Summary
Objective : To summarize recent research and to propose a selection of best papers published in 2018 in the field of Clinical Information Systems (CIS).
Method : Each year a systematic process is carried out to retrieve articles for the CIS section of the IMIA Yearbook of Medical Informatics and to select a set of pest papers for the section. The same query as in the last five years was used. The retrieved articles were categorized in a multi-pass review carried out by the two section editors. The final selection of candidate papers was then peer-reviewed by Yearbook editors and external reviewers. Based on the review results the best papers were then chosen at the selection meeting with the IMIA Yearbook editorial board. Text mining, and term co-occurrence mapping techniques were again used to get an overview of the content of the retrieved articles.
Results : The query was carried out in mid-January 2019, yielding a consolidated, deduplicated result set of 2,264 articles which had been published in 957 different journals. This year, we nominated twelve papers as candidates and three of them were finally selected as best papers in the CIS section. Again, the content analysis of the articles revealed the broad spectrum of topics which is covered by CIS research.
Conclusions : We could observe ongoing trends from our 2017 analysis. The patient increasingly moves in the focus of the research activities and trans-institutional aggregation of data is still an important field of work. The move to use patient and other clinical data directly for the patient and to support data driven process management, the move away from clinical documentation to patient-focused knowledge generation and support of informed decision, is gaining momentum by the application of new or already known but, due to technological advances, now applicable methodological approaches.
Keywords: Medical informatics, International Medical Informatics Association, Yearbook, Clinical Information Systems
Introduction
In our synopsis for the 2018 issue of the IMIA Yearbook of Medical Informatics 1 we concluded that, modern clinical information systems served as backbone for a very complex, trans-institutional information logistics process and that data was more and more reused for multiple purposes. Last year we found a lot of examples showing the benefits and novel approaches to tackle the challenges of such data reuse. And we found that the patient was moving in the focus of interest of CIS research. This year we observed similar results.
About the Paper Selection
The selection process in the CIS section is stable now for five years. It is described in detail in 1 and the full queries are available upon request from the corresponding authors.
The queries were carried out in mid-January 2018. This year the search result set comprised 2,264 unique papers. From these papers 2,160 were retrieved from PubMed and 104 additional publications were found in Web of Science ® . The resulting articles had been published in 957 different journals. Table 1 depicts the top twenty journals with the highest numbers of resulting articles. This year we used RAYYAN, an online systematic review tool 2 to carry out the multipass review by the two section editors (AH, WOH). Both section editors independently reviewed all 2,264 publications. In the first step, ineligible articles were excluded based on their titles and/or abstracts (section editor 1: n=2,184; section editor 2: n=2,140). In this first step, the agreement between the two editors was n=2,089 for “exclude” and n=29 for “not exclude” (i.e. include). The remaining 146 conflicts were solved on mutual consent which resulted in nine additional inclusions. The final candidate selection from the remaining 38 publications was done based on full text review and yielded 12 candidate papers for the CIS section 2019.
This list was then reviewed by the Yearbook editors who checked if any articles had also been selected for other sections. As in the last year, no overlaps were found, and all twelve candidates were sent in the peer-review process for the Yearbook. For each paper at least five independent reviews were collected. During the selection meeting held on April 26, 2019 in Paris where section editors from all Yearbook sections participated, three papers 3 – 5 were finally selected as best papers for the CIS section (Table 2). A content summary of these three best CIS papers can be found in the appendix of this synopsis.
Findings and Trends: Clinical Information Systems Research 2018
During the selection of the best papers we as section editors get a broad overview on the research field of our CIS section. Of course this overview may be biased and to avoid selective perception we again applied our more formal text mining and bibliometric network visualizing approach 6 to summarize the content of titles and abstracts of the articles in our CIS result set as in the previous years 1 , 7 , 8 .
This year we also extracted the 21,264 authors’ keywords from all articles and present their frequency in a tag cloud (cf. figure 1 ). We found 7,250 different key words, of which 5,149 were only used once. Most frequent keywords were “human” (n = 699) followed by “female” (n = 296), “male” (n = 263), “electronic health record(s)” (n = 235), “adult” (n = 230), “child” (n = 191) and “health communication” (n = 164).
Fig. 1.
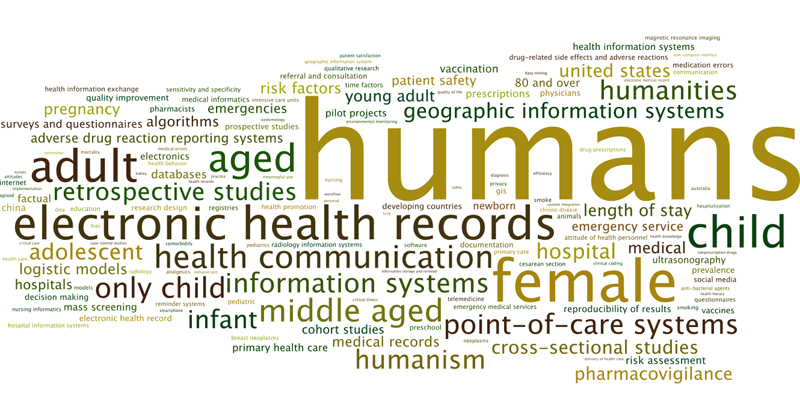
Tag cloud illustrating the frequency authors’ keywords (only top kewords out of n = 7,250 are shown) within the 2,264 papers from the CIS query result set. Font size corresponds to frequency (most frequent keyword was “humans” n = 699)
Figures 2 and 3 depict the resulting co-occurrence maps of the top-100 terms from the titles and of the most relevant terms (top 60 percent, n =524) from the abstracts of the 2,264 papers of the recent CIS result set.
Fig. 2.
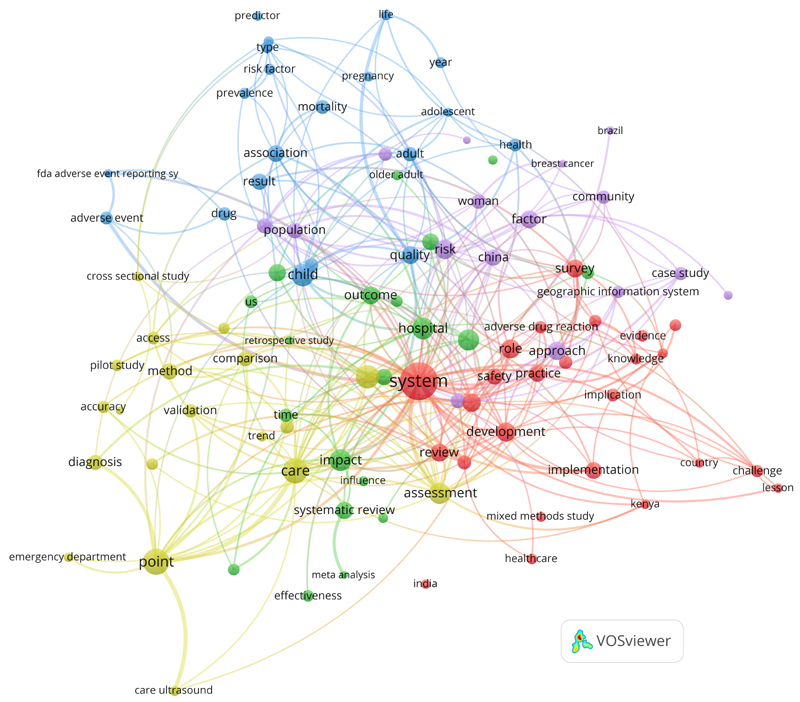
Clustered co-occurrence map of the most relevant terms (top-100) from the titles of the 2,264 papers in the 2019 CIS query result set. Node size corresponds to the frequency of the terms (binary count, once per paper). Edges indicate co-occurrence and distance of nodes corresponds to the association strength of the terms within the titles (only top 200 out of 455 edges are shown). Colors represent the five different clusters. The network was created with VOSviewer 9 .
Fig. 3.
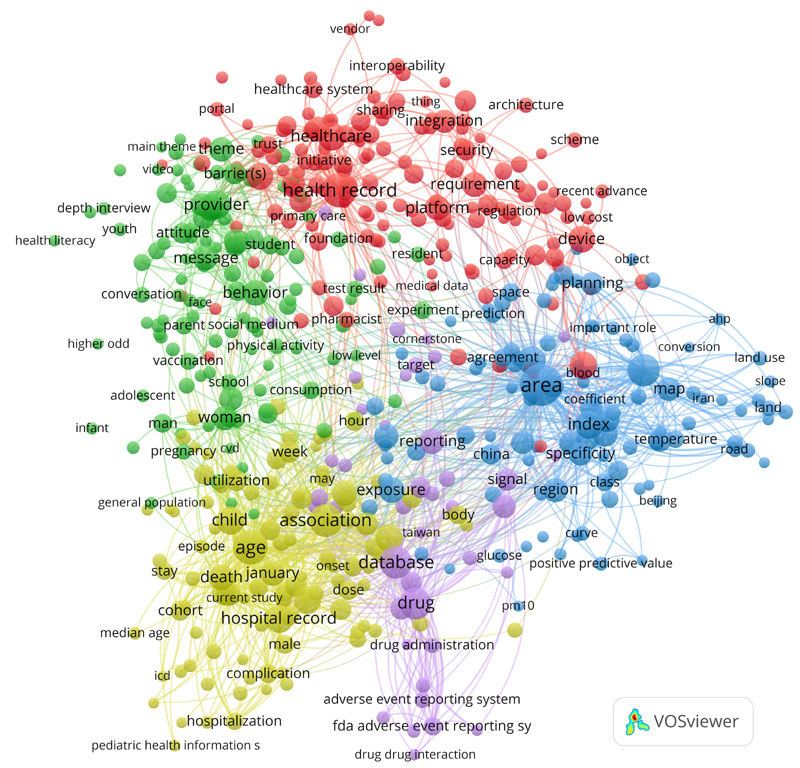
Clustered co-occurrence map of the most relevant terms (top 60 percent, n=524) from the abstracts of the 2,264 papers in the 2019 CIS query result set. Only terms that were found in at least seven different papers were included in the analysis. Node size corresponds to the frequency of the terms (binary count, once per paper). Edges indicate co-occurrence and distance of nodes corresponds to the association strength of the terms within the texts (only top 1,000 of 27,021 edges are shown). Colors represent the five different clusters. The network was created with VOSviewer 9 .
In 2017 we discovered six different clusters of terms within the titles, last year four and this year we found five main clusters. Whereas “human” was the top authors’ keyword in the articles, the analysis of the titles revealed “system” as central hub for these five clusters.
Table 1. Number of retrieved articles for Top-20 journals.
| Journal (Total Number of Journals = 957) | Number of papers |
|---|---|
| PLOS ONE | 51 |
| JOURNAL OF THE AMERICAN MEDICAL INFORMATICS ASSOCIATION | 45 |
| HEALTH COMMUNICATION | 41 |
| APPLIED CLINICAL INFORMATICS | 36 |
| JOURNAL OF MEDICAL INTERNET RESEARCH | 35 |
| INTERNATIONAL JOURNAL OF MEDICAL INFORMATICS | 34 |
| INTERNATIONAL JOURNAL OF ENVIRONMENTAL RESEARCH AND PUBLIC HEALTH | 32 |
| INTERNATIONAL JOURNAL OF CLINICAL PHARMACY | 24 |
| JOURNAL OF MEDICAL SYSTEMS | 24 |
| BMJ OPEN | 23 |
| BMC HEALTH SERVICES RESEARCH | 22 |
| DRUG SAFETY19COMPUTERS | 17 |
| JOURNAL OF HEALTH COMMUNICATION | 15 |
| JOURNAL OF EVALUATION IN CLINICAL PRACTICE | 14 |
| INFORMATICS FOR HEALTH & SOCIAL CARE | 14 |
| ENVIRONMENTAL MONITORING AND ASSESSMENT | 14 |
| BMC PUBLIC HEALTH | 14 |
| PATIENT EDUCATION AND COUNSELING | 13 |
| BMC MEDICAL INFORMATICS AND DECISION MAKING | 12 |
Table 2. Best paper selection of articles for the IMIA Yearbook of Medical Informatics 2019 in the section ‘Clinical Information Systems’. The articles are listed in alphabetical order of the first author’s surname.
| Section Clinical Information Systems |
|---|
| • Eichstaedt JC, Smith RJ, Merchant RM, Ungar LH, Crutchley P, Preoiuc-Pietro D Asch DA, Schwartz HA. Facebook language predicts depression in medical records. Proc Natl Acad Sci U S A 2018;115(44):11203-8. • Parr SK, Shotwell MS, Jeffery AD, Lasko TA, Matheny ME. Automated mapping of laboratory tests to LOINC codes using noisy labels in a national electronic health record system database. J Am Med Inform Assoc 2018;25(10):1292-300. • Xiao C, Ma T, Dieng AB, Blei DM, Wang F. Readmission prediction via deep contextual embedding of clinical concepts. PLoS One 2018;13(4):e01 95024. |
The cluster analysis of the abstracts yielded also five clusters (cf. figure 3 ). The red cluster on top comprises terms connected with electronic health records (EHR) and requirements, data sharing, interoperability, architecture and integration. The blue cluster on the right side is dedicated to geographic information systems and related terms. The violet cluster on the bottom and center represents medication and adverse event related research. The yellow cluster on the bottom left as well as the green cluster on the left side can be seen as containers for context factors mentioned in the abstracts.
As in the previous years, the results of these analyses demonstrate the wide variety of the CIS domain. Again, we found a multitude of well written and highly interesting articles of first-rate quality. Among these we interestingly found a high proportion of articles from Chinese researchers. This is also reflected in our candidate selection where half of the twelve papers come from China or involve Chinese contributors.
From a thematic perspective we identified a growing proportion of contributions showing that the potentials and possibilities due to technological advances, increased computing power, availability of a huge amount of structured data and sophisticated algorithms now can be unlocked.
The first of the best papers in the CIS section 2019 is an example for that. Cao Xiao and colleagues developed a novel deep learning model that learns distributed patient representation from the EHR data and performs prediction for the 30-day readmissions 10 . This inspiring paper is a very well-presented example of how deep learning techniques can be applied to distill new knowledge out from existing EHR data. The approach is highly innovative, and the developed model outperforms state-of-the art approaches in readmission prediction. Another well-presented example of a powerful risk prediction model (1-year incident hypertension) that is based on and validated using a huge amount of patient data by Chengyin Ye and colleagues 11 can be found among the candidate papers.
An “evergreen” within the scope of CIS research is the detection of adverse events within EHR data. This year we chose a contribution by Jiebin Chu and colleagues who used neural attention networks for that 12 . Data Mining also was an important keyword during the last years in CIS research. This year we didn’t find this keyword very often (cf. figure 1 ). However, a very interesting article introducing a novel data or text mining technique termed as optimized swarm search based feature selection 13 coming from Daohui Zeng and colleagues was included in the selection of candidate papers. The “newcomer” Blockchain as identified last year was also a prominent research topic in 2018. We included a contribution by Hongyu Li and colleagues on a blockchain-based data preservation system for medical data in our CIS best paper candidate pool.
The second of the actual best papers tackles an important problem within the domain of clinical information systems in a very inspiring way. Sharidan K. Parr and colleagues developed and applied a machine learning approach that leverages “noisy labels” for automatic mapping of an enormous amount of laboratory data (>6.5 billion lab test results) to LOINC codes 14 . Among the candidate papers another one by Ronald George Hauser and colleagues dealt in a very inspiring way with the standardization of laboratory test results 15 .
An increasing number of research articles deals with process mining or data driven process modeling in the healthcare domain. So for example the contribution from Jingfeng Chen and colleagues on automatic extraction of typical treatment processes from electronic health record data 16 .
On the other hand, clinical processes themselves can also deliver data that can be used for different analyses. Kaat De Pourcq and colleagues present a step-by-step methodology to implement an automated process-oriented performance measurement system for hospitals 17 .
Besides all benefits and possibilities, the use of massive EHR data sets can bring, we have to keep in mind that reality and data not necessarily are congruent. Without careful consideration of the context in which these data are produced many pitfalls and sources of bias may be overlooked. Denis Agniel and colleagues, with their very worth reading candidate paper, remind us of this eternal truth. Another core aspect that may not be overlooked within the CIS domain are the users. Pantelis Natsiavas and colleagues propose a comprehensive user requirements methodology for secure and interoperable health data exchange 18 . This candidate paper is worth reading as it also provides a comprehensive set of user requirements and presents sets of barriers as well as facilitators for health IT solutions.
Last but not least we want to highlight the third of the best papers in the CIS section 2019. In their article which received top ratings from the IMIA Yearbook reviewers Johannes C Eichstaedt and colleagues show that the content shared by consenting users on Facebook can predict a future diagnosis of depression 3 . This study suggests that social media content may point clinicians to specific symptoms of depression. It sheds a new light on privacy and anonymity with regard to health information and vividly indicates that health related information can not only be retrieved from “core” medical and health data, but also from other user provided and publicly available data, such as social media content.
As every year, at the very end of our review of findings and trends for the clinical information systems section, we want to recommend a reading of this year’s survey article in the CIS section 19 .
Conclusions and Outlook
We could observe ongoing trends from our 2017 analysis that the patient increasingly moves in the focus of the research activities and that the trans-institutional aggregation of data is still an important field of work. Common to these efforts is the creation of benefits for the patients. A key differentiator from what we observed in the last years are the methods applied to reach this goal. As already been described in the findings we found a great number of papers - amongst them the best papers for 2018 - that were tackling problems using machine learning approaches combining them with relatively large and sometimes immense clinical data sets or so far uncommon data sets outside the core clinical domain. Apart from these ongoing developments we have also found inspiring work that deals with data driven management of processes and the use of blockchain technology to support data aggregation beyond institutional boundaries. The move to use patient data directly for the patient, to move away from clinical documentation to patient-focused knowledge generation and support of informed decision, is gaining momentum by the application of new or already known but, due to technological advances, now applicable methodological approaches.
Acknowledgements
We would like to acknowledge the support of Lina Soualmia, Adrien Ugon, Brigitte Seroussi, Martina Hutter and the whole Yearbook editorial team as well as the numerous reviewers in the selection process of the best papers.
Footnotes
* Equal Contribution
References
- 1.Hackl W, Hoerbst A. Section Editors for the IMIA Yearbook Section on Clinical Information Systems. On the Way to Close the Loop in Information Logistics: Data from the Patient – Value for the Patient. Yearb Med Inform. 2018;27(01):91–7. doi: 10.1055/s-0038-1667076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ouzzani M, Hammady H, Fedorowicz Z, Elmagarmid A. Rayyan-a web and mobile app for systematic reviews. Syst Rev. 2016;5(01):210. doi: 10.1186/s13643-016-0384-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eichstaedt J C, Smith R J, Merchant R M, Ungar L H, Crutchley P, Preojiuc-Pietro D et al. Facebook language predicts depression in medical records. Proc Natl Acad Sci U S A. 2018;115(44):11203–8. doi: 10.1073/pnas.1802331115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parr S K, Shotwell M S, Jeffery A D, Lasko T A, Matheny M E. Automated mapping of laboratory tests to LOINC codes using noisy labels in a national electronic health record system database. J Am Med Inform Assoc. 2018;25(10):1292–300. doi: 10.1093/jamia/ocy110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao C, Ma T, Dieng A B, Blei D M, Wang F. Readmission prediction via deep contextual embedding of clinical concepts. PLoS One. 2018;13(04):e0195024. doi: 10.1371/journal.pone.0195024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waltman L, van Eck N J, Noyons E CM. A unified approach to mapping and clustering of bibliometric networks. J Informetr. 2010;4(04):629–35. [Google Scholar]
- 7.Hackl W O, Ganslandt T.New Problems - New Solutions: A Never Ending Story Findings from the Clinical Information Systems Perspective for 2015. Yearb Med Inform 2016(1):146–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hackl W O, Ganslandt T. Clinical Information Systems as the Backbone of a Complex Information Logistics Process: Findings from the Clinical Information Systems Perspective for 2016. Yearb Med Inform. 2017;26(01):103–9. doi: 10.15265/IY-2017-023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Eck N J, Waltman L. Software survey: VOSviewer, a computer program for bibliometric mapping. Scientometrics. 2010;84(02):523–38. doi: 10.1007/s11192-009-0146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao C, Ma T, Dieng A B, Blei D M, Wang F. Readmission prediction via deep contextual embedding of clinical concepts. PLoS One. 2018;13(04):e0195024. doi: 10.1371/journal.pone.0195024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye C, Fu T, Hao S, Zhang Y, Wang O, Jin B et al. Prediction of Incident Hypertension Within the Next Year: Prospective Study Using Statewide Electronic Health Records and Machine Learning. J Med Internet Res. 2018;20(01):e22. doi: 10.2196/jmir.9268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu J, Dong W, He K, Duan H, Huang Z.Using neural attention networks to detect adverse medical events from electronic health records J Biomed Inform [Internet] 201887118–30.Available from:https://www.ncbi.nlm.nih.gov/pubmed/30336262 [DOI] [PubMed] [Google Scholar]
- 13.Zeng D, Peng J, Fong S, Qiu Y, Wong R. Medical data mining in sentiment analysis based on optimized swarm search feature selection. Australas Phys Eng Sci Med. 2018;41(04):1087–100. doi: 10.1007/s13246-018-0674-3. [DOI] [PubMed] [Google Scholar]
- 14.Parr S K, Shotwell M S, Jeffery A D, Lasko T A, Matheny M E. Automated mapping of laboratory tests to LOINC codes using noisy labels in a national electronic health record system database. J Am Med Informatics Assoc. 2018;25(10):1292–300. doi: 10.1093/jamia/ocy110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hauser R G, Quine D B, Ryder A. LabRS: A Rosetta stone for retrospective standardization of clinical laboratory test results. J Am Med Inform Assoc. 2018;25(02):121–6. doi: 10.1093/jamia/ocx046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J, Sun L, Guo C, Wei W, Xie Y. A data-driven framework of typical treatment process extraction and evaluation. J Biomed Inform. 2018;83:178–95. doi: 10.1016/j.jbi.2018.06.004. [DOI] [PubMed] [Google Scholar]
- 17.De Pourcq K, Gemmel P, Devis B, Van Ooteghem J, De Caluwe T, Trybou J. A three-step methodology for process-oriented performance: how to enhance automated data collection in healthcare. Informatics Heal Soc Care. 2018;00(00):1–13. doi: 10.1080/17538157.2018.1496087. [DOI] [PubMed] [Google Scholar]
- 18.Natsiavas P, Rasmussen J, Voss-Knude M, Votis K, Coppolino L, Campegiani P et al. Comprehensive user requirements engineering methodology for secure and interoperable health data exchange. BMC Med Inform Decis Mak. 2018;18(01):85. doi: 10.1186/s12911-018-0664-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Combi C, Pozzi G. Clinical Information Systems and Artificial Intelligence: Recent Research Trends. Yearb Med Inform. 2019:83–94. doi: 10.1055/s-0039-1677915. [DOI] [PMC free article] [PubMed] [Google Scholar]


