Summary
Objectives : This survey analyses the latest literature contributions to clinical decision support systems (DSSs) on a two-year period (2017-2018), focusing on the approaches that adopt Artificial Intelligence (AI) techniques in a broad sense. The goal is to analyse the distribution of data-driven AI approaches with respect to “classical" knowledge-based ones, and to consider the issues raised and their possible solutions.
Methods : We included PubMed and Web of Science TM publications, focusing on contributions describing clinical DSSs that adopted one or more AI methodologies.
Results : We selected 75 papers, 49 of which describe approaches in the data-driven AI area, 20 present purely knowledge-based DSSs, and 6 adopt hybrid approaches relying on both formalized knowledge and data.
Conclusions : Recent studies in the clinical DSS area demonstrate a prevalence of data-driven AI, which can be adopted autonomously in purely data-driven systems, or in cooperation with domain knowledge in hybrid systems. Such hybrid approaches, able to conjugate all available knowledge sources through proper knowledge integration steps, represent an interesting example of synergy between the two AI categories. This synergy can lead to the resolution of some existing issues, such as the need for transparency and explainability, nowadays recognized as central themes to be addressed by both AI and medical informatics research.
Keywords: Decision Support System, Clinical; Artificial Intelligence
1 Introduction
Clinical decision-support systems (DSSs) aim to enhance healthcare decision-making, with the final objective to improve the quality of care provided by healthcare organizations 1 , 2 . Since earlier years, clinical DSSs have been often conceived as applications of Artificial Intelligence (AI) methodologies to medical domains 3 . Examples range from the so-called “expert systems” based on rules 4 , to approaches relying on models of deeper and more principled human reasoning, like, e.g., causal reasoning 5 , to more recent proposals exploiting fuzzy logic, Bayesian networks, case-based reasoning (CBR), and other techniques (see, e.g., 6 , 7 ). Al-based DSSs have been used as aids across a wide range of medical tasks among various medical specialties.
According to the Barcelona Declaration for the Proper Development and Usage of Artificial Intelligence in Europe 3 , AI methodologies can be divided into two fundamentally different categories: knowledge-based AI and data-driven AI. Knowledge-based AI consists in “an attempt to model human knowledge in computational terms, starting in a top-down fashion from human self-reporting of what concepts and knowledge individuals use to solve problems or answer queries in a domain of expertise, including common sense knowledge”, and “formalizes and operationalizes this knowledge in terms of software. It rests primarily on highly sophisticated but now quite standard symbolic computing technologies and has already had a huge impact” 3 . Data-driven AI, on the other hand, is characterized by “starting in a bottom-up fashion from large amounts of data of human activity, which are processed with statistical machine learning methods [...] in order to abstract patterns that can then be used to make predictions, complete partial data, or emulate human behaviour in similar conditions in the past. Data-driven AI requires big data and very substantial computing power to reach adequate performance levels” 3 . Knowledge-based methodologies are well established, but less able to exploit large volumes of data, and to automatically build a knowledge model by generalizing from data themselves. In fact, knowledge models often remain human-developed. Knowledge acquisition and formalization is thus a “bottleneck”, which consumes development time and requires a significant initial effort. On the other hand, data-driven methodologies are currently receiving a lot of attention, thanks to the large amount of data available in electronic form, to the availability of powerful computing architectures, and to the significant advancement of machine learning techniques that are able to extract characteristic features and to identify patterns from data with a high level of accuracy.
Interestingly enough, Chen et al. 8 have recently analysed this AI category distinction, referring to the field of clinical DSSs. They have observed that the knowledge-based approach to clinical decision support is limited in scale, due to the lack of evidence in some domains as well as to the cost of human knowledge authoring processes. On the contrary, taking advantage of the accumulated clinical data, stored, e.g., in electronic patient records, powerful data-driven clinical decision support systems can be implemented, possibly leading to more effective outcomes in practice.
Data-driven approaches, however, might lack in transparency and explainability , while, as highlighted by Shortlife et al. 9 , a clinical DSS should enable users to “understand the basis for any advice or recommendations that are offered”.
In this survey, we analyse the latest literature contributions to clinical DSSs (years 2017-2018), focusing on approaches that adopt AI techniques. Our main goal is to understand if “classical” knowledge-based DSSs are still being proposed, or if, as in other domains, earlier AI principles and methods are less represented, in favour of big data analytics and machine learning approaches. The challenges of the surveyed methodological choices are analysed, with a particular focus on the transparency and explainability issues, nowadays recognized as central themes to be addressed by AI and medical informatics research, as mentioned above. Promises deriving from synergies between the two AI categories are also discussed.
2 Methods
We have considered two bibliographic repositories, namely PubMed and Web of Science” (WoS), restricting searches to years 2017 and 2018 (up to November).
Given the focus of our survey, PubMed was queried specifying (in AND) the MeSH terms “Decision Support System, Clinical” and “Artificial Intelligence”. Considering that some contributions may not adopt MeSH terms, we have also searched PubMed by specifying some related keywords to be searched for (one at a time) in the title, namely “machine learning”, “expert system”, “information retrieval”, “cognitive aid”. The search with the keyword “machine learning” actually proved to be too generic (as commented in the Results section below), so we repeated a more focused query that searched for the keywords “machine learning” and “decision” (in AND) in the title. We admitted only papers in English, where the abstract was available. We first assessed the appropriateness of the retrieved papers by reading their abstracts. Those papers that were not excluded in this phase were read in full, by dividing the effort equally between the two co-authors.
WoS does not provide MeSH term filtering; therefore, we indicated “Decision Support” (“Clinical Decision Support” was considered less inclusive) and “Artificial Intelligence” (in AND) as topics, and limited our areas of interest to “Medical Informatics”, “Computer Science Artificial Intelligence”, “Computer Science Information Systems”, and “Computer Science Interdisciplinary Applications”. By reading the title and the abstract, we could focus on the papers of interest. In this case also, papers that were not excluded were read in full, by dividing the effort equally between the two co-authors.
As a general policy, we excluded works that did not describe a DSS, but we kept the papers that (1) were reviews or existing tools comparisons, or (2) coped with related issues, such as the need for data interoperability, or (3) were limited to the presentation of a preliminary design work.
Papers were categorized on the basis of the adopted methodology and task.
3 Results
As regards to PubMed, through the MeSH term search we retrieved 68 papers, 29 of which were excluded after the abstract/full paper reading assessment.
The “cognitive aid” keyword search returned 6 papers, but none of them was devoted to the description of a DSS.
The “machine learning” keyword search returned 1,778 papers; however, this list was too general, since it included aids of very different complexity, typically much simpler than a DSS. We then restricted the search by adding the “decision” keyword in AND. This solution returned 15 papers. After the abstract/full paper reading assessment that led us to identify 8 papers not meeting the study criteria, and after having excluded 3 additional papers already retrieved by the MeSH term search, we kept 4 works.
The “expert system” keyword search provided us with 36 papers, but 19 of them were out of scope (e.g., focused on chemistry/pharmacology), or did not describe a DSS. After having excluded 2 additional papers already retrieved by the MeSH term search, we kept 15 works.
The “information retrieval” keyword search produced 31 papers. Only 2 of them met the selection criteria.
The WoS search returned 113 papers, 17 of which were pertinent and fulfilled our selection criteria. Two of these 17 papers had been retrieved by the PubMed search as well, so we kept 15 works.
In summary, 75 papers were considered in our analysis.
Figure 1 illustrates the overall review flow.
Fig. 1.
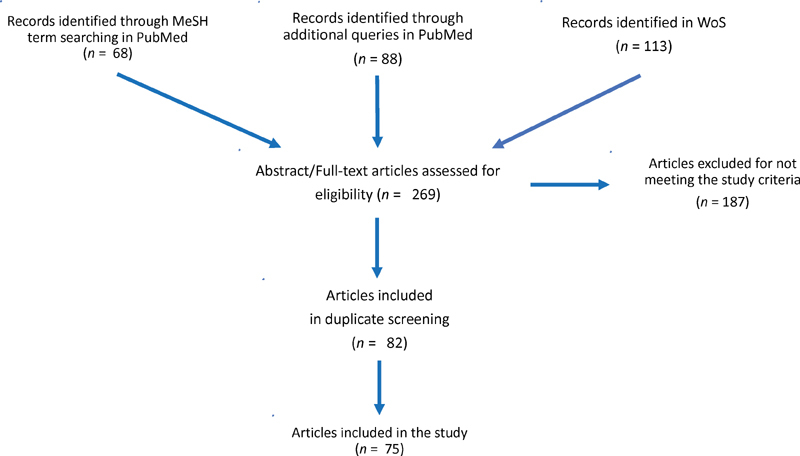
Flow of the review process.
When reading the papers, we distinguished the retrieved works on the basis of the adopted methodology, considering the two main categories (i.e., data-driven vs. knowledge-based DSSs), and refining the distinction by identifying appropriate subgroups. The presence of hybrid approaches was identified as well. We also highlighted the main supported task.
Our results are reported in the following subsections.
3.1 Data-driven Decision Support
Out of the 75 retrieved papers, 49 can be categorized as data-driven decision support approaches 10 – 58 . As regards to the addressed task, a large part of these contributions deals with prediction, intended as classification or regression 10 – 15 , 17 – 19 , 21 – 36 , 38 – 42 , 44 – 46 , 48 , 49 , 52 – 54 , 57 . One work is focused on association rule mining 20 , and one adopts statistics for risk analysis 55 . A set of papers deal with information extraction from natural language texts 16 , 33 , 43 , or more specifically from electronic patient record data 32 , 37 , 44 , 50 , 56 . The extracted information can then be exploited for classification purposes 33 , for statistical correlation and knowledge integration 32 , 43 , or for outlier detection 37 . Retrieval and interpretation of similar complex data, such as time series, images, voice recordings, or radiotherapy plans are addressed in 17 , 30 , 31 , 34 , 38 , 47 , 53 , 58 .
As regards to the adopted methodologies, interestingly, a large part of the classification approaches adopts more than one machine learning techniques 10 – 14 , 16 , 21 , 22 , 24 – 27 , 33 , 39 , 42 , 45 , 48 – 51 , 54 , 57 , such as Support Vector Machines (SVM), Neural Networks (NN), Decision Trees, and Bayesian models, in synergy or in competition. The work in 14 , for instance, adopts machine learning to find out relevant Single Nucleotide Polymorphisms (SNPs) related to Type 2 diabetes, and to predict patient risk. The authors use the Random Forest technique to search for the most important attributes related to diabetes. SVM and Logistic Regression (LR) are also adopted. Their performances have been compared to that achieved by Random Forest. Moreover, the relevance of the attributes obtained through Random Forest has then been used to perform predictions with the k-Nearest Neighbour method, weighting the attributes in the similarity measure according to the relevance provided by Random Forest itself. Working on 677 subjects, Random Forest has outperformed all the other tested machine learning techniques in terms of prediction accuracy (0.853 with respect to 0.835 with LR and 0.825 with SVM on raw data), and in terms of the stability of the estimated relevance of the attributes.
NN deserve a special consideration, since they are the key technique adopted in many of the retrieved papers 15 , 17 – 19 , 28 – 30 , 35 , 40 , 41 , 52 , 58 . Indeed, image interpretation and classification are fields where NN/deep learning approaches work well 59 . As an example, the paper in 30 exploits a convolutional NN for detecting haemorrhage, mass effect, or hydrocephalus at non-contrast-enhanced head computed tomographic examinations, and for identifying suspected acute infarct. In a retrospective study analysing 2,583 representative images, the approach has provided promising results in detecting critical findings at non-contrast-enhanced head computed tomography. Suspected acute infarct detection has shown lower sensitivity (62%) with respect to hydrocephalus detection (80%), but higher specificity (96% with respect to 90%). The authors plan to conduct further investigation in a controlled and prospective clinical setting.
The use of NN requires the choice of proper methods to train the network itself. The work in 17 , for instance, proposes a novel algorithm based on predator-prey particle swarm optimization, to train the weights of a rather simple NN architecture (a single-hidden layer NN). The approach, applied in the field of magnetic resonance image interpretation, has outperformed six state-of-the-art methods.
Papers dealing with information extraction from text, images, time series, or voice recordings, adopt proper methodologies, such as natural language processing techniques (e.g., 32 ), NN (e.g., 58 ), or voice analysis 53 . Table 1 shows the distribution of the surveyed data-driven works by task and by methodology.
Table 1. Distribution of the surveyed data-driven works by task and by methodology.
| Task | Methodology | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Association rules | Decision trees | Hidden Markov Models (HMM) | Neural networks | NLP | Retrieval techniques | Statistics | SVM | Voice analysis | Multiple methods | Total | |
| Image interpretation | 47 | 17, 30, 58 | 34, 38 | 6 | |||||||
| Prediction | 23 | 31 | 15, 17-19, 28-30, 35, 40, 41, 52 | 33, 44 | 34, 38 | 32 | 36, 46 | 53 | 10-14, 21, 22, 24-27, 39, 42, 45, 48, 49, 51, 54,57 | 39 | |
| Risk analysis | 55 | 1 | |||||||||
| Rule mining | 20 | 1 | |||||||||
| Time series interpretation | 31 | 1 | |||||||||
| Voice interpretation | 53 | 1 | |||||||||
| Text interpretation and prediction | 16, 43, 44, 50, 56 | 32, 37, 43 | 16, 33, 50 | 8 | |||||||
| Total | 1 | 2 | 1 | 12 | 6 | 2 | 4 | 2 | 1 | 22 | – |
3.2 Knowledge-based Decision Support
Out of the 75 papers we analysed, 26 reported the adoption of knowledge-based AI methods 60 – 85 , possibly in combination with data- driven methods 61 , 63 , 70 , 71 , 74 , 76 . This later group of 6 hybrid approaches will be specifically detailed in the next subsection.
The knowledge-based methods explicitly represent knowledge as symbols in the form of rules, ontologies, past cases, or other types of knowledge structures. Specifically, the surveyed approaches ranged from the use of ontologies 61 , 62 , 67 – 69 , to rule-based reasoning (including the adoption of fuzzy rules) 64 , 66 – 68 , 72 – 75 , 78 – 80 , 82 – 85 , to the definition of semantic or associative networks 76 , 77 , to temporal reasoning 65 , to probabilistic models such as Bayesian Networks 60 , 63 , 70 , 81 , and to case base reasoning CBR 61 , 71 .
As regards to the addressed task, most works deal with diagnosis 60 , 62 , 64 , 70 , 73 , 74 , 77 – 80 , 82 , 84 , 85 or therapy/treatment support 61 – 63 , 66 , 75 , 81 , while some works address classification 83 , patient’s behaviour tracking or activity scheduling 65 , and training 68 . A set of papers deals with computer-interpretable guidelines (CIGs) 67 , 69 , 71 , 72 .
As an example, the paper in 67 affords the issue of reconciling multiple clinical guidelines for decision support in comor- bid patients. The developed tool exploits semantic web technologies to achieve knowledge modelling and knowledge integration, by aligning multiple ontologi- cally-modelled clinical pathways to develop a unified comorbid patient knowledge model. Guideline execution using reasoning engines to derive guideline-mediated recommendations is also supported. The tool has been analysed in the domain of Atrial Fibrillation and Chronic Heart Failure. Table 2 shows the distribution of the surveyed knowledge-based works by task and by methodology.
Table 2. Distribution of the surveyed knowledge-based works by task and by methodology.
| Task | Methodology | ||||||
|---|---|---|---|---|---|---|---|
| CBR | Ontologies | Probabilistic Models | Rules | Semantic Networks | Temporal Reasoning | Total | |
| Behavior tracking / activity scheduling | 65 | 1 | |||||
| Classification | 83 | 1 | |||||
| Diagnosis | 62 | 60, 70 | 64, 73, 74, 78-80, 82,84,85 | 77 | 13 | ||
| CIG | 71 | 67, 69 | 67, 72 | 4 | |||
| Problem action pattern | 76 | 1 | |||||
| Therapy | 61 | 61, 62 | 63, 81 | 66, 75 | 6 | ||
| Training | 68 | 68 | 1 | ||||
| Total | 2 | 5 | 4 | 15 | 2 | 1 | – |
3.3 Hybrid Approaches
Rather interestingly, among the 26 knowledge-based approaches we analysed in section 3.2, 6 papers report on more properly hybrid approaches, since they are able to take advantage of both formalized knowledge and available data 61 , 63 , 70 , 71 , 74 , 76 . This is a quite natural situation when adopting probabilistic graphical models such as Bayesian Networks, where domain knowledge is typically exploited to build the network structure and/or to identify the key variables (i.e., the qualitative network information), but conditional probability tables (i.e., the quantitative network information) are then learned/ refined from the available data 63 , 70 , 76 . CBR is also intrinsically “hybrid”, since it conjugates the use of formalized knowledge, e.g., the case structure definition, with the exploitation of past operative examples, i.e., of data 61 , 71 . However, we found also other less typical proposals, such as the work in 74 . This specific paper presents a DSS to be used in the domain of Anti-Phospholipid Syndrome diagnosis, which relies on a logic programming approach for knowledge representation and reasoning, complemented with a NN, where neuron connections are learned and tuned from the data, and do not mirror any explicit or easily explainable domain knowledge.
Combinations of several machine learning methods are also frequent, as already mentioned in the “Data-driven decision support” section and can be provided in order to compare classification results obtained through different techniques (see e.g., 11 ), to realize ensemble learning (see e.g., 27 ), or more generally to implement a more complex strategy able to overcome the results of simpler approaches (see e.g., 10 ).
4 Discussion
The main goal of this survey was to identify a possible prevalence of data-driven AI methodologies in the development of clinical DSSs, with respect to knowledge-based approaches. In fact, 65% of the papers that we reviewed in this survey can be categorized in the data-driven methodological area. This finding demonstrates that knowledge-based methods are less represented than statistical and machine learning approaches in the very recent literature. Indeed, machine learning methodologies are particularly appropriate when dealing with high dimensional data, such as time series or medical images. As regards to medical image classification, in particular, deep learning tools are proving particularly suitable. More generally, taking advantage of the data accumulated across the entire care continuum in multiple data sources, medical professionals can use data-driven DSSs to improve patient care more successfully than knowledge-based techniques.
As a matter of fact, the exploitation of all the available patient data is a key element to be considered in a DSS. In fact, in addition to the papers analysed in this survey, using the MeSH keywords described in the “Methods” section, we retrieved from PubMed other works (e.g., 86 , 87 ) that dealt with interoperability, knowledge integration and knowledge management , as pre-requisites towards the development of a clinical DSS. While technically speaking these works could not be included in the survey, as they do not describe a DSS, they show the centrality of these themes. In particular, they suggest that the use of standard terminologies, such as SNOMED-CT, can help to actually implement interoperability. Moreover, developing user-friendly knowledge authoring tools and automatic knowledge acquisition facilities, implementing dimensionality reduction and missing value imputation as data preparation steps, can support the creation of sharable and interoperable knowledge as well.
The paper in 87 also highlights another topic: data-driven AI can be cost-effective, but also potentially beyond human abilities. Building and adopting machine learning techniques can be relatively easy, but understanding the provided outcome is sometimes difficult and obscure, due to the typical black-box nature of machine learning.
The need for transparency and explainability is nowadays being recognized as a central theme to be addressed by AI research. As an example, the DARPA research funding agency in the US highlights this issue through its “Explainable Artificial Intelligence” initiative 88 . Focusing on the field of clinical DSSs, the paper in 9 also clearly states that “black boxes are unacceptable”, since a clinical DSS must enable end users to understand all the generated suggestions.
Very interestingly, DARPA 88 proposes the combination of knowledge-based and data-driven methods as a solution to meet future challenges of AI, beginning with explainability. Even though every system should implement its own explanation module, which may vary on the basis of the application domain and of the adopted methodologies and algorithms, we believe that, in general, the synergy between different knowledge types and AI methodologies can in fact represent a promising strategy to deal with transparency and explainability issues. Indeed, various examples of hybrid systems, combining formalized knowledge and learnt knowledge have been retrieved in this survey. This could be a direction to follow, in order to improve DSS competence, flexibility, and, of course, explainability. Therefore, it is also our opinion that the already mentioned theme of knowledge integration will remain a key research direction for the future.
As a final consideration, it is worth mentioning that this survey has some limitations. In particular, it has a rather narrow focus, namely AI for DSSs. This choice was motivated by the special topic of the current edition of the IMIA Yearbook (2019). As a consequence, the number of examined works is rather limited, and interesting contributions to the field of DSSs may have been left out. Similarly, some works may have been ignored, if not indexed by PubMed or WoS, or not accessible through our queries. Indeed, we are aware of some interesting contributions that were left out, such as, e.g., the work in 89 in the area of deep learning for medical image interpretation, and the work in 90 in the area of CIGs. CIGs, in particular, represent a very active research field 91 , testified in our survey by the works in 67 , 69 , 71 , 72 , where AI is typically adopted for domain knowledge representation, and for automated reasoning as well. In order to avoid the exclusion of further papers, we did not concentrate only on journal publications, as it was done in 92 , and we did not limit our search to papers focusing on a specific study design, as in 93 , or to papers strictly selected on the basis of the quality of the reported study, as in 94 . Finally, it is worth noting that this study covers a much more limited time frame (two years), with respect to more comprehensive and elaborated surveys, such as the ones in 2 , 95 .
5 Conclusion
Recent works in the clinical DSS area demonstrate a prevalence of data-driven AI, with respect to knowledge-based classical approaches. Despite their success, however, data-driven methods may lack transparency in how a conclusion is reached, while the capability of explaining and justifying their outcome to the end user is central in the medical domain 9 . The synergy between different knowledge types and between the two categories of AI methodologies could represent a promising strategy to deal with transparency issues. As the Barcelona Declaration for the Proper Development and Usage of Artificial Intelligence in Europe 3 states, in fact, “the full potential of AI will only be realized with a combination of these two approaches”. An effort towards the definition of hybrid systems, able to integrate knowledge-based and data-driven methods, is already witnessed in the recent literature. Such a strategy also entails data interoperability and knowledge integration issues, to allow the exploitation of different knowledge sources; recent works are approaching this theme as well. We believe that the use of hybrid approaches for DSSs will be a key direction for the future. Indeed, the powerful and promising data-driven DSSs can strongly benefit of methods based on knowledge formalization, and of their generalization and abstraction capabilities, which can be proven particularly helpful to provide a really explainable decision support.
References
- 1.Osheroff J A, Teich J M, Levick D, Saldana L, Velasco F, Sittig Det al. Improving Outcome with Clinical Decision Support: An Implemented Guide, 2nd Ed. Chicago: Healthcare Information and Management Systems Society; 2012
- 2.Peleg M, Tu S.Decision support, knowledge representation and management in medicine. Methods Inf Med 2006;45 Suppl 1:S72-80 [PubMed]
- 3.Barcelona Declaration for the Proper Development and Usage of Artificial Intelligence in Europe. 2017. Mar; 1-4
- 4.Buchanan B G, Shortliffe E H.editors.Rule-based expert systems: The MYCIN experiments of the Stanford heuristic programming project Reading, MA: Addison-Wesley; 1984 [Google Scholar]
- 5.Kulikowski C A, Weiss S M, Szolovits P.Representation of Expert Knowledge for Consultation: The CASNET and EXPERT ProjectsChapter 2 in:editor.Artificial Intelligence in Medicine Boulder Colorado: Westview Press; 1982 [Google Scholar]
- 6.Hatzilygeroudis I, Prentzas J. Integrating (rules, neural networks) and cases for knowledge representation and reasoning in expert systems. Expert Syst Appl. 2004;27(01):63–75. [Google Scholar]
- 7.Shen Y, Colloc J, Jacquet-Andrieu A, Lei K. Emerging medical informatics with case-based reasoning for aiding clinical decision in multi-agent system. J Biomed Inform. 2015;56:307–17. doi: 10.1016/j.jbi.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 8.Chen J H, Alagappan M, Goldstein M K, Asch S M, Altman R B. Decaying relevance of clinical data towards future decisions in data-driven inpatient clinical order sets. Int J Med Inform. 2017;102:71–9. doi: 10.1016/j.ijmedinf.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shortliffe E H, Sepulveda M J. Clinical decision support in the era of artificial intelligence. JAMA. 2017;320(21):2199–200. doi: 10.1001/jama.2018.17163. [DOI] [PubMed] [Google Scholar]
- 10.Parisi L, Rayi Chandran N, Manaog M L. Feature-driven machine learning to improve early diagnosis of Parkinson’s disease. Expert Syst Appl. 2018;15(110):182–90. [Google Scholar]
- 11.Emre Aladag A, Muderrisoglu S, Akbas N B, Zahmacioglu O, Bingol H O. Detecting suicidal ideation on forums: Proof-of-concept study. J Med Internet Res. 2018;20(06):e215. doi: 10.2196/jmir.9840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gil D, Girela J L, Azorin J, De Juan A, De Juan J. Identifying central and peripheral nerve fibres with an artificial intelligence approach. Appl Soft Comput. 2018;67:276–85. [Google Scholar]
- 13.Srividya M, Mohanavalli S, Bhalaji N. Behavioral Modeling for Mental Health using Machine Learning Algorithms. J Med Syst. 2018;42(05):88. doi: 10.1007/s10916-018-0934-5. [DOI] [PubMed] [Google Scholar]
- 14.Lopez B, Torrent-Fontbona F, Vinas R, Fernandez-Real J M. Single Nucleotide Polymorphism relevance learning with Random Forests for Type 2 diabetes risk prediction. Artif Intell Med. 2018;85:43–9. doi: 10.1016/j.artmed.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Orjuela-Canon A D, Camargo Mendoza J E, Awad Garcia C E, Vergara Vela E P. Tuberculosis diagnosis support analysis for precarious health information systems. Comput Methods Programs Biomed. 2018;157:11–7. doi: 10.1016/j.cmpb.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 16.Hernandez-Medrano I, Tello J, Belda C, Urena A, Salcedo I, Espinosa-Anke L et al. Savana: Re-using Electronic Health Records with Artificial Intelligence. Int J Int Mult Art Int. 2017;4(07):8–12. [Google Scholar]
- 17.Wang H, Lv Y, Chen H, Li Y, Zhang Y, Lu Z. Smart pathological brain detection system by predator-prey particle swarm optimization and single-hidden layer neural-network. Multimed Tools Appl. 2018;77(03):3871–85. [Google Scholar]
- 18.Vukicevic A M, Jovicic G R, Jovicic M N, Milicevic V L, Filipovic N D. Assessment of cortical bone fracture resistance curves by fusing artificial neural networks and linear regression. Comput Methods Biomech Biomed Engin. 2018;21(02):169–76. doi: 10.1080/10255842.2018.1431220. [DOI] [PubMed] [Google Scholar]
- 19.Dorado-Moreno M, Pérez-Ortiz M, Gutiérrez P A, Ciria R, Briceño J, Hervás-Martínez C. Dynamically weighted evolutionary ordinal neural network for solving an imbalanced liver transplantation problem. Artif Intell Med. 2017;77:1–11. doi: 10.1016/j.artmed.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 20.Wong B, Ho G TS, Tsui E. Development of an intelligent e-healthcare system for the domestic care industry. Indl Manage Data Syst. 2017;117(07):1426–45. [Google Scholar]
- 21.Yoon J, Zame W R, Banerjee A, Cadeiras M, Alaa A M, van der Schaar M.Personalized survival predictions via Trees of Predictors: An application to cardiac transplantation PLoS One 20181303e0194985eCollection 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moreira M WL, Rodrigues J JPC, Kumar N, Al-Muhtadi J, Korotaev V. Nature-Inspired Algorithm for Training Multilayer Perceptron Networks in e-health Environments for High-Risk Pregnancy Care. J Med Syst. 2018;42(03):51. doi: 10.1007/s10916-017-0887-0. [DOI] [PubMed] [Google Scholar]
- 23.Schetinin V, Jakaite L, Krzanowski W. Bayesian averaging over Decision Tree models for trauma severity scoring. Artif Intell Med. 2018;84:139–45. doi: 10.1016/j.artmed.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Kazemi Y, Mirroshandel S A. A novel method for predicting kidney stone type using ensemble learning. Artif Intell Med. 2018;84:117–26. doi: 10.1016/j.artmed.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez B, Herrero P, Rawson T M, Moore L SP, Evans B, Toumazou C et al. Supervised learning for infection risk inference using pathology data. BMC Med Inform Decis Mak. 2017;17(01):168. doi: 10.1186/s12911-017-0550-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valdes G, Simone C B, 2nd, Chen J, Lin A, Yom S S, Pattison A J et al. Clinical decision support of radiotherapy treatment planning: A data-driven machine learning strategy for patient-specific dosimetric decision making. Radiother Oncol. 2017;125(03):392–7. doi: 10.1016/j.radonc.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 27.Saleh E, Blaszczynski J, Moreno A, Valls A, Romero-Aroca P, de la Riva-Fernandez S et al. Learning ensemble classifiers for diabetic retinopathy assessment. Artif Intell Med. 2018;85:50–63. doi: 10.1016/j.artmed.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 28.Al-Jarrah M A, Shatnawi H. Non-proliferative diabetic retinopathy symptoms detection and classification using neural network. J Med Eng Technol. 2017;41(06):498–505. doi: 10.1080/03091902.2017.1358772. [DOI] [PubMed] [Google Scholar]
- 29.Tang Q, Liu Y, Liu H. Medical image classification via multiscale representation learning. Artif Intell Med. 2017;79:71–8. doi: 10.1016/j.artmed.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 30.Prevedello L M, Erdal B S, Ryu J L, Little K J, Demirer M, Qian S et al. Automated Critical Test Findings Identification and Online Notification System Using Artificial Intelligence in Imaging. Radiology. 2017;285(03):923–31. doi: 10.1148/radiol.2017162664. [DOI] [PubMed] [Google Scholar]
- 31.Gharehbaghi A, Linden M, Babic A. A Decision Support System for Cardiac Disease Diagnosis Based on Machine Learning Methods. Stud Health Technol Inform. 2017;235:43–7. [PubMed] [Google Scholar]
- 32.Zhao C, Jiang J, Xu Z, Guan Y. A study of EMR-based medical knowledge network and its applications. Comput Methods Programs Biomed. 2017;143:13–23. doi: 10.1016/j.cmpb.2017.02.016. [DOI] [PubMed] [Google Scholar]
- 33.Horng S, Sontag D A, Halpern Y, Jernite Y, Shapiro N I, Nathanson L A.Creating an automated trigger for sepsis clinical decision support at emergency department triage using machine learning PLoS One 20171204e0174708eCollection 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu E SF, Wu V WC, Harris B, Foote M, Lehman M, Chan L WC. Vector-model-supported optimization in volumetric-modulated arc stereotactic radiotherapy planning for brain metastasis. Med Dosim. 2017;42(02):85–9. doi: 10.1016/j.meddos.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Lee H, Tajmir S, Lee J, Zissen M, Yeshiwas B A, Alkasab T K et al. Fully Automated Deep Learning System for Bone Age Assessment. J Digit Imaging. 2017;30(04):427–41. doi: 10.1007/s10278-017-9955-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma S, Galatzer-Levy I R, Wang X, Fenyö D, Shalev A Y. A First Step towards a Clinical Decision Support System for Post-traumatic Stress Disorders. AMIA Annu Symp Proc. 2017;2016:837–43. [PMC free article] [PubMed] [Google Scholar]
- 37.Schiff G D, Volk L A, Volodarskaya M, Williams D H, Walsh L, Myers S G et al. Screening for medication errors using an outlier detection system. J Am Med Inform Assoc. 2017;24(02):281–7. doi: 10.1093/jamia/ocw171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nowakova J, Prilepok M, Snasel V.Medical Image Retrieval Using Vector Quantization and Fuzzy Stree J Med Syst. 2017410218. Erratum in: J Med Syst 2018 May; 42(5):98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paydar K, Niakan Kalhori S R, Akbarian M, Sheikhtaheri A. A clinical decision support system for prediction of pregnancy outcome in pregnant women with systemic lupus erythematosus. Int J Med Inform. 2017;97:239–46. doi: 10.1016/j.ijmedinf.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 40.Rajkumar S, Muttan S, Sapthagirivasan V, Jaya V, Vignesh S S. Software intelligent system for effective solutions for hearing impaired subjects. Int J Med Inform. 2017;97:152–62. doi: 10.1016/j.ijmedinf.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 41.Dusenberry M W, Brown C K, Brewer K L. Artificial neural networks: Predicting head CT findings in elderly patients presenting with minor head injury after a fall. Am J Emerg Med. 2017;35(02):260–7. doi: 10.1016/j.ajem.2016.10.065. [DOI] [PubMed] [Google Scholar]
- 42.Finkelstein J, Jeong I C. Machine learning approaches to personalize early prediction of asthma exacerbations. Ann N Y Acad Sci. 2017;1387(01):153–65. doi: 10.1111/nyas.13218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patel T A, Puppala M, Ogunti R O, Ensor J E, He T, Shewale J B et al. Correlating mammographic and pathologic findings in clinical decision support using natural language processing and data mining methods. Cancer. 2017;123(01):114–21. doi: 10.1002/cncr.30245. [DOI] [PubMed] [Google Scholar]
- 44.Yoon J, Davtyan C, van der Schaar M. Discovery and Clinical Decision Support for Personalized Healthcare. IEEE J Biomed Health Inform. 2017;21(04):1133–45. doi: 10.1109/JBHI.2016.2574857. [DOI] [PubMed] [Google Scholar]
- 45.Semenov I, Kopanitsa G. Decision Support System Based on FHIR Profiles. Stud Health Technol Inform. 2018;249:117–21. [PubMed] [Google Scholar]
- 46.Isasi I, Irusta U, Elola A, Aramendi E, Ayala U, Alonso E, Kramer-Johansen J
- 47.Hussain D, Al-Antari M A, Al-Masni M A, Han S M, Kim T S. Femur segmentation in DXA imaging using a machine learning decision tree. J Xray Sci Technol. 2018;26(05):727–46. doi: 10.3233/XST-180399. [DOI] [PubMed] [Google Scholar]
- 48.Shirwaikar R D. Estimation of Caffeine Regimens: A Machine Learning Approach for Enhanced Clinical Decision Making at a Neonatal Intensive Care Unit (NICU) Crit Rev Biomed Eng. 2018;46(02):93–115. doi: 10.1615/CritRevBiomedEng.2018025933. [DOI] [PubMed] [Google Scholar]
- 49.Oude Nijeweme-d’Hollosy W, van Velsen L, Poel M, Groothuis-Oudshoorn C GM, Soer R, Hermens H. Evaluation of three machine learning models for self-referral decision support on low back pain in primary care. Int J Med Inform. 2018;110:31–41. doi: 10.1016/j.ijmedinf.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 50.Jackson R, Kartoglu I, Stringer C, Gorrell G, Roberts A, Song X et al. CogStack - experiences of deploying integrated information retrieval and extraction services in a large National Health Service Foundation Trust hospital. BMC Med Inform Decis Mak. 2018;18(01):47. doi: 10.1186/s12911-018-0623-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng Y, Liu X, Wang Y, Shen F, Liu S, Rastegar-Mojarad M et al. Recommending Education Materials for Diabetic Questions Using Information Retrieval Approaches. J Med Internet Res. 2017;19(10):e342. doi: 10.2196/jmir.7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vaccaro G, Peláez J I, Gil-Montoya J A. A novel expert system for objective masticatory efficiency assessment. PLoS One. 2018;13(01):e0190386. doi: 10.1371/journal.pone.0190386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Montaña D, Campos-Roca Y, Pérez C J. A Diadochokinesis-based expert system considering articulatory features of plosive consonants for early detection of Parkinson’s disease. Comput Methods Programs Biomed. 2018;154:89–97. doi: 10.1016/j.cmpb.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 54.Martín-González S, Navarro Mesa, Juliá-Serdá G, Kraemer J F, Wessel N, Ravelo-García A G. Heart rate variability feature selection in the presence of sleep apnea: An expert system for the characterization and detection of the disorder. Comput Biol Med. 2017;91:47–58. doi: 10.1016/j.compbiomed.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 55.Chung C J, Kuo Y C, Hsieh Y Y, Li T C, Lin C C, Liang W M et al. Subject-enabled analytics model on measurement statistics in health risk expert system for public health informatics. Int J Med Inform. 2017;107:18–29. doi: 10.1016/j.ijmedinf.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 56.Mukhopadhyay A, Maliapen M, Ong V, Jakes R W, Mundy L M, Jialiang Let al. Community-Acquired Pneumonia Case Validation in an Anonymized Electronic Medical Record-Linked Expert System Clin Infect Dis 201764(suppl_2):S141–4. [DOI] [PubMed] [Google Scholar]
- 57.Guidi G, Maffei N, Vecchi C, Gottardi G, Ciarmatori A, Mistretta G M et al. Expert system classifier for adaptive radiation therapy in prostate cancer. Australas Phys Eng Sci Med. 2017;40(02):337–48. doi: 10.1007/s13246-017-0535-5. [DOI] [PubMed] [Google Scholar]
- 58.Haupt F, Berding G, Namazian A, Wilke F, Böker A, Merseburger A et al. Expert System for Bone Scan Interpretation Improves Progression Assessment in Bone Metastatic Prostate Cancer. Adv Ther. 2017;34(04):986–94. doi: 10.1007/s12325-017-0505-z. [DOI] [PubMed] [Google Scholar]
- 59.LeCun Y, Bengio Y, Hinton G E.Deep learning Nature 2015521(7553):436–44. [DOI] [PubMed] [Google Scholar]
- 60.Elkin P L, Schlegel D R, Anderson M, Komm J, Ficheur G, Bisson L. Artificial Intelligence: Bayesian versus Heuristic Method for Diagnostic Decision Support. Appl Clin Inform. 2018;9(02):432–9. doi: 10.1055/s-0038-1656547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brown D, Aldea A, Harrison R, Martin C, Bayley I. Temporal case-based reasoning for type 1 diabetes mellitus bolus insulin decision support. Artif Intell Med. 2018;85:28–42. doi: 10.1016/j.artmed.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 62.Shen Y, Yuan K, Chen D, Colloc J, Yang M, Li Y et al. An ontology-driven clinical decision support system (IDDAP) for infectious disease diagnosis and antibiotic prescription. Artif Intell Med. 2018;86:20–32. doi: 10.1016/j.artmed.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 63.Thanathomwong B. Bayesian-Based Decision Support System for Assessing the Needs for Orthodontic Treatment. Health Inform Res. 2018;24(01):22–8. doi: 10.4258/hir.2018.24.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Segundo U, Aldámiz-Echevarría L, López-Cuadrado J, Buenestado D, Andrade F, Pérez T A et al. Improvement of newborn screening using a fuzzy inference system. Expert Syst Appl. 2017;78:301–18. [Google Scholar]
- 65.Anselma L, Mazzei A, De Michieli F. An artificial intelligence framework for compensating transgressions and its application to diet management. J Biomed Inform. 2017;68:58–70. doi: 10.1016/j.jbi.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 66.Mendez J A, Leon A, Marrero A, Gonzalez-Cava J M, Reboso J A, Estevez J I et al. Improving the anesthetic process by a fuzzy rule based medical decision system. Artif Intell Med. 2018;84:159–70. doi: 10.1016/j.artmed.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 67.Shang Y, Wang Y, Gou L, Wu C, Zhou T, Li J S. Development of a Service-Oriented Sharable Clinical Decision Support System Based on Ontology for Chronic Disease. Stud Health Technol Inform. 2017;245:1153–7. [PubMed] [Google Scholar]
- 68.Nakawala H, Ferrigno G, De Momi E. Development of an intelligent surgical training system for Thoracentesis. Artif Intell Med. 2018;84:50–63. doi: 10.1016/j.artmed.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 69.Abidi S. A Knowledge-Modeling Approach to Integrate Multiple Clinical Practice Guidelines to Provide Evidence-Based Clinical Decision Support for Managing Comorbid Conditions. J Med Syst. 2017;41(12):193. doi: 10.1007/s10916-017-0841-1. [DOI] [PubMed] [Google Scholar]
- 70.Hao S R, Geng S C, Fan L X, Chen J J, Zhang Q, Li L J. Intelligent diagnosis of jaundice with dynamic uncertain causality graph model. J Zhejiang Univ Sci B. 2017;18(05):393–401. doi: 10.1631/jzus.B1600273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hanke S, Kreiner K, Kropf J, Scase M, Gossy C. Reasoning and Data Representation in a Health and Lifestyle Support System. Stud Health Technol Inform. 2017;235:8–12. [PubMed] [Google Scholar]
- 72.Zamborlini V, da Silveira M, Pruski C, Ten Teije A, Geleijn E, van der Leeden M et al. Analyzing interactions on combining multiple clinical guidelines. Artif Intell Med. 2017;81:78–93. doi: 10.1016/j.artmed.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 73.Hossain M S, Ahmed F, Fatema-Tuj Johora, Andersson K. A Belief Rule Based Expert System to Assess Tuberculosis under Uncertainty. J Med Syst. 2017;41(03):43. doi: 10.1007/s10916-017-0685-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vilhena J, Rosario Martins M, Vicente H, Graneda J M, Caldeira F, Gusmao R et al. An Integrated Soft Computing Approach to Hughes Syndrome Risk Assessment. J Med Syst. 2017;41(03):40. doi: 10.1007/s10916-017-0688-5. [DOI] [PubMed] [Google Scholar]
- 75.Khozeimeh F, Alizadehsani R, Roshanzamir M, Khosravi A, Layegh P, Nahavandi S. An expert system for selecting wart treatment method. Comput Biol Med. 2017;81:167–75. doi: 10.1016/j.compbiomed.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 76.Seol J W, Yi W, Choi J, Lee K S. Causality patterns and machine learning for the extraction of problem-action relations in discharge summaries. Int J Med Inform. 2017;98:1–12. doi: 10.1016/j.ijmedinf.2016.10.021. [DOI] [PubMed] [Google Scholar]
- 77.González Aguña A, Díaz Teruel V, Satamaría Pérez A, Gómez González J L, Jiménez Rodríguez M L, Santamaría García J M et al. Dx-Care: A Device to Help in the Diagnosis of Care Problems. Stud Health Technol Inform. 2018;250:256–60. [PubMed] [Google Scholar]
- 78.Braido F, Santus P, Corsico A G, Di Marco F, Melioli G, Scichilone N et al. Chronic obstructive lung disease “expert system”: validation ofa predictive tool for assisting diagnosis. Int J Chron Obstruct Pulmon Dis. 2018;13:1747–53. doi: 10.2147/COPD.S165533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Safdari R, Arpanahi H K, Langarizadeh M, Ghazisaiedi M, Dargahi H, Zendehdel K. Design a Fuzzy Rule-based Expert System to Aid Earlier Diagnosis of Gastric Cancer. Acta Inform Med. 2018;26(01):19–23. doi: 10.5455/aim.2018.26.19-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hassanzad M, Orooji A, Valinejadi A, Velayati A. A fuzzy rule-based expert system for diagnosing cystic fibrosis. Electron Physician. 2017;9(12):5974–84. doi: 10.19082/5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Marquet P, Bedu A, Monchaud C, Saint-Marcoux F, Rérolle J P, Etienne I et al. Pharmacokinetic Therapeutic Drug Monitoring of Advagraf in More Than 500 Adult Renal Transplant Patients, Using an Expert System Online. Ther Drug Monit. 2018;40(03):285–91. doi: 10.1097/FTD.0000000000000503. [DOI] [PubMed] [Google Scholar]
- 82.Başciftci F, Avuclu E. An expert system design to diagnose cancer by using a new method reduced rule base. Comput Methods Programs Biomed. 2018;157:113–20. doi: 10.1016/j.cmpb.2018.01.020. [DOI] [PubMed] [Google Scholar]
- 83.Vijay S AA, Ganesh Kumar P. Fuzzy Expert System based on a Novel Hybrid Stem Cell (HSC) Algorithm for Classification of Micro Array Data. J Med Syst. 2018;42(04):61. doi: 10.1007/s10916-018-0910-0. [DOI] [PubMed] [Google Scholar]
- 84.Paredes R, Tzou P L, van Zyl G, Barrow G, Camacho R, Carmona S et al. Collaborative update of a rule-based expert system for HIV-1 genotypic resistance test interpretation. PLoS One. 2017;12(07):e0181357. doi: 10.1371/journal.pone.0181357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rahmani Katigari M, Ayatollahi H, Malek M, Kamkar Haghighi M. Fuzzy expert system for diagnosing diabetic neuropathy. World J Diabetes. 2017;8(02):80–8. doi: 10.4239/wjd.v8.i2.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dreyer K J, Geis J R. When Machines Think: Radiology’s Next Frontier. Radiology. 2017;285(03):713–8. doi: 10.1148/radiol.2017171183. [DOI] [PubMed] [Google Scholar]
- 87.Ali T, Sungyoung L.Reconciliation of SNOMED CT and domain clinical model for interoperable medical knowledge creation. Conf Proc IEEE Eng Med Biol Soc 2017 Jul; 2654-7 [DOI] [PubMed]
- 88.Launchbury J.A DARPA perspective on artificial intelligence. DARPA Video [Internet]. 2017. Available from:https://www.youtube.com/watch?v=-O01G3tSYpU
- 89.Ehteshami Bejnordi B, Veta M, Johannes van Diest P, van Ginneken B, Karssemeijer N, Litjens G et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women with Breast Cancer. JAMA. 2017;318(22):2199–10. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peleg M, Shahar Y, Quaglini S, Broens T, Budasu R, Fung N et al. Assessment of a personalized and distributed patient guidance system. Int J Med Inform. 2017;101:108–30. doi: 10.1016/j.ijmedinf.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 91.Peleg M. Computer-interpretable clinical guidelines: A methodological review. J Biomed Inform. 2013;46(04):744–63. doi: 10.1016/j.jbi.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 92.Sacchi L, Lanzola G, Viani N, Quaglini S. Personalization and Patient Involvement in Decision Support Systems: Current Trends. Yearb Med Inform. 2015;10(01):106–18. doi: 10.15265/IY-2015-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shay L A, Lafata J E. Where is the evidence? A systematic review of shared decision making and patient outcomes. Med Decis Making. 2015;35(01):114–31. doi: 10.1177/0272989X14551638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.McDermott M S, While A E. Maximizing the healthcare environment: a systematic review exploring the potential of computer technology to promote self-management of chronic illness in healthcare settings. Patient Educ Couns. 2013;92(01):13–22. doi: 10.1016/j.pec.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 95.Middleton B, Sittig D F, Wright A.Clinical Decision Support: a 25 Year Retrospective and a 25 Year Vision Yearb Med Inform 2016. Suppl 1:S103–16. [DOI] [PMC free article] [PubMed] [Google Scholar]


