Abstract
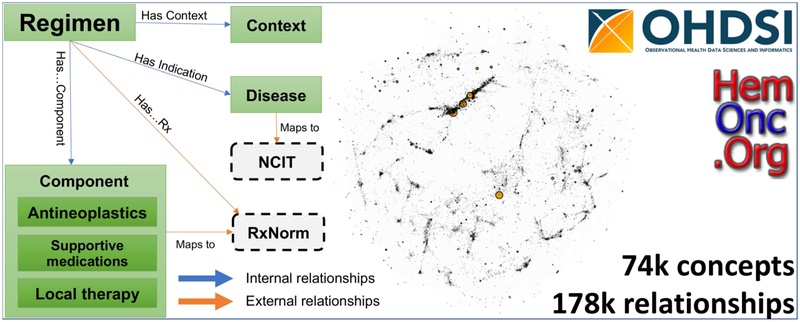
Systematic application of observational data to the understanding of impacts of cancer treatments requires detailed information models allowing meaningful comparisons between treatment regimens. Unfortunately, details of systemic therapies are scarce in registries and data warehouses, primarily due to the complex nature of the protocols and a lack of standardization. Since 2011, we have been creating a curated and semi-structured website of chemotherapy regimens, HemOnc.org. In coordination with the Observational Health Data Sciences and Informatics (OHDSI) Oncology Subgroup, we have transformed a substantial subset of this content into the OMOP common data model, with bindings to multiple external vocabularies, e.g., RxNorm and the National Cancer Institute Thesaurus. Currently, there are >73,000 concepts and >177,000 relationships in the full vocabulary. Content related to the definition and composition of chemotherapy regimens has been released within the ATHENA tool (athena.ohdsi.org) for widespread utilization by the OHDSI membership. Here, we describe the rationale, data model, and initial contents of the HemOnc vocabulary along with several use cases for which it may be valuable.
Keywords: Neoplasms, Ontologies, Knowledge engineering
Introduction
The related fields of hematology and oncology have made a great deal of progress in the treatment of cancer over the past several decades, primarily through the careful application of prospective clinical trials to areas of unmet need.[1] Due to an extensive international network of cooperative study groups, many of these trials have been carried out in a randomized fashion and are thus considered “gold standards” of evidence for cancer care. Despite this, only an estimated 5% of adult cancer patients enroll in clinical trials.[2] For those who do, important details of preceding treatment and subsequent outcomes after the trial is completed are often missing. For example, many trials in heavily pretreated patients merely report a numeric range of “lines” of prior chemotherapies, without any further details about the types of therapies, durations of responses, depth of responses, and toxicities. This obscures the reality that most cancer treatments are given in combination regimens with complex dosing and scheduling, that most cancer drugs are highly toxic and often require additional “supportive” medications to ameliorate side effects, and that reasons for treatment discontinuation are complex and often unrelated to disease progression. Thus, clinical trials only create a glimpse of the deep phenotypes needed to understand the complexity of cancer and its treatment.
For these reasons and due to the large cost of carrying out large prospective randomized controlled trials (RCTs), there is a burgeoning enthusiasm for “real world data” (RWD) to generate “real world evidence” (RWE).[3] These data, primarily scoured from electronic health records, have the promise of revealing in-depth details of cancer treatment history, outcomes, performance status, and comorbidities. A substantial number of public and private institutions are active in this space, but all face a similar major hurdle: the lack of standardization in representing oncology data, particularly chemotherapy regimens and their context-specific disease indications. For example, the Systematized Nomenclature of Medicine – Clinical Terms (SNOMED-CT) only has six chemotherapy regimen concepts. The National Cancer Institute thesaurus (NCIt) has more, 451 in version 19.04f, but these concepts only contain antineoplastic drugs and some disease indications. Here, we present the adaptation of content from HemOnc.org, a community collaborative information resource describing cancer treatment regimens, to work within the Observational Medical Outcomes Partnership (OMOP) common data model (CDM), used by the Observational Health Data Sciences and Informatics (OHDSI) program.[4]
In 2011, we founded the collaborative wiki HemOnc.org, a knowledge base intended primarily for healthcare professionals, built upon the open-source MediaWiki software, and organized primarily by cancer subtype.[5] The site contains information on chemotherapy regimens, antineoplastic and supportive medications, and other topics relevant to the practice of hematology/oncology. As of May 27, 2019 there are a total of 904 content pages with 460,320 lines of content. To consolidate duplicative regimens and to formalize much of the information present on the website, we began to convert portions of the website to the Web Ontology Language (OWL) format, in mid-2017; this work has been described in preliminary form previously.[6]
The OHDSI program is a multi-stakeholder, interdisciplinary collaborative working to make the promise of generating RWE a reality.[7] As the foundational platform for the OHDSI consortium, the OMOP CDM enables the systematic analysis of disparate observational databases. OHDSI’s approach is to transform data contained within observational databases into a common format (data model) with common semantics (terminologies, vocabularies, coding schemes), and perform systematic analyses using a library of standard analytic routines and analytic tools. The OHDSI Oncology Subgroup is tasked with developing extensions of the OMOP CDM/Vocabulary and the OHDSI analytic platform to support observational cancer research.
Although the OWL version of HemOnc has allowed for a degree of formalism previously lacking, the OWL model is not conducive to use in the context of OMOP. In late 2018, we initiated a collaboration with the OHDSI Oncology Subgroup to adapt HemOnc.org content into the more broadly usable OMOP format. This manuscript describes the conversion as well as the current state of the vocabulary.
Methods
The integration of HemOnc.org content with the OMOP CDM involved four key tasks: 1) creation of an extension to the OMOP CDM to handle episodes of care, 2) mapping of content to a relational data model compatible with the OMOP CDM, 3) parsing HemOnc.org to populate the resulting data model, and 4) identification of relevant use cases,
OHDSI Oncology CDM Episode Extension Proposal
Many common oncology scenarios exceed OMOP’s current capabilities. These include: 1) describing cancer treatments at a level of abstraction that matches clinicians’ or researchers’ everyday practice (i.e., a coordinated regimen or complex protocol as opposed to a list of single drugs with or without doses); 2) normalizing regimens that can be referred to in many different ways (e.g., R-CHOP; CHOPR; and cyclophosphamide, doxorubicin, prednisone, rituximab, vincristine all refer to the same regimen); 3) characterizing when an oncology treatment begins and ends (some regimens have a defined duration whereas others are typically given indefinitely until an event such as cancer progression occurs); 4) identifying when a treatment ends and when another begins (e.g., distinguishing between a pre-planned staggered start of combinations of drugs versus an event-triggered change from one set of drugs to another); and 5) determining response to treatment (e.g., determining whether a sequence of treatments was the result of a risk-adapted strategy, a cancer progression event, or a drug intolerance event). These tasks present challenges for modeling within the OMOP framework: as OMOP is oriented toward the representation of low-level clinical events, representation of higher-level abstractions and temporal constraints, as needed to represent clinicians’ and researchers’ view of oncology treatments, must be added through extensions to the model. To address these gaps, we developed an Oncology CDM Extension proposal providing a representation of episodes of care.
Data Model
For the purposes of creating a representation of the HemOnc vocabulary that is compatible with the OMOP CDM, we focused on three types of HemOnc.org pages: 1) intervention content pages; 2) disease-specific content pages; and 3) MediaWiki category[8] pages. Intervention content pages contain details of individual medications or procedures utilized in the practice of hematology/oncology. For medications, this includes mechanism of action, diseases for which the medication is used, history of US Food & Drug Administration (FDA) approvals, and synonyms. Disease-specific content pages are organized by clinical disease subtype, and contain information on treatment guidelines, context-specific treatment plans, prognosis, and drugs under development. The treatment plans conform to a standard structure, which is informed by the data model described below. Finally, category pages contain metatags which are used to develop the class hierarchy of the vocabulary. We analyzed the contents of the pages and both relationships between elements within a page and between pages to develop a relational data model capable of representing all of the relevant HemOnc concepts while maintaining consistency with OMOP conventions.
Parsing and Table Creation
To create the OMOP tables, the content of HemOnc.org was parsed from the HTML pages from the HemOnc.org site. Pages with educational material (e.g., bone marrow biopsy instructions; hematology/oncology fellowship training information) were ignored; the remainder were parsed using R[9] (version 3.5.2). Relationships were instantiated using the nested structure of the HemOnc.org site; e.g., any regimen appearing on the Breast cancer page inherited “Has accepted use” of breast cancer; any regimen under a heading of Adjuvant therapy inherited “Has context” of adjuvant therapy, and so forth. RxNorm codes and MEDLINE date were programmatically accessed using the RxNav[10] application programming interface and the reutils[11] R package, respectively. All concepts are only allowed to appear once in the concept table and are each assigned a unique internal concept code. Concepts selected for public release are then assigned a unique OHDSI concept ID. The tables were developed through an agile and iterative process involving frequent discussions between HemOnc.org representatives and the OHDSI Oncology Subgroup. Simultaneously, the groups had many discussions about which elements to release publicly, balancing complexity with the needs of the OHDSI user community.
Use Cases
As an ongoing process during the development of the HemOnc OMOP model, we identified several use cases illustrating possible applications of the models.
Results
OHDSI Oncology CDM Episode Extension Proposal
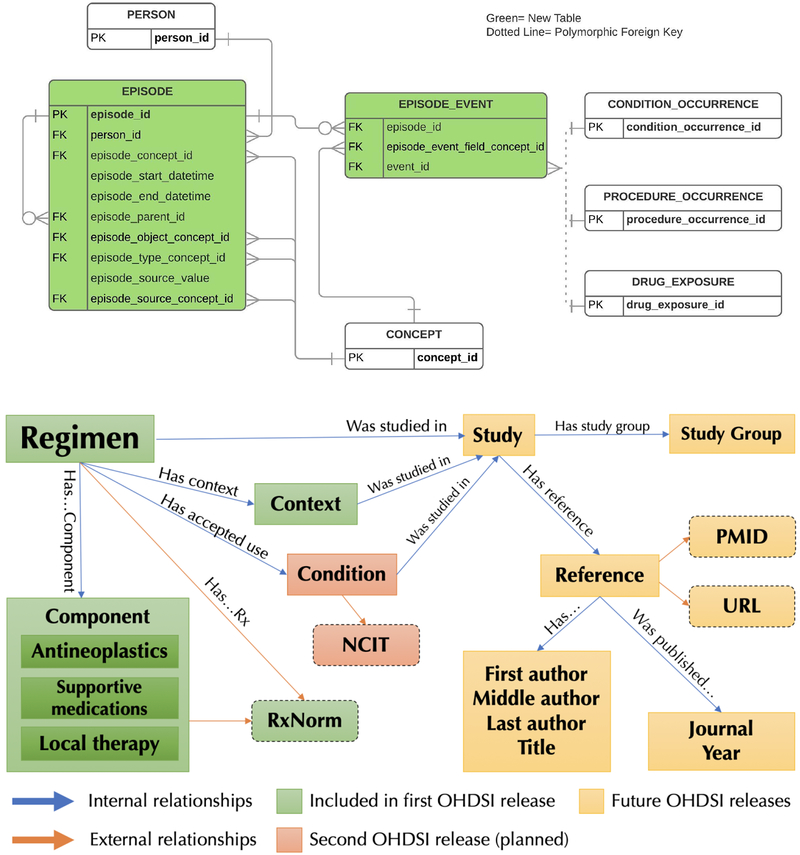
The OMOP CDM Episode extension models oncology treatments as exposures during an episode event. The Extension data model supports the explicit connection between an episode abstraction and the lower level clinical events that implement it (drugs and procedures). The Extension data model is provided in Figure 1.
Figure 1.
OHDSI Oncology CDM Extension Proposal data model. HemOnc oncology drug regimen concepts should be assigned in the episode_object_concept_id column of the EPISODE table. FK: foreign key; PK: primary key
The Extension also recommends the addition of terminologies that support the aggregation of lower-level clinical events into higher-level abstractions. This addition is accomplished through the adoption of the HemOnc chemotherapy regimen ontology as the standard OMOP oncology drug treatment vocabulary. This means that HemOnc oncology drug regimen concepts (as encoded within the OMOP vocabulary) should be assigned to OMOP oncology drug treatment episodes. OMOP developers should use HemOnc’s specification of oncology drug regimens relationships to constituent antineoplastic ingredients/supportive medications, disease context, and detailing of temporal cycles to surface oncology drug regimens from lower-level drug events.
Data Model
Prior to HemOnc.org’s collaboration with the OHDSI Oncology Subgroup, the HemOnc.org content had converged on a semi-formalized standard form. However, a formal concept-relationship model did not exist. In preparation for migration of existing content in the OWL format to the OMOP CDM, as well as to add new content not yet parsed from the website, it was necessary to define a formal model. This process was carried out iteratively with frequent consultation from the OHDSI Oncology Subgroup.
A simplified depiction of the resultant chemotherapy regimen data model is illustrated in Figure 2; the full data model is available in the Supplement. All regimens are tied to a specific condition and to a treatment context, e.g., first-line therapy for ER/PR+ metastatic breast cancer. Regimen concepts contain the specific components comprising the treatment regimen, which are further subdivided into 1) antineoplastics – drugs and/or procedures intended to have a direct or indirect consequence of cancer cell killing; 2) supportive medications – drugs used to ameliorate the side effects of antineoplastics (e.g., antiemetics, growth factor support); 3) local therapies – drugs or other interventions that have a local, non-systemic effect; and 4) immunosuppressives – drugs primarily relevant to regimens used for non-malignant conditions, such as autoimmune hematologic conditions. Study concepts relate to the specific clinical trial that was carried out to evaluate the regimen, almost always within a specific cancer subtype and treatment context. Studies are often organized by a study group, and the primary products of studies cited by HemOnc.org are their reference(s). Finally, references are published by author(s) in a journal at a particular time (anchored to year in the current model).
Figure 2.
HemOnc.org chemotherapy regimen data model (simplified). Dashed boxes represent external vocabularies with instantiated mappings in the HemOnc vocabulary. CC BY-NC-SA 4.0: Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International; NCIt: National Cancer Institute Thesaurus; PMID: PubMed reference number; URL: Uniform Resource Locator.
Each domain of the data model contains a number of attributes that specify the necessary elements of a regimen. For example, regimen-level attributes are shown in Table 1. Classes are bound by binary relationships. For example, a Regimen hasIndicationFor a Condition; a Regimen hasAntineoplastic of Component.
Table 1.
Regimen attributes, with cardinality: [0..n] indicates any number, [1..n] indicates at least one, [0..1] one or zero, etc. Attributes marked with a concept class are currently included in the vocabulary; synonyms are classless. The example is taken from R-CHOP variant #5 used in the treatment of diffuse large B-cell lymphoma (DLBCL).
| Regimen Attribute | Concept Class | Example |
|---|---|---|
| Preferred regimen name [1] | Regimen | R-CHOP |
| Regimen name expansion if acronym [0..n] | Rituximab, Cyclophosphamide, Hydroxydaunorubicin, Oncovin, Predniso(lo)ne | |
| Regimen synonym(s) [0..n] | N/A | CHOP-R; R-CHOP-21; CHOP-R; RCHOP; CHOPR |
| Regimen coded concept(s), if available [0..n] | NCIt ID: C9760 | |
| Regimen type [1..n] | Regimen type | Chemotherapy |
| Regimen schedule [1..2] | 21-day cycle | |
| Regimen duration [1..2] | 6 cycles | |
| If randomized - type [0..1] | Experimental | |
| If experimental - type [0..1] | Escalation | |
| Regimen variant #, if applicable [0..n] | Variant #5 | |
| Regimen variant short description, if applicable [0..n] | prednisone 100 mg, IV rituximab, flat-dose vincristine |
Parsing and Table Creation
As of May 27th, 2019 there were 904 HemOnc.org content pages, of which 728 were parsed. After parsing, there are 24 classes that comprise the vocabulary, with a total of 73,058 unique concept instances. These classes are shown in Table 2, along with a count of each classes’ instances. The standard Regimen class contains 1,546 individuals, which is more than three times as many as are present in NCIt, version 19.04f. Two of the classes are “stub” classes, meaning that the concept does not yet have enough information (e.g., a drug signetur with missing dose information; a regimen with missing drug information). There are 36 relationship types, shown in Table 3, instantiated in 177,268 unique relationships. While the majority of these relationships are internal to HemOnc, 150 are to NCIt disease concepts, 5,792 to RxNorm, and 14 to RxNorm Extension. There are also 3,449 drug and regimen synonyms. For the initial public OHDSI release of the vocabulary, 4,678 concepts from eight concept classes and 24,566 relationships of 17 relationship types are included.
Table 2.
Concept classes and instance counts
| Concept class (Category) | Count | Concept class (Category) | Count |
|---|---|---|---|
| Author | 29,163 | Sig Stub | 858 |
| Reference | 5,626 | Component* | 555 |
| ReferenceTitle | 5,540 | Component Class* | 333 |
| ReferenceURL | 5,485 | Journal | 187 |
| PubMedURL | 5,532 | Condition** | 120 |
| Sig | 3,971 | Year** | 82 |
| Study | 4,869 | Procedure* | 50 |
| Study name short | 4,550 | Condition Class** | 49 |
| Brand name* | 2,131 | Context*+ | 33 |
| Regimen*+ | 1,546 | BioCondition** | 23 |
| Regimen Stub | 1,367 | Regimen type* | 18 |
| Study Group | 958 | Route* | 13 |
| Total unique concepts | 73,058 | ||
These concepts classes are included in the first public OHDSI release
These concepts classes will be included in the second public OHDSI release
These concept classes are standard elements for OHDSI
Table 3.
Relationship types and classes involved in the relationship.
| Plain English Description | Concept 1 Class | Relationship Type | Concept 2 Class | Axioms |
|---|---|---|---|---|
| Generic class hierarchy* | (Multiple) | Is a | (Multiple) | 2,214 |
| Generic external mapping* | (Multiple) | Maps to | (Multiple) | 664 |
| Evidence to support use | (Multiple) | Was studied in | Study | 18,076 |
| Biomarker-specific disease subtype** | BioCondition | Is bio subclass | Condition | 21 |
| Brand name(s)* | Component | Has brand name | Brand Name | 2,140 |
| Drug prescription signetur | Component | Has sig | Sig or Sig Stub | 6,370 |
| Drug route(s)* | Component | May have route | Route | 624 |
| Year of FDA approval | Component | Was FDA approved yr | Year | 390 |
| Middle author | Reference | Has middle author | Author | 68,288 |
| First author | Reference | Has first author | Author | 5,507 |
| Last author | Reference | Has last author | Author | 5,479 |
| Journal of publication | Reference | Was published in | Journal | 5,642 |
| PubMed URL | Reference | Has PMID | PubMedURL | 5,646 |
| Reference title | Reference | Has title | ReferenceTitle | 5,653 |
| Reference URL | Reference | Has URL | ReferenceURL | 5,693 |
| Year of Publication | Reference | Was published year | Year | 5,457 |
| Biomarker-specific regimen** | Regimen | Has bioaccepted use | BioCondition | 167 |
| Antineoplastic interventions* | Regimen | Has antineoplastic | Component | 4,219 |
| Supportive interventions* | Regimen | Has supportive med | Component | 1,258 |
| Immune suppressing interventions* | Regimen | Has immunosuppressor | Component | 165 |
| Local interventions, including CNS therapy* | Regimen | Has local therapy | Component | 134 |
| Episode context of treatment* | Regimen | Has context | Context | 2,487 |
| Disease context of treatment** | Regimen | Has accepted use | Condition | 4,087 |
| Current Regimen** | Regimen | Is current in | Condition | 2,248 |
| Historical Regimen** | Regimen | Is historical in | Condition | 368 |
| Link to preceding treatment(s)** | Regimen | Can be preceded by | Regimen or Regimen Stub | 969 |
| Link to subsequent treatment(s)** | Regimen | Can be followed by | Regimen or Regimen Stub | 868 |
| Direct comparison within an RCT | Regimen | Has been compared to | Regimen or Regimen Stub | 3,872 |
| Regimen type* | Regimen | Has regimen type | Regimen type | 1,688 |
| Antineoplastic interventions (RxNorm)*+ | Regimen | Has antineopl Rx | RxNorm Ingredient | 4,018 |
| Supportive interventions (RxNorm)*+ | Regimen | Has support med Rx | RxNorm Ingredient | 1,001 |
| Immune suppressing interventions (RxNorm)*+ | Regimen | Has immunosuppr Rx | RxNorm Ingredient | 143 |
| Local interventions, including CNS therapy (RxNorm)*+ | Regimen | Has local therap Rx | RxNorm Ingredient | 130 |
| Reference | Study | Has reference | Reference | 5,648 |
| Study group | Study | Has study group | Study Group | 1,147 |
| Study’s short name | Study | Has study short name | Study name short | 4,787 |
| Total relationships: | 177,268 | |||
These relationship types are included in the first public OHDSI release
These relationship types will be included in the second public OHDSI release
These relationship types are named mapping relationships to external vocabularies
As an example, consider the simple two-drug chemotherapy regimen CapeOx.[12] This regimen has 49 relationships in the full vocabulary. As shown in Table 4, this regimen has six treatment contexts and is currently indicated for six disease types. In certain specific disease/context scenarios, it is also part of a larger protocol with preceding or subsequent treatments.
Table 4.
Example of the CapeOx regimen with its relationships. For readability, concept codes have been replaced with concept names. Not shown are 20 studies that evaluated CapeOx alone or as part of an RCT.
| Concept 1 | Relationship Type | Concept 2 | Vocab. 1 | Vocab. 2 |
|---|---|---|---|---|
| CapeOx | Has regimen type | Chemotherapy | HemOnc | HemOnc |
| CapeOx | Has antineoplastic | Capecitabine | HemOnc | HemOnc |
| CapeOx | Has antineoplastic | Oxaliplatin | HemOnc | HemOnc |
| CapeOx | Has antineopl Rx | (RxCUI) 194000 | HemOnc | RxNorm |
| CapeOx | Has antineopl Rx | (RxCUI) 32592 | HemOnc | RxNorm |
| CapeOx | Has context | Adjuvant therapy | HemOnc | HemOnc |
| CapeOx | Has context | Non-curative first-line therapy | HemOnc | HemOnc |
| CapeOx | Has context | Non-curative second-line therapy | HemOnc | HemOnc |
| CapeOx | Has context | Non-curative third-line therapy | HemOnc | HemOnc |
| CapeOx | Has context | Non-curative therapy | HemOnc | HemOnc |
| CapeOx | Has context | Neoadjuvant therapy | HemOnc | HemOnc |
| CapeOx | Has indication | Colon cancer | HemOnc | HemOnc |
| CapeOx | Has indication | Esophageal cancer | HemOnc | HemOnc |
| CapeOx | Has indication | Gastric cancer | HemOnc | HemOnc |
| CapeOx | Has indication | Hepatocellular carcinoma | HemOnc | HemOnc |
| CapeOx | Has indication | Pancreatic cancer | HemOnc | HemOnc |
| CapeOx | Has indication | Rectal cancer | HemOnc | HemOnc |
| CapeOx | Is current in | Colon cancer | HemOnc | HemOnc |
| CapeOx | Is current in | Esophageal cancer | HemOnc | HemOnc |
| CapeOx | Is current in | Gastric cancer | HemOnc | HemOnc |
| CapeOx | Is current in | Hepatocellular carcinoma | HemOnc | HemOnc |
| CapeOx | Is current in | Pancreatic cancer | HemOnc | HemOnc |
| CapeOx | Is current in | Rectal cancer | HemOnc | HemOnc |
| CapeOx | Can be preceded by | Colon cancer surgery | HemOnc | HemOnc |
| CapeOx | Can be preceded by | CAPIRI | HemOnc | HemOnc |
| CapeOx | Can be preceded by | Irinotecan monotherapy | HemOnc | HemOnc |
| CapeOx | Can be preceded by | Gastrectomy | HemOnc | HemOnc |
| CapeOx | Can be preceded by | Capecitabine monotherapy | HemOnc | HemOnc |
| CapeOx | Can be preceded by | Capecitabine and RT | HemOnc | HemOnc |
Use Cases
We suggest several use cases and examples illustrating the potential applications for the HemOnc OMOP CDM model:
For OMOP implementers with source systems that do not natively group drug clinical events into treatment regimen abstractions, the HemOnc regimen vocabulary can be used as the gold-standard oncology drug compendium to aid in the derivation of oncology drug regimens from available low-level clinical events (prescriptions and medical administration records). Example: Patient X has been administered or prescribed Drugs A, B, and C within the same time period. The vocabulary is searched for regimens that only contain Drug A AND Drug B AND Drug C; this narrows the regimen space to either a single regimen or a small group of possible regimens. As a gold-standard, the vocabulary could enable systematic efforts to identify patterns of chemotherapy treatment from structured or unstructured data.[13,14]
For OMOP implementers with source systems that do natively contain oncology drug treatment abstractions, the HemOnc regimen vocabulary can be used as the gold-standard oncology drug compendium for the mapping of oncology drug regimens to a standardized vocabulary. Example: two source systems have native regimen concepts, but they do not share common identifiers; HemOnc can be used as the bridge to join concepts from the two systems.
HemOnc can be used to map regimen acronyms and shorthand found in the natural language of clinical notes to formal regimen concepts. For example, the drug carfilzomib is often written as “carf” in the progress notes of multiple myeloma patients; multidrug regimens such as R-CHOP are rarely, if ever, written out in their constituent components in the medical record. On occasion, regimens are only referred to in clinical notes by the study in which they were evaluated (e.g., “EXTREME” instead of “Carboplatin, Fluorouracil, Cetuximab” [personal communication, Michael Gibson MD, PhD]).
Once the concepts are instantiated and captured at the practice level, patterns of care such as the utilization of pathways can be investigated. At the regional and national levels, patterns of care can be better captured by cancer registries and data aggregators. While the HemOnc vocabulary does not obviate the problem of conflicting information in source systems, it can highlight conflicts such as the many ways that the regimen “FOLFOX” is expressed across systems and practices (personal communication, Robert S. Miller MD, FACP, FASCO).
Discussion
The HemOnc vocabulary represents the most extensive effort in the public domain to date intended to capture the structure of chemotherapy regimens. Most of our effort has been focused on the transformations, resolutions of ambiguities and naming conflicts, and iterative improvements to the data model. As a new vocabulary artifact, HemOnc is a rich source of knowledge representation that could potentially meet multiple cancer phenotyping needs.
Throughout the process of creating the vocabulary, we learned several important lessons that could be broadly applicable to similar efforts of this kind. First: the use of a formal model can provide valuable guidance in clarifying and refining existing data descriptions. The process of developing the HemOnc model led to the identification of a number of inconsistencies and instances of incomplete data on the HemOnc.org website, which we were able to resolve through minor changes. Relatedly, the goals of developing machine-digestible ontologies and user-oriented website content are not necessarily completely in harmony. Mismatches between the capabilities of the MediaWiki software underlying HemOnc.org and the requirements of the ontology development process led to the need for post-processing. More extensive use of tools that explicitly address both semantic modeling and user-oriented content, such as the Semantic MediaWiki extensions, might reduce the need for post-processing. Third, iterative design should be expected. Our design processes iterated over almost 40 candidate tables over the 6+ month project, before arriving at a final set for the first release. Finally, the importance of interdisciplinary expertise cannot be underestimated; this work was done with the collaboration of clinicians, semantic modeling experts, process engineers, database analysts, and human-computer interface experts.
Despite its broad scope, the current vocabulary has several limitations which will inform its future development. Not all concepts on HemOnc.org have been parsed into structured format. For example, the signetur (“signa” or “sig”) for each drug, including dosage as well as route and timing of administration, remain as a complex problem. To date, the regimens listed on HemOnc.org are associated with nearly 5,000 distinct free-text sigs. Each sig is anchored by a structured component concept but is otherwise in free text; therefore, dosage, route, and timing of administration remain incompletely structured. Nevertheless, these details must be incorporated into the ontology so that regimens may be compared in a more granular way. The problem of unstructured sigs may be simplified by classifying all sigs into one of several archetypes and treating each archetype separately. For example, radiation therapy sigs are dissimilar in structure to medication therapy sigs; continuous infusions also have a distinct syntax. Once the basic syntax for an archetype is set, terminology must be standardized across sigs. In addition to syntax, some sigs may be subdivided into two or more sequential components (identified by “, then” or “followed by” or “as follows:”) that each require separate syntactic treatment. The issue of fitting temporal sequences into an ontology structure can then be addressed.[15]
The HemOnc vocabulary is informed primarily by prospective clinical trials and the context in which they are carried out. Notably, many of the unique concepts and relationships that capture these details (e.g., study groups and author names) were not included in the first public OHDSI release. However, these concepts have many applications outside of the retrospective scope of OHDSI. For example, understanding the evidence base that informs standard of care is important to clinicians, insurers, and clinical guideline developers. While some bibliometric data can be accessed directly through MEDLINE, we have augmented this resource e.g., with extensive author name disambiguation; we also chose to instantiate bibliometric relationships such as Reference Was published in Journal for speed and local optimization reasons. Some of these concepts and relationships may not be immediately relevant to the OHDSI user community and the decision on whether to include them in future public releases is the subject of ongoing discussion. Regardless, the full HemOnc vocabulary will be made available upon request by academic and noncommercial users under the existing CC BY-NC-SA 4.0 license [16]; in the future we anticipate that additional classes and relationships will be added to the public OHDSI vocabulary.
Our reliance on published chemotherapy regimens leads to at least two issues: 1) clinicians in the “real-world” may choose to use a regimen in a context other than that for which it was studied, for example using a first-line regimen as second-line; and 2) ad hoc regimens created outside the constraints of a clinical trial are not captured. Furthermore, none of the regimens on HemOnc.org contain investigational drugs, although there are some that contain drugs approved in realms other than the United States; extending the model to incorporate investigational regimens is a focus of future work.
Another challenge is that many complex roles in the vocabulary cannot be adequately expressed using binary relationships. Although the initial goal of the translation of the HemOnc.org regimen information to the OMOP CDM was to build support for key oncology concepts into OHDSI data models and tools, consideration of regimen information in the context of observational research also led to the identification of additional modeling challenges. Given the context and evolving nature of oncology regimens, the introduction of new treatments, the repurposing of existing treatments, and the resulting changes in prescribing practices, rich representations of how and when regimens are used are necessary to address many plausible use cases, particularly those that might address changes in regimen use over time.[17] Appropriate modeling of these complexities may require additional ontological structures capable of representing the richness of relationships between multiple factors.[18]
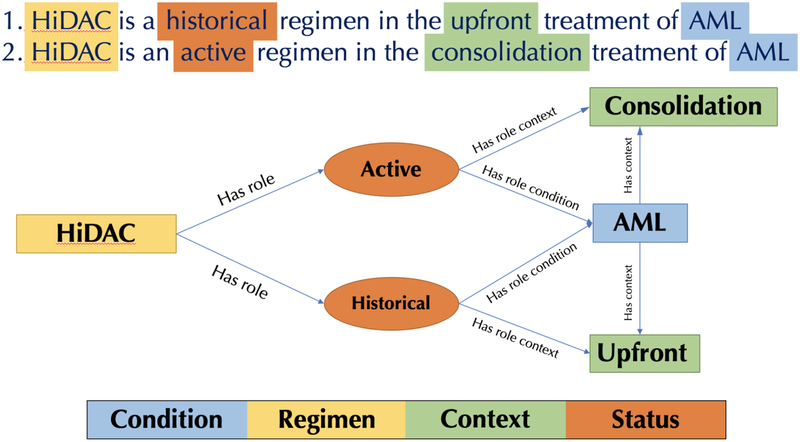
For example, consider a regimen that was used previously for upfront treatment of a given disease, but has been used more recently as a consolidation after (successful) upfront treatment. Representation of these changes will require models capable of indicating that the regimen was used in multiple ways, each of which with differing goals, even if for the same disease (Figure 3). Building on this example, a richer model might associate regimens with episodes (Figure 1) providing (approximate) dates indicating when those regimens were commonly used.
Figure 3.
An example of a ternary relationship in the vocabulary. In this case, the same regimen is either historical (outdated) or current for a single disease – acute myeloid leukemia (AML) – depending on the context in which it is used. One potential solution to this problem is to introduce an additional concept of the “role” that the regimen plays. An attribute of this role taking values of either “active” or “historical” might represent the temporal context of the use of the regimen in a particular context (in this case, consolidation vs. upfront) and disease.
Future extension of the HemOnc OMOP model to accommodate these complexities will be guided by tradeoffs between expressiveness and cost/complexity of modeling familiar to similar knowledge representation efforts. Representations driven by compelling, generalizable use cases will be most likely to be prioritized.
The first release of the HemOnc vocabulary went live on June 10, 2019. Going forward, we anticipate updates on a quarterly basis. Importantly, some concepts will be deprecated, whereas others will be merged or split, as is inevitable for controlled vocabularies. In the future, we plan to map procedures to SNOMED-CT, regimen types to North American Association of Central Cancer Registries (NAACCR) codes, and some regimens to NCIt, which is included in the Unified Medical Language System (UMLS). In conclusion, we look forward to community evaluation and use of this new vocabulary, and we anticipate that it will be a valuable addition towards the normalization and utilization of RWD in the oncology domain.
Supplementary Material
Highlights.
Formal representation of chemotherapeutic regimens is an unmet need
HemOnc.org content is the basis of the largest public regimen vocabulary to date
More than 1,500 regimens have been modeled and represented in OMOP format
A variety of use cases can be addressed with this new standard regimen vocabulary
Public releases will be made available in the ATHENA standardized vocabulary tool
Acknowledgments:
this work was supported in part by National Cancer Institute grants U01CA231840, U24CA184407, and U24CA194215. The funder had no role in the conception or approval of the project.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- [1].Siegel RL, Miller KD, Jemal A, Cancer statistics, 2019, CA Cancer J Clin. 69 (2019) 7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- [2].Kehl KL, Arora NK, Schrag D, Ayanian JZ, Clauser SB, Klabunde CN, Kahn KL, Fletcher RH, Keating NL, Discussions about clinical trials among patients with newly diagnosed lung and colorectal cancer, J Natl Cancer Inst. 106 (2014). doi: 10.1093/jnci/dju216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Khozin S, Blumenthal GM, Pazdur R, Real-world Data for Clinical Evidence Generation in Oncology, J Natl Cancer Inst. 109 (2017). doi: 10.1093/jnci/djx187. [DOI] [PubMed] [Google Scholar]
- [4].Rosenbloom ST, Carroll RJ, Warner JL, Matheny ME, Denny JC, Representing Knowledge Consistently Across Health Systems, Yearb Med Inform. 26 (2017) 139–147. doi: 10.15265/IY-2017-018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Warner JL, Cowan AJ, Hall AC, Yang PC, HemOnc.org: A Collaborative Online Knowledge Platform for Oncology Professionals, J Oncol Pract. 11 (2015) e336–50. doi: 10.1200/JOP.2014.001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Malty AM, Jain SK, Yang PC, Harvey K, Warner JL, Computerized Approach to Creating a Systematic Ontology of Hematology/Oncology Regimens, JCO Clinical Cancer Informatics. (2018) 1–11. doi: 10.1200/CCI.17.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, Suchard MA, Park RW, Wong ICK, Rijnbeek PR, van der Lei J, Pratt N, Norén GN, Li Y-C, Stang PE, Madigan D, Ryan PB, Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers, Stud Health Technol Inform. 216 (2015) 574–578. [PMC free article] [PubMed] [Google Scholar]
- [8].Voss J, Collaborative thesaurus tagging the Wikipedia way, ArXiv:Cs/0604036. (2006). http://arxiv.org/abs/cs/0604036 (accessed March 17, 2019).
- [9].R: The R Project for Statistical Computing, (n.d.). https://www.r-project.org/ (accessed February 25, 2019).
- [10].APIs, (n.d.). https://rxnav.nlm.nih.gov/APIsOverview.html (accessed February 25, 2019).
- [11].Schöfl G, reutils: Talk to the NCBI EUtils, 2016. https://CRAN.R-project.org/package=reutils (accessed February 25, 2019).
- [12].Porschen R, Arkenau H-T, Kubicka S, Greil R, Seufferlein T, Freier W, Kretzschmar A, Graeven U, Grothey A, Hinke A, Schmiegel W, Schmoll H-J, AIO Colorectal Study Group, Phase III study of capecitabine plus oxaliplatin compared with fluorouracil and leucovorin plus oxaliplatin in metastatic colorectal cancer: a final report of the AIO Colorectal Study Group, J. Clin. Oncol 25 (2007) 4217–4223. doi: 10.1200/JCO.2006.09.2684. [DOI] [PubMed] [Google Scholar]
- [13].Savova GK, Tseytlin E, Finan S, Castine M, Miller T, Medvedeva O, Harris D, Hochheiser H, Lin C, Chavan G, Jacobson RS, DeepPhe: A Natural Language Processing System for Extracting Cancer Phenotypes from Clinical Records, Cancer Res. 77 (2017) e115–e118. doi: 10.1158/0008-5472.CAN-17-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Carroll NM, Burniece KM, Holzman J, McQuillan DB, Plata A, Ritzwoller DP, Algorithm to Identify Systemic Cancer Therapy Treatment Using Structured Electronic Data, JCO Clin Cancer Inform. 1 (2017) 1–9. doi: 10.1200/CCI.17.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Styler WF, Bethard S, Finan S, Palmer M, Pradhan S, de Groen PC, Erickson B, Miller T, Lin C, Savova G, Pustejovsky J, Temporal Annotation in the Clinical Domain, Transactions of the Association for Computational Linguistics. 2 (2014) 143–154. doi: 10.1162/tacl_a_00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ontology | HemOnc.org - A Hematology Oncology Wiki, (n.d.). https://hemonc.org/wiki/Ontology (accessed May 22, 2019).
- [17].Abrams TA, Brightly R, Mao J, Kirkner G, Meyerhardt JA, Schrag D, Fuchs CS, Patterns of adjuvant chemotherapy use in a population-based cohort of patients with resected stage II or III colon cancer, J Clin Oncol. 29 (2011) 3255–62. doi: 10.1200/JCO.2011.35.0058. [DOI] [PubMed] [Google Scholar]
- [18].Defining N-ary Relations on the Semantic Web, (n.d.). https://www.w3.org/TR/swbp-naryRelations/ (accessed February 23, 2019).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.