Abstract
Mutualistic symbioses are often a source of evolutionary innovation and drivers of biological diversification1. Widely distributed in the microbial world, particularly in anoxic settings2,3, they often rely on metabolic exchanges and syntrophy2,4. Here, we report a mutualistic symbiosis observed in marine anoxic sediments between excavate protists (Symbiontida, Euglenozoa)5 and ectosymbiotic Deltaproteobacteria biomineralizing ferrimagnetic nanoparticles. Light and electron microscopy observations as well as genomic data support a multi-layered mutualism based on collective magnetotactic motility with division of labour and interspecies hydrogen-transfer-based syntrophy6. The guided motility of the consortia along the geomagnetic field is allowed by the magnetic moment of the non-motile ectosymbiotic bacteria combined with the protist motor activity, which is a unique example of eukaryotic magnetoreception7 acquired by symbiosis. The nearly complete deltaproteobacterial genome assembled from a single consortium contains a full magnetosome gene set8, but shows signs of reduction, with the probable loss of flagellar genes. Based on the metabolic gene content, the ectosymbiotic bacteria are anaerobic sulfate-reducing chemolithoautotrophs that likely reduce sulfate with hydrogen produced by hydrogenosome-like organelles6 underlying the plasma membrane of the protist. In addition to being necessary hydrogen sinks, ectosymbionts may provide organics to the protist by diffusion and predation, as shown by magnetosome-containing digestive vacuoles. Phylogenetic analyses of 16S and 18S ribosomal RNA genes from magnetotactic consortia in marine sediments across the Northern and Southern hemispheres indicate a host–ectosymbiont specificity and co-evolution. This suggests a historical acquisition of magnetoreception by a euglenozoan ancestor from Deltaproteobacteria followed by subsequent diversification. It also supports the cosmopolitan nature of this type of symbiosis in marine anoxic sediments.
Many organisms are able to sense the geomagnetic field to geolocate and navigate9,10. Yet, the genetic, cellular and biophysical processes involved in magnetoreception have remained elusive in sensory biology for decades7,11. So far, magnetoreception has only been well described in magnetotactic bacteria8. These prokaryotes sense magnetic fields owing to ferrimagnetic nanocrystals biomineralized in organelles called magnetosomes12. Their alignment in chains parallel to the cell-motility axis gives a magnetic moment to the bacteria. This ensures navigation of the cells along magnetic field lines using their flagellar apparatus12,13. Despite a 30-year-old report suggesting the existence of magnetotactic microbial algae14, no study has ever proved that microbial eukaryotes (protists) could produce such magnetite-based organelles for magnetoreception.
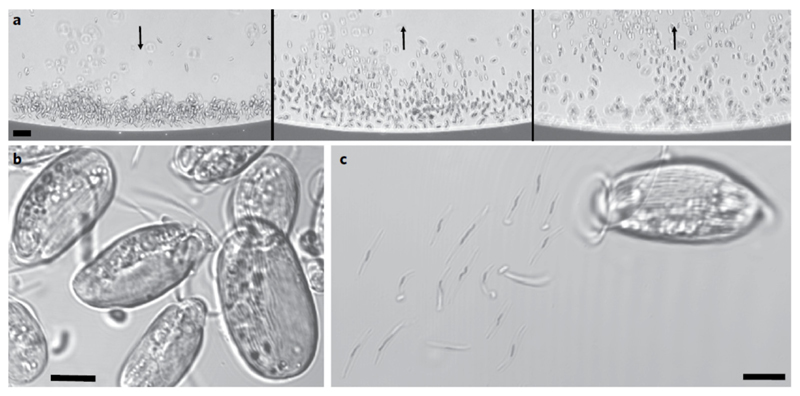
Looking for magnetotactic organisms, we applied a standard magnetic enrichment protocol to marine sediments and water in a dyked area sheltered from major currents in the Mediterranean coast in Carry-le-Rouet, France (see Site description and sample collection in Methods). After magnetic enrichment, populations of north-seeking magnetotactic bacteria, attracted by the south pole of a magnet, were observed as the dominant magnetically responsive organisms (Supplementary Fig. 1). However, we uncovered populations of atypical south-seeking organisms under a light microscope by switching the polarity of the magnetic field (Fig. 1a and Supplementary Video 1). The behaviour and morphological features of these ovoid, striated-looking and flagellated organisms, 16 ± 2 μm in length and 8 ± 2 μm in width (n = 50), were consistent with those of protists (Fig. 1b). The depth profiles of oxygen fugacity and concentrations of magnetotactic cells in the sediment showed that magnetic protists were located below the oxic–anoxic transition zone (Supplementary Fig. 2). These protists were the only organisms with a south-seeking polarity for most of the samples (Supplementary Video 1). When magnetic protists were immobilized between a microscope slide and coverslip under a light microscope, frictions and osmotic shocks led to their mechanical disaggregation and revealed the presence of dozens of 5-μm-long, rod-shaped and slightly curved cells attached to the protist surface (Fig. 1c). Transmission and scanning electron microscopy (TEM and SEM, respectively) analyses confirmed that each magnetic organism was actually an assemblage composed of bacteria biomineralizing magnetosomes and covering entirely a unicellular flagellated protist (Fig. 2a–d). Each of the ectosymbiotic bacteria contained a single chain of 27 ± 9 magnetite particles (n = 100) with a length of 74 ± 8 nm and a width of 67 ± 8 nm (n = 100; Fig. 2b,e and Supplementary Fig. 3). These magnetite particles did not present the morphological features usually observed in magnetite-producing environmental magnetotactic bacteria—that is, the chains were not composed of homogeneously distributed octahedral or bullet-shaped magnetosomes (Supplementary Fig. 1 compared with Supplementary Fig. 3 and Fig. 2e). The bacteria and their magnetosome chains were tightly arranged parallel to the motility axis of the eukaryotic cells (Fig. 2a,d), which optimizes the magnetic moment of the entire consortium. The cohesiveness between the bacteria involved overlapping wing-like structures observed by TEM along each side of the cells (Supplementary Fig. 4a–c). Such wing-like structures were previously described for the ectosymbiotic bacteria of Postgaardi mariagerensis15. Mucilage probably also helped maintain bacterial attachment to the protist cell (Supplementary Fig. 4d). Once detached from the surface of their host, the bacteria remained aligned with the artificial magnetic field but could not swim because of the absence of a flagellum (Supplementary Fig. 5a–e). As a consequence, magnetic ectosymbiotic bacteria (MEB) cannot be considered magnetotactic bacteria, as a magnetotactic bacterium is by definition a motile prokaryote whose motility is influenced by magnetic field lines12. Altogether, these observations indicate that the magnetic orientation of the protist is achieved through the associated magnetic bacteria.
Fig. 1.
Light microscope images of south-seeking magnetic protists sampled in the Mediterranean Sea, Carry-le-Rouet, France. a, The microscope was focused on a point at the edge of the water droplet closest to the north pole of the bar magnet, producing a local field direction indicated by the black arrow (left). Reversing the bar magnet so that the south magnetic pole was closest to the edge of the drop caused south-seeking organisms to rotate and swim in the opposite direction towards the opposite edge of the droplet (middle and right, also indicated by black arrows). b,c, Confocal images of the striated, magnetically responsive protists (b) and their disaggregation 20 min after deposition between a slide and coverslip, showing the presence of rod-shaped bacteria detached from the surface of the protist (c). Scale bars, 20 μm (a) and 5 μm (b,c).
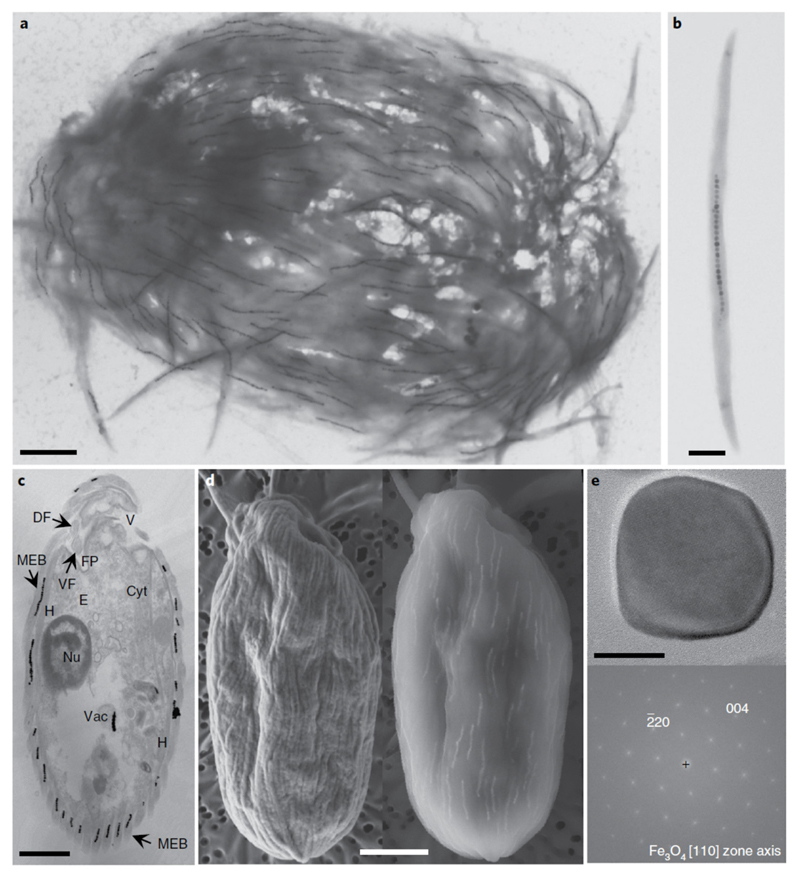
Fig. 2.
Electron microscopy images of the magnetic protist sampled in the Mediterranean Sea, Carry-le-Rouet. a,b, TEM images showing the ultrastructure of a single magnetic consortium containing about 150 magnetosome chains (a) and a magnetic ectosymbiotic bacterium detached from its host (b). c, TEM image of the longitudinal section through a single magnetic consortium showing the general morphological features of the magnetic protist, such as the nucleus (Nu), a battery of extrusomes (E), the vestibulum (V), the cytostome (Cyt), MEB on the extracellular matrix, the flagellar pocket (FP) with the dorsal and ventral flagella (DF and VF, respectively), hydrogenosomes (H) or mitochondria-like organelles in close vicinity to the ectosymbionts, and digestive vacuoles (Vac) in which grazed magnetotactic bacteria and their magnetosomes can be seen. d, Images of a single magnetic consortium observed using a SEM operating at 2 kV (left) or 10 kV (right) showing the presence of magnetosome chains in the bacteria that cover the protist. e, High-resolution TEM image of a single magnetosome biomineralized by an ectosymbiotic bacterium (top) and the corresponding fast Fourier transform (bottom) for which labelled reflexions have been indexed with respect to the magnetite structure. No octahedral or elongated asymmetric shapes are clearly visible (Supplementary Fig. 3). Scale bars, 2 μm (a,c,d), 0.5 μm (b) and 20 nm (e).
We searched for this type of magnetic consortia in other locations to assess their distribution in sediments. Although they were never detected in freshwater sediments and marine areas exposed to major currents, they were routinely observed in the anoxic sediment layers from sheltered marine environments. Protists with similar magnetic behaviour and morphology to those from Carry-le-Rouet were observed in sediments from the Mediterranean Sea at Port Leucate, Port de Boulouris and Cap de Creus, and from the Pacific Ocean at San Francisco Bay, USA and the Akaroa peninsula in New Zealand (Supplementary Fig. 6). Magnetic protists from the Southern Hemisphere also had a swimming direction opposite to that expected for magnetotactic organisms16. All protists shared the same phenotypic traits, exhibiting synapomorphies defined for the Euglenozoa; for example, all have two flagella with reinforced heteromorphic paraxonemal rods5 (that is, one dorsal flagellum with a tubular rod and one ventral flagellum with a lattice structure; Fig. 2c). The morphology and the peculiar association with ectosymbiotic bacteria suggested that the protist host belonged to the Symbiontida group. Symbiontida include few described species (that is, Postgaardi mariagerensis, Bihospites bacati and Calkinsia aureus) that also live in oxygen-deprived marine sediments and harbour ectosymbiotic bacteria17–19.
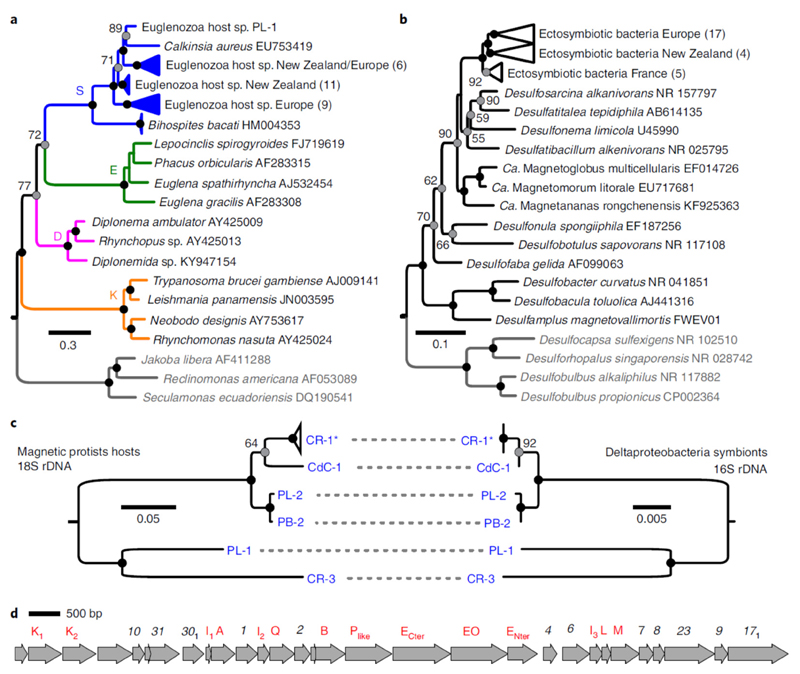
The identity of both the host and symbionts of the magnetic consortia was then investigated by sequencing of the 18S and 16S ribosomal RNA genes amplified from magnetically purified populations and sorted single holobionts (that is, a single protist cell with its MEB). The 18S rRNA phylogenetic analysis confirmed the protists affiliation to the Symbiontida, with Calkinsia aureus being the genetically closest described species (79–84% identity; Fig. 3a). The MEB formed a monophyletic clade affiliated to the Deltaproteobacteria whose members are mostly sulfate-reducing bacteria. They clustered within the order Desulfobacterales (Fig. 3b), which includes the characterized magnetotactic bacteria Candidatus Magnetoglobus multicellularis20 and Desulfamplus magnetovallimortis 21. We report here a magnetosome-producing bacterium affiliated to the Deltaproteobacteria whose magnetite particles are not bullet-shaped22. Fluorescence in situ hybridization using specific fluorescently labelled oligonucleotide probes further confirmed the MEB identity (Supplementary Fig. 7). Only one single deltaproteobacterial 16S rRNA sequence could be retrieved from the cloning and sequencing of individual symbiotic consortia, suggesting the presence of a single ectosymbiotic phylotype per consortium. Accordingly, we searched for any evidence of co-evolution by comparing 16S/18S rRNA gene phylogenies built from hosts and their associated symbionts. The topology of phylogenetic trees of the hosts and their respective MEB were congruent, suggesting that the two symbiotic partners co-evolved and diversified from a single ancestral magnetotactic symbiosis established between Symbiontida and Deltaproteobacteria (Fig. 3c).
Fig. 3.
Diversity of the magnetic protists and their ectosymbionts. a,b, Phylogenetic trees based on 18S rRNA and 16S rRNA gene sequences showing the evolutionary relationships of the magnetic protists with the Euglenozoa (a; S, Symbiontida; E, Euglenida; D, Diplonemida and K, Kinetoplastida) and the MEB with the Desulfobacteraceae (b), respectively. The trees were rooted with other Excavata (protists) and Desulfobacterales (bacteria) families (species in grey). The number of clones obtained are indicated in parenthesis. The numbers next to the grey circles at nodes represent the proportional bootstrap support values. The black circles represent nodes supported by 100% of the replicates. The GenBank accession numbers are also shown. c, Host and symbiont phylogenies built from a subsample of sequences obtained from individual single holobionts only. Topology congruence provides evidence of co-evolution. CR-1* represents a collapsed clade of four single holobionts for which the 16S sequences were identical. CR, Carry-le-Rouet; PL, Port Leucate; PB, Port de Boulouris and CdC, Cap de Creus. d, Chromosomal section containing a magnetosome gene cluster showing the organization of different mam (red) and mad (black) genes of the MEB from a representative holobiont CR-1 isolated from Carry-le-Rouet.
Altogether, the behaviour and structural peculiarities of the magnetotactic holobionts, the existing knowledge about Symbiontida lifestyle and the evidence for host–ectosymbiont co-evolution support that protists and MEB established an obligate mutualistic symbiosis involving magnetoreception. In the absence of available cultures to evaluate a potential fitness gain brought by the symbiosis, we looked for genomic features providing information about mutual beneficial effects. The potential metabolic dependence of ectosymbionts on their protist host was investigated by genome analysis of a single holobiont isolated from Carry-le-Rouet and named CR-1 after whole-genome amplification (WGA) and sequencing. The MEB population CR-1 is considered to be representative of the magnetotactic holobionts as the 16S rRNA gene sequence was at least 92% identical with that of all the 26 MEB sequenced in this study. However, further sequencing will allow assessment of the core genome of these organisms. We assembled a 3.2 Mb long genome of the MEB (98.1% complete), which was significantly smaller than the genome of its free-living closest relatives (Supplementary Table 1), from short and long sequencing reads. Genome size reduction is a process well known in obligate parasites and mutualistic symbionts23, which lose non-essential functions in the symbiotic context by becoming highly specialized and trophically dependent on their host partner24. The functional annotation of the MEB genome in comparison with that of other free-living magnetotactic and nonmagnetotactic Desulfobacteraceae, showed that the proportion of genes involved in signal transduction and motility was up to 3.3 and 7.5 times smaller, respectively (Supplementary Fig. 8). More specifically, none of the genes encoding flagellar proteins or classical chemotaxis pathways were detected (Supplementary Notes). As expected, a magnetosome gene cluster typical of magnetotactic Deltaproteobacteria was fully assembled within a large 366 Kb contig (Fig. 3d and Supplementary Table 2). The magnetotactic consortium phenotype in combination with the ectosymbiont gene content further strengthened the idea that the holobiont magnetotaxis results from an interaction in which the MEB exert magnetic sensing, whereas the protist ensures motility (Supplementary Fig. 5 and Supplementary Video 1). This atypical anterior–posterior orientation of the cells polarity regarding the magnetic moment seems to be restricted only to anaerobes, which seek the surface at the bottom limit of the oxic–anoxic transition zone, instead of the deep. The benefit for the magnetic protist remains to be fully determined but might be similar to that attributed to free-living magnetotactic bacteria with the same unusual magnetotactic behaviour25,26: magnetotaxis could facilitate protist navigation towards chemical and redox optimal niches through sensors that have a different response to chemical gradients. On the bacterial side, several advantages may be hypothesized. For instance, transportation by a predatory protist could reduce the energy cost linked to flagellar-apparatus assembly27 and might also be a way to provide protection for the MEB against predation by other protists. In return, the shell formed by ectosymbionts could maintain an extracellular skeletal structure as described for other symbioses28 and conceal the protist from other predators. However, the most likely benefit for the bacteria is metabolic trade-off with its host.
Elaborated syntrophy, in which metabolic exchange is made possible by the positioning of the magnetotactic consortium in optimal redox conditions, appears the ultimate basis of this symbiosis. The metabolic gene content of MEB strongly suggests that they are strict anaerobic bacteria gaining energy by sulfate reduction coupled with the oxidation of organic or inorganic compounds like other Desulfobacteraceae29. Several sets of genes encoding the major metabolic components involved in sulfate reduction were indeed found in the MEB genome (for example, the dsrAB operon; Supplementary Table 2), while genes involved in hydrogen oxidation (for example, the hynABC operon)29 were also found. Moreover, the MEB genome harboured key enzymes of the Wood–Ljungdahl pathway for CO2 fixation under autotrophic growth conditions (Supplementary Table 2)30. Therefore, the magnetic ectosymbionts are likely anaerobes that are able to grow chemolithoautotrophically with H2 as the electron donor, sulfate as the electron acceptor and CO2 as a carbon source. Consistent with the holobiont biotope, this metabolic potential seems complementary to that assumed for the protist.
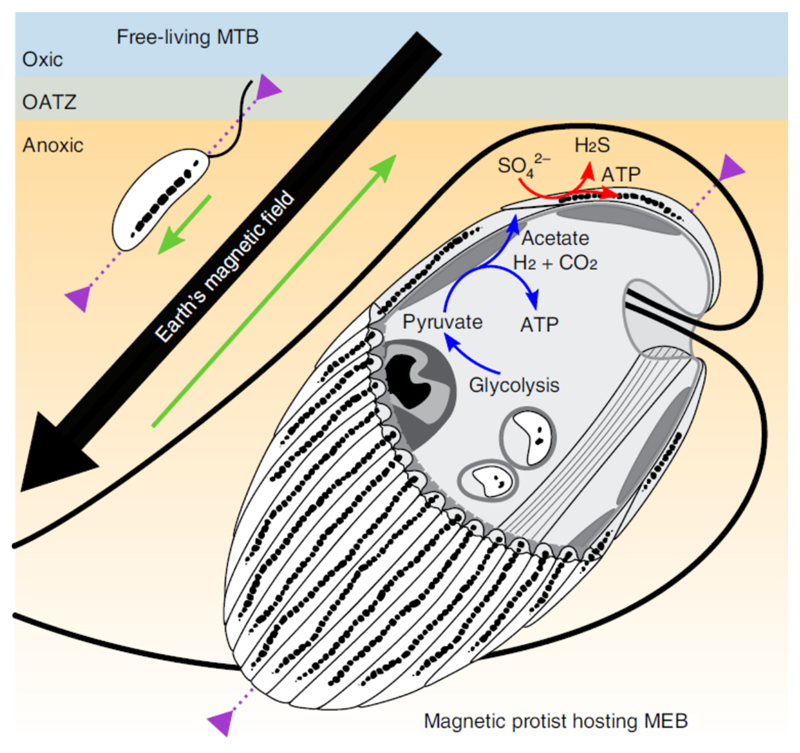
Like other symbiontids found in oxygen-depleted biotopes3,15, the protist harboured a uniform layer of mitochondria-related organelles resembling hydrogenosomes beneath its cell membrane (Fig. 2c and Supplementary Fig. 4). Such mitochondria-related organelles with reduced cristae are known to produce acetate6, molecular hydrogen and CO2 that could diffuse to, and be metabolized by, MEB. Furthermore, the MEB may act as essential hydrogen sinks for the hydrogenosome metabolism to proceed, acting extracellularly very much like endosymbiotic methanogenic archaea or denitrifying bacteria in many hydrogenosome-bearing protists6,31. Hydrogen sulfide produced by MEB sulfate respiration could help maintain a low redox potential surrounding the magnetic holobionts. Although the protist gets an immediate benefit from having a hydrogen sink, it might also benefit from the bacterial primary production. The acquisition of organics might occur via diffusion out of the bacteria coupled to protist osmotrophy, although direct evidence for this is lacking. However, it might also happen by direct predation as in other Euglenozoa5. Several observations indicate that these magnetotactic protists are bacteriovorous phagotrophs capable, among others, of consuming their ectosymbiotic bacteria: (1) the presence of a rod-based feeding apparatus and a tubular cytostome, (2) the predation behaviour observed under the light microscope and (3) the presence of cells with partially dissolved magnetosomes in digestive vacuoles as has already been observed in other grazers32 (Fig. 2c and Supplementary Fig. 9). The morphology and size of the partially degraded magnetosomes that were observed were similar to those biomineralized by the MEB, strongly suggesting that the host feeds on MEB and might cultivate them as a kitchen garden. Based on the MEB gene content and the ultrastructure and behaviour of the consortium, we proposed a model describing protist–MEB symbiosis and the mutual effects relative to several biological functions: navigation and nutrition (Fig. 4). Laboratory culturing assays and the functional annotation of the genome and transcriptome of both partners will enrich this model in the future.
Fig. 4.
Schematic illustration of the magnetotactic consortium showing the magnetotactic behaviour in the Northern Hemisphere and the syntrophic interactions between partners. The green arrows show the anterior–posterior orientation of the organisms, which is parallel and antiparallel to the Earth’s magnetic field lines for the freeliving magnetotactic bacteria (MTB) and the consortium protist-MEB, respectively. The purple arrows show the organism’s motility zone in the sediments. The ATP synthesis by the hydrogenosomes in the protist is symbolized by the blue arrows. Molecular hydrogen, acetate and carbon dioxide are products that could be transported through the plasma and used by the MEB as sources of energy and carbon. The red arrows show the dissimilatory sulfate reduction by MEB, which uses hydrogen as an electron donor and produces hydrogen sulfide outside the consortium. OATZ, oxic–anoxic transition zone.
The discovery of this magnetotactic symbiosis not only challenges our vision of the diversity of magnetically sensitive organisms, but also extends our knowledge of the ecological strategies involved in niche adaptation and symbiosis. Even if the existence of eukaryotic magnetite-based magnetoreceptors was suggested in microalgae (Anisonema, Euglenoidea) 30 years ago14, the lack of further characterization kept the mechanistic basis and the phylogenetic distribution of magnetoreception in unicellular eukaryotes a mystery. Given our data, it is very likely that the original ‘magnetotactic microalga’ was actually a symbiotic association between a symbiontid Euglenozoa and MEB, meaning that no unicellular eukaryote is currently known to produce magnetosomes by itself. The diversity and ecology of this type of symbiosis based on collective magnetotactic motility and interspecies hydrogen-transferbased syntrophy remains to be further characterized. However, the magnetotactic behaviour will undoubtedly facilitate the study of symbiontids and open the way towards the understanding of the mechanistic basis of cooperative sensing and its role in adaptation to anoxic environments.
Methods
Site description and sample collection
Samples were collected by free-diving in the Mediterranean Sea, in Carry-le-Rouet (43.334222° N, 5.175278° E), Port Leucate (42.885295° N, 3.050832° E), Port de Boulouris (43.413902° N, 6.807110° E) and Cap de Creus (42.323277° N, 3.307613° E), and in the Pacific Ocean in San Francisco Bay (37.862580° N, -122.315526° W) and Akaroa, New Zealand (−43.805799° N, 172.965876° E). One-litre plastic bottles were filled to about 0.2–0.3× of their volume with sediment, then filled to capacity with water that overlaid the sediment. Air bubbles were excluded. At least three bottles were collected for each site sampled. Once in the laboratory, the samples were stored under dim light at room temperature (~25 °C). For reasons of ease of access to the sampling site, the detailed ultrastructural characterization and metagenomics were carried out on samples collected from Carry-le-Rouet. The magnetotactic behaviour of the consortia, morphology of the ectosymbiotic cells and their magnetosome shape, and identification of the ectosymbionts and protistan hosts were carried out for all samples. All of these features were very similar between magnetotactic holobionts from the different locations.
Magnetic enrichment and light microscope observation
For each sample, magnetic cells were concentrated by placing two magnetic stirring bars next to the bottles above the sediment–water interface for 2 h, one magnet had its north pole pointing against the bottle (to concentrate south-seeking cells) and the other magnet had its south pole against the bottle (to concentrate north-seeking cells). Examination of magnetically concentrated cells was carried out using the hanging drop technique33 under a Zeiss Primo Star light microscope equipped with phasecontrast and differential interference contrast optics. The local magnetic field used to determine magnetotaxis was reversed by rotating the stirring bar magnet 180° on the microscope stage. Motility and the magnetotactic behaviour of the magnetic protists were also analysed and recorded under the Leica LMD6000 light microscope equipped with a Leica DMC 4500 camera.
Chemical and cell count profiles in microcosms
Dissolved oxygen profiles were measured using a fibre-optic oxygen sensor (50 μm tip diameter; REF OXR50) and a FireStingO2 meter, both from Pyroscience. High-resolution profiles (100 μm steps) were achieved with a motorized micromanipulator (UNISENSE, MM33-2). Sensor calibration was made against saturated air humidity (100%) and a saturated sodium sulfite solution (0%). For cell counts, 40 μl of pore water and sediment was carefully and slowly removed at specific depths in the sample every 2 mm and observed using the hanging drop technique33,34. Magnetic protists accumulating at the edge of the drops due to the magnetic field generated on one side of the drop were counted. The number of cells counted in each drop was multiplied by 25 to obtain the concentration of cells per millilitre. The cell counts were reported as the mean of triplicate counts from the same depth. These measurements, along with dissolved oxygen profiles, were conducted on three different samples collected in Carry-le-Rouet.
Confocal microscopy
Morphological features of the magnetic protists were determined with a Zeiss LSM780 confocal microscope equipped with a Plan-Apochromat ×63/1.40 oil DIC M27 objective. Magnetically concentrated protists were deposited between a slide and coverslip, leading to the detachment of the ectosymbiotic cells from the surface of the euglenozoan after approximately 20 min. Detachment of the bacteria was accelerated by exposure of the sample to 405-nm-wavelength light, the osmotic shock induced by the addition of water next to the coverslip and/or the pressure exerted on the coverslip.
TEM
TEM was used to characterize the ultrastructure of the magnetic protists directly deposited onto TEM copper grids coated with a carbon film, employing an improved technique previously described by Monteil and colleagues32. Electron micrographs were recorded with a Tecnai G2 BioTWIN (FEI Company) equipped with a CCD camera (Megaview III, Olympus Soft imaging Solutions GmbH) with an accelerating voltage of 100 kV. High-resolution TEM, Z-contrast imaging in the high angle annular dark field (STEM-HAADF) mode and X-ray energy dispersive spectroscopy (XEDS) elemental mapping in the STEM-XEDS mode were carried out on a Jeol 2100F microscope. This machine, operating at 200 kV, was equipped with a Schottky emission gun, an ultra-high-resolution pole piece and an ultrathin window JEOL XEDS detector. High-resolution TEM images were obtained with a Gatan US 4000 CCD camera.
Thin-sectioned samples were prepared from magnetically concentrated protists fixed in 2.5% (w/v) glutaraldehyde in sodium cacodylate buffer (0.1 M, pH 7.4) and kept at 4 °C for at least 24 h. Cells were then washed in this same buffer and postfixed for 1 h with 1% (w/v) of osmium tetroxide in cacodylate buffer. Magnetic cells were then dehydrated with successive ethanol baths with increasing concentrations and finally embedded in a monomeric resin (Epon 812). All of the chemicals used for histological preparation were purchased from Electron Microscopy Sciences. Sections (80 nm thick and 3 mm long) were made with the ultramicrotome UCT (Leica Microsystems GmbH), deposited onto TEM copper grids and stained with 1% uranyl acetate for 5 min. Magnetosome and cell sizes were measured from TEM images using ImageJ software (1.48v).
SEM
SEM samples were prepared from magnetically concentrated protists fixed in 2.5% (w/v) glutaraldehyde in sodium cacodylate buffer (0.1 M, pH 7.4) and stored at 4 °C. Samples were filtered on 0.2-μm-diameter isopore Millipore filters (Fig. 2d) or adsorbed on poly-l-lysine-coated coverslips (Supplementary Fig. 5f), then dried and coated with carbon. Images were collected in backscattered and secondary electron modes using a Zeiss Ultra 55 FEG-SEM operating at 1–10 kV, a working distance of 4 mm and an aperture of 10–60 μm.
Cloning and sequencing of the 18S and 16S rRNA genes of magnetically concentrated cells
The 18S and 16S rRNA genes were used to identify magnetically concentrated eukaryotic and prokaryotic cells, respectively, from samples collected in Carry-le-Rouet, Port Leucate, Port de Boulouris, Cap de Creus and Akaroa. Genomic DNA was extracted using the NucleoSpin Soil extraction kit (Macherey-Nagel). DNA was amplified using Phusion Hot Start Flex DNA polymerase following the manufacturer’s recommendations. For eukaryotes, the specific 18S rRNA gene primers EukA 5′-AACCTGGTTGATCCTGCCAGT-3′ and EukB 5′-TGATCCTTCTGCAGGTTCACCTAC-3′ (ref. 35) were used. For bacteria, the primers 27F 5′-AGAGTTTGATCMTGGCTCAG-3′ and 1492R 5′-TACGGHTACCTTGTTACGACTT-3′ (ref. 36) were used. Blunt-end fragments of 16S and 18S rRNA gene sequences were cloned using a Zero Blunt TOPO PCR cloning kit with One Shot TOP10 chemically competent E. coli cells. The inserts of the resulting clones were digested using restriction enzymes to select operational taxonomic units representative of the populations and were sent for sequencing and compared with the NCBI nucleotide database with the Basic Local Alignment Search tool. This first step allowed the taxonomic assignment to the Euglenozoa phylum and Deltaproteobacteria class of the protist host and ectosymbiotic bacteria, respectively, pending further phylogenetic investigation.
Fluorescence in situ hybridization
Fluorescence in situ hybridization was performed according to Pernthaler and colleagues37. A specific ATTO488-labelled probe for the ectosymbiotic bacteria was designed (ECTOp, 5′-CAGTTTCTTCCCACTTGAC-3′) based on the alignment of the most similar 16S rRNA gene sequences of the Desulfobacteraceae family found in GenBank database. The probe specificity was evaluated using the PROBE_MATCH program in the RDP-II38. The nearest non-target hits contain at least one mismatch with the specific probe ECTOp. The nearest non-target hits are from species of the Desulfobacteraceae family along with other taxa. Known multicellular magnetotactic prokaryotes also present in their 16S rRNA gene sequence at least one mismatch with ECTOp. The oligonucleotide probes used in this study were purchased from Eurofins Genomics (Supplementary Table 3). A 20 μl drop of magnetically concentrated cells was deposited on a SuperFrost Plus Gold, Menzel-Gläser (Thermo Scientific) microscope slide. The cells were magnetically directed towards the edge of the drop. After 10 min, the drop was removed and the cells adsorbed on the slide were fixed overnight with 30 μl of a solution of 4% paraformaldehyde. The fixed cells were dehydrated by serial incubation for 5 min each in 50%, 80% and 100% ethanol. The hybridization solution contained 10 ng ml−1 probe, 0.9 M NaCl, 20 mM Tris–HCl (pH 7.4), 1 mM Na2EDTA and 0.01% SDS; the hybridization and washing stringencies recommended for each probe (35% for ECTOp; Supplementary Table 3) were used. Hybridization was performed at 46 °C for 2 h. Slides were mounted with a ProLong Diamond Antifade Mountant (Invitrogen) with or without 4,6-diamidino-2-phenyl-indole (DAPI) and then immediately observed with a Zeiss LSM780 confocal microscope.
Single-consortium sorting and WGA
Single-consortium sorting was carried out on samples collected in Carry-le-Rouet, Port Leucate, Port de Boulouris and Cap de Creus with an InjectMan NI2 micromanipulator and a CellTram vario, hydraulic, manual microinjector from Eppendorf mounted to a Leica DM IL LED microscope equipped with a ×63/0.70 PH objective. The microscope and micromanipulation equipment were placed inside a clean chamber previously exposed for 1 h to ultraviolet germicidal irradiation (wavelength of the lamp: 254 nm). A 10 μl drop of magnetically concentrated cells was gently added to a 30 μl drop of filtered water from the environment on a hydrophobic coverslip to magnetically transfer the magnetic protists towards the edge of the filtered water. A single consortium was transferred with a sterile microcapillary (TransferTip (ES); 15 μm inner diameter) into a 4 μl drop of PBS. This drop containing a single magnetic consortium was stored at 4 °C before WGA. To obtain sufficient gDNA for 16S and 18S rRNA gene and shotgun metagenomic sequencing, WGA was carried out using the multiple displacement amplification technique with the REPLI-g single cell kit (QIAGEN) following the manufacturer’s instructions. The concentration of double-stranded gDNA was measured using the fluorimeter QuBit 4 (ThermoFisher Scientific).
Phylogenetic analysis
Phylogenetic trees based on 18S and 16S rRNA gene sequences were built from the sequences obtained from magnetically concentrated eukaryotic and prokaryotic cells and the closest type strains identified by BLASTN on NCBI (September 2018). For protists, the tree represents the Euglenozoa main groups and was rooted with other Excavata lineages, whereas for bacteria, the tree represents the Desulfobacteraceae family and was rooted with other Desulfobacterales of the Desulfobulbaceae family. A total of 27 complete 18S rRNA gene sequences were used from protists isolated from Carry-le-Rouet (7 sequences), Port Leucate (2 sequences), Port de Boulouris (3 sequences), Cap de Creus (1 sequence) and Akaroa (14 sequences). A total of 26 complete 16S rRNA gene sequences were used from ectosymbiotic bacteria isolated from Carry-le-Rouet (16 sequences), Port Leucate (2 sequences), Port de Boulouris (3 sequences), Cap de Creus (1 sequence) and Akaroa (4 sequences). The multiple sequence alignment was performed with MAFFT39 and trimmed with BMGE40 to get a final alignment containing 1,980 and 1,517 sites, respectively. For both data sets, the tree was built using the maximum-likelihood method implemented in IQ-TREE41 using the TIM3e+R4 and TVMe+R3 substitution models selected by ModelFinder42 with the Bayesian information criterion, respectively. The statistical support of the branches was estimated by the ultrafast bootstrapping method implemented in IQ-TREE with 1,000 replicates. Co-evolution analysis was performed using a subset of sequences obtained after amplification of the 18S and 16S rRNA sequences from WGA products of single consortia. Both maximum-likelihood trees were built using the same approach as described before using the TN+F+R2 and TPM3+F+I models as substitution models for the protist hosts and ectosymbionts, respectively. The statistical support for the branches was estimated by a non-parametric bootstrapping approach implemented in IQ-TREE with 1,000 replicates. Both trees were midpoint rooted.
Shotgun metagenomic sequencing, assembly and functional annotation
One single holobiont, CR-1, was sorted from a sample collected on 30 October 2017 in Carry-le-Rouet using a micromanipulator. Following WGA of this single holobiont, gDNA was first purified on a QIAamp DNA mini kit column (QIAGEN) according to the manufacturer’s protocol. Purified gDNA was then quantified using a QuBit fluorimeter (ThermoFisher Scientific) and quality was evaluated on a 1% TAE agarose gel. For Illumina sequencing, 250 ng WGA DNA was sonicated to a 100–1,000 bp size range using the E210 Covaris instrument (Covaris, Inc.). The fragments were end-repaired, then 3′-adenylated and NextFlex DNA barcodes (Bioo Scientific Corporation) were added using NEBNext DNA modules products (New England Biolabs). After two consecutive clean ups with 1×AMPure XP, the ligated product was amplified by 12 PCR cycles using the Kapa Hifi Hotstart NGS library amplification kit (Kapa Biosystems), followed by purification with 0.6×AMPure XP. After library-profile analysis conducted by a Agilent 2100 Bioanalyzer (Agilent Technologies) and qPCR quantification (MxPro, Agilent Technologies), the library was sequenced using a Illumina MiSeq with MiSeq Reagent Kit v2 (2 × 250 bp; Illumina Inc.). A total of 7.8 × 106 paired-end reads were obtained. The Illumina reads were trimmed with quality control methods before assembly (that is, lowquality nucleotides Q < 20, sequencing adaptors and primer sequences were discarded from the reads and reads shorter than 30 nucleotides after trimming were discarded). For nanopore sequencing, library preparation was done with 2 μg of the same input WGA DNA using the 1D long read Nanopore sequencing kit without BluePippin (SQK-LSK108, Oxford Nanopore). The library was sequenced using nanopore flow cells (R.9.4.1, Oxford Nanopore) and the MinION device with the MinKNOW v.1.10.23 and Albacore v.2.1.10 software. A total of 35,137 reads were obtained with a N50 of 6.8 Kb. An assembly strategy was applied based on SPAdes v.3.12.0 (ref. 43) and Unicycler v.0.4.6 (ref. 44) software. First, two hybrid assemblies (that is, using Illumina and nanopore reads) were launched in parallel with SPAdes (with -k 21,33,55,77,99,127 —only-assembler —sc options) and Unicycler (default options). From the Unicycler assembly graph, the connected component containing the magnetosome gene cluster was detected by alignment with magnetosome protein sequences from other organisms. This component was made of 104 contigs for a total length of 3.2 Mb. In addition, 28 other overlapping contigs (that is, more than 98% identity on more than 1 Kb) from the SPAdes assembly were retrieved by alignment on the 104 contigs of Unicycler. Illumina reads were mapped to these selected contigs with BWA v.0.7.15 (with -k 24 -T 100 options)45 and used as input with nanopore reads to launch a second Unicycler assembly (with —no_correct option and —sc option for SPAdes). Contigs from both Unicycler assemblies were then compared and manually selected by removing inclusions and merging two overlapping contigs. The final assembly resulted in 52 contigs for a total length of 3.2 Mb with a contig N50 of 317 Kb, a GC content of 54% and 9% repeat regions. Assembly completeness and contamination were estimated at 98.1 and 1.3%, respectively, using checkM v.1.0.1146 with a set of 247 Deltaproteobacteria-specific markers. The automatic annotation was performed with the MicroScope platform47. A complete rRNA operon and a total of 48 tRNA (with at least one copy for the 20 canonical amino acids and an additional selenocysteine tRNA) were predicted. We identified 3,013 coding sequences with average coding sequence and intergenic lengths of 966 bp and 145 bp, respectively. Within the MicroScope platform, automatic functional classification of protein-coding genes was made using the eggNOG database v.4.5.1 and the eggNOG-mapper tool v.1.0.3 (with -m diamond option)48. The MEB for which the genome was sequenced undoubtedly represented a distinct genus (Fig. 3b) and based on the phylogeny, what we currently know phenotypically and the potential metabolism of strain CR-1, we propose the name Candidatus Desulfarcum epimagneticum (that is, the magnetic ectosymbiotic sulfate-reducing bacterium with a bow-like morphology).
Supplementary Material
Acknowledgements
This work was supported by a project from the French National Research Agency (ANR Tremplin-ERC, ANR-16-TERC-0025-01). N.M., D.F., C.L.M., D.P. and C.T.L. acknowledge support within the framework of a DFG-ANR project (ANR-14-CE35-0018). R.J.W. and C.T.L. received support from the Dumont d’Urville Science and Technology Programme (grant no. DDU-LVL1501) and New Zealand Ministry for Business, Innovation and Employment (grant no. LVLX1703). D.F. was supported by the Max Planck Society. C.T.L. also received support from the France-Berkeley Fund. P.L.-G. received support from the European Research Council Advanced Grant ‘ProtistWorld’ (grant no. 322669). The SEM facility at IMPMC was supported by funding from Région Ile de France (grant no. SESAME 2006 I-07-593/R); the transmission electron microscopy facility at IMPMC was supported by funding from Region Ile de France (grant no. SESAME 2000 E 1435). We thank A.-L. Monteil and N. Monteil for their help in collecting samples and S. Preveral for her help in confocal observation. Support for the confocal microscope was provided by the Région Provence Alpes Côte d’Azur, Conseil General of Bouches du Rhône, French Ministry of Research, CNRS and Commissariat à l’Energie Atomique et aux Energies Alternatives.
Footnotes
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Author contributions
C.L.M. and C.T.L. designed, performed and analysed most of the experiments. D.V., V.B., S.F. and C.C. designed and performed the genome sequencing and annotation. C.T.L., N.M., K.B. and M.F. performed the electron microscope analyses. E.V. performed the oxygen measurements. G.A. contributed to the identification of microorganisms. N.L. and D.F. contributed to the single-cell sorting. R.J.W. contributed to the sampling. C.L.M. and C.T.L. supervised the project with input from D.P. C.L.M. and C.T.L. wrote the first draft of the manuscript with input from P.L.-G. All authors contributed to data interpretation and editing of the paper.
Competing interests
The authors declare no competing interests.
Data availability
Gene sequences of 18S and 16S rRNA amplified from magnetically purified populations or sorted single holobionts have been assigned to GenBank accession numbers MK131721–MK131747 and MK153697–MK153721, respectively. Sequencing reads and the annotated genome of strain CR-1 were deposited to the European Nucleotide Archive database under the BioProject numbers PRJEB29359 and PRJEB30760, respectively.
References
- 1.Margulis L, Fester R. Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis. MIT Press; 1991. [PubMed] [Google Scholar]
- 2.López-García P, Eme L, Moreira D. Symbiosis in eukaryotic evolution. J Theor Biol. 2017;434:20–33. doi: 10.1016/j.jtbi.2017.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernhard JM, Buck KR, Farmer MA, Bowser SS. The Santa Barbara Basin is a symbiosis oasis. Nature. 2000;403:77–80. doi: 10.1038/47476. [DOI] [PubMed] [Google Scholar]
- 4.Morris BEL, Henneberger R, Huber H, Moissl-Eichinger C. Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev. 2013;37:384–406. doi: 10.1111/1574-6976.12019. [DOI] [PubMed] [Google Scholar]
- 5.Yubuki N, Leander BS. Diversity and evolutionary history of the symbiontida (Euglenozoa) Front Ecol Evol. 2018;6:100. [Google Scholar]
- 6.Müller M, et al. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol Mol Biol Rev. 2012;76:444–495. doi: 10.1128/MMBR.05024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mouritsen H. Long-distance navigation and magnetoreception in migratory animals. Nature. 2018;558:50–59. doi: 10.1038/s41586-018-0176-1. [DOI] [PubMed] [Google Scholar]
- 8.Uebe R, Schüler D. Magnetosome biogenesis in magnetotactic bacteria. Nat Rev Microbiol. 2016;14:621–637. doi: 10.1038/nrmicro.2016.99. [DOI] [PubMed] [Google Scholar]
- 9.Walker MM, et al. Structure and function of the vertebrate magnetic sense. Nature. 1997;390:371–376. doi: 10.1038/37057. [DOI] [PubMed] [Google Scholar]
- 10.Mora CV, Davison M, Wild JM, Walker MM. Magnetoreception and its trigeminal mediation in the homing pigeon. Nature. 2004;432:508–511. doi: 10.1038/nature03077. [DOI] [PubMed] [Google Scholar]
- 11.Nordmann GC, Hochstoeger T, Keays DA. Magnetoreception—a sense without a receptor. PLoS Biol. 2017;15:e2003234. doi: 10.1371/journal.pbio.2003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bazylinski DA, Frankel RB. Magnetosome formation in prokaryotes. Nat Rev Microbiol. 2004;2:217–230. doi: 10.1038/nrmicro842. [DOI] [PubMed] [Google Scholar]
- 13.Popp F, Armitage JP, Schüler D. Polarity of bacterial magnetotaxis is controlled by aerotaxis through a common sensory pathway. Nat Commun. 2014;5 doi: 10.1038/ncomms6398. 5398. [DOI] [PubMed] [Google Scholar]
- 14.Torres de Araujo FF, Pires MA, Frankel RB, Bicudo CEM. Magnetite and magnetotaxis in algae. Biophys J. 1986;50:375–378. doi: 10.1016/S0006-3495(86)83471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson AGB, VandenHoff J, Bernard C, Burton HR, Patterson DJ. The ultrastructure and systematic position of the euglenozoon Postgaardi mariagerensis, Fenchel et al. Arch Protistenkd. 1997;147:213–225. [Google Scholar]
- 16.Blakemore RP, Frankel RB, Kalmijn AJ. South-seeking magnetotactic bacteria in the Southern Hemisphere. Nature. 1980;286:384–385. [Google Scholar]
- 17.Edgcomb VP, et al. Identity of epibiotic bacteria on symbiontid euglenozoans in O2-depleted marine sediments: evidence for symbiont and host co-evolution. ISME J. 2011;5:231–243. doi: 10.1038/ismej.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breglia SA, Yubuki N, Hoppenrath M, Leander BS. Ultrastructure and molecular phylogenetic position of a novel euglenozoan with extrusive episymbiotic bacteria: Bihospites bacati n. gen. et sp. (Symbiontida) BMC Microbiol. 2010;10:145. doi: 10.1186/1471-2180-10-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yubuki N, Edgcomb VP, Bernhard JM, Leander BS. Ultrastructure and molecular phylogeny of Calkinsia aureus: cellular identity of a novel clade of deep-sea euglenozoans with epibiotic bacteria. BMC Microbiol. 2009;9:16. doi: 10.1186/1471-2180-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abreu F, et al. Deciphering unusual uncultured magnetotactic multicellular prokaryotes through genomics. ISME J. 2014;8:1055–1068. doi: 10.1038/ismej.2013.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lefèvre CT, et al. A cultured greigite-producing magnetotactic bacterium in a novel group of sulfate-reducing bacteria. Science. 2011;334:1720–1723. doi: 10.1126/science.1212596. [DOI] [PubMed] [Google Scholar]
- 22.Pósfai M, Lefèvre CT, Trubitsyn D, Bazylinski DA, Frankel RB. Phylogenetic significance of composition and crystal morphology of magnetosome minerals. Front Microbiol. 2013;4:344. doi: 10.3389/fmicb.2013.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323:379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- 24.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2011;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 25.Leão P, et al. North-seeking magnetotactic Gammaproteobacteria in the Southern Hemisphere. Appl Environ Microbiol. 2016;82:5595–5602. doi: 10.1128/AEM.01545-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simmons SL, Bazylinski DA, Edwards KJ. South-seeking magnetotactic bacteria in the Northern Hemisphere. Science. 2006;311:371–374. doi: 10.1126/science.1122843. [DOI] [PubMed] [Google Scholar]
- 27.Bray D, editor. Cell Movements: From Molecules to Motility. 2nd edn. Garland Science; 2001. pp. 3–60. [Google Scholar]
- 28.Leander BS, Keeling PJ. Symbiotic innovation in the oxymonad Streblomastix strix. J Eukaryot Microbiol. 2004;51:291–300. doi: 10.1111/j.1550-7408.2004.tb00569.x. [DOI] [PubMed] [Google Scholar]
- 29.Rabus R, Hansen TA, Widdel F. In: The Prokaryotes. Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. Springer; 2013. pp. 309–404. [Google Scholar]
- 30.Schauder R, Eikmanns B, Thauer RK, Widdel F, Fuchs G. Acetate oxidation to CO2 in anaerobic bacteria via a novel pathway not involving reactions of the citric acid cycle. Arch Microbiol. 1986;145:162–172. [Google Scholar]
- 31.Hamann E, et al. Environmental Breviatea harbour mutualistic Arcobacter epibionts. Nature. 2016;534:254–258. doi: 10.1038/nature18297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Monteil CL, et al. Accumulation and dissolution of magnetite crystals in a magnetically responsive ciliate. Appl Environ Microbiol. 2018;84:e02865–17. doi: 10.1128/AEM.02865-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schüler D. The biomineralization of magnetosomes in Magnetospirillum gryphiswaldense . Int Microbiol. 2002;5:209–214. doi: 10.1007/s10123-002-0086-8. [DOI] [PubMed] [Google Scholar]
- 34.Jogler C, et al. Cultivation-independent characterization of ‘Candidatus Magnetobacterium bavaricum’ via ultrastructural, geochemical, ecological and metagenomic methods. Environ Microbiol. 2010;12:2466–2478. doi: 10.1111/j.1462-2920.2010.02220.x. [DOI] [PubMed] [Google Scholar]
- 35.Medlin L, Elwood HJ, Stickel S, Sogin ML. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene. 1988;71:491–499. doi: 10.1016/0378-1119(88)90066-2. [DOI] [PubMed] [Google Scholar]
- 36.Lane DJ. In: Nucleic Acid Techniques in Bacterial Systematics. Stackebrandt E, Goodfellow M, editors. John Wiley & Sons; 1991. pp. 115–175. [Google Scholar]
- 37.Pernthaler J, Glockner FO, Schonhuber W, Amann R. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Methods Microbiol. 2001;30:207–226. [Google Scholar]
- 38.Cole JR, et al. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 2003;31:442–443. doi: 10.1093/nar/gkg039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katoh K, Standley DM. MAFFT multiple sequence alignment software versio 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Antipov D, Korobeynikov A, McLean JS, Pevzner PA. hybridSPAdes: an algorithm for hybrid assembly of short and long reads. Bioinformatics. 2016;32:1009–1015. doi: 10.1093/bioinformatics/btv688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vallenet D, et al. MicroScope in 2017: an expanding and evolving integrated resource for community expertise of microbial genomes. Nucleic Acids Res. 2017;45:D517–D528. doi: 10.1093/nar/gkw1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huerta-Cepas J, et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 2017;34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene sequences of 18S and 16S rRNA amplified from magnetically purified populations or sorted single holobionts have been assigned to GenBank accession numbers MK131721–MK131747 and MK153697–MK153721, respectively. Sequencing reads and the annotated genome of strain CR-1 were deposited to the European Nucleotide Archive database under the BioProject numbers PRJEB29359 and PRJEB30760, respectively.