Abstract
Purpose:
Small cell lung cancer (SCLC) has been treated clinically as a homogeneous disease, but recent discoveries suggest that SCLC is heterogeneous. Whether metabolic differences exist among SCLC subtypes is largely unexplored. In this study, we aimed to determine whether metabolic vulnerabilities exist between SCLC subtypes that can be therapeutically exploited.
Experimental Design:
We performed steady state metabolomics on tumors isolated from distinct GEMMs representing the MYC and MYCL-driven subtypes of SCLC. Using genetic and pharmacological approaches, we validated our findings in chemo-naive and resistant human SCLC cell lines, multiple GEMMs, four human cell line xenografts, and four newly-derived PDX models.
Results:
We discover that SCLC subtypes driven by different MYC family members have distinct metabolic profiles. MYC-driven SCLC preferentially depends on arginine-regulated pathways including polyamine biosynthesis and mTOR pathway activation. Chemo-resistant SCLC cells exhibit increased MYC expression and similar metabolic liabilities as chemo-naive MYC-driven cells. Arginine depletion with pegylated arginine deiminase (ADI-PEG 20) dramatically suppresses tumor growth and promotes survival of mice specifically with MYC-driven tumors, including in GEMMs, human cell line xenografts, and a PDX from a relapsed patient. Finally, ADI-PEG 20 is significantly more effective than the standard of care chemotherapy.
Conclusion:
These data identify metabolic heterogeneity within SCLC and suggest arginine deprivation as a subtype-specific therapeutic vulnerability for MYC-driven SCLC.
Keywords: small cell lung cancer, mouse models, MYC, arginine
Introduction
Small cell lung cancer (SCLC) is a major subtype of lung cancer accounting for ~15% of all lung cancer cases with a five-year survival rate of less than 6% (1). Surgery for SCLC is rare due to highly metastatic disease. The standard treatment regimen is platinum-based chemotherapy usually combined with the topoisomerase II inhibitor etoposide (2). First-line treatment in SCLC elicits responses in 60–80% of patients, but almost all patients relapse within 6–12 months and second-line therapy provides minimal survival benefit (2). This treatment approach has remained largely unchanged for ~40 years. New approaches are urgently needed to identify more effective therapeutic strategies in chemo-naive and resistant SCLC.
It has become increasingly appreciated that SCLC is a heterogeneous disease exhibiting intratumoral and intertumoral heterogeneity (3–9). Nearly all SCLCs have loss-of-function alterations in RB1 and TP53 (10–12). MYC family (MYC, MYCL, and MYCN) amplifications and overexpression however, are mutually exclusive (10, 11). Human SCLC cell lines have been categorized as classic or variant based on their morphology, neuroendocrine gene expression pattern, and status of MYC family member expression (13–15). Specifically, MYCL and ASCL1 expression are associated with classic SCLC, whereas MYC and NEUROD1 expression are associated with variant SCLC. Previous studies identified Mycl and Ascl1 as key drivers of tumorigenesis in classic SCLC that are required for tumor growth (3, 16, 17). The variant morphology was not observed in genetically engineered mouse models (GEMMs) until recently when our group showed that MycT58A overexpression in Rb1fl/fl;Trp53fl/fl mice promotes SCLC that recapitulates variant characteristics (13–15, 5, 18). Importantly, these molecular subtypes are therapeutically relevant as MYC-driven SCLC is particularly sensitive to inhibition of Aurora A/B kinases or CHK1 (5, 4, 19, 20). Indeed, a recent clinical trial with Aurora A inhibitor Alisertib in relapsed SCLC appeared to be a failure until patient samples were stratified based on MYC status (6). Together these studies suggest that SCLC can be defined based on MYC family member expression with unique therapeutic vulnerabilities.
Metabolic changes accompanying cell transformation are necessary to meet the metabolic demands of malignant cells, which include changes in energy formation, biosynthesis and redox homeostasis (21). MYC is one of the most frequently deregulated oncogenes in cancer and is a master regulator of glycolysis, glutamine metabolism, nucleotide biosynthesis and other metabolic processes (22). Mammalian Target of Rapamycin (mTOR) is a serine/threonine kinase that regulates cell growth, protein translation and a network of metabolic changes including lipid and nucleotide biosynthesis (23). mTOR is stimulated by growth factors via the PI3K/AKT pathway and/or amino acids including arginine, leucine or glutamine via the Ragulator complex (24). mTOR inhibitors in combination with either BCL2 inhibitors, BH3 mimetics or chemotherapy have shown efficacy in SCLC cell lines and xenografts, although these studies did not evaluate MYC status or the chemo-resistant setting (25–27). In SCLC clinical trials, mTOR inhibitors did not demonstrate a significant improvement in outcome either in the first-line setting combined with chemotherapy or in the second-line setting as a monotherapy (28–30). However, these studies did not determine whether MYC status could stratify patient response.
In addition to promoting mTOR activity, arginine regulates nitric oxide generation via nitric oxide synthase (NOS) and polyamine biosynthesis via ornithine decarboxylase 1 (ODC1) (31). Nitric oxide (NO) can exhibit both anti- and pro-tumor effects, and has been shown to regulate angiogenesis, apoptosis, cell cycle, invasion and metastasis (32). Polyamines are highly regulated organic cations that are elevated in proliferating tissues including various cancers (31). While high polyamine levels are associated with increased cancer cell proliferation, reduced apoptosis and increased expression of metastasis genes, the mechanisms underlying these effects have not been well defined (31). Previous work demonstrated that a single variant SCLC cell line was dependent on polyamine biosynthesis, but it is not clear whether classic SCLC cells are also dependent (33, 34). Since arginine is the precursor for NO generation, polyamine biosynthesis, and mTOR pathway activation, depleting arginine in tumors has been proposed as a therapeutic strategy for cancer. ADI-PEG 20 is a pegylated version of arginine deiminase (ADI) that depletes peripheral blood arginine levels and is currently in clinical trials for multiple cancers including SCLC (35). Argininosuccinate synthase 1 (ASS1) catalyzes the generation of argininosuccinate, a precursor in arginine biosynthesis. While ASS1 is a relatively ubiquitous enzyme, loss of ASS1 causes tumors to be highly auxotrophic for arginine, and this is correlated with chemo-resistance and poor clinical outcomes (36). Accordingly, tumors and cell lines that lack ASS1 have been shown to be more sensitive to ADI-PEG 20 (36). In a recent clinical trial of ADI-PEG 20 in patients with relapsed sensitive or refractory SCLC, most SCLCs did not demonstrate tumor regression, but 18% (4/22) of patients exhibited stable disease (). This study did not evaluate MYC status so it is currently unknown whether SCLC subtypes have differential responses to arginine depletion.
Here, we used an unbiased metabolomic approach with mouse and human model systems to define novel metabolic liabilities that can be therapeutically exploited in MYC-driven SCLC.
Methods
Mice
RPM (JAX stock #029971), RPR2, RPP, and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) (JAX stock #005557) mice were housed in an environmentally controlled room and experiments were performed in accordance with University of Utah’s Institutional Animal Care and Use Committee (IACUC). RPM mice were generated as previously described (5). RPP and RPR2 mice were provided by D. MacPherson and J. Johnson, respectively (3, 37). At 6–8 weeks of age, anesthetized mice were infected with 108 plaque-forming units of Ad5-Cgrp-Cre (RPM and RPP) or Ad5-CMV-Cre (RPR2) viruses (University of Iowa) by intratracheal instillation as described elsewhere (38). Viruses were administered in a Biosafety Level 2+ room according to Institutional Biosafety Committee guidelines.
For drug treatment studies, mice were given freshly prepared cisplatin (5 mg/kg, Sigma #P4394) in PBS on Day 1, etoposide [8 mg/kg (RPP) or 10 mg/kg (RPM), Sigma #E1383] in 70% PEG in water on Day 2 by intraperitoneal (i.p.) injection and repeated on a weekly basis until sacrifice. Control mice received PBS with equivalent volumes based on body weight. From the 5th round of chemotherapy onwards, mice were treated with etoposide only for toxicity reasons. For RPP treatments, mice received a one week break following two rounds of treatment and were treated on alternate weeks following the 5th round of treatment to ameliorate toxicity. Freshly prepared AZD2014 (20 mg/kg, AstraZeneca) in 1% Tween 80 in deionized water was administered orally on a 2 day on/5 day off regimen from Day 1 until sacrifice. ADI-PEG 20 (5 IU, Polaris) was administered via i.p. injections once a week. Due to their differences in tumor growth rate, RPM mice were imaged twice a week whereas RPP mice were imaged once weekly during the course of treatment. For survival studies, endpoints include but are not limited to: difficulty breathing, eating or moving, obvious signs of pain, or weight loss > 20% of initial body weight. For the xenograft experiments, 3–5 ×106 cells were suspended in a 1:1 mixture of Matrigel (BD Bioscience #356237) and RPMI, and then implanted into the flanks of NSG mice. Treatment was initiated once tumor volume reached 100–200 mm3 with an identical treatment regimen to the RPM mice. Mice were sacrificed once the tumor volume reached 2000 mm3 or when mice exhibited any sign of distress.
Patient derived xenograft (PDX) derivation
All patients were consented for the collection of human specimens, and approved by the University of Utah Institutional Review Board (IRB_00010924 or IRB_00089989) in accordance with the U.S. Common Rule. PDX implantation was conducted under a protocol approved by the University of Utah’s IACUC. PDXCTC (HCISCLC002, 003, 008, 009, 010, 011, and 015): Circulating tumor cells (CTCs) were enriched from 10–30 ml of whole blood collected in EDTA vacutainers (BD# 366643). White blood cells and red blood cells (RBCs) were crosslinked using RosetteSep™ Cocktail (StemCell Technologies, Cat#15127) and CTCs were separated by density gradient centrifugation using Lymphoprep™ (StemCell Technologies, Cat# 7801). The enriched CTCs were resuspended in 100 µl of Matrigel (Corning, Cat# 47743–715) and injected subcutaneously into NSG mice. PDXEBUS (HCISCLC004): Endobronchial ultrasound-guided transbronchial needle aspirate (EBUS-TBNA) specimens were obtained according to standard clinical protocols. The EBUS-TBNA specimen was processed by adding ice-cold, sterile PBS to a total volume of 500 µl, centrifuged, and resuspended in 5 ml of PBS supplemented with 2% FBS and placed on ice. To remove RBCs, the specimen was incubated with 25 ml of 1xACK (Ammonium-Chloride-Potassium) lysing buffer for 5 min at room temperature, centrifuged, and washed with 5 ml of PBS with 2% FBS. Subsequently, the specimen was centrifuged, PBS was removed, and tissue was combined with 100 µl volume of ice cold Matrigel and allowed to solidify for 5 min at 37°C. To generate the PDX, EBUS-TBNA derived tissue embedded in Matrigel was implanted subcutaneously into the flank of NSG mice. PDXSURGERY (HCISCLC0012): Tumor tissue was harvested into serum-free DMEM and kept at 4°C. The tumors were cut into 3×3 mm fragments and implanted subcutaneously into the flank of NSG mice. Successfully engrafted PDX tumors were further passaged into NSG mice for expansion and analysis. Confirmation that PDX tumors are of human origin was verified with human-specific anti-mitochondrial IHC (Abcam, Cat#ab92824). Xenograft-associated lymphoproliferative disorders were ruled out with human-specific CD45 IHC (Santa Cruz Biotechnology, Cat#sc-18901). SCLC histopathology was confirmed by pathologists. For drug studies involving PDX, treatment was initiated once the tumor volume reached 100–200 mm3 with an identical treatment regimen to the RPM mice. Mice were sacrificed once the tumor volume reached 2000 mm3 or when mice exhibited any signs of distress.
Cell lines
Cell lines were cultured in RPMI or HITES supplemented with 10% FBS and 1% pen/strep. GLC1, GLC8, NCI-H1092, NCI-H2141 and SBC4 were kindly provided by M. Sos (Cologne, Germany). NCI-H82, NCI-H524, NCI-H446, NCI-H889 and NCI-H69 were obtained from ATCC. NCI-H1963 was kindly provided by R. Govindan (St. Louis, MO, USA) and H1048 and DMS53 were kindly provided by D. MacPherson (Seattle, WA, USA). Cell lines were tested for Mycoplasma contamination using e-Myco PCR detection kit (Bulldog Bio: 25233) in March, 2019. GLC1, GLC8, H69, H82, H446, H524, H1092, H2141 and SBC4 were authenticated by short tandem repeat (STR) profiling in June, 2017. DMS53, H1048, H889 and H1963 were validated by STR profiling in February, 2018. Multiple vials of cell lines were cryopreserved upon acquisition and the cells were passaged for no more than 6 months in culture.
Nutrient deprivation assays
For amino acid withdrawal assays, 2.5–5 × 103 cells were seeded in a flat-bottom 96-well plate overnight with 5–6 technical replicates. The following day, the culture medium was changed either to complete RPMI or arginine, leucine, or glutamine-depleted RPMI. Cell viability was measured using Cell Titer Glo® (CTG) (Promega) 72 hrs post depletion. For chemo-resistant cell lines, 10–15 ×104 cells were seeded in 6-well plates and counted at 0 and 72 hrs using Countess II (AMQAX1000) and results depicted as relative viability. Arginine or leucine-depleted RPMI was prepared by using RPMI-1640 medium without L-arginine, L-leucine and L-lysine powder, supplemented with additional nutrients to obtain complete RPMI media or arginine/leucine-depleted RPMI (US Biologicals R8999–03A). Glutamine-depleted RPMI was prepared using RPMI-1640 medium without L-glutamine powder (Corning 90–022), supplemented with or without glutamine to obtain complete RPMI or glutamine-depleted RPMI, respectively. L-arginine (BP-370), L-leucine (BP-385) and L-lysine hydrochloride (BP-386) were acquired from Fisher Scientific, L-glutamine (25030–81) from Invitrogen, putrescine dihydrochloride (P-5780) and L-citrulline (C7629) from Sigma-Aldrich.
Immunoblot antibodies
Primary antibodies for immunoblot include: MYC (CST #13987, 1:1,000), MYCN (Santa Cruz sc-791, 1:200), ODC1 (EMD Millipore, MABS36, 1:100), ASCL1 (BD Bioscience, BDB556604, 1:300), phospho-4EBP1 (CST #2855, 1:1,000), 4EBP1 (CST #9644, 1:2,000), phospho-S6 (CST #2211, 1:1,000), S6 (CST #2217, 1:1,000), phospho-H2AX (CST #9718, 1:1000), PARP (CST #9532, 1:1000), ASL (Abcam#201026), ASS1 for mouse tumors (Abcam #170952, 1:1,000), ASS1 for human PDX and cell lines (Polaris Pharmaceuticals, 1:1000), and HSP90 (CST #4877, 1:1,000).
Cell viability assays
2.5–5 × 103 cells were seeded per well in triplicate in white, flat-bottom 96-well plates. The next day, cells were treated to generate 8-point dose response curves with increasing doses of L-NAME (Sigma #N5751), DFMO (Santa Cruz #252762), AZD8055 (Selleckchem #S1555) or AZD2014. After 96 hrs of treatment, cell viability was measured using CTG reagent on a luminometer. For assays involving chemotherapy, the cells were treated with cisplatin or etoposide. AZD2014, cisplatin and etoposide used for in vitro studies were identical to the those used for in vivo studies. After 48 hrs of treatment, cell viability was measured using CTG reagent on a luminometer. Normalized, transformed dose response curves were generated and analyzed using GraphPad Prism (GraphPad) to determine EC50 for each compound. Non-targeting siRNAs (D-001810) or those targeting ODC1 (L-006668–00) or MYC (L-003282–02) were acquired from Dharmacon. X-treme gene siRNA transfection reagent (4476093001) was acquired from Sigma-Aldrich. siRNAs were used at 100 nM concentration and cells were transfected following manufacturer’s guidelines.
Statistics
GraphPad Prism was used to perform statistical analyses. Survival studies were analyzed using log-rank (Mantel-Cox) test. Error bars represent mean ± SD unless otherwise indicated. For the statistical analysis of the in vitro drug treatments or tumor burden, column analysis was performed using Student’s unpaired t-test with p value < 0.05 considered statistically significant. For tumor growth in vivo over time, two-way ANOVA was performed with Greenhouse-Geisser correction with or without Sidak’s multiple comparison test as indicated in figure legends.
Other methods
The details of other methods including metabolomics, immunoblot, ChIP-seq, RNA-seq, immunohistochemistry, drug screen bioinformatic data, plasmids and microCT imaging are given in the Supplementary Materials and Methods.
Results
MYC-driven SCLC is metabolically distinct from MYCL-driven SCLC with enrichment of arginine biosynthetic pathways
Our laboratory recently developed a MYC-driven GEMM of SCLC that is molecularly distinct from MYCL-associated GEMMs (5). We sought to determine whether these distinct subtypes of SCLC have unique metabolic profiles that could imply new therapeutic vulnerabilities. To test this, we analyzed the abundance of ~120 metabolites by steady state metabolomics comparing MYC-driven tumors from Rb1fl/fl;p53fl/fl;MycT58ALSL/LSL (RPM) mice to MYCL-driven tumors from Rb1fl/fl;p53fl/fl;Ptenfl/fl (RPP) mice (16, 5, 37). We previously showed that RPM tumors express higher levels of MYC and NEUROD1 and exhibit variant morphology, whereas RPP tumors express higher levels of Mycl and ASCL1 and exhibit classic morphology (5). Since RPP mice also develop NSCLC (18), we restricted our analysis to tumors with SCLC histology verified by H&E sections from the corresponding tumor. Principal Component Analysis (PCA) revealed that RPM tumors are metabolically distinct from RPP tumors (n=9 tumors per genotype in technical triplicate) (Fig. 1A). Supervised analysis identified significantly altered metabolites (VIP score >1) in each subtype (Fig. 1B). Approximately the same number of metabolites were relatively accumulated vs depleted in the RPM tumors, indicating complex metabolic changes between the genotypes. We further identified metabolites significantly upregulated in RPM tumors compared to RPP tumors and these included multiple components of arginine biosynthesis and nucleotide metabolism pathways (Supplementary Fig. S1A). Metabolite Set Enrichment Analysis (MSEA) of this dataset identified “Nucleotide metabolism”, “Arginine and proline metabolism”, and “Urea cycle” as some of the top potentially altered pathways in RPM tumors (Fig. 1C). We recently showed that nucleotide metabolism is differentially altered in MYC-driven SCLC (39), and thus focus on arginine metabolism here. Next, we explored whether these patterns were reflected at the gene expression level. Using publicly available datasets (5), we performed Gene Set Enrichment Analysis (GSEA) comparing RPM tumors to Rb1fl/fl;p53fl/fl;Rbl2fl/fl (RPR2) tumors, a second MYCL-driven model of SCLC that also expresses high Mycl and ASCL1 (40). According to GSEA, genes involved in “Arginine and proline metabolism” were significantly upregulated in RPM compared to RPR2 tumors (Supplementary Fig. S1B). Together these data demonstrate that SCLC subtypes driven by different MYC family members have distinct metabolic profiles with altered arginine biosynthetic pathways.
Fig. 1. MYC-driven SCLC is metabolically distinct from MYCL-driven SCLC with enrichment of arginine biosynthetic pathways.
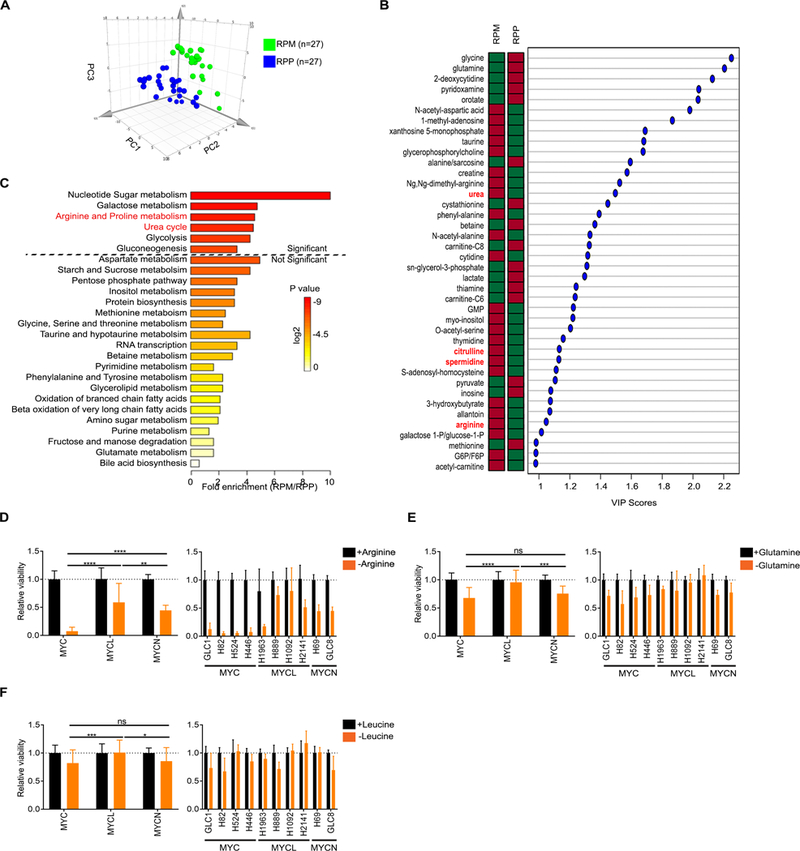
A, Principal component (PC) analysis of the metabolic signatures from RPM and RPP murine lung tumors. Nine individual tumors of each genotype were divided into three samples for technical replicates. B, Significantly altered metabolites between RPM and RPP tumors from panel A with variable importance in projection (VIP) score (VIP >1). Metabolites involved in arginine biosynthesis are marked in red. Relative metabolite abundance is indicated in the bar, with red representing relative accumulation and green representing relative depletion. C, MSEA of metabolites significantly upregulated in RPM compared to RPP tumors derived from panel B. D-F, Relative cell viability measured by CTG assay in response to depletion of arginine (D), glutamine (E), or leucine (F) in human SCLC cell lines (individual cell lines and grouped by genotype) measured in 4–6 replicates, 72 hrs post amino acid withdrawal. Mean ± SD of n = 2–5 experiments. Two-tailed unpaired t-test, ns = not significant, *p = 0.013, **p = 0.0045, ***p ≤ 0.0004, ****p < 0.0001.
Based on the metabolite and gene expression data, we tested whether human cell lines representing MYC (n = 4), MYCL (n = 3–4) or MYCN (n = 2)-driven subsets of SCLC (Supplementary Fig. S1C) exhibit differential responses to arginine withdrawal in vitro. Human SCLC cells were grown in complete media overnight. The following day, cells were changed to either complete media or amino-acid depleted media and assessed for proliferation after 72 hours. Interestingly, MYC-driven cell lines were much more sensitive to arginine depletion than MYCL- or MYCN-driven cell lines (Fig. 1D). To determine whether this dependency was specific to arginine, we depleted cells of either glutamine or leucine. While glutamine and leucine depletion both caused a reduction in cell growth in MYC-driven cells compared to MYCL-driven cells, the magnitude of this effect was minimal compared to arginine (Fig. 1E and F).
MYC-driven SCLC cells are sensitive to inhibition of polyamine biosynthesis and the mTOR pathway
Since MYC-driven SCLC cells exhibited a strong dependency on arginine, we sought to determine the function of arginine that cells may be reliant upon. Arginine regulates nitric oxide synthesis through NOS, polyamine biosynthesis through ODC1 (31), and mTOR pathway activation through the Ragulator complex (41) (Fig. 2A). In an attempt to separate these functions of arginine, we treated cells of each SCLC subtype with established inhibitors against NOS, ODC1 or the mTOR pathway (Fig. 2A). MYC-driven cell lines were technically more sensitive than MYCL- and MYCN-driven cells to NOS inhibition with L-NG-nitroarginine methyl ester (L-NAME) (Fig. 2B). However, this required extremely high doses of L-NAME (> 4,000 µM) that are not physiologically relevant. Similarly, MYC-driven cell lines were more sensitive to ODC1 inhibition by D, L-alpha-difluoromethylornithine (DFMO) than MYCL- or MYCN-driven cell lines, although these doses were also relatively high (100–600 µM) (Fig. 2C). ODC1 appears to be the major target of DFMO as cell viability upon DFMO treatment is rescued by addition of putrescine (Fig. 2D). To further test the role of ODC1 in MYC-driven SCLC, we knocked down ODC1 using pooled short interfering RNAs (siRNAs). Upon treatment with ODC1 siRNAs, MYC-driven cell lines exhibited reduced proliferation at 48 and 72 hrs post treatment while there was no effect on a MYCL-driven cell line (Fig. 2E and F) suggesting that MYC-driven cells are preferentially dependent on polyamine biosynthesis.
Fig. 2. MYC-driven SCLC cells are sensitive to inhibition of polyamine biosynthesis and the mTOR pathway.

A, Schematic of arginine biosynthetic pathways with indicated pharmacological inhibitors used near red bars. B-C, EC50 values of SCLC cell lines (individual and grouped by genotype) treated with L-NAME (B) or DFMO (C) in triplicate for 96 hrs. Mean ± SD for n = 2–6 experiments. D, Relative cell viability in response to 1 mM DFMO with or without 1 mM putrescine in MYC-driven H82 (left) and H446 (right) cells seeded in triplicate for 96 hrs. Mean ± SD, n = 2 experiments. E, Relative cell viability following treatment with non-targeting (CTRL) or ODC1 siRNAs at indicated timepoints. Mean ± SD, n = 2 experiments. F, ODC1 protein levels following treatment with non-targeting CTRL (–) or ODC1 siRNAs (+) at 72 hrs assessed by immunoblot. HSP90 serves as loading control. G–H, EC50 values of SCLC cell lines (individual and grouped by genotype) treated with AZD8055 (G) or AZD2014 (H) in triplicate for 96 hrs. Mean ± SD for n = 2–6 experiments. # = verified mutation in the PI3K/AKT pathway in the indicated cell lines. H446 cells harbor loss of PTEN and mTOR mutation (missense), while H69 cells have an activating PIK3CA mutation. Two-tailed unpaired t-test, ns = not significant, *p = 0.0182, **p < 0.0013, ****p < 0.0001.
To test the cells reliance on the mTOR pathway, we used the mTORC1/2 ATP-competitive inhibitors AZD8055 and AZD2014, which are close analogues of each other. MYC- and MYCN-driven cell lines displayed a striking sensitivity to mTOR pathway inhibition with low nanomolar EC50s (50–750 nM) compared to micromolar EC50s in MYCL-driven cell lines (Fig. 2G and H). We did not observe any subtype-specific differences in mTOR pathway activation as assessed by immunoblot for p4EBP1 and pS6 (Supplementary Fig. S2A), suggesting that the basal state of the pathway does not explain this result. To test whether arginine directly regulates the mTOR pathway in these cells, we analyzed cells at multiple time points following arginine depletion. While arginine depletion strikingly reduced mTOR pathway levels in MYC-driven cell lines, the effects were not as pronounced in MYCL- and MYCN-driven cell lines (Supplementary Fig. S2B). Additionally, we also observed an increase in DNA damage and apoptosis markers in MYC-driven cells compared to MYCL- and MYCN-driven cells following arginine depletion (Supplementary Fig. S2B). We further validated the specificity of our findings by withdrawing leucine from our media and testing its role in mTOR pathway regulation. We observed that, unlike arginine depletion, leucine depletion did not exhibit a striking reduction in mTOR pathway levels in MYC-driven cells (Supplementary Fig. S2C).
A recent NCI drug screen profiled the effects of 526 small molecule inhibitors on the proliferation of 65 SCLC cell lines with associated gene expression data (42). To confirm our findings in an independent dataset, we classified these cell lines based on high or low MYC expression and analyzed drug responses. We found that human cell lines with high MYC expression demonstrated an increased sensitivity to multiple mTORC1/2 and dual PI3K/mTOR inhibitors including AZD8055 and AZD2014 (Supplementary Fig. S2D). We did not find known NOS or ODC1 inhibitors included in this drug screen. Together, these results indicate that MYC-driven SCLC cells have increased dependency on arginine and its regulated pathways including polyamine biosynthesis and mTOR.
To determine whether these vulnerabilities were simply associated with proliferation rates, we measured cell doubling times by manually counting the cells at multiple timepoints. While MYC-driven cells had significantly increased proliferation rates in general (Supplementary Fig. S2E), MYC-high H524 cells exhibited drug sensitivity despite relatively slow proliferation. Furthermore, when cells were treated with a different metabolic inhibitor 2-Deoxy-D-glucose (2-DG), we did not observe genotype-specific sensitivity (Supplementary Fig. S2F). Together, this suggests that MYC status, not necessarily proliferation rate per se, correlates with sensitivity to arginine withdrawal, polyamine biosynthesis and mTOR pathway inhibition.
Chemo-resistant SCLC cells are metabolically distinct from chemo-naive cells and depend on arginine, polyamine biosynthesis and the mTOR pathway
Chemo-resistance is one of the major barriers to SCLC treatment in the clinic. To determine whether metabolic changes occur during chemo-resistance, we established two matched pairs of human chemo-resistant cell lines: DMS53 and H1048 (43). DMS53 is considered to be a MYC+/ASCL1-high cell line that clusters with classic SCLC cell lines exhibiting neuroendocrine features (5) consistent with our immunoblot results (Fig. 3A). While a published report suggests that H1048 is a MYC-low cell line (44), RNA-Seq from another published report describes H1048 as having similar MYC expression as DMS53 (42). More recent reports suggest that H1048 expresses the POU2F3 transcription factor and is negative for ASCL1 and NEUROD1, representing a variant form of SCLC associated with the tuft cell lineage (45). Both cell lines were treated with effective concentrations at 20% viability (EC20) doses of cisplatin, etoposide, or both (the standard of care in SCLC) repeatedly until they acquired stable resistance. While H1048 acquired stable resistance against cisplatin and etoposide, DMS53 failed to acquire stable resistance to etoposide even after 16 doses, consistent with the notion that these cells are inherently etoposide-resistant (46). In general, once the cell lines became resistant to one agent, they exhibited cross-resistance to the other chemotherapy—mimicking trends seen in the human disease (Supplementary Fig. S3A). To analyze metabolic differences between chemo-naive and chemo-resistant cell lines, we performed unbiased metabolite profiling as in Fig. 1. We used etoposide/cisplatin-resistant cells (ECR) for H1048, and the cisplatin-resistant cells (CR) for DMS53 since we could not generate ECR for this line. PCA revealed that resistant cells are metabolically distinct from parental cells (Fig. 3B and Supplementary Fig. S3B–S3C). MSEA using significantly upregulated metabolites (VIP score > 1) revealed that some of the potentially altered pathways in chemo-resistant cells are protein and amino acid biosynthesis pathways including “Arginine and proline metabolism” and “Urea cycle” (Fig. 3C and D). Together this suggests that chemo-resistant SCLC cells are metabolically distinct from chemo-naive cells.
Fig. 3. Chemo-resistant SCLC is metabolically distinct from chemo-naive SCLC and exhibits dependency on arginine, polyamine biosynthesis, and the mTOR pathway.
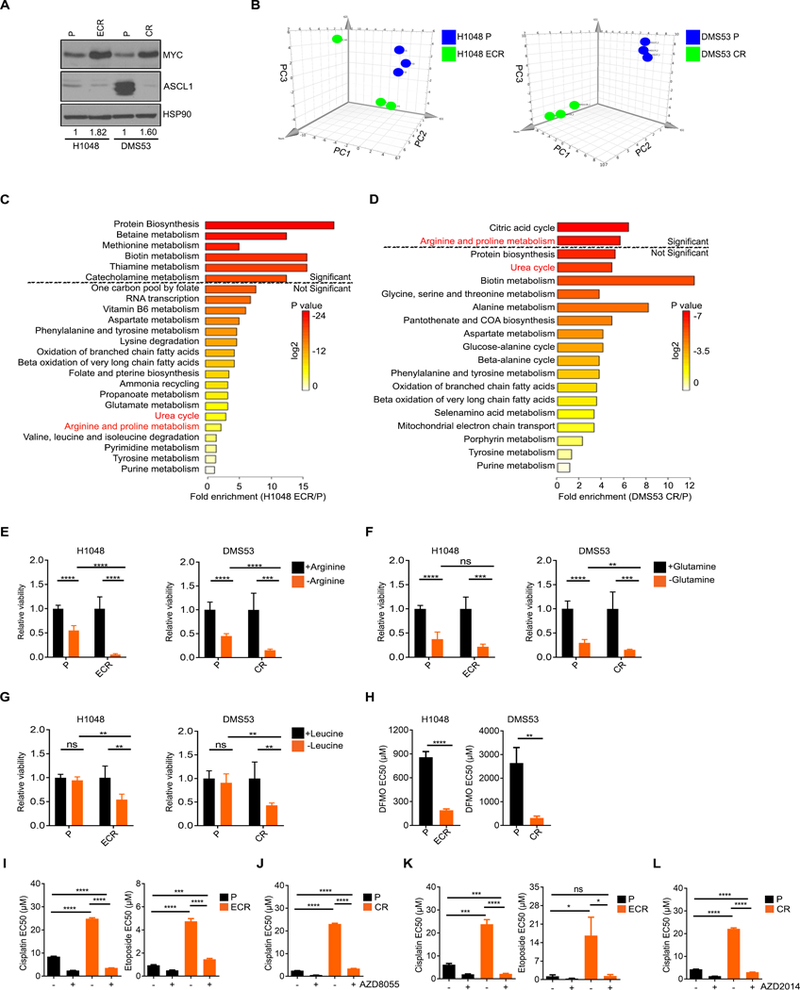
A, MYC and ASCL1 protein levels in H1048 parental (P) and etoposide/cisplatin-resistant (ECR) H1048 and DMS53 cisplatin-resistant CR cell lines as assessed by immunoblot. MYC protein levels in chemo-resistant cell lines relative to HSP90 are normalized to parental cells and indicated under the blots. B, Principal component (PC) analysis of the metabolic signatures from H1048 P vs H1048 ECR (left) and DMS53 vs DMS53 CR (right) cell lines. n = 3 biological replicates per cell line. C and D, MSEA of metabolites significantly upregulated in H1048 ECR (C) or DMS53 CR (D) compared to chemo-naive cells (P) of each line. E-G, Relative cell viability in response to depletion of arginine (E), glutamine (F), or leucine (G) in H1048 P vs ECR (left) and DMS53 P vs CR (right) measured in duplicate, 72 hrs post amino acid withdrawal. Mean ± SD for n = 2–4 experiments. H, EC50 values for H1048 P vs ECR and DMS53 P vs CR cell lines treated with DFMO. Mean ± SD for n = 2 experiments. I-L, EC50 values for cisplatin and etoposide in combination with AZD8055 or AZD2014. H1048 P vs H1048 ECR (I and K) and DMS53 vs DMS53 CR (J and L) in triplicate treated for 48 hrs. Mean ± SD for n = 2 experiments. Two-tailed unpaired t-test, ns = not significant, *p < 0.02, **p < 0.002, ***p < 0.0008, ****p < 0.0001.
To test whether chemo-resistant cells are also dependent on exogenous arginine, we depleted arginine from parental and chemo-resistant cell lines. As observed in MYC-driven cells, chemo-resistant SCLC cells were much more sensitive to arginine withdrawal compared to chemo-naive cells (Fig. 3E). Chemo-resistant cells also exhibited preferential sensitivity to glutamine and leucine withdrawal but not to the same extent as arginine (Fig. 3F and G). The differences in amino acid responses observed with H1048 and DMS53 parental cells compared to cells used in Figure 1 might be attributed to the different molecular subtypes of the cell lines used (9). Overall, these data indicate that chemo-resistant cells demonstrate an altered metabolic profile and increased arginine dependency similar to chemo-naive MYC-driven cells.
Since chemo-resistant cells displayed an increased dependency on arginine, we sought to explore the role of arginine using inhibitors to NO generation, polyamine biosynthesis and the mTOR pathway. Chemo-resistant cells were not more sensitive than parental cells to NOS inhibition with L-NAME, suggesting that NOS is not involved in chemo-resistance (Supplementary Fig. S3D). In contrast, ODC1 inhibition by DFMO significantly inhibited growth of chemo-resistant cells (Fig. 3H), indicating that chemo-resistant cells are dependent on polyamine biosynthesis similar to chemo-naive MYC-driven SCLC cells. We did not observe differences in ODC1 levels across parental and chemo-resistant lines, indicating that the need for polyamines rather than ODC1 levels may dictate DFMO sensitivity in chemo-resistant cells in culture (Supplementary Fig. S3E).
Next, we tested whether chemo-resistant cells demonstrate preferential sensitivity to mTOR pathway inhibition using AZD8055 and AZD2014. Compared to the chemo-naive MYC-driven SCLC cells, mTOR inhibitors exhibited a relatively modest inhibition of cell growth in chemo-resistant cells (Supplementary Fig. S3F). Parental H1048 cells harbor an activating PIK3CA mutation that may explain their lack of increased sensitivity to mTOR inhibitors upon chemo-resistance (47). We hypothesized that mTOR activation may be particularly important during the stress response of chemotherapy. To test this, we treated parental and chemo-resistant cells with mTOR inhibitors combined with cisplatin or etoposide. Combining mTOR pathway inhibition with either cisplatin or etoposide dramatically sensitized chemo-resistant cells to chemotherapy, often to levels comparable to the chemo-naive cells (Fig. 3I–L). mTOR pathway activity was upregulated in both sets of chemo-resistant cells as indicated by phosphorylated 4EBP1, while pS6 levels did not change (Supplementary Fig. S3G). These results suggest that mTOR pathway activity protects chemo-resistant cells from chemotherapy. Given that the chemo-resistant cells demonstrated similar metabolic liabilities as the MYC-driven cell lines, we analyzed chemo-naive and resistant cells for MYC protein levels. Immunoblotting revealed that the chemo-resistant cells had acquired increased levels of MYC (Fig. 3A). Together, this suggests that MYC expression and its associated metabolic vulnerabilities may be enriched during chemotherapy resistance, although we did not determine that the metabolic vulnerabilities depend on MYC alone.
MYC directly regulates the metabolic dependencies observed in MYC-driven SCLC cells
Since MYC-driven SCLC cells exhibited a strong dependency on arginine, polyamine biosynthesis and mTOR pathway, we decided to investigate if MYC had a causal role in regulating these dependencies. We used doxycycline-inducible short hairpin RNA (shRNA) to silence MYC gene expression. In MYC-driven H82 and H446 cells, we observed that MYC shRNA led to a partial reduction in MYC protein levels in a time-dependent manner (Fig. 4A). MYC knockdown reversed the metabolic dependency on arginine similar to levels seen in MYCL-driven lines (Fig. 4B). The observed effects were specific to MYC as we did not observe reduction in MYC protein levels or a reversal in arginine dependency upon knockdown with a non-targeting RENILLA shRNA (Fig. 4C and D). Next, we assessed whether MYC knockdown altered the sensitivity of cells to DFMO or mTOR inhibitor treatment. MYC knockdown led to a partial reversal in DFMO sensitivity in both H82 and H446 cells (Fig. 4E and F). MYC knockdown led to a modest but significant increase in EC50 values for mTOR inhibitors in H82 but not H446 cells (PTEN loss, missense mutation in mTOR) (Fig. 4G–J). Together, these results suggest that MYC at least partially regulates the metabolic dependencies observed in MYC-driven SCLC cells.
Fig. 4. The metabolic vulnerabilities observed in MYC-driven SCLC cells depend on MYC.

A, Immunoblot of MYC levels following doxycycline (doxy)-inducible MYC shRNA knockdown in H82 (left) and H446 (right) cells at indicated time points compared to doxy-treated uninfected cells. B, Relative cell viability in response to arginine depletion in uninfected (doxy-treated) or H82-shMYC cells (left) and H446-shMYC cells (right) measured in 4–6 replicates, 72 hrs post-withdrawal, compared to H1092 (MYCL-associated) cells at the indicated time points of doxy treatment. Mean ± SD, n = 2–3 experiments. C, Immunoblot of MYC levels following doxy-inducible RENILLA shRNA knockdown in H82 (left) and H446 (right) cells at indicated time points. D, Relative cell viability in response to arginine depletion in H82-shRENILLA cells (left) and H446-shRENILLA cells (right) measured in 4–6 replicates, 72 hrs post-withdrawal as in panel B. Mean ± SD, n = 2–3 experiments. E-J, EC50 values or relative cell viability of H82-shMYC (top row) and H446-shMYC (bottom row) cells treated with DFMO (E and F), AZD8055 (G and H), or AZD2014 (I and J) in triplicate for 96 hrs. Mean ± SD, n = 2–3 experiments. K, MYC and ODC1 protein levels by immunoblot in human cells grouped by MYC status. L, MYC protein levels following treatment with non-targeting CTRL (–) or MYC (+) siRNAs at indicated time points assessed by immunoblot. H1048 ECR (left) and DMS53 CR (right). M, Relative cell viability in response to arginine depletion following treatment with non-targeting CTRL (–) or MYC (+) siRNAs, measured in 2–5 replicates, 72 hrs post-withdrawal. Mean ± SD, n = 3 experiments. H1048 ECR (left) and DMS53 CR (right). A-J, H82, H446 and MYCL (H1092) cells were treated with doxy corresponding to the longest timepoint in the assay. HSP90 serves as loading control for all immunoblots. Two-tailed unpaired t-tests. *p = 0.0248, **p < 0.0045, ***p < 0.0001, ****p < 0.0001.
To better understand the mechanism of these therapeutic liabilities, we assessed levels of the key proteins involved in each pathway including ASS1, ODC1 and markers of mTOR pathway activation. Both ASS1 and ODC1 have previously been implicated as MYC targets (48, 49). Given the dependency of MYC-driven cells on exogenous arginine, we reasoned that they may have low levels of ASS1 and thereby be unable to generate sufficient arginine in its absence. In contrast to our expectations, MYC-driven cell lines expressed ASS1, while MYCL-driven cells had substantially less (Supplementary Fig. S4A). Arginine-deprived cells were partially or fully rescued upon citrulline addition consistent with ASS1’s ability to convert citrulline back to arginine (Supplementary Fig. S4B). We were surprised that MYCL-driven cells were not more sensitive to arginine withdrawal given their low ASS1 levels, so we questioned whether MYCL-driven cells could induce ASS1 in culture. To test this, we depleted arginine in MYCL-driven H1963 cells and assessed ASS1 at multiple time points. Arginine depletion significantly induced ASS1 as early as 8 hours and this continued to increase over 48 hours (Supplementary Fig. S4C). These results demonstrate that arginine availability can regulate ASS1 levels, and that basal ASS1 expression in cell culture may not be sufficient to predict arginine dependency.
Next, we assessed ODC1 levels across the panel of human cell lines. We did not observe subtype-specific differences in ODC1 levels as assessed by immunoblot, indicating that the need for polyamines rather than ODC1 levels may dictate DFMO sensitivity in MYC-driven cell lines in culture (Fig. 4K). To assess this further in tumors, we analyzed chromatin immunoprecipitation-sequencing (ChIP-Seq) data from MYC-driven SCLC mouse tumors. As expected from published studies (48), MYC bound the promoter region of Odc1 in vivo (Supplementary Fig. S4D). Consistent with Odc1 being a transcriptional target of MYC, Odc1 expression was significantly higher in RPM tumors compared to RPR2 tumors (Supplementary Fig. S4E). Furthermore, gene expression data from human SCLC tumors revealed that MYC and ODC1 levels are positively correlated (Supplementary Fig. S4F). In contrast, markers of mTOR pathway activity did not differ according to MYC status in human cell lines (Supplementary Fig. S2A). MYC knockdown led to a subtle reduction in ODC1 levels in H82 cells, but not H446 cells, in a time-dependent manner, but had no effect on mTOR pathway levels as measured by phospho-S6 and phospho-4EBP1 in either cell line (Supplementary Fig. S4G–S4H). Thus, it is possible that increased polyamine biosynthesis through ODC1 levels and/or activity in MYC-driven tumors may explain the increased dependency on polyamines. This is further supported by the observation that cell viability upon ODC1 inhibition with DFMO is rescued by addition of putrescine (Fig. 2D). In addition, supplementation of putrescine reversed mTOR inhibitor sensitivity in MYC-driven cell lines indicated by the increase in EC50 values (Supplemental Fig. S4I), suggesting a possible role of the mTOR pathway in regulating polyamine biosynthesis in SCLC. Together this suggests that MYC promotes metabolic dependencies in SCLC and this is at least partially through its regulation of polyamine biosynthesis genes such as ODC1.
Finally, we investigated if MYC had a causal role in regulating the metabolic dependencies observed upon chemo-resistance. We used pooled siRNAs to knockdown MYC in our chemo-resistant cell lines since these cells were difficult to infect. Upon treatment with MYC siRNAs, we observed a partial reduction in MYC protein levels over time (Fig. 4L). MYC knockdown in chemo-resistant cells led to a partial but statistically significant reversal in their arginine dependency (Fig. 4M). Altogether, these results suggest at least a partial role for MYC in regulating arginine dependency in relapsed SCLC.
MYC-driven mouse tumors are highly sensitive to arginine depletion in vivo
We next sought to determine whether our in vitro findings could be recapitulated in the preclinical RPM (MYC-driven) and RPP (MYCL-driven) mouse models of SCLC. We decided to test the mTOR inhibitor AZD2014 as it is currently undergoing phase 1 and 2 clinical trials for SCLC ( and ). To determine the efficacy of AZD2014 in vivo, RPM and RPP mice were infected with Ad5-Cgrp-Cre to initiate tumors in neuroendocrine cells, and then monitored for tumor development by microCT imaging. Consistent with published studies, RPM mice developed tumors approximately six weeks post-infection, whereas RPP mice developed tumors ~4–5 months post-infection. Upon tumor detection, mice were randomly assigned to one of four treatment groups: control (PBS), chemotherapy (5 mg/kg cisplatin and 8–10 mg/kg etoposide, i.p.), AZD2014 (20 mg/kg, p.o), or AZD2014 plus chemotherapy. Etoposide at 10 mg/kg in combination with 5 mg/kg cisplatin led to toxicity in RPP mice, and was adjusted to 8 mg/kg etoposide. Etoposide at 8 mg/kg with 5 mg/kg cisplatin lacked efficacy in RPM mice (Supplementary Fig. S5A), so etoposide was adjusted to 10 mg/kg in RPM mice. In RPP mice, combination chemotherapy significantly prolonged survival as expected (Supplementary Fig. S5B). AZD2014 monotherapy, however, did not improve overall survival of RPP mice compared to PBS control (Supplementary Fig. S5B). RPP mice treated with AZD2014 plus chemotherapy did not demonstrate an improvement in overall survival when compared to mice treated with chemotherapy alone (Supplementary Fig. S5B), suggesting that mTOR inhibition is ineffective in RPP mice. In RPM mice, chemotherapy significantly improved overall survival similar to and consistent with our previous studies (Supplementary Fig. S5C). Unlike in RPP mice, AZD2014 monotherapy caused a subtle but significant improvement in median survival of RPM mice compared to controls (Supplementary Fig. S5C). Furthermore, the combination of AZD2014 and chemotherapy modestly but significantly improved the overall survival of RPM mice beyond that of chemotherapy alone (Supplementary Fig. S5C). As measured by microCT imaging, control and AZD2014-treated mice exhibited a rapid increase in lung tumor volume following tumor detection (Supplementary Fig. S5D–S5E). Mice receiving AZD2014 plus chemotherapy demonstrated statistically-significant tumor stasis compared to chemotherapy-treated mice (Supplementary Fig. S5D). Over the course of 19 days of treatment, AZD2014 alone did not reduce tumor growth compared to control, but ~50% of the AZD2014 plus chemotherapy-treated mice exhibited stable disease compared to only ~12% of mice treated with chemotherapy alone (Supplementary Fig. S5F). Following Day 19, even tumors treated with combination chemotherapy and AZD2014 had rapidly rebounded.
While these studies demonstrate that AZD2014 with chemotherapy is more effective than chemotherapy in first-line treatment of MYC-driven SCLC, it is not clear whether this strategy is effective in relapsed disease. While tumors in RPM mice exhibit resistance following chemotherapy (5), their extremely rapid growth rate makes it difficult to address this question in this model.
Since arginine is required for polyamine biosynthesis and mTOR pathway activation, strategies to deplete arginine such as ADI-PEG 20 could represent an effective therapy for MYC-driven SCLC. To test this, MYC-driven RPM and MYCL-driven RPP and RPR2 mice were randomly assigned to treatment groups upon tumor detection. RPR2 and RPP mice were assigned to control (PBS) or ADI-PEG 20 (5 IU, i.p.). RPM mice were assigned to control, ADI-PEG 20, chemotherapy (5 mg/kg cisplatin and 10 mg/kg etoposide, i.p.), or ADI-PEG 20 plus chemotherapy. ADI-PEG 20 monotherapy did not improve overall survival of RPP or RPR2 mice compared to PBS control (Fig. 5A and B), suggesting that arginine depletion is not effective in MYCL-driven tumors. Remarkably, in RPM mice, ADI-PEG 20 monotherapy led to a dramatic improvement in overall survival beyond that of combination chemotherapy (Fig. 5C). ADI-PEG 20 treatment led to an additional 20 days of increased survival beyond that of combination chemotherapy (42 days vs 22.5 days) and as such, almost doubled the median survival rate compared to the current standard of care. As a result, ADI-PEG 20 is the most effective therapy administered to RPM mice in our hands thus far, surpassing Alisertib (5), AZD2014 (Supplementary Fig. S5C), Mizoribine (39), and combination chemotherapy. Interestingly, the addition of chemotherapy to ADI-PEG 20 treatment did not increase the overall survival of mice beyond that of ADI-PEG 20 alone (44 days vs 42 days) (Fig. 5C), suggesting that the survival benefit in the combination group was due entirely to ADI-PEG 20.
Fig. 5. MYC-driven mouse tumors are preferentially sensitive to arginine depletion in vivo.

A-C, Kaplan-Meier survival curve of RPP mice (A), RPR2 (B) or RPM (C) mice treated with indicated agents. ADI-PEG 20 treatments marked by vertical dashed lines. Chemotherapy treatments (weekly cisplatin and etoposide) marked by ticks on the x-axis in panel C. Log rank (Mantel-Cox) test. ns = not significant, ****p < 0.0001. Number of mice per group indicated in figure. D, Fold change in tumor burden (total tumor volume) in indicated treatment groups of RPM mice at indicated days post-treatment initiation. Mean ± SD. Two-way ANOVA with Greenhouse-Geisser correction, **p = 0.0015, ***p = 0002. E, Representative microCT images from RPM mice pseudo-colored with tumors (yellow) and normal tissue and airway (purple) at indicated time points post-treatment initiation. F, Total tumor volume change from Day 0 to Day 19 (or last scan before death) by waterfall plot in individual RPM mice. Gray shading on x axis indicates partial responses or stable disease. G, Immunoblot for ASS1 and ASL protein levels in individual RPM, RPP or RPR2 tumors. HSP90 serves as loading control. Two-tailed unpaired t-test, ns = not significant, **p < 0.0072, ***p = 0.0009.
As an additional measure of therapeutic impact, tumor volume in RPM mice was calculated by microCT imaging (5). Over a course of 19 days of treatment, mice receiving ADI-PEG 20 with or without chemotherapy demonstrated statistically-significant tumor regression followed by tumor stasis when compared to chemotherapy-treated mice (Fig. 5D and E). We also analyzed the percent change in total tumor volume at day 19 (or time of death if sooner) compared to day 0 in each treatment group by waterfall plot. Remarkably, 20 of 22 mice treated with ADI-PEG 20 with or without chemotherapy exhibited stable disease or regression, compared to mice treated with chemotherapy alone where only 2/17 mice exhibited stable disease or regression at this time point (Fig. 5F). Additionally, 4/12 mice treated with ADI-PEG 20, and 7/10 mice treated with the combination of ADI-PEG 20 and chemotherapy, displayed tumor regression compared to 1/17 mice treated with chemotherapy alone (Fig. 5F). Altogether, this suggests MYC-driven SCLC is highly sensitive to arginine depletion in vivo.
We examined the status of arginine biosynthetic enzymes ASS1 and ASL in RPM, RPP, and RPR2 tumors by immunoblot. While ASL was expressed across all subtypes, ASS1 levels were undetectable in RPM tumors while a majority of RPP and RPR2 tumors expressed ASS1 (Fig. 5G), providing a potential mechanistic explanation for their sensitivity to arginine depletion in vivo. Whether the discrepancy with ASS1 levels in human cell lines compared to mouse tumors reflects a mouse/human or in vitro/in vivo-specific phenomenon is currently unclear.
MYC-driven human SCLC is preferentially sensitive to arginine depletion in vivo
To determine the impact of arginine depletion in human xenografts, we implanted NSG mice subcutaneously with two variant (H82 and GLC1) or two classic (H1092 and H69) SCLC cell lines. Compared to PBS control, ADI-PEG 20 significantly impeded the growth of both MYC-high variant cell lines, but neither of the MYCL/N-high classic cell lines (Fig. 6A–D). These results further validate the efficacy of ADI-PEG 20 in MYC-high SCLC.
Fig. 6. MYC-driven human SCLC is preferentially sensitive to arginine depletion in vivo.

A–D, Tumor volume of xenografts in NSG mice injected with GLC1 (A), H82 (B), H69 (C), or H1092 (D) and treated with PBS control or ADI-PEG 20. Weekly ADI-PEG 20 treatments marked by dashed lines. E, Immunoblot for MYC, ASS1 and ASCL1 protein levels in individual PDX tumors. HSP90 serves as loading control. F-G, Tumor volume of xenografts in NSG mice injected with PDX#2 (F), or PDX#8, 9, and 10 depicted together in one graph (G). See Supplemental Figure 6D–F for individual PDX responses. Number of mice treated indicated in figures. Mean ± SD. Two-way ANOVA with Greenhouse-Geisser correction followed by Sidak’s multiple comparison test, *p < 0.03, **p < 0.008, ***p < 0.0008, ****p < 0.0001.
To determine the efficacy of arginine deprivation in an uncultured human tumor, we analyzed ASS1 and MYC status in a novel cohort of nine SCLC patient-derived xenograft (PDX) models that were generated from circulating tumor cells and/or tumor tissue using previously described methods (50)(Supplementary Figure S6A–B). Patients #2 and 3 received radiation therapy prior to PDX generation. Additionally, patients #2, 3 and 11 were also treated with carboplatin/etoposide prior to PDX generation. Immunoblotting revealed that 8 of 9 tumors were ASCL1+/MYC- (Figure 6E). Of these, 7/8 ASCL1+/MYC- PDX were either lowly or strongly positive for ASS1 either by immunoblotting or IHC with intratumoral variability (Fig. 6E and Supplementary Fig. S6C). One PDX (#2) was MYC+/ASCL1-/ASS1- and derived from a patient following relapse from chemotherapy. Consistently, treatment of PDX #2 with combination chemotherapy (cisplatin and etoposide) led to modest inhibition of tumor growth (Fig. 6F). Treatment of PDX #2 with ADI-PEG 20, however, significantly impeded tumor growth beyond that of combination chemotherapy (Fig. 6F). In contrast, MYC-/ASS1+ PDXs (#8, 9 and 10) were resistant to arginine depletion, similar to our observations with MYCL/N-high cell line xenografts and MYCL-driven GEMMs (Fig. 6G and Supplementary Fig. S6D–F). Together, MYC-driven human SCLC cell lines, xenografts, mouse tumors, and a human PDX demonstrate enhanced dependency on arginine.
Discussion
Despite numerous clinical trials and years of research, therapeutic options for SCLC remain limited. However, recent studies suggest that molecular subtypes of SCLC exist with distinct biology and therapeutic responses (9, 3, 5, 4, 6, 7). Here, we found that MYC and MYCL-driven SCLC tumors have distinct metabolic profiles with a dependency on arginine in MYC-driven tumors and cell lines. MYC-driven SCLC cells are highly dependent on arginine-regulated pathways including polyamine biosynthesis and mTOR pathway activation. Importantly, chemo-resistant SCLC displayed increased MYC expression and increased dependency on arginine, polyamine biosynthesis and mTOR pathway activity. Using GEMMs of distinct SCLC subtypes, we found that arginine depletion using ADI-PEG 20 was highly effective as a monotherapy in MYC-driven SCLC in mouse and human tumors, surpassing the efficacy of the standard of care chemotherapy.
While MYC-driven SCLC was preferentially sensitive to ODC1 inhibition, we did not test ODC1 inhibitors in vivo. There is still an incomplete understanding of polyamine function, and inhibitors have had modest efficacy as single agents in clinical trials (31, 51). ODC1 is a known MYC target, and we found Odc1 is a consistently-upregulated polyamine pathway enzyme in MYC-driven SCLC, so further studies warrant determining whether ODC1 is the critical target promoting polyamine dependency. Likewise, the mechanisms by which MYC may promote mTOR dependency in SCLC are unknown. mTOR inhibition can alter a vast array of metabolic networks and a recent report in prostate cancer found that the mTOR pathway regulates polyamine biosynthesis (52) and can regulate ODC1 mRNA translation (53), so future studies warrant understanding the relationship between these pathways. This is consistent with our data where we observe rescue of mTOR inhibition with putrescine. While our studies suggest that MYC-driven SCLC cell lines are likely dependent on arginine for polyamine biosynthesis, further studies are warranted to completely understand the function of arginine in MYC-driven SCLC. Recent advances in isotope tracing could help to shed light on the functions of arginine and polyamines in SCLC.
It was recently shown that tumors with activated mTOR signaling have increased dependency on the guanine nucleotide biosynthesis enzyme, IMPDH. IMPDH inhibition in the context of activated mTOR signaling leads to nucleotide depletion and an anabolic imbalance promoting cell death (54). We also observed an enrichment of nucleotide biosynthesis pathways in MYC-driven tumors (Fig. 1C) and MYC-driven tumors are also highly sensitive to IMPDH inhibition (39). Both MYC and mTOR regulate common targets such as ribosome biogenesis and nucleotide metabolism (22). Thus, it is possible that a similar anabolic imbalance may explain why MYC-driven tumors are sensitive to inhibition of both mTOR and polyamine biosynthesis.
While previous clinical trials with mTOR inhibitors in SCLC have not shown favorable outcomes, patients in those studies were not stratified based on MYC status (28–30). Based on our findings, it is possible that selecting patients based on expression of MYC family members may improve outcomes in the clinic. Additionally, all of the previous clinical trials with mTOR inhibitors used Rapalogs, which fail to inhibit mTORC2 in contrast to the dual mTORC1/2 inhibitors used here. While MYC-driven human cells demonstrated increased sensitivity to mTOR inhibition compared to MYCL-driven cells, mTOR inhibition alone was not particularly effective in our aggressive GEMM of MYC-driven SCLC. mTOR inhibition in combination with chemotherapy, however, significantly inhibited tumor growth and prolonged survival of mice with MYC-driven, but not MYCL-driven, SCLC. Thus, we predict that clinical trials with dual mTORC1/2 inhibitors combined with chemotherapy may be a more effective therapeutic strategy for first-line or second-line SCLC specifically for those with MYC-high tumors.
One of the major barriers to clinical progress in SCLC is the rapid emergence of chemotherapy resistance. Using unbiased metabolomic analyses, we found that chemo-resistant SCLC cells are metabolically distinct from chemo-naive SCLC. Our data implicate arginine and polyamine biosynthesis as regulators of chemo-resistance, similar to our results in MYC-driven SCLC. Interestingly, MYC expression was increased in two independent human cell lines following chemotherapy resistance. MYC activity was also reportedly upregulated in chemo-resistant PDX models and human SCLC tumors and cell lines (15, 55, 56). Pharmacological inhibition of the mTOR pathway in combination with chemotherapy reversed chemo-resistance in vitro, and chemo-resistant cells were also dependent upon arginine. This suggests mTOR inhibition with chemotherapy, or arginine deprivation, as potential second-line strategies in patients with MYC-high relapsed SCLC.
The striking sensitivity of murine MYC-driven SCLC to ADI-PEG 20 monotherapy warrants further investigation of ADI-PEG 20 as a first-line therapy for SCLC. Additionally, our results in MYC-high human cell line xenografts and a relapsed PDX suggest that MYC-driven human SCLC is preferentially sensitive to arginine withdrawal in vivo compared to MYCL/N-driven SCLC. While a Phase II clinical trial of relapsed SCLC patients treated with ADI-PEG 20 did not demonstrate significant tumor regression, 18% (4/22) of patients exhibited stable disease (). Our data suggest it will be important to determine whether the patients with stable disease were enriched for MYC-high tumors. Other arginine-modulating agents such as Pegzilarginase are also in clinical trials (), highlighting the need to assess MYC status in these trials as well. Given that chemotherapy did not improve the response to ADI-PEG 20 in our GEM models, it raises the possibility of identifying new combination strategies that synergize with arginine depletion for greater efficacy.
It is not yet clear why human cell lines and mouse tumors differ in their expression patterns of ASS1. It has been observed that ASS1 is silenced in multiple cancers to meet their requirements for purines and pyrimidines via aspartate, which otherwise could be used for the synthesis of arginine through ASS1 (57, 58). We speculate that MYC-driven mouse tumors silence ASS1 in order to divert aspartate to meet the metabolic demand for nucleotides. In contrast, cell lines are grown in nutritional excess and may not need to silence ASS1 to divert aspartate to nucleotide biosynthesis. We have observed an enrichment of nucleotide biosynthesis pathways in MYC-driven tumors (Fig. 1C) and shown that MYC-driven tumors exhibit an increased dependency on de novo guanine nucleotide synthesis (39). Further study is warranted to fully understand the regulation of ASS1 in SCLC.
In summary, SCLC subsets driven by different MYC family members have distinct metabolic programs that can be exploited to uncover subtype-specific therapies. Furthermore, chemo-resistant SCLC also acquires metabolic changes leading to new therapeutic vulnerabilities. Further studies into the unique metabolic liabilities among SCLC subtypes and their chemo-resistant counterparts may ultimately lead to more personalized diagnosis and treatment strategies for SCLC patients.
Supplementary Material
Translational relevance.
Small cell lung cancer (SCLC) is a highly aggressive form of lung cancer with poor overall survival. The standard of care consisting of combination platinum-based chemotherapy has remained similar for the last ~40 years. Recent work suggests that SCLC is a heterogeneous disease comprised of distinct molecular subtypes. Using a metabolomic approach, we discover that the MYC-driven subset of SCLC is highly dependent on arginine biosynthetic pathways. This metabolic dependency can be exploited using a clinically relevant agent to deplete arginine (ADI-PEG 20). In genetically engineered mouse models (GEMMs), the efficacy of ADI-PEG 20 substantially exceeds that of combination chemotherapy specifically in MYC-driven SCLC. ADI-PEG 20 is also effective in a MYC-high patient-derived xenograft (PDX) from a relapsed patient and in human MYC-high cell line xenografts. These results suggest arginine depletion as a potential therapeutic strategy for MYC-high SCLC.
Statement of Significance.
Using a metabolomic approach, we discover that the MYC-driven subset of SCLC is highly dependent on arginine biosynthetic pathways that can be exploited using clinically relevant agents for arginine depletion (ADI-PEG 20). These results suggest arginine depletion as a potential new therapeutic strategy specifically for MYC-high SCLC.
Acknowledgments:
We acknowledge support from the National Cancer Institute (NCI) of the National Institutes of Health (NIH) under award P30CA042014 awarded to Huntsman Cancer Institute (HCI) for the use of core facilities including Preclinical Research Resource, Biorepository and Molecular Pathology, and High-Throughput Genomics and Bioinformatics Analysis. Pilot funding was provided by the Huntsman Cancer Foundation and the Cell Response and Regulation program at HCI. TGO was supported in part by the American Lung Association (LCD-506758), Lung Cancer Research Foundation, and the NIH NCI (R21-1R21CA216504-01A1). RJD was supported by a V Foundation Clinical Investigator Award and NCI R35-CA220449. Thanks to K. Sutherland and A. Berns for permission to use Cgrp-Cre viruses, RPP mice from D. MacPherson, RPR2 tumors from J. Sage and J. Johnson, and NSG mice from S. Holmen. We appreciate technical assistance from members of the Oliver Lab including C. Lin, P. Ballieu, I. Can, D. Hansen and C. Whitney for assistance with mouse work, B. Anderson and K. Gligorich for histological services, B. Dalley and T. Mosbruger for bioinformatics support, and R. Olsen and R. Dahlgren for administrative support.
Abbreviations:
- ADI-PEG
20 Pegylated arginine deiminase
- DFMO
D, L-alpha-difluoromethylornithine
- GEMM
Genetically engineered mouse model
- L-NAME
L-NG-nitroarginine methyl ester
- MSEA
Metabolite set enrichment analysis
- PCA
Principal component analysis
- PDX
Patient-derived xenograft
- SCLC
Small cell lung cancer
- STR
Short tandem repeat
Footnotes
Conflicts of interest:
R.J. DeBerardinis is a scientific advisor for Agios Pharmaceuticals. N.T. Ingolia is an inventor on a patent for ribosomal profiling.
References
- 1.Gazdar AF, Bunn PA, Minna JD. Small-cell lung cancer: what we know, what we need to know and the path forward. Nat Rev Cancer 2017;17:725–37. [DOI] [PubMed] [Google Scholar]
- 2.William WN, Glisson BS. Novel strategies for the treatment of small-cell lung carcinoma. Nat Rev Clin Oncol 2011;8:611–9. [DOI] [PubMed] [Google Scholar]
- 3.Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, et al. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep 2016;16:1259–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardnell RJ, Li L, Sen T, Bara R, Tong P, Fujimoto J, et al. Protein expression of TTF1 and cMYC define distinct molecular subgroups of small cell lung cancer with unique vulnerabilities to aurora kinase inhibition, DLL3 targeting, and other targeted therapies. Oncotarget 2017;8:73419–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mollaoglu G, Guthrie MR, Böhm S, Brägelmann J, Can I, Ballieu PM, et al. MYC drives progression of small cell lung cancer to a variant neuroendocrine subtype with vulnerability to Aurora kinase inhibition. Cancer Cell 2017;31:270–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Owonikoko TK, Nackaerts K, Csoszi T, Ostoros G, Baik C, Mark Z, et al. Randomized phase 2 study of investigational aurora A kinase (AAK) inhibitor alisertib (MLN8237) + paclitaxel (P) vs placebo + P as second line therapy for small-cell lung cancer (SCLC). Abstr ESMO Congr Copenhagen 2016.
- 7.Poirier JT, Gardner EE, Connis N, Moreira AL, de Stanchina E, Hann CL, et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene 2015;34:5869–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lim JS, Ibaseta A, Fischer MM, Cancilla B, O’Young G, Cristea S, et al. Intratumoural heterogeneity generated by Notch signalling promotes small-cell lung cancer. Nature 2017;545:360–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, et al. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 2019;19:289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.George J, Lim JS, Jang SJ, Cun Y, Ozretić L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peifer M, Fernández-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 2012;44:1111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carney DN, Gazdar AF, Bepler G, Guccion JG, Marangos PJ, Moody TW, et al. Establishment and identification of small cell lung cancer cell lines having classic and variant features. Cancer Res 1985;45:2913–23. [PubMed] [Google Scholar]
- 14.Gazdar AF, Carney DN, Nau MM, Minna JD. Characterization of variant subclasses of cell lines derived from small cell lung cancer having distinctive biochemical, morphological, and growth properties. Cancer Res 1985;45:2924–30. [PubMed] [Google Scholar]
- 15.Johnson BE, Russell E, Simmons AM, Phelps R, Steinberg SM, Ihde DC, et al. MYC family DNA amplification in 126 tumor cell lines from patients with small cell lung cancer. J Cell Biochem 1996;24:210–7. [DOI] [PubMed] [Google Scholar]
- 16.McFadden DG, Papagiannakopoulos T, Taylor-Weiner A, Stewart C, Carter SL, Cibulskis K, et al. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell 2014;156:1298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim D-W, Wu N, Kim Y-C, Cheng PF, Basom R, Kim D, et al. Genetic requirement for Mycl and efficacy of RNA Pol I inhibition in mouse models of small cell lung cancer. Genes Dev 2016;30:1289–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gazdar AF, Savage TK, Johnson JE, Berns A, Sage J, Linnoila RI, et al. The comparative pathology of genetically engineered mouse models for neuroendocrine carcinomas of the lung. J Thorac Oncol 2015;10:553–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sos ML, Dietlein F, Peifer M, Schöttle J, Balke-Want H, Müller C, et al. A framework for identification of actionable cancer genome dependencies in small cell lung cancer. Proc Natl Acad Sci U S A 2012;109:17034–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sen T, Tong P, Stewart CA, Cristea S, Valliani A, Shames DS, et al. CHK1 inhibition in small-cell lung cancer produces single-agent activity in biomarker-defined disease subsets and combination activity with Cisplatin or Olaparib. Cancer Res 2017;77:3870–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv 2016;2:e1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov 2015;5:1024–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dibble CC, Manning BD. Signal integration by mTORC1 coordinates nutrient input with biosynthetic output. Nat Cell Biol 2013;15:555–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell 2017;168:960–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potter DS, Galvin M, Brown S, Lallo A, Hodgkinson CL, Blackhall F, et al. Inhibition of PI3K/BMX cell survival pathway sensitizes to BH3 mimetics in SCLC. Mol Cancer Ther 2016;15:1248–60. [DOI] [PubMed] [Google Scholar]
- 26.Tsurutani J, West KA, Sayyah J, Gills JJ, Dennis PA. Inhibition of the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin pathway but not the MEK/ERK pathway attenuates laminin-mediated small cell lung cancer cellular survival and resistance to imatinib mesylate or chemotherapy. Cancer Res 2005;65:8423–32. [DOI] [PubMed] [Google Scholar]
- 27.Marinov M, Ziogas A, Pardo OE, Tan LT, Dhillon T, Mauri FA, et al. AKT/mTOR pathway activation and BCL-2 family proteins modulate the sensitivity of human small cell lung cancer cells to RAD001. Clin Cancer Res 2009;15:1277–87. [DOI] [PubMed] [Google Scholar]
- 28.Besse B, Heist RS, Papadmitrakopoulou VA, Camidge DR, Beck JT, Schmid P, et al. A phase Ib dose-escalation study of everolimus combined with cisplatin and etoposide as first-line therapy in patients with extensive-stage small-cell lung cancer. Ann Oncol 2014;25:505–11. [DOI] [PubMed] [Google Scholar]
- 29.Tarhini A, Kotsakis A, Gooding W, Shuai Y, Petro D, Friedland D, et al. Phase II study of everolimus (RAD001) in previously treated small cell lung cancer. Clin Cancer Res 2010;16:5900–7. [DOI] [PubMed] [Google Scholar]
- 30.Pandya KJ, Dahlberg S, Hidalgo M, Cohen RB, Lee MW, Schiller JH, et al. A randomized, phase II trial of two dose levels of temsirolimus (CCI-779) in patients with extensive-stage small-cell lung cancer who have responding or stable disease after induction chemotherapy: a trial of the Eastern Cooperative Oncology Group (E1500). J Thorac Oncol 2007;2:1036–41. [DOI] [PubMed] [Google Scholar]
- 31.Gerner EW, Meyskens FL. Polyamines and cancer: old molecules, new understanding. Nat Rev Cancer 2004;4:781–92. [DOI] [PubMed] [Google Scholar]
- 32.Keshet R, Erez A. Arginine and the metabolic regulation of nitric oxide synthesis in cancer. Dis Model Mech 2018;11:33332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luk GD, Goodwin G, Marton LJ, Baylin SB. Polyamines are necessary for the survival of human small-cell lung carcinoma in culture. Proc Natl Acad Sci U S A 1981;78:2355–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luk GD, Abeloff MD, Griffin CA, Baylin SB. Successful treatment with DL-alpha-difluoromethylornithine in established human small cell variant lung carcinoma implants in athymic mice. Cancer Res 1983;43:4239–43. [PubMed] [Google Scholar]
- 35.Ensor CM, Holtsberg FW, Bomalaski JS, Clark MA. Pegylated arginine deiminase (ADI-SS PEG20,000 mw) inhibits human melanomas and hepatocellular carcinomas in vitro and in vivo. Cancer Res 2002;62:5443–50. [PubMed] [Google Scholar]
- 36.Delage B, Fennell DA, Nicholson L, McNeish I, Lemoine NR, Crook T, et al. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. Int J cancer 2010;126:2762–72. [DOI] [PubMed] [Google Scholar]
- 37.Cui M, Augert A, Rongione M, Conkrite K, Parazzoli S, Nikitin AY, et al. PTEN is a potent suppressor of small cell lung cancer. Mol Cancer Res 2014;12:654–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 2001;15:3243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang F, Ni M, Chalishazar MD, Huffman KE, Kim J, Cai L, et al. Inosine monophosphate dehydrogenase dependence in a subset of small cell lung cancers. Cell Metab 2018;28:369–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaffer BE, Park K-S, Yiu G, Conklin JF, Lin C, Burkhart DL, et al. Loss of p130 accelerates tumor development in a mouse model for human small-cell lung carcinoma. Cancer Res 2010;70:3877–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang S, Tsun Z-Y, Wolfson RL, Shen K, Wyant GA, Plovanich ME, et al. Lysosomal amino acid transporter SLC38A9 signals arginine sufficiency to mTORC1. Science 2015;347:188–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Polley E, Kunkel M, Evans D, Silvers T, Delosh R, Laudeman J, et al. Small cell lung cancer screen of oncology drugs, investigational agents, and gene and microRNA expression. J Natl Cancer Inst 2016;108:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner AH, Devarakonda S, Skidmore ZL, Krysiak K, Ramu A, Trani L, et al. Recurrent WNT pathway alterations are frequent in relapsed small cell lung cancer. Nat Commun 2018;9:3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suda K, Rozeboom L, Yu H, Ellison K, Rivard CJ, Mitsudomi T, et al. Potential effect of spliceosome inhibition in small cell lung cancer irrespective of the MYC status. PLoS One 2017;12:e0172209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Y-H, Klingbeil O, He X-Y, Wu XS, Arun G, Lu B, et al. POU2F3 is a master regulator of a tuft cell-like variant of small cell lung cancer. Genes Dev 2018;32:915–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Owonikoko TK, Zhang G, Deng X, Rossi MR, Switchenko JM, Doho GH, et al. Poly (ADP) ribose polymerase enzyme inhibitor, veliparib, potentiates chemotherapy and radiation in vitro and in vivo in small cell lung cancer. Cancer Med 2014;3:1579–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cardnell RJ, Feng Y, Mukherjee S, Diao L, Tong P, Allison Stewart C, et al. Activation of the PI3K/mTOR pathway following PARP inhibition in small cell lung cancer. PLoS One 2016;11:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bello-Fernandez C, Packham G, Cleveland JL. The ornithine decarboxylase gene is a transcriptional target of c-Myc. Proc Natl Acad Sci U S A 1993;90:7804–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai W-B, Aiba I, Long Y, Lin H-K, Feun L, Savaraj N, et al. Activation of Ras/PI3K/ERK pathway induces c-Myc stabilization to upregulate argininosuccinate synthetase, leading to arginine deiminase resistance in melanoma cells. Cancer Res 2012;72:2622–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hodgkinson CL, Morrow CJ, Li Y, Metcalf RL, Rothwell DG, Trapani F, et al. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med 2014;20:897–903. [DOI] [PubMed] [Google Scholar]
- 51.Casero RA, Marton LJ. Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat Rev Drug Discov 2007;6:373–90. [DOI] [PubMed] [Google Scholar]
- 52.Zabala-Letona A, Arruabarrena-Aristorena A, Martín-Martín N, Fernandez-Ruiz S, Sutherland JD, Clasquin M, et al. mTORC1-dependent AMD1 regulation sustains polyamine metabolism in prostate cancer. Nature 2017;547:109–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dai N, Rapley J, Angel M, Yanik MF, Blower MD, Avruch J. mTOR phosphorylates IMP2 to promote IGF2 mRNA translation by internal ribosomal entry. Genes Dev 2011;25:1159–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Valvezan AJ, Turner M, Belaid A, Lam HC, Miller SK, McNamara MC, et al. mTORC1 couples nucleotide synthesis to nucleotide demand resulting in a targetable metabolic vulnerability. Cancer Cell 2017;32:624–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brennan J, O’Connor T, Makuch RW, Simmons AM, Russell E, Linnoila RI, et al. myc family DNA amplification in 107 tumors and tumor cell lines from patients with small cell lung cancer treated with different combination chemotherapy regimens. Cancer Res 1991;51:1708–12. [PubMed] [Google Scholar]
- 56.Drapkin BJ, George J, Christensen CL, Mino-Kenudson M, Dries R, Sundaresan T, et al. Genomic and functional fidelity of small cell lung cancer patient-derived xenografts. Cancer Discov 2018;8:600–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rabinovich S, Adler L, Yizhak K, Sarver A, Silberman A, Agron S, et al. Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis. Nature 2015;527:379–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, Vander Heiden MG. Supporting aspartate biosynthesis is an essential function of respiration in proliferating cells. Cell 2015;162:552–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


