667. doi: 10.1161/01.cir.101.6.660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matsui T., Tao J., del Monte F., Lee K.H., Li L., Picard M., Force T.L., Franke T.F., Hajjar R.J., Rosenzweig A. Akt activation preserves cardiac function and prevents injury after transient cardiac ischemia in vivo. Circulation. 2001;104:330–335. doi: 10.1161/01.cir.104.3.330. [DOI] [PubMed] [Google Scholar]
- 102.Khan M.A., Reckman Y.J., Aufiero S., van den Hoogenhof M.M., van der Made I., Beqqali A., Koolbergen D.R., Rasmussen T.B., van der Velden J., Creemers E.E., Pinto Y.M. RBM20 Regulates Circular RNA Production From the Titin Gene. Circ. Res. 2016;119:996–1003. doi: 10.1161/CIRCRESAHA.116.309568. [DOI] [PubMed] [Google Scholar]
- 103.Pan R.-Y., Liu P., Zhou H.-T., Sun W.-X., Song J., Shu J., Cui G.J., Yang Z.J., Jia E.Z. Circular RNAs promote TRPM3 expression by inhibiting hsa-miR-130a-3p in coronary artery disease patients. Oncotarget. 2017;8:60280–60290. doi: 10.18632/oncotarget.19941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zheng C., Niu H., Li M., Zhang H., Yang Z., Tian L., Wu Z., Li D., Chen X. Cyclic RNA hsa-circ-000595 regulates apoptosis of aortic smooth muscle cells. Mol. Med. Rep. 2015;12:6656–6662. doi: 10.3892/mmr.2015.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dang R.Y., Liu F.L., Li Y. Circular RNA hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186/HIF-1α axis. Biochem. Biophys. Res. Commun. 2017;490:104–110. doi: 10.1016/j.bbrc.2017.05.164. [DOI] [PubMed] [Google Scholar]
- 106.Boeckel J.N., Jaé N., Heumüller A.W., Chen W., Boon R.A., Stellos K., Zeiher A.M., John D., Uchida S., Dimmeler S. Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA. Circ. Res. 2015;117:884–890. doi: 10.1161/CIRCRESAHA.115.306319. [DOI] [PubMed] [Google Scholar]
- 107.Yang P., Qiu Z., Jiang Y., Dong L., Yang W., Gu C., Li G., Zhu Y. Silencing of cZNF292 circular RNA suppresses human glioma tube formation via the Wnt/β-catenin signaling pathway. Oncotarget. 2016;7:63449–63455. doi: 10.18632/oncotarget.11523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Holdt L.M., Stahringer A., Sass K., Pichler G., Kulak N.A., Wilfert W., Kohlmaier A., Herbst A., Northoff B.H., Nicolaou A. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016;7:12429. doi: 10.1038/ncomms12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Burd C.E., Jeck W.R., Liu Y., Sanoff H.K., Wang Z., Sharpless N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Devaux Y., Vausort M., McCann G.P., Zangrando J., Kelly D., Razvi N., Zhang L., Ng L.L., Wagner D.R., Squire I.B. MicroRNA-150: a novel marker of left ventricular remodeling after acute myocardial infarction. Circ Cardiovasc Genet. 2013;6:290–298. doi: 10.1161/CIRCGENETICS.113.000077. [DOI] [PubMed] [Google Scholar]
- 111.Zhao Z., Li X., Gao C., Jian D., Hao P., Rao L., Li M. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci. Rep. 2017;7:39918. doi: 10.1038/srep39918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yu R.Z., Grundy J.S., Geary R.S. Clinical pharmacokinetics of second generation antisense oligonucleotides. Expert Opin. Drug Metab. Toxicol. 2013;9:169–182. doi: 10.1517/17425255.2013.737320. [DOI] [PubMed] [Google Scholar]
- 113.Piwecka M., Glažar P., Hernandez-Miranda L.R., Memczak S., Wolf S.A., Rybak-Wolf A., Filipchyk A., Klironomos F., Cerda Jara C.A., Fenske P. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357 doi: 10.1126/science.aam8526. eaam8526. [DOI] [PubMed] [Google Scholar]
- 114.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., Verdine V., Cox D.B.T., Kellner M.J., Regev A. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cox D.B.T., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Barrett S.P., Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–1847. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Meganck R.M., Borchardt E.K., Castellanos Rivera R.M., Scalabrino M.L., Wilusz J.E., Marzluff W.F., Asokan A. Tissue-Dependent Expression and Translation of Circular RNAs with Recombinant AAV Vectors In Vivo. Mol. Ther. Nucleic Acids. 2018;13:89–98. doi: 10.1016/j.omtn.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jaski B.E., Jessup M.L., Mancini D.M., Cappola T.P., Pauly D.F., Greenberg B., Borow K., Dittrich H., Zsebo K.M., Hajjar R.J., Calcium Up-Regulation by Percutaneous Administration of Gene Therapy In Cardiac Disease (CUPID) Trial Investigators Calcium upregulation by percutaneous administration of gene therapy in cardiac disease (CUPID Trial), a first-in-human phase 1/2 clinical trial. J. Card. Fail. 2009;15:171–181. doi: 10.1016/j.cardfail.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jessup M., Greenberg B., Mancini D., Cappola T., Pauly D.F., Jaski B., Yaroshinsky A., Zsebo K.M., Dittrich H., Hajjar R.J., Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID) Investigators Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID): a phase 2 trial of intracoronary gene therapy of sarcoplasmic reticulum Ca2+-ATPase in patients with advanced heart failure. Circulation. 2011;124:304–313. doi: 10.1161/CIRCULATIONAHA.111.022889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kaczmarek J.C., Kowalski P.S., Anderson D.G. Advances in the delivery of RNA therapeutics: from concept to clinical reality. Genome Med. 2017;9:60. doi: 10.1186/s13073-017-0450-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wesselhoeft R.A., Kowalski P.S., Anderson D.G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 2018;9:2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Petkovic S., Müller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43:2454–2465. doi: 10.1093/nar/gkv045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dieterich C., Papantonis A. Springer Nature; 2018. Circular RNAs: Methods and Protocols. [Google Scholar]