Abstract
The simultaneous delivery of multiple therapeutics to a single site has shown promise for cancer targeting and treatment. However, because of the inherent differences in charge and size between drugs and biomolecules, new approaches are required for colocalization of unlike components in one delivery vehicle. In this work, we demonstrate that triblock copolymers containing click nucleic acids (CNAs) can be used to simultaneously load a prodrug enzyme (cytosine deaminase, CodA) and a chemotherapy drug (doxorubicin, DOX) in a single polymer nanoparticle. CNAs are synthetic analogs of DNA comprised of a thiolene backbone and nucleotide bases that can hybridize to complementary strands of DNA. In this study, CodA was appended with complementary DNA sequences and fluorescent dyes to allow its encapsulation in PEG-CNA-PLGA nanoparticles. The DNA-modified CodA was found to retain its enzyme activity for converting prodrug 5-fluorocytosine (5-FC) to active 5-fluorouracil (5-FU) using a modified fluorescent assay. The DNA-conjugated CodA was then loaded into the PEG-CNA-PLGA nanoparticles and tested for cell cytotoxicity in the presence of the 5-FC prodrug. To study the effect of coloading DOX and CodA within a single nanoparticle, cell toxicity assays were run to compare dually loaded nanoparticles with nanoparticles loaded only with either DOX or CodA. We show that the highest level of cell death occurred when both DOX and CodA were simultaneously entrapped and delivered to cells in the presence of 5-FC.
Graphical Abstract
INTRODUCTION
Nanotechnology-based approaches have led to a large progress in delivering therapeutic agents to cancerous sites in the body1 while reducing off-target side effects.1,2 To increase efficacy, combination therapies have been proposed to eradicate tumors before they develop problematic drug resistance.3-5 One such approach is to load multiple agents in the same nanoparticle, including chemically unlike moieties such as hydrophilic nucleic acids or proteins and hydrophobic chemotherapy drugs, such as doxorubicin or paclitaxel.3,6-9 This encapsulation approach is especially important for biomolecules that require protection from proteolytic degradation of the body’s immune system.10,11 Entrapping both hydrophobic and hydrophilic agents in a single carrier is difficult due to their different solvent partition coefficients. While poly(lactic-co-glycolic acid) (PLGA) particles produced using oil-in-water emulsions have been used, these often suffer in vivo due to difficulties encountered in loading proteins, the overall long-term stability during circulation as well as difficulties with producing them at diameters less than 200 nm.12 Because of this, proteins are more often conjugated with polymers such as polyethylene glycol (PEG) or entrapped within a cross-linked or stimuli-responsive nanoparticle.13 However, due to the inherently large differences in size and polarity between proteins and hydrophobic drugs and despite numerous advances in bioconjugation techniques,14,15 coentrapment within a single delivery vehicle has only been achieved through particle formulations16-19 that rely on cationic polymers,20,21 which can suffer due to rapid elimination in vivo or show a limited degree of success.22-31
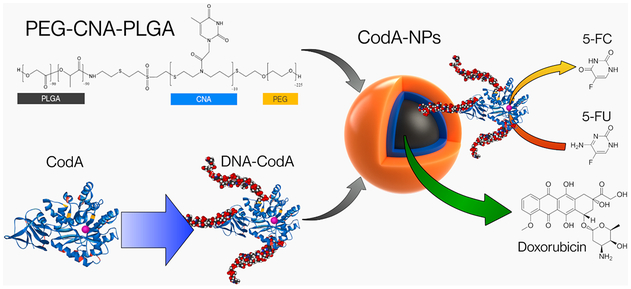
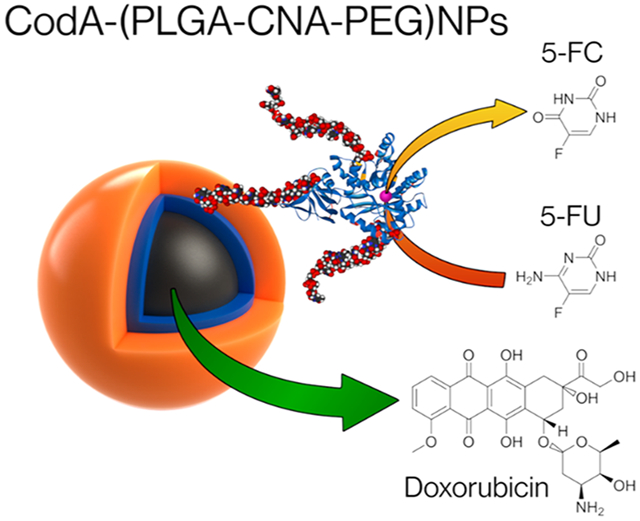
In this work we show the use of synthetic nucleic acids, namely, click nucleic acids or CNAs, to enable the simultaneous entrapment and delivery of a prodrug enzyme, cytosine deaminase (CodA), and the chemotherapy drug doxorubicin (DOX) in a single nanoparticle by using a PEG-CNA-PLGA block polymer. Click nucleic acids (CNAs) are a synthetic analog of DNA comprised of a thiolene backbone and nucleotide bases.32 We have recently shown that CNA in the appropriate solvent environment can hybridize to complementary strands of DNA.33 In addition, we recently demonstrated the successful synthesis and production of PEG-CNA-PLGA nanoparticles and their utility for loading both complementary strands of DNA along with pyrene as a preliminary study.34 In the work shown here, we have advanced our previous findings by coentrapping protein therapeutics, along with chemotherapy drugs in PEG-CNA-PLGA nanoparticles. To load CodA into the PEG-CNA-PLGA nanoparticles, we first attached complementary DNA sequences and dyes to the enzyme; work by Mirkin et al. using DNA-modified proteins showed the structural integrity and functionality was maintained after similar conjugations.35 To ensure this continued to be true, the DNA-modified CodA was tested for enzymatic activity with the prodrug 5-fluorocytosine (5-FC) using a modified fluorometric assay based on the o-phthaldehyde amino acid detection assay.36-38 The DNA-conjugated CodA was then loaded into the PEG-CNA-PLGA nanoparticles and tested for cell cytotoxicity in the presence of 5-FC (Scheme 1). The effect of coloading DOX and CodA within a single nanoparticle was also studied and compared to using nanoparticles loaded with either DOX or CodA alone, showing that the highest level of cell death occurs when both are simultaneously entrapped and delivered to cells in the presence of 5-FC (Scheme 1).
Scheme 1.
Graphic Representation of PEG-CNA-PLGA Nanoparticles Co-Loaded with Hydrophilic DNA-Conjugated Prodrug Activating Enzymes and Hydrophobic Chemotherapy Drugs
MATERIALS AND METHODS
Materials.
AlexaFluor488 NHS ester (Invitrogen), DNA sequences (Integrated DNA Tech.), ammonium chloride (Fisher), amphicillin (Sigma), biuret reagent (Sigma), coomassie stain (simplyblue safestain; Invitrogen), DBCO-NHS (Sigma), doxorubicin (DOX, Tokyo chem. Indus.), Dulbecco’s Modified Eagle Medium (Gibco), fetal bovine serum (Gibco), fluorocytosine (5-FC, Sigma), fluorouracil (5-FU, Sigma), folin, and coicalteu phenol reagent (Sigma), gel cassettes (NuPage 4–12%; Life Tech.), imidazole (Chem-Impex int’l Inc.), isopropyl β-d-thiogalactopyranoside (IPTG, Gold Biotech.), l-glutamine-penicillin-streptomycin solution (Sigma), loading buffer (LDS NuPage; Invitrogen), MDA-MB-468 cell line (ATCC), Ni-NTA Beads (HisPur; Thermo), o-phtalaldehyde (Sigma), poly(lactic-co-glycolic acid) (RG502H; PLGA, Sigma), thiolated poly(ethylene glycol) (PEG-SH, Lysan Bio Inc.), trypsin-EDTA (GenDEPOT), tryptone (Becton Dickinson), and yeast extract (Becton Dickinson).
Cytosine Deaminase (CodA) Expression.
E. coli strain BL21(DE3) was used as the template for amplifying CodA gene using the forward and reverse primer sequence given below. The primers incorporate a HindIII site at the 5′ position and an XhoI site at the 3′ position of the amplified sequence. The amplification was done using a standard PCR technique with a 65 °C annealing temperature.
5 ′ −3′: aagcttGGCGGTGGCTCGAATAACGCTTTACAAA-CAATTATTAAC
3′ −5′: ctcgagACGTTTGTAATCGATGGCTTCT
Pet21B vector was digested for 1 h at 37 °C with the mentioned restriction enzymes and the primers treated using a standard ligation mixture supplied by NEB. Finally the ligated plasmid was then transformed into the electrocompetent E. coli DH5α using an Eppendorf eporator at 1300 V. The transformed E. coli was then incubated overnight on an agar plate supplemented with Ampicillin as selection marker in order to create a 50% glycerol stock solution stored at —21 °C.
CodA Production.
CodA producing bacteria was pricked from the described glycerol stock and incubated in 5 mL of LB supplemented with 1 mg/mL Ampicillin overnight at 37 °C in an orbital shaker. A total of 1 mL of the activated bacteria was added to an Erlenmeyer flask containing 100 mL of LB with the same antibiotic supplementation and allowed to grow in the same incubation conditions until an absorbance of 0.7 at 600 nm was reached, the point at which 100 μL of a 1 M IPTG solution was added and allowed to express the protein in the same incubation conditions for 2.5 h. The solutions were centrifuged 5 min at 10000 g, the supernatant was discarded, and bacterial pellets were resuspended and collected in a total of 25 mL Equilibration Buffer. The bacterial suspension was sonicated at 25% output for 4 min in 1 min on/off cycles for 20 min and centrifuged at 12000 g and the supernatant was collected for protein purification.
Protein Purification.
Ni-NTA beads were cleaned following company instructions. To collect the CodA proteins, the bacterial supernatants were reacted with the cleaned Ni beads and washed 2– 3X with wash buffer. Any absorbed proteins were released by mixing the cleaned beads in 200 μL of elution buffer for 30 min in an end to end shaker and a last 2 min centrifugation at 700 g to obtain the purified CodA. Excess imidazole was then removed with a desalting column, final product concentration was assessed by its absorbance and stored at 4 °C in PBS.
CodA DNA and Dye Modification.
Varying concentrations of the purified CodA in PBS was mixed with equal volume of Bicarbonate buffer to slightly increase the pH and reacted in a 1:5 CodA to DBCO-NHS, to increase the solubility DMSO was added to the mixture so that final DMSO concentration was of 30%. This was allowed to proceed 4 h at 4 °C and without further purification further reacted with 5 mol equiv of NHS-AlexaFluor488 at 4 °C overnight. This was followed by 20 kDa MWCO Dialysis; it was found to be necessary to perform this step at 4 °C to avoid protein degradation (data not reported). Dialysis was performed against 1 L of PBS changed at 2, 4, 8, and then approximately every 12 h. Samples of the dialysate were periodically taken in order to monitor the process up to a total dialysis time of 72 h. The purified DBCO and dye modified enzyme was then quantified by Ohnishi and Barr’s Lowry’s method, calibrated with raw CodA quantified by its extinction coefficient (Figure S1), and then mixed with azido-DNA in a 1:5 ratio and left to react overnight at room temperature in an end to end shaker protected from light. Final product was then again followed by a dialysis purification and quantification, as described previously.
OPAME Assay.
The developing solution was made by dissolving 270 mg (2 mmol) of o-phthalaldehyde, 105 μL (1.5 mmol) of mercaptoethanol in 2.5 mL of ethanol and diluted to 50 mL with PBS. The solution was stored protected from light at room temperature and an aliquot filtered through a 0.22 μm PTFE filter before use. When an assay was to be performed equal volumes of the sample and the filtered developing reagent was mixed in an Eppendorf tube and left reacting in the dark for 30 min before fluorometrically assessing the concentration of an aliquot with 410 nm source excitation and a 473 nm read. Calibration was performed by varying concentrations of NH4Cl finding a linear response below 400 nM NH4Cl The assay brightness was found to have a time response (Figure S2) so it was deemed necessary to run the assay 30–60 min after adding the reagent and for every run to include a calibration in parallel.
Protein Quantification.
Protein was quantified by either an estimated 55550 cm−1 M-1 extinction coefficient39 at 380 nm or by an adapted Ohnishi and Barr modification of Lowry’s method.40 Briefly, 110 μL of Biuret reagent was mixed with 10 μL of protein solution and left reacting 10 min after which 5 μL of Folin and Coicalteu phenol reagent was added and mixed thoroughly. 100 μL of the final solution was used for the spectrometric analysis measuring the absorbance at 750 nm. Calibration was performed by using purified CodA protein quantified by abs as described above so that the results reported are self-consistent.
PEG-CNA(T10)-PLGA Polymer Nanoparticles (NPs).
The copolymer was synthesized as previously reported and prior to use, anhydrously stored in a concentrated 50 mg/mL DCM solution at –21 °C. Next, polymer NPs were formulated by dissolving PEG-CNA-PLGA in methylene chloride (DCM) to a final concentration of 50 mg/mL. A total of 100 μL of the polymer solution was then added to a 1/2 dram glass vial containing 500 μL of PBS and 100 μL of either clean DCM or a 250 μg/mL Doxorubicin (DOX) in DCM was further added according to the type of NPs being made. After vigorous shaking, the solution was heated up to 37 °C in an aluminum heating block and maintained uncapped for 2 h; the temperature was then increased to 50 °C and maintained 1 h to ensure evaporation of all remaining DCM. While stirring the solution was left to equilibrate to room temperature (RT) before centrifugation at 22000 g for 20 min. Supernatant was discarded and the nanoparticles were resuspended in the appropriate volume of buffer for the next step. Particle sizes were determined by nanoparticle tracking analysis, the results of which are shown in Figure S3.
CodA Entrapment in Polymer Nanoparticles.
In a similar way to the previously reported DNA conjugation,34 DNA conjugated 100 pmol of CodA was loaded per mg of nanoparticles by mixing a 50 μL aliquot of the polymer nanoparticles, 20 μL of the modified enzyme, and 30 μL of DMSO. The reactions were left overnight at 4 °C, diluted to 1 mL with PBS and centrifuged for 20 min at 22000 g. After three washes, the final pellets were resuspended in 100 μL of PBS. The amounts of encapsulated protein and DOX were determined using calibration curves shown in Figures S4-S6.
Cell Culture.
MDA-MB-468 breast cancer cells were seeded at 37 °C in a 5% CO2-enriched atmosphere with Dulbecco’s Modified Eagle Medium supplemented with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin-streptomycin. When cell density approached confluence, the cells were incubated with 5 mL of trypsin for 5–15 min and once detached centrifuged at 150 g for 5 min and resuspended in fresh media. Cell concentration was estimated by a dual chamber hemocytometer. A total of 0.5–1 million cells were reseeded in a flask and 96-well plates were prepared by adding 100 μL at 105 cells/mL to each well needed for the assay. Cells were left overnight after seeding before any further operation was performed.
Cell Viability Assay.
To the cells seeded in a 96-well plate, 90 μL of either fresh media or media supplemented with 5-FC was added. To that 10 μL of the appropriate concentration of nanoparticles to yield the desired final concentration was added followed by incubation for 24 h at 37 °C. Next, the cells were washed with PBS and replaced with 100 μL of new media supplemented with MTT stock solution to contain 0.5 mg/mL of MTT. Results were obtained by an absorbance reading at 560 nm and a reference reading at 650 nm.
Confocal imaging.
Confocal microscopy slides were prepared analogously to the cell viability assay by first seeding 1 mL of MDA-MB-468 at 5 X 104 cells/mL in a 24-well plate preprepared to contain a glass slide. Cells where left to attach overnight followed by incubation for 24 h then incubated 24 h with 1 mg/mL of NPs. Right before the 24 h mark 10 mL of cell media was warmed up to room temperature and 10 μL of a previously prepared 0.2 mg/mL DAPI in DMSO (stored at –21 °C) was added together with 100 μL of DiD 1 mg/mL in DMSO. DAPI and DiD staining was used to image the cell nuclei and membrane, respectively. The staining media was discarded after 10 min of incubation with the cells and washed twice more with PBS before immediately preparing the samples for microscopy.
RESULTS AND DISCUSSION
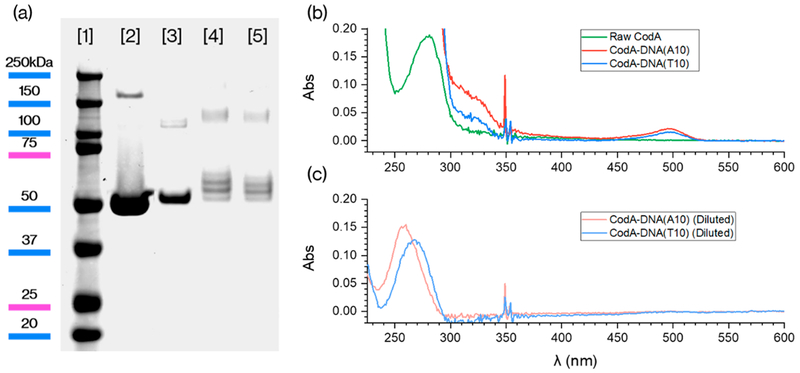
In this study, we first produced the prodrug enzyme cytosine deaminase (CodA), which catalyzes the deamination of cytosine to uracil in microorganisms in the pyrimidine synthetic pathway.41 For cancer therapy, CodA has been used to convert the prodrug fluorocytosine (5-FC) to the active drug fluorouracil (5-FU), which is believed to inhibit nucleotide synthetic enzyme thymidylate synthase.42 To produce CodA, we first expressed the protein in BL21(DE3) (E. coli) by inserting the gene in a Pet21B vector using HindIII and Xhol as restriction enzymes followed by transfection through electroporation. Cells were subsequently stored as a glycerol stock and then the enzyme was produced by growing the cells in LB at 37 °C and inducing with IPTG for 2.5 h when a 0.7 OD at 600 nm was reached. Cells were lysed and CodA proteins were collected and purified using Ni-NTA beads. The purity of the product was assessed by polyacrylamide gel electrophoresis (SDS-PAGE; Figure 1a) where a single band of ~50 kDa was observed.
Figure 1.
(a) SDS-PAGE gel of [1] protein ladder, [2] unmodified CodA, and [3] CodA after DBCO and AlexaFluor488 conjugation and after reacting with [4] A10 and [5] T10 DNA. (b, c) UV–vis absorbance of the dye and DNA-conjugated CodA.
The low toxicity of the prodrug 5-FC makes it an attractive therapy agent to target cancer cells, provided that the enzyme can be site-specifically delivered.24 In order to load CodA into our previously reported PLGA-CNA(T10)-PEG nanoparticles,34 we first conjugated the protein with polyadenine DNA strands so that the enzyme could specifically bind to the polythymine CNA domain. In addition, to track protein loading within the polymer nanoparticles, the enzyme was further modified with the dye AlexaFluor488. To attach both DNA and dye to each protein, we reacted CodA first with five molar equivalents of NHS-DBCO at pH 8.2 in 30% DMSO for 4 h. Next, NHS-AlexaFluor488 was directly added to the protein reactions using a 1:5 protein/dye molar ratio for 4 h. To prevent protein degradation all of the reactions were performed at 4 °C. Removing excess dye and NHS-DBCO from the proteins was done by dialysis at 4 °C for 72 h. The products of each step were characterized by SDS-PAGE and as shown in Figure 1a, specific bands that correspond to each modification step (dye attachment followed by T10 or A10 conjugation) were detected. UV-vis absorbance of the purified protein (Figure 1b) also confirms the successful conjugation of both the AlexaFluor488 dye and DNA oligonucleotides and based on calibration curves (Figure S4) we were able to determine ~1 dye molecule and ~3—4 DNA strands were attached per protein. In addition, a gel run with no Coomassie stain (Figure S7) showed the expected fluorescence from the successful addition of the AlexaFluor488 dye.
Next, the dye and DNA-conjugated CodA enzymes were tested for converting 5-FC to 5-FU to determine if modifying the protein either inhibited or hindered activity. For this, while the traditional method is to detect a shift in the UV absorbance peak at 286 nm as 5-FC is converted to 5-FU,43 the strong absorbance at 260 nm of the DNA attached to the enzyme (Figure 1b) obscured our ability to use UV-vis to monitor activity. Therefore, a common fluorescence technique that detects the presence of primary amines was employed since ammonia is a byproduct of the 5-FC deamination to 5-FU (Figure 2a). o-Phthalaldehyde becomes fluorescent only when reacted with both a sulfhydryl group and a primary amine and thus can be used to assess the amount of 5-FC reacted with CodA and spectrometrically quantify37 the progress of the reaction.
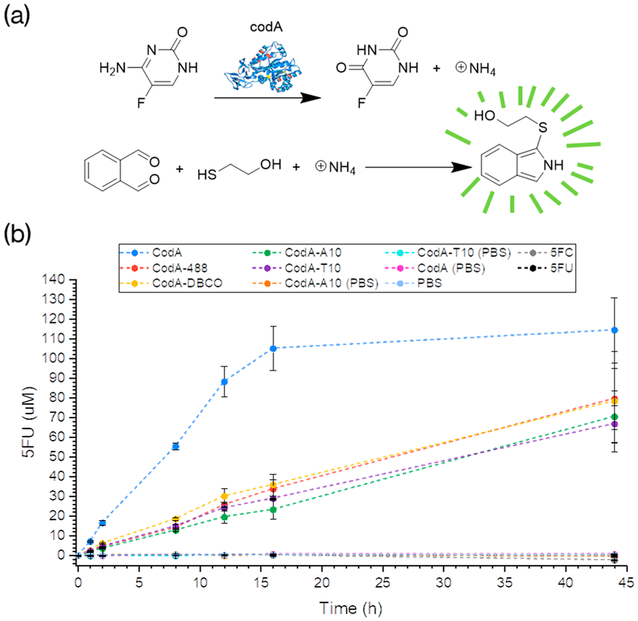
Figure 2.
(a) Scheme of the OPAME assay for determining 5-FC deamination rates. (b) Plots showing the amount of 5-FC converted to 5-FU over time as determined by the OPAME assay. Samples were run with using 500 μM 5-FC or 5-FU in the absence of the presence of 10 nM CodA. All reactions were run in triplicate with each point showing the average value obtained and subsequent error bars.
To ascertain that the presence of 5-FC, CodA alone, or DNA and dye-conjugated CodA does not yield a fluorescent signal in the presence of o-phthalaldehyde and mercaptoethanol (OPAME), we reacted 60 μL of each of the control substrates with 60 μL of the OPAME solution (2 mmol o-phthalaldehyde, 1.5 mmol mercaptoethanol, and 2.5 mL EtOH) diluted to a final volume of 50 mL with PBS. Next, the reactions were incubated at room temperature in the dark for 30 min followed by measuring fluorescence at 473 nm (405 nm excitation). As shown in Figures 2b and S2, little to no measurable fluorescence could be detected indicating that the presence of amine groups on the enzyme or prodrugs alone could not generate any fluorescence in the OPAME assay.
Next, 10 nM CodA (native, dye, DNA, and dye conjugated) enzymes were reacted with 500 μM 5-FC and incubated in PBS at 37 °C while being continuously vigorously mixed in an orbital shaker. At varying time points, aliquots were removed and assayed using the OPAME test as described above. In order to determine the amount of ammonia being produced over time which should be in equimolar amounts to the amount of 5-FC converted to 5-FU, calibration curves with NH4Cl were used (Figure S8) for each collected time point. Because the intensity of the fluorescence response was found to decrease after 30–60 min, fluorescence measurements were run at a set point of 45 min after starting the OPAME tests (Figure S2).
As shown in Figure 2a, modifying the enzyme decreased the rate of 5-FC conversion, regardless of whether the modification was with dye alone or DNA-dye. Because of the nonspecific decrease in enzyme activity, we believe that the CodA activity is related in part to the denaturation of the protein during the first reaction with NHS-DBCO as the reactions were run at pH 8.2 and 30% DMSO, which is also supported by the data shown in Figure S9. Despite this decrease in activity, however, the enzyme is still capable of activating a prodrug for cancer therapy.
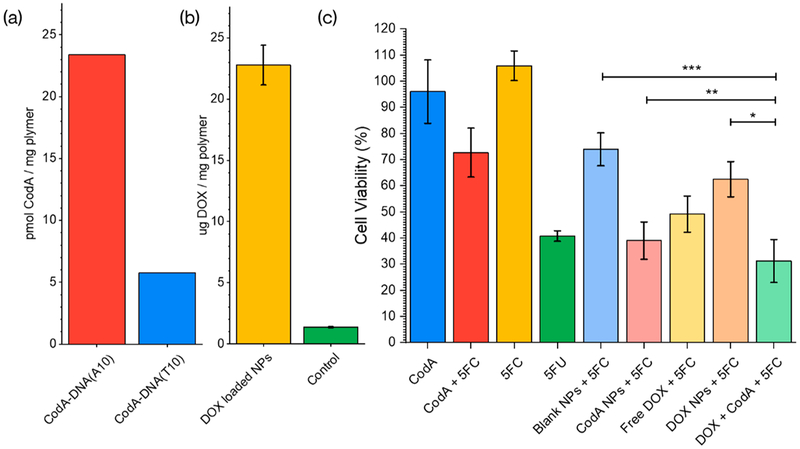
Next, to encapsulate both CodA and a chemotherapy drug such as DOX in a single carrier, the triblock copolymer PEG-CNA(T10)-PLGA was synthesized as reported in our previous work.18 PEG-CNA(T10)-PLGA nanoparticles were then formulated by emulsifying a solution of the polymer dissolved in methylene chloride with water and increasing the temperature slowly under high stirring until all the organic solvent is evaporated, leaving a nanoparticle mixture that could be collected and cleaned by high speed centrifugation. Next, 200 pmol of the DNA and dye conjugated CodA proteins were reacted with 2 mg of the PEG-CNA(T10)-PLGA nanoparticles at 30/70 DMSO/water overnight at 4 °C followed by 3 consecutive washes to remove any free, nonencapsulated protein. An aliquot was lyophilized, weighed, and dissolved in pure DMSO to fluorometrically quantify the amount of CodA entrapped for a given mass of PEG-CNA(T10)-PLGA nanoparticles. As shown in Figure 3a, using these methods we were able to discern that ~46 pmol (23%) of the added DNA(A10)-CodA protein was encapsulated in ~2 mg of polymer nanoparticles. In contrast, when the CodA protein was conjugated with either dye alone or T10 DNA, only ~12 pmol (6%) could be encapsulated, showing again that specific DNA-CNA hybridization was needed to load the enzyme in the PEG-CNA-PLGA nanoparticles. In addition to loading the CodA enzyme in the polymer nanoparticles, the chemotherapy drug DOX could also be simultaneously entrapped by simply adding it in during the polymer nanoparticle synthesis. Since the fluorescence emission of DOX (610 nm) is separate from that of the AlexaFluor 488 dye on the enzyme, we were able to determine ~23 μg DOX could be loaded per mg of polymer nanoparticle (Figure 3b).
Figure 3.
(a) Estimated entrapment of DNA modified CodA for both a polyadenine and polythymine-modified enzyme. (b) Amount of Doxorubicin. (c) MTT results of incubating polymer nanoparticles with cells. All samples contained 500 μM of 5-FC except for the first column (CodA) and the fourth one (5-FU). CodA and CodA+5FU samples contained 10 nM of the enzyme and 500 μM 5-FU. All nanoparticle samples contained ~1 mg/mL NPs. The DOX alone test contained 23 μg/mL of the drug in solution -the estimated amount encapsulated in the NPs. Blank and DOX NPs contained no enzyme. Error bars represent the standard error calculated from a n = 6, we estimated a *p < 0.006 of the coloaded NPs being less toxic than Doxorubicin NPs, a **p < 0.031 for being more toxic than CodA-loaded NPs and a ***p < 10−7 of being less toxic than blank NPs for a normal distribution.
The CodA and DOX loaded polymer nanoparticles were next evaluated for cell killing and compared to that of nanoparticles loaded with CodA or DOX alone. In this instance, breast cancer MDA-MB-468 cell lines were used in an MTT assay, and all tests were done in the presence of 5-FC. As controls, polymer nanoparticles were added with no DOX loaded or no addition of 5-FC; 5-FU was also tested alone as a comparison. For all tests, MDA-MB-468 cells were seeded and incubated overnight in 96-well plates at a cell density of 104 cells per well. Next, the various therapy agents (polymer nanoparticles, CodA, DOX, 5-FC, 5-FU) were added and incubated with the cells for 24 h at 37 °C. Next, the media was removed, the cells were washed 2X with PBS, and MTT assays were performed. As shown in Figure 3c, compared to blank PLGA-CNA-PEG nanoparticles in the presence of the prodrug 5-FC, the CodA-loaded NPs show a significant increase in cell toxicity, which can be attributed to the enzyme activation of the prodrug. In comparing the CodA-loaded nanoparticles and 5-FC with 5-FU alone, highly comparable results in cell viability were obtained, demonstrating that although 5-FU can diffuse into cells immediately upon adding to cells, the enzyme loaded polymer nanoparticles did not cause lower cell killing capabilities as compared to the drug alone, despite the need for the prodrug 5-FC to enter the polymer corona, diffuse to the enzyme embedded and react to produce the active drug 5-FU. The polymer nanoparticles loaded with enzyme seemed to also show higher cell killing as compared to enzyme alone with 5-FC, but this behavior may in part be attributed to an inherent cell toxicity the PEG-CNA(T10)-PLGA nanoparticles seem to show (“Blank NPs + 5-FC” as compared to the control of 5-FC alone) at the polymer nanoparticle concentrations used (Figure S10). Lastly, the polymer nanoparticles loaded with both CodA and DOX were tested with the MDA-MB-468 cells in the presence of the prodrug 5-FC. As shown in Figure 3c, comparing either DOX alone or DOX-loaded polymer nanoparticles, activating prodrugs in the vicinity of the cells along with DOX delivery showed significant enhancement of cell killing, supporting the hypothesis that delivering and activating multiple therapy agents using a single carrier can enhance cancer cell eradication.
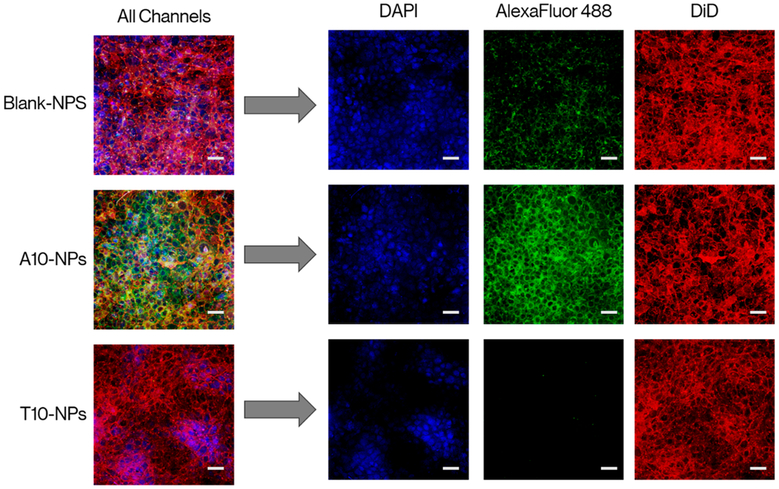
Finally, confocal imaging was run to visually assess cellular uptake of the enzyme-loaded (no DOX) nanoparticles. For this, MDA-MB-468 cells were seeded in 24-well plates that contained a glass slide in the bottom of each well. Cells were left to attach overnight followed by incubating the cells with 1 mg/mL of the CodA loaded polymer nanoparticles for 24 h. The slides where then washed with PBS and stained for 10 min at 37 °C with DMEM media containing 0.2 μg of DAPI and 10 μg of DiD per mL acting as a nuclear and membrane stains, respectively.
As shown in Figure 4, only cells incubated with PEG-CNA(T10)-PLGA nanoparticles loaded with DNA(A10)-AlexaFluor488 conjugated CodA showed any marked fluorescence in the green, showing not only that CodA is indeed entrapped in the polymer nanoparticles but also that the enzyme is being delivered intracellularly. Cells incubated with polymer nanoparticles reacted with DNA(T10)-AlexaFluor488 conjugated CodA showed nearly zero green emission supporting the earlier results that enzyme loading in the PEG-CNA(T10)-PLGA nanoparticles is dependent on specific CNA–DNA hybridization. Lastly, cell uptake kinetic analyses were performed by reacting seeded MDA-MB-468 cells with DOX-loaded PEG-CNA(T10)-PLGA nanoparticles and at various time points, removing the media and measuring intracellular delivery of the nanoparticles by flow cytometry. As shown in Figure S11, nanoparticle uptake appeared to reach a maximum value after ~5 h of incubation with cells with little to no change up to 10 h of measurement.
Figure 4.
Confocal imaging of MDA-MB-468 incubated for 24h with 1 mg/mL of unconjugated NPs as well as particles reacted with AlexaFluor 488 modified cytosine deaminase conjugated with either polyadenine or polythymine DNA. As shown, green fluorescence from the AlexaFluor488 dye was only detected from cells incubated with PEG-CNA(T10)-PLGA nanoparticles reacted with poly-A conjugated CodA, demonstrating the specificity of CNA-DNA hybridization. Scale bars represent 50 μm.
CONCLUSION
In this work we have shown the successful simultaneous entrapment of the prodrug activating enzyme cytosine deaminase (CodA) with the chemotherapy drug doxorubicin (DOX) within a single polymer nanoparticle. This result was achieved by conjugating the protein with polyadenine strands, then using complementary base pairing to PEG-CNA(T10)-PLGA to incorporate the enzyme into preformed polymer nanoparticles that could also be loaded with DOX. To test for prodrug activation by the DNA conjugated CodA, a fluorometric assay based on o-phthalaldehyde was developed, which showed that despite some nonspecific decrease in activity, the DNA-modified CodA retained the ability to convert prodrug 5-FC to active drug 5-FU. After encapsulating the DNA-CodA in PEG-CNA-PLGA nanoparticles, cell toxicity assays were performed against MDA-MB-468 breast cancer cells. These results showed low toxicity with particles alone in the presence of 5-FC, but a significant increase in cell killing upon loading with DNA-CodA. Furthermore, the toxic effects were compounded when the hydrophobic drug doxorubicin was also encapsulated, showing potential for adapting this technique in the codelivery of chemotherapy agents. Lastly, confocal imaging was used to demonstrate successful targeting of the cells with the enzyme loaded PEG-CNA-PLGA nanoparticles and that complementary base pairing was required to associate the CNA strands of the polymer with the DNA oligonucleotides on the enzyme.
Supplementary Material
ACKNOWLEDGMENTS
A.H. gratefully acknowledges the initial support of the Balsells fellowship program.
Funding
We gratefully acknowledge financial support from NSF-MRSEC (DMR1420736) and the National Institutes of Health Award # DP2EB020401.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-mac.9b00040.
Fluorescently scanned SDS-PAGE gel showing dye conjugation on the studied proteins (Figure S7), calibration lines used in protein quantification in the studied solvents in all the mentioned methods described in the main text (S1, S4, S5, and S6), the resulting calibration for the developed OPAME assay (S8) as well as its time stability (S2), a brief study on the inherent toxicity of the described blank NPs (S10), an assay on denaturing effects of the reaction conditions on codA (S9), NTA of the studied NPs (S3) and finally a cytometry test on the delivered DOX on cells with the studied NPs (S11) (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Miller KD; Siegel RL; Lin CC; Mariotto AB; Kramer JL; Rowland JH; Stein KD; Alteri R; Jemal A Cancer Treatment and Survivorship Statistics, 2016. Ca-Cancer J. Clin 2016, 66 (4), 271–289. [DOI] [PubMed] [Google Scholar]
- (2).Krishnan V; Rajasekaran AK Clinical Nanomedicine: A Solution to the Chemotherapy Conundrum in Pediatric Leukemia Therapy. Clin. Pharmacol. Ther. 2014, 95 (2), 168–178. [DOI] [PubMed] [Google Scholar]
- (3).Kemp JA; Shim MS; Heo CY; Kwon YJ “Combo” Nanomedicine: Co-Delivery of Multi-Modal Therapeutics for Efficient, Targeted, and Safe Cancer Therapy. Adv. Drug Delivery Rev. 2016, 98, 3–18. [DOI] [PubMed] [Google Scholar]
- (4).Morton SW; Lee MJ; Deng ZJ; Dreaden EC; Siouve E; Shopsowitz KE; Shah NJ; Yaffe MB; Hammond PT A Nanoparticle-Based Combination Chemotherapy Delivery System for Enhanced Tumor Killing by Dynamic Rewiring of Signaling Pathways. Sci. Signaling 2014, 7 (325), ra44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Burrell RA; McGranahan N; Bartek J; Swanton C The Causes and Consequences of Genetic Heterogeneity in Cancer Evolution. Nature 2013, 501 (7467), 338–345. [DOI] [PubMed] [Google Scholar]
- (6).Li H; Chen L; Shi Y; Yuan B; Ma Y; Wei H; Zhao G Design of Block Copolymer Micellar Aggregates for Co-Delivery of Enzyme and Anticancer Prodrug. Chem. - Asian J 2017, 12 (2), 176–180. [DOI] [PubMed] [Google Scholar]
- (7).Chen L; Liu Z; Jin R; Yang X; Bai Y; Liu S; Chen X Stepwise Co-Delivery of an Enzyme and Prodrug Based on a Multi-Responsive Nanoplatform for Accurate Tumor Therapy. J. Mater. Chem. B 2018, 6 (39), 6262–6268. [DOI] [PubMed] [Google Scholar]
- (8).Zheng C; Zheng M; Gong P; Deng J; Yi H; Zhang P; Zhang Y; Liu P; Ma Y; Cai L Polypeptide Cationic Micelles Mediated Co-Delivery of Docetaxel and SiRNA for Synergistic Tumor Therapy. Biomaterials 2013, 34 (13), 3431–3438. [DOI] [PubMed] [Google Scholar]
- (9).Qu MH; Zeng RF; Fang S; Dai QS; Li HP; Long JT Liposome-Based Co-Delivery of SiRNA and Docetaxel for the Synergistic Treatment of Lung Cancer. Int. J. Pharm. 2014, 474 (1–2), 112–122. [DOI] [PubMed] [Google Scholar]
- (10).Zhang Y; Satterlee A; Huang L In Vivo Gene Delivery by Nonviral Vectors: Overcoming Hurdles? Mol. Ther. 2012, 20 (7), 1298–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Harris JM; Chess RB Effect of Pegylation on Pharmaceuticals. Nat. Rev. Drug Discovery 2003, 2 (3), 214–221. [DOI] [PubMed] [Google Scholar]
- (12).Hanson JA; Chang CB; Graves SM; Li Z; Mason TG; Deming TJ Nanoscale Double Emulsions Stabilized by Single Component Block Copolypeptides. Nature 2008, 455 (7209), 85–88. [DOI] [PubMed] [Google Scholar]
- (13).Tseng YC; Mozumdar S; Huang L Lipid-Based Systemic Delivery of SiRNA. Adv. Drug Delivery Rev. 2009, 61 (9), 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Soppimath KS; Aminabhavi TM; Kulkarni AR; Rudzinski WE Biodegradable Polymeric Nanoparticles as Drug Delivery Devices. J. Controlled Release 2001, 70 (1–2), 1–20. [DOI] [PubMed] [Google Scholar]
- (15).Mahon E; Salvati A; Baldelli Bombelli F; Lynch I; Dawson KA Designing the Nanoparticle-biomolecule Interface for “Targeting and Therapeutic Delivery. J. Controlled Release 2012, 161 (2), 164–174. [DOI] [PubMed] [Google Scholar]
- (16).Saad M; Garbuzenko OB; Minko T Co-Delivery of SiRNA and an Anticancer Drug for Treatment of Multidrug-Resistant Cancer. Nanomedicine 2008, 3 (6), 761–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Lee ALZ; Wang Y; Cheng HY; Pervaiz S; Yang YY The Co-Delivery of Paclitaxel and Herceptin Using Cationic Micellar Nanoparticles. Biomaterials 2009, 30 (5), 919–927. [DOI] [PubMed] [Google Scholar]
- (18).Huang Y; Yu F; Park Y-S; Wang J; Shin M-C; Chung HS; Yang VC Co-Administration of Protein Drugs with Gold Nanoparticles to Enable Percutaneous Delivery. Biomaterials 2010, 31 (34), 9086–9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Jiang T; Mo R; Bellotti A; Zhou J; Gu Z Gel-Liposome-Mediated Co-Delivery of Anticancer Membrane-Associated Proteins and Small-Molecule Drugs for Enhanced Therapeutic Efficacy. Adv. Funct. Mater. 2014, 24 (16), 2295–2304. [Google Scholar]
- (20).Lv H; Zhang S; Wang B; Cui S; Yan J Toxicity of Cationic Lipids and Cationic Polymers in Gene Delivery. J. Controlled Release 2006, 114 (1), 100–109. [DOI] [PubMed] [Google Scholar]
- (21).Choi CHJ; Hao L; Narayan SP; Auyeung E; Mirkin CA Mechanism for the Endocytosis of Spherical Nucleic Acid Nanoparticle Conjugates. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (19), 7625–7630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Gaumet M; Vargas A; Gurny R; Delie F Nanoparticles for Drug Delivery: The Need for Precision in Reporting Particle Size Parameters. Eur. J. Pharm. Biopharm. 2008, 69 (1), 1–9. [DOI] [PubMed] [Google Scholar]
- (23).Champion JA; Katare YK; Mitragotri S Particle Shape: A New Design Parameter for Micro- and Nanoscale Drug Delivery Carriers. J. Controlled Release 2007, 121 (1–2), 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Kulkarni SA; Feng S Effects of Particle Size and Surface Modification on Cellular Uptake and Biodistribution of Polymeric Nanoparticles for Drug Delivery. Pharm. Res. 2013, 30 (10), 2512–2522. [DOI] [PubMed] [Google Scholar]
- (25).Bazile DV; Ropert C; Huve P; Verrecchia T; Mariard M; Frydman A; Veillard M; Spenlehauer G Body Distribution of Fully Biodegradable [14C]-Poly(Lactic Acid) Nanoparticles Coated with Albumin after Parenteral Administration to Rats. Biomaterials 1992, 13 (15), 1093–1102. [DOI] [PubMed] [Google Scholar]
- (26).Cheng J; Teply B. a; Sherifi I; Sung J; Luther G; Gu FX; Levy-Nissenbaum E; Radovic-Moreno AF; Langer R; Farokhzad OC Formulation of Functionalized PLGA-PEG Nanoparticles for in Vivo Targeted Drug Delivery. Biomaterials 2007, 28 (5), 869–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Danhier F; Ansorena E; Silva JM; Coco R; Le Breton A; Préat V PLGA-Based Nanoparticles: An Overview of Biomedical Applications. J. Controlled Release 2012, 161 (2), 505–522. [DOI] [PubMed] [Google Scholar]
- (28).Dziubla TD; Karim A; Muzykantov VR Polymer Nanocarriers Protecting Active Enzyme Cargo against Proteolysis. J. Controlled Release 2005, 102 (2), 427–439. [DOI] [PubMed] [Google Scholar]
- (29).Hosseininasab S; Pashaei-Asl R; Khandaghi AA; Nasrabadi HT; Nejati-Koshki K; Akbarzadeh A; Joo SW; Hanifehpour Y; Davaran S Synthesis, Characterization, and in Vitro Studies of PLGA-PEG Nanoparticles for Oral Insulin Delivery. Chem. Biol. Drug Des. 2014, 84 (3), 307–315. [DOI] [PubMed] [Google Scholar]
- (30).Crotts G; Park TG Protein Delivery from Poly(Lactic-Co-Glycolic Acid) Biodegradable Microspheres: Release Kinetics and Stability Issues. J. Microencapsulation 1998, 15 (6), 699–713. [DOI] [PubMed] [Google Scholar]
- (31).Zheng C-H; Gao J-Q; Zhang Y-P; Liang W-Q A Protein Delivery System: Biodegradable Alginate-chitosan-poly(Lactic-Co-Glycolic Acid) Composite Microspheres. Biochem. Biophys. Res. Commun. 2004, 323 (4), 1321–1327. [DOI] [PubMed] [Google Scholar]
- (32).Xi W; Pattanayak S; Wang C; Fairbanks B; Gong T; Wagner J; Kloxin CJ; Bowman CN Clickable Nucleic Acids: Sequence-Controlled Periodic Copolymer/Oligomer Synthesis by Orthogonal Thiol-X Reactions. Angew. Chem. Int. Ed 2015, 54 (48), 14462–14467. [DOI] [PubMed] [Google Scholar]
- (33).Han X; Domaille DW; Fairbanks BD; He L; Culver HR; Zhang X; Cha JN; Bowman CN New Generation of Clickable Nucleic Acids: Synthesis and Active Hybridization with DNA. Biomacromolecules 2018, 19, 4139–4146. [DOI] [PubMed] [Google Scholar]
- (34).Harguindey A; Domaille DW; Fairbanks BD; Wagner J; Bowman CN; Cha JN Synthesis and Assembly of Click-Nucleic-Acid-Containing PEG-PLGA Nanoparticles for DNA Delivery. Adv. Mater. 2017, 29 (24), 1700743. [DOI] [PubMed] [Google Scholar]
- (35).Brodin JD; Sprangers AJ; McMillan JR; Mirkin CA DNA-Mediated Cellular Delivery of Functional Enzymes. J. Am. Chem. Soc. 2015, 137 (47), 14838–14841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Roth M Fluorescence Reaction for Amino Acids. Anal. Chem. 1971, 43 (7), 880–882. [DOI] [PubMed] [Google Scholar]
- (37).Simons SS; Johnson DF The Structure of the Fluorescent Adduct Formed in the Reaction of O-Phthalaldehyde and Thiols with Amines. J. Am. Chem. Soc. 1976, 98 (22), 7098–7099. [Google Scholar]
- (38).Long YF; Liao QG; Huang CZ; Ling J; Li YF Conformational Change Detection of DNA with the Fluorogenic Reagent of O-Phthalaldehyde-β-Mercaptoethanol. J. Rhys. Chem. B 2008, 112 (6), 1783–1788. [DOI] [PubMed] [Google Scholar]
- (39).Chazan A Peptide Property Calculator; http://biotools.nubic.northwestern.edu/proteincalc.html. [Google Scholar]
- (40).Ohnishi ST; Barr JK A Simplified Method of Quantitating Protein Using the Biuret and Phenol Reagents. Anal. Biochem. 1978, 86 (1), 193–200. [DOI] [PubMed] [Google Scholar]
- (41).Sakai T; Yu T. s.; Tabe H; Omata S Purification of Cytosine Deaminase from Serratia Marcescens. Agric. Biol. Chem. 1975, 39 (8), 1623–1629. [Google Scholar]
- (42).Longley DB; Harkin DP; Johnston PG 5-Fluorouracil: Mechanisms of Action and Clinical Strategies. Nat. Rev. Cancer 2003, 3 (5), 330–338. [DOI] [PubMed] [Google Scholar]
- (43).Porter DJT; Austin EA Cytosine Deaminase: The Roles of Divalent Metal Ions in Catalysis. J. Biol. Chem. 1993, 268 (32), 24005–24011. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.