Abstract
Optical induction of intracellular signaling by membrane-associated and integral membrane proteins allows spatiotemporally precise control over second messenger signaling and cytoskeletal rearrangements that are important to cell migration, development, and proliferation. Optogenetic membrane recruitment of a protein-of-interest to control its signaling by altering subcellular localization is a versatile means to these ends. Here, we summarize the signaling characteristics and underlying structure-function of RGS-LOV photoreceptors as single-component membrane recruitment tools that rapidly, reversibly, and efficiently carry protein cargo from the cytoplasm to the plasma membrane by a light-regulated electrostatic interaction with the membrane itself. We place the technology-relevant features of these recently described natural photosensory proteins in context of summarized protein engineering and design strategies for optically controlling membrane protein signaling.
Graphical Abstract
Introduction
Over the nearly past two decades, optogenetics [1,2] and optochemical [3,4] approaches to controlling the excitability and signaling of genetically targeted cells and cell-like systems [5,6] have transitioned from specialized upstart technologies to core techniques in cell biology. This prominence has driven the discovery of natural proteins that push the boundaries of known photosensory signaling mechanisms [7–10], the creation of innovative photoreceptor engineering strategies [11–14], and the solving of high-resolution structures of natural and engineered light-activated proteins [15–17] - all of which efforts have been aimed at informing how to construct photoinducible signaling tools from natural proteins.
Outside of neuroscience and muscle biology where electrogenic control over excitable cell spiking dominates, technology development and application have extensively focused on mammalian transcriptional activation (as well-summarized by others [18]) or of primary focus here, intracellular signaling by membrane receptors [19–22] and membrane-associated proteins [23–30] for controlling the effector functions of kinases, G-protein coupled receptors (GPCRs), and small GTPases that are involved with second messenger signaling, environmental sensing, and cytoskeletal rearrangements in cell migration, development, and proliferation. Because the signaling of these effectors and the phenotypic cellular behaviors they regulate occur on the timescale of ~ 100 – 103 seconds, which is similar to the typical photocycle of a non-electrogenic photosensory protein, the spatiotemporal dynamics of their signaling and information encoding/decoding schemes [22–25] are well-suited for optogenetic analyses.
Here, we will first briefly describe approaches for optical control over intracellular signaling by membrane proteins, with an emphasis on optogenetic approaches with fully genetically encoded tools in mammalian cells that alter subcellular localization of membrane-associated proteins (Figure 1). This emphasis reflects the current prevalence in reports, not an implied importance of these model/expression systems or signaling pathways/applications over others. The contextual summary highlights general signal induction strategies at a high-level, as opposed to detailing the myriad specific tools reported to date – for which we point to these recent and exhaustive reviews by others [1,2,18]. We will also summarize the signaling structure-function of very recently reported light-oxygen-voltage (LOV) photoreceptors that directly bind the plasma membrane by a light-regulated protein-lipid electrostatic interaction [7,8], in context of guiding the design and engineering of single-component optogenetic membrane recruitment tools.
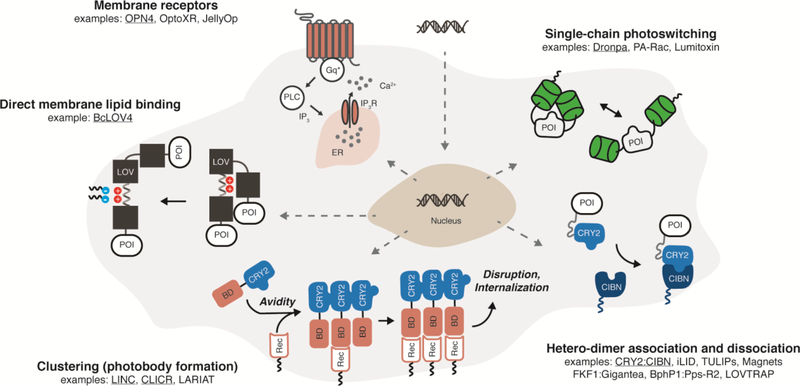
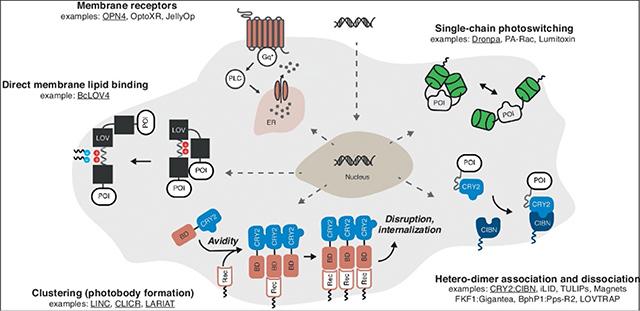
Figure 1. Fully genetically encoded strategies for optically inducible intracellular signaling by integral membrane and membrane-associated proteins in mammalian cells.
Examples schematized are underlined. Abbreviations: POI = protein of interest. BD = binding domain, CIBN = cryptochrome interacting binding partner, CRY2 = cryptochrome, ER = endoplasmic reticulum, Gq* = activated Gαq, IP3 = inositol triphosphate, IP3R = IP3 receptor, LOV = light-oxygen-voltage, PLC = phospholipase C, Rec = membrane receptor.
Allosteric switching of effector biochemical function
There are numerous ways to classify optogenetic tools: by photoreceptor from which they are derived, by induction wavelength, and by application, to name a few. For the sake of simplicity, we organize the approaches for optical induction of membrane protein intracellular signaling into two effector-centric modes: (i) photoswitching of effector biochemical activity (described in this section), and (ii) photocontrol over subcellular localization of a constitutively active effector to regulate signaling (as described in the next section). Here, photo-switching implies that the protein-level enzymatic/binding efficiency of the signaling effector domain/segment is altered between the photoactivated and dark-adapted states of the photosensory domain. These photo-switching tools are typically single-chain proteins since they do not rely on engineered binding partners.
Co-opting the natural signaling function of photosensory membrane receptors is conceptually straightforward. For example, human melanopsin (OPN4), a light-activated Gαq-coupled GPCR normally found in non-vision-forming retinal ganglion cells, initiates calcium release from the endo/sarcoplasmic reticulum and downstream calcium-dependent transcription and excitability [22,31]. Invertebrate opsins from diverse organisms also engage mammalian signaling pathways [21,32], such as a Gas-coupled opsin from jellyfish (JellyOp). Chimeric opsins created by substituting the signaling-responsible cytoplasmic loops with those of ligand-responsive GPCRs (often termed “OptoXRs”) have been derived from both mammalian opsins [20] and more recently, microbial opsins [33].
The most common component for constructing single-chain, chimeric photo-switches is the light-oxygen-voltage (LOV) sensor domain from Phototropin 1 of Avena sativa (AsLOV2), a non-dimerizing monomer with existing high-resolution structures of its active signaling state by NMR [34] and x-ray crystallography [35] (PDB code 2v1a). LOV signal transmission is mediated by flavin photocycling-initiated protein conformational changes that disrupt a β-sheet interaction with a C-terminal Jα-helix [34], to which effector proteins-of-interest (POI) can be fused such as small GTPases as in photoactivatable Rac (PA-Rac) [27], (extracellular) surface-displayed peptide toxins (Lumitoxins) to antagonize endogenous channels [36], and small peptide tags [12,37–40]. The signaling function of the POI is presumably diminished in the dark by steric hindrance or occlusion of its binding site by the LOV domain fused to the terminus, although the structured molecular contacts between these domains are seldom reported (assuming they exist). A recent report describes how effector loop regions can be computationally designed to interact with the AsLOV2 flavin binding pocket, such that light-induced conformational changes introduce “extrinsic disorder” to these loop regions that disrupts effector signaling in a structurally principled manner [15]; importantly, theses mechanistic assertions were confirmed by structure determination and correlated signaling assays [15].
Monomeric photoswitches can also be constructed by engineering single-chain proteins from light-activated homodimers, such as the β-barrel green fluorescent protein-based Dronpa [28]. Dronpa dimerization is bi-directional or photo-switchable with two different colors of light (ultraviolet and blue), a beneficial feature because its shutoff can be independent of its thermal reversion between states. Beyond allosteric switching, homodimer dissociation can be effective in disrupting oligomerization-dependent signaling activity of membrane-associated proteins, as demonstrated with receptor tyrosine kinases (RTKs) fused to bacterial cobalamin binding domains [19]. Although this latter system requires cofactor supplementation in mammalian systems, natural LOV that photo-dissociate (PDB code 4hj6) [10] exist that bind mammalian-endogenous flavins, and have been used to engineer chimeric Cas9 DNA-binding domains in bacteria [41].
Oligomerization systems to alter subcellular localization of effector cargo
Whereas allosteric systems switch the effector protein-level activity, optically induced signaling by a constitutively active POI merely requires a change in the subcellular localization of the POI that consequently changes its local concentration or availability to its partner. These signaling systems are ostensibly easier to design and engineer than allosteric photoswitches because effector signaling is largely decoupled in structure-function from the conformational changes of the photosensory domain, thus requiring only a modest amount of linker engineering (in length and rigidity) between the photosensor and its passive cargo, the POI, to work in principle.
Nature has already “engineered” several photosensory heterodimerization pairs suitable for membrane signaling in mammalian cells (using endogenous cofactors) when stimulated by blue light (e.g. cryptochrome CRY2 with CIBN, and the LOV domain FKF1 with Gigantea) or near-infrared light (e.g. bacteriophytochrome BphP1 with Pps-R2) [29,42,43]. In the most common design configuration, an interaction domain is non-specifically localized to the plasma membrane by prenylation at a C-terminal “CAAX” motif, and the dark-adapted photosensor initially sequesters a fused POI in the cytoplasm, unavailable to signal at the membrane; the POI is recruited to the membrane to increase signaling upon light-activated heterodimerization.
Despite the ready availability of natural heterodimer pairs, many engineered heterodimer pairs have also been reported. These artificial heterodimerization pairs can be derived from natural homodimerizers by creating a highly charged and electrostatically asymmetric dimerization interface that promotes electrostatic stabilization between heterodimers and repulsion between homodimers, as done with the Magnet [13] system derived from the LOV protein Vivid (VVD, PDB code 3rh8). Several reported AsLOV2-based photoswitches expose the binding regions of short peptides [12,37,38] with known binding partners to drive heterodimer association for the downstream purpose of altering the subcellular localization of another effector, as is the case with iLID (SsrA peptide binding to SsrB) (PDB code 4wf0) and TULIPs (epitope binding to a PDZ domain). Thus, it is important to note that the signaling mode classification used here is organizational, not fundamental in nature or mutually exclusive.
Optically induced dissociation of a thermally stable heterodimer is rarer. When the naturally fused regulatory and effector domains of cyanobacterial orange carotenoid protein (OCP) are split, the resulting engineered domains form a stable heterodimer in the dark that is disrupted by blue light stimulation ([44], see also the contribution of Kerfeld to this issue). While the ketocarotenoid cofactor bound by OCP is not endogenously biosynthesized in mammalian cells, carotenoids are often bioavailable by supplementation. To the best of our knowledge though, Nature has not provided such a heterodimerization pair involving a photoreceptor that binds an endogenous mammalian cofactor; even though bacteriophytochromes (and phytochromes) optically dissociate from their interaction partners from the photoactivated state, the dark-adapted photoreceptor is unbound. However, an artificial pair has been cleverly created in the LOVTRAP system [14] (PDB code 5efw), in which the binding of Zdark (an engineered Z subunit of Protein A) to the critical Jα-helix of dark-adapted AsLOV2 is disrupted when the latter photocycles.
While these modular systems are fairly “plug-and-play,” beyond linker engineering they do require protein expression level tuning, in relative expression level between heterodimerization partners and total expression level of the POI, for suitable dynamic range to ensure a strong signaling change upon illumination without permanent association in the dark-adapted state. These expression level setpoints can be reasonably locked in with proper clonal selection of stably transducing cell lines [23,24], but heterogenous expression across a cell population can lead to inconsistent function for applications in primary cells and transiently transfected cell lines.
Beyond dimers, oligomerization state can be grossly controlled to alter subcellular localization using cryptochromes, which are known to cluster into large (~10−7 m) internal bodies [45] of unknown colloidal structure (as do phytochromes [46] that bind mammalian-exogenous phycocyanobilin). Homo-oligomeric clustering at the membrane can disrupt membrane protein signaling by internalization as reported with LINC [47] and the related LARIAT (hetero-oligomers of multimeric proteins) [48] systems. Conversely, photobody formation can activate signaling by enhancing the overall avidity of the fused POI to an endogenous membrane receptor (CLICR, [49]). This latter approach is promising because it requires a single transgene. However, the aggregates are large and the indirect membrane recruitment is a multi-step process.
A more straightforward single-component approach to membrane recruitment, in which a single-transgene tool carries a POI cargo directly to the membrane, without aggregation and a multi-step binding process, would be highly useful. Next, we will discuss a recent advance to this end: the discovery of natural LOV photoreceptors that are directly recruited by the plasma membrane itself in a blue light-dependent manner.
Single-component membrane recruitment by BcLOV4
Recently, we and colleagues reported the identification and characterization of a class of LOV photoreceptors that directly associate with the plasma membrane inner leaflet by a light-switched and high-affinity electrostatic interaction with anionic phospholipids [7,8] (Figure 2). Their photocycle-coupled signal transmission is mediated by an unmasking of a membrane-interacting polybasic amphipathic helical linker that couples the LOV sensor Jα-helix to a highly structured C-terminal DUF domain (domain of unidentified function) (Figure 2a–b); an N-terminal RGS domain (regulator of G-protein signaling) inhibits the electrostatic interaction in the dark-adapted state, but has no detectable interaction with mammalian proteins. Of relevance here, these RGS-LOV (RGS-associated LOV) proteins function as single-component optogenetic tools for dynamic membrane recruitment from the cytoplasm by binding the plasma membrane itself (Figure 2c–d). To date, we have most thoroughly characterized BcLOV4 from Botrytis cinerea, but the dynamic membrane association phenomenon is general (see Figure S5 of reference [7]) to its fungal RGS-LOV homologs [7,8]. Here, we will expand upon the discussion of BcLOV4 as a single-component optogenetic tool with a contextual emphasis on mammalian signaling.
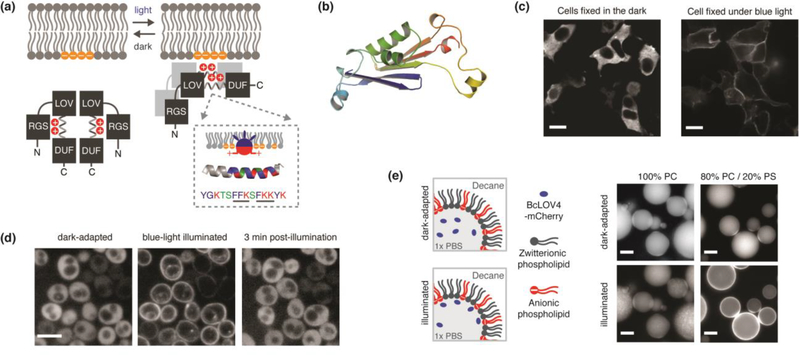
Figure 2. BcLOV4, a membrane-interacting photoreceptor and single-component system for dynamic membrane recruitment across diverse model systems.
(a) Schematized signaltransmission mode of membrane recruitment by a directly light-regulated and high-affinity electrostatic interaction with anionic phospholipids, which are largely enriched in the plasma membrane in mammalian cells. The interaction is inhibited by an N-terminal RGS (Regulator of G-protein signaling). A critical membrane binding site exists in a polybasic amphipathic helix in the linker region between the LOV sensor and a C-terminal DUF (domain of unidentified function). Inset: Schematized helix-membrane interaction and amphipathic helix sequence from BcLOV4, with known membrane binding motifs underlined. Blue = hydrophobic. Red = Basic. Green = Polar. (b) Pymol rendered model of predicted DUF structure by de novo energy minimization modeling in Rosetta. The DUF has a PAS-like mixed α-helix / anti-parallel β-sheet topology, suggesting that the LOV-DUF interaction is an evolutionary conserved PAS-PAS interaction. (c-e) Fluorescence micrographs of light-activated membrane recruitment in diverse contexts: (c) Expressed as FLAG epitope-tagged protein in mammalian HEK cells, fixed in the dark/light, and visualized by immunocytochemistry with Alexa488-conjugated anti-FLAG antibody. Scale = 10um. (d) Expressed in fungal S. cerevisiae cells and visualized by mCherry-fusion tag. Scale = 5 um. (e) In vitro as purified recombinant mCherry-tagged protein that binds lipid-stabilized water-in-oil emulsion interfaces. BcLOV4 does not bind purely zwitterionic interfaces of phosphatidylcholine (PC), but binds anionic interfaces with 20% phosphatidylserine (PS) that emulates mammalian plasma membrane inner leaflets. Scale = 25 um. Images modified from Reference 7. Copyright 2018 National Academy of Sciences.
BcLOV4 membrane recruitment is fast (τon ~ 1 second) because it has a high affinity for interfaces of mammalian plasma membrane-like composition (Kd ~ 130 nM for 80% phosphatidylcholine (PC) / 20% phosphatidylserine (PS)). This timescale is near diffusion-limited, such that it associates with the plasma membrane near instantaneously upon diffusing to it. It also undocks quickly from the membrane in the dark (τoff ~ 1–1.5 minutes), and mutants with much longer residence times of ~ 10 minutes have been engineered by lengthening the photocycle (see Figure S8 of [7]). These undocking timescales are ~one minute longer than the respective photocycles, and thus, the photocycle is partially rate-limiting in the overall dissociation kinetics of the system, with other rate-determining factors still to be elucidated (as discussed further below).
While its high-affinity lipid interaction in the photoactivated state ensures robust membrane recruitment, its low affinity in the dark-adapted state (Kd ~ low micromolar) keeps it well- sequestered in the cytoplasm even when over-expressed in eukaryotic cells. Because its mammalian binding “target” is the inner leaflet itself, which is a giant endogenous sink for activestate protein, the relative stoichiometric tuning of interaction partners is unnecessary to achieve a high on:off ratio for the interaction in the light vs. dark. This feature distinguishes it from heterodimerization systems that may require the selection of clonal cell lines with optimized expression levels to overcome the inherent expression level heterogeneity of transiently transfected cells in order to ensure robustness [23]. Accordingly, its single-component operation simplifies transgene delivery and cell line development, and also frees optical bandwidth by eliminating a second fluorescent protein tag needed in heterodimerization systems to visualize individual components in live cells.
BcLOV4 is versatile. Beyond robust performance in mammalian cells (Figure 2c), it also functions when expressed in yeast (Figure 2d) and in vitro as purified recombinant protein in lipid-stabilized water-in-oil emulsions (Figure 2e), the latter related to optochemical control in droplet-based in vitro-compartmentalized signaling systems [5]. Because its primary membrane interaction site is internal (in linear polypeptide space), it tolerates protein fusions to its N- and/or C-terminus when engineering chimeras. As stated, optogenetic chimeras for membrane recruitment are commonly employed in mammalian cytoskeletal biology applications, and indeed, BcLOV4 can be effectively applied to that end as demonstrated here (Figure 3).
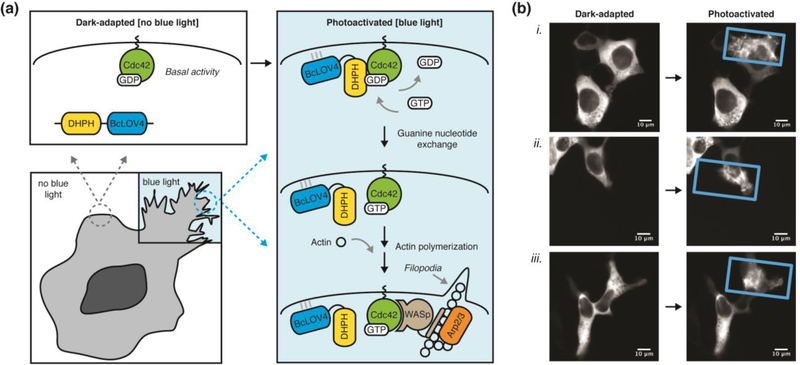
Figure 3. Spatially localized and optically induced cytoskeletal rearrangements by BcLOV4-derived opto-DHPH.
(a) Schematic overview of spatially precise opto-DHPH membrane recruitment and consequent activation of Cdc42 by the DHPH domain of the Intersectin GEF, which drives downstream actin polymerization. Cdc42 = Cell division control protein 42. DHPH = Diffuse B-cell lymphoma homology, Pleckstrin homology domain. GEF = Guanine exchange factor. Arp2/3 = Actin-related protein-2/3. WASp = Wiskott-Aldrich Syndrome protein. (b) Optically induced filopodia formation in HEK cells visualized by fluorescence imaging of a C-terminal mCherry tag. Only the blue light-illuminated (rectangle) regions show pronounced protusions, which are induced with very little stimulation (duty cycle = 0.8% = 0.5 sec per minute, λ = 450nm, 15 mW/cm2; spatially patterned by a digital micromirror device). Post-illumination times (i) 300 sec, (ii, iii) 500 sec Supplementary Movie 1 corresponds to cell (i).
Opto-DHPH is a chimera of BcLOV4 (mammalian codon-optimized and with C-terminal mCherry visualization tag) with an N-terminal DHPH (Dbl-homology, Pleckstrin-homology) domain of the Intersectin1 (ITSN) guanine exchange factor (GEF) that activates the Cdc42 small GTPase and downstream actin polymerization (Figure 3a). This particular Cdc42-GEF signaling pathway has been manipulated by numerous optogenetic heterodimerization systems based on iLID, CRY2, and BphP1 [26,29,50]. Spatially patterned illumination (using a digital micromirror device [51]) of opto-DHPH in transfected HEK293 cells causes pronounced filopodia formation that is restricted to the blue light-illumination field (Figure 3b and Supplementary Video 1). Optogenetic induction of the cytoskeletal change is very efficient; cells robustly respond to very sparse optical stimulation (< 1% duty cycle or 0.5 sec per minute) at experimental light levels (photocycling ED50 = 15 mW/cm2 at λ = 450 nm). Thus, BcLOV4 is clearly promising as a single-component tool for optogenetic membrane recruitment and signaling of its fusion partner as cargo.
Optogenetic implications of RGS-LOV signaling structure-function
The RGS-LOV signal transmission mode is distinct from known PAS-superfamily (Per-Arnt-Sim [52], to which LOV belong) lipid-binding proteins, which are integral membrane proteins (e.g. PhoQ: PDB code 3bq8, LuxQ: PDB code 2hje), and we are unaware of other photosensory proteins that are directly recruited to the membrane in response to light. However, despite the novelty of the photosensory signal transmission mode, de novo structural predictions by energy minimization modeling in Rosetta [53] suggest that the highly structured DUF domain is PAS-like [7] (Figure 2b), which would bring RGS-LOV in line with evolutionarily conserved tandem PAS proteins [52].
Importantly, RGS-LOV signaling follows known determinants of canonical LOV protein signaling. Optical membrane recruitment can be largely abolished by a cysteine-to-alanine mutation that prevents canonical photoadduct formation, and conversely, BcLOV4 can be made constitutively active or permanently membrane localized in the dark by mutating a conserved glutamine at the Jα helix terminus to structurally mimic an active “lit” state [54]. These mutants are useful as experimental controls for implicating photocycle involvement in signaling and accounting for the non-LOV role of blue-light alone in assays [7,27]. Likewise, a kinetic mutant that approaches functional bi-stability (membrane undocking timescale of ~10 minutes) has been rationally engineered by lengthening the photocycle [7].
The membrane-interacting amphipathic helix, which was initially identified through bioinformatics based on sequence conservation across 66 RGS-LOV homologs and secondary structure predictions [7], contains known plasma membrane interaction motifs rich in phenylalanine and lysine (“FFK” and “FKK”), which are found in other membrane-associated proteins (e.g. BAD: PDB code 1g5m, M2 proton channel of Influenza A: PDB code 2rlf). Its hydrophobic residues embed into the phospholipid bilayer while the surrounding cationic residues bind to the anionic membrane phospholipid headgroups such that the helix sits on top (not across) of the inner leaflet (Figure 2a inset). This non-stereospecific lipid-interaction motif is thus nonselective amongst lipid headgroups beyond their anionic charge density [7]. Thus, why do RGS-LOV preferentially bind the plasma membrane? The simplest explanation is that the mammalian plasma membrane is the most enriched subcellular structure for anionic phospholipids, especially for phosphatidylserine [55–57].
Membrane geometry and mechanical properties may also play a role in its subcellular preference profile, as amphipathic helices are known to “sense” membrane curvature and defects (and/or induce them). The inner leaflet is not only a densely anionic membrane, but also a largely defect-free and “flat” one due to its high sterol content [55–57]. Such membrane physical characteristics are well recognized by amphipathic helices with cationic residues surrounding bilayer-partitioning residues with large sidechains (like that of RGS-LOV), and support long-range electrostatic interactions between them [55,56]. These long-range electrostatic interactions may be critical to the signaling response in mammalian cells. For example, high-salinity prevents BcLOV4 from associating with in vitro membrane interfaces, and RGS-LOV that distribute to the nucleus in the dark-adapted state do not bind the inner nuclear membrane, the latter presumably due to the presence of a thick and dense nuclear lamina (see Figures S5 and S6 of [7]).
While it is possible that BcLOV4 binds a plasma membrane-associated partner that could influence subcellular localization selectivity, the interaction is likely transient if such a mammalian partner exists; multiple attempts at (AP/MS) affinity purification mass spectrometry-based interactome analyses of stably transducing BcLOV4-HEK293 cells showed no preferential partner in a light-dependent manner in our hands. One consequence of this preliminary insight is that BcLOV4 may be fairly “inert” as an optogenetic tool, devoid of spurious protein-protein interactions with basal levels of mammalian proteins. The membrane undocking kinetics of BcLOV4, though, which lags the photoadduct thermal reversion timescale by typically ~one minute in cells and in lipid-stabilized emulsions, does suggest the existence of unidentified lipid-interaction(s) across the multi-domain protein once it is membrane-localized (or possibly a slower photoadduct reversion for the lipid-bound state than in the bulk solution). In other non-mammalian cellular and in vitro model systems, the distribution profile and determinants will certainly be influenced by the respective membrane compositions (lipids, proteins, matrices, etc.), membrane structural properties (curvature, defect-density, fluidity, etc.), intracellular milieu (salinity, oxidation-reduction environment, pH, molecular crowding, etc.), and post-translational modifications, and thus, it is important to note that the commonalities of RGS-LOV signaling characteristics observed across model systems may be coincidental.
A high-resolution structure of the lipid-bound state is likely needed to conclusively determine what governs its plasma membrane preference and to refine our proposed multi-domain rearrangement and signal transmission mode. A high-resolution structure would also inform the rational design of a monomeric form of the native dimeric BcLOV4 (although naturally monomeric RGS-LOV may exist), a truncated or “minimal” BcLOV4 that preserves only the essential structural elements required for light-induced membrane binding, or variants with decreased membrane undocking timescales to improve overall temporal precision as a tool. However, given that the high-affinity protein-membrane interaction is near diffusion-limited, it is unlikely that meaningfully faster membrane recruitment times can be achieved by engineered RGS-LOV or any other optogenetic tool.
Conclusion and Future Directions: Is it a feature or bug?
Despite the comparative advantages respectively reported for existing optogenetic systems, there is no single solution for an ideal tool for membrane recruitment and signaling. Shortcomings of any particular system are typically attributable to inherent biophysical arguments or signaling responses downstream of optical induction, not to poor protein engineering. For example, speed comes at the expense of optical induction efficiency; fast undocking benefits temporal resolution of a tool but necessitates more light to sustain signaling because the activated protein thermally reverts quickly post-induction, whereas a “slow” tool is more efficient because it better sustains signaling post-induction [58], even if the chromophore extinction coefficient is the same between the two scenarios. In our own work in studying calcium signaling dynamics, the intrinsic “slowness” of the CRY2:CIBN system confers quasi bi-stability that is helpful in limiting photobleaching when performing second messenger signaling assays with fluorescent reporters [23].
In another example of contextual dichotomy, because no transition between biological states is infinitely steep, high-affinity light-activated binding that promotes rapid membrane recruitment and robust association between partners places an upper bound on protein expression level, in order to avoid permanent association in the dark (i.e. when expression level exceeds the binding affinity in the dark-adapted state), as described with iLID [38]. Similarly, the strong intrinsic preference of RGS-LOV for the inner leaflet is advantageous for rapid plasma membrane recruitment, but could hinder applications that target other subcellular structures.
The proverbial quip from computer engineering of “It’s not a bug, it’s a feature” is perhaps apropos for any characteristic of any optogenetic tool. Accordingly, it is useful to consider an application-specific operating window in optogenetics, or a useful dynamic range in analogy to the therapeutic index in pharmacology. The successful implementation of optogenetic tools for membrane recruitment and hetero-dimerization often necessitates extensive side-by-side comparisons of multiple technologies in context of the specific end-application to identify such windows; some examples of thorough characterization on application-specific kinetics [59,60] and expression level-based performance [38] in subcellular optogenetics can be found in works by others.
The BcLOV4 protein discussed here possesses functional windows well-suited for optically inducible membrane recruitment-based signaling in mammalian cells with respect to temporal precision, signaling induction efficiency by sparse illumination, and signaling contrast ratio between the photoactivated and dark-adapted states. It is possible that these characteristics were evolutionarily optimized for membrane recruitment-based signaling as natural proteins, but as engineered optogenetic tools, the RGS-LOV proteins studied to date were not intentionally designed so, beyond mammalian codon optimization to increase expression level and the rational engineering of the aforementioned bi-stable mutant by lengthening the flavin photocycle duration. Thus, further engineering, structure-function analyses, and/or experimental characterization of other homologous RGS-LOV proteins [7,8] will likely prove to be fruitful endeavors with exciting and valuable outcomes.
Supplementary Material
Highlights.
Diverse protein engineering strategies for optically induced membrane signaling.
Design strategies include allosteric switching and subcellular localization changes.
Light-regulated protein-lipid electrostatic interaction by RGS-LOV proteins.
RGS-LOV as single-component optogenetic tools for membrane recruitment.
Acknowledgments
P.H-A., S.T.G., and B.Y.C. prepared the manuscript. P.H-A. and B.Y.C. constructed hardware and conducted assays for the primary data reported here. This work was funded by: National Science Foundation / Systems and Synthetic Biology (MCB 1652003), National Institutes of Health / National Institute on Drug Abuse (R21 DA040434), National Institutes of Health / National Institute of Neurological Disorders and Stroke (R01 NS101106).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
* Special interest; ** Outstanding interest
- 1.Losi A, Gardner KH, Möglich A: Blue-Light Receptors for Optogenetics. Chemical Reviews 2018, 118:10659–10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rost BR, Schneider-Warme F, Schmitz D, Hegemann P: Optogenetic Tools for Subcellular Applications in Neuroscience. Neuron 2017, 96:572–603. [DOI] [PubMed] [Google Scholar]
- 3.Ankenbruck N, Courtney T, Naro Y, Deiters A: Optochemical Control of Biological Processes in Cells and Animals. Angewandte Chemie International Edition 2018, 57:2768–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kienzler MA, Isacoff EY: Precise modulation of neuronal activity with synthetic photoswitchable ligands. Current Opinion in Neurobiology 2017, 45:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caldwell RM, Bermudez JG, Thai D, Aonbangkhen C, Schuster BS, Courtney T, Deiters A, Hammer DA, Chenoweth DM, Good MC: Optochemical Control of Protein Localization and Activity within Cell-like Compartments. Biochemistry 2018, 57:2590–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartelt SM, Steinkühler J, Dimova R, Wegner SV: Light-Guided Motility of a Minimal Synthetic Cell. Nano Letters 2018, 18:7268–7274. [DOI] [PubMed] [Google Scholar]
- 7. **.Glantz ST, Berlew EE, Jaber Z, Schuster BS, Gardner KH, Chow BY: Directly light-regulated binding of RGS-LOV photoreceptors to anionic membrane phospholipids. Proceedings of the National Academy of Sciences 2018, 115:E7720–E7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. *.Glantz ST, Carpenter EJ, Melkonian M, Gardner KH, Boyden ES, Wong GK, Chow BY: Functional and topological diversity of LOV domain photoreceptors. Proc Natl Acad Sci U S A 2016, 113:E1442–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Govorunova EG, Sineshchekov OA, Rodarte EM, Janz R, Morelle O, Melkonian M, Wong GKS, Spudich JL: The Expanding Family of Natural Anion Channelrhodopsins Reveals Large Variations in Kinetics, Conductance, and Spectral Sensitivity. Scientific reports 2017, 7:43358–43358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conrad KS, Bilwes AM, Crane BR: Light-Induced Subunit Dissociation by a LOV domain Photoreceptor from Rhodobacter sphaeroides. Biochemistry 2013, 52:378–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohlendorf R, Schumacher CH, Richter F, Möglich A: Library-Aided Probing of Linker Determinants in Hybrid Photoreceptors. ACS Synthetic Biology 2016, 5:1117–1126. [DOI] [PubMed] [Google Scholar]
- 12. *.Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, Bear JE, Kuhlman B: Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proceedings of the National Academy of Sciences 2015, 112:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawano F, Suzuki H, Furuya A, Sato M: Engineered pairs of distinct photoswitches for optogenetic control of cellular proteins. Nature Communications 2015, 6:6256. [DOI] [PubMed] [Google Scholar]
- 14. **.Wang H, Vilela M, Winkler A, Tarnawski M, Schlichting I, Yumerefendi H, Kuhlman B, Liu R, Danuser G, Hahn KM: LOVTRAP: an optogenetic system for photoinduced protein dissociation. Nature methods 2016, 13:755–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. **.Dagliyan O, Tarnawski M, Chu P-H, Shirvanyants D, Schlichting I, Dokholyan NV, Hahn KM: Engineering extrinsic disorder to control protein activity in living cells. Science 2016, 354:1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diensthuber RP, Bommer M, Gleichmann T, Möglich A: Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure 2013, 21:1127–1136. [DOI] [PubMed] [Google Scholar]
- 17.Kim YS, Kato HE, Yamashita K, Ito S, Inoue K, Ramakrishnan C, Fenno LE, Evans KE, Paggi JM, Dror RO, et al. : Crystal structure of the natural anion-conducting channelrhodopsin GtACR1. Nature 2018, 561:343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polesskaya O, Baranova A, Bui S, Kondratev N, Kananykhina E, Nazarenko O, Shapiro T, Nardia FB, Kornienko V, Chandhoke V, et al. : Optogenetic regulation of transcription. BMC neuroscience 2018, 19:12–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kainrath S, Stadler M, Reichhart E, Distel M, Janovjak H: Green-Light-Induced Inactivation of Receptor Signaling Using Cobalamin-Binding Domains. Angewandte Chemie (International ed. in English) 2017, 56:4608–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K: Temporally precise in vivo control of intracellular signalling. Nature 2009, 458:(7241)–1029. [DOI] [PubMed] [Google Scholar]
- 21.Bailes HJ, Zhuang L-Y, Lucas RJ: Reproducible and Sustained Regulation of Gαs Signalling Using a Metazoan Opsin as an Optogenetic Tool. PLOS ONE 2012, 7:e30774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hannanta-Anan P, Chow BY: Optogenetic Control of Calcium Oscillation Waveform Defines NFAT as an Integrator of Calcium Load. Cell Syst 2016, 2:283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hannanta-Anan P, Chow BY: Optogenetic Inhibition of Gαq Protein Signaling Reduces Calcium Oscillation Stochasticity. ACS Synth Biol 2018, 7:1488–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. *.Bugaj LJ, Sabnis AJ, Mitchell A, Garbarino JE, Toettcher JE, Bivona TG, Lim WA: Cancer mutations and targeted drugs can disrupt dynamic signal encoding by the Ras-Erk pathway. Science 2018, 361:eaao3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson MZ, Ravindran PT, Lim WA, Toettcher JE: Tracing Information Flow from Erk to Target Gene Induction Reveals Mechanisms of Dynamic and Combinatorial Control. Molecular Cell 2017, 67:757–769.e755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Neill PR, Kalyanaraman V, Gautam N, Van Haastert P: Subcellular optogenetic activation of Cdc42 controls local and distal signaling to drive immune cell migration. Molecular Biology of the Cell 2016, 27:1442–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman BH, K.M.: A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 2009, 461:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou XX, Chung HK, Lam AJ, Lin MZ: Optical control of protein activity by fluorescent protein domains. Science 2012, 338:810–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. *.Kaberniuk AA, Shemetov AA, Verkhusha VV: A bacterial phytochrome-based optogenetic system controllable with near-infrared light. Nat Methods 2016, 13:591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou XX, Fan LZ, Li P, Shen K, Lin MZ: Optical control of cell signaling by single-chain photoswitchable kinases. Science 2017, 355:836–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye H, Baba MD-E, Peng R-W, Fussenegger M: A Synthetic Optogenetic Transcription Device Enhances Blood-Glucose Homeostasis in Mice. Science 2011, 332:1565–1568. [DOI] [PubMed] [Google Scholar]
- 32.Zemelman BV, Lee GA, Ng M, Miesenbock G: Selective photostimulation of genetically chARGed neurons. Neuron 2002, 33:15–22. [DOI] [PubMed] [Google Scholar]
- 33.Zheng W, Zhou J, Luan Y, Yang J, Ge Y, Wang M, Wu B, Wu Z, Chen X, Li F, et al. : Spatiotemporal Control of GPR37 Signaling and Its Behavioral Effects by Optogenetics. Frontiers in Molecular Neuroscience 2018, 11:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. **.Harper SM, Neil LC, Gardner KH: Structural basis of a phototropin light switch. Science 2003, 301:1541–1544. [DOI] [PubMed] [Google Scholar]
- 35.Halavaty AS, Moffat K: N- and C-Terminal Flanking Regions Modulate Light-Induced Signal Transduction in the LOV2 Domain of the Blue Light Sensor Phototropin 1 from Avena sativa. Biochemistry 2007, 46:14001–14009. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt D, Tillberg PW, Chen F, Boyden ES: A fully genetically encoded protein architecture for optical control of peptide ligand concentration. Nat Commun 2014, 5:3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sosnick TR, Weiss EL, Glotzer M: TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods 2012, 9:379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. *.Zimmerman SP, Hallett RA, Bourke AM, Bear JE, Kennedy MJ, Kuhlman B: Tuning the Binding Affinities and Reversion Kinetics of a Light Inducible Dimer Allows Control of Transmembrane Protein Localization. Biochemistry 2016, 55:5264–5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renicke C, Schuster D, Usherenko S, Essen L-O, Taxis C: A LOV2 Domain-Based Optogenetic Tool to Control Protein Degradation and Cellular Function. Chemistry & Biology 2013, 20:619–626. [DOI] [PubMed] [Google Scholar]
- 40.Niopek D, Wehler P, Roensch J, Eils R, Di Ventura B: Optogenetic control of nuclear protein export. Nature Communications 2016, 7:10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richter F, Fonfara I, Bouazza B, Schumacher CH, Bratovič M, Charpentier E, Möglich A: Engineering of temperature- and light-switchable Cas9 variants. Nucleic acids research 2016, 44:10003–10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. *.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL: Rapid blue-light-mediated induction of protein interactions in living cells. Nat Meth 2010, 7:973–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pudasaini A, Shim JS, Song YH, Shi H, Kiba T, Somers DE, Imaizumi T, Zoltowski BD: Kinetics of the LOV domain of ZEITLUPE determine its circadian function in Arabidopsis. eLife 2017, 6:e21646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lechno-Yossef S, Melnicki MR, Bao H, Montgomery BL, Kerfeld CA: Synthetic OCP heterodimers are photoactive and recapitulate the fusion of two primitive carotenoproteins in the evolution of cyanobacterial photoprotection. The Plant Journal 2017, 91:646–656. [DOI] [PubMed] [Google Scholar]
- 45.Yu X, Sayegh R, Maymon M, Warpeha K, Klejnot J, Yang H, Huang J, Lee J, Kaufman L, Lin C: Formation of nuclear bodies of Arabidopsis CRY2 in response to blue light is associated with its blue light-dependent degradation. The Plant cell 2009, 21:118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Buskirk EK, Decker PV, Chen M: Photobodies in Light Signaling. Plant Physiology 2012, 158:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taslimi A, Vrana JD, Chen D, Borinskaya S, Mayer BJ, Kennedy MJ, Tucker CL: An optimized optogenetic clustering tool for probing protein interaction and function. Nat Commun 2014, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee S, Park H, Kyung T, Kim NY, Kim S, Kim J, Heo WD: Reversible protein inactivation by optogenetic trapping in cells. Nature Methods 2014, 11:633. [DOI] [PubMed] [Google Scholar]
- 49.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV: Optogenetic protein clustering and signaling activation in mammalian cells. Nat Meth 2013, 10:249–252. [DOI] [PubMed] [Google Scholar]
- 50.de Beco S, Vaidžiulyté K, Manzi J, Dalier F, di Federico F, Cornilleau G, Dahan M, Coppey M: Optogenetic dissection of Rac1 and Cdc42 gradient shaping. Nature communications 2018, 9:4816–4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trojanowski NF, Fang-Yen C: Simultaneous Optogenetic Stimulation of Individual Pharyngeal Neurons and Monitoring of Feeding Behavior in Intact C. elegans In C. elegans: Methods and Applications. Edited by Biron D, Haspel G: Humana Press; 2015:105–119. 10.1007/978-1-4939-2842-2_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Möglich A, Ayers RA, Moffat K: Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 2009, 17:1282–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. *.Dou J, Vorobieva AA, Sheffler W, Doyle LA, Park H, Bick MJ, Mao B, Foight GW, Lee MY, Gagnon LA, et al. : De novo design of a fluorescence-activating β-barrel. Nature 2018, 561:485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ganguly A, Thiel W, Crane BR: Glutamine Amide Flip Elicits Long Distance Allosteric Responses in the LOV Protein Vivid. Journal of the American Chemical Society 2017, 139:2972–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jackson Catherine L, Walch L, Verbavatz J-M: Lipids and Their Trafficking: An Integral Part of Cellular Organization. Developmental Cell 2016, 39:139–153. [DOI] [PubMed] [Google Scholar]
- 56.Drin G, Antonny B: Amphipathic helices and membrane curvature. FEBS Letters 2010, 584:1840–1847. [DOI] [PubMed] [Google Scholar]
- 57.van Meer G, Voelker DR, Feigenson GW: Membrane lipids: where they are and how they behave. Nature Reviews Molecular Cell Biology 2008, 9:112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Taslimi A, Zoltowski B, Miranda JG, Pathak GP, Hughes RM, Tucker CL: Optimized second-generation CRY2-CIB dimerizers and photoactivatable Cre recombinase. Nature chemical biology 2016, 12:425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. *.van Bergeijk P, Adrian M, Hoogenraad CC, Kapitein LC: Optogenetic control of organelle transport and positioning. Nature 2015, 518:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benedetti L, Barentine AES, Messa M, Wheeler H, Bewersdorf J, De Camilli P: Light-activated protein interaction with high spatial subcellular confinement. Proceedings of the National Academy of Sciences 2018, 115:E2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.