SUMMARY
Triple-negative breast cancer (TNBC) is an aggressive and heterogeneous disease that lacks clinically actionable genetic alterations that limit targeted therapies. Here we explore a systems pharmacology approach that integrates drug-target networks and large-scale genomic profiles of TNBC and identify wogonoside, one of the major active flavonoids, as a potent angiogenesis inhibitor. We validate that wogonoside attenuates cell migration, tube formation, and rat aorta microvessel outgrowth, and reduces formation of blood vessels in chicken chorioallantoic membrane and TNBC cell-induced matrigel plugs. In addition, wogonoside inhibits growth and angiogenesis in TNBC-cell xenograft models. This network-based approach predicts, and we empirically validate, wogonoside’s anti-angiogenic effects resulting from VEGF secretion. Mechanistically, wogonoside inhibits Gli1 nuclear translocation and transcriptional activities associated with Hedgehog signaling, by promoting Smoothened degradation in a proteasome-dependent mechanism. This study offers a powerful, integrated, systems pharmacology-based strategy for oncological drug discovery and identifies wogonoside as a potential TNBC angiogenesis inhibitor.
Keywords: angiogenesis, hedgehog signaling, Smoothened, triple-negative breast cancer, wogonoside, systems pharmacology
Graphical Abstract
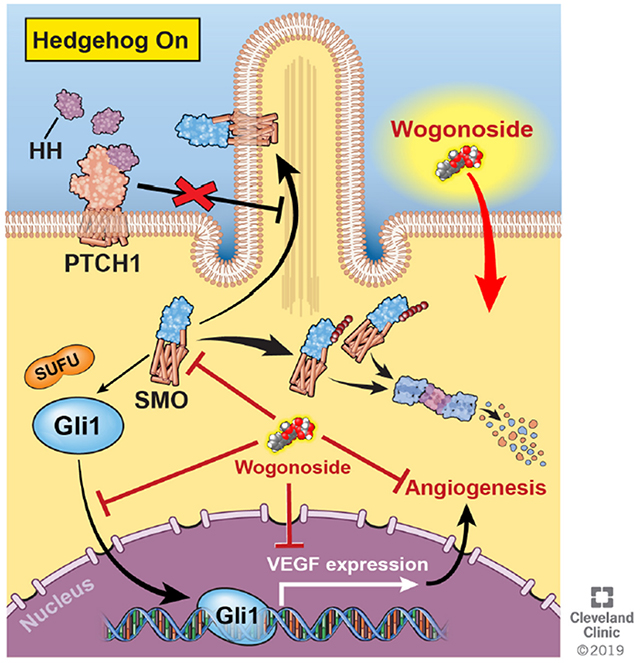
eTOC Blurb
Huang et al., introduce a systems pharmacology approach and identify wogonoside as an effective angiogenesis inhibitor in triple-negative breast cancer (TNBC) in vitro and in vivo. Mechanistically, the authors demonstrate that wogonoside inhibits the Gli1 nuclear translocation and transcription activities of Hedgehog signaling via specifically promoting ubiquitination-dependent degradation of Smoothened.
INTRODUCTION
Triple-negative breast cancer (TNBC), characterized by absence or minimal expression of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2), is an aggressive and heterogeneous subtype that constitutes 15-20% of breast cancer patients (Penault-Llorca and Viale, 2012). The development of targeted therapies for TNBC has been challenging due to the unexpected genomic heterogeneity and lack of consistently recurrent genetic alterations (Bareche et al., 2018; Bianchini et al., 2016). Neoadjuvant chemotherapy including a combination of Taxanes (mitotic inhibitors) and anthracyclines (DNA intercalators) remains standard of care in non-metastatic TNBC, since neither endocrine therapies nor HER2-targeted agents are effective (Bianchini et al., 2016; Foulkes et al., 2010; Lehmann and Pietenpol, 2014). While chemotherapy is only effective in a small population of TNBC patients, long-term survival is poor due to high rate of relapse and 30-50% of patients acquiring resistance (Foulkes et al., 2010). Therefore, there is an urgent need for effective therapeutic strategies for TNBC, ones which surmount the genomic heterogeneity of patients with TNBC (Cheng et al., 2019b).
Angiogenesis is an essential step for tumor development, which is principally initiated by chemoattractant and proliferative cytokines such as vascular endothelial growth factor (VEGF) (Carmeliet and Jain, 2011). Previous studies have shown that TNBC possesses high microvessel density (Mohammed et al., 2011) and VEGF amplification compared to non-TNBC (Andre et al., 2009) such that patients with TNBC have significantly higher intratumoral VEGF levels and shorter recurrence-free survival (Linderholm et al., 2009). So far, 11 anti-VEGF drugs were approved for multiple cancer types, either alone or in combination with various cytotoxic chemotherapies or targeted therapies (Zirlik and Duyster, 2018). For example, anti-VEGF monoclonal antibody (mAb) bevacizumab has improved the response rate in several clinical trials (Earl et al., 2015; von Minckwitz et al., 2012; von Minckwitz et al., 2014a). However, antibody-based anti-angiogenic therapies have a limited effect on overall survival of cancer patients and inhibition of the VEGF signaling pathway is not effective in most cancer types. Thus, development of anti-angiogenic therapies, especially small molecules with low toxicity, is a pressing need for highly targeted therapies in TNBC. While identification of agents (e.g., angiogenesis inhibitors) with ideal pharmacokinetics/pharmacodynamics is important, traditional experimental approaches are costly and often show lack efficacy in vivo.
The Hedgehog signaling pathway plays a crucial role in cell proliferation and differentiation by regulating vascular formation in early embryonic development (Pak and Segal, 2016). The binding of Hedgehog ligands to Patched (PTCH), a 12 pass transmembrane protein inhibiting membrane translocation of Smoothened (SMO), results in the activation of glioma-associated oncogene homolog proteins (Gli), a key protein regulating cellular cycle and apoptosis (Hui and Angers, 2011). Recently, regulation of the mesenchymal marker expression and epithelial-mesenchymal transition of TNBC cells showed that the expression of SMO and Gli1 is significantly elevated in TNBC (Tao et al., 2011). In addition, increased blood vessel density in breast cancer is observed upon activation of the Hedgehog signaling pathway (Harris et al., 2012), and Gli enhances the vascular endothelial growth factor A (VEGFA) gene promoter resulting in the upregulation of VEGFA in breast cancer cells (Cao et al., 2012). Thus, Hedgehog signaling inhibitors present opportunities for potential therapeutic strategies in TNBC.
Recent advances in systems biology and omics technologies have enabled the development of in silico - network-based (Cheng et al., 2018; Cheng et al., 2019a; Cheng et al., 2012b), genomics-based (Jahchan et al., 2013; Jin et al., 2012; Lee et al., 2012), and systems pharmacology-based approaches to drug discovery (Fang et al., 2017a). In this study, we applied a systems pharmacology-based methodology, in order to identify consistent, therapeutic agents to treat TNBC. Using this integrated computational and experimental approach, we predicted wogonoside, a bioactive flavonoid extracted from the root of Scutellaria baicalensis Georgi, as an effective angiogenesis inhibitor in TNBC. We experimentally validated the prediction, and critically, then via a network-based approach predicted and tested its mechanism in TNBC, showing that wogonoside blocks Hedgehog signaling in vitro and in vivo.
RESULTS
Systems Pharmacology-based Prediction of anti-TNBC Agents from Natural Products
To identify potential TNBC therapeutic agents, we employed a systems pharmacology approach that quantifies the therapeutic potential of natural products for TNBC by integrating the known drug-target interactions, as well as disease genomic and genetic profiles. We hypothesized that a natural promiscuous (“multi-target”) drug is a candidate to treat TNBC if its targets are more likely to be functional gene products (proteins) of TNBC (Fang et al., 2017c; Jiang et al., 2018). The null hypothesis asserts that natural products randomly target TNBC functional proteins across the human proteome. A permutation testing was performed to calculate the statistical significance of a natural product to be prioritized in treating TNBC. Then, the nominal P-values from the permutation tests were corrected as adjusted P-values (q) using the Benjamini-Hochberg approach (Benjamini and Hochberg, 1995). Subsequently, a Z-score (z) was calculated for each natural product to be prioritized in treating TNBC during permutation testing:
| (1) |
where x is the observed number of TNBC proteins targeted by a given natural product, μ is the mean number of TNBC proteins targeted by a given natural product during 100,000 permutations, and σ is the standard deviation.
We computed a list of candidate natural products (see Methods) ranked by increasing z-score through searching 150 known TNBC proteins (Data S1) derived from large-scale genetic and genomic studies within a network of 38,220 compound-protein interactions (Data S2). To evaluate the performance of the computational model, we retrieved published indications from PubMed for 706 natural products predicted to be anti-TNBC (Data S1). The area under the receiver operating characteristic curve is 71.9%, suggesting a reasonable accuracy in predicting therapeutic agents in treating TNBC (Figure 1A).
Figure 1. A diagram illustrating systems pharmacology-based prediction of anti-tumor effects of wogonoside in triple-negative breast cancer (TNBC).
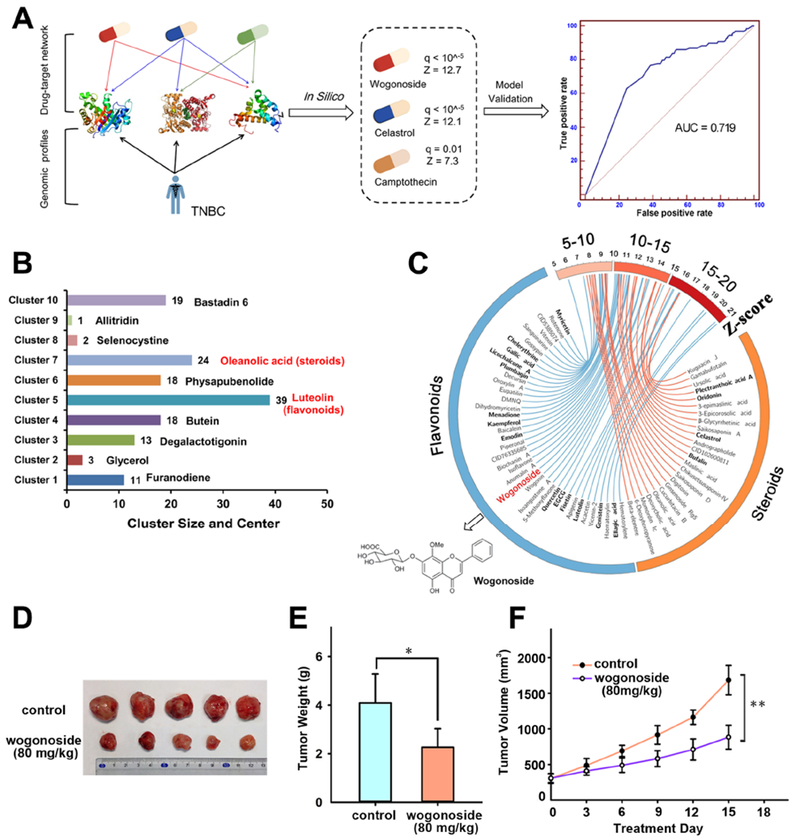
(A) An in silico model for predicting anti-TNBC indications for natural products by integrating the drug-target network and experimentally validated functional genes in TNBC. The performance of the in silico model was evaluated using Receiver Operating Characteristic (ROC) curve. The area under ROC curve (AUC) was shown. (B) The chemical structure clustering analysis for 148 natural products having the significantly predicted anti-TNBC indications (q<10−5). (C) Circos plot (Circo v0.69) representing the predicted anti-TNBC indications (q<10−5) for 24 steroids and 39 flavonoids. The predicted q values with corresponding Z-score are exhibited as connected lines (edges). Natural products with previously published experimental data in TNBC are highlighted in bold font. Wogonside was selected for experimental validation using subject matter expertise based on a combination of factors (see Data S1 for references). (D) Effect of wogonoside on MDA-MB-231 tumor growth in nude mice. (E) The tumor weight of control and wogonoside treatment group (n=7) was measured after 15 days treatment. (F) The tumor volume of control and wogonoside treatment group (n=7) was measured every three days. The comparisons were made relative to the control group and the significance of the difference is indicated as *P value < 0.05 and **P value < 0.01.
Among the 706 natural products, we computationally identified 148 as significantly anti-TNBC (adjusted p-value [q] <10−5). Chemical clustering groups them into 10 clusters. Cluster 5 (named flavonoids, n=39) is the largest (Figure 1B). Figure 1C displays the predicted anti-TNBC data for 39 flavonoids. We next used subject matter expertise based on: (i) removing flavonoids with potential cytochrome p450 (CYP450) inhibition (CYP450 inhibitors involving in the high risk of adverse drug-drug interactions (Lynch and Price, 2007) or having preclinical toxicity predicted by admetSAR (Cheng et al., 2012a); (ii) novelty of prediction through exclusion of flavonoids with literature-reported anti-TNBC effects; and (iii) strength of the systems pharmacology-based prediction (a higher z-score in Data S1). Applying these criteria resulted in wogonoside (q<10−5, z=12.7), one of the major active flavones in Scutellaria baicalensis Georgi, as the best candidate (Data S1). We thus selected wogonoside and tested its anti-tumor effect in TNBC.
A nude mouse model bearing transplanted MDA-MB-231 tumor was used to evaluate anti-TNBC effect of wogonoside (Figure 1D) in vivo. The resected tumor weight was reduced to 55.47% by wogonoside (80 mg/kg) compared with the untreated group (Figure 1E). In addition, wogonoside significantly suppresses growth of MDA-MB-231 cell xenografts (Figure 1F), suggesting that wogonoside restrains the growth of transplanted MDA-MB-231 tumor in nude mouse model.
Wogonoside Inhibits the Expression of VEGF
To explore the mechanism-of-action of wogonoside in TNBC, MTT cell viability assay was performed. However, we found that wogonoside had no effect on the viability of both MDA-MB-231 and MDA-MB-468 cells after 24 h pretreatment (Figure S1). To further investigate the potential molecular mechanism, we integrated 13 literature-reported TNBC gene-encoding proteins targeted potentially by wogonoside in breast-specific co-expressed human protein-protein interactome network (see Methods). The network was derived from RNA-sequencing profiles across 32 human tissues in the Genotype-Tissue Expression (GTEx) database (Consortium, 2015). Network analysis (Cheng et al., 2018) shows that wogonoside potentially targets VEGFA in TNBC (Figure 2A) consistent with our previous study that wogonoside decreases the secretion of VEGF in MCF-7, an estrogen positive breast cancer cell line (Huang et al., 2016). RNA-sequencing data from the GTEx database (Consortium, 2015) indicates that VEGFA is highly expressed in breast tissue.
Figure 2. Effects of wogonoside on VEGF secretion, protein and mRNA expression and transcriptional activity in MDA-MB-231 and MDA-MB-468 cell lines.
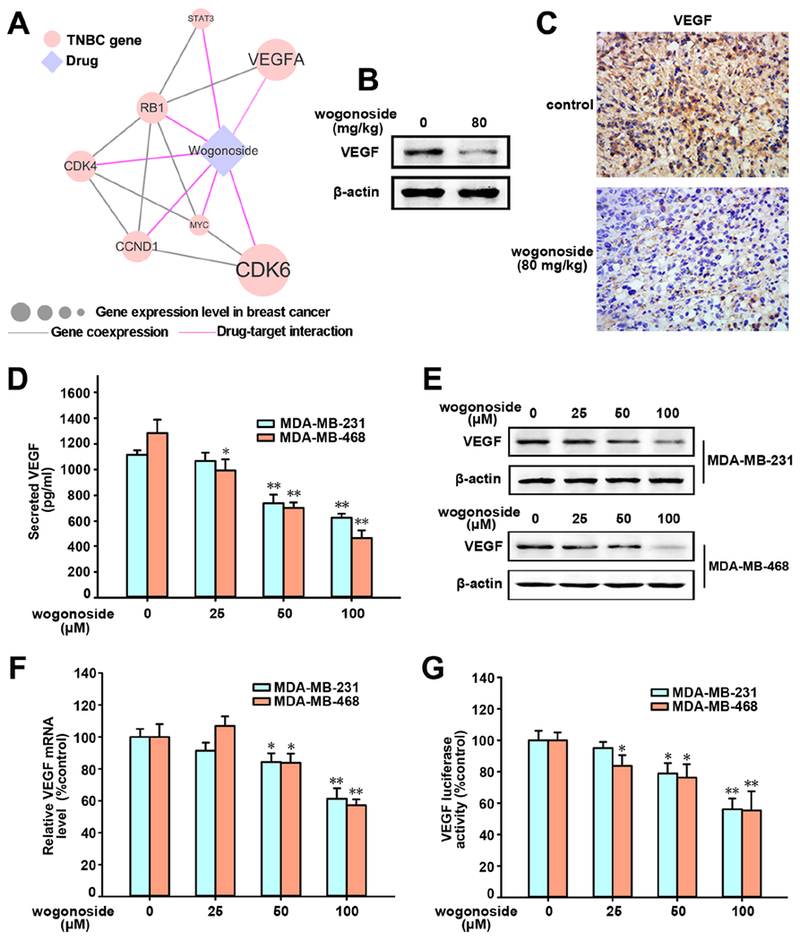
(A) Network analysis highlighting the inferred mechanism-of-action for wogonoside in TNBC. The potential molecular mechanisms of wogonoside against TNBC were investigated via integration of known drug targets and experimentally validated TNBC genes into tissue-specific co-expressed protein-protein interactome network (see Methods). Node size indicates the protein-coding gene expression level in breast comparing to other 31 tissues from GTEx database (Consortium, 2015). Larger size highlighting the high expression level in breast comparing to other tissues. Co-expression denotes the co-expressed gene pairs (p-value < 0.05, F-statistics) encoding protein-protein interactions in TNBC RNA sequencing data from The Cancer Genome Atlas database (Cancer Genome Atlas, 2012). (B) Effect of wogonoside on VEGF expression in xenograft model of MDA-MB-231 cells in nude mice (n=5) was detected by Western blot analysis. (C) The expression of VEGF in xenograft model was detected by immunochemistry using specific antibody. (D) The concentration of VEGF in MDA-MB-231 and MDA-MB-468 CM were measured by ELISA kits. (E) VEGF expression was detected by western blot analysis using specific antibodies. (F) The mRNA level of VEGF was investigated by RT-PCR. (G) VEGF transcriptional activity was tested by Dual-Luciferase reporter assay. The comparisons were made relative to the control group and the significance of the difference is indicated as *P value < 0.05 and **P value < 0.01. See also Figures S1 and S4.
We next inspected whether wogonoside inhibits VEGF expression in TNBC using the transplanted MDA-MB-231 tumors. Western blot analysis (Figure 2B) reveals that wogonoside reduces VEGF expression of mouse tumor tissues, which is further confirmed by the immunohistochemical (IHC) staining assay (Figure 2C). We further evaluated the effect of wogonoside on VEGF expression in TNBC cells by ELISA (enzyme-linked immunosorbent assay). Figure 2D reveals that VEGF secretion in MDA-MB-231 and MDA-MB-468 cells are significantly reduced by wogonoside. Western blot assays show that wogonoside inhibits VEGF protein expression in a concentration-dependent manner (Figure 2E) as well. In addition, RT-PCR (Figure 2F) and luciferase reporter assays (Figure 2G) reveal that VEGF mRNA level and VEGF promoter activity are suppressed by wogonoside in both MDA-MB-231 and MDA-MB-468 cell lines. Altogether, wogonoside inhibits VEGF expression in both mouse model (Figures 2B and 2C) and human MDA-MB-231 and MDA-MB-468 cell lines (Figures 2D–2G).
Wogonoside Inhibits the Hedgehog Signaling Pathway
The activation of the Hedgehog/Gli signaling pathway is significantly increased in TNBC cells and Gli1 regulates the VEGFA gene promoter directly (Cao et al., 2012; Tao et al., 2011). We therefore examined the effects of wogonoside on Hedgehog/Gli signaling. The immunohistochemical (IHC) staining assay reveals that the nuclear translocation of Gli1 is inhibited by wogonoside (Figure 3A) in vivo. Western blot analysis further confirms that wogonoside reduces Gli1 expression in the nucleoplasm of mouse tumor tissues (Figure 3B).
Figure 3. Effects of wogonoside on Hedgehog signaling pathway.
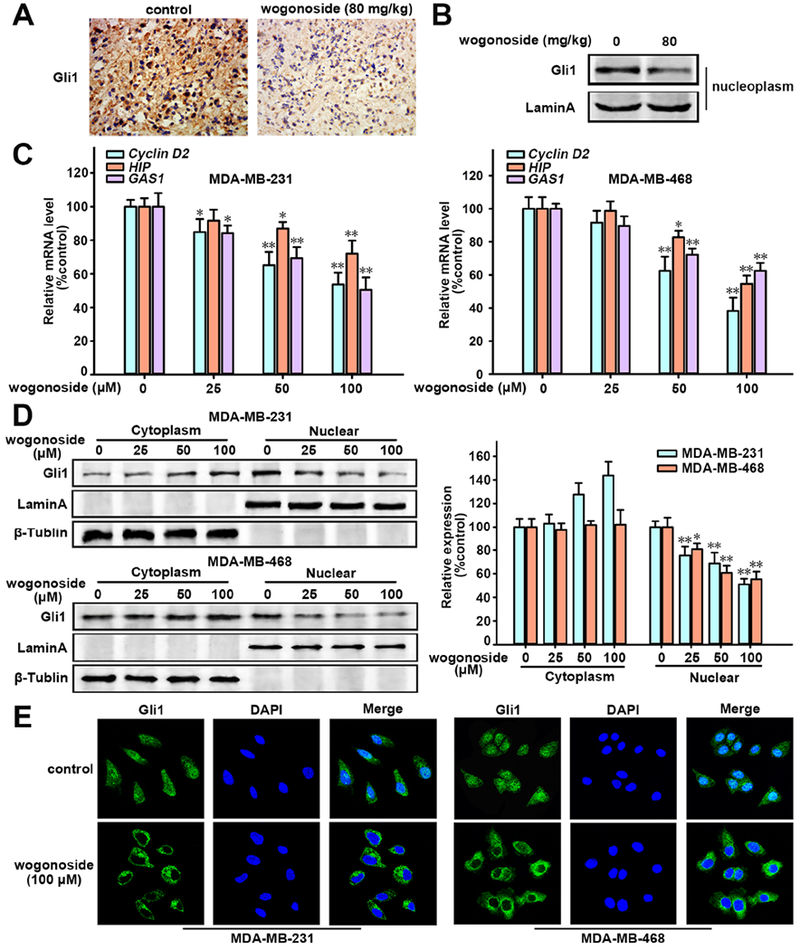
(A) Effect of wogonoside on Gli1 expression in xenograft tissue of MDA-MB-231 cells in nude mice was detected by immunochemistry. (B) Gli1 expression in nucleoplasm of MDA-MB-231 cell xenograft tissue was detected by Western blot analysis using specific antibodies and Lamin A was used as nuclear marker. (C) The mRNA level of Cyclin D2, HIP and GAS1 was detected by RT-PCR. (D) Western blot analysis of Gli1 expression in cytosolic and nuclear lysates. Lamin A and β-tublin were used as nuclear and cytoplasmic markers, respectively. (E) Gli1 nuclear tanslocation was analyzed by immunofluorescence confocal microscopy. The comparisons were made relative to the control group and the significance of the difference is indicated as *P value < 0.05 and **P value < 0.01. See also Figure S2.
Furthermore, the Gli luciferase reporter assay shows that the transcriptional activity of Gli signaling is inhibited by wogonoside (IC50 = 22.17 μM, Figure S2A) in both MDA-MB-231 and MDA-MB-468 cell lines. In addition, the mRNA levels of three main target genes of Gli1 (including Cyclin D2, HIP and GAS1) are decreased by wogonoside (Figure 3C). As the cytoplasmic accumulation and intracellular localization of Gli1 are essential for activating the transcription of target genes (Briscoe and Therond, 2013), we tested Gli1 expression in cytosolic and nuclear lysates by western blot analysis. We found that wogonoside decreased Gli1 levels in both MDA-MB-231 and MDA-MB-468 cell nucleus in a concentration-dependent manner (Figure 3D). In addition, immunofluorescence (Figure 3E) shows that the nuclear translocation of Gli1 in MDA-MB-231 and MDA-MB-468 cell lines is inhibited by wogonoside. Collectively, wogonoside inhibits Gli1 nuclear translocation and transcriptional activity of the Hedgehog signaling pathway.
Wogonoside Inhibits the Expression of SMO via Promoting its Ubiquitination
SMO is a key upstream activator involved in the Hedgehog signaling pathway (Cannonier and Sterling, 2015; Stanton and Peng, 2010). To explore how wogonoside affect Gli1 nuclear translocation, we tested protein expression of SMO in whole cell lysate. In vivo assay reveals that wogonoside reduces the SMO expression, confirmed by immunohistochemical (IHC) staining (Figure 4A) and Western blot assay (Figure 4B). Moreover, we found that wogonoside down-regulated the expression of SMO in a concentration-dependent manner (Figure 4C), without affecting Gli1 in both MDA-MB-231 and MDA-MB-468 cell lines. To explore the mechanism of the down-regulating of SMO expression by wogonoside in depth, we examined the transcription level of SMO. Interestingly, the mRNA levels of SMO in both MDA-MB-231 and MDA-MB-468 cell lines are not changed by wogonoside (Figure S2B). We therefore turned to inspect wogonoside’s effects by focusing on SMO protein expression.
Figure 4. Effects of wogonoside on the expression of SMO.
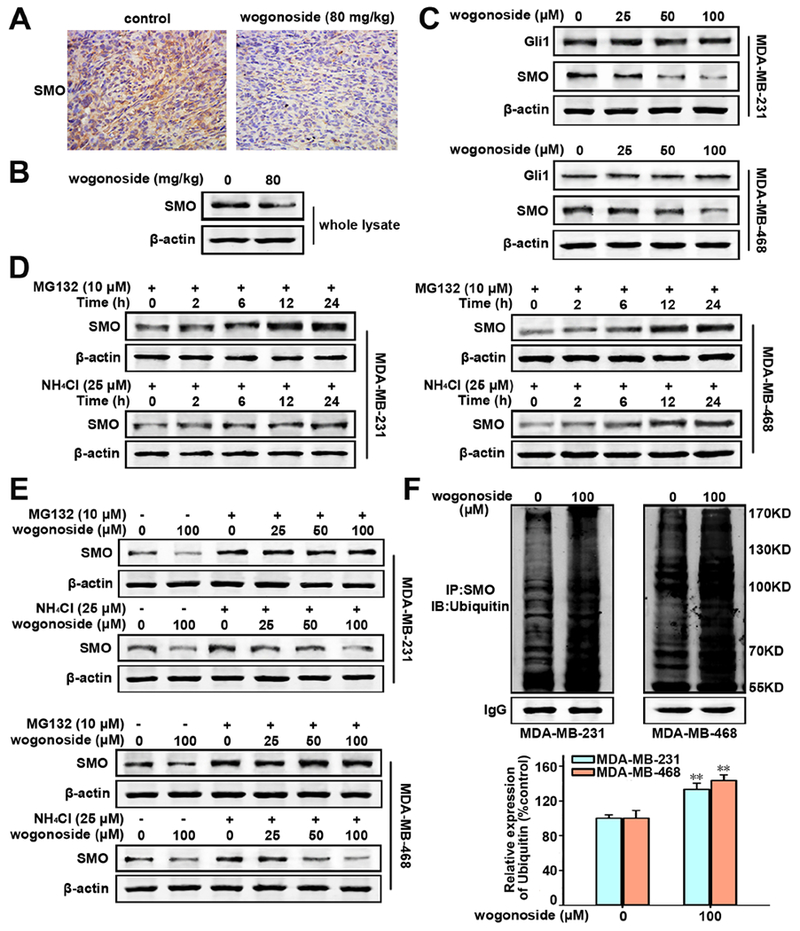
(A) Effect of wogonoside on SMO expression in xenograft tissue of MDA-MB-231 cells in nude mice was detected by immunochemistry. (B) The total SMO expression in the whole lysate of MDA-MB-231 cell xenograft tissue was tested by Western blot. (C) The total Gli1 and SMO expression in MDA-MB-231 and MDA-MB-468 cell lines was tested by Western blot. (D) The expression of SMO in MDA-MB-231 and MDA-MB-468 cells treated with MG132 (10 μM) or NH4Cl (25 mM) for different time points (0, 2, 6, 12 and 24 h) by Western blot. (E) The expression of SMO in cells treated with MG132 (0 or 10 μM) or NH4Cl (0 or 25 mM) and wogonoside (0, 25, 50 and 100 μM) as indicated was investigated by western blot. (F) SMO ubiquitination was tested by protein immunoprecipitation assay. The comparisons were made relative to the control group and the significance of the difference is indicated as *P value < 0.05 and **P value < 0.01.
It has been reported that Drosophila SMO is degraded via both lysosome- and proteasome-dependent mechanisms (Li et al., 2012; Zhou et al., 2017). As expected, the protein level of SMO is significantly increased when both MDA-MB-468 and MDA-MB-231 cell lines were treated with either MG132, a proteasome inhibitor, or NH4Cl, a lysosome inhibitor, in a time-dependent manner (Figure 4D). These observations suggest that SMO is also degraded by both lysosome- and proteasome-dependent manner in TNBC cells. We then treated TNBC cells by wogonoside with MG132 (10 μM) or NH4Cl (25 mM) at the same time. Figure 4E shows that the SMO reduction by wogonoside in MDA-MB-231 and MDA-MB-468 cell lines were both abrogated by adding MG-132. However, the expression of SMO still decreased at different wogonoside concentrations (25, 50 and 100 μM), when NH4Cl was added. In addition, wogonoside (100 μM) promotes the ubiquitination of SMO in both MDA-MB-231 and MDA-MB-468 cell lines, as indicated by multiple co-immunoprecipitation experiments followed by Western blot assays (Figure 4F). In summary, wogonoside down-regulates SMO expression by promoting its proteasome degradation.
Wogonoside Binding to SMO Promotes its Ubiquitination
It has been reported that Smurf-family of E3 ubiquitin ligases and Cul4-DDB1 are involved in the ubiquitination of SMO in Drosophila melanogaster (Li et al., 2018a; Li et al., 2018b). We therefore investigated whether Cul4-DDB1 is involved in human SMO degradation triggered by wogonoside. Firstly, we silenced the expression of Cul4A in MDA-MB-231 cells by specific siRNAs (Figure 5A). Endogenous SMO protein was then immunoprecipitated and detected by ubiquitin antibody using western blotting. We found that wogonoside significantly increased the SMO ubiquitination (Figure 5B). Notably, siRNA-mediated downregulation of Cul4A effectively impairs wogonoside-induced SMO ubiquitination in MDA-MB-231 cells. These observations indicate that Cullin4 is a potential E3 ligase for human SMO, consistent with ubiquitination regulation of SMO in Drosophila melanogaster.
Figure 5. A physical interaction between wogonoside and SMO.
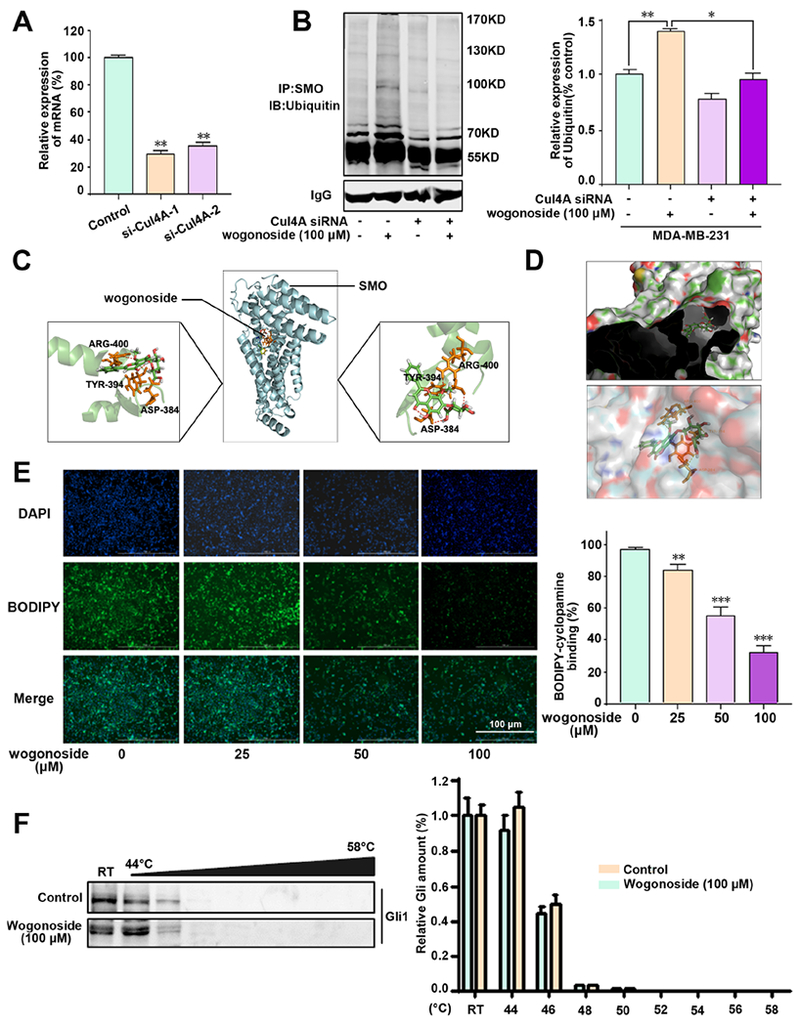
(A) The relative expression of mRNA of Cul4A in MDA-MB-231 cells following specific siRNA treatment. (B) SMO ubiquitination was determined by protein immunoprecipitation assay. Cul4A was silenced in MDA-MB-231 cells by a specific siRNA. MDA-MB-231 cells were then treated with wogonoside (100 μM) for 24 h and SMO protein was immunoprecipitated and detected by ubiquitin antibody using western blotting. Relative expression of SMO ubiquitin (% control) was shown in right. (C) The binding mode of wogonoside with SMO by molecular docking simulation (see Methods). (D) The aromatic ring branched chain of wogonoside stretched into the hydrophobic pocket consisted of Asp384, Ser385, Val386, Ser387, Gly388, Ile389, Cys390, Phe391, Val392, Gly393, Tyr394 and Arg477. (E) A physical interaction of wogonoside and SMO was detected by BODIPY-cyclopamine by SMO binding assay. The quantitative data of BODIPY-cyclopamine binging was shown in right of panel E. (F) MDA-MB-231 cells were treated with DMSO or wogonoside (100 μM) for 12 h and then subjected to Cellular Thermal Shift Assay (CETSA). CETSA shows that wogonoside has no effect on stabilizing Gli protein in MDA-MB-231 cells. The comparisons were made relative to the control group and the significance of the difference is indicated as **P value < 0.01 and ***P value < 0.001. See also Figure S2.
Small molecule modulators inhibit the Hedgehog signaling pathway by binding to Gli directly, or by targeting SMO to suppress Gli indirectly (Hui et al., 2013; Wang et al., 2013). To explore how wogonoside promotes the proteasome degradation of SMO, we first performed molecular docking simulation to investigate potential interactions of wogonoside with SMO or Gli1. This suggested a strong interaction between wogonoside and SMO, including hydrogen bonds involving Arg515 and Ser456 (Figure 5C), and aromatic ring docking into the hydrophobic pocket consisting of Glu380, Cys381, Ala382, Trp383, Leu384, Glu385, Ser456, Gly457, Tyr459, Thr460, Arg515, Ser518, Asn519 and Met522 (Figure 5D), while a weak interaction between wogonoside and Gli1 (Figure S2C).
To investigate the interaction between wogonoside and SMO/Gli further, SMO binding and cellular thermal shift assays (CETSA) were performed. BODIPY-cyclopamine, a fluorescent derivative of cyclopamine, specifically binding SMO-expressing cells, was used for SMO binding assay (Chen et al., 2002). We found that wogonoside significantly reduced BODIPY-cyclopamine binding to human SMO in a dose-dependent manner (Figure 5E), indicating a direct interaction between wogonoside and SMO by binding with the cyclopamine pocket. The CETSA assay monitors and quantifies the extent to which a drug reaches and directly binds to a protein of interest within cells by quantifying the changes in the thermal stability of proteins upon ligand binding (Jafari et al., 2014; Martinez Molina et al., 2013). In CETSA, the heat challenge results in Gli protein degradation due to unfolding, while wogonoside has no effect on Gli protein stability in MDA-MB-231 cells (Figure 5F). Taken together, wogonoside is a potential, direct SMO inhibitor in TNBC cells, by specifically promoting ubiquitination-dependent degradation of SMO.
Wogonoside Inhibits Angiogenesis Induced by TNBC Cells
It has been reported that SMO overexpression is involved in the development of TNBC and the Hedgehog/Gli signaling pathway regulates the expression of VEGF (Cao et al., 2012; Tao et al., 2011) by accelerating the proliferation, migration and invasion of vascular endothelial cells (Ferrara et al., 2003). We therefore examined the effects of wogonoside on angiogenesis induced by TNBC cells. We incubated human umbilical vein endothelial cells (HUVECs) in TNBC cell-conditioned medium (MDA-MB-231 CM or MDA-MB-468 CM). The matrigel invasion assay indicates that CM collected from MDA-MB-231 or MDA-MB-468 cells without wogonoside treatment stimulated apparent migration and invasion of HUVECs (Figure S3A). Upon exposure to CM collected from TNBC cells treated with wogonoside (25, 50 and 100 μM), the number of invasive HUVECs decreased in a concentration-dependent manner. Thus, wogonoside down-regulates the expression and transcription of VEGF in TNBC cells, as well as the migration and invasion ability of HUVECs induced by TNBC cell CM.
We next examined the effect of wogonoside on angiogenesis in TNBC cells. The tube formation assay shows that elongated and robust tube-like structures and the number of tubes of HUVECs are markedly inhibited by wogonoside compared with MDA-MB-231 CM or MDA-MB-468 CM stimulated group (Figure 6A). In addition, the rat aortic ring assay, which could mimic several key stages of angiogenesis, including endothelial cell proliferation, migration and tube formation, was also performed. We found that the vessel sprouting of rat aortic ring was significantly stimulated by MDA-MB-231 CM or MDA-MB-468 CM compared with the control group (Figure 6B). As a result, the new microvessel growth from aortic rings was inhibited by wogonoside in both MDA-MB-231 and MDA-MB-468 cell lines. After injecting MDA-MB-231 cells or MDA-MB-468 cells in the chicken embryos, the CAM assay indicated that more new blood vessels formed in the TNBC cell group compared with the control group (Figure 6C). We found that wogonoside (50, 100 and 200 ng/CAM) inhibited the growth of the newly formed blood vessels (Figure 6C). In summary, wogonoside inhibits angiogenesis induced by MDA-MB-231 and MDA-MB-468 cells.
Figure 6. Effects of wogonoside on TNBC cell-induced angiogenesis in vitro.
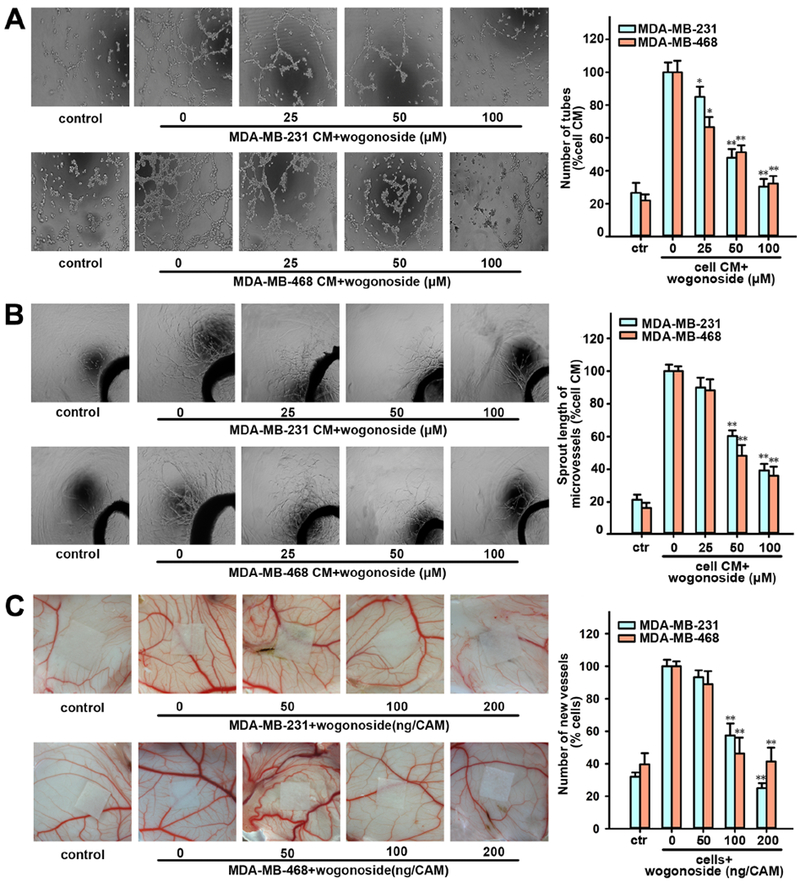
(A) The tube formation ability of HUVECs cultured by CM collected from MDA-MB-231 or MDA-MB-468 pretreated with wogonoside (0, 25, 50 and 100 μM) for 24 h and 1% FBS medium in control group as indicated was tested by the endothelial cell tube formation assay. (B) The rat aortic ring microvessel sprouting induced by CM collected from MDA-MB-231 or MDA-MB-468 pretreated with wogonoside (0, 25, 50 and 100 μM) for 24 h and 1% FBS M199 medium in the control group as indicated was tested by rat aortic ring assay. (C) Effect of wogonoside on the angiogenesis of chicken chorioallantoic membrane. Data are presented as mean±SD. The comparisons were made relative to MDA-MB-231 or MDA-MB-468 CM group and significance of difference is indicated as *P value < 0.05 and **P value < 0.01. See also Figure S3.
Wogonoside Inhibits Angiogenesis of TNBC in vivo
We further investigated the effect of wogonoside on TNBC cell-induced angiogenesis in vivo. The matrigel plug assay shows that wogonoside (80 mg/kg) suppresses the formation of blood vessels compared to the MDA-MB-231 or MDA-MB-468 cells-injected group (Figure 7A). Furthermore, the whole-mount of CD31 staining shows that wogonoside significantly reduces the vascular density in matrigel plugs compared with MDA-MB-231 or MDA-MB-468 cells-injected group as well (Figure 7B). Specifically, the hemoglobin concentration in matrigel plugs is reduced by 66.3±1.3% and 57.3±5.3% with the treatment of wogonoside in MDA-MB-231 and MDA-MB-468 cell-injected groups, respectively (Figure 7C). In addition, histological analysis of sections stained with CD31 (an endothelial-specific marker) shows that wogonoside reduces the vascular density in tumors compared with the control group (Figure 7D). Taken together, wogonoside inhibits angiogenesis induced by MDA-MB-231 and MDA-MB-468 cells in vivo.
Figure 7. Effects of wogonoside on angiogenesis in vivo.
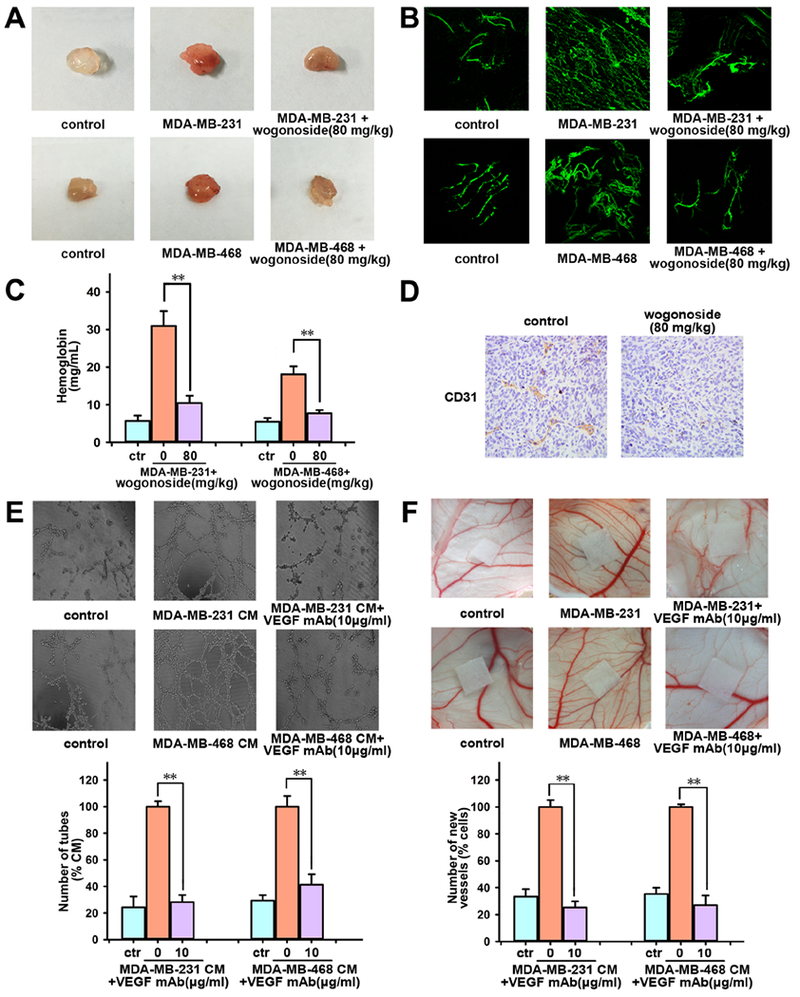
Matrigel containing saline injection (control group) or MDA-MB-231 cells and MDA-MB-468 cells was injected subcutaneously to assess angiogenesis in vivo. (A) Macroscopic appearance of matrigel plugs isolated from each group of mice (n=5). (B) The whole-mount of CD31 staining was viewed by laser scanning confocal microscope. (C) The hemoglobin content in matrigel plugs was determined. (D) The expression of CD31 in MDA-MB-231 cells xenograft in nude mice was tested by immunochemistry. (E) Effect of VEGF neutralizing antibody (10 μg/ml) on the tube formation of HUVECs induced by CM from MDA-MB-231 or MDA-MB-468 cells was tested by endothelial cell tube formation assay. (F) Effect of VEGF neutralizing antibody (10 μg/ml) on the enhanced angiogenesis induced by MDA-MB-231 cells or MDA-MB-468 cells (1×106 cells/CAM) was tested by chicken chorioallantoic membrane (CAM) assay. The comparisons were made relative to MDA-MB-231 or MDA-MB-468 cells group and the significance of the difference is indicated as *P value < 0.05 and **P value < 0.01.
We next used VEGF neutralizing antibody (VEGF mAb) to determine whether the VEGF inhibition by wogonoside contributes to suppressing angiogenesis in TNBC. VEGF mAb (10 μg/ml) markedly reduces the number of HUVECs tubes stimulated by MDA-MB-231 CM or MDA-MB-468 CM (Figure 7E) compared with the CM treated group. In addition, the newly formed blood vessels in CAM decreased after adding VEGF mAb as well (Figure 7F). VEGF is primarily secreted by tumor cells and specifically activates VEGFR2 (vascular endothelial growth factor receptor 2), a membrane receptor mainly expressed on endothelial cells (Yancopoulos et al., 2000). When cultured with MDA-MB-231 CM or MDA-MB-468 CM, the expression of p-VEGFR2 in HUVECs was much higher compared with the control group (Figure S3B–C). In summary, the inhibitory effect of wogonoside on VEGF contributes to the reduction of angiogenesis induced by TNBC cells.
DISCUSSION
Considering that tumor growth of breast cancer is dependent on angiogenesis, the recent addition of angiogenesis inhibitors (i.e., bevacizumab, Avastin) to neoadjuvant chemotherapy significantly improve the progression-free survival of patients with TNBC compared with chemotherapy alone (Cella et al., 2011; Robert et al., 2011; von Minckwitz et al., 2014b). Among a variety of proangiogenic factors, VEGF and the activation of its VEGFR that specifically stimulate endothelial cells (ECs) proliferation and migration during angiogenesis, have served as primary targets for anti-angiogenic therapy (Potente et al., 2011). However, the existing antibody-based VEGF blockers (e.g., bevacizumab and ramucirumab) used in TNBC have undesirable side effects and most patients develop resistance to anti-VEGF/VEGFR therapy (Giovannini et al., 2010). Hence, effective anti-angiogenic agents with fewer side effects and wider application in chemotherapy are urgently needed.
In this study, we applied a systems pharmacology methodology to identify effective therapeutic agents for treating TNBC and wogonoside was significantly predicted to have potential to treat TNBC. Using subject matter expertise based on a combination of factors (Data S1), including pharmacokinetics/pharmacodynamics assessment, novelty and strength of the prediction, we selected wogonoside and validated it as a potential angiogenesis inhibitor in TNBC in vitro and in vivo. Specifically, the inhibitory effect of wogonoside on TNBC was subsequently validated by attenuation of tumor growth in nude mouse model bearing transplanted MDA-MB-231 cells. To investigate the potential molecular mechanisms of wogonoside against TNBC, we further integrated drug targets of wogonoside and known TNBC genes into the breast-specific human protein-protein interactome network. Network analysis prompts us to explore whether wogonoside targets the VEGF pathway (Figure 2A). In line with this, we indeed observed that wogonoside inhibited the transcription and expression of VEGF rather than influencing the viability of TNBC cells, even when treated with high concentration for 24 h (Figure S1). These observations suggest that wogonoside inhibits TNBC angiogenesis rather than having direct cytotoxic effects on TNBC cells. The tube formation and rat aortic ring experiments, together with the CAM assays, all point to wogonoside inhibiting TNBC cell-induced angiogenesis in vitro. The matrigel plug and xenograft models further indicate that wogonoside inhibits angiogenesis in vivo. This is further supported by the VEGF mAb experiment and the expression of p-VEGFR2 in HUVECs co-cultured with TNBC cells, suggesting that VEGF plays a key role in wogonoside’s reduction of angiogenesis in TNBC, consistent with network analysis (Figure 2A). We also tested wogonin (Data S1), a wogonoside analog, in TNBC cell lines. We found that wogonin cannot consistently inhibit VEGF secretion of MDA-MB-231 cells at 60 μM (Figure S4A–B) and had no effect on the migration of HUVECs co-cultured by MDA-MB-468 CM as compared with wogonoside (Figures S4C–F). In summary, we have identified wogonoside as an effective angiogenesis inhibitor for treating TNBC in vitro and in vivo.
Emerging data suggest the involvement of Hedgehog signaling in tumor-associated angiogenesis: (i) increased blood vessel density in breast cancer was observed upon Hedgehog signaling activation (Harris et al., 2012); (ii) Hedgehog promotes neovascularization by regulating Ang-1 in bone-marrow derived proangiogenic cells (Nakamura et al., 2010); and (iii) canonical Hedgehog signaling augments tumor angiogenesis by induction of VEGF (Chen et al., 2011). tGli1, an alternative splicing form of Gli1, enhances the hVEGF gene promoter resulting in upregulation of VEGF in breast cancer cells (Cao et al., 2012; Carpenter and Lo, 2012). To explore the potential mechanism of wogonoside’s impact on VEGF and angiogenesis in TNBC further, we examined Hedgehog/Gli signaling by the Gli luciferase reporter assay and observed that micromolar wogonoside inhibits the transcriptional activity. In addition, Gli1 nuclear translocation was decreased by wogonoside in a concentration-dependent manner. Importantly, we demonstrated that wogonoside directly inhibited SMO (a key upstream activator involving in the nuclear translocation and transcriptional activity of Gli1 (Cannonier and Sterling, 2015)) protein expression by promoting its proteasome degradation, which contributes to its pharmacologic inhibition of TNBC growth in vitro and in vivo.
Ubiquitination of SMO regulates its trafficking and cell surface expression (Li et al., 2012). We investigated whether Cul4-DDB1 is involved in human SMO degradation triggered by wogonoside. We observed that siRNA-mediated downregulation of Cul4A effectively impairs the wogonoside-induced SMO ubiquitination in MDA-MB-231 cells, indicating that Cullin4 is potential E3 ligases for human SMO in wogonoside-triggered SMO ubiquitination. In the light of known mechanism-of-action of existing small molecule inhibitors on Hedgehog signaling pathway (Hui et al., 2013; Wang et al., 2013), we tested the direct interaction of wogonoside with SMO or Gli1. Via SMO binding and the cellular thermal shift assays, we found that wogonoside was a direct SMO inhibitor, while it may not directly interact with Gli in MDA-MB-231 cells. In addition, we inspected the effects of wogonoside on the ubiquitination level of three key proteins in TNBC cells, including c-Jun, EGFR and STAT-3. Figures S5 reveal that wogonoside has no effect on the ubiquitination of these proteins, although further proteome-wide off-target effects are warranted to detect the specific ubiquitination status by wogonoside. Altogether, wogonoside directly binds to SMO and specifically promotes ubiquitination-dependent degradation of SMO.
We acknowledged several potential limitations of the current systems pharmacology approach. First, although our sizeable efforts assembled large-scale, experimentally reported compound-protein interactions for natural products from publicly available databases, the network data may be incomplete and its quality is also not assured. For example, some compound-protein interactions for natural products are obtained from functional assays, not physical binding studies, which may lead to false positive predictions. Integration of large-scale, unbiased drug-induced transcriptome (Subramanian et al., 2017) or drug-induced proteome (Litichevskiy et al., 2018) data may help in further improving our systems pharmacology-based model. Second, our current systems pharmacology model cannot separate therapeutic anticancer effects from those observed due to lack of detailed functional effects of drug targets and unknown biological consequences of mutations on TNBC genes/proteins. Drug targets representing nodes within cellular networks are often intrinsically coupled with both therapeutic and adverse profiles (Bedi et al., 2016), as drugs can inhibit or activate protein functions (including antagonists versus agonists). TNBC alleles from genetic or genomic studies include gain-of-function and loss-of-function. A unique integration of functional genomic assays, along with state-of-art pharmaco-epidemiologic analysis on patient-level data (e.g., health insurance claims data) and in vitro or in vivo mechanistic studies, may accelerate systems pharmacology-based drug discovery in the future (Cheng et al., 2018; Skrott et al., 2017).
In conclusion, wogonoside inhibits angiogenesis in TNBC by reducing VEGF expression and inhibiting the Hedgehog signaling pathway in vitro and in vivo. Specifically, wogonoside inhibits the nuclear translocation and transcription activity of Gli1 by binding to SMO directly and specifically promoting ubiquitination-dependent degradation of SMO in TNBC cells. These findings offer compelling preclinical evidence in support of wogonoside development as a potential angiogenesis inhibitor for the treatment of TNBC.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will fulfilled by the Lead Contact, Feixiong Cheng (chengf@ccf.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Lines
Human breast cancer MDA-MB-231 cells and MDA-MB-468 cells were originally obtained from the Cell Bank of Shanghai Institute of Cell Biology, Chinese academy of sciences (CAS). The primary human umbilical vein endothelial cells (HUVECs) were obtained from LONZA and NIH3T3 cells were obtained from American Type Culture Collection (ATCC, Manassas, VA). MDA-MB-231 and NIH3T3 cells were maintained in DMEM medium with 10% fetal bovine serum (FBS) and MDA-MB-468 cells were maintained in L-15medium containing 10% FBS, 100 U/ml penicillin and 100 μg/ml streptomycin, in a stable environment with 5% CO2 at 37 °C. HUVECs were grown in M199 medium containing endothelial cell growth supplement (ECGS, 30 mg/ml), epidermal growth factor (EGF, 10 ng/ml), 20% fetal bovine serum (FBS), 100 U/ml penicillin and 100 U/ml streptomycin. After three to five passages, HUVECs were collected for use in all experiments.
Animal Studies
Five to six-week-old female BALB/c-nude mice (Slaccas Shanghai Laboratory Animal Co., Ltd., Shanghai, China) were maintained in a pathogen-free environment (23 ± 2 °C, 55 ± 5% humidity) on a 12 h light/12 h dark cycle with food and water supplied adlibitum throughout the experimental period. Each mouse was subcutaneously inoculated with injections of 1×107 MDA-MB-231 cells into its left axilla. After initial growth of the MDA-MB-231 xenografts, mice with similar tumor volume (eliminate mice with tumors that are too large or too small) were randomly assigned to two groups (eight mice/group): a negative group (0.9% normal saline, gavage) and a wogonoside-treated group (80 mg/kg, gavage, once every other day). Mice were weighed, and tumor width (W) and length (L) were measured every day. Tumor volume was estimated according to the standard formula: V=1/2×L×W2 Fifteen days later, the nude mice were killed and the tumor xenografts were extracted, weighed and excised for immunoblot analysis of protein markers (VEGF, CD31, SMO and Gli1). Animal study and euthanasia were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of the China Pharmaceutical University.
METHODS DETAILS
Manual curation of functional genes for triple-negative breast cancer (TNBC)
We manually collected 150 functional genes for TNBC from over 30 research articles or reviews (see Data S1 for references). These functional genes included known TNBC targets in clinical trials, known TNBC driver genes (also called significantly mutated genes) reported by large-scale cancer genome sequencing projects, and experimentally validated TNBC genes identified by siRNA or gene knockdown studies. For example, 50 most significantly mutated genes in TNBC were identified based on re-analysis of the 77 patients with TNBCs from The Cancer Genome Atlas (TCGA) (Pareja et al., 2016).
Reconstruction of Drug-Target Network for Natural Products
We firstly collected chemical structure information for natural products by integrating data from six data sources as described in previous studies (Fang et al., 2017a; Fang et al., 2017c). In total, 259,547 unique natural products were obtained. Compound-protein interaction (DPI) mapping of natural products were performed using three types of data sources: i) direct CPI databases including ChEMBL(Gaulton et al., 2017) and BindingDB (Gilson et al., 2016); ii) indirect CPI databases including STITCH 5 (Szklarczyk et al., 2016), Herbal Ingredients’ Targets Database (HIT) (Ye et al., 2011), and traditional Chinese medicine integrated database (TCMID) (Xue et al., 2013); and iii) manually curated CPIs collected from literatures (see Data S2 for references).
We downloaded compound-protein binding affinity data (Ki, Kd, or IC50) from ChEMBL (v21) and BindingDB (accessed in Sep 2017). We filtered data items that met the criteria as described in a previous study (Fang et al., 2017c). Subsequently all chemical structures were standardized through removing salt ions and standardizing dative bonds using Open Babel toolkit (v2.3.2) (O’Boyle et al., 2011). Then 8,015 direct CPIs were obtained after mapping 259,547 unique natural products to data items filtered.
Indirect CPIs were integrated via the below criteria. For STITCH source (accessed in September 2017), the thickness of each CPI stands for the confidence score of the association. We only kept CPIs from Homo sapiens, and retained CPIs with experimental evidence score higher than 0.7 (Szklarczyk et al., 2016) in this study. Furthermore, we extracted CPIs from HIT and TCMID using a web crawler approach (Fang et al., 2017a). In total, we obtained 22,620 indirect CPIs (Data S2) for natural products from available databases after removing the duplicated CPIs.
We further manually extracted indirect CPIs from natural product-specific pharmacological references covering over 2,000 publications dating from 2009 to 2017. We collected 10,640 indirect CPIs (Data S2) connecting 726 natural products to 1,300 human proteins. After merging the three data sources, we obtained 38,220 CPIs connecting 3,882 unique natural products to 5,643 human proteins (Data S2).
In Silico Prediction of Anti-TNBC Indications for Natural Products
We used an integrated statistical framework to predict potential anti-TNBC indication for natural products by incorporating their drug-target (compound-protein) network into the curated functional genes/proteins of TNBC. We hypothesized that a natural promiscuous (“multi-targets”) drug shows a high possibility to treat TNBC if its targets are more likely to be functional proteins in TNBC. The null hypothesis asserts that the targets of a natural product randomly target TNBC across the human proteome. We calculated statistical significance of a natural product to be prioritized for TNBC using permutation testing as below:
| (2) |
A nominal P was computed for each natural product by calculating the number of observed TNBC functional proteins (Sm (p)) greater than the permutations (Sm).
Here we repeated 100,000 times of permutations by randomly selecting 150 genes (the same number of TNBC functional genes) from the genome-wide simulation (20,462 human protein-coding genes from the NCBI database) (Coordinators, 2017). Subsequently, the nominal P-values from the permutation tests were corrected as adjusted P-values (q) using R based on the Benjamini-Hochberg approach (Benjamini and Hochberg, 1995). Finally, the second index (Z-score, equation (1)) was calculated for each natural product to be prioritized for anti-TNBC indication during permutation testing.
Chemical structure clustering analysis
To investigate the chemical feature of natural products having significant anti-TNBC indications (q<10<−5), we performed clustering analysis for 148 significantly predicted natural products. The clustering analysis was performed by computing the root-mean-square-difference (RMSD) of the Tanimoto distance of pairwise compounds using FCFP_4 fingerprint as described previously (Fang et al., 2017a; Fang et al., 2017b).
Tissue-specific Human Protein-Protein Interactome Network Analysis
We downloaded the RNA-seq data (RPKM value) of 32 tissues from GTEx V6 release (accessed on April 01,2016, https://gtexportal.org/home/). For each tissue (e.g., breast), we regarded those genes with RPKM ≥ 1 in more than 80% of samples as tissue-expressed genes and the remaining genes as tissue-unexpressed. To quantify the expression significance of tissue-expressed gene i in tissue t, we calculated the average expression ⟨E(i)⟩ and the standard deviation δE(i) of a gene’s expression across all considered tissues. The significance of gene expression in tissue t is defined:
| (3) |
Herein, we performed a breast-specific co-expressed protein-protein interactome network analysis by comparing genome-wide expression profiles of breast to 31 other tissues from GTEx database (Consortium, 2015). Specifically, we assembled 15 commonly used databases with various types of experimental evidence and an unbiased, systematic human protein-protein interactions (PPIs) we have recently utilized (Cheng et al., 2018): (i) binary PPIs tested by high-throughput yeast-two-hybrid (Y2H) systems in which we combined binary PPIs from two publicly available high-quality Y2H datasets (Rolland et al., 2014; Rual et al., 2005); (ii) kinase-substrate interactions; (iii) signaling networks from SignaLink2.0 (Fazekas et al., 2013); (iv) binary PPIs from three-dimensional (3D) protein structures reported in Instruct (Meyer et al., 2013); and (v) carefully literature-curated PPIs identified by low-throughput experiments. All inferred data (e.g., evolutionary analysis and gene co-expression data) are excluded. The detailed descriptions for building human protein-protein interactome are provided in our previous studies (Cheng et al., 2018; Cheng et al., 2014; Cheng et al., 2015).
We next reconstructed the breast-specific co-expressed PPI network by mapping RNA sequencing data from TCGA (Cancer Genome Atlas, 2012). Normalized RNA sequencing data (RPKM) were extracted using the R package from TCGA-Assembler (Zhu et al., 2014). We computed the co-expression (Pearson correlation coefficient) and p-value (F-statistics) for each PPI based on RNA sequencing data in TNBC. Finally, we only used the p-value (P) < 0.05 as co-expressed PPIs to build the TNBC-specific co-expressed human protein-protein interactome network for network analysis in Figure 2A.
Compound Preparation
Wogonoside (>98% purity; Langze Pharmaceutical Co, Ltd., Nanjing, China) was dissolved in dimethylsulfoxide (DMSO) as stock solution at 0.1 M and stored at −20 °C. Wogonoside stock solution was fre shly diluted with medium to the final concentration before each experiment in vitro. The final DMSO concentration did not exceed 0.1%. In the in vivo study, wogonoside was prepared as intragastric administration (0.5% Sodium Carboxymethyl Cellulose, CMC). Chemicals and reagents used in this study were described in key resources table.
KEY RESOURCES TABLE
| chemicals and reagents | Catalog# | Source |
|---|---|---|
| MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) | M5655 | Sigma |
| DMSO (dimethylsulfoxide) | D2650 | Sigma |
| CMC (Sodium Carboxymethyl Cellulose) | 419273 | Sigma-Aldrich |
| DMEM Medium | 10566016 | Gibco |
| L-15 Medium | 11415056 | Leibovitz’s |
| M199 Medium | 11150067 | Gibco |
| FBS (Fetal Bovine Serum) | 10099141 | Gibco |
| ECGS (endothelial cell growth supplement) | B211-GS | Sigma |
| EGF (epidermal growth factor) | E5036 | Sigma |
| Penicillin-Streptomycin | Vetec-V900929 | Sigma-Aldrich |
| BSA (bovine serum albumin) | A1933 | Sigma |
| VEGF ELISA kit | DY293B-05 | R&D Systems |
| HiScript® II Q RT SuperMix for qPCR | R222-01 | Vazyme |
| Hiscript® II Reverse Transcriptase | R201-02 | Vazyme |
| ExFect®2000 Transfection Reagent | T202-02 | Vazyme |
| Matrigel Basement Membrane Matrix | 356234 | BD Bioscience |
| Hematoxylin and Eosin Staining Kit | C0105 | Beyotime |
| ACA (ε-amino-n-caproic acid) | 145815 | J&K Scientific Ltd. |
| Thrombin | T4393 | Sigma |
| Fibrinogen | F4883 | Sigma |
| 4’,6-Diamidino-2-phenylindole dihydrochloride (DAPI) | #4083 | Cell Signaling Technology |
| Triton X-100 | T8787 | Sigma |
| MG132 | 474791 | EMD Millipore |
| NH4Cl (Ammonium chloride) | A9434 | Sigma |
| Dual Luciferase Reporter Assay Kit | DL101-01 | Vazyme |
| Nuclear and Cytoplasmic Protein Extraction Kit | P0028 | Beyotime |
| RNA isolater® Total RNA Extraction Reagent | R401-01 | Vazyme |
| Immunohistochemistry Application Solutions Kit | #13079 | Cell Signaling Technology |
| Lipofectamine 2000 | 11668-019 | invitrogen |
| Bodipy-Cyclopamine | 2160-50 | BioVision |
| The antibodies used in this study. | ||
| Antibodies | Catalog# | Source |
| anti-VEGF | ab69479 | Abcam |
| anti-β-actin | sc-47778 | Santa Cruz |
| anti-CD31 | ab28364 | Abcam |
| anti-EGFR | #2085 | Cell Signaling Technology |
| anti-Gli1 | ab49314 | Abcam |
| anti-c-Jun | ab32137 | Abcam |
| anti-LaminA | sc-293162 | Santa Cruz |
| anti-β-Tublin | sc-5274 | Santa Cruz |
| anti-SMO | ab72130 | Abcam |
| anti-STAT3 | #9139 | Cell Signaling Technology |
| anti-Ubiquitin | #3936 | Cell Signaling Technology |
| anti-VEGFR2 | #9698 | Cell Signaling Technology |
| anti-p-VEGFR2 | #2478 | Cell Signaling Technology |
| anti-Gli1 | #2553 | Cell Signaling Technology |
| anti-IgG | sc-2025 | Santa Cruz |
| Experimental Models: Cell Lines. | ||
| Name of cell line | Catalog# | Source |
| MDA-MB-231 | TCHu227 | Cell Bank of CAS |
| MDA-MB-468 | TCHu136 | Cell Bank of CAS |
| HUVECs | C2517A | LONZA |
| NIH3T3 | CRL-1658 | ATCC |
| The primers used in this study for various PCR assays. | ||
| Genes | Forward primer (5’-3’) | Reverse primer (5’-3’) |
| VEGF | GGTGGACATCTTCCAGAGTA | GGCTTGTCACATCTGCAAGTA |
| β-actin | CTGTCCCTGTATGCCTCT | ATGTCACGCACGATTTCC |
| Smo | TCTCGGGCAAGACATCCT | TAGCCTCCCACAATAAGCA |
| Cyclin D2 | TGGAGCTGCTGTGCCACG | GTGGCCACCATTCTGCGC-3 |
| HIP | CCTGGGTTCTGACCACTGTT | TGGTCACTGGAGCTTGTGAG |
| GAS1 | CGGAGCTTGACTTCTTGGAC | CCCAACCCTTCAAATTGCTA |
| Cul4A | ACCTCGCACAGATGTACCAG | AGGTTGACGAACCGCTCATTC |
Immunohistochemistry
Tumor xenografts of mice in control and wogonoside (80 mg/kg) treated groups were immersed in 4% formaldehyde (pH 7.4) for 24 h, embedded in paraffin, cut into sections 4 mm thick using standard histological techniques to prepare paraffin sections. The expression of CD31, VEGF, SMO and Gli1of the tissues from control and wogonoside (80 mg/kg) treated groups were assessed using Immunohistochemistry Application Solutions Kit with specific antibodies (1:100).
Cell Viability Assay
The MTT assay was used to evaluate the viability of human breast cancer cells. MDA-MB-231 and MDA-MB-468 cells were plated at a density of 2 × 105 cells per well into 96-well plates in medium with 10% FBS. After overnight growth, cells were treated with various concentrations of wogonoside (0, 25, 50 and 100 μM) for 24 h in 5% CO2 incubator at 37 °C. Then 20 μl of 0.5% MTT were added into the medium and incubated for 4 h. The supernatant was removed and 100 μl DMSO was added to dissolve the precipitate. Absorbance was measured at 570 nm.
Enzyme-Linked ImmunoSorbent Assay (ELISA)
The concentration of human VEGF protein in MDA-MB-231 CM or MDA-MB-468 CM was determined by a human VEGF Duo-set enzyme linked immune sorbent assay kit in accordance with the manufacture instructions.
VEGF-Luciferase Reporter Assay
The hVEGF-pGL2 promoter reporter was provided by Dr. Barbara K. Vonderhaar (Mammary Biology and Tumorigenesis Laboratory, National Cancer Institute, National Institutes of Health, Bethesda, MD). MDA-MB-231 or MDA-MB-468 cells were cotransfected with hVEGF-pGL2 with Renilla luciferase Reporter (as internal control) and then maintained in medium with 0.5% FBS and various concentrations of wogonoside (0, 25, 50 and 100 μM) for 24 h. The luciferase activity of cell lysate was determined by the Dual-Luciferase Reporter kit. Luciferase signals were collected by DualLuciferase assay system (Thermo Fisher Scientific, Rockford, IL).
Western Blot Analysis
After treatment with wogonoside (0, 25, 50 and 100 μM) for 24 h, cell extracts were obtained by lysis with RIPA buffer. Protein samples were separated by SDS-PAGE and transfered to a polyvinylidene difluoride (PVDF) membrane (Millipore, Billerica, MA). Membranes were incubated with the primary antibodies (key resources table) specific for target proteins overnight at 4°C, and then incubated with the secondary antibody which was coupled to infrared (IR) dyes for one hour at room temperature. Detection was performed by the Odyssey Infrared Imaging System (LI-COR Inc., USA) using a fluorescent readout and quantified using the Image Studio 5.2. software (LI-COR, USA).
Real-Time PCR Analysis
Total RNA was extracted after treated with wogonoside (0, 25, 50 and 100 μM, 24 h) using Total RNA Extraction Reagent (Vazyme). cDNA was made using 500 ng of total RNA with Hiscript® II Reverse Transcriptase (Vazyme). qPCR incubations were run with 200 nM of gene-specific primers and HiScript® II Q RT SuperMix for qPCR (Vazyme). The qPCR primer sequences and protocol were described in key resources table. qPCR data was analyzed by the ΔΔCt method.
Gene Knockdown
The target siRNA sequences against Cul4A (siCul4A-1,5’-GCACAGAUCCUUCCGUUUA-3’; siCul4A-2. 5’-GGUUUAUCCACGGUAAAGA-3’), and the scrambled siRNA (siControl) were synthesized by Gima Company. Cells were transfected with 25 nmol/l siCul4A using lipofectamine 2000 (invitrogen). The siRNA-transfected MDA-MB-231 cells were used for subsequent experiments after 24 hours. Knockdown efficiency of Cul4A was verified by RT-PCR.
Molecular docking studies
Molecular docking simulations were used to explore the potential interaction of wogonoside on Gli1 or SMO. The crystal structure of Gli1 (pdb code: 5ED2) and SMO (pdb code: 4N4W) were prepared by the Protonate 3D tool in MOE (version 2010.10, Chemical Computing Group Inc. Montreal, Quebec, Canada, 2010) and all the water molecules were removed. Hydrogen atoms were added using MOE. The structure of wogonoside was modeled and minimized in MOE. Docking simulations were carried out in the CDOCKER module implemented in Discovery Studio 4.0 (version 2.5, Accelrys Inc., San Diego, CA, 2009).
BODIPY-cyclopamine Based Fluorescence Binding Assay
293 cells were transfected with a human SMO expression vector in 6-well plates. Transfected cells were washed with phosphate-buffered saline (PBS) cantaining 1% fetal bovine serum, fixed in 4% paraformaldehyde in PBS and treated with 10 nM BODIPY-cyclopamine and different concentrations of the wogonoside at 37°C for 3 h. Cell nuclei was also stained with DAPI. The fluorescence images were captured and analyzed with Cytation 5 (Biotek, Winooski, VT).
Cellular Thermal Shift Assay (CETSA)
Cellular thermal shift assay was conducted as described previously (Wu et al., 2018). In briefly, MDA-MB-231 cells were treated with DMSO or 100 μM wogonoside for 12 h, and then cells were collected and resuspended in phosphate buffered saline (PBS). Each aliquot (3×106 cells) were heated at indicated temperatures for 3 min on PCR instrument (Bio-Rad, CA) and frozen twice in liquid nitrogen. The samples were centrifuged and the supernatants were subjected to Western Blotting detection.
Gli-luciferase Reporter Assay
NIH3T3 cells were transfected with 8xGli firefly luciferase reporter plasmid (kindly provided by Aykut Uren at Georgetown University) and were seeded into 96-well plates (20,000 cells/well) and cultured to confluency. The cells were then maintained in DMEM medium with 0.5% FBS and different concentrations of wogonoside in the presence of SAG (20 nM). After the cells were treated for additional 48 h, the luciferase activities were detected by adding luciferase substrate D-luciferin and a Cystation 5 plate reader (BioTek, Winooski, VT).
Conditioned Media Collection
MDA-MB-231 or MDA-MB-468 cells (5×106 cells/culture flask) were treated with various concentrations of wogonoside (0, 25, 50 and 100 μM) for 24 h, then washed with PBS (pH 7.4) twice to remove the unreacted drug. All the cells pretreated with wogonoside were cultured for another 12 h with low serum media containing 1% FBS, and the supernatant was collected and referred to as MDA-MB-231or MDA-MB-468 derived conditioned medium (MDA-MB-231 CM or MDA-MB-468 CM).
Endothelial Cell Invasion Assay
A transwell chamber (6.5 mM in diameter, 8 mM pore-size, CorningCostar, Cambridge, MA) was used to test the invasion ability of HUVECs. Firstly, transwell chambers were loaded with 0.1 ml matrigel at 37°C for 1 h. HUVECs were incubated in regular medium or regular medium containing 50% MDA-MB-231 or MDA-MB-468 CM for 24 h, then trypsinized and suspended at a final concentration of 5×105 cells/ml in serum-free M199. Cell suspension was loaded into each of the upper wells and regular medium or regular medium containing 50% CM was added into in the lower compartment. Following incubation at 37°C in 5% CO 2 for 24 h, the non-migratory cells on the upper surface were removed by a cotton swab. The invasive cells on the lower surface were fixed with 100% methanol and stained with hematoxylin and eosin. The migrated cells were quantified by manual counting and five randomly chosen fields were analyzed for each group.
Tube Formation Assay
Matrigel mixed with an equal volume of non-FBS medium, and then transferred into a 96-well plate to solidify and polymerize at 37°C for 45 min. After pretreated with regular medium (control) or regular medium containing 50% CM, HUVECs were harvested with trypsin, suspended in 1% FBS medium and seeded onto matrigel. Following incubated for 8 h, the plate was examined for capillary tube formation under an inverted microscope. Tubular structures were photographed and quantified by manual counting of tube numbers. Five randomly chosen fields were analyzed for each well.
Rat Aortic Ring Assay
The thoracic aorta was harvested from male Sprague Dawley rats (six weeks old) and cut into 1 mm slices and set in a 24-well plate. Prepare the clotting media containing M199+ (M199 with 200 U/ml penicillin and 200 μg/ml streptomycin), 0.3% fibrinogen and 0.5% ACA. Then the growth media consisting of M199+ with 20% FBS and 0.5% ACA was added to each well. One to two days later, cells started to sprout from the explants, forming microvessel-like structures. After three days of growth, six rings were used as a group and fed with 1 ml of M199+ alone or M199+ containing 50% CM with various concentrations of wogonoside (0, 25, 50 and 100 μM) as indicated. Plates were then stored in the incubator at 37°C. S even days later, the sprouting microvessels in five randomly chosen fields were measured and photographed under a microscope for each group.
Chicken Chorioallantoic Membrane (CAM) Assay
Fertilized chicken eggs were incubated at 37°C for 9 days. After this incubation, a small hole was punched on the broad side of the egg, and a window was carefully created through the egg shell. MDA-MB-231or MDA-MB-468 cells (1×106 cells/embryo) and sterilized filter paper disks (5 mm×5 mm) saturated with wogonoside (0, 50, 100 and 200 ng/CAM) were placed on the CAMs, which were incubated at 37°C for another 2 days. Inject appropriate volume of 10% fat emulsion was into the embryo chorioallantois for observing the density and length of vessels toward the CAM face. Neovascular zones under the filter paper disks were observed and photographed by a digital camera at ×5 magnification. The number of newly growth vessels was counted on digitalized pictures.
In Vivo Angiogenesis Study
3 to 4-wk-old BALB/c-nude mice (Slaccas Shanghai Laboratory Animal Co., Ltd., Shanghai, China) were maintained in a pathogen-free environment (23 ± 2°C, 55 ± 5% humidity) on a 12 h light/12 h dark cycle with food and water supplied adlibitum throughout the experimental period. Mice were divided into three groups: control group (injected subcutaneously with 600 μl matrigel containing 200 μl saline), model group (600 μl matrigel containing 200 μl MDA-MB-231or MDA-MB-468 cell suspension) and wogonoside treated group, which were treated with wogonoside (80 mg/kg, gavage, everyday) for ten days. Mice were killed and the gel plugs were excised, photographed, viewed whole-mount of CD31 staining and homogenized in 1 ml PBS buffer, centrifuged to test the content of hemoglobin in the supernatant was by Drabkin’s reagent (Sigma-Aldrich).
Preparation of Cytosolic and Nuclear Extracts
MDA-MB-231 or MDA-MB-468 cells were treated with various concentrations of wogonoside (0, 25, 50 and 100 μM) for 24 h. Nuclear and cytosolic protein extracts were prepared according to the user guide of Nuclear and Cytoplasmic Protein Extraction Kit.
Immunofluorescence Staining
Cells were grown on coverslips and pretreated with wogonoside (100 μM) for 24 h, fixed with 4% paraformaldehyde, permeabilized in 0.2% Triton X-100 and incubated with 3% BSA. After incubated with primary antibodies (1:100), cells were exposed to secondary antibodies (1:1000) and stained with DAPI. Cells were observed and photographed with a confocal laser scanning microscope (Fluoview FV 1000, Olympus, Tokyo, Japan).
Quantification and statistical analysis
The data shown in the study were obtained from at least three independent experiments and all data in different experimental groups were expressed as the mean± standard deviation (SD). Statistical analyses were performed using a One-Way ANOVA, with post-hoc analysis. Details of each statistical analysis are provided in the figure legends. Differences with P values < 0.05 were considered statistically significant.
DATA AND SOFTWARE AVAILABILITY
The code written for and used in this study is available from the Lead Contact (Feixiong Cheng, chengf@ccf.org) upon reasonable request. Additional data supporting the findings of this study are available within the supplemental information files or from the Lead Contact upon reasonable request.
Supplementary Material
SIGNIFICANCE.
In this study, we applied a systems pharmacology approach that integrates known drug-target network and large-scale genomic profiles of TNBC for identification of effective therapeutic agents in treating TNBC. We found that wogonoside, one of the major active flavonoids derived from Scutellaria baicalensis Georgi, exhibited an effective angiogenesis inhibitor of TNBC in vitro and in vivo. In summary, we demonstrated that wogonoside inhibited the Gli1 nuclear translocation and transcription activities of Hedgehog signaling by promoting SMO degradation via a proteasome-dependent manner. This study offers a powerful, integrated computational and experimental pharmacology approaches for oncological drug discovery and identifies wogonoside as a potential angiogenesis inhibitor to treat TNBC in preclinical studies. Our approach can minimize the translational gap between pre-clinical testing results in animal models and clinical outcomes, which is a significant problem in the development of efficient therapeutic strategies for TNBC. From a translational perspective, if broadly applied, the computational and experimental pharmacology tools developed here could help develop effective treatment strategies for other types of cancer and complex diseases in clinic.
Highlights.
A systems pharmacology approach uncovers wogonoside as a TNBC angiogenesis inhibitor.
Network analysis identifies that wogonoside inhibits VEGF expression in TNBC.
Wogonoside inhibits the Gli1 nuclear translocation and transcription activities.
Wogonoside promotes Smoothened (SMO) degradation in a proteasome-dependent mechanism.
ACKNOWLEDGEMENTS
This work was supported by the National Heart, Lung, and Blood Institute of the National Institutes of Health under Award Number K99HL138272 and R00HL138272 to F.C. This work has been also funded in whole or in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E. This research was supported (in part) by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organizations imply endorsement by the US Government. C.E. is the Sondra J and Stephen R Hardis Endowed Chair in Cancer Genomic Medicine at the Cleveland Clinic, and an ACS Clinical Research Professor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information including 5 Supplemental figures (Figures S1–S5) and 2 Supplemental databases (Data S1 [Related to Figures 1 and 2] and Data S2 [Related to Figures 1 and 2]) can be found with this article online.
DECLARATION OF INTERESTS
F.C. and C.E. have a pending patent application related to this study. The other authors declare no conflict of interest.
REFERENCES
- Andre F, Job B, Dessen P, Tordai A, Michiels S, Liedtke C, Richon C, Yan K, Wang B, Vassal G, et al. (2009). Molecular characterization of breast cancer with high-resolution oligonucleotide comparative genomic hybridization array. Clin. Cancer Res 15, 441–451. [DOI] [PubMed] [Google Scholar]
- Bareche Y, Venet D, Ignatiadis M, Aftimos P, Piccart M, Rothe F, and Sotiriou C (2018). Unravelling triple-negative breast cancer molecular heterogeneity using an integrative multiomic analysis. Ann. Oncol 29, 895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedi O, Dhawan V, Sharma PL., and Kumar P (2016). Pleiotropic effects of statins: new therapeutic targets in drug design. Naunyn-Schmiedeberg’s Arch. Pharmacol 389, 695–712. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, and Hochberg Y (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Royal Stat. Soc 57, 289–300. [Google Scholar]
- Bianchini G, Balko JM, Mayer IA, Sanders ME, and Gianni L (2016).Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol 13, 674–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe J, and Therond PP (2013). The mechanisms of Hedgehog signalling and its roles in development and disease. Nat. Rev. Mol. Cell Biol 14, 416–429. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas N (2012). Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannonier SA, and Sterling JA (2015). The role of Hedgehog signaling in tumor induced bone disease. Cancers 7, 1658–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, Geradts J, Dewhirst MW, and Lo HW (2012). Upregulation of VEGF-A and CD24 gene expression by the tGLI1 transcription factor contributes to the aggressive behavior of breast cancer cells. Oncogene 31, 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, and Jain RK (2011). Molecular mechanisms and clinical applications of angiogenesis. Nature 473, 298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter RL, and Lo HW (2012). Hedgehog pathway and GLI1 isoforms in human cancer. Discov. Med 13, 105–113. [PMC free article] [PubMed] [Google Scholar]
- Cella D, Wang M, Wagner L, and Miller K (2011). Survival-adjusted health-related quality of life (HRQL) among patients with metastatic breast cancer receiving paclitaxel plus bevacizumab versus paclitaxel alone: results from Eastern Cooperative Oncology Group Study 2100 (E2100). Breast Cancer Res. Treat 130, 855–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JK, Taipale J, Cooper MK, and Beachy PA (2002). Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev 16, 2743–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Tang T, Eastham-Anderson J, Dunlap D, Alicke B, Nannini M, Gould S, Yauch R, Modrusan Z, DuPree KJ, et al. (2011). Canonical hedgehog signaling augments tumor angiogenesis by induction of VEGF-A in stromal perivascular cells. Proc. Natl. Acad. Sci. USA 108, 9589–9594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Desai RJ, Handy DE, Wang R, Schneeweiss S, Barabasi AL, and Loscalzo J (2018). Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun 9, 2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Kovacs IA, and Barabasi AL (2019a). Network-based prediction of drug combinations. Nat. Commun 10, 1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Jia P, Wang Q, Lin CC, Li WH, and Zhao Z (2014). Studying tumorigenesis through network evolution and somatic mutational perturbations in the cancer interactome. Mol. Biol. Evol 31, 2156–2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, Lee PW, and Tang Y (2012a). admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties. J. Chem. Inf. Model 52, 3099–3105. [DOI] [PubMed] [Google Scholar]
- Cheng F, Liang H, Butte AJ, Eng C, and Nussinov R (2019b). Personal mutanomes meet modern oncology drug discovery and precision health. Pharmacol. Rev 71, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Liu C, Jiang J, Lu W, Li W, Liu G, Zhou W, Huang J, and Tang Y (2012b). Prediction of drug-target interactions and drug repositioning via network-based inference. PLoS Comput. Biol 8, e1002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GT (2015). The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coordinators NR (2017). Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 45, D12–d17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl HM, Hiller L, Dunn JA, Blenkinsop C, Grybowicz L, Vallier AL, Abraham J, Thomas J, Provenzano E, Hughes-Davies L, et al. (2015). Efficacy of neoadjuvant bevacizumab added to docetaxel followed by fluorouracil, epirubicin, and cyclophosphamide, for women with HER2-negative early breast cancer (ARTemis): an open-label, randomised, phase 3 trial. Lancet Oncol 16, 656–666. [DOI] [PubMed] [Google Scholar]
- Fang J, Cai C, Wang Q, Lin P, Zhao Z, and Cheng F (2017a). Systems pharmacology-based discovery of natural products for precision oncology through targeting cancer mutated genes. CPT Pharmacometrics Syst. Pharmacol 6, 177–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Gao L, Ma H, Wu Q, Wu T, Wu J, Wang Q, and Cheng F (2017b). Quantitative and Systems Pharmacology 3. Network-Based Identification of New Targets for Natural Products Enables Potential Uses in Aging-Associated Disorders. Frontiers Pharmacol 8, 747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J, Wu Z, Cai C, Wang Q, Tang Y, and Cheng F (2017c). Quantitative and Systems Pharmacology. 1. In silico prediction of drug-target interactions of natural products enables new targeted cancer therapy. J. Chem. Inf. Model 57, 2657–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazekas D, Koltai M, Turei D, Modos D, Palfy M, Dul Z, Zsakai L, Szalay-Beko M, Lenti K, Farkas IJ, et al. (2013). SignaLink 2 - a signaling pathway resource with multi-layered regulatory networks. BMC Syst. Biol 7, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara N, Gerber HP, and LeCouter J (2003). The biology of VEGF and its receptors. Nat. Med 9, 669–676. [DOI] [PubMed] [Google Scholar]
- Foulkes WD, Smith IE, and Reis-Filho JS (2010). Triple-negative breast cancer. N. Engl. J. Med 363, 1938–1948. [DOI] [PubMed] [Google Scholar]
- Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D, Mutowo P, Atkinson F, Bellis LJ, Cibrian-Uhalte E, et al. (2017). The ChEMBL database in 2017. Nucleic Acids Res 45, D945–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, and Chong J (2016). BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res 44, D1045–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannini M, Aldrighetti D, Zucchinelli P, Belli C, and Villa E (2010). Antiangiogenic strategies in breast cancer management. Crit. Rev. Oncol./Hematol 76, 13–35. [DOI] [PubMed] [Google Scholar]
- Harris LG, Pannell LK, Singh S, Samant RS, and Shevde LA (2012). Increased vascularity and spontaneous metastasis of breast cancer by hedgehog signaling mediated upregulation of cyr61. Oncogene 31, 3370–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Zhao K, Hu Y, Zhou Y, Luo X, Li X, Wei L, Li Z, You Q, Guo Q, et al. (2016). Wogonoside inhibits angiogenesis in breast cancer via suppressing Wnt/beta-catenin pathway. Mol. Carcinog 55, 1598–1612. [DOI] [PubMed] [Google Scholar]
- Hui CC, and Angers S (2011). Gli proteins in development and disease. Annu. Rev. Cell Dev. Biol 27, 513–537. [DOI] [PubMed] [Google Scholar]
- Hui M, Cazet A, Nair R, Watkins DN, O’Toole SA, and Swarbrick A (2013). The Hedgehog signalling pathway in breast development, carcinogenesis and cancer therapy. Breast Cancer Res 15, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafari R, Almqvist H, Axelsson H, Ignatushchenko M, Lundback T, Nordlund P, and Martinez Molina D (2014). The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc 9, 2100–2122. [DOI] [PubMed] [Google Scholar]
- Jahchan NS, Dudley JT, Mazur PK, Flores N, Yang D, Palmerton A, Zmoos AF, Vaka D, Tran KQ, Zhou M, et al. (2013). A drug repositioning approach identifies tricyclic antidepressants as inhibitors of small cell lung cancer and other neuroendocrine tumors. Cancer Discov 3, 1364–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Lu W, Shen X, Wang Q, Lv J, Liu M, Cheng F, Zhao Z, and Pang X (2018). Repurposing sertraline sensitizes non-small cell lung cancer cells to erlotinib by inducing autophagy. JCI Insight 3, e98921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin G, Fu C, Zhao H, Cui K, Chang J, and Wong ST (2012). A novel method of transcriptional response analysis to facilitate drug repositioning for cancer therapy. Cancer Res 72, 33–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, MacBeath G, and Yaffe MB (2012). Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell 149, 780–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann BD, and Pietenpol JA (2014). Identification and use of biomarkers in treatment strategies for triple-negative breast cancer subtypes. J. Pathol 232, 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Chen Y, Shi Q, Yue T, Wang B, and Jiang J (2012). Hedgehog-regulated ubiquitination controls smoothened trafficking and cell surface expression in Drosophila. PLoS Biol 10, e1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Cho YS, Wang B, Li S, and Jiang J (2018a). Regulation of Smoothened ubiquitylation and cell surface expression through a Cul4-DDB1-Gbeta E3 ubiquitin ligase complex. J. Cell Sci 131, jcs218016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Li S, Wang B, and Jiang J (2018b). Hedgehog reciprocally controls trafficking of Smo and Ptc through the Smurf family of E3 ubiquitin ligases. Sci. Signal 11, eaan8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderholm BK, Hellborg H, Johansson U, Elmberger G, Skoog L,Lehtio J, and Lewensohn R (2009). Significantly higher levels of vascular endothelial growth factor (VEGF) and shorter survival times for patients with primary operable triple-negative breast cancer. Ann. Oncol 20, 1639–1646. [DOI] [PubMed] [Google Scholar]
- Litichevskiy L, Peckner R, Abelin JG, Asiedu JK, Creech AL, Davis JF, Davison D, Dunning CM, Egertson JD, Egri S, et al. (2018). A library of phosphoproteomic and chromatin signatures for characterizing cellular responses to drug perturbations. Cell Syst 6, 424–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch T, and Price A (2007). The effect of cytochrome P450 metabolism on drug response, interactions, and adverse effects. Am. Fam. Physician 76, 391–396. [PubMed] [Google Scholar]
- Martinez Molina D, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, and Nordlund P (2013). Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87. [DOI] [PubMed] [Google Scholar]
- Meyer MJ, Das J, Wang X, and Yu H (2013). INstruct: a database of high-quality 3D structurally resolved protein interactome networks. Bioinformatics 29, 1577–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed RA, Ellis IO, Mahmmod AM, Hawkes EC, Green AR, Rakha EA, and Martin SG (2011). Lymphatic and blood vessels in basal and triple-negative breast cancers: characteristics and prognostic significance. Modern Pathol 24, 774–785. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Sasajima J, Mizukami Y, Sugiyama Y, Yamazaki M, Fujii R, Kawamoto T, Koizumi K, Sato K, Fujiya M, et al. (2010). Hedgehog promotes neovascularization in pancreatic cancers by regulating Ang-1 and IGF-1 expression in bone-marrow derived pro-angiogenic cells. PLoS One 5, e8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, and Hutchison GR (2011). Open Babel: An open chemical toolbox. J. Cheminformat 3, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak E, and Segal RA (2016). Hedgehog signal transduction: Key players, oncogenic drivers, and cancer therapy. Dev. Cell 38, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pareja F, Geyer FC, Marchio C, Burke KA, Weigelt B, and Reis-Filho JS (2016). Triple-negative breast cancer: the importance of molecular and histologic subtyping, and recognition of low-grade variants. NPJ Breast Cancer 2, 16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penault-Llorca F, and Viale G (2012). Pathological and molecular diagnosis of triple-negative breast cancer: a clinical perspective. Ann. Oncol 23, vi19–22. [DOI] [PubMed] [Google Scholar]
- Potente M, Gerhardt H, and Carmeliet P (2011). Basic and therapeutic aspects of angiogenesis. Cell 146, 873–887. [DOI] [PubMed] [Google Scholar]
- Robert NJ, Dieras V, Glaspy J, Brufsky AM, Bondarenko I, Lipatov ON, Perez EA, Yardley DA, Chan SY, Zhou X, et al. (2011). RIBBON-1: randomized, double-blind, placebo-controlled, phase III trial of chemotherapy with or without bevacizumab for first-line treatment of human epidermal growth factor receptor 2-negative, locally recurrent or metastatic breast cancer. J. Clin. Oncol 29, 1252–1260. [DOI] [PubMed] [Google Scholar]
- Rolland T, Tasan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, et al. (2014). A proteome-scale map of the human interactome network. Cell 159, 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. (2005). Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178. [DOI] [PubMed] [Google Scholar]
- Skrott Z, Mistrik M, Andersen KK, Friis S, Majera D, Gursky J, Ozdian T, Bartkova J, Turi Z, Moudry P, et al. (2017). Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature 552, 194–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton BZ, and Peng LF (2010). Small-molecule modulators of the Sonic Hedgehog signaling pathway. Mol. BioSyst 6, 44–54. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Narayan R, Corsello SM, Peck DD, Natoli TE, Lu X, Gould J, Davis JF, Tubelli AA, Asiedu JK, et al. (2017). A next generation Connectivity Map: L1000 platform and the first 1,000,000 profiles. Cell 171, 1437–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, and Kuhn M (2016). STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res 44, D380–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Mao J, Zhang Q, and Li L (2011). Overexpression of Hedgehog signaling molecules and its involvement in triple-negative breast cancer. Oncol. Lett 2, 995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Minckwitz G, Eidtmann H, Rezai M, Fasching PA, Tesch H, Eggemann H, Schrader I, Kittel K, Hanusch C, Kreienberg R, et al. (2012). Neoadjuvant chemotherapy and bevacizumab for HER2-negative breast cancer. N. Engl. J. Med 366, 299–309. [DOI] [PubMed] [Google Scholar]
- von Minckwitz G, Loibl S, Untch M, Eidtmann H, Rezai M, Fasching PA, Tesch H, Eggemann H, Schrader I, Kittel K, et al. (2014a). Survival after neoadjuvant chemotherapy with or without bevacizumab or everolimus for HER2-negative primary breast cancer (GBG 44-GeparQuinto)dagger. Ann. Oncol 25, 2363–2372. [DOI] [PubMed] [Google Scholar]
- von Minckwitz G, Puglisi F, Cortes J, Vrdoljak E, Marschner N, Zielinski C, Villanueva C, Romieu G, Lang I, Ciruelos E, et al. (2014b). Bevacizumab plus chemotherapy versus chemotherapy alone as second-line treatment for patients with HER2-negative locally recurrent or metastatic breast cancer after first-line treatment with bevacizumab plus chemotherapy (TANIA): an open-label, randomised phase 3 trial. Lancet Oncol 15, 1269–1278. [DOI] [PubMed] [Google Scholar]
- Wang C, Wu H, Katritch V, Han GW, Huang XP, Liu W, Siu FY, Roth BL, Cherezov V, and Stevens RC (2013). Structure of the human smoothened receptor bound to an antitumour agent. Nature 497, 338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Wang W, Chen W, Lian F, Lang L, Huang Y, Xu Y, Zhang N, Chen Y, Liu M, et al. (2018). Pharmacological inhibition of dihydroorotate dehydrogenase induces apoptosis and differentiation in acute myeloid leukemia cells. Haematologica 103, 1472–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue R, Fang Z, Zhang M, Yi Z, Wen C, and Shi T (2013). TCMID: Traditional Chinese Medicine integrative database for herb molecular mechanism analysis. Nucleic Acids Res 41, D1089–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos GD, Davis S, Gale NW, Rudge JS, Wiegand SJ, and Holash J (2000). Vascular-specific growth factors and blood vessel formation. Nature 407, 242–248. [DOI] [PubMed] [Google Scholar]
- Ye H, Ye L, Kang H, Zhang D, Tao L, Tang K, Liu X, Zhu R, Liu Q, Chen YZ, et al. (2011). HIT: linking herbal active ingredients to targets. Nucleic Acids Res 39, D1055–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Yao X, Pang S, Chen P, Jiang W, Shan Z, and Zhang Q (2017). The deubiquitinase UCHL5/UCH37 positively regulates Hedgehog signaling by deubiquitinating Smoothened. J. Mol. Cell Biol 10, 243–257. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Qiu P, and Ji Y (2014). TCGA-assembler: open-source software for retrieving and processing TCGA data. Nat. Methods 11, 599–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirlik K, and Duyster J (2018). Anti-angiogenics: current situation and future perspectives. Oncol. Res. Treat 41, 166–171. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code written for and used in this study is available from the Lead Contact (Feixiong Cheng, chengf@ccf.org) upon reasonable request. Additional data supporting the findings of this study are available within the supplemental information files or from the Lead Contact upon reasonable request.


