Abstract
DNA-mediated assembly of core-satellite structures composed of Zr(IV)-based porphyrinic metal-organic framework (MOF) and NaYF4,Yb,Er upconverting nanoparticles (UCNPs) for photodynamic therapy (PDT) is reported. MOF NPs generate singlet oxygen (1O2) upon photoirradiation with visible light without the need for additional small molecule, diffusional photosensitizers such as porphyrins. Using DNA as a templating agent, well-defined MOF-UCNP clusters are produced where UCNPs are spatially organized around a centrally located MOF NP. Under NIR irradiation, visible light emitted from the UCNPs is absorbed by the core MOF NP to produce 1O2 at significantly greater amounts than what can be produced from simply mixing UCNPs and MOF NPs. The MOF-UCNP core-satellite superstructures also induce strong cell cytotoxicity against cancer cells, which are further enhanced by attaching epidermal growth factor receptor targeting affibodies to the PDT clusters, highlighting their promise as theranostic photodynamic agents.
1. Introduction
Extensive research in recent years has focused on the development of new materials for detecting diseased locations in the body and inducing site-specific therapy. Photodynamic therapy (PDT) has been employed because of its minimal invasiveness and ability to localize irradiation at tumor sites.[1,2] Because PDT relies on photogenerated single oxygen (1O2), effective therapy requires a combination of light, photosensitizer, and tissue oxygenation. However, typical photosensitizers are porphyrin derivatives whose hydrophobicity complicates dispersion and transport through aqueous environments.[3] To address this, different nanoparticle carriers have been studied for loading such photosensitizers, including porous silica or polymer nanoparticles due to their biocompatibility and large surface area.[4,5] Furthermore, in order to take advantage of the increased tissue penetration of NIR light as compared to UV or visible, upconverting nanoparticles (UCNPs) have been utilized to excite the photosensitizers due to their ability to absorb in the NIR and emit visible light.[2,5-11] For example, we recently showed that UCNPs coated with porous silica shells can be loaded with zinc phthalocyanine to produce 1O2 upon NIR irradiation and induce cell cytotoxicity in vitro.[6] However, photosensitizer loading in the silica-coated UCNPs was very low, reaching a maximum of 0.34 wt%, dramatically limiting singlet oxygen production and making it difficult to apply for effective cancer treatment.[2] Aside from low loading yields, the small molecule photosensitizers can also diffuse out of the porous carriers during the delivery process, leading to losses and further lowering efficacy for PDT.[8,12,13]
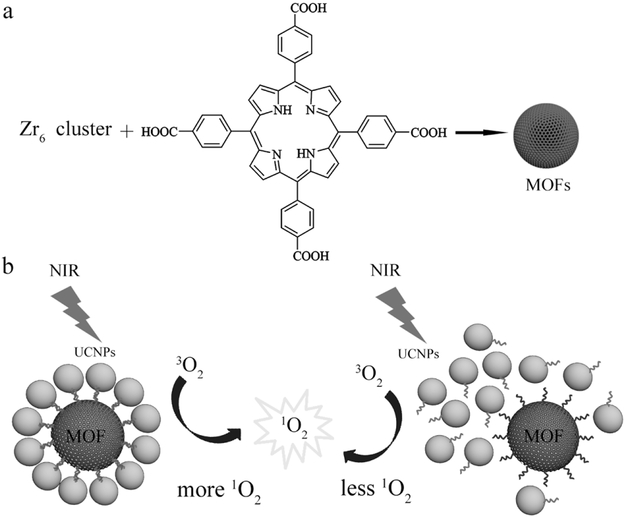
An alternative strategy would be to incorporate the porphyrins directly into metal-organic frameworks (MOFs) to yield structures that can efficiently generate 1O2 without needing small molecule photosensitizers.[3,14-16] MOFs typically possess uniform pores and channels, structural adaptivity and flexibility, and multiple coordination sites that originate from a near-limitless choice of building blocks.[17-21] In addition, nanoscale MOFs have been incorporated into sensing and catalysis applications.[22-28] For example, Lin and co-workers recently showed that Hf-porphyrin MOF nanoparticles (MOF NPs) were effective PDT agents for treating head and neck cancers with 640 nm light,[14,16,29] while Zhou and co-workers showed a size-dependent cellular uptake of porphyrin Zr-MOF PCN-224 nanoparticles (porous coordination network).[3] In these studies however, the need for visible light limits the utility of the MOF nanoparticles for in vivo applications due to the tissue transparency window. Therefore, to utilize the MOF NPs for targeted therapy in vivo, we show here methods to fabricate well-ordered MOF-UCNP core-satellite superstructures through DNA interactions that could generate high amounts of 1O2 upon NIR excitation (Scheme 1). We also demonstrate the therapeutic efficacy of the MOF-UCNP clusters in vitro as a function of assembly and the MOF:UCNP molar ratios used.
Scheme 1.
a) Illustration of PCN-224 MOF NPs synthesis. b) Self-assembled MOF–UCNP core–satellite superstructures (left) and random mixtures of MOFs and UCNPs (right) for PDT.
2. Results and Discussion
First, Zr(IV)-based porphyrinic MOFs were synthesized. These MOFs were chosen for their biocompatibility and structural robustness due to strong electrostatic interactions between the Zr cations and carboxylate groups on the porphyrin.[3,21,30] In particular, PCN-224, developed by Zhou and c-workers, was studied because of its large stability and porosity, which allows efficient diffusion of molecular oxygen.[3] More importantly, PCN-224 can be easily prepared with nanoscale dimensions, and the free carboxyl groups on the periphery are convenient for conjugation of biomolecules such as targeting ligands or DNA.[3] Here, PCN-224 MOF nanoparticles were synthesized by combining 50 mg tetrakis (4-carboxyphenyl) porphyrin (H2TCPP), 150 mg zirconylchloride octahydrate (ZrOCl2.8H2O) with a solution of benzoic acid dissolved in N,N-dimethylformamide (DMF), and stirring at 90 °C for 6 h.[3] The benzoic acid is used to control the nucleation rate of the MOF NP by competing for the Zr6 clusters.[3] As shown in Figure S1 (Supporting Information), the as-prepared PCN-224 MOF NPs were spherical with an average diameter of 52.1 ± 9.8 nm. X-ray diffraction (XRD) confirmed the crystalline nature of the MOF NPs and no other impurities were detected (Figure S2, Supporting Information). UV–vis absorption spectroscopy of the H2TCPP molecule showed a Soret band at 419 nm and four Q-bands at 513, 548, 589, and 645 nm. The PCN-224 MOF NPs showed slight redshifts for all Q-bands with the peaks appearing at 515, 550, 591, and 646 nm (Figure S3, Supporting Information), which probably results from the coordination of the carboxylate groups of TCPP to Zr6 clusters.[31] The PCN-224 MOF NPs also showed fluorescence emission under visible light irradiation with an emission maximum of ~650 nm (Figure S4, Supporting Information).
Next, the PCN-224 MOF NPs were conjugated to DNA oligonucleotides to fabricate the MOF–UCNP clusters, with the aim of using two complementary DNA strands (DNA1 and DNA2; Table S1, Supporting Information) for the assembly. For this, the MOF nanoparticles were first reacted with Nα,Nα-bis(carboxymethyl)-l-lysine hydrate, which has three carboxylic acid groups that can coordinate with the unsaturated Zr coordination sites, leaving amine groups exposed on the surface of the MOF, which was confirmed by the UV–vis and Fourier transform infrared spectroscopy (FTIR) shown in Figure S5 (Supporting Information). The amine-modified MOF NPs were then reacted with dibenzocyclooctyne-N-hydroxysuccinimidyl ester (NHS-DBCO), followed by DNA1 strands whose 5′ ends were modified with azide units (Table S1, Supporting Information). The DNA-conjugated MOF NPs were next purified by centrifuge filtration. To determine the concentration of MOF and DNA-conjugated MOF NPs in the stock solutions, a standard absorption curve was first determined by weighing out the as-synthesized MOFs, dispersing them in water, and measuring UV–vis absorbance (Figure S6, Supporting Information). UV–vis analysis of the filtrates was run to show that there are ≈260 strands of DNA per MOF NP (Figure S7, Supporting Information).
Herein, NaFY4:Yb/Er UCNPs were synthesized using a thermal decomposition method to produce monodisperse particles 23.8 ± 1.4 nm in diameter (Figure S8, Supporting Information).[6,32] The powder XRD pattern of the as-prepared NaYF4,Yb,Er UCNPs and selected area electron diffraction pattern further confirmed the UCNPs have a β-phase crystalline structure (Figure S8 and S9, Supporting Information). In order to transfer the as-prepared UCNPs into water, C18-PEG-NH2 was added to intercalate with the oleic acid surfactants on the UCNP surface.[6,28] Next, DNA2 strands complementary to the DNA1 sequence on the MOF NPs were conjugated to the UCNPs by first reacting the C18-PEG-NH2 coated UCNPs with NHS-DBCO for 4–5 h. As with the MOF NPs, azide-terminated DNA2 strands were then added to conjugate the UCNPs through copper-free click reaction (Table S1). Finally, the well-dispersed DNA2-UCNPs were purified through 30K microcentrifuge filters. Transmission electron microscopy (TEM) images showed the DNA conjugated UCNPs to be well-dispersed with little aggregation (Figure S10, Supporting Information), and analysis of UV–vis spectra of the filtrate showed that ≈78 DNA2 strands were attached per UCNP (Figure S11, Supporting Information).[33]
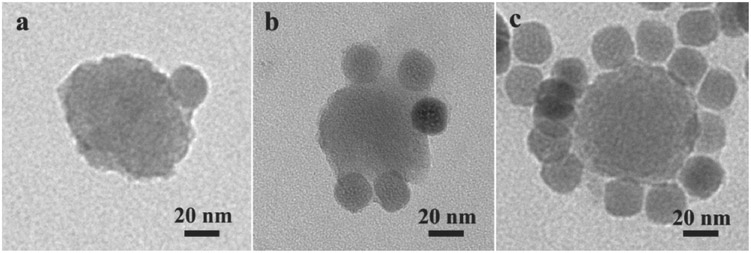
The core–satellite MOF–UCNP superstructures were then fabricated by annealing the DNA1-modified PCN-224 NPs (DNA1-MOFs) with the DNA2-modified UCNPs (DNA2-UCNPs) at 60 °C for 10 min followed by slow cooling. This procedure was done using molar ratios of MOF:UCNP that ranged from 1:1 to 1:24. As shown in Figure 1, TEM imaging revealed well-defined core–satellite assemblies with a single PCN-224 MOF NP forming the core and UCNPs as the satellites. Almost all the DNA2-UCNPs were conjugated with a centrally located DNA1-MOF up to 1:12 molar ratios of MOF: UCNP before and after 980 nm laser irradiation (Figures S12 and S13, Supporting Information). With increasing UCNP loadings beyond 1:12, many free UCNPs were found detected that were not conjugated to MOF NPs (Figure S14, Supporting Information). As controls, MOF NPs and UCNPs bound with noncomplementary DNA were also heated and cooled; as shown in Figure S15 (Supporting Information) only random distributions of nanoparticles were observed.
Figure 1.
TEM images of assembled MOF–UCNP superstructures with DNA1-MOF and DNA2-UCNPs using MOF:UCNP ratios of a) 1:1, b) 1:6, and c) 1:12.
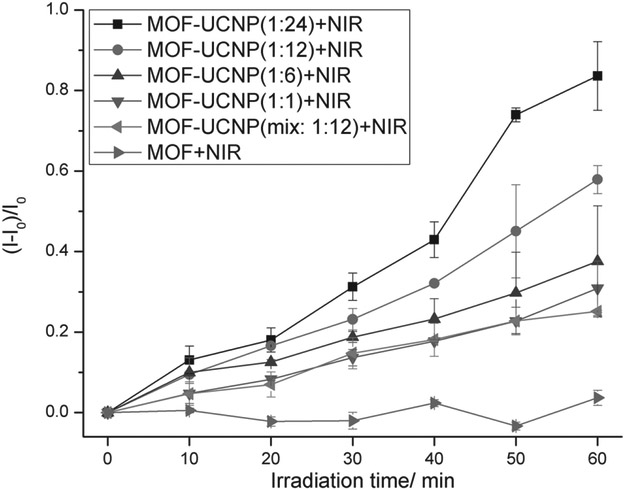
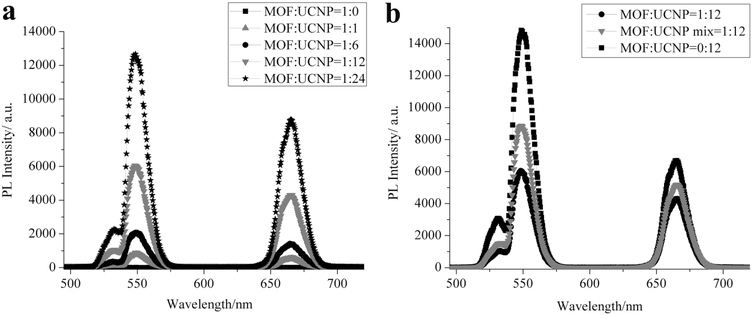
Due to the overlap of the MOF NP absorbance with the emissive wavelengths of the UCNPs upon NIR excitation (Figure S16, Supporting Information), it was reasoned that singlet oxygen (1O2) could be produced from the MOF–CNP clusters with 980 nm irradiation, well within the tissue transparency window. To study this, Singlet Oxygen Sensor Green (SOSG) was used to measure the production of 1O2 from the MOF–UCNP core–satellite superstructures upon 980 nm laser excitation (laser spot size: 1 mm in diameter; laser power: 500 mW; and laser confluence: 15.9 W cm−2).[3,14] The reagent SOSG reacts with 1O2 to generate green fluorescence that can be quantified with a fluorimeter. As shown in Figure 2, at a constant concentration of ~1.9 × 10−9 M MOF NPs, 1O2 production increased with UCNPs loading, reaching 57.9% from 1:12 MOF:UCNP core–satellite structures. Although a maximum value of 83.6% 1O2 was detected from 1:24 MOF:UCNP assemblies, some of this may be attributed to the presence of free UCNPs in addition to the core–satellite structures. As a comparison, mixtures of MOF NPs and UCNPs (1:12 MOF:UCNP) with noncomplementary DNA led to significantly less 1O2 (25.1%) than for the DNA assembled core–satellite structures at the same MOF:UCNP ratios. This result demonstrates that positioning the UCNPs directly at the surface of the MOF NPs clearly improves energy transfer from the UCNPs to the MOFs for 1O2 production (Scheme 1). In addition to 1O2 measurements, photoluminescence studies of the 1:12 MOF–UCNP core–satellite clusters showed an ≈61% and ≈35% decrease in green and red emission, respectively, as compared to UCNPs alone (Figure 3b and Table S2, Supporting Information). The larger loss in green emission may be attributed to the higher absorption overlap between the MOF NP and the UCNPs in the region of 515–580 nm (Figure S16, Supporting Information). As a comparison, random distributions of MOF NPs and UCNPs (1:12 MOF:UCNP) showed only ≈43% and ≈21% decrease in green and red emission, respectively.
Figure 2.
Singlet oxygen generated from MOF NPs, mixtures of MOF NPs and UCNPs, and DNA assembled MOF–UCNP core–satellite superstructures at different MOF:UCNP molar ratios under 980 nm laser irradiation.
Figure 3.
Photoluminescence spectra of a) DNA assembled MOF–UCNP core–satellite superstructures made at different MOF:UCNP molar ratios. b) Comparison of photoluminescence from MOF NPs alone and with UCNPs (mixed or DNA assembled).
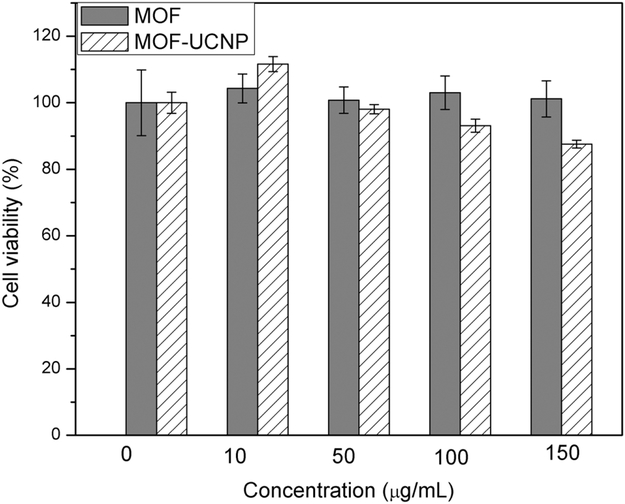
Next, the MOF–UCNP superstructures were tested for photodynamic therapy in vitro. For this, thiol-polyethylene glycol (HS-PEG) chains were first conjugated with the MOF–UCNP clusters to impart biocompatibility and reduce the nonspecific binding of blood proteins and macrophages to the nanoparticles.[6,34-37] Conjugation was performed by reacting the MOF–UCNP superstructures with 10 mg thiolated-PEG2000 overnight in water, followed by centrifugation to remove excess pEG.[6,15,38,39] The PEG-coated nanoparticles were next tested in 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) cell viability assays by first incubating MDA-MB-468 cells with either MOF NPs or the MOF–UCNP clusters at different concentrations (0, 10, 50, 100, and 150 μg mL−1) for 4 h. Any free particles not taken up by cells were removed by washing with phosphate buffered saline (PBS). Filtered MTT solutions were then added to the wells and incubated for a further 4 h, after which the media was carefully removed and mixed with 100 μL acidic isopropyl alcohol (40 × 10−3 M HCl). The wells were gently agitated at room temperature overnight to completely dissolve formazan crystals in each well, after which the wells were measured at 570 and 750 nm by a microplate reader. As shown in Figure 4, the MOF–UCNP superstructures exhibited negligible cytotoxicity when nanoparticle concentrations lower than 150 μg mL−1 were used (based on PEGylated MOFs).
Figure 4.
In vitro cell viability of MDA-MB-468 cells incubated with PEGylated MOF NPs and MOF–UCNP core–satellite superstructures at different concentrations for 4 h.
To specifically target the MOF–UCNP superstructures to MDA-MB-468 cells, which overexpress epidermal growth factor receptors (EGFRs), we next coupled EGFR binding affibodies to the UCNP nanocrystals. Affibodies are small, 56 amino acid long proteins that form stable triple alpha helix bundles and can be engineered to bind molecular targets with high specificity and tunable affinity. EGFR affibodies have been commonly used as ligands for targeting tumor cells such as breast and ovarian, and they can be produced and modified more easily than antibodies.[40-42] EGFR affibodies were produced by modifying the ZEGFR:l907 plasmid to express a cysteine at the C-terminus, followed by a 6× His tag for metal affinity purification.[43] This modified affibody was recombinantly expressed and purified in Escherichia coli. The affibody was then modified with NHS-fluorescein to install about approximately three dye molecules per affibody, as quantified by UV–vis spectroscopy. Next the Cys-affibody was conjugated to the free amine groups on the UCNPs using succinimidyl 4-(N-maleimidomethyl) cyclohexane-1-carboxylate (SMCC). Again, analysis of UV–vis spectra revealed that approximately eight affibodies were successfully conjugated to each DNA2-UCNP.
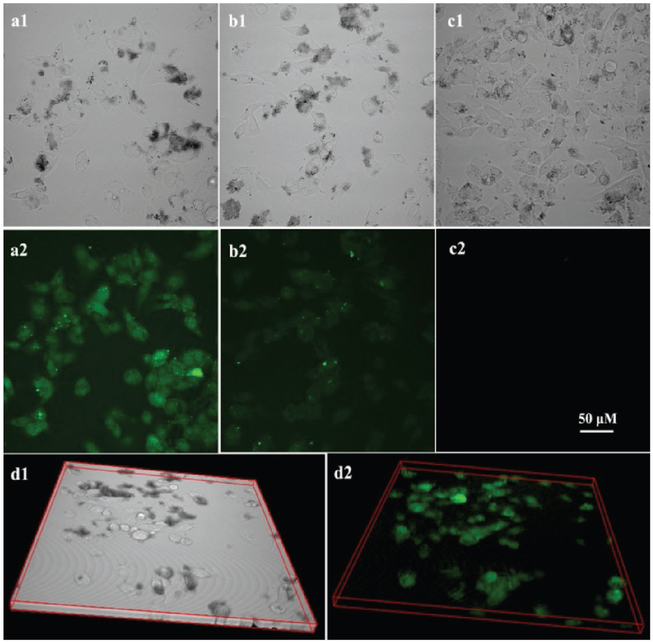
First, to test cell binding and uptake of the MOF–UCNP clusters with and without affibody, the affibody-UCNPs (with free amine group) were reacted with N-hydroxysuccinimide-fluorescein to track the targeting efficiency. As shown in Figure 5, confocal imaging showed significant more cell binding from the affibody-conjugated clusters as opposed to those coated with PEG alone, demonstrating the potential use of the EGFR affibody for cancer cell targeting. The 3D confocal imaging in Figure 5d and Figure Sl7 (Supporting Information) showed that the MOF–UCNP core–satellite superstructures were uptaken by the MBD-MB-468 cells and the superstructures were dispersed in the whole cells. Next, the efficacy of the MOF–UCNPs for cell killing was studied in vitro. For this, MOF–UCNP and affibody-bound MOF–UCNP clusters at 100 μg mL−1 were incubated with MDA-MB-468 cells for 4 h. The MDA-MB-468 cells were then subjected to irradiation with 980 nm light at 15.9 W cm−2 intensity for 10 and 20 min, followed by incubating for an additional 12 h at 37 °C in the dark, followed by MTT viability assays. As shown in Figure 6, 10 and 20 min 980 nm light irradiation had not affected MDA-MB-468 cells without nanoparticles. Moreover, no acute cell death was observed even after 20 min irradiation with 980 nm light for MOFs without UCNPs. However, a decrease in cell viability of 63.7% was observed after 20 min irradiation with the MOF–UCNP structures. Cells incubated with unassociated MOFs and UCNPs showed only a 48.1% decrease in cell viability. Most significantly, the affibody-conjugated MOF–UCNP superstructures showed the highest efficacy, resulting in more than ≈82% cell death after 20 min photoirradiation.
Figure 5.
Confocal luminescence imaging of MDA-MB-468 cells incubated with a) MOF–UCNPaffibody, fluorescence core–satellite superstructures, b) MOF–UCNPfluorescence core–satellite superstructures, and c) MOF–UCNPaffibody core–satellite superstructures. d) 3D confocal bright field and luminescence images of MDA-MB-468 cells incubated with MOF–UCNPaffibody, fluorescence core–satellite superstructures. (All the figures have the same scale bar. Images were collected using a Nikon A1 microscope with a 0.2% setting of a maximum 70 mW laser power.)
Figure 6.
Quantitative analysis of MDA-MB-468 cell viabilities with MOF NPs, mixtures of MOF NPs and UCNPs, and DNA assembled MOF–UCNP core–satellite superstructures after 980 nm laser irradiation for 10 and 20 min. Data are averages ± one SD (n > 3 experimental replicates). *p < 10−4, **p < 10−6 relative to control (black). ***p < 10−5, ****p < 10−3.
3. Conclusions
In summary, core–satellite MOF–UCNP assemblies were constructed and evaluated as agents for PDT treatment at NIR wavelengths. By using DNA sequences, well-ordered core–satellite MOF–UCNP superstructures were synthesized. Due to the association of UCNPs and the MOF by DNA hybridization, large quantities of singlet oxygen were obtained under 980 nm laser irradiation, which also showed enhanced cell cytotoxicity as compared to when UCNPs were simply mixed with the MOF NPs. Furthermore, when the core–satellite MOF–UCNP superstructures were further conjugated with EGFR affibodies, significant gains in targeted photodynamic therapy were observed, resulting in ≈82% cell death after 20 min NIR excitation. It is expected that this work will facilitate the creation of new multifunctional PDT materials based on assembled UCNP and MOF nanostructures.
4. Experimental Section
Materials:
H2TCPP was purchased from Frontier Scientific. Dulbecco’s Modified Eagle’s Medium (DMEM, high glucose with L-glutamine sterile filtered) and fetal bovine serum were purchased from Sigma-Aldrich. Singlet Oxygen Sensor Green was purchased from Life Technologies (USA). The DNA was purchased from Integrated DNA Technologies Inc. Other chemicals were of at least analytical grade and were used as received. All aqueous solutions were prepared with Milli-Q water. All other materials (not mentioned) and solvents were used as received without further purification from the following suppliers (Alfa Aesar, Sigma-Aldrich, and Fisher Scientific).
Synthesis of PCN-224 MOF NPs:
50 mg H2TCPP, 150 mg ZrOCl2.8H2O, and benzoic acid was mixed in 70 mL DMF solution. The reaction mixture solution was heated at 90 °C for 6 h. After the reaction was done, the products were collected by centrifugation at 15 000 rpm for 20 min followed by washing with fresh DMF three times.
Synthesis of NaYF4,Yb,Er UCNPs. NaYF4:Yb, Er UCNPs:
The UCNPs used herein were synthesized using previously described methods.[27] In a typical procedure, 0.1562 g YCl3, 0.0503 g YbCl3, and 0.0055 g ErCl3 were mixed with 6 mL oleic acid and 15 mL octadecene. The solution was slowly heated to 160 °C with vigorous stirring under argon for 50 min to form a homogeneous transparent solution, and then allowed to cool to room temperature. Then, a 10 mL methanol solution of NaOH (0.1 g) and NH4F (0.148 g) was added drop by drop and stirred for another 40 min. The solution was then slowly heated and degassed at 110 °C for 20 min and then refilled with argon and degased three times in total. After that the solution was heated to 300 °C within 10 min and reacted for 70 min under argon. After the solution was cooled back to room temperature, the products were precipitated from the solution with ethanol and washed with ethanol and water (1:1) three times.
Modifying PCN-224 MOF NPs with DNA1 or DNA 2:
Typically, 100 μL ≈41 × 10−9 M of the as-prepared MOFs (in DMF, contains 0.42 × 10−3 m Zr) was mixed with 100 μL water and 10.5 μL 20 × 10−3 m ligand (Nα,Nα-bis(carboxymethyl)-l-lysine hydrate) (MOF:ligand = 1:5). After reacting for 20–30 min, the products were purified by 30K centrifuge filtration and re-dispersed in water. Then, 3 μL 2 × 10−3 m NHS-DBCO was added and reacted for 4–5 h. After purifying through 30K filters, 16.4 μL of 100 × 10−6 m of azide-terminated DNA1 was added and reacted overnight. The number of DNA1 sequences bound to each MOF NP was determined by UV–vis analysis.
Modifying UCNPs with DNA2:
In order to transfer the as-prepared oleic acid stabilized UCNPs into water, 6.0 mg was first dispersed in 500 μL chloroform. Next, 6.0 mg of C18-PEG-NH2 dissolved in 1 mL water was added to the UCNP solution drop wise. Then, the entire mixture was vigorously stirred overnight to evaporate. Next, the amine-UCNPs were collected by centrifugation and reacted with NHS-DBCO for 4–5 h followed by reacting with DNA2 overnight (UCNP:NHS-DBCO:DNA = 1:200:200). The samples were purified by 30K filters and kept at 4 °C. The number of DNA2 sequences bound to each UCNP was determined by UV–vis analysis.
Preparation of Core-Satellite MOF-UCNP Superstructures:
To prepare MOF–UCNP core-satellite assemblies (taking the molar ratio of MOF:UCNP = 1:12 as an example), 10 μL of 41 × 10−9 m DNA1-MOF NPs was mixed with 49.2 μL DNA2-UCNPs (100 × 10−9 m) and NaCl solution was added to keep the final salt concentration at 50 × 10−3 m. The mixture was heated to 60 °C for 10 min followed by a slow cool down to room temperature for at least 5 h. In order to prepare EGFR affibody-conjugated MOF–UCNP superstructures, the DNA2-UCNPs were first conjugated with EGFR affibody by thiol-maleimide reaction (see below the “Affibody-Fluorescein Conjugated with DNA2-UCNPs” heading). Next, the affibody-DNA2-UCNPs were hybridized to the DNA1-MOFs. For the samples used for cytotoxicity assays, 100 μL of 2 mg mL−1 MOF NPs, MOF–UCNP core–satellite, and mixtures were reacted with 10 mg HS-PEG Molecular Weight (M.W) 2000) overnight. The PEGylated samples were then purified by centrifugation and re-dispersed in DMEM medium for cytotoxicity studies.
Singlet Oxygen Generation Test:
SOSG was employed for detecting singlet oxygen. Typically, 200 μL 9.73 × 10−9 m DNA-MOFs, DNA2-UCNP, and NaCl solution were mixed at different MOF:UCNP ratios to prepare the MOFs, MOF–UCNP superstructures, and MOF–UCNP mixture samples. Then, the samples were heated to 60 °C and then cooled down to room temperature. Before 980 nm laser irradiation, 10 μL 5 × 10−3 m SOSG DMSO solution was added. Then, the samples were irradiated with 980 nm laser light for 0, 10, 20, 30, 40, 50, and 60 min. Every 10 min, 100 μL solution was taken out and the fluorescence intensity was obtained on a fluorimeter using 504 nm excitation and reading emission at 525 nm.
EGFR Affibody Production:
The affibody ZEGFR:1907 sequence was modified by including a cysteine followed by six histidine residues at the N-terminus for site-specific conjugation and affinity purification, respectively. This sequence was cloned into a pZE21 vector and transformed into E. coli strain DH5a (New England Biolabs). Transformants were grown overnight in 5 mL of lysogeny broth (LB) media containing 50 μg mL−1 kanamycin. Cells were then centrifuged for 2 min at 2000 g and resuspended into 50 mL of fresh media and grown to an optical density (OD) of 0.5 at which time protein expression was induced through the addition of 60 ng mL−1 of anhydrotetracycline (Sigma-Aldrich). Expression was allowed to proceed for 4 h, followed by cell lysis via sonication and purification of the affibody from the soluble lysate fraction with Ni-NTA agarose resin (Thermo Fisher Scientific).
Affibody Conjugated with NHS-Fluorescein:
In this part, 10 μL 34 × 10−6 m affibody solution was mixed with NHS-fluorescein at the ratio of 1:100. After reaction for few hours, the product was purified by dialysis with a 2000 MWCO dialysis tube for 1 h. After dialysis the solution volume was 37 μL. Then the product was characterized by UV–vis spectra. The concentration of the fluorescein (28.97 × 10−6 m) could be calculated by the UV–vis spectra. So there was 28.97/9.19 = 3.2 fluorescein attached to each affibody molecule.
Affibody-Fluorescein Conjugated with DNA2-UCNPs:
The DNA2-UCNPs (with free amine groups) were first reacted with SMCC at the ratio of 1:250 for 5 h. Then it was purified by centrifugation. At the same time, the affibody (with fluorescein) was reduced by tris-(2-carboxyethyl)phosphine (TCEP) and then purified by dialysis. Next, the reduced affibody-fluorescein was mixed with SMCC-treated DNA2-UCNPs at the ratio of 100:1. After overnight reaction, the product was purified by centrifugation and checked by UV–vis analysis. Through this method, it was determined that there were approximately eight affibodies attached to each UCNP.
Cell Culture:
The MDA-MB-468 cells were grown to confluence at 37 °C and with 5% CO2 in DMEM containing 10% fetal bovine serum and 1% penicillin/streptomycin.
Cytotoxicity Assay:
The cell viability of MDA-MB-468 cells was determined by using MTT assay. MDA-MB-468 cells were seeded into a 96-well cell culture plate at 104 per well and incubated for 24 h at 37 °C under 5% CO2. Then, MOF–UCNP core–satellite superstructures dispersed in DMEM were added to each well to give final particle concentrations of 0, 10, 50, 100, and 150 μg mL−1. The cells were incubated for another 4 h at 37 °C under 5% CO2. After incubation, particle containing media was removed, cell wells were washed using PBS to remove the nonuptake particles, and 90 μL of fresh medium was added. Then 20 μL of filter-sterilized MTT reagent (5 mg mL−1 in PBS) was added to each well, and the plates were incubated at 37 °C for further 4 h. After incubation, medium was removed and the precipitated formazan crystals were dissolved by adding acidic isopropyl alcohol (40 × 10−3 m HCl). Absorption values of the dissolved formazan crystals in each well were measured at 570 and 750 nm using a microplate reader. All the samples were prepared in triplicate.
Photodynamic Test:
MDA-MB-468 cells were incubated with MOF–UCNP core–satellite superstructures at a final concentration of 100 μg mL−1 for 4 h, excess core–satellite MOF–UCNP superstructures were removed by PBS washing, and 100 μL of fresh medium was added. Then, 980 nm laser (laser spot size: 1 mm in diameter; laser power: 500 mW; and laser confluence: 15.9 W cm−2) was used to irradiate each cell well in the 96-well plate for 10 or 20 min and then replaced with fresh culture medium and cultivated for another 12 h at 37 °C in order to determine the effects of the 980 nm laser irradiation and laser induced 1O2 on cell viability. Cell viability was determined by MTT assay. All the samples were prepared in triplicate.
Supplementary Material
Acknowledgements
The research was primarily supported by the National Institute of Health (1R21EB020911-01), NIH (DP2EB020401), Army Research Office under Award No. W911NF-14-1-0211, and the National Science Foundation Award No. DMR 1420736. The authors acknowledge the Light Microscopy Facility located in the Department of Molecular, Cellular, and Developmental Biology and Dr. James Orth for assistance.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Liangcan He, Department of Chemical and Biological Engineering, University of Colorado, Boulder, CO 80303, USA.
Michael Brasino, Department of Chemical and Biological Engineering, University of Colorado, Boulder, CO 80303, USA.
Chenchen Mao, Department of Electrical, Computer and Energy Engineering, University of Colorado, Boulder, CO 80303, USA.
Suehyun Cho, Department of Electrical, Computer and Energy Engineering, University of Colorado, Boulder, CO 80303, USA.
Wounjhang Park, Department of Electrical, Computer and Energy Engineering, University of Colorado, Boulder, CO 80303, USA; Materials Science and Engineering Program, University of Colorado, Boulder, CO 80303, USA.
Andrew P. Goodwin, Department of Chemical and Biological Engineering, University of Colorado, Boulder, CO 80303, USA; Materials Science and Engineering Program, University of Colorado, Boulder, CO 80303, USA
Jennifer N. Cha, Department of Chemical and Biological Engineering, University of Colorado, Boulder, CO 80303, USA; Materials Science and Engineering Program, University of Colorado, Boulder, CO 80303, USA
References
- [1].Dolmans DEJGJ, Fukumura D, Jain RK, Nat. Rev. Cancer 2003, 3, 380. [DOI] [PubMed] [Google Scholar]
- [2].Idris NM, Gnanasammandhan MK, Zhang J, Ho PC, Mahendran R, Zhang Y, Nat. Med. 2012, 18, 1580. [DOI] [PubMed] [Google Scholar]
- [3].Park J, Jiang Q, Feng D, Mao L, Zhou H-C, J. Am. Chem. Soc. 2016, 138, 3518. [DOI] [PubMed] [Google Scholar]
- [4].Chen Q, Wang C, Cheng L, He W, Cheng Z, Liu Z, Biomaterials 2014, 35, 2915. [DOI] [PubMed] [Google Scholar]
- [5].Dong H, Du S-R, Zheng X-Y, Lyu G-M, Sun L-D, Li L-D, Zhang P-Z, Zhang C, Yan C-H, Chem. Rev. 2015, 115, 10725. [DOI] [PubMed] [Google Scholar]
- [6].He L, Dragavon J, Cho S, Mao C, Yildirim A, Ma K, Chattaraj R, Goodwin AP, Park W, Cha JN, J. Mater. Chem. B 2016, 4, 4455. [DOI] [PubMed] [Google Scholar]
- [7].Ai X, Ho CJH, Aw J, Attia ABE, Mu J, Wang Y, Wang X, Wang Y, Liu X, Chen H, Gao M, Chen X, Yeow EKL, Liu G, Olivo M, Xing B, Nat. Commun. 2016, 7, 10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu Y, Liu Y, Bu W, Cheng C, Zuo C, Xiao Q, Sun Y, Ni D, Zhang C, Liu J, Shi J, Angew. Chem., Int. Ed. 2015, 54, 8105. [DOI] [PubMed] [Google Scholar]
- [9].Tian G, Ren W, Yan L, Jian S, Gu Z, Zhou L, Jin S, Yin W, Li S, Zhao Y, Small 2013, 9, 1929. [DOI] [PubMed] [Google Scholar]
- [10].Wang C, Tao H, Cheng L, Liu Z, Biomaterials 2011, 32, 6145. [DOI] [PubMed] [Google Scholar]
- [11].Li Y, Tang J, He L, Liu Y, Liu Y, Chen C, Tang Z, Adv. Mater. 2014, 27, 4075. [DOI] [PubMed] [Google Scholar]
- [12].Zhou J, Liu Z, Li F, Chem. Soc. Rev. 2012, 41, 1323. [DOI] [PubMed] [Google Scholar]
- [13].Lu S, Tu D, Hu P, Xu J, Li R, Wang M, Chen Z, Huang M, Chen x., Angew. Chem., Int. Ed. 2015, 54, 7915. [DOI] [PubMed] [Google Scholar]
- [14].Lu K, He C, Lin W, J. Am. Chem. Soc. 2014, 136, 16712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liu J, Yang Y, Zhu W, Yi X, Dong Z, Xu X, Chen M, Yang K, Lu G, Jiang L, Liu Z, Biomaterials 2016, 97, 1. [DOI] [PubMed] [Google Scholar]
- [16].Lu K, He C, Lin W, J. Am. Chem. Soc. 2015, 137, 7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu Y, Tang Z, Adv. Mater. 2013, 25, 5819. [DOI] [PubMed] [Google Scholar]
- [18].Liu X, He L, Zheng J, Guo J, Bi F, Ma X, Zhao K, Liu Y, Song R, Tang Z, Adv. Mater. 2015, 27, 3273. [DOI] [PubMed] [Google Scholar]
- [19].Furukawa H, Cordova KE, O’Keeffe M, Yaghi OM, Science 2013, 341, 1230444. [DOI] [PubMed] [Google Scholar]
- [20].Cohen SM, Chem. Rev. 2012, 112, 970. [DOI] [PubMed] [Google Scholar]
- [21].Gao W-Y, Chrzanowski M, Ma S, Chem. Soc. Rev. 2014, 43, 5841. [DOI] [PubMed] [Google Scholar]
- [22].He L, Liu Y, Liu J, Xiong Y, Zheng J, Liu Y, Tang Z, Angew. Chem., Int. Ed. 2013, 52, 3741. [DOI] [PubMed] [Google Scholar]
- [23].Dhakshinamoorthy A, Garcia H, Chem. Soc. Rev. 2012, 41, 5262. [DOI] [PubMed] [Google Scholar]
- [24].Liu J, Chen L, Cui H, Zhang J, Zhang L, Su C-Y, Chem. Soc. Rev. 2013, 43, 6011. [DOI] [PubMed] [Google Scholar]
- [25].He C, Liu D, Lin W, Chem. Rev. 2015, 115, 11079. [DOI] [PubMed] [Google Scholar]
- [26].Li Z, Yu R, Huang J, Shi Y, Zhang D, Zhong X, Wang D, Wu Y, Li Y, Nat. Commun. 2015, 6, 8248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhao S, Wang Y, Dong J, He C-T, Yin H, An P, Zhao K, Zhang X, Gao C, Zhang L, Lv J, Wang J, Zhang J, Khattak AM, Khan NA, Wei Z, Zhang J, Liu S, Zhao H, Tang Z, Nat. Energy 2016, 1, 16184. [Google Scholar]
- [28].Zhao M, Yuan K, Wang Y, Li G, Guo J, Gu L, Hu W, Zhao H, Tang Z, Nature 2016, 539, 76. [DOI] [PubMed] [Google Scholar]
- [29].Lu K, He C, Guo N, Chan C, Ni K, Weichselbaum RR, Lin W, J. Am. Chem. Soc. 2016, 138, 12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bai Y, Dou Y, Xie L-H, Rutledge W, Li J-R, Zhou H-C, Chem. Soc. Rev. 2016, 45, 2327. [DOI] [PubMed] [Google Scholar]
- [31].Fateeva A, Chater PA, Ireland CP, Tahir AA, Khimyak YZ, Wiper PV, Darwent JR, Rosseinsky MJ, Angew. Chem., Int. Ed. 2012, 51, 7440. [DOI] [PubMed] [Google Scholar]
- [32].He L, Mao C, Cho S, Ma K, Xi W, Bowman CN, Park W, Cha JN, Nanoscale 2015, 7, 17254. [DOI] [PubMed] [Google Scholar]
- [33].Li Z, Zhu Z, Liu W, Zhou Y, Han B, Gao Y, Tang Z, J. Am. Chem. Soc. 2012, 134, 3322. [DOI] [PubMed] [Google Scholar]
- [34].He Q, Zhang J, Shi J, Zhu Z, Zhang L, Bu W, Guo L, Chen Y, Biomaterials 2010, 31, 1085. [DOI] [PubMed] [Google Scholar]
- [35].Chou LYT, Zagorovsky K, Chan WCW, Nat. Nano 2014, 9, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sykes EA, Chen J, Zheng G, Chan WCW, ACS Nano 2014, 8, 5696. [DOI] [PubMed] [Google Scholar]
- [37].Albanese A, Walkey CD, Olsen JB, Guo H, Emili A, Chan WCW, ACS Nano 2014, 8, 5515. [DOI] [PubMed] [Google Scholar]
- [38].He F, Yang G, Yang P, Yu Y, Lv R, Li C, Dai Y, Gai S, Lin J, Adv. Funct. Mater. 2015, 25, 3966. [Google Scholar]
- [39].Xing H, Bu W, Zhang S, Zheng X, Li M, Chen F, He Q, Zhou L, Peng W, Hua Y, Shi J, Biomaterials 2012, 33, 1079. [DOI] [PubMed] [Google Scholar]
- [40].Antaris AL, Chen H, Cheng K, Sun Y, Hong G, Qu C, Diao S, Deng Z, Hu X, Zhang B, Zhang X, Yaghi OK, Alamparambil ZR, Hong X, Cheng Z, Dai H, Nat. Mater. 2016, 15, 235. [DOI] [PubMed] [Google Scholar]
- [41].Miao Z, Ren G, Liu H, Jiang L, Cheng Z, J. Biomed. Opt. 2010, 15, 036007. [DOI] [PubMed] [Google Scholar]
- [42].Jokerst JV, Miao Z, Zavaleta C, Cheng Z, Gambhir SS, Small 2011, 7, 625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Friedman M, Orlova A, Johansson E, Eriksson TLJ, Hoiden-Guthenberg I, Tolmachev V, Nilsson FY, Stahl S, J. Mol. Biol. 2008, 376, 1388. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.