SUMMARY
Identification of RNA-interacting pharmacophores could provide chemical probes and, potentially, small molecules for RNA-based therapeutics. Herein, using a high-throughput differential scanning fluorimetry assay, we identified small molecule natural products with the capacity to bind the discrete stem-looped structure of pre-miR-21. The most potent compound identified was a prodiginine-type compound, butylcylcoheptyl prodiginine (bPGN), with the ability to inhibit Dicer-mediated processing of pre-miRNA-21 in vitro and in cells. Time dependent RT-qPCR, western blot, and transcriptomic analyses showed modulation of miR-21 expression and its target genes such as PDCD4 and PTEN upon treatment with bPGN, supporting on-target inhibition. Consequently, inhibition of cellular proliferation in HCT-116 colorectal cancer cells was also observed when treated with bPGN. The discovery that bPGN can bind and modulate the expression of regulatory RNAs such as miR-21 helps set the stage for further development of this class of natural product as a molecular probe or therapeutic agents against miRNA-dependent diseases.
Graphical Abstract
eTOC Blurb:
Matarlo et al. describes the identification of a natural product using a biophysical screen for small-molecule compounds that bind and modulate the stability of an oncogenic microRNA. The compound, butylcylcoheptyl prodiginine, specifically binds to precursor microRNA-21 to inhibit its processing into the mature oncogenic miR-21, and selectively arrest growth of colon cancer cells.
INTRODUCTION
MicroRNAs (miRNAs) play pivotal roles for maintaining cellular homeostasis and are involved in virtually all developmental, physiological, and disease processes such as proliferation, migration, cell cycle, and apoptosis (Asangani et al., 2008; Connolly et al., 2010; Krichevsky and Gabriely, 2009). The fundamental principles of the biogenesis of miRNAs are generally well understood (Lin and Gregory, 2015). First, primary-miRNA (pri-miR) transcripts are processed into short stem-looped precursor-miRNAs (pre-miRs) in the nucleus by Drosha-DGCR8 proteins. Pre-miRs are then transported into the cytoplasm by Exportin 5-RAN-GTP where they are further processed into mature ~22 nucleotide double stranded miRNAs by the Dicer-TRBP complex. Once formed, a double stranded miRNA is incorporated into the RNA-induced silencing complex (RISC) after which the passenger strand is degraded, forming the mature silencing complex (Gregory et al., 2006). Functionally, miRNAs mediate translational repression or transcript degradation through antisense interactions with target mRNAs and a single mRNA can be regulated by numerous miRNAs (Bartel, 2018; Bartel and Chen, 2004; Lin and Gregory, 2015). Studies have shown that miRNA-mediated post-transcriptional regulation requires high levels of specific miRNAs and thus, aberrant over- or under- expression of miRNAs has been linked to disease pathologies such as cancer (Denzler et al., 2016). A prime example, miRNA-21–5p (miR-21), is a small 22-nucleotide regulatory oncogenic miRNA that is overexpressed in most cancers including colorectal cancer (CRC). miR-21 regulates numerous driver genes, oncogenes, and tumor suppressor genes such as PDCD4, PTEN, STAT3, and MYC (Asangani et al., 2008; Krichevsky and Gabriely, 2009; Ma et al., 2013). In addition to cancer, dysregulation of miR-21 expression has been implicated in other diseases including obesity (Seeger et al., 2014), cardiovascular diseases (Bauersachs, 2012; Gryshkova et al., 2018), and diabetes (Jiang et al., 2017; Zhong et al., 2013). Thus, modulation of dysregulated miR-21 processing and function could provide a promising strategy for future drug discovery with broad potential therapeutic applications.
In recent years, there has been an increase in RNA-based therapeutic research that has focused on delivering short sequence-oligonucleotides (RNAi) to silence target mRNAs in cancer cells (Crooke et al., 2018; MacLeod and Crooke, 2017). Our efforts and others’ have centered on the identification of chemical pharmacophores that bind specifically to discrete RNA structures as potential starting points for developing chemical probes and/or therapeutic agents. To that end, a growing body of evidence shows that miRNAs and pre-miRNAs are potential targets for small-molecule modulators (Connelly et al., 2017; Jiang et al., 2015; Lorenz et al., 2015; Naro et al., 2015). For example, Disney et al. developed a method called Inforna to identify small molecule binding partners for discrete RNA secondary structures (Disney et al., 2016). In addition, the small molecule AC1MMYR2 (2,4-diamino-1,3-diazinane-5-carbonitrile) has also been shown to specifically target miR-21 biogenesis in cells and in vivo despite non-selective inhibition of other miRNAs in vitro (Ren et al., 2015; Shi et al., 2013). Furthermore, Garner et al. developed a high-throughput assay to screen small-molecule ligands that may bind pre-miRNAs to inhibit Dicer processing (Lorenz et al., 2015). These studies have demonstrated that specific small-molecule inhibitors of miRNA processing are obtainable from suitably controlled assay systems and that structured RNAs can indeed be druggable targets.
Here, we use a RNA-based differential scanning fluorimetry (DSF) assay to identify modulators of miRNA biogenesis and processing. This effort resulted in the identification of the natural product butylcylcoheptyl prodiginine (bPGN, 1) (Figure 1A) which can regulate the expression level of miR-21 in HCT-116 colorectal cancer cells. 1 belongs to a family of bioactive tripyrrole natural product compounds called prodiginines produced by bacteria with diverse reported biological activities including antibacterial (Gondil et al., 2017), antimalarial (Castro, 1967), antifungal (Woodhams et al., 2018), and anticancer (Perez-Tomas and Vinas, 2010). Well-known derivatives from this family include obatoclax (Gariboldi et al., 2015) and prodigiosin (Perez-Tomas et al., 2003) (Figure 1A). Obatoclax has been tested in clinical trials and shows promise against hematological malignancies (Joudeh and Claxton, 2012; Schimmer et al., 2008; Schimmer et al., 2014) but has displayed limited efficacy in solid tumors (Arellano et al., 2014; Goard and Schimmer, 2013). Despite the clinical development of obatoclax, the reported molecular mechanism of prodiginine anti-cancer activity is still not fully understood (Chen et al., 2014; Xie et al., 2015), thus further elucidation of its molecular mechanism may provide important data toward improving its therapeutic applications. Here, we report for the first time that 1 can bind to pre-miR-21 and inhibit Dicer-mediated processing at non-cytotoxic concentration which results in cellular proliferation arrest by downregulating miR-21 expression. The previously unreported interaction of a prodiginine-type natural product with regulatory non-coding RNA to mediate downregulation of cancer-associated genes broadens the chemical classes identified as RNA-interacting pharmacophores for the development of molecular probes and provides avenues for potential therapeutic applications.
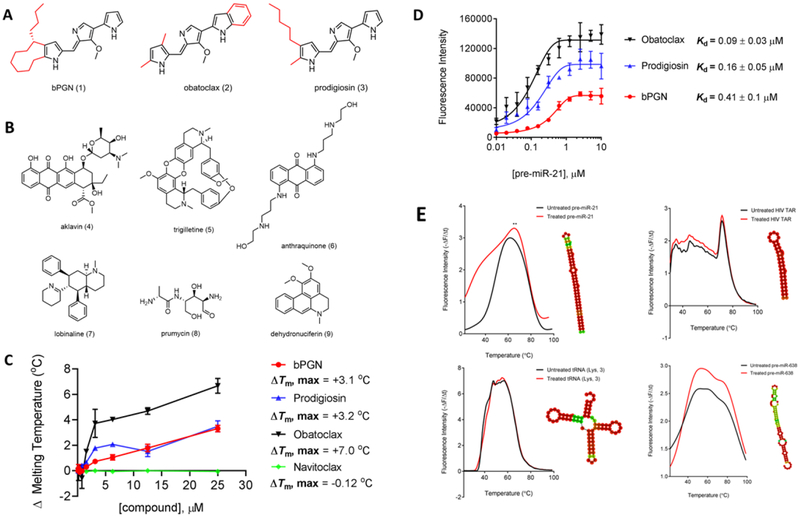
Figure 1: Identification of RNA-binding compound, bPGN (1).
(A) The chemical structures of prodiginine-class compounds: butylcycloheptyl prodiginine (1), obatoclax (2), and prodigiosin (3). (B) The chemical structures of active natural product compounds identified through a DSF-based HTS against pre-miR-21. (C) Titration of 1, 2, 3, and navitoclax ranging from 0 – 25 μM to pre-miR-21 using DSF shows melting temperature (Tm) changes. (D) Direct binding affinity of 1, 2, 3, was determined by titrating compounds from 0 – 10 μM with pre-miR-21. Fluorescence was monitored at 545/580 nm. Kd was assessed using the non-linear regression fit (Sigmoidal, 4PL) function in GraphPad Prism. (E) Pre-miR-21, tRNA (Lys, 3), HIV TAR, and pre-miR-638 were incubated with 5 μM of 1 for 10 minutes followed by a Tm shift measurement by DSF. Only pre-miR-21 showed significant Tm shift. Interestingly, pre-miR-628 showed significant fluorescence intensity change, but no Tm shift was observed. All experiments were performed in triplicate.
RESULTS
Screening for Pre-miR-21 Thermal Modulators Identifies bPGN as a RNA Binding Molecule
Compounds that modulated the thermal stability of pre-miR-21 in vitro were identified using a high-throughput differential scanning fluorimetry (DSF)-based screen. Briefly, we performed a screen of 3682 pure natural products against an annealed pre-miR-21 oligonucleotide and identified which compounds could modulate its thermal stability in vitro (Figure S1). This screen resulted in the identification of 32 compounds that either increased or decreased the thermal stability of pre-miR-21 (Table S1). In a single specificity screen using another stem-looped RNA structure, these compounds did not significantly affect the in vitro thermal stability of the 5’-UTR stem-loop A (SLA) RNA fragment from the Dengue-II virus genome (data not shown). Based on a literature search, 7 of the 32 active compounds have not been previously identified as RNA-binding molecules and thus were tested for their ability to inhibit growth in cells (Figure 1B, compounds 1, 4 - 9). Previous studies have shown the significance of miR-21 in growth and proliferation of HCT-116 cells (Chen et al., 2017), thus we chose to treat these cells with the compounds ranging from 0 – 25 μM and assessed for viability 24 hours post-treatment (Table 1). Among these compounds, aklavin (AKL, 4), trigilletine (TRG, 5), and an anthraquinone (ANT, 6) all showed high nanomolar half maximal growth inhibitory concentrations (GI50), close to the value of 5-fluorouracil (5-FU) positive control (0.72 μM). Lobinaline (LOB, 7), prumycin (PRU, 8), and dehydronuciferin (DHY, 9) all showed ≥ 10 μM GI50 (Table 1). The most active compound bPGN, 1, showed a GI50 as low as 0.035 μM in HCT-116 cells. Concomitantly, we also tested 2 other prodiginine compounds, obatoclax and prodigiosin, and determined their GI50 to be 0.10 μM and 0.24 μM, respectively against HCT-116 cancer cells. Significantly, the GI50 of 1, obatoclax, and prodigiosin in normal colon cells were all determined to be ≥ 10 μM, signifying a selective mechanism of these compounds against cancer cells.
Table 1: Differential scanning fluorimetry and cytotoxicity results for pre-miR-21 stability modulating compounds.
PubChem CID # is the compound’s identifier (https://pubchem.ncbi.nlm.nih.gov/). The ΔTm MAX was calculated by taking the melting temperature shift difference of pre-miR-21 treated with the compound vs untreated. aThe apparent Kd (Kdapp) was measured by direct binding assay using intrinsic fluorescence max 545/580 nm. bGI50 was determined using the XTT cell viability assay. Calculations were performed using the non-linear regression fit (Sigmoidal, 4PL) function in GraphPad Prism. cNormal colon cells used were ATCC CRL-1790. d1,5-bis[3-(2-hydroxyethylamino)propylamino]anthracene-9,10-dione hydrochloride. All experiments were repeated in triplicate.
| Compound number | PubChem CID | Short Name | ΔTm MAX (°C) | aKd (μM)app | GI50 (μM) HCT-116b | GI50 (μM) normal colonc |
|---|---|---|---|---|---|---|
| 1 | 54601748 | bPGN | +3.1 | 0.41 ± 0.04 | 0.035 ± 0.003 | ≥ 10 |
| 2 | 16681698 | Obatoclax | +3.2 | 0.09 ± 0.02 | 0.10 ± 0.01 | ≥ 10 |
| 3 | 5351169 | Prodigiosin | +7.0 | 0.16 ± 0.06 | 0.24 ± 0.04 | ≥ 10 |
| 4 | 5701988 | Aklavin | −17.1 | nd | 0.57 ± 0.06 | 1.7 ± 0.4 |
| 5 | 301935 | Trigilletine | +5.48 | nd | 3.5 ± 0.5 | 1.0 ± 0.2 |
| 6 | 5351330 | Anthraquinoned | +8.70 | nd | 3.2 ± 0.6 | 1.1 ± 0.8 |
| 7 | 5351224 | Lobinaline | −14.0 | nd | > 10 | > 10 |
| 8 | 5458561 | Prumycin | +8.41 | nd | > 10 | 5.1 ± 0.1 |
| 9 | 821347 | Dehydronuciferin | −15.7 | nd | > 10 | > 10 |
| 10 | 3385 | 5-FU | nd | nd | 0.72 ± 0.02 | 2.0 ± 0.2 |
In vitro, a dose dependent titration of 1, obatoclax, and prodigiosin (up to 20 μM) afforded a maximum melting temperature shift (ΔTm, max) of +3.1, +7.0, and +3.2°C, respectively against pre-miR-21 (Figure 1C). Using the intrinsic fluorescence of the highly conjugated tripyrrol backbone, we measured the direct binding affinity (Kd) of 1, obatoclax, and prodigiosin to pre-miR-21 and calculated an affinity of 0.41, 0.09, and 0.16 μM, respectively (Figure 1D). Navitoclax (structure shown in Figure S3) was used as a control for these experiments as navitoclax and obatoclax have been shown to share a common molecular mechanism as BH3-mimetics in cells (Chen et al., 2011; Koehler et al., 2014). Our results showed no significant interaction between pre-miR-21 and Navitoclax (Figure 1C). Using fluorescence displacement assay, Navitoclax did not show apparent binding affinity (Kdapp) to pre-miR-21 (data not shown), consistent with the lack of molecular interaction to pre-miR-21 in our DSF assay. Taken together, these results suggest that the tripyrrolic backbone is indeed responsible for RNA binding by the prodiginine family of compounds. Furthermore, we posit that the disconnect between the binding affinity (410 nM) and cellular growth inhibitory activity (35nM) of 1 may be due to an additive effect between bPGN inhibition of pre-miR-21 processing and other previously reported mechanisms for this class of compounds (i.e. as a BH3 mimetic) which would certainly enhance cellular activities.
Finally, to discern specificity of 1 in vitro, we tested the binding of 1 against other structured RNAs such as tRNA (Lys,3), HIV TAR, and pre-miR-638, and showed that 1 did not significantly change the Tm of these RNAs up to 5-fold molar excess of 1 compared to modulation of pre-miR-21 Tm (Figure 1E). Furthermore, results using differential scanning calorimetry (DSC), a dye-independent measurement of thermal stability, show similar stabilizing effects on pre-miR-21 structure in the presence of 1 consistent with DSF (Figure S4). These results directed us to focus our additional studies on elucidating the mechanism of action of 1 in modulating the expression and function of oncogenic miR-21.
bPGN Rapidly Modulates miR-21 but not Pre-miR-21 Expression in Cells
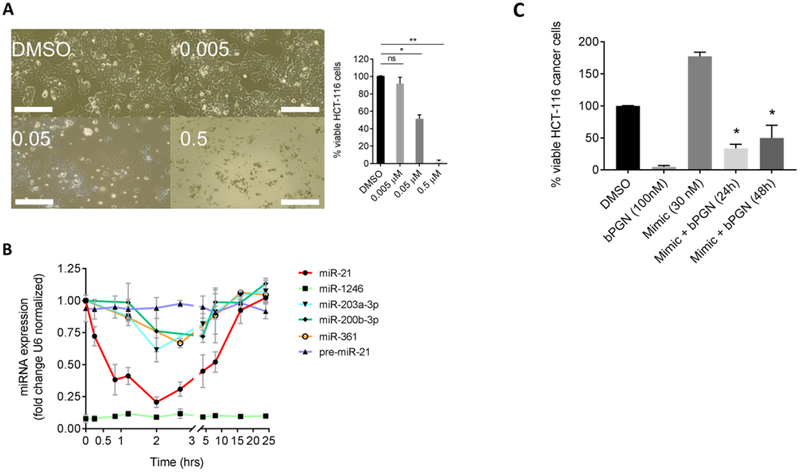
In addition to its pro-apoptotic effect, obatoclax have been shown to inhibit proliferation of leukemia and colorectal cancer cells by inhibiting cell cycle progression, but the exact mechanism remained unclear and targets remain unidentified (Konopleva et al., 2008; Or et al., 2016). Here, a concentration dependent reduction in cell proliferation and viability was also observed when HCT-116 cells were treated with compound 1 at 0 (DMSO only), 0.005, 0.05, and 0.5 μM (Figure 2A). This static effect resulted in 55% reduction of cell number 24-hours post-treatment at 0.05 μM (p < 0.01), but complete cell death at concentration > 0.5 μM (p < 0.001). To determine if the proliferation arrest we observed is indeed caused by the inhibition of pre-miR-21 processing by 1, HCT-116 cells were treated with 50 nM of 1 and the expression levels of pre-miR-21 and miR-21 were evaluated via quantitative RT-PCR (RT-qPCR) at time points ranging from 0 to 24 hours post-treatment. GAPDH and U6 snRNA were used as reference control genes and miRs −1246, −203a-3p, −200b-3p, and −361 expression levels were used to assess selectivity. Figure 2B shows that 1 rapidly reduced miR-21 levels by ~80% (p < 0.001) within 2 hours post-treatment but was less effective in downregulating the expression level of the other tested miRs (up to ~30% reduction). miR-1246 is known to be only highly expressed in HCT-116 spheroid cells (Yamada et al., 2014), and thus remained at low levels up to 24 hours post-treatment. Interestingly, miR-21 expression level gradually recovered up to 24 hours post-treatment, suggesting involvement of a compensatory mechanism due to its pivotal role in cell survival (i.e. the miR-21:STAT3 feed-back loop (Krichevsky and Gabriely, 2009)). Notably, pre-miR-21 expression level did not significantly change up to 24 hours post-treatment indicating that Drosha-mediated processing of pri-miRNA to pre-miRNA was unaffected by 1 (Figure 2B). Furthermore, transfection of exogenous miR-21 mimic 4 hours before treatment of 1, afforded greater percentage of cells 24 and 48 hours post-treatment (Figure 2C), signifying that the proliferation arrest caused by 1 can be mitigated by the addition of exogenous miR-21.
Figure 2: sub-cytotoxic concentration of 1 induces cellular proliferation arrest by modulating miR-21 expression in HCT-116 cells.
(A) HCT-116 cells were treated with DMSO, 0.005, 0.05, or 0.5 μM of 1 and incubated for 24 hr. Cells were then imaged, and viability was determined using the XTT cell-viability assay. Cell proliferation was inhibited at 0.05 μM and cell death was observed at 0.5 μM. ns = not significant, *p < 0.01, ** p < 0.001. (B) HCT-116 cells were treated with 50 nM 1 and total RNA was extracted from various time points (0–24 hr) for RT-qPCR analysis. Expression levels of miRs −21, −1246, −203a-3p, −200b-3p, and −361 and pre-miR-21 were analyzed using U6 snRNA as reference gene. miR-1246 did not show significant amplification up to 24 hr. (C) To show if miR-21 can rescue bPGN treated cells, HCT-116 cells were transfected with miR-21 mimic 4–5 hr before the cells were treated with 100 nM bPGN (2× GI50). Cell viability was measured at 24 and 48 hr by XTT assay. *p < 0.05. All experiments were performed in triplicate.
bPGN can Inhibit Dicer Activity in Vitro and in Vivo
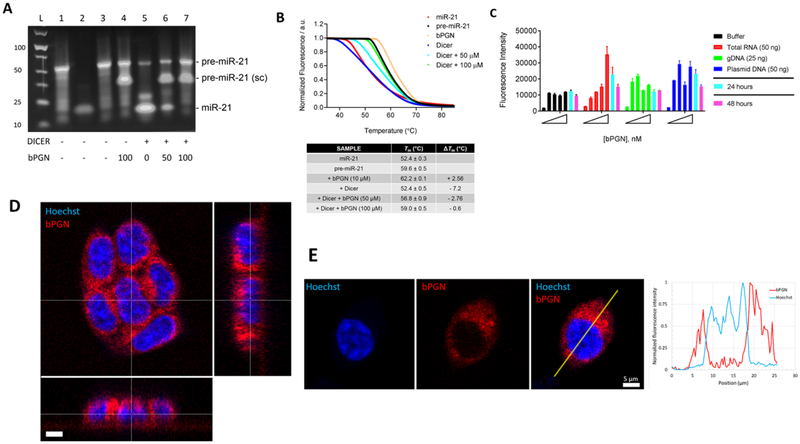
To determine if 1 can inhibit Dicer-mediated pre-miR-21 processing, we performed two in vitro Dicer activity assays. First, we showed by gel shift assay that Dicer activity can be inhibited by 1 in a dose dependent manner (Figure 3A). Lanes 1, 2, and 3 are controls showing pure pre-miR-21, miR-21, and pre-miR-21 with DMSO only, respectively. Major bands, at ~55 and ~22 bp corresponding to pre-miR-21 and miR-21 respectively, are observed. Lane 4 shows pre-miR-21 with 100 μM (4-fold excess) of 1 without Dicer. Here, two bands were observed at ~55 and ~35 bp, suggesting that two major distinct structured species of pre-miR-21 exist in the presence of 1. We hypothesize that 1 induces a supercoiling structural change because of the lower bp band similar to previous reports shown for other compounds binding to RNA (Jain et al., 2013). Supercoiling of RNA can have a thermal stabilizing effect as more intramolecular interactions are made. This result is also consistent with our DSC results where we observe two thermal transitions (Tm1 = 42.3°C; Tm2 = 68.0°C) for pre-miR-21 in the presence of 1 (Figure S4), where presumably the higher melting temperature (trace 2) corresponds to the supercoiled species. Lanes 5–7 show the effect of increasing 1 concentration on pre-miR-21 processing by Dicer. In Lane 5, depletion of the ~55 bp band and the concomitant increase in ~22 bp band is observed in the absence of 1. In Lanes 6 and 7 a dose dependent decrease in the ~22 bp band is observed in the presence of 50, and 100 μM of 1, suggesting a dose dependent inhibition of Dicer processing of pre-miR-21 in vitro.
Figure 3: bPGN localizes in the cytoplasm and inhibits Dicer-mediated pre-miR-21 processing.
(A) Gel-shift assay with or without 1 was performed by incubating 10U Dicer per 10 μM pre-miR-21 (for up to 36 hr in 37°C incubator) after which the RNA was purified and loaded onto a 4% agarose gel. Samples in each lane are as follows: pre-miR-21 (lane 1), miR-21 (lane 2) and pre-miR-21 with DMSO (lane 3), pre-miR-21 with 100 μM 1 (lane 4), pre-miR-21 with Dicer (lane 5), pre-miR-21 with Dicer and 50/100 μM of 1 (lanes 6 and 7, respectively). sc = supercoiled RNA. (B) From the same reaction above, 1 μM of the purified RNA was mixed with 1× SYBR Green dye and the thermal melt of the mixture was analyzed using DSF. Tm is the average melting temperature calculated from experimental triplicates and ΔTm is the difference between samples and DMSO only (control). Tm was obtained using the non-linear regression fit (Sigmoidal, 4PL) function in GraphPad Prism where y = fluorescence, x = temperature. (C) a concentration of 0, 12.5, 25, 50, and 100 nM of 1 was incubated with buffer (black), 50 ng/μL purified total RNA from HCT-116 cells (red), 25 ng/μL purified genomic DNA from HCT-116 cells (green), or 50 ng/μL of plasmid DNA (blue). Fluorescence intensity was measured 4 hr post incubation at max 545/580 nm. The 50 nM sample was also measured after 24 (cyan) and 48 (magenta) hr of incubation. Our data show that bPGN fluorescence was not quenched below background level during any time points measured. (D) 3D laser scanning confocal image of a cluster of live HCT-116 cells treated with 50 nM bPGN (red) for 24 hr and stained with Hoechst (blue) reveals minimal presence of bPGN within the nuclei of the cells. The large image is the xy plane, and x-z and y-z orthogonal slices (thin white lines in the main image) through the cluster are depicted in the bottom and right images, respectively. Scale bar is 5 μm. (E) Laser scanning confocal images of a live HCT-116 cell stained with Hoechst (blue) and treated with 50 nM bPGN (red) for 24 hr. The merged image shows little overlap between the red and blue signals. A profile plot of normalized fluorescence in each channel as a function of position along the yellow line in the merged image reveals minimal bPGN presence in the nucleus. Scale bar is 5 μm. Confocal images are representative of global response in HCT-116 cells.
To further support these results, we also used DSF as a more sensitive method to determine if Dicer processing of pre-miR-21 was inhibited by 1. Here, under Dicer reaction conditions, the Tm of pre-miR-21 was 62.2°C and 59.6°C in the presence (beige) and absence (black) of 10 μM 1, respectively (Figure 3B). In the presence of Dicer and only DMSO (blue), we observe a shift in the Tm by −7.2°C, but a −2.8 and −0.6°C shift in the presence of 50 (aqua) and 100 (green) μM 1, respectively (Figure 3B). These results suggest that the presence of 1 can reduce the global residency of pre-miR-21 on Dicer in vitro. Pure miR-21 (red) was also tested as a control and had a Tm of 52.4°C, strongly correlating with the sample of pre-miR-21 incubated with Dicer with only DMSO (blue). As an assay control, we used the same method to evaluate known Dicer inhibitors: hexachlorophene (Lorenz et al., 2015) and regorafenib (Chen et al., 2017). Indeed, high concentration of hexachlorophene or regorafenib strongly inhibited Dicer activity consistent with our results with 1 (Figure S5). Taken together, these data strongly support that the binding of 1 to pre-miR-21 can lead to inhibition of Dicer-mediated processing of pre-miR-21 in vitro which may correspond to its activity in cells.
Finally, work by Su et al. showed that prodigiosin induces cytotoxicity in hepatocellular carcinoma cells via intercalation into DNA grooves in a copper-mediated manner (Su et al., 2015). To determine if this is true for 1, we assessed its cellular localization using fluorescence microscopy (max 545/580 nm) by treating cells with either DMSO or 50 nM of 1. Importantly, 1 is intrinsically fluorescent due to the highly conjugated tripyrrolic backbone as also previously reported for prodigiosin (Darshan and Manonmani, 2016; Han et al., 2014), thus no chemical modification was necessary. As an additional control, we also show that incubation of 1 with purified total RNA and DNA (gDNA or plasmid DNA) does not quench fluorescence of 1 up to 48 hours of incubation in vitro (Figure 3C). 1 hour prior to imaging, cells were treated with 0.5 μg/mL Hoechst nuclear stain. 3D laser scanning confocal imaging shows that 1 predominantly accumulated at the cytoplasmic space up to 24 hours post-treatment (Figure 3D). Analyses of the x-y plane as well, as the subsequent x-z and y-z orthogonal slices, reveal minimal presence of the 1 in the nucleus. Furthermore, quantification of fluorescence intensity shows minimal overlap between bPGN and Hoechst signals (Figure 3E). Taken together, our data indicate that at 50 nM, 1 does not induce cellular proliferation arrest via intercalating DNA. It also supports our hypothesis that 1 most likely inhibits Dicer-mediated processing of pre-miR-21 which occurs in the cytoplasm, but not the inhibition of Drosha-mediated processing which takes place in the nucleus.
bPGN-Induced miR-21 Downregulation Causes Upregulation of Pdcd4 and PTEN
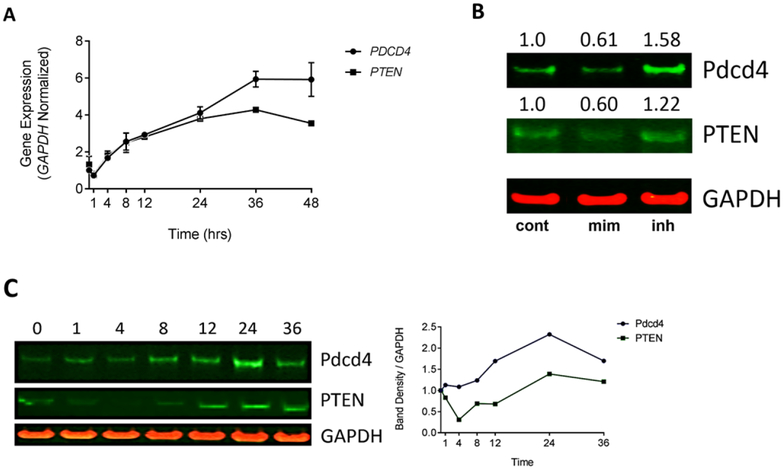
Overexpression of miR-21 in colorectal cancer cells downregulates expression of tumor suppressor protein, programmed cell death 4 (Pdcd4) (Asangani et al., 2008), which is known to inhibit cell proliferation in other cancer cells (Wang et al., 2016a). In addition, it has been shown that aberrant PI3K-Akt pathway signaling plays an important role in tumorigenesis and is often associated with the absence (or mutation) of PTEN (Vivanco and Sawyers, 2002). PTEN, a highly regulated miR-21 target, negatively regulates the PI3K-Akt pathway via dephosphorylation of PIP3, ultimately inhibiting activation of Akt and consequently inhibiting cellular proliferation signaling. To determine if the reduction of miR-21 by treatment of 1 influences the expression of PDCD4 and PTEN in cells, we performed time dependent RT-qPCR and western blot analyses. RT-qPCR showed that the PDCD4 and PTEN transcripts were upregulated 2-fold by 8 hours post-treatment with 1 and continuously increased up to 2 – 4-fold by 36 hours post-treatment (Figure 4A). To see if Pdcd4 and PTEN protein expression could be modulated by miR-21, siRNA miR-21 mimic or inhibitor were transfected in HCT-116 cells. Results showed by western blot analysis that protein expression was decreased by ~40% when treated with the miR-21 mimic and increased by 22–58% when treated by the miR-21 inhibitor 48 hours post-transfections (Figure 4B). Consistent with the siRNA results, cells treated with 50 nM of 1 showed a time dependent increase in Pdcd4 and PTEN protein levels within 24 hours post-treatment (Figure 4C). Taken together, our data reveal that on-target inhibition of miR-21 expression in HCT-116 cells leads to the upregulation of the PDCD4 and PTEN transcripts and protein levels in a time dependent manner. Importantly, upregulation of these proteins is sufficient to induce the anti-proliferation effect observed upon treatment with 1.
Figure 4: HCT-116 cells treated with 1 upregulate miR-21 target genes PDCD4 and PTEN.
(A) PDCD4 and PTEN mRNA expression was measured by RT-qPCR and showed significant upregulation up to 36 hr post-treatment with 1. GAPDH was used as an internal control. (B) HCT-116 cells were treated with DMSO (lane 1) or 30 nM miR-21 mimic (lane 2) or inhibitor (lane 3) and were analyzed for Pdcd4 and PTEN expression level 48 hr post-transfection. Lipofectamine 2000 was used as the transfection agent. Band density was normalized to GAPDH. Fold change (value shown) was determined by double normalizing to GAPDH and the DMSO control. (C) HCT-116 cells were treated with 50 nM 1 and cells were harvested for protein extraction at time points ranging from 0–36 hours post-treatment. Anti-Pdcd4/anti-PTEN was used to detect expression levels of Pdcd4 and PTEN and GAPDH was used as an internal control. Band density was quantified and double normalized GAPDH and time point 0. All experiments were performed in triplicate, western blots are representative of 3 replicates.
bPGN Inhibits Proliferation by Upregulating miR-21 Target Genes
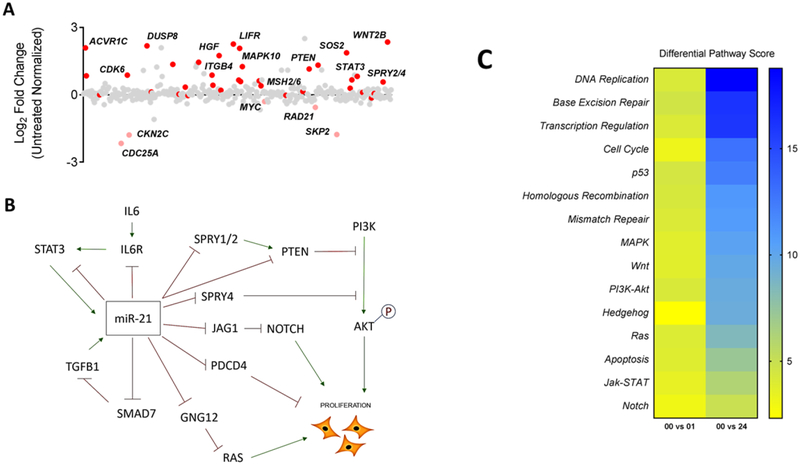
To gain greater insight into the differential effects of 1 to miR-21 target genes, we performed a time course transcriptomic study using the NanoString Technologies platform to collectively identify which genes were modulated. TargetScan (targetscan.org, (Agarwal et al., 2015)) and miRDB (mirdb.org, (Wong and Wang, 2015)) both predict over 200 target genes for miR-21[−5p], many of which have been experimentally validated (Buscaglia and Li, 2011; Krichevsky and Gabriely, 2009). In our analysis, we found 28 of 41 predicted genes that were significantly differentiated from untreated control cells (p < 0.05) (Figure 5A, Table S2). Most notably, miR-21 target genes that are reported to play a role in suppressing cellular proliferation, such as PTEN, STAT3, and TGFB1/TGFBR2, were significantly upregulated. In addition, a differential pathway analysis using our NanoString data reveals multiple pathways important for cell proliferation as highly modulated upon treatment with 1 including modulation of the cell cycle, DNA replication, and PI3K-Akt pathways (Figure 5B). Taken together, our data indicate that the inhibition of cellular proliferation observed when cells are treated with 50 nM of 1 can be due to modulation of miR-21 expression and function which leads to the upregulation of target genes such as PTEN, which could then lead to the subsequent inhibition of cell proliferation. Figure 5C summarizes the connection between PTEN and other target genes and their effect on cellular proliferation.
Figure 5: HCT-116 cells treated with bPGN show global miR-21 target gene upregulation.
(A) Of the 800 genes analyzed using NanoString Technologies gene expression platform, 41 are predicted miR-21 targets by TargetScan and miRDB. 28 of these genes were upregulated (red, p < 0.05) up to 24 hr post-treatment compared to untreated control. Red are predicted miR-21 target genes. (B) Differential pathway score heat map of modulated pathways. Scores were calculated using nSolver Analysis software (NanoSting) and were obtained from HCT-116 cells 1 and 24 hr post-treatment with 1. (C) A representative connection pathway map model of significantly upregulated miR-21 target genes and how they can function to inhibit cellular proliferation upon treatment with 1.
DISCUSSION
As an increasing number of important cell functions are determined to be regulated, at least in part, by small regulatory RNAs, the identification of pharmacophores that interact with miRNAs are of more importance. Thus, development of methodologies to identify these pharmacophores will be very useful for the growth of the field. Here, for the first time, we employ a RNA-based high-throughput DSF-based screen and identified butylcycloheptyl prodiginine (bPGN, 1), a derivative from the class of tripyrrolic natural product called prodiginines, as an active RNA-binding small molecule. Currently, numerous evidences suggest that at clinical concentrations of prodiginines such as obatoclax or prodigiosin, proapoptotic Bcl-2 family proteins are activated in a p53-independent manner. This has been proposed to be the primary mechanism of tumor cytotoxicity for this class of compounds (Soto-Cerrato et al., 2004). Other observed mechanisms of anticancer activities for prodiginines include intracellular acidification, copper-dependent DNA cleavage, modulation of signal transduction and MAPKs, cell cycle arrest, and most recently, the inhibition of Wnt/β-catenin signaling pathway (Perez-Tomas and Vinas, 2010; Wang et al., 2016b). In this report, we add another previously unreported mechanism and show for the first time that 1, and derivatives thereof, can bind at nanomolar concentration and stabilize the discrete stem-looped structure of pre-miR-21. Importantly, we show that binding of 1 can interfere with Dicer-mediated processing of pre-miR-21, which leads to rapidly downregulates mature miR-21 expression in cells. We extended these findings by showing that 1 can consequently induce transcript and protein expression of Pdcd4 and PTEN, which can then lead to the observed reduced proliferation state of HCT-116 colorectal cancer cell. Indeed, the complexity of the mode of action(s) of prodiginines may account for the disconnect between our in vitro binding assay with pre-miR-21 (~410 nM) and cellular activity (~35nM) as we could be observing a more potent cellular activity by intrinsic additive effects. Nevertheless, the anti-proliferative, a static rather than cytotoxic effect of 1, by binding regulatory RNAs represents yet another apoptosis-independent mechanism by which prodiginines elicit anticancer activity.
Indeed, other studies have observed the same antiproliferative and reduced cell viability effect with other prodiginines, but the underlying mechanisms and/or targets in these studies remained unidentified. For example, Hsieh et al. showed that dephosphorylated Akt could lead to GSK-3β activation when treated with prodigiosin, but the molecular basis for how Akt was dephosphorylated was not elucidated (Hsieh et al., 2012). Here, our functional gene analysis shows significant upregulation of PTEN transcript (increased by 121 %). Consistently, studies have shown that PTEN is a direct target of miR-21 (Luo et al., 2017; Meng et al., 2007), but can also be indirectly downregulated by miR-21 through the suppression of SPRY genes (Chai et al., 2018; Feng et al., 2011). Thus, it was not surprising that inhibition of pre-miR-21 processing by 1 caused an increase in PTEN and, to a lesser degree, SPRY2 transcripts (Figure 5A), which could consequently lead to inhibition of PI3K-Akt signaling and ultimately inhibit cellular proliferation, as previously observed.
Prodigiosin have been shown to stabilize topoisomerase I/II-DNA complex in lymphocyte cells which may lead to DNA-damage-induced cytotoxicity in these cells (Montaner et al., 2005). It is hypothesized that the selective cytotoxicity of prodigiosin to cancer cells is due to the higher copper levels in cancer cells compared with normal cells. However, our live cell imaging studies revealed that 1 accumulates in the cytoplasmic space for up to 24 hours post-treatment, suggesting that the proliferation arrest we observed upon treatment of HCT-116 cells with 1 at sub-cytotoxic concentration is independent of DNA-damage and must be due to increased levels of miR-21 targets such as Pdcd4 and PTEN. Consequently, we posit that selective growth inhibitory effect towards cancer cells observed here is mediated by the inhibition of overexpressing miR-21 which increases its target genes, leading to reduced cell proliferation of HCT-116 cells. It is also important to note that Figure 2B shows downregulation of other miRNAs, albeit not as significant as miR-21, but in the same rapid manner. Previous reports have shown that the half-life of miRNAs is measured in days rather than hours (Gantier et al., 2011), but rapid degradation of stable miRNAs is not unusual and have been observed in many studies (Hwang et al., 2007; Krol et al., 2010; Ruegger and Grosshans, 2012). Unlike the biogenesis of miRNAs however, turnover and regulation thereof, are still poorly understood. Enzymes that play a role in miRNA turnover are slowly being identified (Chatterjee and Grosshans, 2009; Ramachandran and Chen, 2008) such as PNPT1 which have been shown to degrade mature miRNAs without affecting pri-/pre-miRs (Das et al., 2010). Here, future endeavors to identify factors responsible for rapid miRNA turnover and how 1 can activate rapid degradation of miRNAs is of great interests but requires further investigation.
Finally, recent work by Disney et al. has shown the importance of fully understanding the effects of clinical agents on regulatory RNAs or the transcriptome, beyond their initially-described protein targets (Velagapudi et al., 2018). Our studies here show that 1 can have substantial effect on pre-miR-21 stability through direct binding which leads to a reduction in Dicer processing and subsequent down-regulation of miR-21 expression. This data provides yet another example of a unstudied chemotype that can bind pre-miRNA. In contrast to pre-miR-21, we did not observe significant thermal modulation of other stem-looped structured RNAs (Figure 1E), thus we posit that 1 may confer selectivity to specific pre-miRNAs (e.g. via specific loop sequences or alternating nucleotide sequence as seen with prodigiosin binding to DNA). It is also possible that 1 may have additional direct effects on the actions of miR-21, which will require further structural and functional studies to deduce. To this end, further investigation of the global binding of 1 to other pre-miRNAs to identify vulnerable pre-miRNA targets are currently ongoing. In addition, structural insights of the molecular interactions of 1 to pre-miR-21 will also enhance our knowledge of the selectivity of this chemical pharmacophore.
STAR* METHODS
Detailed methods are provided and include the following:
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Barry O’Keefe (okeefeba@mail.nih.gov)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
HCT-116 and was purchased from American Type Culture Collection (ATCC) and was established from an adult human colorectal cancer. HCT-116 was cultured in Roswell Park Memorial Institute (RPMI) media with 10% Fetal bovine serum (FBS). CCD 841 CoN was purchased from ATCC and was established from a normal colon epithelial cells of a 21 weeks gestation fetus. CCD 841 CoN cell line was cultured in Eagle’s Minimum Essential Medium (EMEM) with 10% FBS. Both cell lines were subject to 1:5 passaging every 3 days and incubated in 37°C with 5% CO2. Cells were purchased directly from ATCC but were not authenticated in house.
METHOD DETAILS
RNA and Biochemicals
Unmodified, desalted, and HPLC purified pre-miR-21 was purchased from Eurofins Scientific (Louisville, KY, USA). RNA was folded via resuspending with buffer (10 mM Tris-HCl pH 7.5 with 20 mM NaCl), and heated to 95°C for 90 sec, and allowed to cool to room temperature overnight. Folded RNA was aliquoted and stored in −20°C until assayed. The following sequence of pre-miR-21 was purchased:
5’-UGUCGGGUAGCUUAUCAGACUGAUGUUGACUGUUGAAUCUCAUGGCAACACCAGUCGAUGGGCU GUCUGACA-3’
Butylcycloheptyl prodiginine (bPGN) was supplied by Developmental Therapeutics Program (DTP), Division of Cancer Treatment and Diagnosis in the National Cancer Institute (NCI). Plates for high-throughput screens were provided by NCI, Molecular Targets Program (MTP). Compounds such as 5-FU, obatoclax, prodigiosin, navitoclax, spermidine, methoctramine, hexachlotophene, and regorafenib were purchased from Sigma Aldrich or selleckchem.com.
Differential Scanning Fluorimetry (DSF)
The high throughput DSF-screen was performed in a total reaction volume of 6 μL containing 1 μM pre-miR-21, 1× SYBR Green Dye (ThermoFisher, Cat#S7567), 2% DMSO, and buffer (0.25 mM Tris-HCl and 0.25 mM NaCl) in a 384-well white polypropylene reaction plate (Roche, Cat# 04729749001) and was treated with 3 μM concentration of compounds from the NCI Molecular Targets Program (MTP) pure compound library using an automated liquid handler. Spermidine and methoctramine were used as a stabilizing or destabilizing control, respectively. In a specificity screen, pre-miR-21, Lys3 tRNA, HIV TAR, and pre-miR-638 were treated with 5 μM bPGN to determine ΔTm. To obtain maximum change in Tm, DSF dose response was performed from a master mix containing 1.0 μM pre-miR-21, 1× SYBR Green, 2% DMSO in a reaction buffer (0.25 mM Tris-HCl and 0.25 mM NaCl). Then, bPGN, obatoclax, prodigiosin, or navitoclax was titrated ranging from 0 – 20 μM in each well at final volume of 6 μL. All DSF analysis was performed using Roche LightCycler 480 Instrument II using the following parameters: manual mode with integration time set at 0.75 seconds, continuous acquisition mode up to 99°C, ramp rate of 0.09°C/s and 5 acquisitions per °C. Raw data was imported into GraphPad Prism 7.03 for analysis and high-resolution figures. From the raw data, the temperature which gave half-denatured state (Log GI50) of pre-miR-21 was calculated (labeled as Tm) using the non-linear regression fit (Sigmoidal, 4PL) function, y = fluorescence, x = temperature. The exact Tm of premiR-21 with or without bPGN can be extrapolated using differential scanning calorimetry (below).
Differential Scanning Calorimetry (DSC)
DSC experiments were performed using VP-DSC (Malvern Instruments). 30 μM Pre-miR-21, with or without 300 μM bPGN, was prepared in 1.5% DMSO adjusted buffer (0.25 mM Tris-HCl pH 7.5 with 0.25 mM NaCl), degassed, and added into the sample cell. Cell compartments were capped, adjusted for positive pressure (30 psi), and allowed to progress through a series of 8 alternating up and down scans, all scans running at a rate of 60°C/hr with a filter period of 16 seconds. The completed sample thermogram was first corrected for buffer-DMSO effects (subtracting the first up scan) followed by a normalized correction of the absolute baseline, the cubic approximation of the pre-transition (structured) and the post-transition (unstructured) regions of the thermogram. The baseline-corrected thermogram was fitted to a non-2-state unfolding model according to manufacturer’s protocol, the fitting error evaluated for single versus multiple unfolding units. From this model-fitting, values for melting temperature (Tm) and enthalpy of unfolding (ΔHcal) were extrapolated for each transition. Analyzed data were imported into GraphPad Prism 7.03 to render figures.
RNA/DNA Quenching and Direct Binding (Kd) Assays
Total RNA (50 ng/μL) and gDNA (25 ng/μL) from HCT-116 or 50 ng/μL plasmid were incubated with 0, 12.5, 25, 50, and 100 nM of bPGN in buffer (0.25 mM Tris-HCl and 0.25 mM NaCl). Fluorescence intensity was measured using SpectraMax i3x (Molecular Devices) at excitation 545 nm and emission 580 nm. Direct binding assay was performed in a 40 μL reaction volume containing 1.0 μM pre-miR-21 and compounds (bPGN, obatoclax, and prodigiosin) ranging from 0 – 10 μM in a reaction buffer (0.25 mM Tris-HCl and 0.25 mM NaCl) and the fluorescence intensity was using the same parameters. Raw fluorescence data were imported into GraphPad Prism 7.03 for analysis and high-resolution figures. Kd was calculated using the non-linear regression fit (Sigmoidal, 4PL) function.
Cell Culture, XTT cell viability assay, and bPGN treatment
HCT-116 cells (ATCC CCL-247) were cultured in RPMI with 10% FBS. Normal colon cells (CoN ATCC CRL-1790) were cultured in EMEM with 10% FBS. For dose response experiments, 2 × 103 cells were seeded in 96-well plates and incubated for 24 hours in a 5% CO2 / 37°C incubator. Cells were then treated with compounds ranging from 0 – 10 μM and incubated for another 24 hours. The XTT (2,3-Bis-(2-Methoxy-4-Nitro-5-Sulfophenyl)-2H-Tetrazolium-5-Carboxanilide) cell viability assay was performed according to manufacturer’s protocol (ThermoFisher, Cat#X6493). Absorbance (450 nm) was read using Tecan-Magellan plate reader (Tecan Trading AG, Switzerland). Cells for RT-qPCR, NanoString mRNA expression, and protein analyses were cultured in 100 mm cell culture plates until 50–60% confluence before treatment. Cells were treated with 50 nM bPGN and collected for time point studies at 0, 1, 4, 8, 16, 24, 36, 48, and 72 hours post-treatment and subjected to protein and total RNA extraction following manufacturer’s protocol for mirVana PARIS (ThermoFisher). For siRNA targeting miR-21, cells were treated with DMSO (control), 30 nM mimic (Ambion #4464066), or 30 nM inhibitor (Ambion #4464084) using Lipofectamine 2000 as the lipid carrier and incubated for 48 hours prior to mirVana PARIS extraction. All experiments were performed in experimental triplicate.
RT-qPCR
Materials for RT-qPCR for mRNA and miRNA expression profiling were EXIQON ExiLERATE (#303402) and miRCURY (#203351) LNA reagents and primers (Exiqon, Woburn MA, USA). Standard designed primers include hsa-miR-21–5p, hsa-miR-1246, hsa-miR-203a-3p, hsa-miR-200b-3p, hsa-miR-361, UniSp6 (loading control), U6 snRNA (reference gene), and GAPDH (reference gene). Pre-miR-21 and PDCD4 were ordered using the following primers:
pre-miR-21: forward – TGTCGGGTAGCTTATCAGAC, reverse – TTCAGACAGCCCATCGACTG
PDCD4: forward – TAGATTGTGTGCAGGCTA, reverse – GTTCAGCTTCAGATATGTCTC
RT-qPCR was performed according to manufacturer’s protocol using the following conditions: Denature – 95°C for 10 min. Then Cycle 45 times of 10 sec at 95°C, 1 min at 60°C, and 10 sec at 95°C. End by extension – 55°C for 1 min and 4°C hold. PDCD4 fold change expression was determined using the following equation: 2−ΔΔCP, where ΔΔCP = ΔCPtreated – ΔCPuntreated and ΔCP is the difference in the CP values of PDCD4 and GAPDH (i.e. ΔCP = PDCD4 – GAPDH).
Western Blot Analysis
Cell lysates were subjected to standard trichloroacetic acid (TCA)/acetone precipitation. The isolated protein fraction pellet was dried, reconstituted in 8 M urea, and the total protein concentration was determined by UV spectrophotometry. For SDS-PAGE, a quantity of 20 μg protein was loaded per lane of NuPAGE 4–12% Bis-Tris gels (Invitrogen) and separated at 200 V constant voltage. Gels were briefly washed in 20 % ethanol and proteins were transferred to PVDF membranes using the iBlot 2 dry transfer device (Invitrogen) using the following method: 20 V for 1 minute, 23 V for 4 minutes, and 25 V for 3 minutes. Membranes were blocked in Odyssey Blocking Buffer in PBS (LiCor) and probed with mouse anti-PDCD4 or PTEN (Santa Cruz Biotechnology) and rabbit anti-GAPDH (Cell Signaling Technology) primary antibodies. Mulitplex detection was accomplished using goat anti-mouse IRDye 800CW and goat anti-rabbit IRDye 680RD secondary antibodies (LI-COR Biotechnology).
Dicer Processing Assay
Dicer reaction assays were set up according to manufacturer’s protocol (Genlantis, San Diego CA, Cat#T510001). 10 μM of pre-miR-21 was incubated with 10U of DICER with or without bPGN (or other compounds) up to 36 hours at 37°C. The reaction was quenched, and RNA was purified by ethanol precipitation (or an RNA purification kit). 1 μM of the RNA is used for DSF assay (see above for protocol) and the rest was loaded into a pre-stained, pre-casted 4% agarose get for the gel-shift electrophoresis assay (Invitrogen, E-Gel Power Snap Electrophoresis System, Cat#G8342ST). Experiments were performed in experimental duplicates.
Live Cell Imaging
2 × 105 HCT-116 cells were cultured in a 35 mm glass bottom culture dish (Cellvis, CA, USA, #D35–141.5N) and treated with 50 nM bPGN and incubated for 24 hours. Cell were then treated with 0.5 μg/mL Hoechst stain 1 hours prior to imaging. A Nikon TI-Eclipse microscope outfitted with 20 X objective (0.8 NA) an Andor Zyla camera (Belfast, UK) and a Tokai Hit (Shizuoka-ken, Japan) stage-top incubator was used for wide field fluorescence imaging. A Leica SP8 laser scanning confocal microscope with a 40X (0.85 NA) air objective and a Tokai Hit stage-top incubator collected 3D images of the bPGN and Hoechst fluorescence using HyD detectors in photon counting mode. Cells were maintained at 37°C and 5% CO2 during the entire period of imaging, and bPGN fluorescence was measured using the tetramethylrhodamine channel. Experiments were performed in duplicates and on 3 separate days. Images were minimally processed (cropping) and analyzed with FIJI (intensity profiles; NIH) and Imaris (othogonal slicing; Bitplane, Concord, MA) software.
NanoString Analysis
NanoString analysis was performed by the National Cancer Institute (NCI), Center for Cancer Research(CCR) Genomics Core Facility. The NanoString nCounter Human Pan-Cancer Pathway Panel assay kit (http://www.nanostring.com/) was employed to identify altered mRNAs with or without 50 nM treatment of bPGN at time points: 0, 1, and 24 hours post-treatment. 100 ng of total RNA was used for nCounter sample preparation according to manufacturer’s instructions (NanoString Technologies). Raw data was normalized to 40 Housekeeping genes using the NanoString nSolver software. For our analysis, a 5 fM signal threshold was implemented as statistically significant, thus we excluded 227 genes because of low signals. Furthermore, in addition to the 40 housekeeping genes used as internal controls during analysis, all data were normalized to untreated control (time point 0). P-values were calculated using Pearson correlation as a distance metric and pairwise complete-linkage as a clustering method. nSolver Pathway Analysis was used to obtain differential pathway scores. Hierarchal clustering figure was made using GraphPad Prism from the normalized values.
QUANTIFICATION AND STATISTICAL ANALYSIS
All plots show means with error bars representing S.E.M., unless otherwise noted. Experiments were completed at least in triplicate except for nanoString mRNA analysis which are two replicates. Data were plotted in Graphpad Prism 7.03.
DATA AND SOFTWARE AVAILABILITY
nSolver and Advanced Analysis software are available in nanoString Technologies website (https://www.nanostring.com/). FIJI and Imaris are available from NIH and Oxford Instruments, respectively.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti-PDCD4 | Santa Cruz Biotechnology | sc-376430 |
| Mouse anti-PTEN | Santa Cruz Biotechnology | sc-7974 |
| rabbit anti-GAPDH | Cell Signaling Technology | 2118 |
| goat anti-mouse IRDye 800CW | LI-COR Biotechnology | 926–32210 |
| goat anti-rabbit IRDye 680RD | LI-COR Biotechnology | 926–68071 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Butylcycloheptyl prodiginine (bPGN) | Developmental Therapeutics Program (DTP) of the National Cancer Institute (NCI). | -N/A- |
| 5-Fluorouracil | Sigma | F6627 |
| obatoclax | selleckchem | S1057 |
| Prodigiosin hydrochloride | Sigma | P0103–100UG |
| Navitoclax | selleckchem | S1001 |
| spermidine | Sigma | S2626–1G |
| Methoctramine hydrate | Sigma | M105–10MG |
| Hexachlorophene | selleckchem | S4632 |
| Regorafenib | selleckchem | S1178 |
| SYBR Green Dye | ThermoFisher | S7567 |
| Critical Commercial Assays | ||
| XTT | ThermoFisher | X6493 |
| mirVana PARIS | ThermoFisher | AM1556 |
| Dicer reaction assays | Genlantis | T510001 |
| NanoString nCounter Human Pan-Cancer Pathway Panel assay kit | nanoString Technology | XT-CSO-PATH1–12 |
| Deposited Data | ||
| NanoString_Matarlo_mRNA Time point studies of bPGN treated HCT-116 cell line |
This paper |
GSM3731921 GSM3731922 GSM3731923 GSM3731924 GSM3731925 GSM3731926 |
| Experimental Models: Cell Lines | ||
| HCT-116 colorectal cancer cell line, human | ATCC | ATCC CCL-247 |
| CoN - Normal colon cell line, human | ATCC | ATCC CRL-1790 |
| Oligonucleotides | ||
| pre-miR-21, UGUCGGGUAGCUUAUCAGACUGAUGUUGACUGUUGAAUCUCAUGGCAACACCAGUCGAUGGGCUGUCUGACA | Eurofins Scientific | https://www.eurofinsgenomics.com/en/products/products.aspx |
| miR-21, UAGCUUAUCAGACUGAUGUUGA | Eurofins Scientific | https://www.eurofinsgenomics.com/en/products/products.aspx |
| HIV TAR, CUGGUUAGACCAGAUCUGAGCCUGGGAGCUCUCUGGCUAACUAG | Eurofins Scientific | https://www.eurofinsgenomics.com/en/products/products.aspx |
| Pre-miR-638, GUGAGCGGGCGCGGCAGGGAUCGCGGGCGGGUGGCGGCCUAGGGCGCGGAGGGCGGACCGGGAAUGGCGCGCCGUGCGCCGCCGGCGUAACUGCGGCGCU | Eurofins Scientific | https://www.eurofinsgenomics.com/en/products/products.aspx |
| Lys3 tRNA, GCCCGGAUAGCUCAGUCGGUAGAGCAUCAGACUUUUAAUCUGAGGGUCCAGGGUUCAAGUCCCUGUUCGGGCGCCA | Miller JT et al., 2004 | N/A |
| miR-21 siRNA mimic | ThermoFisher | 4464066 |
| miR-21 siRNA inhibitor | ThermoFisher | 4464084 |
| qPCR primers, pre-miR-21: forward – TGTCGGGTAGCTTATCAGAC, reverse – TTCAGACAGCCCATCGACTG PDCD4: forward – TAGATTGTGTGCAGGCTA, reverse – GTTCAGCTTCAGATATGTCTC PTEN: default Exiqon construct GAPDH: default Exiqon construct |
EXIQON ExiLERATE | 303402 |
| Software and Algorithms | ||
| GraphPad Prism 7.03 | Graphpad (NIH) | https://www.graphpad.com/scientific-software/prism/ |
| FIJI | NIH | N/A |
| Imaris | Bitplane | N/A |
| NanoString nSolver | nanoString Technologies | https://www.nanostring.com/products/analysis-software/nsolver |
| nSolver Pathway Analysis; Advanced Analysis | nanoString Technologies | https://www.nanostring.com/products/analysis-software/advanced-analysis |
SIGNIFICANCE.
As the concept of druggable RNA targets expand beyond viral and bacterial systems (e.g. riboswitch and ribosomal RNA), our results strongly support that small molecules can indeed be used as modulators of functional oncogenic regulatory RNAs in cells. Regulatory non-coding RNAs are an emerging target for small molecules. Development of methods to identify small molecule modulators of RNA stability is a useful strategy for discovering unstudied chemical probes to elucidate biological functions and potential therapeutics. Towards that end, we apply a high-throughput DSF-based RNA screen to identify small molecule natural products that can modulate the stability of discrete structured RNAs. From this method, we report a prodiginine-type compound as an active pharmacophore for RNA-binding. Our studies detail how such compound can downregulate miR-21 processing and diminish the function of oncogenic miR-21 in colon cancer cells. The identification of RNA-binding pharmacophores such as the natural product butylcylcoheptyl prodiginine (bPGN) is important for the continued development of the field and this study will lead to further research of this compound class, and natural products in general, to better discern their potential as pharmacophores for RNA-binding therapeutic agents against relevant diseases including cancer.
Highlights:
A screen to identify natural products that can bind regulatory non-coding RNAs
Identification of natural pharmacophores that can bind precursor microRNA-21
Butylcylcoheptyl prodiginine inhibits microRNA-21 and reduces growth of cancer cells
Regulatory non-coding RNAs can be potential targets for small molecule therapeutics
ACKNOWLEDGEMENTS
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Funding support also from National Cancer Institute Director’s Innovation Award to JSM. We thank Mary McNally, Nina Bubunenko, and Xiaolin Wu (Fredrick National Laboratory for Cancer Research, Genomics Laboratory) for performing the NanoString experiments. We thank Jennifer Miller (Stuart Le Grice’s Lab) for providing the tRNA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes 5 figures and 2 tables.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Agarwal V, Bell GW, Nam JW, and Bartel DP (2015). Predicting effective microRNA target sites in mammalian mRNAs. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano ML, Borthakur G, Berger M, Luer J, and Raza A (2014). A phase II, multicenter, open-label study of obatoclax mesylate in patients with previously untreated myelodysplastic syndromes with anemia or thrombocytopenia. Clin Lymphoma Myeloma Leuk 14, 534–539. [DOI] [PubMed] [Google Scholar]
- Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, and Allgayer H (2008). MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 27, 2128–2136. [DOI] [PubMed] [Google Scholar]
- Bartel DP (2018). Metazoan MicroRNAs. Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, and Chen CZ (2004). Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 5, 396–400. [DOI] [PubMed] [Google Scholar]
- Bauersachs J (2012). miR-21: a central regulator of fibrosis not only in the broken heart. Cardiovasc Res 96, 227–229; discussion 230–223. [DOI] [PubMed] [Google Scholar]
- Buscaglia LE, and Li Y (2011). Apoptosis and the target genes of microRNA-21. Chin J Cancer 30, 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro AJ (1967). Antimalarial activity of prodigiosin. Nature 213, 903–904. [DOI] [PubMed] [Google Scholar]
- Chai C, Song LJ, Han SY, Li XQ, and Li M (2018). MicroRNA-21 promotes glioma cell proliferation and inhibits senescence and apoptosis by targeting SPRY1 via the PTEN/PI3K/AKT signaling pathway. CNS Neurosci Ther 24, 369–380. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Chatterjee S, and Grosshans H (2009). Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461, 546–549. [DOI] [PubMed] [Google Scholar]
- Chen J, Jin S, Abraham V, Huang X, Liu B, Mitten MJ, Nimmer P, Lin X, Smith M, Shen Y, et al. (2011). The Bcl-2/Bcl-X(L)/Bcl-w inhibitor, navitoclax, enhances the activity of chemotherapeutic agents in vitro and in vivo. Mol Cancer Ther 10, 2340–2349. [DOI] [PubMed] [Google Scholar]
- Chen S, Wang G, Niu X, Zhao J, Tan W, Wang H, Zhao L, and Ge Y (2014). Combination of AZD2281 (Olaparib) and GX15–070 (Obatoclax) results in synergistic antitumor activities in preclinical models of pancreatic cancer. Cancer Lett 348, 20–28. [DOI] [PubMed] [Google Scholar]
- Chen X, Xie B, Cao L, Zhu F, Chen B, Lv H, Fan X, Han L, Bie L, Cao X, et al. (2017). Direct binding of microRNA-21 pre-element with Regorafenib: An alternative mechanism for anti-colorectal cancer chemotherapy? J Mol Graph Model 73, 48–53. [DOI] [PubMed] [Google Scholar]
- Connelly CM, Boer RE, Moon MH, Gareiss P, and Schneekloth JS Jr. (2017). Discovery of Inhibitors of MicroRNA-21 Processing Using Small Molecule Microarrays. ACS Chem Biol 12, 435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EC, Van Doorslaer K, Rogler LE, and Rogler CE (2010). Overexpression of miR-21 promotes an in vitro metastatic phenotype by targeting the tumor suppressor RHOB. Mol Cancer Res 8, 691–700. [DOI] [PubMed] [Google Scholar]
- Crooke ST, Witztum JL, Bennett CF, and Baker BF (2018). RNA-Targeted Therapeutics. Cell Metab 27, 714–739. [DOI] [PubMed] [Google Scholar]
- Darshan N, and Manonmani HK (2016). Prodigiosin inhibits motility and activates bacterial cell death revealing molecular biomarkers of programmed cell death. AMB Express 6, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SK, Sokhi UK, Bhutia SK, Azab B, Su ZZ, Sarkar D, and Fisher PB (2010). Human polynucleotide phosphorylase selectively and preferentially degrades microRNA-221 in human melanoma cells. Proc Natl Acad Sci U S A 107, 11948–11953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzler R, McGeary SE, Title AC, Agarwal V, Bartel DP, and Stoffel M (2016). Impact of MicroRNA Levels, Target-Site Complementarity, and Cooperativity on Competing Endogenous RNA-Regulated Gene Expression. Mol Cell 64, 565–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Disney MD, Winkelsas AM, Velagapudi SP, Southern M, Fallahi M, and Childs-Disney JL (2016). Inforna 2.0: A Platform for the Sequence-Based Design of Small Molecules Targeting Structured RNAs. ACS Chem Biol 11, 1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng YH, Wu CL, Tsao CJ, Chang JG, Lu PJ, Yeh KT, Uen YH, Lee JC, and Shiau AL (2011). Deregulated expression of sprouty2 and microRNA-21 in human colon cancer: Correlation with the clinical stage of the disease. Cancer Biol Ther 11, 111–121. [DOI] [PubMed] [Google Scholar]
- Gantier MP, McCoy CE, Rusinova I, Saulep D, Wang D, Xu D, Irving AT, Behlke MA, Hertzog PJ, Mackay F, et al. (2011). Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res 39, 5692–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gariboldi MB, Taiana E, Bonzi MC, Craparotta I, Giovannardi S, Mancini M, and Monti E (2015). The BH3-mimetic obatoclax reduces HIF-1alpha levels and HIF-1 transcriptional activity and sensitizes hypoxic colon adenocarcinoma cells to 5-fluorouracil. Cancer Lett 364, 156–164. [DOI] [PubMed] [Google Scholar]
- Goard CA, and Schimmer AD (2013). An evidence-based review of obatoclax mesylate in the treatment of hematological malignancies. Core Evid 8, 15–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondil VS, Asif M, and Bhalla TC (2017). Optimization of physicochemical parameters influencing the production of prodigiosin from Serratia nematodiphila RL2 and exploring its antibacterial activity. 3 Biotech 7, 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, and Shiekhattar R (2006). MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol Biol 342, 33–47. [DOI] [PubMed] [Google Scholar]
- Gryshkova V, Fleming A, McGhan P, De Ron P, Fleurance R, Valentin JP, and Nogueira da Costa A (2018). miR-21–5p as a potential biomarker of inflammatory infiltration in the heart upon acute drug-induced cardiac injury in rats. Toxicol Lett 286, 31–38. [DOI] [PubMed] [Google Scholar]
- Han L, Zhou Y, Huang X, Xiao M, Zhou L, Zhou J, Wang A, and Shen J (2014). A multi-spectroscopic approach to investigate the interaction of prodigiosin with ct-DNA. Spectrochim Acta A Mol Biomol Spectrosc 123, 497–502. [DOI] [PubMed] [Google Scholar]
- Hsieh HY, Shieh JJ, Chen CJ, Pan MY, Yang SY, Lin SC, Chang JS, Lee AY, and Chang CC (2012). Prodigiosin down-regulates SKP2 to induce p27(KIP1) stabilization and antiproliferation in human lung adenocarcinoma cells. Br J Pharmacol 166, 2095–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, and Mendell JT (2007). A hexanucleotide element directs microRNA nuclear import. Science 315, 97–100. [DOI] [PubMed] [Google Scholar]
- Jain SS, Anderson CM, DiRienzo F, Taylor IR, Jain K, Guha S, and Hoque N (2013). RNA binding and inhibition of primer extension by a Ru(III)/Pt(II) metal complex. Chem Commun (Camb) 49, 5031–5033. [DOI] [PubMed] [Google Scholar]
- Jiang CS, Wang XM, Zhang SQ, Meng LS, Zhu WH, Xu J, and Lu SM (2015). Discovery of 4-benzoylamino-N-(prop-2-yn-1-yl)benzamides as novel microRNA-21 inhibitors. Bioorg Med Chem 23, 6510–6519. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Lyu XM, Yuan Y, and Wang L (2017). Plasma miR-21 expression: an indicator for the severity of Type 2 diabetes with diabetic retinopathy. Biosci Rep 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joudeh J, and Claxton D (2012). Obatoclax mesylate : pharmacology and potential for therapy of hematological neoplasms. Expert Opin Investig Drugs 21, 363–373. [DOI] [PubMed] [Google Scholar]
- Koehler BC, Scherr AL, Lorenz S, Elssner C, Kautz N, Welte S, Jaeger D, Urbanik T, and Schulze-Bergkamen H (2014). Pan-Bcl-2 inhibitor obatoclax delays cell cycle progression and blocks migration of colorectal cancer cells. PLoS One 9, e106571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopleva M, Watt J, Contractor R, Tsao T, Harris D, Estrov Z, Bornmann W, Kantarjian H, Viallet J, Samudio I, et al. (2008). Mechanisms of antileukemic activity of the novel Bcl-2 homology domain-3 mimetic GX15–070 (obatoclax). Cancer Res 68, 3413–3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, and Gabriely G (2009). miR-21: a small multi-faceted RNA. J Cell Mol Med 13, 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schubeler D, et al. (2010). Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell 141, 618–631. [DOI] [PubMed] [Google Scholar]
- Lin S, and Gregory RI (2015). MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 15, 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz DA, Song JM, and Garner AL (2015). High-throughput platform assay technology for the discovery of pre-microrna-selective small molecule probes. Bioconjug Chem 26, 19–23. [DOI] [PubMed] [Google Scholar]
- Luo M, Tan X, Mu L, Luo Y, Li R, Deng X, Chen N, Ren M, Li Y, Wang L, et al. (2017). MiRNA-21 mediates the antiangiogenic activity of metformin through targeting PTEN and SMAD7 expression and PI3K/AKT pathway. Sci Rep 7, 43427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Choudhury SN, Hua X, Dai Z, and Li Y (2013). Interaction of the oncogenic miR-21 microRNA and the p53 tumor suppressor pathway. Carcinogenesis 34, 1216–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod AR, and Crooke ST (2017). RNA Therapeutics in Oncology: Advances, Challenges, and Future Directions. J Clin Pharmacol 57 Suppl 10, S43–S59. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, and Patel T (2007). MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 133, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montaner B, Castillo-Avila W, Martinell M, Ollinger R, Aymami J, Giralt E, and Perez-Tomas R (2005). DNA interaction and dual topoisomerase I and II inhibition properties of the anti-tumor drug prodigiosin. Toxicol Sci 85, 870–879. [DOI] [PubMed] [Google Scholar]
- Naro Y, Thomas M, Stephens MD, Connelly CM, and Deiters A (2015). Aryl amide small-molecule inhibitors of microRNA miR-21 function. Bioorg Med Chem Lett 25, 4793–4796. [DOI] [PubMed] [Google Scholar]
- Or CR, Chang Y, Lin WC, Lee WC, Su HL, Cheung MW, Huang CP, Ho C, and Chang CC (2016). Obatoclax, a Pan-BCL-2 Inhibitor, Targets Cyclin D1 for Degradation to Induce Antiproliferation in Human Colorectal Carcinoma Cells. Int J Mol Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Tomas R, Montaner B, Llagostera E, and Soto-Cerrato V (2003). The prodigiosins, proapoptotic drugs with anticancer properties. Biochem Pharmacol 66, 1447–1452. [DOI] [PubMed] [Google Scholar]
- Perez-Tomas R, and Vinas M (2010). New insights on the antitumoral properties of prodiginines. Curr Med Chem 17, 2222–2231. [DOI] [PubMed] [Google Scholar]
- Ramachandran V, and Chen X (2008). Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321, 1490–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Zhou X, Yang JJ, Liu X, Zhao XH, Wang QX, Han L, Song X, Zhu ZY, Tian WP, et al. (2015). AC1MMYR2 impairs high dose paclitaxel-induced tumor metastasis by targeting miR-21/CDK5 axis. Cancer Lett 362, 174–182. [DOI] [PubMed] [Google Scholar]
- Ruegger S, and Grosshans H (2012). MicroRNA turnover: when, how, and why. Trends Biochem Sci 37, 436–446. [DOI] [PubMed] [Google Scholar]
- Schimmer AD, O’Brien S, Kantarjian H, Brandwein J, Cheson BD, Minden MD, Yee K, Ravandi F, Giles F, Schuh A, et al. (2008). A phase I study of the pan bcl-2 family inhibitor obatoclax mesylate in patients with advanced hematologic malignancies. Clin Cancer Res 14, 8295–8301. [DOI] [PubMed] [Google Scholar]
- Schimmer AD, Raza A, Carter TH, Claxton D, Erba H, DeAngelo DJ, Tallman MS, Goard C, and Borthakur G (2014). A multicenter phase I/II study of obatoclax mesylate administered as a 3- or 24-hour infusion in older patients with previously untreated acute myeloid leukemia. PLoS One 9, e108694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger T, Fischer A, Muhly-Reinholz M, Zeiher AM, and Dimmeler S (2014). Long-term inhibition of miR-21 leads to reduction of obesity in db/db mice. Obesity (Silver Spring) 22, 2352–2360. [DOI] [PubMed] [Google Scholar]
- Shi Z, Zhang J, Qian X, Han L, Zhang K, Chen L, Liu J, Ren Y, Yang M, Zhang A, et al. (2013). AC1MMYR2, an inhibitor of dicer-mediated biogenesis of Oncomir miR-21, reverses epithelial-mesenchymal transition and suppresses tumor growth and progression. Cancer Res 73, 5519–5531. [DOI] [PubMed] [Google Scholar]
- Soto-Cerrato V, Llagostera E, Montaner B, Scheffer GL, and Perez-Tomas R (2004). Mitochondria-mediated apoptosis operating irrespective of multidrug resistance in breast cancer cells by the anticancer agent prodigiosin. Biochem Pharmacol 68, 1345–1352. [DOI] [PubMed] [Google Scholar]
- Su JC, Chang JH, Huang JW, Chen PP, Chen KF, Tseng PH, and Shiau CW (2015). Copper-obatoclax derivative complexes mediate DNA cleavage and exhibit anti-cancer effects in hepatocellular carcinoma. Chem Biol Interact 228, 108–113. [DOI] [PubMed] [Google Scholar]
- Velagapudi SP, Costales MG, Vummidi BR, Nakai Y, Angelbello AJ, Tran T, Haniff HS, Matsumoto Y, Wang ZF, Chatterjee AK, et al. (2018). Approved Anti-cancer Drugs Target Oncogenic Non-coding RNAs. Cell Chemical Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco I, and Sawyers CL (2002). The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer 2, 489–501. [DOI] [PubMed] [Google Scholar]
- Wang W, Zhao L, Wei X, Wang L, Liu S, Yang Y, Wang F, Sun G, Zhang J, Ma Y, et al. (2016a). MicroRNA-320a promotes 5-FU resistance in human pancreatic cancer cells. Sci Rep 6, 27641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Li B, Zhou L, Yu S, Su Z, Song J, Sun Q, Sha O, Wang X, Jiang W, et al. (2016b). Prodigiosin inhibits Wnt/beta-catenin signaling and exerts anticancer activity in breast cancer cells. Proc Natl Acad Sci U S A 113, 13150–13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong N, and Wang X (2015). miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 43, D146–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhams DC, LaBumbard BC, Barnhart KL, Becker MH, Bletz MC, Escobar LA, Flechas SV, Forman ME, Iannetta AA, Joyce MD, et al. (2018). Prodigiosin, Violacein, and Volatile Organic Compounds Produced by Widespread Cutaneous Bacteria of Amphibians Can Inhibit Two Batrachochytrium Fungal Pathogens. Microb Ecol 75, 1049–1062. [DOI] [PubMed] [Google Scholar]
- Xie C, Edwards H, Caldwell JT, Wang G, Taub JW, and Ge Y (2015). Obatoclax potentiates the cytotoxic effect of cytarabine on acute myeloid leukemia cells by enhancing DNA damage. Mol Oncol 9, 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada N, Tsujimura N, Kumazaki M, Shinohara H, Taniguchi K, Nakagawa Y, Naoe T, and Akao Y (2014). Colorectal cancer cell-derived microvesicles containing microRNA-1246 promote angiogenesis by activating Smad 1/5/8 signaling elicited by PML down-regulation in endothelial cells. Biochim Biophys Acta 1839, 1256–1272. [DOI] [PubMed] [Google Scholar]
- Zhong X, Chung AC, Chen HY, Dong Y, Meng XM, Li R, Yang W, Hou FF, and Lan HY (2013). miR-21 is a key therapeutic target for renal injury in a mouse model of type 2 diabetes. Diabetologia 56, 663–674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
nSolver and Advanced Analysis software are available in nanoString Technologies website (https://www.nanostring.com/). FIJI and Imaris are available from NIH and Oxford Instruments, respectively.