Abstract
We sought to determine whether preeclampsia, delivery of a small for gestational age infant or spontaneous preterm birth were associated with the presence of bacterial DNA in the human placenta. Here we show that there was no evidence for the presence of bacteria in the large majority of placental samples, from both complicated and uncomplicated pregnancies. Almost all signals were related either to acquisition of bacteria during labour and delivery or contamination of laboratory reagents with bacterial DNA. The exception was Streptococcus agalactiae (Group B Streptococcus), where non-contaminant signals were detected in ~5% of samples collected prior to the onset of labour. We conclude that bacterial infection of the placenta is not a common cause of adverse pregnancy outcome and that the human placenta does not have a microbiome, but it does represent a potential site of perinatal acquisition of S. agalactiae, a major cause of neonatal sepsis.
Keywords: pre-eclampsia, preterm birth, fetal growth restriction, small for gestational age, bacteria, microbiome, 16S rRNA, shotgun metagenomics
Placental dysfunction is associated with common adverse pregnancy outcomes which determine a substantial proportion of the global burden of disease1. However, the cause of placental dysfunction in the majority of such cases is unknown. A number of studies have employed sequencing-based methods for bacterial detection (metagenomics and 16S rRNA gene amplicon sequencing) and have concluded that the placenta is physiologically colonized with a diverse population of bacteria (the “placental microbiome”) and that the nature of this colonization may differ between healthy and complicated pregnancy2–4. This contrasts with the view in the pre-sequencing era that the placenta was normally sterile5. However, several studies which applied sequencing-based methods informed by the potential for false positive results due to contamination6–8 have failed to detect a placental microbiome9–12. The aim of the present study was to determine whether preeclampsia (PE), delivery of a small for gestational age (SGA) infant and spontaneous preterm birth (PTB) were associated with the presence or a pattern of bacterial DNA in the placenta and to determine whether there was evidence to support the existence of a placental microbiome. We employed samples from a large, prospective cohort study of nulliparous pregnant women,13 applying an experimental approach informed by the potential for false positive results14.
Experimental approach
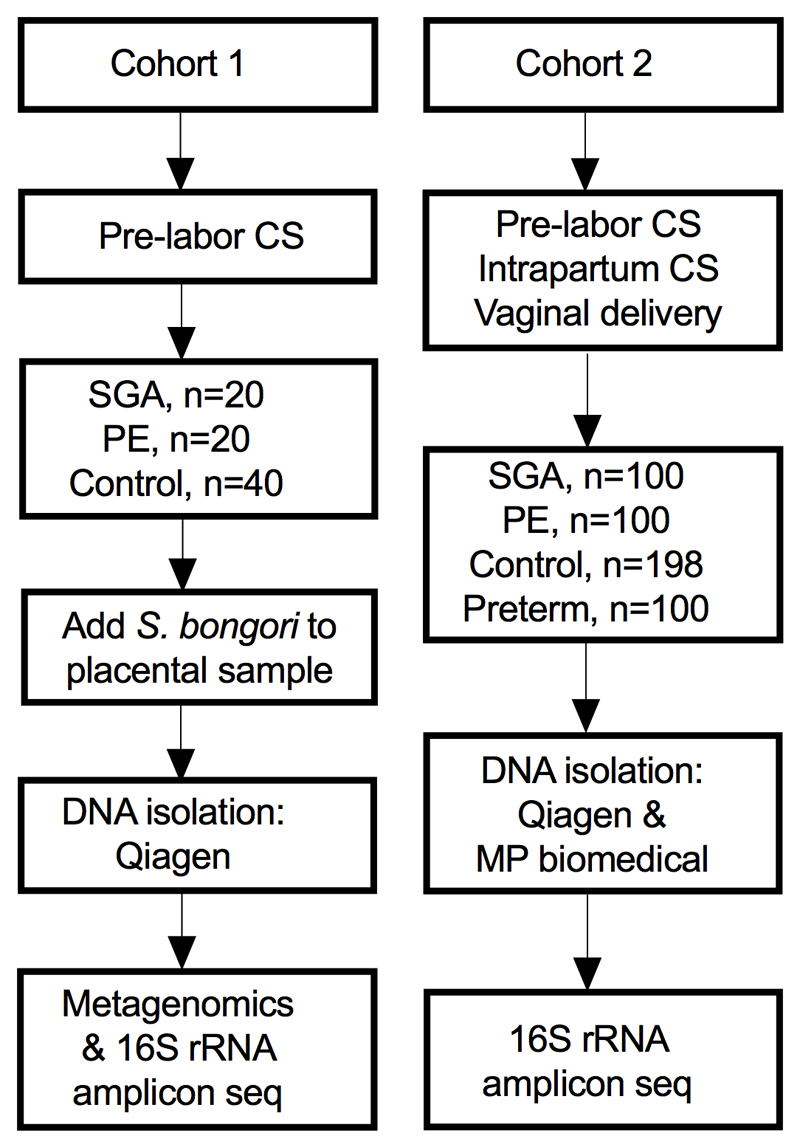
We studied two cohorts of patients (Extended Data Fig. 1 and Supplementary Tables 1 and 2). Cohort 1 were all delivered by pre-labour Caesarean section (CS) and included 20 PE, 20 SGA and 40 matched controls. The placental biopsies were spiked with ~1,100 colony forming units (CFU) of Salmonella bongori (positive control) and samples were analysed using both deep metagenomic sequencing of total DNA (424 million reads on average per sample) and 16S rRNA gene amplicon sequencing. Cohort 2 included 100 PE, 100 SGA, 198 matched controls (two controls were used twice) and 100 preterm births. All these samples were analysed twice using 16S rRNA gene amplicon sequencing from DNA extracted by two different kits.
Cohort 1: Metagenomics and 16S rRNA
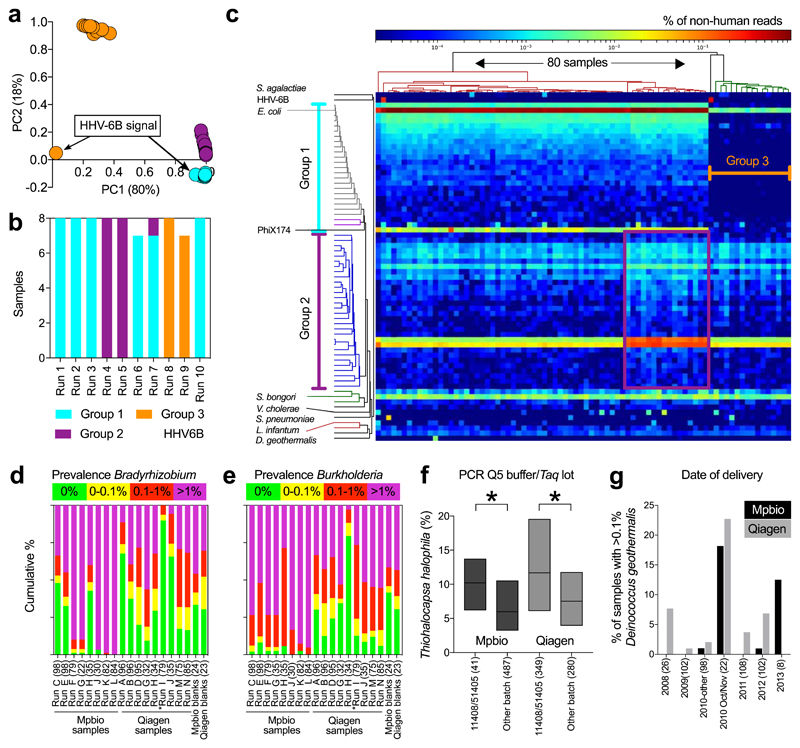
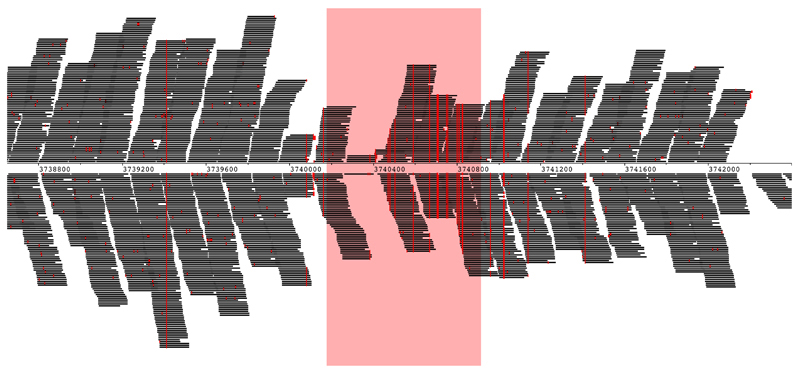
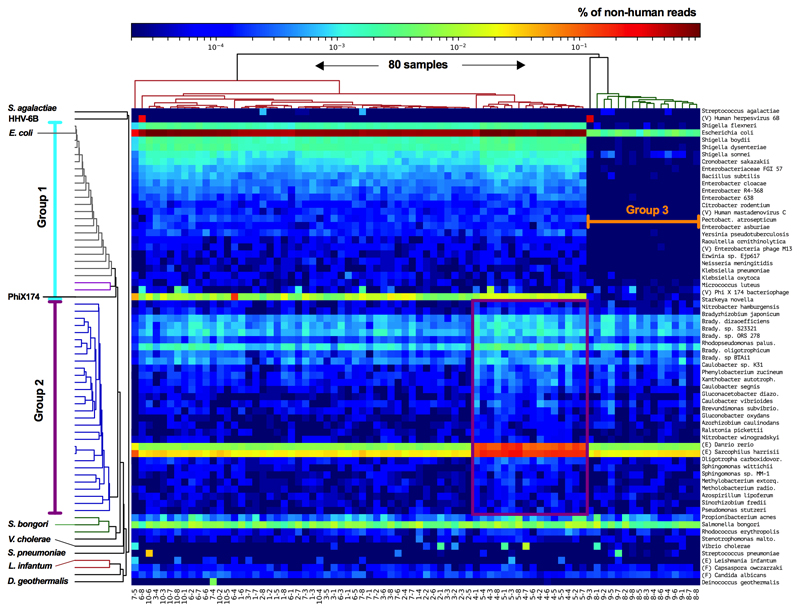
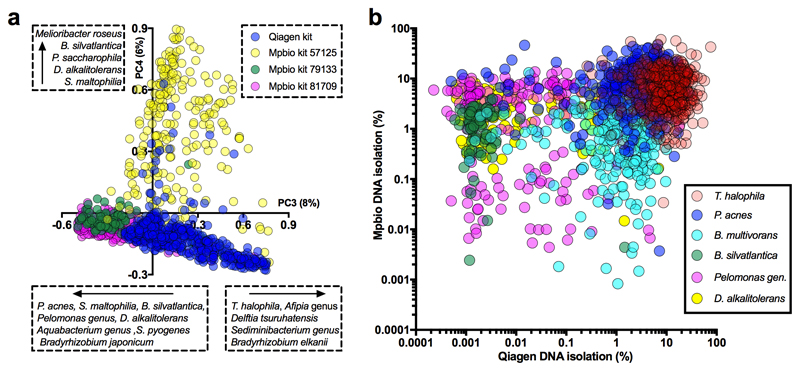
The positive control (S. bongori, average 180 reads per sample, Extended Data Fig. 2a) was detected in all samples. Multiple other bacterial signals were also observed. Principal component analysis (PCA; Fig. 1a) demonstrated that almost all of the variation in the metagenomics data (98%), was represented by principal components 1 (80%) and 2 (18%). This variation was driven by batch effects and not case/control status (Fig. 1b). Any variation that is associated with processing batches, and not the sampling framework, must be due to contamination. A heatmap (Fig. 1c) showed that eight out of the ten runs had a pronounced Escherichia coli signal (>20,000 reads in 64 samples and 50-150 reads in 16 samples), a large collection of additional bacterial signals, and high levels of PhiX174 reads (Group 1; Fig. 1c). Additional analyses mapping all E. coli reads from all samples together against the closest reference genome (WG5) showed that all E. coli reads belonged to the same strain (Extended Data Fig. 3) and are, therefore, due to contamination. All samples belonging to runs 4 and 5 (Fig. 1b) also had strong Bradyrhizobium and Rhodopseudomonas palustris signals (Group 2 in PCA analysis). Runs 8 and 9 (Group 3) lacked these strong signals. Two samples had strong Human Herpesvirus 6B (HHV-6B) signals (>10,000 read pairs; Fig. 1a-c) which reflected inheritance of the chromosomally integrated virus, which affects 0.5-1% of individuals in western populations15.
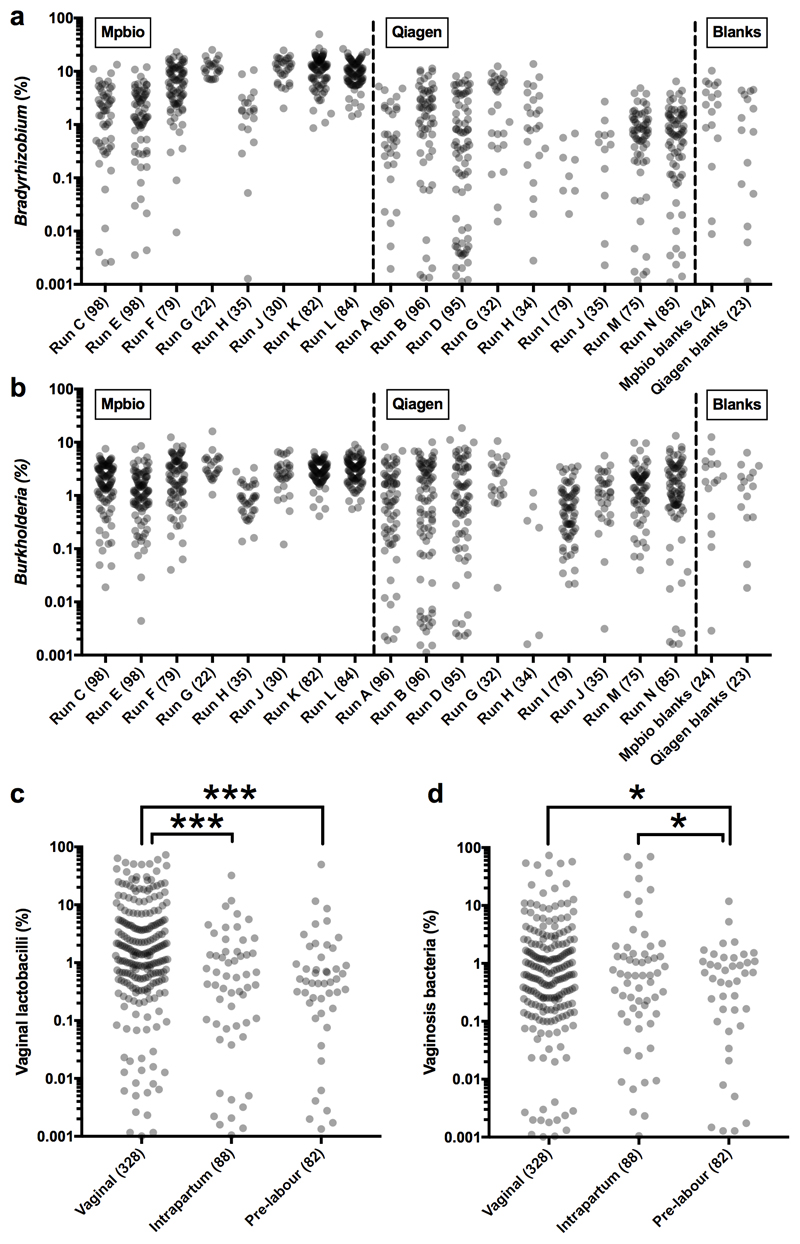
Fig. 1. Batch effect detection in metagenomic and 16S rRNA amplicon sequencing data, Cohort 1 samples.
a-c) Summary of metagenomics data. a) PCA of summarized genus level Kraken output. b) MiSeq sequencing runs (n=8 per run). c) Heatmap of all non-human read abundance (see Extended Data Figure 4). d,e) Read abundance by run and DNA isolation method (Mpbio or Qiagen) in chronological order, (d) Bradyrhizobium, and (e) Burkholderia. Scatterplots are shown in Extended Data Fig. 6. f) Associations between Thiohalocapsa halophila and Q5 Buffer or Taq polymerase. Interquartile range is shown. * P < 0.001. g) D. geothermalis detection (>0.1% reads) by year of delivery. Number of samples in each group (n).
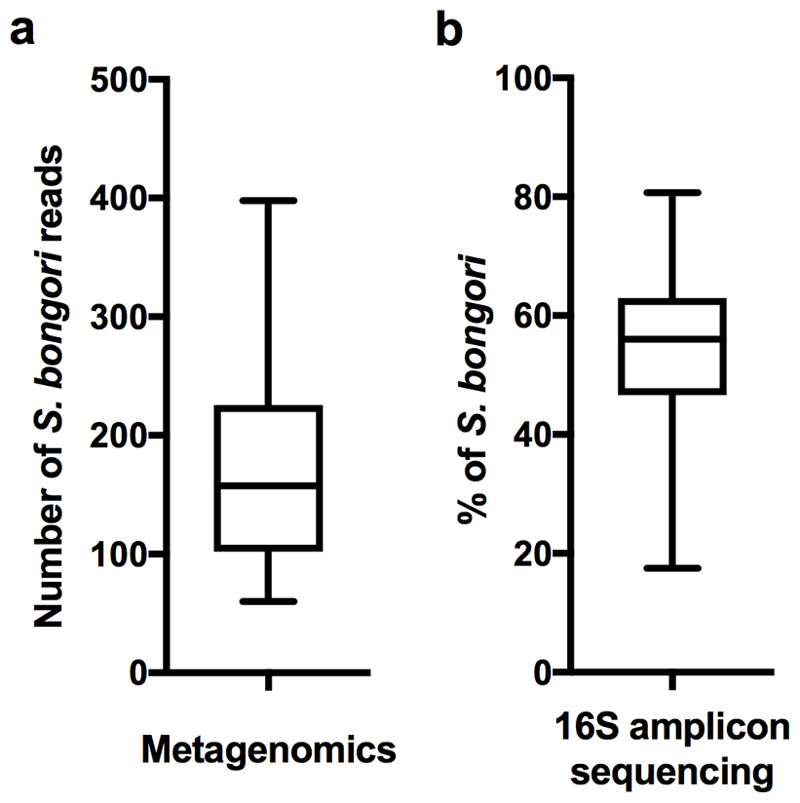
We analysed the concordance between metagenomics and 16S rRNA gene amplicon sequencing in 79 samples from Cohort 1 (Table 1, one 16S primer pair failed). The only signal consistently detected using both methods was S. bongori. An average of ~33,000 S. bongori reads (~54% of total reads) were found by 16S rRNA amplicon sequencing (Extended Data Fig. 2b). S. bongori was not detected in the 16S negative controls (DNA extraction blanks; Table 1). The level of agreement between metagenomics and 16S rRNA for the other bacterial signals was assessed using the kappa statistic, scaled from 0 (no agreement) to 1 (perfect agreement). Only two signals demonstrated agreement (moderate-substantial) between the two methods: Streptococcus agalactiae and Deinococcus geothermalis (Table 1). The results were consistent when using different definitions of positive (Supplementary Table 3) and neither signal was detected in negative controls. The number of positive samples was too small for informative comparison of cases and controls.
Table 1. Comparison of main signals using metagenomics (MG) with 16S rRNA amplicon sequencing.
| Species | Positive signals MG and 16S (79 = max) |
Avg. no. of MG reads in positive samples | Avg. % of 16S reads in positive samples* | Concordance MG and 16S kappa score (P value)† | Part of an MG batch effect‡ | Presence 16S in neg. controls Absent/Weak/Strong (n=5)t§ | |||
|---|---|---|---|---|---|---|---|---|---|
| Both* | MG only | 16S only* | Neither | ||||||
| Salmonella bongori | 79 | 0 | 0 | 0 | 178 | 54 % | N/A | No | 5/0/0 |
| Escherichia coli | 1 | 78 | 0 | 0 | 18602 | 1.2 % | 0 (-) | Gr. 1&2 | 4/1/0 |
| Shigella (genus) | 0 | 75 | 0 | 4 | 254 | N/A | 0 (-) | Gr. 1&2 | 5/0/0 |
| Salmonella enterica | 0 | 75 | 0 | 4 | 33 | N/A | 0 (-) | Gr. 1&2 | 5/0/0 |
| Cronobacter sakazakii | 0 | 65 | 0 | 14 | 21 | N/A | 0 (-) | Gr. 1&2 | 5/0/0 |
| Bacillus subtilis | 0 | 63 | 0 | 16 | 13 | N/A | 0 (-) | Gr. 1&2 | 5/0/0 |
| Y. pseudotuberculosis | 0 | 59 | 0 | 20 | 3 | N/A | 0 (-) | Gr. 1&2 | 5/0/0 |
| Neisseria meningitidis | 0 | 44 | 0 | 35 | 2 | N/A | 0 (-) | Gr. 1&2 | 5/0/0 |
| Bradyrhizobium (genus) | 0 | 79 | 0 | 0 | 125 | N/A | 0 (-) | Gr. 2 | 5/0/0 |
| R. palustris | 0 | 79 | 0 | 0 | 45 | N/A | 0 (-) | Gr. 2 | 5/0/0 |
| Caulobacter (genus) | 12 | 67 | 0 | 0 | 14 | 1.4 % | 0 (-) | Gr. 2 | 1/3/1 |
| Methylobacterium (genus) | 9 | 69 | 0 | 1 | 8 | 2.4 % | 0.003 (0.36) | Gr. 2 | 1/4/0 |
| Burkholderia (genus) | 21 | 57 | 0 | 1 | 7 | 1.9 % | 0.009 (0.27) | Gr. 2 | 1/4/0 |
| Propionibacterium acnes | 66 | 13 | 0 | 0 | 20 | 4.8 % | 0 (-) | No | 0/3/2 |
| S. pneumoniae | 0 | 11 | 0 | 68 | 115 | N/A | 0 (-) | No | 5/0/0 |
| Vibrio cholerae | 0 | 14 | 0 | 65 | 46 | N/A | 0 (-) | No | 5/0/0 |
| Thiohalocapsa halophila | 0 | 0 | 71 | 8 | N/A | 4.2 % | 0 (-) | No | 0/0/5 |
| S. maltophilia | 5 | 51 | 1 | 22 | 2 | 1.9 % | 0.03 (0.24) | No | 2/3/0 |
| Acinetobacter baumanii | 1 | 26 | 0 | 52 | 2 | 2.4 % | 0.05 (0.08) | No | 4/1/0 |
| Micrococcus luteus | 1 | 46 | 0 | 32 | 15 | 2.0 % | 0.02 (0.20) | No | 4/1/0 |
| Gardnerella vaginalis | 0 | 5 | 0 | 74 | 1 | N/A | 0 (-) | No | 4/1/0 |
| Lactobacillus crispatus | 0 | 4 | 0 | 75 | 1 | N/A | 0 (-) | No | 5/0/0 |
| Deinococcus geothermalis | 1 | 1 | 0 | 77 | 68 | 33 % | 0.66 (<0.0001) | No | 5/0/0 |
| Streptococcus agalactiae | 3 | 4 | 0 | 72 | 8 | 13 % | 0.58 (<0.0001) | No | 5/0/0 |
16S rRNA amplicon sequencing signals higher than 1% are defined as positive.
One-sided P values
See Fig. 1 for definition of groups 1 and 2.
Strong signals are defined as more than 1%
A number of bacterial signals associated with PC2, including the Caulobacter, Methylobacterium, and Burkholderia genera, were also detected by 16S rRNA gene sequencing. However, the kappa statistics were low and these signals were also detected in negative controls (Table 1). Vibrio cholerae and Streptococcus pneumoniae signals were detected using metagenomics in 14 and 11 samples, respectively. However, neither was detected using 16S rRNA (Table 1). Assembly and analysis of these reads demonstrated that the closest matches were isolates from Bangladesh (PRJEB14661 V. cholerae) and the Global Pneumococcal Sequence project (PRJEB31141 S. pneumoniae), which had been sequenced on the same pipelines at the Sanger Institute, indicating that these signals are due to cross contamination during library preparation or sequencing (the same explanation applies for Leishmania infantum, Fig. 1c).
Cohort 2: duplicate 16S rRNA
By combining the data from two independent DNA isolation methods (MP Biomedical kit, hereafter “Mpbio”, or Qiagen kit) we were able to visualize batch effects using PCA (Extended Data Fig. 5a) or of species individually (Figure 1d-g) and to analyse signal reproducibility. For example, Bradyrhizobium was detected nearly ubiquitously and in high abundance in some 16S rRNA sequencing runs but was less frequently detected and in lower abundance in others (compare runs K and L with runs I and J; Fig. 1d). The Burkholderia genus, which has been suggested to have a role in PTB3, had a higher signal in samples isolated using the Mpbio DNA isolation reagents than with the Qiagen kit and in addition showed pronounced run-to-run variation (Fig. 1e). Furthermore, both Bradyrhizobium and Burkholderia were commonly detected in the negative controls. Batch effects based on the use of particular PCR reagent lots can similarly be visualized. For example, the association of Thiohalocapsa halophila with either the PCR reagent 5x Q5 Buffer (lot# 11408) or the Q5 Taq polymerase (lot# 51405), which were both used to process the same 390 samples, is shown in Fig. 1f.
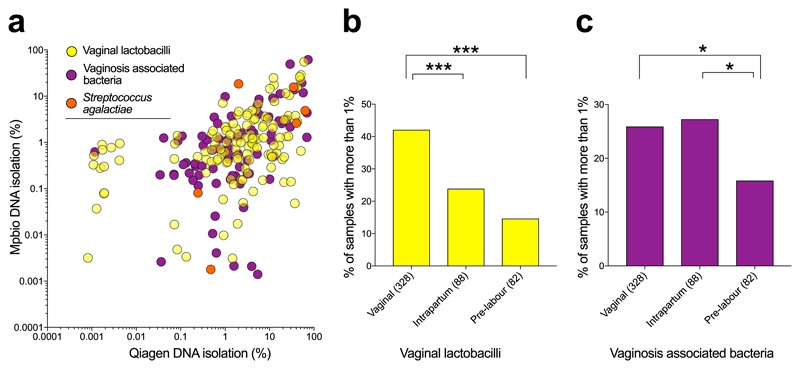
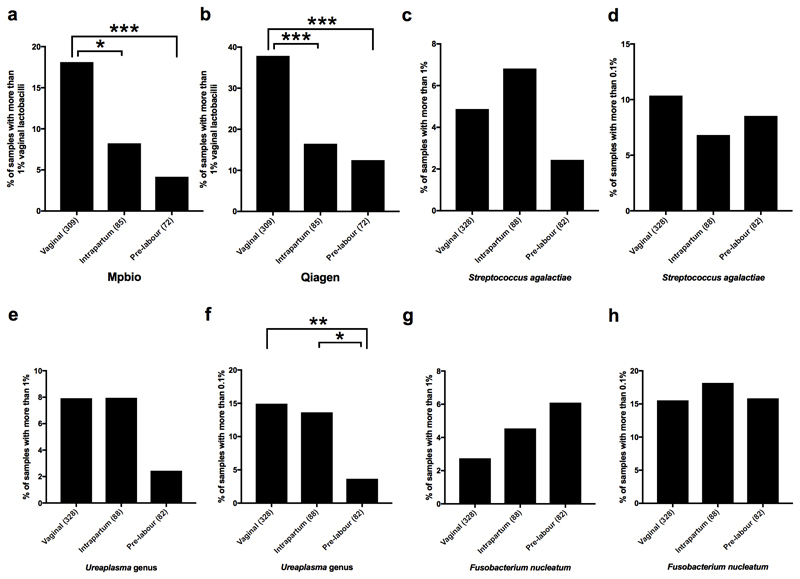
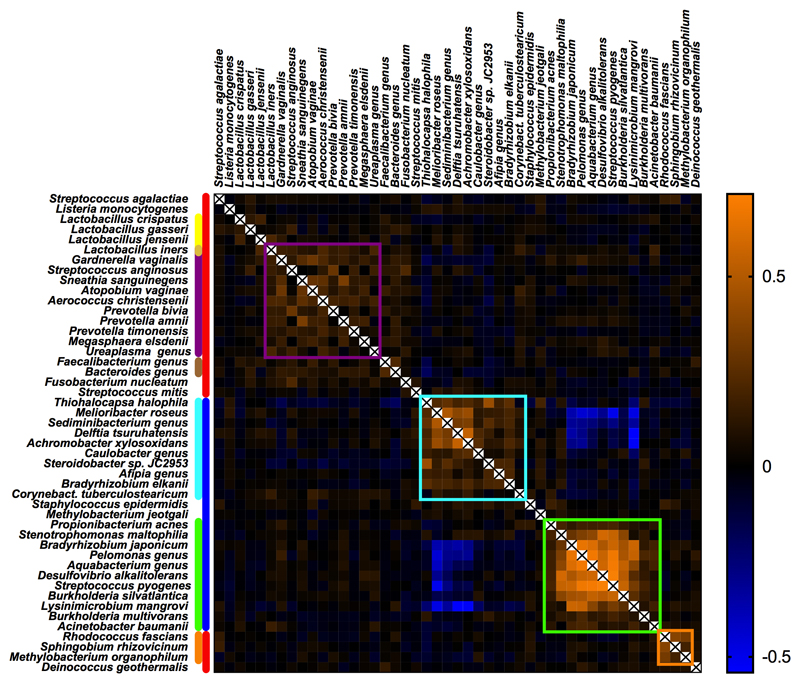
We used the kappa statistic to quantify the level of agreement between 16S rRNA amplicon sequencing of two DNA samples from the same patient extracted using the two different kits (Supplementary Table 4). The majority of the most prevalent bacterial groups had low kappa scores and there was a low correlation between the magnitude of the signals comparing the two DNA extraction methods (Extended Data Fig. 5b). Moreover, these signals also demonstrated striking batch effects using PCA (Extended Data Fig. 5a). Intriguingly, four ecologically unexpected bacterial groups of high prevalence exhibited a fair level of concordance (Rhodococcus fascians, Sphingobium rhizovicinum, Methylobacterium organophilum and D. geothermalis). Further analysis demonstrated a temporal pattern of these signals (Fig. 1g). All placental samples were washed in sterile PBS to remove surface contamination, such as maternal blood, and the temporal pattern of these bacterial signals is consistent with them being derived from batches of this reagent. Some ecologically plausible species, such as S. agalactiae and Listeria monocytogenes, vaginal lactobacilli, vaginosis associated bacteria, faecal bacteria and some bacteria of likely oral origin had modest to high kappa scores, indicating that they were sample associated. In contrast to the laboratory contaminants, the signals for these bacterial groups correlated when comparing the two DNA extractions (Fig. 2a) and were not associated with batch effects identifiable using PCA. Sample-associated signals (non-reagent contaminants) of a few species not typically associated with a vaginal or rectal habitat but with the oral habitat were detected, such as Streptococcus mitis, Streptococcus vestibularis and Fusobacterium nucleatum. However, it was only a very small minority of samples which exhibited these signals (below that of S. agalactiae) and none of these oral signals were identified by metagenomic analysis of pre-labour CS samples (Cohort 1).
Fig. 2. Mode of delivery and detection of vaginal bacteria by 16S rRNA amplicon sequencing.
a) Concordant detection of vaginal lactobacilli and a combination of all vaginosis associated bacteria by both Qiagen (x-axis) and Mpbio (y-axis) results in Spearman’s rho correlation coefficients of 0.37 and 0.59, respectively when analysing the upper right quadrant only (>0.1%). b,c) Comparisons with vaginally associated bacteria and mode of delivery. Mann-Whitney U tests were used where values below 1% are regarded as 0%. * P < 0.05, *** P < 0.001. Scatterplots are in Extended Data Fig. 6. Percent read count based on the higher value for given species using Qiagen or Mpbio DNA isolation kit (using all 498 samples).
Delivery-associated signals
Vaginal organisms (lactobacilli and vaginosis associated bacteria) were more abundant than S. agalactiae in Cohort 2 (vaginal, intrapartum and pre-labour CS deliveries) but less abundant than S. agalactiae in Cohort 1 (pre-labour CS deliveries only). Hence, we next examined the relationship between mode of delivery and the 16S rRNA signal. Vaginal lactobacilli (Lactobacillus iners, Lactobacillus crispatus, Lactobacillus gasseri and Lactobacillus jensenii) were found more frequently and in higher numbers in vaginally delivered placentas than in placentas delivered via intrapartum or pre-labour CS (Fig. 2b), irrespective of DNA isolation method (Extended Data Figs. 7a-b). Vaginosis associated bacteria were found at approximately the same frequency in vaginal and intrapartum CS samples but significantly less frequently in pre-labour CS samples (Fig. 2c). A heatmap generated using the Spearman rho correlation coefficients of all abundant and relevant bacterial groups generated a cluster of vaginally associated bacteria, representative of vaginal community group IV16, which reflects sample contamination during labour and delivery (Extended Data Fig. 8). The other clusters represented the contamination signatures of the two different DNA extraction kits and a fourth cluster reflected contamination associated with the date of collection of the placental biopsies (2012-2013).
Genuine signals and pregnancy outcome
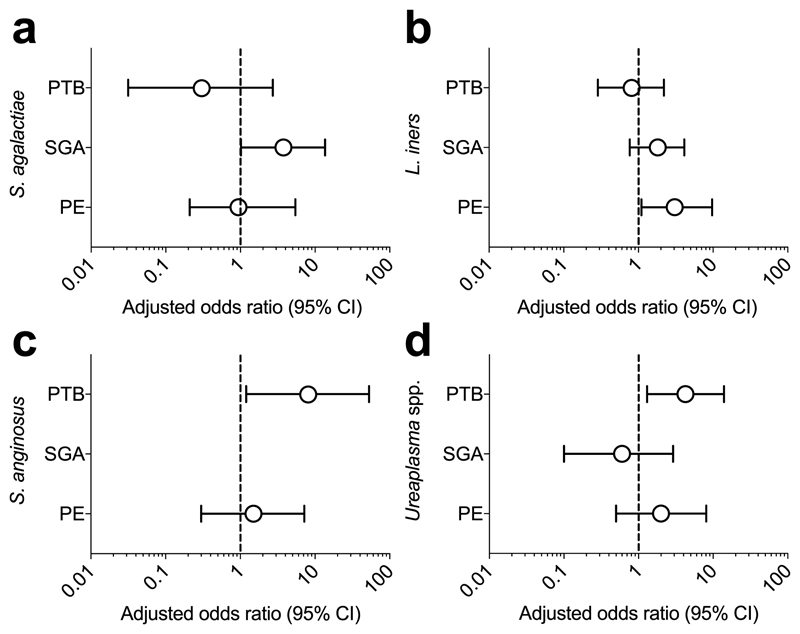
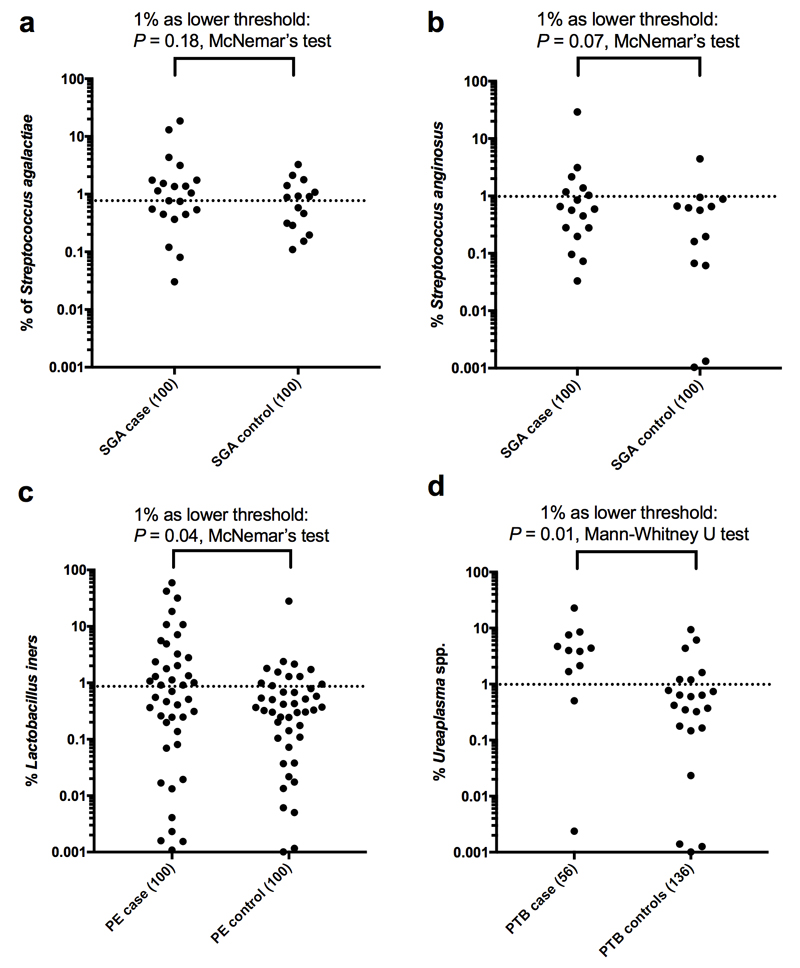
The presence of S. agalactiae was analysed with respect to clinical outcome (SGA, PE, PTB) as it was the only organism that met all the criteria of a genuine placenta-associated bacterial signal (Table 2). There was a non-significant (P=0.06, n=100) trend for an association with SGA (Fig. 3) but no association with PE or PTB. Exploratory analysis of the 16S amplicon sequencing data of all sample-associated signals, including delivery-associated bacteria, showed that S. mitis and F. nucleatum were not associated with adverse pregnancy outcome (Supplementary Table 5). Of note however were the significant associations of the delivery-associated bacteria L. iners with PE and Streptococcus anginosus and the Ureaplasma genus with PTB (Fig. 3, Supplementary Table 5 and Extended Data Fig. 9). In one placental sample from a preterm birth, a strong L. monocytogenes signal was found (7% and 52% of all reads with Mpbio and Qiagen respectively).
Table 2. Simplified overview on the nature of bacterial findings.
| Signals | ||||||
|---|---|---|---|---|---|---|
|
|
||||||
| Independent of: | Not in negative controls‡ | Sample-associated§ | verified meta-genomics‖ | |||
|
|
||||||
| DNA extraction batch* | Date of delivery† | Mode of delivery | ||||
| Capable pathogens | ||||||
| Streptococcus agalactiae | ✔ | ✔ | ✔ | ✔ | ✔ | ✔ |
| Listeria monocytogenes e | ✔ | ✔ | ✔ | ✔ | ✔ | - |
| Vaginal lactobacilli | ||||||
| Lactobacillus crispatus | ✔ | ✔ | - | ~ | ✔ | ~ |
| Lactobacillus iners | ✔ | ✔ | - | ~ | ✔ | - |
| Lactobacillus gasseri | ✔ | ✔ | - | ✔ | ✔ | - |
| Lactobacillus jensenii | ✔ | ✔ | - | ~ | ✔ | - |
| Vaginosis associated bacteria | ||||||
| Gardnerella vaginalis | ✔ | ✔ | - | ~ | ✔ | - |
| Atopobium vaginae | ✔ | ✔ | - | ~ | ✔ | - |
| Ureaplasma genus | ✔ | ✔ | - | ✔ | ✔ | - |
| Prevotella bivia | ✔ | ✔ | - | ~ | ✔ | - |
| Prevotella amnii | ✔ | ✔ | - | ✔ | ✔ | - |
| Prevotella timonensis | ✔ | ✔ | - | ~ | ✔ | - |
| Aerococcus christensenii | ✔ | ✔ | - | ✔ | ✔ | - |
| Streptococcus anginosus | ✔ | ✔ | - | ~ | ✔ | - |
| Sneathia sanguinegens | ✔ | ✔ | - | ✔ | ✔ | - |
| Megasphaera elsdenii | ✔ | ✔ | - | ~ | ✔ | - |
| Faecal associated bacteria | ||||||
| Bacteroides genus | ✔ | ✔ | - | ~ | ✔ | - |
| Faecalibacterium prausnitzii | ✔ | ✔ | - | ~ | ✔ | - |
| Roseburia faeces | - | ✔ | - | ~ | ✔ & - | - |
| Coriobacterium sp. | ✔ | ✔ | - | ~ | ✔ | - |
| Collinsella intestinalis | ✔ | ✔ | - | + | ✔ | - |
| Suspected oral origin | ||||||
| Fusobacterium nucleatum | ✔ | ✔ | ✔ | ~ | ✔ | - |
| Streptococcus mitis | ✔ | ✔ | ✔ | ~ | ✔ | - |
| Streptococcus vestibularis | - | ✔ | ✔ | ~ | ✔ & - | - |
| Genuine reagent contaminants | ||||||
| Acinetobacter baumanii e | - | ✔ | ✔ | ~ | - | ~ |
| Thiohalocapsa halophila | - | ✔ | ✔ | - | - | - |
| Propionibacterium acnes | - | ✔ | ✔ | - | - | - |
| S. maltophilia | - | ✔ | ✔ | - | - | - |
| Bradyrhizobium japonicum | - | ✔ | ✔ | - | - | - |
| Melioribacter roseus | - | ✔ | ✔ | - | - | - |
| Pelomonas genus | - | ✔ | ✔ | - | - | - |
| Methylobacterium genus | - | ✔ | ✔ | - | - | - |
| Aquabacterium genus | - | ✔ | ✔ | - | - | - |
| Sediminibacterium genus | - | ✔ | ✔ | - | - | - |
| Desulfovibrio alkalitolerans | - | ✔ | ✔ | - | - | - |
| Delftia tsuruhatensis | - | ✔ | ✔ | - | - | - |
| Streptococcus pyogenes | - | ✔ | ✔ | ~ | - | - |
| Burkholderia multivorans | - | ✔ | ✔ | - | - | - |
| Caulobacter genus | - | ✔ | ✔ | - | - | - |
| Steroidobacter sp. JC2953 | - | ✔ | ✔ | - | - | - |
| Afipia genus | - | ✔ | ✔ | - | - | - |
| Burkholderia silvatlantica | - | ✔ | ✔ | - | - | - |
| Lysinimicrobium mangrove | - | ✔ | ✔ | - | - | - |
| Bradyrhizobium elkanii | - | ✔ | ✔ | - | - | - |
| Achromobacter xylosoxidans | - | ✔ | ✔ | - | - | - |
| C. tuberculostearicum | - | ✔ | ✔ | - | - | - |
| Rhodococcus fascians | ✔ | - | ✔ | ~ | ✔ | - |
| Sphingobium rhizovicinum | ✔ | - | ✔ | ~ | ✔ | - |
| Methylobac. organophilum | ✔ | - | ✔ | ~ | ✔ | - |
| Deinococcus geothermalis e | ✔ | - | ✔ | ✔ | ✔ | ✔ |
Includes batch effects caused by different DNA isolation kits, PCR reagents and MiSeq run.
A ✔ indicates absence, ~ indicates detection (any %) in less than 20% of negative controls.
Detection of signal in corresponding Qiagen and Mpbio DNA isolations. “✔ & -” indicates that signals from these OTUs are sample-associated in most 16S runs, but reagent contaminants in others. See Supplementary Table 4 for details.
See Table 1 and Supplementary Table 3. A ~ indicates some level of concordance was detected using a different 16S threshold. Presence or absence of verification should be interpreted with caution, as indicated by examples.
Fig. 3. Bacterial signals and adverse pregnancy outcome.
a-d) Adjusted odds ratios for the association of S. agalactiae, L. iners, S. anginosus and Ureaplasma spp. with PTB, SGA and PE. PE and SGA both had 100 matched cases and controls. The PTB analysis included 56 preterm cases and 136 unmatched controls (all vaginally delivered). Odds ratios were adjusted for clinical characteristics by logistic regression. The odds ratio and its confidence interval cannot be calculated for S. anginosus and SGA because one of the discordant values is zero. Additional details are in Supplementary Table 5.
Validating Streptococcus agalactiae
A nested PCR/qPCR approach, targeted towards the sip gene, encoding the surface immunogenic protein (SIP) of S. agalactiae, was used to verify its presence in 276 placental samples where a 16S sequencing result was available. Seven of 276 samples were positive using PCR/qPCR and all seven were also positive (>1%) by 16S analysis. 14 samples were positive by 16S sequencing but not by PCR/qPCR, no sample was positive using PCR/qPCR and negative by 16S, and 255 samples were negative by both methods. This yielded a kappa statistic of 0.48, indicating moderate agreement and a P value of 9.7 x 10-21. We conclude that the detection of S. agalactiae by 16S rRNA amplification was verified by two further independent methods (metagenomics and PCR/qPCR) and the level of agreement in both cases was well above what could be expected by the play of chance. It remains to be determined why some samples were positive for S. agalactiae by 16S sequencing but negative by the PCR/qPCR method. Generally, the latter would be considered more sensitive, particularly in samples with a higher microbial biomass, due to the complex amplification kinetics when a large number of diverse 16S template molecules are present. However, in the absence of other significant bacterial signals, it is possible that 16S sequencing is more sensitive for detecting very small numbers of S. agalactiae, as the organism’s genome has seven copies of the 16S rRNA gene, but only one copy of sip 17.
Discussion
We studied placental biopsies from a total of 537 women, including 318 cases of adverse pregnancy outcome and 219 controls using multiple methods of DNA extraction and detection, and drew a number of important conclusions. First, we found that the biomass of bacterial sequences in DNA extracted from human placenta was extremely small. Second, the major source of bacterial DNA in the samples studied was contamination from laboratory reagents and equipment. Third, both metagenomics and 16S amplicon sequencing were capable of detecting a very low amount of a spiked-in signal. Fourth, samples of placental tissue become contaminated during the process of labour and delivery, even when they were dissected from within the placenta. Finally, the only organism for which there was strong evidence that it was present in the placenta prior to the onset of labour was S. agalactiae. It was not part of any batch effect, it was detected by three methods, there was a statistically highly significant level of agreement between 16S amplicon sequencing and both metagenomics (P=1.5 x 10-8) and a targeted PCR/qPCR assay (P=9.7 x 10-21), none of 47 negative controls analysed by 16S sequencing were positive for S. agalactiae, and there was no association with mode of delivery (Extended Data Fig. 7). However, there was no significant association between the presence of the organism and PE, SGA or PTB. Exploratory analysis of other signals did demonstrate an association between PTB and the presence of Ureaplasma reads (>1%), consistent with previous studies,13,18 but this was likely the result of ascending uterine infection. We conclude that bacterial placental infection is not a major cause of placentally-related complications of human pregnancy and that the human placenta does not have a resident microbiome.
The finding of S. agalactiae in the placenta before labour could be of considerable clinical importance. Perinatal transmission of S. agalactiae from the mother’s genital tract can lead to fatal sepsis in the infant. It is estimated that routine screening of all pregnant women for the presence of S. agalactiae and targeted use of antibiotics prevents 200 neonatal deaths per year in the USA19. Our findings identify an alternative route for perinatal acquisition of S. agalactiae. Further studies will be required to determine the association between the presence of the organism in the placenta and fetal or neonatal disease. However, if such a link was identified, rapid testing of the placenta for the presence of S. agalactiae might allow targeting of neonatal investigation and treatment. Our work also sheds light on the possible routes of fetal colonization. While we see no evidence of a placental microbiome, the frequency of detection of vaginal bacteria in the placenta increased after intrapartum CS suggesting ascending or haematogenous spread. Similarly, haematogenous spread as the result of transient bacteraemia could potentially explain the presence of the small number of sample-associated oral bacterial signals16. Such spread could lead to fetal colonization immediately before delivery.
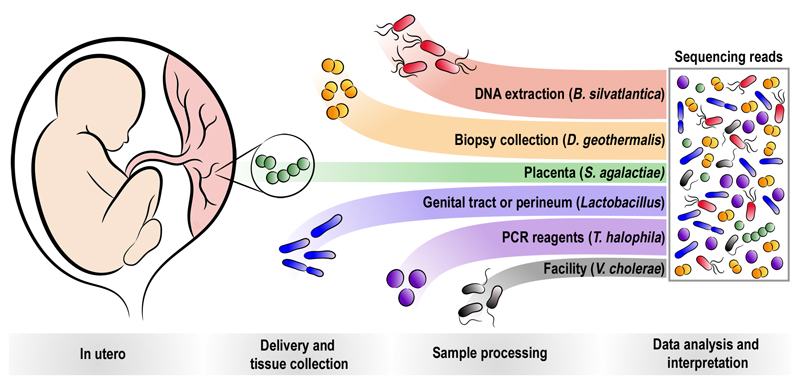
We identified five different patterns of contamination (Fig. 4), namely, contamination of the placenta with real bacteria during the process of labour and delivery (Fig. 2), contamination of the biopsy when it was washed with PBS, contamination of DNA during the extraction process, contamination of reagents used to amplify the DNA prior to sequencing, and contamination from the reagents or equipment used for sequencing. Using 16S rRNA amplicon sequencing, the positive control (S. bongori) accounted for more than half of the reads, indicating that the method is highly sensitive. However, when the method is applied to samples with little or no biomass, these sources of contamination can lead to apparent signals, hence it is critical to employ a methodological approach which allows differentiation between true bacterial signals and these sources of contamination. Additional technical discussion is in Supplementary Information File 1.
Fig. 4. Sources of bacterial signals detected in human placental samples.
Bacteria may sometimes be present in utero, such as S. agalactiae. Bacteria or bacterial DNA also frequently contaminate the placenta during labour and delivery (e.g. Lactobacillus), during sample collection (e.g. D. geothermalis), and always during sample processing (e.g. B. silvatlantica and T. halophila). Contamination may also occur during library preparation or sequencing from other projects carried out at the facility (e.g. V. cholera in the metagenomic sequencing).
In conclusion, in a study of 537 placentas carefully collected, processed and analysed to detect real bacterial signals, we found no evidence to support the existence of a placental microbiome and no significant relationship between placental infection with bacteria and the risk of preeclampsia, SGA and preterm birth. However, we identified an important pathogen, S. agalactiae, in the placenta of approximately five percent of women prior to the onset of labour.
Methods
Ethics
This study is in compliance with all relevant ethical regulations. The Pregnancy Outcome Prediction study (POPs) was approved by the Cambridgeshire 2 Research Ethics Committee (reference number 07/H0308/163). The study and the characteristics of the eligible and participating women have been previously described in detail14,20. In brief, 4,212 nulliparous women with a singleton pregnancy were followed through from their first ultrasound scan to delivery. At the time of delivery, placental samples were obtained using a standardized protocol by a team of trained technicians, where the majority of samples were obtained within 3 hours of delivery (IQR: 0.3-8.4 h). All participants gave written informed consent for the study and for subsequent analysis of their samples.
Patient selection
For Cohort 1, cases of SGA (≤5th percentile based on customized birth weight21; n=20) or pre-eclampsia (according to the 2013 ACOG (The American College of Obstetricians and Gynecologists) Guidelines22; n=20) were matched one-to-one with healthy controls (n=40). Only deliveries by pre-labour CS were included in this cohort. The cases and controls were matched as closely as possible for maternal body mass index (BMI), maternal age, gestational age, sample collection time, maternal smoking, and fetal sex. Clinical characteristics are presented in Supplementary Table 1.
For Cohort 2, cases of SGA (≤5th customized birth weight percentile21; n=100) or preeclampsia (2013 ACOG guidelines22; n=100) were selected. The cases were matched one-to-one with healthy controls (n=198, two controls were used twice). All deliveries were at term (≥37 week’s gestation). The same matching criteria as in the first cohort were used with the addition of an absolute match for mode of delivery. Placentas from 100 preterm births (<37 week’s gestation) deliveries were also included in the study (clinical characteristics in Supplementary Table 2). Flowcharts describing the two cohorts as well as subsequent sample processing and analysis steps are presented in Extended Data Fig. 1.
Placenta collection
Placentas were collected after delivery and the procedure has previously been described in detail21. We confined our sampling to the placental terminal villi (fetal tissue). We chose this as the villi are the site of exchange, across the vasculosyncytial membrane, between the fetus and mother. This location is the closest interface between the fetus with the mother’s blood and tissues. If the placenta was colonized, one would expect bacteria to ascend the genital tract (local infiltration) or to come from the mother’s blood (haematogenous). Hence, we believe that this would be the most plausible site for bacteria to be found. Villous tissue was obtained from four separate lobules of the placenta after trimming to remove adhering decidua from the basal plate. The tissue in the selected areas had no visible damage, hematomas, or infarctions. To remove maternal blood, the selected tissue samples were rinsed in chilled sterile phosphate-buffered saline (Oxoid Phosphate Buffered Saline Tablets, Dulbecco A; Thermo Fisher Scientific) dissolved in ultrapure water (ELGA Purelab Classic 18MΩ.cm). After initial collection all placental samples were frozen in liquid nitrogen and stored at -80°C until further processing. For DNA isolation, approximately 25 mg of villous tissue (combined weight obtained from fragments of all four biopsy collection points) was cut from the stored tissue. In order to reduce the risk of environmental contamination of the samples the entire experimental procedure was carried out in a Class 2 biological safety cabinet (tissue cutting, DNA isolation, setting up PCR reactions). The tissue was cut with single-use sterile forceps and scalpel. Each matched case-control pair was processed in parallel on the same day for each step of the entire experimental procedure (tissue cutting, DNA isolation, setting up PCR reactions). Also, the same lot of laboratory reagents were used for each pair. For each lot of laboratory reagents negative controls were included (described in detail below).
DNA isolation from Cohort 1
DNA was isolated from placental tissue with the Qiagen Qiaamp DNA mini kit (cat# 51304; Qiagen, Manchester, UK) according to the manufacturer’s instructions with the addition of a freeze/thaw cycle after the overnight tissue lysis. Prior to DNA isolation, intact S. bongori was added to the placental tissue (1,100 CFU, described in detail below). The placental tissue with added S. bongori was lysed in a proteinase K based solution (100 µl Buffer ALT, 80 µl of S. bongori, 20 µl proteinase K) overnight (18 hours at 56°C) and thereafter freeze/thawed once. After the thawed samples were brought to room temperature, RNA was removed with the addition of 4 µl RNase A (Qiagen, cat#19101) and incubated at room temperature for 2 minutes. Spin filtering and washing of the DNA was carried out according to the manufacture’s instruction. The DNA was eluted from the spin column with 200 µl Buffer AE after a 5 minute incubation (the elution step was repeated once with another 200 µl Buffer AE and 5 minute incubation). In order to prevent accidental cross-contamination between samples, gloves were changed between handling each sample. Throughout the protocol (DNA extraction, primer aliquoting, 16S rRNA gene amplification and library preparation), nuclease-free plastics were used (unless supplied with kit): PCR clean 2.0 and 1.5 ml DNA LoBind Tubes (Eppendorf, Hamburg, Germany), and nuclease-free filter tips (TipONE sterile filter tips, STARLAB (UK), Ltd, Milton Keynes, UK). For each box of DNA isolation kit used, extraction blanks were carried out. These DNA extraction blanks, or negative controls, contained only the reagents from each DNA isolation kit (no added biological material) and were subjected to the complete DNA extraction procedure: tissue homogenization, matrix binding, spin filtering, washing, and elution of nucleic acids. The negative controls were subjected to the entire analysis protocol alongside the placental samples: DNA isolation, 16S rRNA gene PCR amplification, sequencing and data analysis.
Positive control
As a positive control, a known amount of intact S. bongori (strain NCTC-12419) was added to each of the placental tissue samples in Cohort 1 prior to DNA isolation (n=80). S. bongori was incubated with shaking overnight at 37°C in LB broth. When the OD600 reached 0.9 (approximately equivalent to 7.2 × 108 bacteria/ml, measured with a Ultrospec™ 10 Cell Density Meter, GE Healthcare, Little Chalfont, Buckinghamshire, UK) the culture was chilled on ice. To minimize bacterial growth outside of the shaking incubator, all cultures and dilutions were kept on ice. In order to increase the proportion of live bacteria added as positive controls, 1 ml of the S. bongori suspension was diluted in 14 ml fresh LB broth (OD600 was 0.06) and incubated with shaking (1.5 hours at 37°C; OD600 was 0.8). The S. bongori culture was then serially diluted to an estimated concentration of 1,000 S. bongori per 80 µl which was used to spike the placental samples. To determine the actual number of colony forming units (CFU) added to the placental samples, the S. bongori suspension was further diluted and aliquots cultured on LB plates overnight (37°C). The number of colonies was counted. Based on 3 plates with distinct individual colonies (between 29 and 205 colonies/plate), the number of S. bongori added to each placental tissue sample was calculated to be 1,100 CFU.
DNA isolation from Cohort 2
DNA was isolated twice from each placenta using two different extraction kits. The DNA isolations were carried out in accordance with respective manufacturers instructions, with the addition of two extra washes in the MP Biomedical kit.
For the Qiagen Qiaamp DNA mini kit (cat# 51304; Qiagen), the placental tissue was digested in a Proteinase K based solution (100 µl Buffer ALT, 80 µl PBS, 20 µl proteinase K) for at least 3 hours. Four µl RNase A (Qiagen, cat#19101) was added to the tissue lysate and incubated at room temperature for 2 minutes. Spin filtering and washing of the DNA was carried out according to the manufacture’s instruction. The DNA was eluted from the spin column with 200 µl Buffer AE after a 5 minute incubation (the elution step was repeated once with another 200 µl Buffer AE and 5 minute incubation).
For the MP Biomedical Fast DNA Spin kit (cat#116540600; MP Biomedical, Santa Ana, CA, USA), the placental tissue was homogenized in 1.0 ml of CLS-TC solution by bead-beating (Lysing Matrix A tubes, 40 sec, speed 6.0 on a FastPrep-24, MP Biomedical). After spinning the samples, equal volumes of the supernatant were combined with Binding Matrix. The mixture was transferred to a spin filter, after spin filtering the DNA was washed three times with SEWS-M. The DNA was eluted by re-suspending the Binding Matrix in 100 µl DES buffer, incubating the tubes at 55°C for 5 minutes before recovering the DNA by centrifugation.
The same measures to prevent contamination of the samples as described in the Cohort 1 DNA isolation section were taken. Extraction blanks were generated for each box/lot of both DNA isolation kits in a similar manner as was done for Cohort 1. DNA concentrations were determined by Nanodrop Lite (Thermo Fisher Scientific, Waltham, MA, USA).
Metagenomic sequencing
Sample processing for the metagenomics analysis was performed exactly as described by Lager et al.23. In brief the NEB Ultra II custom kit (NEB, Ipswich, Massachusetts, USA) was used for library generation which were then sequenced on the Illumina HiSeq X Ten platform (150 base, paired end) in 10 runs (flowcells) of 8 samples (lanes) each. The sequencing coverage was designed to generate >30-fold coverage of the human chromosomal DNA in each sample.
16S rRNA gene amplification
For detection of the bacterial 16S rRNA gene, PCR amplification of the V1-V2 region was performed using V1 primers with 4 degenerate positions to optimize coverage as recommended by Walker et al24. The V1-V2 amplicon is relatively short (~260bp) and, with paired end reads, almost all of the amplified product is sequenced on both stands and thus at higher accuracy. This is not the case with the longer V1-V3 amplicon. This region has also been used in other studies of the placental microbiome10–12. The following barcoded primers were used Forward-27: 5’-AATGATACGGCGACCACCGAGATCTACACnnnnnnnnnnnnACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNAGMGTTYGATYMTGGCTCAG-3’ and Reverse-338: 5’-CAAGCAGAAGACGGCATACGAGATnnnnnnnnnnnnGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNGCTGCCTCCCGTAGGAGT. The n-string represents unique 12-mer barcodes used for each sample studied and distinct indexes were used at both the 5’ and 3’ends of the amplicons. The primers were purchased from Eurofins Genomics (Ebersberg, Germany). Before aliquoting, the cabinet and pipettes were cleaned with DNA AWAY Surface Decontaminant. The primers were diluted in Tris-EDTA buffer (Sigma-Aldrich Company Ltd., Gillingham, UK) in PCR clean nuclease-free DNA LoBind Tubes (Eppendorf) with nuclease-free filter tips (TipONE sterile filter tips, STARLAB). The PCR amplification was carried out in quadruplicate reactions for each sample on a SureCycler 8800 Thermal Cycler (Agilent Technologies, Stockport, UK) with high-fidelity Q5 polymerase (cat# M0491L; New England Biolabs, Hitchin, UK), dNTP solution mix (cat#N0447L, New England Biolabs), and UltraPure DNase/RNase-Free Water (Thermo Fisher Scientific) in 0.2 ml PCR strips (STARLAB). Amplification was performed with 500 ng DNA per reaction, and the final primer concentration was 0.5 µM. The PCR amplification profile was an initial step of 98°C for 2 min followed by 10 cycles of touch-down (68 to 59°C; 30 sec), and 72°C (90 sec), followed by 30 cycles of 98°C (30 sec), 59°C (30 sec), and 72°C (90 sec). After completion of cycling, the reactions were incubated for 5 min at 72°C. After completion of the PCR, the four replicates of each sample were pooled, cleaned up with AMPure XP beads (cat# A63881; Beckman Coulter, High Wycombe, UK) and eluted in Tris-EDTA buffer (Sigma-Aldrich). DNA concentration was determined by Qubit Fluorometric Quantitation (cat# Q32854; Invitrogen, Carlsbad, CA, USA). Equimolar pools of the PCR amplicons were run on 1% agarose/TBE gels and ethidium bromide used to visualize the DNA. The DNA bands were excised and cleaned up with a Wizard SV Gel and PCR Clean-Up System (Promega UK, Southampton, UK). The equimolar pools were sequenced on the Illumina MiSeq platform using paired-end 250 cycle MiSeq Reagent Kit V2 (Illumina, San Diego, CA, USA).
Bioinformatic analysis of metagenomics data
Bioinformatic analysis first required removal of human reads followed by identification of the species of non-human reads. KneadData (http://huttenhower.sph.harvard.edu/kneaddata) is a tool designed to perform quality control on metagenomic sequencing data, especially data from microbiome experiments, and we used this to remove the human reads. Forward and reverse reads from each sample were filtered using KneadData (v0.6.1) with the following trimmomatic options: HEADCROP9, SLIDINGWINDOW:4:20, MINLEN: 100. A custom Kraken25 reference database (v0.10.6) was built, using metagm_build_kraken_db and -max_db_size 30, in order to detect any bacterial, viral and potential non-human eukaryotic signals. This custom Kraken reference database included both the default bacterial and viral libraries, and an accessions.txt file was supplied (via -ids_file) containing a diverse array of organisms chosen from all sequenced forms of eukaryotic life (see Supplementary Table 6 for accession numbers). This wide array was chosen to both detect potentially relevant unknown organisms, but also to identify additional human reads which had not been mapped to the human reference genome. In the metagenomic data, various non-human eukaryotic signals were identified by Kraken in every placental sample at a similar percentage, and were mostly assigned to Pan paniscus (Supplementary Table 6). As a verification, reads mapping to eukaryotic species were extracted (Supplementary Information File 1) and contigs were assembled. These were analysed using BLASTN and were indeed identified as human. This indicates that these (often lower quality or repetitive) eukaryotic reads are in fact human reads that were not removed by mapping against the human reference genome. An exception to this was that in 17 samples an elevated number of reads were assigned to Danio rerio and Sarcophilus harrisii (Zebrafish and Tasmanian devil respectively, both of which had been sequenced on the Sanger Institute pipeline). Kraken was run using the metagm_run_kraken option. All human derived signals (Eukaryotic non-fungal reads found in every placental sample at a similar percentage) were removed prior to further analysis. See Source Data of Figure 1a-c for abundance information. The origins of Streptococcus pneumonia and Vibrio cholerae reads were analysed by extracting their respective reads as identified by the Kraken using custom scripts (Supplementary Information File 1), performing an assembly on these reads using Spades (v3.11.0)26 and by using BLAST (blastn, database: others)27 to find the closest match. The first step of the strain level analysis of E. coli reads in order to find the closest E. coli reference genome match was identical to the steps described above. Subsequently, E. coli reads were mapped against E. coli WG5 (Genbank: CP02409.1) using BWA (v0.7.17-r1188)28 and visualized using Artemis (v.16.0.0)29. E. coli reads were both analysed per sample and by combining all E. coli reads from all samples together.
Bioinformatic analysis of 16S rRNA gene amplicon data
In order to analyse all fourteen 16S rRNA amplicon data together using the MOTHUR (v1.40.5) MiSeq SOP30 and the Oligotyping (v2.1) pipeline31 the data from each individual run were initially individually processed in the MOTHUR pipeline as described below. All the reads need to be aligned together as a requirement of the Oligotyping pipeline so after the most memory intensive filtering steps had been performed, they were combined and processed again. Modifications to the MOTHUR MiSeq SOP are as follows: the "make.contigs" command was used with no extra parameters on each individual run. The assembled contigs were taken out from the MOTHUR pipeline and the four poly NNNNs present in the adapter/primer sequences were removed using the "-trim_left 4" and "-trim_right 4" parameters in the PRINSEQ-lite (v0.20.3) program32. The PRINSEQ trimmed sequences were used for the first "screen.seqs" command to remove ambiguous sequences and sequences containing homopolymers longer than 6 bp. In addition, any sequences longer than 450 bp or shorter than 200 bp were removed. Unique reads (“unique.seqs”) were aligned (“align.seqs”) using the Silva bacterial database "silva.nr_v123.align"33 with flip parameter set to true. Any sequences outside the expected alignment coordinates ("start=1046”, “end=6421") were removed. The correctly aligned sequences were subsequently filtered ("filter.seqs") with "vertical=T" and "trump=.". The filtered sequences were de-noised by allowing three mismatches in the "pre.clustering" step and chimeras were removed using Uchime with the dereplicate option set to "true". The chimera free sequences were classified using the Silva reference database "silva.nr_v123.align" and the Silva taxonomy database "silva.nr_v123.tax" and a cut off value of 80%. Chloroplast, Mitochondria, unknown, Archaea, and Eukaryota sequences were removed. All reads from each sample were subsequently renamed, placing the sample name of each read in front of the read name. The “deunique.seqs” command, which creates a redundant fasta file from a fasta and name file, was performed prior to concatenating all the data of all fourteen 16S runs together using the “merge.files” command which was done on both the fasta and the group files. The “unique.seqs” command was again used before again aligning all reads as described previously before finishing the MOTHUR pipeline with the “deunique.seqs” command.
Oligotyping and species identification
After the MOTHUR pipeline, the redundant fasta file, which now only contains high quality aligned fasta reads, was subsequently used for oligotyping using the unsupervised "Minimum Entropy Decomposition" (MED) for sensitive partitioning of high-throughput marker gene sequences31. A minimum substantive abundance of an oligotype (-M) was defined at 1000 reads and a maximum variation allowed (-V) was set at 3 using the command line "decompose 14runs.fasta -M 1000 -V 3 -g –t”. The node representative sequence of each oligotype (OTP) was used for species profiling using the ARB program - A Software Environment for Sequence Data (version 5.5-org-9167)34. For ARB analysis we used a customized version of the SILVA SSU Ref database (NR99, release 123) that was generated by removing uncultured taxa. Oligotype abundances are provided in Supplementary Information File 2 and additional metadata, for e.g. contamination identification via PCA (Extended Data Figure 3), is provided in the Source Data files.
Sensitivity analysis
To compare 16S rRNA amplicon sequencing and metagenomics sensitivity the S. bongori signals (positive control) spiked into Cohort 1 were analysed (Extended Data Figs. 2a-b). In 16S rRNA amplicon sequencing analysis 1,100 colony forming units of S. bongori resulted in an average of 33,000 S. bongori reads (~54%). Thus, the remaining bacterial signal (reagent contamination background + other signals) contributes the remaining 46% of the reads. This is approximately equivalent to another 937 S. bongori colony forming units (1,100/(54/46)). Thus, if there are 937 bacteria in the sample (everything except the spike) this should produce a signal of 100% when there are no spiked-in bacteria present. Thus, the sensitivity of this assay in Cohort 2, which did not contain a spike is 0.106% of sequencing reads per CFU (100%/937CFU). However, while an average of 54% S. bongori reads were detected in all spiked samples it can be reasoned that samples with the highest S. bongori percentages only have reagent contamination DNA to compete with during the PCR step and not any other sample-associated signals. S. bongori percentages in the top 20th percentile on average account for 71% of all reads which would correspond to a sensitivity limit of ~0.2% of reads / CFU (100/(1100/(71/29)). A threshold of 1%, as utilized by Lauder et al.10, can however be considered a more reliable cut-off for determining whether a signal should be considered biologically relevant. A threshold of 1% would be indicative of multiple replication events (more than 2) and thus metabolic activity or repeated invasion of the tissue by the respective organism. In addition, a 1% threshold for the 16S rRNA data is comparable with the sensitivity of metagenomics as on average 180 S. bongori read pairs were detected with metagenomics (Extended Data Fig. 2a). In contrast to 16S analysis, the S. bongori spike has no meaningful effect on quantification in metagenomics as microorganisms only represent a very small fraction of the total amount of reads (the vast majority of reads are human). Hence 6 CFU are required on average per metagenomics read pair and 6 CFU would result in a signal of approximately 1% of 16S amplicon reads in Cohort 2 using the Qiagen kit.
Nested PCR
We developed a nested PCR assay to sensitively detect the S. agalactiae sip gene. 276 placental DNA samples (isolated with the Qiagen kit as described above) were used of which 226 had no (0%) S. agalactiae reads detected by 16S rRNA gene sequencing, while S. agalactiae reads were detected in 50 samples (range 0.002-63.37% of 16S rRNA reads). The first-round PCR was performed using the DreamTaq PCR Master Mix (2X) (cat# K1071; Thermo Fisher Scientific) and the following primers for the sip gene at a final concentration of 0.5 µM: Forward 5’-TGAAAATGAATAAAAAGGTACTATTGACAT-3’ and Reverse 5’-AAGCTGGCGCAGAAGAATA-3’. Amplification was performed in 50 µl and using 500 ng of placental DNA per reaction. Genomic S. agalactiae DNA (ATCC cat# BAA-611DQ) was used as positive control at 20 or 2 copies/reaction. One reaction was set up with H2O instead of gDNA as negative control. The PCR amplification profile had an initial step of 95°C for 3 min followed by 15 cycles of 95°C (30 sec), 48°C (30 sec), and 72°C (60 sec). After completion of cycling, the reactions were incubated for 3 min at 72°C. The second-round qPCR was performed using the TaqMan Multiplex Master Mix (cat# 4461882; Thermo Fisher Scientific) and two TaqMan Assays (Thermo Fisher Scientific): Ba04646276_s1 (Gene Symbol: SIP; Dye Label, Assay Concentration: FAM-MGB, 20X) at a final 1X concentration; RNase P TaqMan assay (ABY dye/QSY probe Thermo Fisher Scientific cat# 4485714) at a final 0.5X concentration, added as a positive control for the human DNA. In each well, 6 µl of the first-round PCR (or H20 in the No Template Control/blank wells) was used as the reaction substrate in a total volume of 15 µl. The PCR amplification profile had an initial step of 95°C for 20 sec followed by 40 cycles of 95°C (5 sec) and 60°C (20 sec).
Statistics
The inter-rater agreement kappa scores35 and P values were computed by DAG_Stat36. Comparison of cases and controls was performed using multivariable logistic regression, with conditional logistic regression employed for paired comparisons, using Stata v15.1 (Statacorp, College Station, TX, USA). Other statistical calculations were performed in GraphPad Prism 7 (GraphPad Software, Inc., La Jolla, CA, USA). Principal component analyses (PCA) were performed with the prcomp function from the R package in RStudio (v0.99.902) with all settings, where applicable, set to TRUE. As the effect size was not known in advance we performed power calculations with varying prevalence and effect sizes (OR) for 100 case-control pairs (pre-eclampsia and growth restriction) used in the 16S rRNA amplicon sequencing study. These showed that a 5% prevalence in controls and OR=5 gives 82% power to detect the signal at significance level 0.05. The bioinformatic analysis and the setting of the minimum detection thresholds were performed in a blinded fashion in respect to adverse pregnancy outcome status. All reported P values are two-sided except for concordance calculations, as indicated.
Data availability
The 16S rRNA gene sequencing datasets generated and analyzed in this study are publicly available under European Nucleotide Archive (ENA) accession no. ERP109246 (https://www.ebi.ac.uk/ena/data/view/PRJEB27192). The metagenomics data sets, which primarily contain human sequences, are available with managed access in the European Genome-phenome Archive (EGA) accession no. EGAD00001004198. (https://ega-archive.org/datasets/EGAD00001004198)
Extended Data
Extended Data Fig. 1. Two cohorts of placental samples were analysed.
Cohort 1 (n=80) contained only samples from pre-labour CS and S. bongori was added to the samples before DNA isolation as a positive control. The samples in cohort 1 was analysed by both metagenomics as well as by 16S rRNA amplicon sequencing. Cohort 2 (n=498) contained placental samples from CS and vaginal deliveries. DNA was isolated twice from each placental sample with two different DNA extraction kits. The samples were analysed by 16S rRNA amplicon sequencing. CS = Caesarean section, SGA = small for gestational age (birth weight <5th percentile) using a customized reference, PE = preeclampsia using the ACOG 2013 definition, Preterm= birth at <37 week’s gestation.
Extended Data Fig. 2. Positive control experiment comparison between metagenomics and 16S amplicon sequencing.
Adding approximately 1,100 CFU of S. bongori to the placental tissue before DNA isolation resulted in a) an average of 180 reads (SD: 90 reads) by metagenomic sequencing (n=80) or b) on average 54% of all 16S rRNA amplicon sequencing reads (~33,000 reads) being identified as S. bongori (SD: 13%; n=79). Box represents the interquartile range. Whiskers represent Max/Min in both figures.
Extended Data Fig. 3. Strain analysis of E. coli reads found by metagenomics.
All reads identified in all 80 samples by Kraken as E. coli were extracted and mapped together against the closest E. coli reference genome (Genbank: CP02409.1). Single Nucleotide Polymorphisms (SNPs), shown in red, were consistent for all samples across the genome. SNPs were rare, except in the fimbrial chaperone protein gene (EcpD) indicated in light red. Sequence differences which appear as short sporadic red lines represent sequencing errors. Strain variation would have resulted in dashed vertical lines.
Extended Data Fig. 4. Detailed heatmap metagenomic data.
Heatmap showing the abundance of all non-human reads as detected by metagenomics. Human reads remaining after filtering (89.8%, SD: 1.5%) are not shown for scaling purposes. The majority of taxa (shown on the right) are found in higher abundance within groups 1 and/or 2 (indicated on the left with light blue and purple, respectively). The purple box highlights the samples and species associated with group 2. The lane ID of each sample is represented by the first number (x-axis). All samples from lanes 4 and 5 form Group 2 and all samples from lanes 8 and 9 form Group 3 (see Figs. 1a-b).
Extended Data Fig. 5. Species associated with batch effects visualized by PCA also do not show signal reproducibility.
a) Principal component analyses of selections of samples from Cohort 2 (16S), or of all Cohort 2 samples as shown here, allows for the identification of batch effects and allows for the identification of contaminating species associated with the use of specific DNA isolation methods/kits and/or other reagents. An analysis of all samples shows that principal components 3 (x-axis) and 4 (y-axis) are strongly correlated with the use of Qiagen or specific Mpbio DNA isolation kits. b) Examples of bacteria detected in high abundance and frequency when processed with the Qiagen (x-axis) and/or Mpbio (y-axis) DNA isolation kit. Patterns lacking positive correlation (compare with Fig. 2a) demonstrate that signals are not sample but batch associated.
Extended Data Fig. 6. Scatterplot representations of the abundance of a) Bradyrhizobium and b) Burkholderia in respect to sequencing run batch effects and c) vaginal lactobacilli and d) vaginosis bacteria in respect to the mode of delivery found during 16S amplicon sequencing.
In a & b Numbers in brackets indicate the number of samples sequenced in a given run. Values of zero are not shown on the logarithmic axis. c,d) Comparisons between modes of delivery were performed by Mann-Whitney U tests, where values below 1% are regarded as 0% (not biologically relevant). * P < 0.05, *** P < 0.001.
Extended Data Fig. 7. Mode of delivery and the detection of bacterial signals.
a, b) The association of vaginal lactobacilli with the mode of delivery, as determined by the analysis of 466 samples by 16S amplicon sequencing which were successfully sequenced twice using Mpbio (a) and Qiagen (b) DNA isolation. Comparisons of the Mpbio and Qiagen DNA isolations highlight that the same patterns are observed in the associations with mode of delivery. Comparisons also show that the Qiagen DNA isolation was more sensitive, resulting in twice as many signals above the 1% threshold. Figures c-h were generated using all 498 placental samples using the highest value of either DNA isolation method for each bacterial group per sample. c, d) S. agalactiae was not associated with the mode of delivery irrespective of whether a 0.1% threshold was used (the 16S detection limit, relevant for detecting traces of contamination during delivery) or whether a 1% threshold was used (the minimum percentage considered to be potentially ecologically relevant). e, f) The Ureaplasma genus was associated with the mode of delivery, comparable to Figure 2c which describes the combination of all vaginosis associated bacteria. g, h) F. nucleatum was not associated with the mode of delivery, irrespective of threshold. Comparisons between modes of delivery were performed by Mann-Whitney U tests. * P < 0.05, ** P < 0.01, *** P < 0.001.
Extended Data Fig. 8. Heatmap of Spearman’s rho correlation coefficients of bacterial signals as found by 16S rRNA amplicon sequencing.
Sample-associated signals (red bar), are typically identified by elevated kappa scores as shown in Supplementary Table 4. Reagent contaminants are indicated with a blue bar. Vaginosis associated bacteria (purple bar) show positive correlations (purple square) with each other, Lactobacillus iners and fecal bacteria (brown bar). Lactobacilli (yellow bar) show limited positive correlation with fecal bacteria. Reagent contaminants mainly associated with the Qiagen (light blue) or the Mpbio kit (green) form distinct clusters. Species which are strongly associated with sample collection contamination in 2012-2013 are indicated in orange. For each species the highest value (%) found using either the Qiagen or the Mpbio DNA isolation kit, was used as input (using all 498 samples).
Extended Data Fig. 9. Bacterial signals and adverse pregnancy outcome.
Scatterplot representations of the association of a) S. agalactiae with SGA, b) S. anginosus with SGA, c) L. iners with preeclampsia, and d) Ureaplasma with PTB. Samples with 0% signal are not shown. Signals above 1% (dotted line) are regarded as positive for the McNemar’s test (a-c) and signals below 1% are considered 0% (d).
Supplementary Material
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Supplementary Information file 1 contains all Supplementary Tables, a Supplementary Discussion and Supplementary Methods. Supplementary Information file 2 contains detailed oligotype abundance information.
Acknowledgements
The work was supported by the Medical Research Council (United Kingdom; MR/K021133/1) and the National Institute for Health Research (NIHR) Cambridge Comprehensive Biomedical Research Centre (Women’s Health theme). We would like to thank Leah Bibby, Samudra Ranawaka, Katrina Holmes, Josephine Gill, Ryan Millar and Leonor Sánchez Busó for technical assistance during the study. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care.
Footnotes
Author contributions
G.C.S.S, D.S.C-J, J.P, and S.J.P conceived the experiments. G.C.S.S, D.S.C-J, J.P, S.J.P, and S.L. designed the experiments. S.L. and M.C.G optimized the experimental approach. S.L and F.G. performed the experiments. M.C.G. analysed all the sequencing data. U.S. matched cases and controls, performed statistical analyses and provided logistical support for patient and sample metadata. EC managed sample collection and processing and the biobank in which all sample were stored. All authors contributed in writing the manuscript and approved the final version.
Competing interests
JP reports grants from Pfizer, personal fees from Next Gen Diagnostics Llc, outside the submitted work; SJP reports personal fees from Specific, personal fees from Next Gen Diagnostics, outside the submitted work; DSC-J reports grants from GlaxoSmithKline Research and Development Limited, outside the submitted work and non-financial support from Roche Diagnostics Ltd, outside the submitted work; GCSS reports grants and personal fees from GlaxoSmithKline Research and Development Limited, personal fees and non-financial support from Roche Diagnostics Ltd, outside the submitted work; DSC-J and GCSS report grants from Sera Prognostics Inc, non-financial support from Illumina inc, outside the submitted work. MCG, SL, US, FG and EC have nothing to disclose.
Reprints and permissions information is available at http://www.nature.com/reprints.
References
- 1.Brosens I, Pijnenborg R, Vercruysse L, Romero R. The “Great Obstetrical Syndromes” are associated with disorders of deep placentation. Am J Obstet Gynecol. 2011;204:193–201. doi: 10.1016/j.ajog.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aagaard K, et al. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6 doi: 10.1126/scitranslmed.3008599. 237ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antony KM, et al. The preterm placental microbiome varies in association with excess maternal gestational weight gain. Am J Obstet Gynecol. 2015;212:653.e1–16. doi: 10.1016/j.ajog.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. Human gut colonization may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep. 2016;6:23129. doi: 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez-Muñoz ME, Arrieta MC, Ramer-Tait AE, Walter J. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome. 2017;5:48. doi: 10.1186/s40168-017-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salter SJ, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jervis-Bardy J, et al. Deriving accurate microbiota profiles from human samples with low bacterial content through post-sequencing processing of Illumina MiSeq data. Microbiome. 2015;3:19. doi: 10.1186/s40168-015-0083-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Goffau MC, et al. Recognizing the reagent microbiome. Nat Microbiol. 2018;3:851–853. doi: 10.1038/s41564-018-0202-y. [DOI] [PubMed] [Google Scholar]
- 9.Lauder AP, et al. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome. 2016;4:29. doi: 10.1186/s40168-016-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leiby JS, et al. Lack of detection of a human placenta microbiome in samples from preterm and term deliveries. Microbiome. 2018;6:196. doi: 10.1186/s40168-018-0575-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Theis KR, et al. Does the human placenta delivered at term have a microbiota? Results of cultivation, quantitative real-time PCR, 16S rRNA gene sequencing, and metagenomics. Am J Obstet Gynecol. 2019;220:267. doi: 10.1016/j.ajog.2018.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leon LJ, et al. Enrichment of clinically relevant organisms in spontaneous preterm delivered placenta and reagent contamination across all clinical groups in a large UK pregnancy cohort. Appl Environ Microbiol. 2018;84:e00483–18. doi: 10.1128/AEM.00483-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sovio U, White IR, Dacey A, Pasupathy D, Smith GCS. Screening for fetal growth restriction with universal third trimester ultrasonography in nulliparous women in the Pregnancy Outcome Prediction (POP) study: a prospective cohort study. Lancet. 2015;386:2089–2097. doi: 10.1016/S0140-6736(15)00131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hornef M, Penders J. Does a prenatal bacterial microbiota exist? Mucosal Immunol. 2017;10:598–601. doi: 10.1038/mi.2016.141. [DOI] [PubMed] [Google Scholar]
- 15.Leong HN, et al. The prevalence of chromosomally integrated human herpesvirus 6 genomes in the blood of UK blood donors. J Med Virol. 2007;79:45–51. doi: 10.1002/jmv.20760. [DOI] [PubMed] [Google Scholar]
- 16.Ravel J, et al. Vaginal microbiome of reproductive-age women. Proc Natl Acad Sci USA. 2011;108:4680–4687. doi: 10.1073/pnas.1002611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glaser P, et al. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 2002;45:1499–1513. doi: 10.1046/j.1365-2958.2002.03126.x. [DOI] [PubMed] [Google Scholar]
- 18.Abele-Horn M, Scholz M, Wolff C, Kolben M. High-density vaginal Ureaplasma urealyticum colonization as a risk factor for chorioamnionitis and preterm delivery. Acta Obstet Gynecol Scand. 2000;79:973–978. [PubMed] [Google Scholar]
- 19.Schrag SJ, et al. Group B streptococcal disease in the era of intrapartum antibiotic pylaxis. N Engl J Med. 2000;342:15–20. doi: 10.1056/NEJM200001063420103. [DOI] [PubMed] [Google Scholar]
- 20.Pasupathy D, et al. Study protocol. A prospective cohort study of unselected primiparous women: the pregnancy outcome prediction study. BMC Pregnancy Childbirth. 2008;8:51. doi: 10.1186/1471-2393-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gardosi J, Mongelli M, Wilcox M, Chang A. An adjustable fetal weight standard. Ultrasound Obstet Gynecol. 1995;6:168–174. doi: 10.1046/j.1469-0705.1995.06030168.x. [DOI] [PubMed] [Google Scholar]
- 22.American College of Obstetricians and Gynecologists; Task Force on Hypertension in Pregnancy. Hypertension in pregnancy. Report of the American College of Obstetricians and Gynecologists’ Task Force on Hypertension in Pregnancy. Obstet Gynecol. 2013;122:1122–1131. doi: 10.1097/01.AOG.0000437382.03963.88. [DOI] [PubMed] [Google Scholar]
- 23.Lager S, et al. Detecting eukaryotic microbiota with single-cell sensitivity in human tissue. Microbiome. 2018;6:151. doi: 10.1186/s40168-018-0529-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker AW, et al. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome. 2015;3:26. doi: 10.1186/s40168-015-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nurk S, et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol. 2013;20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson M, et al. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. Artemis: and integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–469. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eren AM, et al. Oligotyping: Differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol. 2013;4:1111–1119. doi: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmieder R, Edwards R. Quality control and preprocessing of metagenomics datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ludwig W, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viera AJ, Garrett JM. Understanding interobserver agreement: the kappa statistic. Fam Med. 2005;37:360–363. [PubMed] [Google Scholar]
- 36.Mackinnon A. A spreadsheet for the calculation of comprehensive statistics for the assessment of diagnostic tests and inter-rater agreement. Comput Biol Med. 2000;30:127–134. doi: 10.1016/s0010-4825(00)00006-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene sequencing datasets generated and analyzed in this study are publicly available under European Nucleotide Archive (ENA) accession no. ERP109246 (https://www.ebi.ac.uk/ena/data/view/PRJEB27192). The metagenomics data sets, which primarily contain human sequences, are available with managed access in the European Genome-phenome Archive (EGA) accession no. EGAD00001004198. (https://ega-archive.org/datasets/EGAD00001004198)