Summary
Objectives : There exists a communication gap between the biomedical informatics community on one side and the computer science/artificial intelligence community on the other side regarding the meaning of the terms “semantic integration" and “knowledge representation“. This gap leads to approaches that attempt to provide one-to-one mappings between data elements and biomedical ontologies. Our aim is to clarify the representational differences between traditional data management and semantic-web-based data management by providing use cases of clinical data and clinical research data re-representation. We discuss how and why one-to-one mappings limit the advantages of using Semantic Web Technologies (SWTs).
Methods : We employ commonly used SWTs, such as Resource Description Framework (RDF) and Ontology Web Language (OWL). We reuse pre-existing ontologies and ensure shared ontological commitment by selecting ontologies from a framework that fosters community-driven collaborative ontology development for biomedicine following the same set of principles.
Results : We demonstrate the results of providing SWT-compliant re-representation of data elements from two independent projects managing clinical data and clinical research data. Our results show how one-to-one mappings would hinder the exploitation of the advantages provided by using SWT.
Conclusions : We conclude that SWT-compliant re-representation is an indispensable step, if using the full potential of SWT is the goal. Rather than providing one-to-one mappings, developers should provide documentation that links data elements to graph structures to specify the re-representation.
Keywords: Semantic Web, artificial intelligence, knowledge management, common data model
1 Introduction
1.1 A Communication Gap in Biomedical Informatics
The technical means that enable data sharing and data integration are a key problem in biomedical data management. Integration of data can happen at multiple levels, and semantic integration is the second to last integration level, only followed by shared business process, according to Blobel and Oemig 1 . Semantic integration aims to preserve “the detail, uncertainty, and above all the context of the data involved” 2 . Ontologies are an integral part of current semantic integration approaches. To achieve computer-assisted integration solutions, ontologies should be machine-interpretable and thus, need to provide information about details, uncertainty, and context in a computer-interpretable language 2 (as opposed to textual definitions written in any given natural language). Ontologies are an increasingly popular and successful tool for encoding and sharing machine-interpretable knowledge, such as background information about an area of biomedicine or other domains, as well as general information about the structure of the world such as is provided by upper- level ontologies like the BFO (Basic Formal Ontology) or the SUMO (Suggested Upper Merged Ontology) 3 . These ontologies are usually implemented and distributed as OWL (Web Ontology Language) files 4 , 5 containing logical definitions.
In a 2018 paper, Brochhausen et al. indicated a communication gap in biomedical informatics regarding the interpretation of the term “semantic integration” and, more generally, “semantics” 6 . They showed how common data models (CDMs) were cited as fostering semantic integration or providing “semantics” despite their lack of representation of detail-oriented contextual information expressing levels of diagnostic confidence (suspected vs. confirmed, etc.) provided in a machine-interpretable language. This shows that the interpretation of the terms “sematic interpretation” and “semantics” differs between the biomedical informatics community and the computer science/big data community, which is found in the publication by Cheatham and Pesquita 2 . To help mitigate that situation and address issues of the ability of resources (such as ontologies, controlled vocabularies, and terminologies) to contribute to semantic integration, Brochhausen et al. proposed “computable semantics” as a baseline to establish whether a resource is capable of supporting semantic integration 6 . For a resource to provide computable semantics means there must be an effective method that could assign or validate the meaning of the symbols and expressions. In logic and mathematics, an effective method (sometimes also called mechanical method) is a method that allows to compute the answer to a given problem in a finite number of steps, and is logically bound to give the correct answer (and no wrong answers) 7 .
Utecht et al. 8 have shown one way of demonstrating that an ontology-driven system entails the capability to provide computable semantics. For a project managing drug-drug interaction evidence information, they created an ontology that represented 44 different evidence types (such as longitudinal studies, observational studies, etc.) completed with necessary and sufficient conditions for class inclusion. In a pilot test, the team retrieved 30 evidence items (e.g. a journal paper), that had previously been assigned to one of the evidence types manually. In the test, a person had to answer five questions about each evidence item filling in a web-based form. As they were captured, the answers about these evidence items were used to generate Resource Description Framework (RDF) representations. Based on the information entered and the axiomatic definitions of the evidence types, running the OWL reasoner HermiT ( http://www.hermit-reasoner.com ) sorted all evidence items correctly into evidence types.
Providing the ability to automatically sort data items, e.g. diagnoses based on properties such as anatomical location, has inspired developments in clinical vocabularies, specifically the SNOMED Clinical Terms (SNOMED CT), over the last years. For example, the SNOMED CT representation of “Herpes simplex iridocyclitis” (SCTID: 13608004) specifies in a machine interpretable language 9 that the finding site for this disorder is the “ciliary body structure” or “iris structure”, the causative agent is “human herpes simplex virus”, the associated morphology is “inflammation”, and the pathological process is “infection”. This means that any instance of a disorder that does not fulfill these criteria would not be sorted in that category. These specifications would potentially also allow the validation of clinical coding by checking whether the finding site, causative agent, associated morphology, and pathological agent specified elsewhere in the medical record are consistent with the code. Miñarro-Giménez et al. have made the point that an increase in formal logical axioms to SNOMED CT would help to overcome still existent low code coincidence between annotators 10 .
A recent paper that aimed to assess knowledge representation of clinical data across health systems demonstrated the existence of a communication gap regarding the term “knowledge representation”, in particular in distinction to “data representation”. Rosenbloom et al. 11 assessed three commonly used standards for sharing clinical data: Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) 12 , 13 , PCORNet (National Patient-Centered Clinical Research Network) CDM 14 , 15 , and Health Level Seven International (HL7) Fast Healthcare Interoperability Resource (FHIR) 16 . While we agree with the authors that the resources reviewed in their paper contribute to “a recent growth in high-impact efforts to support quality-assured and standardized clinical data sharing across different institutions and EHR (Electronic Health Record) systems”, we do not agree with those resources contributing to better knowledge representation and, thus, fostering semantic integration.
Rosenbloom et al. do not provide a definition of knowledge representation, but use “knowledge representation” as a search term for their review. According to Davis et al. 17 , knowledge representation is best understood by looking at five distinct “roles” that it plays: (i) it is based on a surrogate, a representation of the entities in the world; (ii) it consists of a set of ontological commitments; (iii) it provides a fragmentary theory of intelligent reasoning, including rules of inference; (iv) it acts as a medium of pragmatically efficient computation; and (v) it is a medium of human expression. Knowledge representation or knowledge representation and reasoning (KRR), as it is sometimes called, is a subfield of artificial intelligence and has a long history dating back to the early days of symbolic Artificial Intelligence (AI). The reasoning aspect of KRR, i.e. , the capability of a computer system to automatically draw inferences based on a set of inference rules, is what allows for filling the roles (iii) and (iv) of Davis’ definition of knowledge representation.
Brochhausen et al. showed that neither OMOP CDM, nor PCORNet CDM exhibit roles (iii) and (iv) defining knowledge representation 6 . Hence, they do not provide knowledge representation in the sense of computer/information science. FHIR does provide avenues to fill roles (iii) and (iv). While an extensive review of FHIR’s knowledge representation capabilities is out of the scope of this paper, Martinez-Costa and Schulz have pointed out from the perspective of knowledge representation that despite the fact that FHIR at the time of their writing (2017) required some manual effort, there are feasible strategies to use FHIR for knowledge representation and semantic integration 18 .
A rich corpus of literature about the lack of reliability in coding clinical data 10 , 19 20 21 22 23 demonstrates the reason why axiomatic definitions, even for tasks or databases that do not (yet) explicitly require reasoning, are relevant. Without the capability of using an effective method to ascertain the correctness, consistency, and reliability of coding, semantic integration will not be possible in a way that can be validated.
1.2 Using Semantic Web Technologies for Biomedical Data - an Engineering-oriented Perspective
The initial motivation for semantic Web technologies (SWTs) was to enable computers to play a more active role in handling, organizing, and managing data on the Internet:
“The concept of machine-understandable documents does not imply some magical artificial intelligence allowing machines to comprehend human mumblings. It relies solely on the machine’s ability to solve well-defined problems by performing well-defined operations on well-defined data. So, instead of asking machines to understand people’s language, the new technology, like the old, involves asking people to make some extra effort, in repayment for which they get major new functionality–just as the extra effort of producing HTML mark-up is outweighed by the benefit of having content searchable on the web” 24 .
SWT include numerous key methodologies, but at its core is the Resource Description Framework (RDF) 25 . In RDF, information such as the fact that “Hydrogen potassium ATPase” is a “proton pump”, is captured by a statement that identifies the two entities about which the statement is made, and by specifying the relation that holds between the two. These statements are referred to as triples, because they consist of three parts: subject, predicate, and object 26 , 27 . The bold rectangle in Fig. 1 shows a representation of a triple. RDF uses unique resource identifiers (URIs) to refer to the entities and relationships in a domain 28 . Using URIs for the entities in the domain, such as “Hydrogen potassium ATPase” in the example in Figure 1, allows us to build complex and large graphs based on the simple triple structure. Due to the use of URIs, the two triples that contain Hydrogen potassium ATPase, [Omeprazole, inhibits, Hydrogen potassium ATPase] and [Hydrogen potassium ATPase, is a, Proton pump] get connected to build a small graph consisting of three nodes and two edges.
Fig. 1.
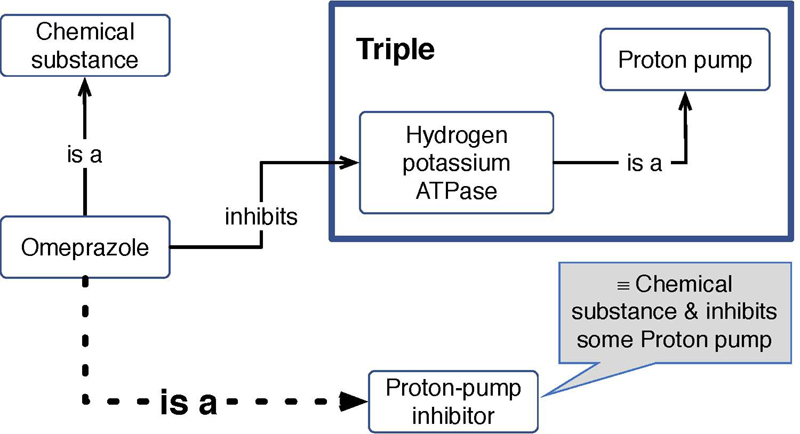
Example of using RDF triples and axioms to represent knowledge in the pharmacology domain. The transparent boxes represent RDF subjects and objects, the lines represent predicates between subject and object. The gray box shows a necessary and sufficient condition for being a member of the class 'Proton-pump inhibitor'. The dotted line represents an inferred relationship.
Through RDF together with languages to define controlled vocabularies and ontologies, such as RDF Schema (RDFS) 29 and OWL 30 , we can present the SWT knowledge representation strategy 26 , 27 . At the core of this representation strategy is the possibility to use formal logic to draw inferences from premises and axioms to make implicit information explicit. Figure 1 shows the example of a reasoner using the RDF statements and a necessary and sufficient condition for the class “Proton pump inhibitor” to infer the statement that “Omeprazole is a proton pump inhibitor”. The use of such axioms and rules of inference, for example in ontologies 6 , marks one of the key features of using SWTs.
The goal of this paper is not to claim that sharing and integration of clinical data requires SWTs, but the considerations presented above and in Brochhausen et al. clearly demonstrate that semantic integration requires a knowledge representation approach that is absent from both OMOP and PCORNet CDM. Those resources, of course, still provide value in biomedical informatics, but previous research indicates a number of use cases that require or benefit from using SWTs:
From the material presented above it is obvious that semantic web technologies are useful tools for all use cases where we seek to validate coding or automate a classification of cases into different categories 8 . Axiomatically rich ontologies have been shown to support a number of medically relevant functionalities such as automatic sorting of entities based on axiomatic definitions. Utecht et al. have shown that studies reporting evidence regarding drug-drug interactions can be sorted automatically into a complex system of study types using the Drug-drug Interaction and Drug-drug interaction Evidence Ontology (DIDEO) based on a six questions about the studies 8 .
SWTs are used to allow integration of structured, but uncoded data for clinical and clinical research purposes. Mate et al. demonstrated an ontology driven system to manage extract, transform, and load (ETL) procedures to reuse standard care data from electronic medical record (EMR) to answer research questions 31 .
Integrating heterogeneous uncoded but structured data describing instances of the same types of medical phenomena to allow either query data in a truth preserving manner using the biomedical context. Brochhausen et al. demonstrated that ontology-based representation was able to fix problems in querying biobank data from different biobanks at the same institution, by using RDF and the Ontology of Biobanking (OBIB) 32 .
SWTs have shown great promise in improving the curation and usage of drug- drug interaction information 33 34 35 36 .
These are, of course, only a few examples, illustrating the type of problems and the scope of applying semantic web technologies in the biomedical arena. A PubMed query for “’semantic web technologies’ OR ’semantic web technology’ OR SWT [all]” retrieved 560 hits in December 2018.
2 Objectives
In the daily practice of an SWT specialist working with clinical data and clinical research data, requests to map or annotate existing clinical data and clinical research data with “ontology terms” are quite common. One reason for these requests is an understandable lack of awareness on the consumer side that using ontologies productively is an effort that goes beyond coding or re-coding existing data, but that it requires transforming (mostly) tabular data into a graph data format. The results of such approaches have been reported in the literature 37 38 39 . Previous works showed that using terms from OWL ontologies to annotate biomedical data that is not graph data may yield some results, such as assessing the domain coverage of the ontology or semantic integration based on the taxonomy that is part of the OWL file 40 , 41 . However, utilizing the artificial intelligence capabilities linked to KRR requires the data is transformed to graph- based data representation.
Our aim is to provide use cases for using preexistent ontologies and SWTs to map clinical data and clinical research data in a way that realizes the Artificial Intelligence capabilities of those technologies. In our ontological representation, we follow the best practice of reusing existing ontologies where possible 42 , 43 .
Our focus in this effort is to promote awareness and understanding of the level of re-representation necessary to enable true knowledge representation based on this data. As such, we present conceptualizations of what the data is about, to help alleviate the communication gap between medical researchers and biomedical informaticians on the one side and computer scientists and the artificial intelligence community on the other side. Researchers and data curators in biomedical informatics are encouraged to embrace pre-existing tools for restructuring tabular data as graph data, such as W3C CSV2RDF 44 or RDB2RDF 45 . Our aim is to foster understanding of the re-structuring of the data that is useful for those curating it, especially those with medical domain knowledge, to ensure automatic transformation delivers correct and meaningful results. We point to ontology resources that foster orthogonal and consistent ontology development and demonstrate reuse of OWL entities from ontologies following those strategies. We demonstrate the semantic ambiguity of terms from clinical and clinical research standards and Common Data Elements (CDEs). Modeling ontologies to be one-to-one mappable to those artifacts leads to diminishing the advantages of the SWT approach by creating a multitude of study-specific classes in a pre-coordination approach.
3 Knowledge Representation Applied to Medical Data
3.1 General Approach
In our reuse of pre-existing ontologies we have embraced the collaborative, community-driven development paradigm in the biomedical ontologies community led by the Open Biological and Biomedical Ontologies (OBO) Foundry 46 . The OBO Foundry is a collaborative effort to build a library of orthogonal ontologies for both biomedical and biological domains following a core set of principles. In addition, the OBO Foundry provides a consistent way to manage naming and identifiers 46 . The results of the OBO Foundry are made available through the OBO Foundry website 47 and through the Ontobee service 48 , which allows users to explore many OBO Foundry ontologies using one term search 49 .
Smith and Ceusters stressed that the need for a shared upper ontology is a practical consequence of the need for collaborative ontology development in science 50 . They pointed out the relevance of the ontological realist methodology in building the OBO Foundry. The advantage of adopting a realist stance for collaborative ontology development is that the appropriateness and correctness of the ontological representation (and thus the ontological commitments) is linked to scientific research, including experimentation and scientific arguments 50 . The linkage between the ontological realist methodology and the individual OBO Foundry ontologies is ensured by the fact that the Basic Formal Ontology (BFO) is the upper ontology of most OBO Foundry ontologies 51 52 53 54 . According to Arp et al. , all OBO Foundry domain ontologies have adopted BFO as their upper level ontology 53 .
As a collective of open biological and biomedical ontologies collaborating around shared design principles, OBO Foundry has broad and expanding term coverage for entities that one might need to model when creating semantic representation for data in the biomedical domain. However, even with this broad coverage, it is not unusual to encounter phenomena for which there are no adequate terms existing in OBO Foundry ontologies. We generally approach this issue in our projects by working with the developers of the relevant ontology, proposing terms and their definitions, and requesting their addition to the ontology 55 . Because this process is not immediate, it is sometimes necessary to create a small application ontology that has placeholders for the desired terms that we can use as we proceed with crafting our representations. Of course, having already defined and implemented a draft for the required term makes the term request easier to discuss and fulfill.
In some cases, there may not be an existing ontology that is a natural fit for the term or terms needed. Depending on the scale of this gap and the nature of the terms in question, including how generally useful they are likely to be for the larger community, it may make sense either to simply develop our own terms for internal use in an application ontology, or to initiate the development of a new OBO ontology that covers the relevant subdomain. Note that because BFO provides a full upper- level theory that is shared by all OBO ontologies, and because there are already existing interoperable OBO ontologies for many areas of biology and medicine, even in the case where we develop new terms without the intention of releasing them as part of a new ontology, these terms are not built in isolation but are developed in the context of existing OBO ontologies, with logical definitions that capture their relations to those resources.
3.2 The Cancer Imaging Archive
The Cancer Imaging Archive (TCIA) is the National Cancer Institute’s primary resource for acquiring, curating, managing, and distributing images and related data to support cancer research 56 57 58 . TCIA hosts over 36 million de-identified medical images of cancer (28 distinct cancer types) organized into 96 distinct collections 59 . TCIA was created to support research reproducibility and research reuse.
We are developing PRISM (Platform for Imaging in Precision Medicine) as the future basis of TCIA and offering this advanced informatics platform as an open source, easily deployable resource to support other research communities. Within the PRISM platform, we are developing state-of-the-art technologies for semantic integration of clinical and research information drawn from multiple sources. Identified near-term goals and challenges include: uniform management of non-image data; semantic query mechanisms and enhanced data exploration; and automatic curation of current and new data types. Many TCIA collections include non-image data in a variety of formats, often as downloadable spreadsheet files, which makes them difficult to combine or query. Further complicating this is the use of different representation schemes for similar information in different collections.
Our ongoing work to make these diverse non-image data more accessible and usable transforms them into shared semantic representations in OWL that use OBO Foundry resources, and will allow for queries that span collections to answer questions such as:
Which patients in lung cancer collections have been diagnosed with metastatic colon cancer, and how was that diagnosis obtained?
Which patients in head and neck cancer collections have tumors specifically in their oropharynx, and have been diagnosed with human papillomavirus, and how were those diagnoses obtained?
Our semantic representations based on these data use OBO Foundry ontologies including the Human Disease Ontology and The Uber Anatomy Ontology (Uberon). Instances for individual entries in TCIA collection data are linked to ontology classes to explicitly represent locations, disease types, diagnosis methods, etc.
Figure 2 shows excerpts of similar data contained in two different head and neck cancer collections in the TCIA: the Head-Neck-PET-CT collection 60 , which contains non-image data, including diagnostic and treatment information for patients with head and neck cancer, and the HNSCC (Head and Neck Squamous Cell Carcinoma) collection 61 , which contains much of the same information. Though using different notation, these collections overlap significantly in their contents, including patient sex and other demographic data, tumor staging, HPV status, and an indication of the primary tumor location. Figure 3 shows our semantically-enhanced representation of positive HPV status for a patient in a head and neck cancer collection, which provides a unique contextually-rich and axiomatically-defined representation for what the different values (“positive”, “negative”, “+”, “-”, “N/A”) represent. Note that while in a conversation or even in written documentation one might say, “this patient’s HPV status is positive,” an “HPV status” per se is a fairly nebulous entity from the realist perspective, which strives to represent things as they actually are. Our goal is to represent, in a form even a computer can understand, the relevant portions of reality that the authors of this collection were trying to describe by creating a column named “HPV status” and populating it with entries like “positive” or “+”. The best description we can extract based on that information is that at some point a “diagnostic process” occurred that involved the infected human, and involved some “HPV assay”. “HPV assay” is a subclass of OBI: assay, defined as “A planned process with the objective to produce information about the HPV status of the human that is the evaluant, by physically examining the human or samples taken from their body”. That diagnostic process produced a “diagnosis” as its output, and if there is some instance of the “papillomavirus infectious disease” that inheres in that human, the diagnosis is about that instance of the HPV infectious disease. For the pilot described here, mapping rules for the value sets to an ontological representation were specified manually and then automatically executed in transforming the source data into RDF.
Fig. 2.
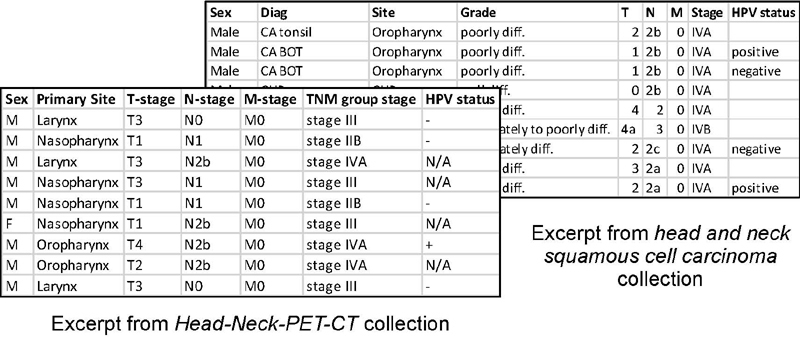
Data excerpts from two head and neck cancer collections
Fig. 3.
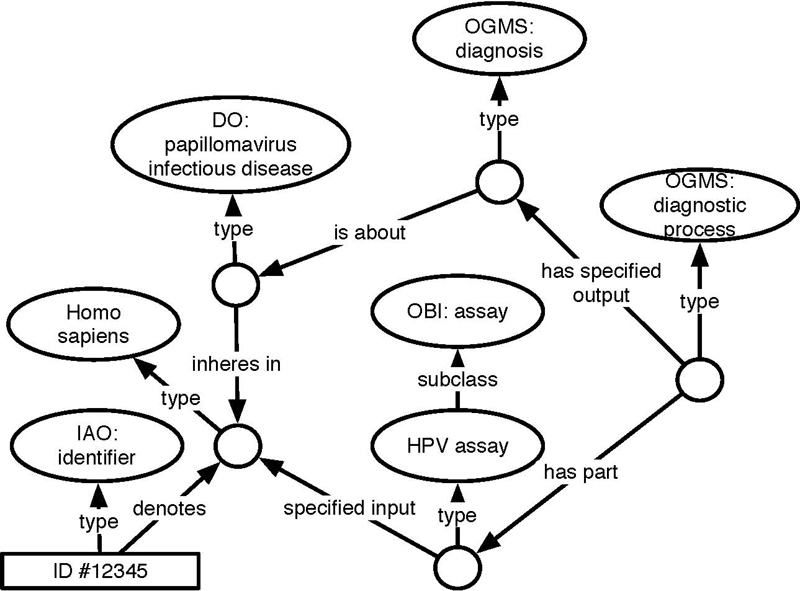
RDF representation of positive HPV status for head and neck cancer records. “Positive” or “+” map to this representation. “Negative” or “-” will map to a representation with a human undergoing the HPV assay, not establishing the existence of a papillomavirus infectious disease.
These representation patterns are used to transform tabular non-image data associated with TCIA collections into OWL/ RDF instance data linked and annotated with the corresponding ontology terms. These data are then loaded into a triple store database for reasoning and querying. The resulting triple store contains assertions linking patient identifiers to RDF instances representing patients, affected body parts, diagnoses, relations among those, etc. We are able to query this database using SPARQL (SPARQL Protocol and RDF Query Language) to identify patient records matching criteria based on fields that were previously not queryable in TCIA collections, as well as queries that retrieve results spanning collections. Work is ongoing to implement this approach for additional data types and additional existing TCIA collections, with the eventual goal of a shared representation for all TCIA non-image data, including the ability to automatically curate new incoming data as it is submitted.
3.3 The Data Coordinating and Operations Center
The IDeA (Institutional Development Award) States Pediatric Clinical Trials Network (ISPCTN) is a research network with the goal to provide medically underserved and rural populations with access to state- of-the-art clinical trials, apply findings from relevant pediatric cohort studies to children in IDeA Program state locations, and build pediatric research capacity at a national level 62 . It is part of the Environmental influences on Child Health Outcomes (ECHO) program, which is funded by the National Institutes of Health (NIH) 63 64 65 . The University of Arkansas for Medical Sciences serves as the Data Coordinating and Operations Center (DCOC) for the ISPCTN. One study undertaken by the ISPCTN deals with Neonatal Opioid Withdrawal Syndrome (NOWS), aiming to characterize current clinical practice in opioid withdrawal in newborns. The study data collection form includes patient demographics, facility characteristics, maternal and fetal exposure, maternal history, pharmacologic and non-pharmacologic treatment, and discharge disposition. As part of the DCOC mission to provide reliable and innovative data coordination and management, one of the authors (MB) was asked to provide an overview of how the NOWS data dictionary could be translated into a graph-based data representation using Semantic Web Technologies.
The NOWS data dictionary consists of 267 elements that are closely linked to questionnaire items to be completed by study representatives. Examples for those questions are: “Was the infant > 36 weeks of gestational age?”, “Was there a maternal history of opioid use?”, “Did the infant need major surgical intervention ?”, and “What lactation interventions were employed?”. The data dictionary assigns an item name, a description label, a response type, and a response label to each data element. There is additional information for each element, such as information about the questionnaire order and logic.
In Table 1 , we present three examples illustrating that a one-to-one mapping between a data element and an OWL/RDF class would be suboptimal. We elaborate the shortcomings of one-to-one mapping and demonstrate how such mapping would lead to losing the advantage of SWT.
Table 1. An excerpt from the DCOC’s NOWS data dictionary.
| Item name | Description label | Response type | Response label | Response options | Data type |
|---|---|---|---|---|---|
| INGAGE | Was the infant ≥ 36 weeks gestational age? | radio | YN | Yes, No | INT |
| BRTHDTC | Date of birth | [empty] | [empty] | [empty] | DATE |
| INMEDSUSD | Indicate the medication(s) used to treat NOWS for this infant at the transferring hospital | checkbox | MMBCPUO | Morphine,Phenobarbital,Methadone,Buprenorphine,Clonidine,Unknown, Other | ST |
The INGAGE data element is one example of NOWS data elements that has a yes/ no answer. Though study administrators frequently employ this type of questions, the information captured that way is particularly sparse from the perspective of semantics. In a case like this capturing only the response (yes, no) does not provide any machine-interpretable semantic information on what that piece of data means. Obviously, changing the text of the answer to reverse the meaning of the answer, if, for example, one changed the description label to “Was the infant <36 weeks gestational age?” would not be discernible by representing the answer alone. Thus, our first aim is to provide semantically-rich data that is machine-interpretable. Doing so requires capturing the semantics in the form of a question and maintaining the association between the question and the answer. Figure 4 shows the semantic presentation we have chosen for this INGAGE data element.
Fig. 4.
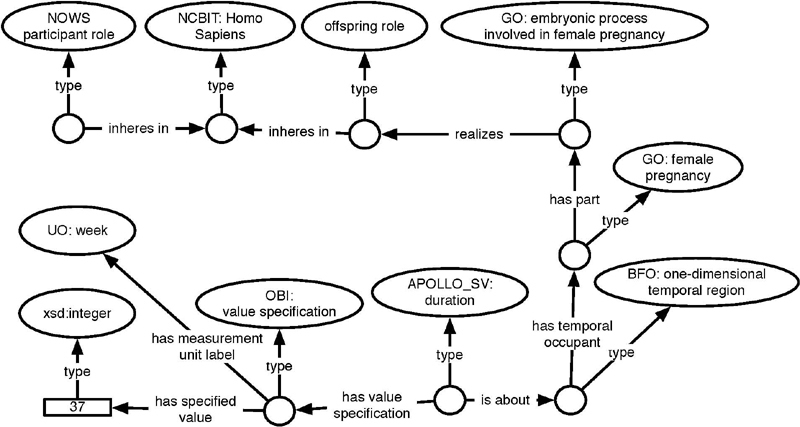
SWT representation of INGAGE from Table 1
Using a one-to-one mapping approach to the INGAGE data element would mean creating a class of potential participants in an OWL file that were all born with at least a gestational age of 36 weeks. Doing so would indeed be completely possible and, of course, also possible in RDF. If the class created was also axiomatically defined, we would lose less semantics than with the yes/no answer option. However, doing so is not advisable. Following that strategy, we would need one added class for every study that needs a different gestational age as an inclusion criterion. If those classes are axiomatically defined, we would add a lot of reasoning to our ontology, without much gain, except for individual studies. From an SWT perspective, it is much more advisable to ensure that all elements that we need to define the inclusion criterion do exist in our RDF data. For the example at hand, this means we can capture the integer value for the gestational age for all participants or patients regardless of the inclusion criteria of one specific study, e.g. NOWS. Using SPARQL, we are then able to query for all participants and patients that are at least 36 weeks of gestational age or at least 34 weeks of gestational age, depending on the requirements of the study at hand. We do not need to deal with numerous predefined classes in our ontology that slow down reasoning. The numerical values can now be extracted along with the units of measurement and used in calculations, such as analyses. For operations that go beyond the capabilities of SPARQL, it is advisable to run these calculations in tools external to the SWT suite. SWTs are, at their core, not analysis tools but knowledge management tools that can help to feed better and more meaningful data into our analysis cycles.
Date of birth is an extremely common data element in clinical data and clinical research data. It is also a data element the meaning of which is highly contextual. While some data repositories specify that this is the patient’s or the participant’s date of birth, we still regularly find “date of birth” as the form or question prompts. Strictly speaking, this practice is semantically ambiguous, since we can only know contextually that what is meant is the date of birth of the patient and not, say, the date of birth of the healthcare provider. However, typically the context is sufficient to elucidate that situation. In the data for the NOWS study, it is relevant to specify that this is the date of birth of the infant, which is the NOWS participant and not the date of birth of the mother. The latter is also relevant, as NOWS collects numerous data elements related to the mother’s medical history, such as history of opioid use. The RDF presented in Figure 5 shows how these issues are disambiguated.
Fig. 5.
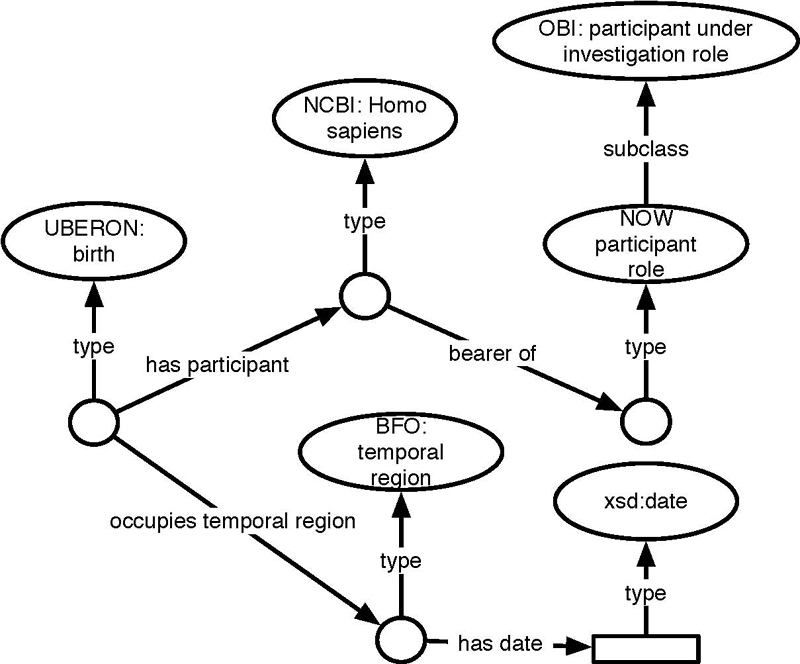
SWT representation of BRTHDTC from Table 1 .
Regarding the mapping of data, we also considered the same route rejected for the previous example (INGAGE), i.e., creating a class that captures a NOWS participant’s date of birth. For the same reasons as explained previously, we chose not to do so. Instead, we wanted to ensure that all elements necessary to retrieve that kind of information using a SPARQL query to match the corresponding pattern of triples were present.
Figure 6 shows a representation of information about drugs being used in the transferring hospital to manage the infant’s neonatal opioid withdrawal symptom. The way this data element is set up, with discrete answer options, except the ubiquitous “other” and “unknown,” provides data that can easily be semantically enriched linking the information to existing controlled vocabularies and terminologies. The advantage of doing a re-representation like the one above lies in a better chance of maintaining semantic integrity, if the data is integrated with data from other sources, using a different level of granularity regarding drug information or using a different terminology or controlled vocabulary. Using ChEBI identifiers (Chemical Entities of Biological Interest) 66 for the active ingredients allows the integration of data from the NOWS study with data that reports drug products using the Drug Ontology (DRON) 67 68 69 as a bridge.
Fig. 6.
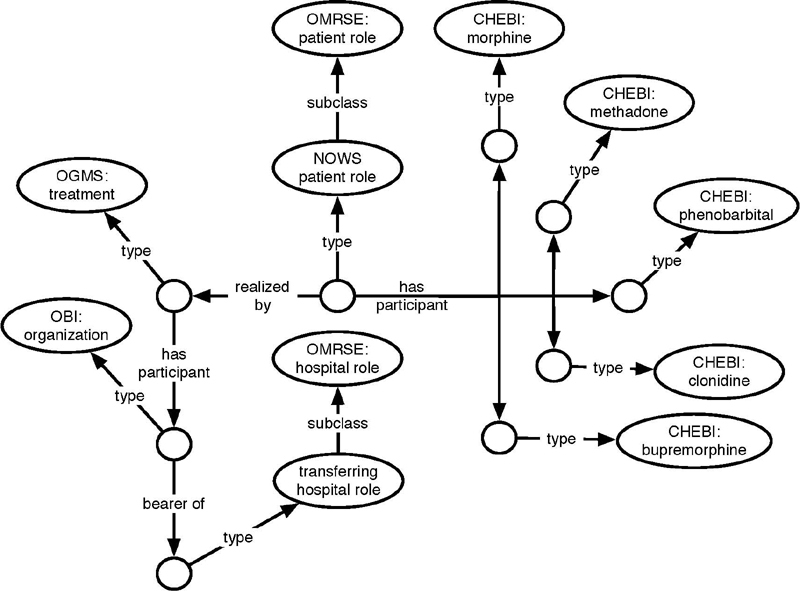
WT representation of INMEDSUSD from Table 1 .
4 Discussion
The projects described above aim to enhance pre-existing data by crafting detailed semantic representations based on axiomatically rich ontologies, and using those to re-represent these data. By building the semantics directly into our representations of the data using freely available open biomedical ontologies, we make these data understandable and useable, both to researchers and software, including software that performs automated reasoning to support producing new inferences about the data.
In the PRISM case, this work made available key information about TCIA collections that was previously not retrievable. Additionally, it supports combining similar information across collections, for instance clinical data about imaging subjects, that provides essential context for understanding and analyzing the disease depicted in these images. Figure 7 shows a SPARQL query and results illustrating this. This query retrieves identifiers across two head-and-neck cancer collections for records whose subjects have a “positive HPV diagnosis” and have also been “diagnosed with cancer of the oropharynx”. Prior to this effort, a researcher interested in investigating HPV diagnoses and tumor images in head and neck cancer cases would have had to navigate a wiki page, download separate spreadsheets, and figure out how to interpret and how to query each of those spreadsheets in order to make combined use of these data. This is already a huge advantage for cohort identification including clinical data in the TCIA. In order to further facilitate this type of investigation using these data, work is ongoing within this project to produce a user-friendly interface that will allow investigators to search and access this semantically integrated data without requiring any knowledge of ontologies, query languages, or other semantic web technologies.
Fig. 7.
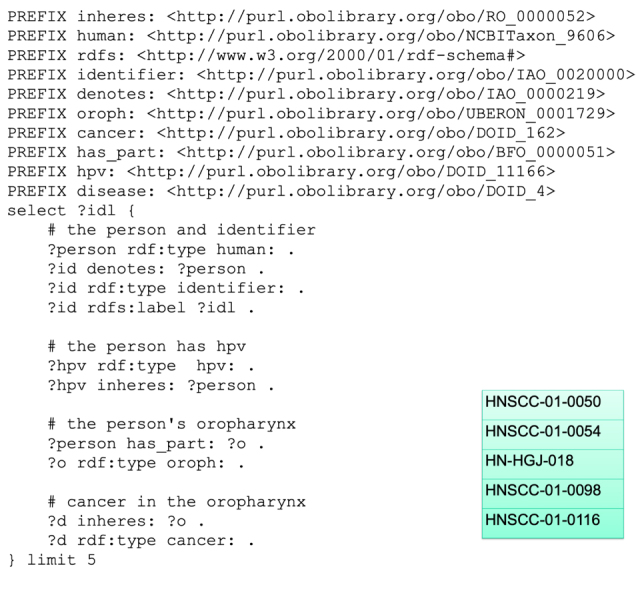
SPARQL query with results across head and neck cancer collections for individuals with HPV and cancer of the oropharynx
Regarding the Neonatal Opiate Withdrawal project, our effort was exploratory. Using the study form as an example, the goal of exploring an SWT-based knowledge management approach is to assess:
1) The feasibility of representing, curating, and extracting all information relevant to reporting;
2) The scope to which pre-existing ontologies provide coverage for the representations necessary;
3) Reusability of representation patterns across studies;
4) Flexibility and maintainability of knowledge representation against evolving needs and objectives of studies.
The project to date has shown that SWT-oriented data representation is able to adequately represent the information and data at hand. In comparison to an approach that rests exclusively on definition of common data elements, the SWT-enhanced approach provides knowledge representation capabilities, such as representing context. The coverage of pre-existing ontologies has been very good. Most of the concepts that did not already exist in OBO Foundry ontologies were study-specific. At this point we cannot make a statement about 3) and 4) with any certainty, but we can report that regarding 3) the generic representation of many aspects and the addition of few study specific entities suggest that re-use will be an option across studies and will create synergies for data management.
5 Conclusion
As illustrated by the examples discussed above from our ongoing projects working with semantic representations of biomedical data, mapping data elements directly to ontology terms is often not a feasible solution for representing the meaning of data in a useful way. Even when in some cases such one- to-one mappings of data elements to newly created ontology terms may be feasible, doing so comes at the additional cost of increased reasoning over the ontology. Furthermore, doing so puts an unnecessary burden on developers to pre-coordinate information that could instead be easily aggregated from the knowledge graph at the time a query is run.
For these reasons, we argue that progress in the practice of representing biomedical data with ontologies requires a shift in thinking about how these resources are to be used: rather than mapping data elements directly to classes or individuals in an ontology, we work to always provide a full graph representation of the patterns of elements involved in relaying the meaning behind the data elements. This allows developers to begin with a set of data elements to identify the elements needed in their ontology and allows straightforward creation of RDF based on instance data coming from tabular and other less knowledge-structured formats. Toward that end, one of the authors’ ongoing projects establishes a web repository of ontology use patterns built on SWT to promote open sharing and discussion of applying such patterns to represent biomedical instance data.
Acknowledgements
The PRISM project presented in this paper has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Under this contract, the University of Arkansas is funded by Leidos Biomedical Research subcontract 16X011. Funding was also provided by U24CA215109 and 3U24CA215109-02S1.
The DCOC work presented in this paper was funded by grant number U24OD024957 from the National Institutes of Health Office of the Director through the ECHO program.
Sarah Bost participated in literature search, writing, and technical editing of this paper.
Tracy S. Nolan assisted with data retrieval and data interpretation in the PRISM research presented in this paper. The authors thank the editors and external reviewers for their valuable input.
References
- 1.Blobel B, Oemig F.Why Do We Need an Architectural Approach to Interoperability? Eur J Biomed Inform 2016 May 20;12(1)
- 2.Cheatham M, Pesquita C, Zomaya A Y, Sakr S.Semantic Data IntegrationIn:editors.Handbook of Big Data Technologies Cham: Springer International Publishing; 2017263–305. [Google Scholar]
- 3.Niles I, Pease A.Towards a standard upper ontology. In: Proceedings of the International Conference on Formal Ontology in Information Systems (FOIS) - Volume 2001. New York, NY, USA: ACM; 2001. p. 2-9
- 4.Hitzler P, Krötzsch M, Parsia B, Patel-Schneider P F, Rudolph S.OWL 2 Web Ontology Language Primer (Second Edition). 2012 Dec 11;65
- 5.OWL 2 Web Ontology Language Document Overview (Second Edition). 2012 Dec 11;7
- 6.Brochhausen M, Bona J, Blobel B. The role of axiomatically-rich ontologies in transforming medical data to knowledge. Stud Health Technol Inform. 2018;249:38–49. [PubMed] [Google Scholar]
- 7.Hunter G
- 8.Utecht J, Brochhausen M, Judkins J, Schneider J, Boyce R D. Formalizing evidence type definitions for drug-drug interaction studies to improve evidence base curation. Stud Health Technol Inform. 2017;245:960–4. [PMC free article] [PubMed] [Google Scholar]
- 9.International Health Terminology Standards Development Organisation. SNOMED CT Compositional Grammar v2.3.1 Specification and Guide [Internet]. 2016 [cited 2019 Apr 24]. Available from:https://confluence.ihtsdotools.org/display/DOCSCG
- 10.Minarro-Gimenez J A, Martinez-Costa C, Karlsson D, Schulz S, Goeg K R.Qualitative analysis of manual annotations of clinical text with SNOMED CTPLoS One 2018;13(12) [DOI] [PMC free article] [PubMed]
- 11.Rosenbloom S T, Carroll R J, Warner J L, Matheny M E, Denny J C. Representing Knowledge Consistently Across Health Systems. Yearb Med Inform. 2017;26(01):139–47. doi: 10.15265/IY-2017-018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Overhage J M, Ryan P B, Reich C G, Hartzema A G, Stang P E. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc. 2012;19(01):54–60. doi: 10.1136/amiajnl-2011-000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitz Henry F, Resnic F S, Robbins S L, Denton J, Nookala L, Meeker D et al. Creating a Common Data Model for Comparative Effectiveness with the Observational Medical Outcomes Partnership. Appl Clin Inform. 2015;6(03):536–47. doi: 10.4338/ACI-2014-12-CR-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collins F S, Hudson K L, Briggs J P, Lauer M S. PCORnet: turning a dream into reality. J Am Med Inform Assoc. 2014;21(04):576–7. doi: 10.1136/amiajnl-2014-002864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleurence R L, Curtis L H, Califf R M, Platt R, Selby J V, Brown J S. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc. 2014;21(04):578–82. doi: 10.1136/amiajnl-2014-002747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bender D, Sartipi K.HL7 FHIR: An Agile and RESTful approach to healthcare information exchange. In: Proceedings of the 26th IEEE International Symposium on Computer-Based Medical Systems; 2013. p. 326-31
- 17.Davi R, Shrobe H, Szolovits P. What Is a Knowledge Representation? AI Mag. 1993;14(01):17. [Google Scholar]
- 18.Martinez-Costa C, Schulz S. HL7 FHIR: Ontological Reinterpretation of Medication Resources. Stud Health Technol Inform. 2017;235:451–5. [PubMed] [Google Scholar]
- 19.Chiang M F, Hwang J C, Yu A C, Casper D S, Cimino J J, Starren J. Reliability of SNOMED- CT Coding by Three Physicians using Two Terminology Browsers. AMIA Annu Symp Proc. 2006;2006:131–5. [PMC free article] [PubMed] [Google Scholar]
- 20.Andrews J E, Richesson R L, Krischer J. Variation of SNOMED CT Coding of Clinical Research Concepts among Coding Experts. J Am Med Inform Assoc. 2007;14(04):497–506. doi: 10.1197/jamia.M2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Czwikla J, Domhoff D, Giersiepen K. [ICD coding quality for outpatient cancer diagnoses in SHI claims data] Z Evidenz Fortbild Qual Im Gesundheitswesen. 2016;118-119:48–55. doi: 10.1016/j.zefq.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Gologorsky Y, Knightly J J, Chi J H, Groff M W. The Nationwide Inpatient Sample database does not accurately reflect surgical indications for fusion. J Neurosurg Spine. 2014;21(06):984–93. doi: 10.3171/2014.8.SPINE131113. [DOI] [PubMed] [Google Scholar]
- 23.McKenzie K, Enraght-Moony El, Waller G, Walker S M, Harrison J E, McClure R J. Causes of injuries resulting in hospitalisation in Australia: assessing coder agreement on external causes. Inj Prev. 2009;15(03):188–96. doi: 10.1136/ip.2008.020479. [DOI] [PubMed] [Google Scholar]
- 24.Berners-Lee T, Hendler J. Publishing on the semantic web. Nature. 2001;410:1023–4. doi: 10.1038/35074206. [DOI] [PubMed] [Google Scholar]
- 25.RDF - Semantic Web Standards [Internet]. [cited 2019 Jan 17]. Available from:https://www.w3.org/RDF/
- 26.Ruttenberg A, Clark T, Bug W, Samwald M, Bodenreider O, Chen Het al. Advancing translational research with the Semantic Web BMC Bioinformatics 20078(Suppl 3):S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Antoniou G, Groth P, van Harmelen F, Hoekstra R. MIT Press; 2012. A Semantic Web primer. [Google Scholar]
- 28.Berners-Lee T, Fielding R, Masinter L.RFC 3986 Uniform Resource Identifier (URI): Generic Syntax [Internet]. 2005 [cited 2019 Jan 17]. Available from:http://www.rfc-editor.org/rfc/rfc3986.txt
- 29.RDFS - Semantic Web Standards [Internet]. [cited 2019 Jan 17]. Available from:https://www.w3.org/2001/sw/wiki/RDFS
- 30.OWL - Semantic Web Standards [Internet]. [cited 2019 Jan 17]. Available from:https://www.w3.org/OWL/
- 31.Mate S, Köpcke F, Toddenroth D, Martin M, Prokosch H U, Bürkle Tet al. Ontology-Based Data Integration between Clinical and Research Systems. PLoS One 2015 Jan 14;10(1) [DOI] [PMC free article] [PubMed]
- 32.Brochhausen M, Zheng J, Birtwell D, Williams H, Masci A M, Ellis H J et al. OBIB-a novel ontology for biobanking. J Biomed Semant. 2016;7(01):23. doi: 10.1186/s13326-016-0068-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noor A, Assiri A, Ayvaz S, Clark C, Dumontier M. Drug-drug interaction discovery and demystification using Semantic Web technologies. J Am Med Inform Assoc. 2017;24(03):556–64. doi: 10.1093/jamia/ocw128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herrero-Zazo M, Segura-Bedmar I, Hastings J, Martinez P. DINTO: Using OWL Ontologies and SWRL Rules to Infer Drug-Drug Interactions and Their Mechanisms. J Chem Inf Model. 2015;55(08):1698–707. doi: 10.1021/acs.jcim.5b00119. [DOI] [PubMed] [Google Scholar]
- 35.Judkins J, Tay-Sontheimer J, Boyce R D, Brochhausen M.Extending the DIDEO ontology to include entities from the natural product drug interaction domain of discourse. J Biomed Semant 2018 May 9;9(15) [DOI] [PMC free article] [PubMed]
- 36.Herrero-Zazo M, Segura-Bedmar I, Martinez P. Conceptual models of drug-drug interactions: A summary of recent efforts. Knowl-Based Syst. 2016;114:99–107. [Google Scholar]
- 37.Shah N H, Jonquet C, Chiang A P, Butte A J, Chen R, Musen M A.Ontology-driven indexing of public datasets for translational bioinformatics BMC Bioinformatics 200910(Suppl 2):S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsu W, Gonzalez N R, Chien A, Pablo Villablanca J, Pajukanta P, Vinuela F et al. An integrated, ontology-driven approach to constructing observational databases for research. J Biomed Inform. 2015;55:132–42. doi: 10.1016/j.jbi.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cinaglia P, Veltri P, Cannataro M.eMiRo: an ontology-based system for clinical data integration and analysis. In: Proceeding of the 25th Italian Symposium on Advanced Database System (SEBD); 2017. CEUR Workshop Proceedings Vol. 2037
- 40.Shah N H, Rubin D L, Supekar K S, Musen M A. Ontology-based annotation and query of tissue microarray data. AMIA Annu Symp Proc. 2006;2006:709–13. [PMC free article] [PubMed] [Google Scholar]
- 41.Brochhausen M, Fransson M N, Kanaskar N V, Eriksson M, Merino-Martinez R, Hall R A et al. Developing a semantically rich ontology for the biobank-administration domain. J Biomed Semant. 2013;4(01):23–23. doi: 10.1186/2041-1480-4-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simperl E. Reusing ontologies on the Semantic Web: a feasibility study. Data Knowl Eng. 2009;68(10):905–25. [Google Scholar]
- 43.Lonsdale D, Embley D W, Ding Y, Xu L, Hepp M. Reusing ontologies and language components for ontology generation. Data Knowl Eng. 2010;69(04):318–30. [Google Scholar]
- 44.CSV2RDF [Internet]. W3C. 2014 [cited 2019 Apr 29]. Available from:https://www.w3.org/2013/csvw/wiki/CSV2RDF
- 45.RDB2RDF Working Group. RDB2RDF [Internet]. W3C. 2012 [cited 2019 Apr 29]. Available from:https://www.w3.org/2001/sw/wiki/RDB2RDF
- 46.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat Biotechnol. 2007;25(11):1251–5. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.The OBO Foundry [Internet]. [cited 2019 Jan 17]. Available from:http://www.obofoundry.org/
- 48.Ontobee [Internet]. [cited 2019 Jan 17]. Available from:http://www.ontobee.org/
- 49.Xiang Z, Mungall C, Ruttenberg A, He Y, Bodenreider O, Martone M, Ruttenberg A.Ontobee: a linked data server and browser for ontology termsIn:editors.Proceedings of the 2nd International Conference on Biomedical Ontology Buffalo, NY: 2011279–81. [Google Scholar]
- 50.Smith B, Ceusters W.Ontological realism: A methodology for coordinated evolution of scientific ontologies Appl Ontol 20105(3-4):139–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grenon P, Smith B, Goldberg L. Biodynamic ontology: applying BFO in the biomedical domain. Stud Health Technol Inform. 2004;102:20–38. [PubMed] [Google Scholar]
- 52.Grenon P, Smith B. SNAP and SPAN: towards dynamic spatial ontology. Spat Cogn Comput. 2004;4(01):69–104. [Google Scholar]
- 53.Arp R, Smith B, Spear A D. MIT Press; 2015. Building ontologies with Basic Formal Ontology. [Google Scholar]
- 54.Smith B, Brochhausen M. Putting biomedical ontologies to work. Methods Inf Med. 2010;49(02):135–40. doi: 10.3414/ME9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.FAQ: Request an ontology term [Internet]. The OBO Foundry. [cited 2019 Apr 29]. Available from:http://www.obofoundry.org/faq/how-do-i-request-a-term.html
- 56.Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P et al. The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J Digit Imaging. 2013;26(06):1045–57. doi: 10.1007/s10278-013-9622-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kalpathy-Cramer J, Freymann J B, Kirby J S, Kinahan P E, Prior F W. Quantitative Imaging Network: data sharing and competitive algorithm validation leveraging The Cancer Imaging Archive. Transl Oncol. 2014;7(01):147–52. doi: 10.1593/tlo.13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chennubhotla C, Clarke L P, Fedorov A, Foran D, Harris G, Helton E et al. An assessment of imaging informatics for precision medicine in cancer. Yearb Med Inform. 2017;26(01):110–9. doi: 10.15265/IY-2017-041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.The Cancer Imaging Archive (TCIA) - A growing archive of medical images of cancer [Internet]. The Cancer Imaging Archive (TCIA). [cited 2019 Jan 17]. Available from:http://www.cancerimag-ingarchive.net/
- 60.Vallieres M, Kay-Rivest E, Perrin L J, Liem X, Furstoss C, Khaouam Net al. Data from Head-Neck-PET-CT [Internet]. The Cancer Imaging Archive; 2017 [cited 2019 Jan 15]. Available from:https://doi.org/10.7937/K9/TCIA.2017.8oje5q00
- 61.Grossberg A, Mohamed A, Elhalawani H, Bennett W, Smith K, Nolan Tet al. Data from Head and Neck Cancer CT Atlas [Internet]. The Cancer Imaging Archive; 2017 [cited 2019 Jan 15]. Available from:https://doi.org/10.7937/K9/TCIA.2017.umz8dv6s
- 62.Snowden J, Darden P, Palumbo P, Saul P, Lee J.The institutional development award states pediatric clinical trials network: building research capacity among the rural and medically underserved Curr Opin Pediatr 20183002297–302.(6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ward R M, Benjamin D K, Davis J M, Gorman R L, Kauffman R, Kearns G L et al. The Need for Pediatric Drug Development. J Pediatr. 2018;192:13–21. doi: 10.1016/j.jpeds.2017.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gillman M W, Blaisdell C J. Environmental influences on Child Health Outcomes, a research program of the National Institutes of Health. Curr Opin Pediatr. 2018;30(02):260–2. doi: 10.1097/MOP.0000000000000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith B, Knox S, Benjamin D K. Coordination of the Environmental influences on Child Health Outcomes program: so the whole is greater than the sum of its parts. Curr Opin Pediatr. 2018;30(02):263–8. doi: 10.1097/MOP.0000000000000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hastings J, Owen G, Dekker A, Ennis M, Kale N, Muthukrishnan Vet al. ChEBI in 2016: Improved services and an expanding collection of metabolites Nucleic Acids Res 201644(Database issue):D1214–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hanna J, Joseph E, Brochhausen M, Hogan W R. Building a drug ontology based on RxNorm and other sources. J Biomed Semant. 2013;4(44):9. doi: 10.1186/2041-1480-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hanna J, Bian J, Hogan W R. An accurate and precise representation of drug ingredients. J Biomed Semant. 2016;7(07):9. doi: 10.1186/s13326-016-0048-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hogan W R, Hanna J, Hicks A, Amirova S, Bramblett B, Diller Met al. Therapeutic indications and other use-case-driven updates in the drug ontology: anti-malarials, anti-hypertensives, opioid analgesics, and a large term request. J Biomed Semant 2017 Mar 3;8 [DOI] [PMC free article] [PubMed]


