Abstract
We investigated three bovine respiratory pathobionts in healthy cattle using qPCR optimised and validated to quantify Histophilus somni, Mannheimia haemolytica and Pasteurella multocida over a wide dynamic range. A longitudinal study was conducted to investigate the carriage and density of these bacteria in the nasal passages of healthy beef calves (N = 60) housed over winter in an experimental farm setting. The three pathobiont species exhibited remarkably different carriage rates and density profiles. At housing, high carriage rates were observed for P. multocida (95%), and H. somni (75%), while fewer calves were positive for M. haemolytica (13%). Carriage rates for all three bacterial species declined over the 75-day study, but not all individuals became colonised despite sharing of environment and airspace. Colonisation patterns ranged from continuous to intermittent and were different among pathobiont species. Interval-censored exponential survival models estimated the median duration of H. somni and P. multocida carriage at 14.8 (CI95%: 10.6–20.9) and 55.5 (CI95%: 43.3–71.3) days respectively, and found higher density P. multocida carriage was associated with slower clearance (p = 0.036). This work offers insights into the dynamics of pathobiont carriage and provides a potential platform for further data collection and modelling studies.
Subject terms: Infectious-disease epidemiology, Bacteriology
Introduction
Respiratory microbiota are diverse communities in humans and livestock1. In humans, Staphylococcus aureus and Streptococcus pneumoniae are frequently carried in the nasal passages as commensals but are also significant pathogens on occasion2. Bacteria with this dual commensal-pathogen behaviour have been dubbed pathobionts, although key drivers between commensal and pathogenic states remain to be identified.
Similarly, healthy cattle carry pathobionts in their nasal passages, most commonly members of the Pasteurellaceae family: Histophilus somni, Mannheimia haemolytica and Pasteurella multocida3,4. In the UK, these bacteria are frequently identified in clinical cases of bovine respiratory disease (BRD), along with Mycoplasma bovis and viruses including bovine respiratory syncytial virus (BRSV), parainfluenza virus-3 (BPI3-V), bovine herpesvirus-1 (BHV-1) and bovine diarrhoea virus (BVDV)5. Although the relative importance for disease causation of these various microbial species and their capacity to transit from harmless colonisation of the upper respiratory tract to harmful infection of the lower respiratory tract certainly varies, transition may also be associated with possession of specific virulence factors such as leukotoxin in M. haemolytica6–8.
Pasteurellaceae carriage appears to be benign without predisposing factors such as respiratory viral infection or environmental stressors (e.g. temperature, humidity, transport, mixing of groups of animals); these are thought to be associated with bacterial proliferation and migration into the lower respiratory tract, resulting in pneumonia9–11. Respiratory disease can affect cattle of any age, but is most commonly seen in calves aged under 6 months, typically while housed or shortly after transportation, hence the clinical terminology ‘enzootic pneumonia’ or ‘shipping fever’12. Significant economic losses are incurred, resulting from decreased production, increased morbidity and mortality, and through increased costs of husbandry, veterinary care and preventive measures13. Control of BRD is directed at improving management conditions and preventing viral and bacterial infections through vaccination and antimicrobial therapy14,15. However, reports of resistance to antimicrobials including newer compounds are common and of increasing concern16–18.
Bacterial carriage in the bovine upper respiratory tract has generally been detected by culture19–23, but this is problematic due to the fastidious nature of the organisms and overgrowth by faster-growing species24. Advances in molecular techniques have allowed in-depth investigation of organisms that are currently difficult or impossible to culture, for example by PCR25 and sequencing3,26,27. Real-time quantitative PCR (qPCR) is a rapid and specific method to reliably detect and measure density of bacterial species in a range of clinical samples. TaqMan qPCR uses a 5′ hydrolysis probe alongside oligonucleotide primers to enhance assay specificity28. Density of carriage of bovine respiratory pathobionts has not previously been investigated using qPCR to determine organism load, which may be an important predictor or determinant of disease.
Bacterial carriage is often assessed over time by repeated sampling and assaying for the presence or absence of target organisms. Inherently, this longitudinal approach gives rise to interval-censored data, when an event of interest is known to occur within an interval29. In humans, upper respiratory tract pneumococcal carriage has been modelled using exponential interval-censored survival models to determine the rate of clearance and duration of carriage30, but these approaches are rarely used in veterinary research29.
In the studies reported here, we optimised three TaqMan qPCR assays for detection and quantification of Histophilus somni, Mannheimia haemolytica and Pasteurella multocida in bovine nasal swabs, and applied these assays to the investigation of rates and densities of carriage in the nasal passages of healthy beef calves. Using interval-censored exponential survival analyses, we modelled the time to bacterial clearance and investigated whether sex and density of carriage influenced the duration of nasal bacterial carriage. This approach offers novel insights into Pasteurellaceae carriage patterns in healthy livestock. Characterising bacterial colonisation dynamics in healthy animals should improve our understanding of the biology of carriage, including transmission dynamics, and in turn help inform prevention and control strategies.
Results
Growth curves in liquid culture
Pure cultures were maintained throughout growth curve studies for each bacterial species (H. somni, M. haemolytica and P. multocida), as confirmed by well-isolated colonies and pure Gram stain of overnight plate cultures. Ten-fold serial dilutions yielded appropriate colony counts for all three bacterial species. Growth rates of H. somni, M. haemolytica and P. multocida determined by optical density measurements can be seen in Supplementary Fig. S1.
Real-time PCR standard curves and assay performance
Cultures of H. somni, M. haemolytica and P. multocida in liquid medium were used to construct standard curves for each organism (values given in Table 1 and shown in Supplementary Fig. S2) using the same PCR conditions used to amplify DNA targets in nasal swabs stored in skim milk-tryptone-glucose-glycerol (STGG). Linear regression of Cq value versus observed and extrapolated log10 mean colony count/ml for corresponding 10-fold serial dilutions of bacterial broth cultures provided equations for conversion of Cq values to CFU/ml31. These equations were later applied to Cq values obtained from nasal swabs to obtain genome copies/ml. For all standard curve dilutions, bacteriophage T4 internal amplification controls gave Cq values as expected, indicating successful DNA extraction and providing no evidence of PCR inhibition.
Table 1.
Evaluation of Histophilus somni, Mannheimia haemolytica and Pasteurella multocida qPCR assays on pure log-phase broth cultures of reference strains.
| Assay characteristic | Histophilus somni | Mannheimia haemolytica | Pasteurella multocida | |
|---|---|---|---|---|
| Standard Equation | Slope (standard error) | −0.295 (0.00610) | −0.292 (0.00688) | −0.264 (0.00332) |
| Intercept | 11.3 | 12.4 | 11.2 | |
| r2 | 0.995 | 0.991 | 0.998 | |
| Biological replicates | 2 | 3 | 3 | |
| Degrees of freedom (total) | 13 | 17 | 14 | |
| Efficiency (%)† | 97.5 | 96.0 | 83.5 | |
| Linear dynamic range (log10) | 8 | 7 | 6 | |
| Cq cut-off value‡ | 35 cycles | 34 cycles | 35 cycles |
Linear regression of log10 colony count/ml against cycle quantification (Cq) value to generate standard equation for conversion of swab Cq value to genome copies/ml.
†PCR Amplification Efficiency (%) = [(10−slope) − 1] × 100.
‡The Cq value corresponding to the endpoint dilution of the standard curve at which samples tested positive.
All assays were highly reproducible and repeatable for all bacterial species, producing consistent Cq values at each dilution of the standard curve for biological replicates tested on separate days (coefficient of variation, CV < 6%), and for technical replicates performed on the same day of testing (CV < 4%). For all standard curves, coefficient of determination values were high (r2 > 0.99). Amplification efficiency was > 95% for both M. haemolytica and H. somni and lower for P. multocida at 84% (Table 1). For all three assays, DNA extracts from three dilutions of all heterologous bacterial strains comprising a specificity panel (Supplementary Table S1) yielded negative results.
Re-confirmation of PCR results by culture and sequencing
All nasal swabs collected at day 0 (N = 60) were cultured for the presence of H. somni, M. haemolytica and P. multocida. Presumptive and phenotypically confirmed isolates were stored at −70 °C. As only one swab was determined positive for H. somni by culture, PCR products from H. somni PCR positive nasal swabs (N = 5) were sequenced to confirm identity. In all five, the sequences were representative of H. somni as evident following Basic Local Alignment Search Tool (BLAST) searches: 100% homology to H. somni was seen for 3 of 5 queried sequences, 99% to the fourth and 98% to the fifth. For all 5 queried swab sequences there was homology between 96 and 100% for sequences deposited as ‘uncultured bacterium clone’, probably representing H. somni. One swab had 93% homology to one sequence deposited as Actinobacillus capsulatus. The majority of these swabs (43/60) were determined positive for P. multocida by both culture and PCR, with a smaller number PCR positive and culture negative (14/60). Fewer swabs were determined positive for M. haemolytica (4/60) by both culture and PCR. Similarly, a number (5/60) were PCR positive and culture negative. No swabs were culture positive but negative in the PCR for corresponding species (Supplementary Table S2).
Characteristics of the study animals and signs of respiratory disease
Animal characteristics (sex, sire breed and age) are shown in Table 2. Animals were monitored for signs of respiratory disease using the Wisconsin calf respiratory scoring system. Nasal discharges were observed on three occasions (scored between 1 and 3), two of which were in the same animal. Coughing was observed in one animal on one occasion. No Dictyocaulus viviparous larvae were detected in faeces of any animal.
Table 2.
Sex, sire breed and age of calves in the study (N = 60).
| Green Barn (N = 30) | Red Barn (N = 30) | Total | |
|---|---|---|---|
| Sex | |||
| Heifer | 15/30 | 14/30 | 29/60 |
| Steer | 15/30 | 16/30 | 31/60 |
| Sire Breed | |||
| Charolais | 20/30 | 21/30 | 41/60 |
| Hereford | 4/30 | 3/30 | 7/60 |
| Limousin | 6/30 | 6/30 | 12/60 |
| Median Age (day 0) | |||
| Days (range) | 305 (276–319) | 307 (258–324) | 305 (258–324) |
Carriage rates and patterns
A total of 299 nasal swabs obtained during winter housing of calves were analysed by qPCR to determine carriage rates and densities of H. somni, M. haemolytica and P. multocida; 238 swabs were positive for at least one of these three target bacteria. P. multocida was most frequently detected (227/299; 75.9%), followed by H. somni (80/299; 26.8%) and M. haemolytica (17/299; 5.7%). The overall proportions of swabs positive in each barn were similar for P. multocida (Green Barn: 118/150; 78.7%, Red Barn: 109/149; 73.2%; p = 0.327) and M. haemolytica (Green Barn: 7/150; 4.7%, Red Barn: 10/149; 6.7%; p = 0.608). The proportion of swabs positive for H. somni was lower in the Green (20/150; 13.3%) than in the Red Barn (60/149; 40.3%; p < 0.001). In all cases, the T4 internal amplification control assay Cq values were as expected, indicating successful DNA extraction with no evidence of PCR inhibition.
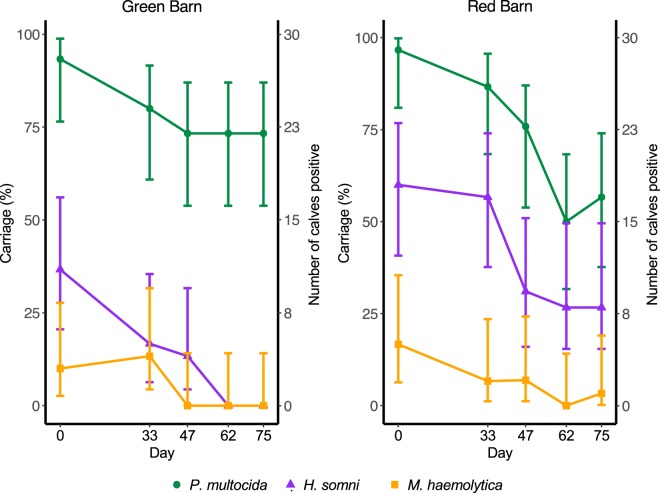
Target bacterial nucleic acid was detected in swabs from each of the sixty animals on at least one sampling occasion. Carriage rates were highest on the first occasion of sampling (day 0), and declined for all bacterial species thereafter, with similar trends observed in both barns. P. multocida was carried at a higher rate than H. somni and M. haemolytica on all sampling occasions in the Green Barn, and on days 0 and 47 in the Red Barn. Clearance of H. somni and M. haemolytica was observed in the Green barn from days 62 and 47, respectively (Fig. 1). Log-linear models provided no evidence to suggest any difference in carriage rates between barns and pens (see Supplementary Information, Method and Data S1).
Figure 1.
Nasal carriage of Pasteurella multocida (green circles), Histophilus somni (purple triangles) and Mannheimia haemolytica (orange squares) in healthy beef calves determined by qPCR on nasal swabs collected on five occasions from sixty calves housed in two identical barns (N = 30 Green Barn, N = 30 Red Barn). Error bars represent Newcombe 95% confidence intervals for the single proportion.
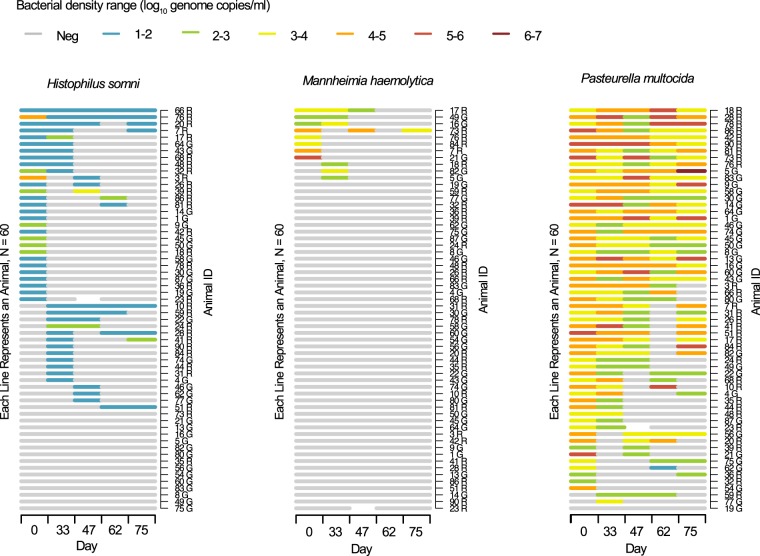
A carriage episode was defined as a period of carriage detected in consecutive samples without interruption. Forty-five (75%) of the 60 calves were positive for H. somni on at least one occasion: 36 of these calves had a single carriage episode and 9 had two episodes. H. somni was detected at the first visit in 29 calves, of which two remained positive at all subsequent study visits. Details of individual carriage trajectories are shown in Fig. 2.
Figure 2.
Carriage patterns and density of Histophilus somni, Mannheimia haemolytica and Pasteurella multocida determined by qPCR on nasal swabs collected on five sampling days (0, 33, 47, 62 and 75) from sixty calves. Animals housed in Green and Red barns are identified by G and R following ID numbers respectively. Density is represented as log10 genome copies/ml. One animal (ID 23 R) had a missing sample at day 47.
Carriage of P. multocida was detected in 57 (95%) of the 60 calves at day 0: 38 of these calves had a single carriage episode and 19 had two episodes. Carriage was detected in 26 calves on all study visits. Of three calves negative for P. multocida on day 0, carriage was detected in two at the subsequent visit, while the third remained negative throughout the study.
Carriage of M. haemolytica was detected in 8 (13%) of the 60 animals at the first visit, of which 7 had one carriage episode and one had three. In three further calves, M. haemolytica was detected at the second visit, while this species of pathobiont was not detected in the remaining 49 animals at any visit.
Co-carriage was defined as detection of more than one bacterial species in the same sample and co-carriage of two and three species was found in 47 and three animals respectively. Co-carriage with H. somni and P. multocida occurred most frequently and M. haemolytica was co-carried with P. multocida more commonly than with H. somni. No evidence of association between pairs of species was apparent using Fisher’s exact test (p > 0.221 for all nine pairwise comparisons; six pairwise comparisons were not performed because neither of the relevant pair of bacterial species were present; p-values may be underestimated as we have not controlled for multiple comparisons).
Carriage density
Individual animal density data are shown in Fig. 2. For each bacterium, density values for all positive samples were also plotted as histograms revealing distinct density profiles (Fig. 3). The modal value for H. somni carriage density accounting for the vast majority (82.5%) of swabs was 1–2 log10 genome copies/ml with fewer samples (13.8%) between 2 and 3 logs. Only two samples in two calves were in the 4–5 log range and one sample in the 3–4 log range. Although carriage rates were low for M. haemolytica, carriage density ranged between 2 and 6 logs when it occurred. Pasteurella multocida was also carried over a wide range of densities, most commonly between 3 and 5 log10 genome copies/ml, extending up to 6.08 (ID: 5 G) logs in one sample. Although density of P. multocida carriage was also dynamic within and between calves, some animals maintained carriage at high densities in the 4–6 log range over several sampling visits, and only one sample in one calf (ID: 62 G) was in the 1–2 log range (Fig. 2).
Figure 3.
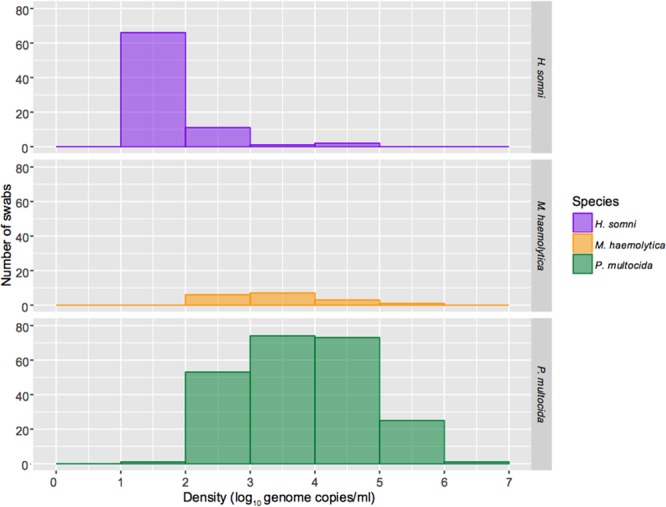
Histograms for all positive swabs summarising density distribution profiles of Histophilus somni (N = 80), Mannheimia haemolytica (N = 17) and Pasteurella multocida (N = 227). Note these data include up to 5 positive swabs from single animals, as shown in Fig. 2.
Duration of carriage and hazard of clearance
Interval-censored exponential survival models for carriage (without inclusion of carriage density, host animal sex, barn or pen as covariates) estimated the median duration of H. somni carriage to be 14.8 days (CI95%: 10.6–20.9) and the hazard of clearance to be 0.0467 per day (CI95%: 0.0326–0.0634). For P. multocida, the median carriage duration was estimated to be 55.5 days (CI95%: 43.3–71.3) and hazard of clearance to be 0.0125 per day (CI95%: 0.00969–0.0159, Fig. 4). For either bacterium, the animals’ sex (H. somni: p = 0.414; P. multocida: p = 0.311), barn (H. somni: p = 0.060; P. multocida: p = 0.106) or pen (H. somni: 0.119; P. multocida: p = 0.449) did not significantly influence carriage duration in survival models. Carriage rates of M. haemolytica were too low for meaningful survival modelling. The effects of carriage density on carriage duration was modelled in univariable analyses. Log hazard ratio and density data were suggestive of a non-linear trend for both H. somni and P. multocida, therefore we modelled the effect of density using categories based on density quartiles rather than continuously. For P. multocida, density of carriage significantly influenced subsequent carriage duration (p = 0.036, Table 3). Categories 2–4 of P. multocida density were significantly associated with increased carriage duration compared to the reference category (1); these trends were non-linear with categories 2 and 4 associated with a longer duration (lower hazard) compared to category 3, and category 4 (highest density) associated with the longest subsequent carriage duration (Table 3). Density was not seen to affect H. somni.
Figure 4.
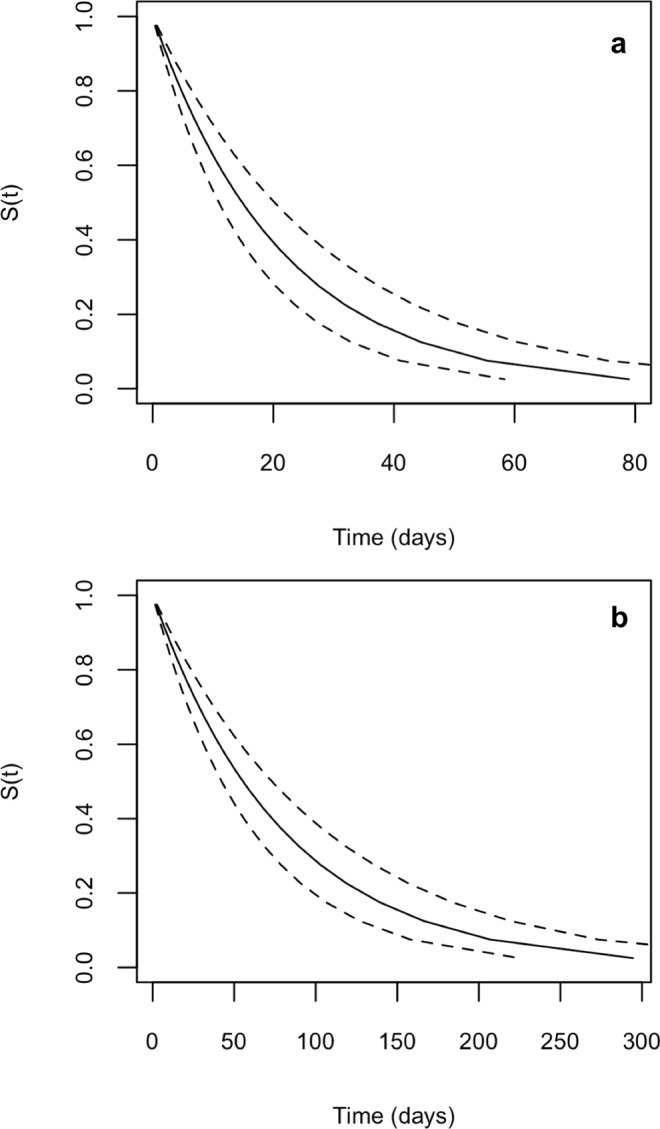
Survival curves: proportion of carriage episodes still ongoing by time for Histophilus somni (panel a) and Pasteurella multocida (panel b). Dotted lines represent 95% confidence interval.
Table 3.
Interval-censored exponential survival models.
| Species | Model | Covariate | Hazard ratio | CI95% of hazard ratio | Hazard ratio p-value§ | Log-likelihood |
|---|---|---|---|---|---|---|
| H. somni (N = 54) | M0: Unconditional | 0.0467* | 0.0333–0.0655 | −55.8 | ||
| M1: Density† | Category 1 (N = 14)1 | 0.0556* | 0.0261–0.112 | −55.0 | ||
| Category 2 (N = 13)2 | 0.967 | 0.383–2.44 | 0.943 | |||
| Category 3 (N = 13)2 | 0.610 | 0.219–1.69 | 0.342 | |||
| Category 4 (N = 14)2 | 0.850 | 0.266–2.72 | 0.784 | |||
| M2: Sex | Heifer1 | 0.0411* | 0.0260–0.0617 | −55.5 | ||
| Steer2 | 1.29 | 0.635–2.61 | 0.484 | |||
| P. multocida (N = 78) | M0: Unconditional | 0.0125* | 0.00950–0.0164 | −109 | ||
| M1: Density‡ | Category 1 (N = 20)1 | 0.0280* | 0.0145–0.0524 | −104 | ||
| Category 2 (N = 19)2 | 0.393 | 0.189–0.814 | 0.0119 | |||
| Category 3 (N = 19)2 | 0.321 | 0.153–0.674 | 0.00267 | |||
| Category 4 (N = 20)2 | 0.414 | 0.197–0.868 | 0.0196 | |||
| M2: Sex | Heifer1 | 0.0108* | 0.00791–0.0146 | −108 | ||
| Steer2 | 1.33 | 0.464–1.23 | 0.255 |
H. somni log-likelihood ratio test statistic: (M0 vs. M1), χ2 (3 df) = 1.62, p = 0.655; (Model 0 vs. Model 2), χ2 (1 df) = 0.668, p = 0.414.
P. multocida log-likelihood ratio test statistic: (M0 vs. M1), χ2 (3 df) = 8.52, p = 0.036; (Model 0 vs. Model 2), χ2 (1 df) = 1.028, p = 0.311.
*Hazard for unconditional model and baseline category.
†H. somni density categories based on quartiles (log10 genome copies/ml): <1.26; ≥1.26 to 1.45; ≥1.45 to 1.96; ≥1.96.
‡P. multocida density categories based on quartiles (log10 genome copies/ml): <3.40; ≥3.40 to 3.99; ≥3.99 to 4.77; ≥4.77.
§P-values shown are for the null hypothesis of no difference between each of the category levels2 in M1 (categories 2, 3 and 4) and M2 (steer) compared to their respective baseline categories1 (category 1 and heifer).
Discussion
We present the first longitudinal investigation into nasal carriage patterns and densities of common bovine respiratory pathobionts in healthy animals using molecular detection tools. The detailed quantitative profiles we obtained over a typical housing period demonstrate marked differences between the bacteria studied in terms of frequency, duration of carriage and microbial density. These results provide methodological and biological information that will be important for future study of bacterial carriage and transmission in housed animals, and the impact of vaccines and viral infections upon respiratory disease.
PCR performed on nasal swabs may give a rapid and reliable indication of both the current colonisation status and, in the case of PCR-positive culture-negative samples, perhaps the recent footprint of colonisation as compared to conventional bacteriological culture which can only reflect the former. To validate detection of target bacteria using the qPCR assays presented in this study we cultured a subset of nasal swabs collected on day 0 (N = 60). In all cases, swabs negative by PCR were also negative by culture and detection of all species was enhanced using PCR, probably reflecting swabs with target DNA but too few viable organisms for successful culture. In addition, we experienced consistent difficulty in isolating H. somni. Selective agars (Haemophilus selective and chocolate with bacitracin) supported growth of type strain ATCC 43625 but no field isolates were obtained from nasal swabs by this method; instead H. somni was cultured from only one swab using non-selective chocolate agar. Reassuringly amplicons from five nasal swabs determined PCR positive (one of which was culture-positive) had sequence homology to H. somni. This highlights the difficulty with culture which requires specialist skills and can yield values for bacterial density only over a narrow range, unless very labour-intensive serial dilutions are prepared for each sample.
The sodA gene of M. haemolytica and 16S rRNA gene sequences of H. somni and P. multocida were proven to be qPCR primer targets capable of separating these pathobionts from other closely-related and common non-pathogenic bacterial species (Supplementary Table S1). Hitherto, discrimination between M. haemolytica and M. glucosida by PCR has generally relied either on analysis of melting curves following SYBR green qPCR, or on the assumption that PCR positive samples are M. haemolytica because M. glucosida has not been implicated in clinical cases of bovine respiratory disease32,33. We were able to discriminate successfully between M. haemolytica and all other members of the Mannheimia genus using a TaqMan probe. A well-known limitation of PCR is that it does not distinguish viable from non-viable organisms. We used cells harvested from liquid cultures in the exponential phase of growth to ensure that the bacterial cell populations for production of calibration curves were highly likely to be viable31.
Animals remained healthy and no animals were diagnosed with respiratory disease throughout the study; on only a few occasions were very slight nasal discharge or coughing observed, but these were mild and transient, and no treatment was necessary. No Dictyocaulus viviparous (bovine lung worm) larvae were detected on any occasion. Surveillance data on diagnostic submissions to Veterinary Investigation Centres confirm bacterial species involved in bovine pneumonia in the UK; most frequently Mannheimia spp. closely followed by P. multocida5. The prevalence of P. multocida as a commensal in the healthy bovine respiratory tract varies21,24,34; the high carriage rates by PCR (61.7–95.0%) we report here in healthy animals suggest it may function as part of the core nasal microbiota. By contrast, we rarely detected M. haemolytica (carriage rates of up to 13.3%), although when carriage did occur it was over a wide range of densities (2–6 logs). It has been suggested M. haemolytica is carried preferentially deeper in the nasopharynx at the palatine tonsil35,36, but our detection of this species in short nasal swabs suggests it may also be shed in nasal secretions. Using culture, nasal carriage of H. somni has been reported in healthy calves (6.6%) and those with respiratory disease (11.9%)21. We observed higher rates by PCR (13.3% to 48.3%). This may reflect the increased sensitivity of detection by qPCR compared to culture, suggesting that H. somni may have been underreported previously.
Patterns of carriage for all three pathobiont species were similar between barns but not identical (clearance of H. somni was observed in the Green barn only). This suggests epidemiology may vary somewhat in different but overtly similar groups of animals. Furthermore, carriage rates for all species declined over the study period in both barns. This observation may reflect animals becoming older. Older animals which are immunologically more mature, or those which have developed appropriate immune responses following exposure may be better equipped to clear bacteria than younger animals.
We defined an episode of carriage as a period when an animal was positive for any one bacterial species in consecutive samples without interruption. One animal experienced three such episodes of carriage with M. haemolytica and carriage was often transient for all three species. While such apparent transient carriage episodes may reflect genuine clearance and re-acquisition, negative results could also be due to intermediate and other sample(s) below the limit of detection of the assay, in the context of apparent large fluctuations in density over time (Fig. 2).
We do not know whether and to what extent the bacteria are evenly distributed within the upper respiratory tract of colonised animals. Conversely, apparent continuous episodes of carriage in our study could also represent repeated acquisition and successive carriage episodes with the same or different strains of H. somni, M. haemolytica or P. multocida. This is a limitation of repeated sampling studies which can be partially addressed by taking more frequent samples or undertaking more detailed characterisation of the bacteria detected. Approaches such as pulsed-field gel electrophoresis, multilocus sequence typing37 and whole genome sequencing can be used to study transmission events; however, their success is dependent on adequate genetic diversity38.
We did not find any evidence of interspecies competition between these three bacterial species which might have influenced their carriage rates. It is possible that carriage rates were too low for us to detect an association between bacterial species which has been reported by others39,40 with the number of animals in our study. Serotype displacement and replacement is known to occur following dysbiosis of the respiratory niche, for example pneumococcal carriage following universal vaccination in childhood41. In cattle, although differences in serotype prevalence have been suggested between healthy animals and those suffering with respiratory disease, this area of microbiology is relatively unexplored42,43.
Interval-censoring occurs when the time-to-event is not observed precisely but is known to have occurred within a particular interval. This commonly occurs in monitoring for infectious diseases when discrete time points are chosen to monitor carriage/infection status29. Biased estimation can result if this imprecision is ignored and the midpoint or right end point of the observed interval is taken as the exact event/failure time44. Using interval-censored exponential survival analysis we estimated the median duration of H. somni carriage at 14.8 days (hazard; 0.0467 per day) and at 55.5 days for P. multocida (hazard; 0.0125 per day), concordant with the increased prevalence observed for P. multocida. We found increasing P. multocida carriage density was significantly associated with increased carriage duration. One explanation for reduced carriage duration at lower densities could be the requirement of a certain number of bacteria for colonisation45, however at higher densities (over a certain threshold) the opportunity for naïve individuals to become exposed increases. Incidentally, H. somni was predominately carried at lower densities (1–2 logs) and was not significantly associated with carriage duration (Fig. 3). M. haemolytica carriage episodes were too few (prevalence 1.7–13.3%) to model using survival analysis but carriage density did extend transiently up to 6 log10 genome copies/ml. Given the infrequency of observed second episodes for both bacterial species, it was not possible to generate precise estimates for the median duration of carriage per episode type. Only two animals had sustained carriage of H. somni for the entire study, but twenty-six had sustained carriage of P. multocida. It would be of interest to define factors which determine persistence of carriage with these pathobionts.
A greater incidence of respiratory disease has been reported in male calves than in female calves46, suggesting sex may influence respiratory pathobiont colonisation dynamics. We included sex as a covariate in exponential survival models but did not find that it was significantly associated with carriage duration for either H. somni or P. multocida. Furthermore, when carriage rates observed at each occasion were stratified by sex there was no difference between the two proportions carrying on any occasion (data not presented). This may indicate that reported differences in disease rates may not be due to marked differences in carriage biology but instead to other factors41.
This study uses qPCR to quantify and track carriage of the target bacterial species over time in the nasal airways. A limitation of PCR is that it will detect DNA from inviable organisms that persist in the respiratory tract even when viable organisms are no longer present or cannot be detected by culture. It can be argued that specific detection of DNA from these unculturable organisms provides evidence of either current or recent colonisation and is thus of biological interest. In studies in humans we have often found samples positive by culture and negative by PCR31 perhaps reflecting that, on occasion, culture has the potential to be more sensitive than PCR when there are very small numbers of organisms present. Interestingly, we did not encounter this phenomenon in this study, perhaps reflecting the poor sensitivity of culture for these bacterial species. In common with all studies of mucosal colonisation, our study is subject to sampling error – we will have failed to detect colonisation on occasion by obtaining the swabs from the wrong place, at the wrong time or insufficiently frequently and, indeed, our choice of the nasal cavity may not have been optimal for detection for one or more of these organisms. Accordingly, our results are likely to be underestimates and there is a need to develop sufficiently non-invasive techniques which will permit more frequent or even continuous sampling with due consideration to animal welfare. More broadly, this is a study in healthy cattle, and we cannot be certain whether and to what extent the strains of bacteria they carry in the nose are associated with outbreaks of disease or how specific virulence factors may vary among the same bacterial species. Moreover, our failure to detect statistical significance, for instance in relation to possible associations among organisms, may be explained by the relatively small sample size.
For the three bacterial species studied, we observed unexpected marked differences in prevalence, carriage duration and density. These observed differences provide evidence for genuine differences in the carriage biology of these organisms and do not reflect sampling artefact. Quantitative molecular techniques have identified increased nasal bacterial carriage and density following respiratory viral infection in the upper respiratory tract of healthy mice and children47,48. We hypothesise density may be important for transmission of bovine pathobionts between herd members and may affect the likelihood of invasive disease. Moreover, respiratory viral infection may influence bacterial colonisation dynamics. The methodological approaches and carriage patterns presented in this report lay the foundation for well powered studies exploring transmission models of bovine respiratory disease.
Materials and Methods
Cattle and husbandry
Beef calves, either Charolais, Hereford or Limousin crosses, were bred at Rothamsted Research, North Wyke Farm, Devon, UK. In November 2015 at weaning, 60 calves aged 5 to 7 months were transferred to the Biotechnology and Biological Sciences Research Council (BBSRC) North Wyke Farm Platform National Capability49 and allocated to one of two identical, physically adjacent purpose-built cattle housing facilities, designated the ‘Green Barn’ and the ‘Red Barn’. Barns are highly standardised and representative of typical modern commercial winter housing facilities for beef cattle in the UK: solid concrete sidewalls to animal height (~1.5 m) and ventilated naturally above animal height with space boarding. Barns are orientated SW-NE such that the prevailing winds assist with their ventilation through end apertures (solid gates approximately 2 m high with openings above and a space-boarded gable end). Windbreaks closing the end apertures are used to provide further protection in adverse weather (strong winds, rain or low temperatures). Animals were allocated to housing, ensuring bodyweight, sire breed and sex were balanced between the two barns. In January 2016, within each barn, calves were separated into six pens each of five animals, nose-to-nose contact between animals within adjacent pens was permitted. Calves were housed on deep litter straw bedding with access to water and silage ad libitum. Animals were observed daily throughout all studies by animal technicians and any cattle showing abnormal behaviour that might have indicated poor health were examined in more detail by one of us (AT) using the Wisconsin scoring system for signs of respiratory disease (cough, nasal and ocular discharge, abnormal ear/head tilt and rectal temperature)50. However, no signs indicative of BRD or any other clinical problem were observed. Animals received no vaccines prior to or during the study, nor were antimicrobials administered to any animal during the course of the study.
Sample collection
Nasal swabs
Nasal swabs were collected from each calf on five separate visits between January 2016–April 2016, on days: 0, 33, 47, 62 and 75 as follows: any excessive debris on nares was cleaned with a disposable paper towel. A swab 15 cm in length with breakable polyester tip (MW821 HydraFlock, Medical Wire & Equipment, Corsham, UK) was inserted to approximately 10 cm depth and rotated 360° against the mucous membranes, then withdrawn carefully, avoiding contact with other areas of the nasal cavity. The swab tip was aseptically broken off into 1.5 ml skim milk-tryptone-glucose-glycerol (STGG) medium. Swab samples were marked with a random number from 1–90 using pre-printed labels, maintained at 4 °C for no more than 3 hours and then vortexed to release bacteria into the STGG and frozen at −70 °C until further analysis31,51. Gloves were changed between handling each animal. On days 33 and 47 a dual-tipped polyester swab (MW821DC Dual Hydraflock, Medical Wire & Equipment, Corsham, UK) was collected; one tip was transferred to STGG and the other to RNAlater stabilisation solution (Thermo Fisher Scientific, UK) for gene expression studies (to be reported later). A nasal swab was not collected on day 47 from one animal (ID: 23 R) as difficulty was experienced collecting a sample from this individual.
Parasitological sampling
Shortly after housing in November 2015, composite samples of 10 freshly deposited faecal pats collected from the floor of each barn were tested for the presence of respiratory nematode larvae (Dictyocaulus viviparus) using the Baermann technique. Following parasitological testing, all calves were treated once routinely with 200 mg ivermectin (Noromectin®, Norbrook) pour-on (40 ml of product) to control respiratory and gastrointestinal nematodes and external parasites.
Bacterial culture
H. somni American Type Culture Collection (ATCC) 43625, M. haemolytica ATCC 33396 and P. multocida ATCC 43137 were used as positive control strains for culture and PCR assay development. All reference strains used in the study (Supplementary Table S1) for PCR assay optimisation and specificity testing were cultured on Columbia blood agar supplemented with 5% sheep blood (CBA; Thermo Fisher Scientific, Basingstoke, UK) overnight at 37 °C with 5% CO2, except for H. somni and Haemophilus influenzae, which were cultured on Chocolate agar (E&O laboratories, UK) at 37 °C, for 24–48 hours with 5% CO2. All cultures were sub-cultured and Gram-stained to ensure purity.
For isolation of Pasteurellaceae, nasal swabs were thawed on ice, vortexed and a 50 µl STGG broth aliquot was spread over the entire surface of an agar plate52. For detection of M. haemolytica and P. multocida swabs were cultured on CBA, Pasteurella selective agar and MacConkey agar with salt (Thermo Fisher Scientific, Basingstoke, UK). Haemophilus selective agar (Thermo Fisher Scientific, Basingstoke, UK) and Chocolate agar plates (E&O laboratories, UK) were used for the enumeration of H. somni. Plates were cultured in an atmosphere containing 5% CO2 at 37 °C for 16–72 hours. Plates were examined after 16 hours and all colonies phenotypically resembling M. haemolytica or P. multocida were subcultured onto CBA, while presumptive H. somni colonies were subcultured onto Chocolate agar. Bacterial identification was done according to standard microbiological techniques for H. somni53, M. haemolytica54 and P. multocida55. For long term storage, bacterial cells were harvested into Brain Heart Infusion broth or Haemophilus Test Medium (H. somni only) supplemented with 20% glycerol (Media Services, School of Cellular and Molecular Medicine, University of Bristol, UK) and stored at −70 °C.
DNA extraction
Frozen nasal swab samples and bacterial reference strains were thawed on ice and then vortexed. Each sample (300 µl) was aliquoted into a 2 ml extraction tube. Automated extraction of nucleic acid from samples was carried out in a QIAsymphony SP instrument (QIAGEN, CA, USA) using QIAsymphony DSP Virus/Pathogen Mini Kit (QIAGEN, CA, USA). An elution volume of 140 µl was produced from 200 µl of the 300 µl aliquot and stored at −70 °C. To determine successful DNA extraction and absence of PCR inhibition the bacteriophage T4 was used as an internal amplification control31.
Real-time PCR for bacteria
Real-time TaqMan PCR (qPCR) assays targeting sodA for M. haemolytica and the 16S rRNA region of the bacterial genome for H. somni and P. multocida were performed, using previously published primers and TaqMan probes32,56. For M. haemolytica a novel TaqMan probe was designed to work with previously published primers (Table 4).
Table 4.
Primer and probe (P/P) sequences used for qPCR assays in this study.
| Target Species | Target | P/P Name | P/P Sequence (5′–3′) | Length | Melting Point (°C) | Reference |
|---|---|---|---|---|---|---|
| M. haemolytica | sodA | Mh-SGF | AGCAGCGACTACTCGTGTTGGTTCAG | 26 | 65.6 | Guenther et al.32 |
| M. haemolytica | sodA | Mh-SGR | AAGACTAAAATCGGATAGCCTGAAACGCCTG | 31 | 68.7 | Guenther et al.32 |
| M. haemolytica | sodA | Mh-BV1P* | TTCAACCGCTAACCAGGACAACCCAC | 26 | 68.4 | This study |
| P. multocida | 16S rRNA | Pm-TMF | CGCAGGCAATGAATTCTCTTC | 21 | 58.5 | Mahony and Horwood56 |
| P. multocida | 16S rRNA | Pm-TMR | GGCGCTCTTCAGCTGTTTTT | 20 | 58.3 | Mahony and Horwood56 |
| P. multocida | 16S rRNA | Pm-TMP* | ACTGCACCAACAAATGCTTGCTGAGTTAGC | 30 | 69.2 | Mahony and Horwood56 |
| H. somni | 16S rRNA | Hs-TMF | AGGAAGGCGATTAGTTTAAGAGATTAATT | 29 | 58.8 | Mahony and Horwood56 |
| H. somni | 16S rRNA | Hs-TMR | TCACACCTCACTTAAGTCACCACCT | 25 | 60.0 | Mahony and Horwood56 |
| H. somni | 16S rRNA | Hs-TMP* | ATTGACGATAATCACAGAAGAAGCACCGGC | 30 | 69.7 | Mahony and Horwood56 |
*Probe fluorophore and quencher: 5′ FAM, 3′ BHQ-1.
Primers and probes were assessed and optimised using Primer Express Software v.3.0 (Life Technologies, USA), and specificity was assessed in silico by BLAST searches using the National Center for Biotechnology Information database (NCBI).
Extracts were thawed and centrifuged prior to PCR for 1 minute at 1000 × g. MicroAmp optical 384-well reaction plates (Life Technologies, USA) were prepared using a QIAgility pipetting robot and software (QIAGEN, CA, USA). The qPCRs were performed in a 20 µl volume consisting of 10 µl TaqMan® (Applied Biosystems), 5 µl Primer/Probe Mix (Sigma Aldrich) and 5 µl nucleic acid template. Working concentrations of primers and probe were 300 nM and 100 nM respectively. Cycling conditions on a ViiA7 real-time PCR instrument (Thermo Fisher Scientific) for P. multocida, H. somni and T4 (internal amplification control) assays were as follows: 95 °C for 20S hold stage, followed by 50 cycles of 95 °C for 3S and 60 °C for 60S. Cycling conditions on a QS7 real-time PCR instrument (Thermo Fisher Scientific) for M. haemolytica were employed as follows: 95 °C for 20S hold stage, followed by 50 cycles of 95 °C for 3S and 69 °C for 60S. Cross-reactivity was observed in this assay with M. glucosida CCUG 38457 T under the same cycling conditions used for P. multocida, H. somni and T4 as described above. A thermal gradient PCR was conducted and determined an alternative annealing temperature at 69 °C where no cross-reactivity was observed (data not presented). Fluorescence emission was measured at the end of the elongation step. No template and positive template controls were included in every run.
Real-time PCR data collected for P. multocida, H. somni and T4 were analysed using ViiA7 software (v1.1) and data collected for M. haemolytica were analysed using QuantStudio™ Real-Time PCR Software (v1.3). For all assays, auto baseline settings were employed, and threshold values manually set after all PCR runs were completed. Thresholds were set above any background amplification and approximately halfway through the exponential phase. Results were exported as csv files for further analysis.
Specificity panel
To determine the specificity of the three assays, a panel of 40 bacterial strains (Supplementary Table S1) were selected based either on genetic relatedness to target organisms, known involvement with respiratory disease in ruminants, or being common bacteria of different genera. Specificity panel bacteria were cultured overnight as described above and diluted 10−3, 10−4, 10−5 in L6 lysis buffer (Public Health England, Bristol, UK), followed by automated DNA extraction using a QIAsymphony SP instrument (as described above).
Growth curves in liquid broth culture
Log-phase liquid cultures of H. somni, M. haemolytica and P. multocida were used to construct a 10-fold dilution series for each organism, quantified at each dilution by culture and colony counting. Growth curves were performed in triplicate on separate days for M. haemolytica and P. multocida and in duplicate for H. somni (biological replicates). M. haemolytica and P. multocida reference strains were grown in liquid medium as described in39,40. In brief, 4–6 colonies from pure plate cultures were used to inoculate 20 ml Brain Heart Infusion (BHI) broth (Media Services, School of Cellular and Molecular Medicine, University of Bristol) and incubated overnight at 37 °C with shaking at 200 rpm. An aliquot of overnight inoculum was transferred to fresh BHI broth the next morning to achieve a starting optical density (OD) measured at 600 nm (Thermo Spectronic Genesys 6, Thermo Electron Scientific Instruments LLC, WI, USA) of 0.05 (approximately 107 CFU/ml). After 1 hour, and at subsequent regular intervals 1 ml aliquots were taken for further OD measurements until stationary phase was reached. A final reading was taken at 24 hours. For H. somni, Veterinary Fastidious Medium (VFM) (Oxoid, Basingstoke, UK) was directly inoculated with 4–6 colonies from pure plate culture (starting OD of 0.05, approximately 107 CFU/ml) and incubated at 37 °C, with shaking at 200 rpm and 5% CO257. For all bacterial species, at late logarithmic phase, a 100 µl aliquot of liquid culture was taken and 10-fold serial dilutions prepared in 900 µl STGG. For M. haemolytica and P. multocida 50 µl aliquots of dilutions 10−3 to 10−8 of the series were plated out in triplicate onto BHI agar. For H. somni, 100 µl aliquots from dilutions 10−3 to 10−8 were plated onto Chocolate agar in triplicate. All plates were incubated for 12–24 hours at 37 °C, in 5% CO2 for colony counts. Counts over 750 were considered too numerous to count and where necessary were extrapolated from higher dilutions yielding lower counts. Bacterial counts were performed in triplicate and the mean expressed as log10 colony count/ml. After plating, the dilution series was immediately cooled and held frozen at −70 °C.
Real-time PCR standard curves
Liquid broth cultures of each bacterial species were used to generate qPCR standard curves31 which were used to evaluate assay performance and to quantify template from bovine nasal swabs.
Dilutions of liquid cultures for each organism (described above) were thawed on ice and a 300 µl aliquot of each dilution (10−1 to 10−10) was inactivated at 100 °C for 10 minutes using a digital heat block (Grant Boekel, BBD, Grant Instruments, Cambridge, UK)31. Successful inactivation was confirmed through appropriate plate cultures for each species. Nucleic acid was extracted from all dilutions and PCR runs were conducted for each bacterial species as described above.
Standard curves were generated by linear regression of cycle quantification (Cq) values versus log10 CFU/ml values for corresponding 10-fold serial dilutions of broth cultures. Five technical replicates of Cq values were performed at each dilution. The linear operating range, Cq cut-off value and amplification efficiency were determined for each assay. The amplification efficiency (E) was calculated based on the slope of the standard curve as follows: E (%) = (10−slope − 1) × 100. The endpoint dilution of the standard curve at which tested samples were positive was used to determine Cq cut-off values.
DNA sequencing
As culture confirmation proved technically difficult for this bacterial species, products generated from the H. somni PCR assay performed on nasal swabs were purified using a QIAquick PCR purification kit (Qiagen, CA, USA) according to the manufacturer’s instructions and sequenced (Eurofins Genomics, Ebersberg, Germany) with identification based on BLAST analysis of the target 16S rRNA gene.
Standard curves
A linear regression model was fitted to colony counts and Cq values obtained from liquid culture as described above. The log10 mean colony count/ml was considered as the response variable. Mean Cq value (N = 5) was an explanatory variable. A parallel lines model was fitted to allow for differences in the intercept between biological replicates of the growth curves, obtaining the best estimate of the slope by assuming an additive effect of biological replicate. After fitting a parallel lines model, a predictive model was produced for each species using the mean intercept across the biological replicates. Cq values obtained from collected nasal swabs were converted to genome copies/ml by interpolation using the predictive models. In vivo, clinical samples are likely to contain both viable and non-viable bacterial cells. Accordingly, colony forming units/ml (CFU/ml) and genome copies/ml cannot be used interchangeably for these assays, and genome copies/ml are presented.
Carriage and co-carriage
Rates of carriage of H. somni, M. haemolytica and P. multocida were calculated as the numbers of animals positive by qPCR for each agent divided by the numbers of animals sampled on each occasion, unless stated otherwise. Carriage estimates for each bacterial species were calculated for each barn (Green and Red) and as an overall average. Co-carriage rate, defined as carriage of either two or three of the bacterial species on the same occasion, was calculated as an overall average for the two barns. Confidence intervals for proportions and differences between proportions were calculated using the Wilson score58 and Newcombe-Wilson hybrid score59 methods respectively60. Levels of co-carriage of H. somni, M. haemolytica and P. multocida were assessed at each visit using Fisher’s exact test (because of expected values lower than 5) to assess for the independence of the carriage of pairs of different bacterial species, based on the counts of calves with different combinations of positive/negative qPCR results.
Interval-censored survival analysis
Interval-censored survival analysis was used to estimate the rate (‘hazard’) of clearance of bacterial carriage. A parametric proportional hazards regression model for interval-censored data with an exponential distribution was fitted to carriage episodes from all animals (N = 60). An episode was defined as a period of uninterrupted H. somni or P. multocida carriage. Further details on rules defining carriage episodes and on dataset construction are provided in Supplementary Information, Method S2. The number of carriage episodes for each bacterium contributing to the survival analysis was not equal to the number of animals observed; some animals experienced multiple carriage episodes whilst others were carriage-free. Adjustment for correlation between recurrent episodes of carriage within the same animal was made by including animal identity as a clustering variable and calculating cluster robust standard errors61. The effects of sex and density of carriage when first positive were considered as covariates in univariable analyses. To assess the relationship between log hazard ratio and density (log10 genome copies/ml) we grouped the observations into categories based on the density quartiles. Parameter estimates for each density category were plotted to assess the fit. The hazard for each sex and density category was estimated and expressed as a hazard ratio relative to the baseline category. Median carriage duration was calculated as −loge(0.5) divided by the estimated hazard value. Improvement in model fit between the unconditional model (no covariates specified) and the model with the covariate of interest was assessed by calculating the log-likelihood test ratio statistic. The strength of the relationship between categorical covariates (sex/carriage density) and hazard of clearance was assessed using the hazard ratio p-value. All analyses were performed in R version 3.5.0, using base functions and the following supplementary packages: ggplot2 (version 2.2.1), longCatEDA (version 0.3162), icenReg (version 2.0.763).
Ethics statement
All animal studies were approved by the Rothamsted Research Ethical Review Board and animal care and use protocol protocols adhered to the Animals (Scientific Procedures) Act 1986 as revised 1 January 2013 to comply with European Directive 2010/63/EU, permit number (3003338).
Supplementary information
Supplementary Information: Insights into Pasteurellaceae carriage dynamics in the nasal passages of healthy beef calves
Acknowledgements
The authors thank Rothamsted Research (BBS/E/C/000J0100) for hosting animal work; Hannah Fleming, Bruce Griffith and Simon White for excellent support and animal care during sample collection periods (Rothamsted Research); the late Robert Orr for initial contributions towards study conceptualisation and extensive knowledge of the North Wyke Farm Platform operations (Rothamsted Research); Debbie Langton for sequencing support (University of Bristol). The authors acknowledge the support from the NIHR Health Protection Research Unit in Evaluation of Interventions at University of Bristol.
Author Contributions
M.E., A.F. and A.T. conceptualised the study. A.T., M.E., A.F., A.M., R.R., B.V. and B.M.A. developed methodology. A.T. and M.E. performed and validated the experiments. A.T., M.E., A.F. and M.B. visualised data. A.T., R.R., A.M. and M.E. conducted formal analysis. M.E., A.F., M.B. and M.L. supervised. A.T. wrote the initial manuscript draft, managed the project and curated data. A.T., M.E., A.F., A.M., R.R., M.B., M.L., B.V. and B.M.A. reviewed and edited the manuscript. M.L. provided resources. M.E. and M.L. acquired funding. M.E. oversaw project administration.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A. Finn and M. C. Eisler contributed equally.
Change history
10/29/2019
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Contributor Information
A. C. Thomas, Email: amyc.thomas@bristol.ac.uk
M. C. Eisler, Email: mark.eisler@bristol.ac.uk
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-48007-5.
References
- 1.Lima SF, Teixeira AGV, Higgins CH, Lima FS, Bicalho RC. The upper respiratory tract microbiome and its potential role in bovine respiratory disease and otitis media. Sci Rep. 2016;6:12. doi: 10.1038/srep29050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brugger, S. D., Bomar, L. & Lemon, K. P. Commensal-pathogen interactions along the human nasal passages. PLoS Pathog12, 10.1371/journal.ppat.1005633 (2016). [DOI] [PMC free article] [PubMed]
- 3.Gaeta NC, et al. Deciphering upper respiratory tract microbiota complexity in healthy calves and calves that develop respiratory disease using shotgun metagenomics. J. Dairy Sci. 2017;100:1445–1458. doi: 10.3168/jds.2016-11522. [DOI] [PubMed] [Google Scholar]
- 4.Pass DA, Thomson RG. Wide distribution of Pasteurella haemolytica type-1 over nasal mucosa of cattle. an J Comp Med. 1971;35:181. [PMC free article] [PubMed] [Google Scholar]
- 5.Agency), A. A. P. H. Veterinary Investigation Diagnosis Analysis (VIDA) report. Yearly trends 2009 to 2016: Cattle, https://www.gov.uk/government/publications/veterinary-investigation-diagnosis-analysis-vida-report-2016 (2016).
- 6.Singh K, Ritchey JW, Confer AW. Mannheimia haemolytica: Bacterial-Host Interactions in Bovine Pneumonia. Vet Pathol. 2011;48:338–348. doi: 10.1177/0300985810377182. [DOI] [PubMed] [Google Scholar]
- 7.Saadati M, Gibbs HA, Parton R, Coote JG. Characterisation of the leukotoxin produced by different strains of Pasteurella haemolytica. J Med Microbiol. 1997;46:276–284. doi: 10.1099/00222615-46-4-276. [DOI] [PubMed] [Google Scholar]
- 8.Larsen, J. et al. Evolution of the leukotoxin promoter in genus Mannheimia. Bmc Evolutionary Biology9, 10.1186/1471-2148-9-121 (2009). [DOI] [PMC free article] [PubMed]
- 9.Panciera RJ, Confer AW. Pathogenesis and pathology of bovine pneumonia. Vet Clin North Am Food Anim Pract. 2010;26:191–214. doi: 10.1016/j.cvfa.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor JD, Fulton RW, Lehenbauer TW, Step DL. & Confer, A. W. The epidemiology of bovine respiratory disease: What is the evidence for predisposing factors? Can Vet J. 2010;51:1095–1102. [PMC free article] [PubMed] [Google Scholar]
- 11.EBLEX. Better management of bovine respiratory disease (BRD/Pneumonia), http://beefandlamb.ahdb.org.uk/wp/wp-content/uploads/2016/03/BRP-plus-BRD-Pneumonia-080316.pdf (2013).
- 12.Dabo SM, Taylor JD, Confer AW. Pasteurella multocida and bovine respiratory disease. Anim Health Res Rev. 2007;8:129. doi: 10.1017/S1466252307001399. [DOI] [PubMed] [Google Scholar]
- 13.Griffin D. Economic impact associated with respiratory disease in beef cattle. Vet Clin North Am Food Anim Pract. 1997;13:367–377. doi: 10.1016/S0749-0720(15)30302-9. [DOI] [PubMed] [Google Scholar]
- 14.Salt JS, Thevasagayam SJ, Wiseman A, Peters AR. Efficacy of a quadrivalent vaccine against respiratory diseases caused by BHV-1, PI3V, BVDV and BRSV in experimentally infected calves. Veterinary Journal. 2007;174:616–626. doi: 10.1016/j.tvjl.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Rice JA, Carrasco-Medina L, Hodgins DC, Shewen PE. Mannheimia haemolytica and bovine respiratory disease. Anim Health Res Rev. 2007;8:117–128. doi: 10.1017/s1466252307001375. [DOI] [PubMed] [Google Scholar]
- 16.Portis E, Lindeman C, Johansen L, Stoltman G. A ten-year (2000–2009) study of antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex Mannheimia haemolytica, Pasteurella multocida, and Histophilus somni in the United States and Canada. J Vet Diagn Invest. 2012;24:932–944. doi: 10.1177/1040638712457559. [DOI] [PubMed] [Google Scholar]
- 17.Klima CL, et al. Pathogens of bovine respiratory disease in North American feedlots conferring multidrug resistance via integrative conjugative elements. J Clin Microbiol. 2014;52:438–448. doi: 10.1128/jcm.02485-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anholt, R. M. et al. Antimicrobial susceptibility of bacteria that cause bovine respiratory disease complex in Alberta, Canada. Front Vet Sci4, 10.3389/fvets.2017.00207 (2017). [DOI] [PMC free article] [PubMed]
- 19.DeRosa DC, Mechor GD, Staats JJ, Chengappa MM, Shryock TR. Comparison of Pasteurella spp. simultaneously isolated from nasal and transtracheal swabs from cattle with clinical signs of bovine respiratory disease. J Clin Microbiol. 2000;38:327–332. doi: 10.1128/jcm.38.1.327-332.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Magwood SE, Barnum DA, Thomson RG. Nasal bacterial flora of calves in healthy and in pneumonia prone herds. an J Comp Med. 1969;33:237–243. [PMC free article] [PubMed] [Google Scholar]
- 21.Allen JW, et al. The microbial flora of the respiratory tract in feedlot calves: associations between nasopharyngeal and bronchoalveolar lavage cultures. Can J Comp Med. 1991;55:341–346. [PMC free article] [PubMed] [Google Scholar]
- 22.Catry B, et al. Detection of tetracycline-resistant and susceptible Pasteurellaceae in the nasopharynx of loose group-housed calves. Vet Res Commun. 2006;30:707–715. doi: 10.1007/s11259-006-3347-8. [DOI] [PubMed] [Google Scholar]
- 23.Noyes NR, et al. Mannheimia haemolytica in feedlot cattle: prevalence of recovery and associations with antimicrobial use, resistance, and health outcomes. J Vet Intern Med. 2015;29:705–713. doi: 10.1111/jvim.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hotchkiss EJ, et al. Prevalence of Pasteurella multocida and other respiratory pathogens in the nasal tract of Scottish calves. Vet Rec. 2010;167:555–560. doi: 10.1136/vr.c4827. [DOI] [PubMed] [Google Scholar]
- 25.Angen O, Ahrens P, Tegtmeier C. Development of a PCR test for identification of Haemophilus somnus in pure and mixed cultures. Vet Microbiol. 1998;63:39–48. doi: 10.1016/s0378-1135(98)00222-3. [DOI] [PubMed] [Google Scholar]
- 26.Holman DB, McAllister TA, Topp E, Wright A-DG, Alexander TW. The nasopharyngeal microbiota of feedlot cattle that develop bovine respiratory disease. Vet Microbiol. 2015;180:90–95. doi: 10.1016/j.vetmic.2015.07.031. [DOI] [PubMed] [Google Scholar]
- 27.Timsit E, et al. Evolution of the nasopharyngeal microbiota of beef cattle from weaning to 40 days after arrival at a feedlot. Vet Microbiol. 2016;187:75–81. doi: 10.1016/j.vetmic.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 28.Biesbroek G, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190:1283–1292. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 29.Radke BR. A demonstration of interval-censored survival analysis. Prev Vet Med. 2003;59:241–256. doi: 10.1016/s1067-5877(03)00103-x. [DOI] [PubMed] [Google Scholar]
- 30.Abdullahi O, et al. Rates of acquisition and clearance of Pneumococcal serotypes in the nasopharynges of children in Kilifi district, Kenya. J Infect Dis. 2012;206:1020–1029. doi: 10.1093/infdis/jis447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thors V, et al. Population density profiles of nasopharyngeal carriage of 5 bacterial species in pre-school children measured using quantitative PCR offer potential insights into the dynamics of transmission. Hum Vaccin Immunother. 2016;12:375–382. doi: 10.1080/21645515.2015.1090069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guenther S, et al. Real-time PCR assay for the detection of species of the genus Mannheimia. J Microbiol Methods. 2008;75:75–80. doi: 10.1016/j.mimet.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 33.Angen O, et al. Respiratory disease in calves: Microbiological investigations on trans-tracheally aspirated bronchoalveolar fluid and acute phase protein response. Vet Microbiol. 2009;137:165–171. doi: 10.1016/j.vetmic.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Autio T, et al. Etiology of respiratory disease in non-vaccinated, non-medicated calves in rearing herds. Vet Microbiol. 2007;119:256–265. doi: 10.1016/j.vetmic.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frank, G. H., Briggs, R. E. & Debey, B. M. In Workshop on Pasteurellosis in ProductionAnimals. 83–88.
- 36.Frank GH, Briggs RE, Gillette KG. Colonization of the nasal passages of calves with Pasteurella haemolytica serotype 1 and regeneration of colonization after experimentally induced viral infection of the respiratory tract. Am J Vet Res. 1986;47:1704–1707. [PubMed] [Google Scholar]
- 37.Hotchkiss EJ, Hodgson JC, Schmitt-van de Leemput E, Dagleish MP, Zadoks RN. Molecular epidemiology of Pasteurella multocida in dairy and beef calves. Vet Microbiol. 2011;151:329–335. doi: 10.1016/j.vetmic.2011.03.018. [DOI] [PubMed] [Google Scholar]
- 38.Campbell, F., Strang, C., Ferguson, N., Cori, A. & Jombart, T. When are pathogen genome sequences informative of transmission events? PLoS Pathog14, 10.1371/journal.ppat.1006885 (2018). [DOI] [PMC free article] [PubMed]
- 39.Bavananthasivam J, et al. Proximity-dependent inhibition of growth of Mannheimia haemolytica by Pasteurella multocida. Appl Environ Microbiol. 2012;78:6683–6688. doi: 10.1128/aem.01119-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dassanayake RP, et al. Bibersteinia trehalosi inhibits the growth of Mannheimia haemolytica by a proximity-dependent mechanism. Appl Environ Microbiol. 2010;76:1008–1013. doi: 10.1128/aem.02086-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bogaert D, et al. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet. 2004;363:1871–1872. doi: 10.1016/s0140-6736(04)16357-5. [DOI] [PubMed] [Google Scholar]
- 42.Katsuda K, et al. Serotyping of Mannheimia haemolytica isolates from bovine pneumonia: 1987-2006. Veterinary Journal. 2008;178:146–148. doi: 10.1016/j.tvjl.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 43.Klima CL, Alexander TW, Hendrick S, McAllister TA. Characterization of Mannheimia haemolytica isolated from feedlot cattle that were healthy or treated for bovine respiratory disease. Can J Comp Med. 2014;78:38–45. [PMC free article] [PubMed] [Google Scholar]
- 44.Wang LM, McMahan CS, Hudgens MG, Qureshi ZP. A flexible, computationally efficient method for fitting the proportional hazards model to interval-censored data. Biometrics. 2016;72:222–231. doi: 10.1111/biom.12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kenkre, V. M. & Kuperman, M. N. Applicability of the Fisher equation to bacterial population dynamics. Phys Rev E67, 10.1103/PhysRevE.67.051921 (2003). [DOI] [PubMed]
- 46.Sanderson MW, Dargatz DA, Wagner BA. Risk factors for initial respiratory disease in United States’ feedlots based on producer-collected daily morbidity counts. Can J Vet Res. 2008;49:373–378. [PMC free article] [PubMed] [Google Scholar]
- 47.Mina, M. J., McCullers, J. A. & Klugman, K. P. Live attenuated influenza vaccine enhances colonization of Streptococcus pneumoniae and Staphylococcus aureus in mice. MBio5, 10.1128/mBio.01040-13 (2014). [DOI] [PMC free article] [PubMed]
- 48.Thors V, et al. The effects of live attenuated influenza vaccine on nasopharyngeal bacteria in healthy 2 to 4 year olds a randomized controlled trial. Am J Respir Crit Care Med. 2016;193:1401–1409. doi: 10.1164/rccm.201510-2000OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McAuliffe GA, Takahashi T, Orr RJ, Harris P, Lee MRF. Distributions of emissions intensity for individual beef cattle reared on pasture-based production systems. J Clean Prod. 2018;171:1672–1680. doi: 10.1016/j.jclepro.2017.10.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McGuirk SM. Disease management of dairy calves and heifers. Vet Clin North Am Food Anim Pract. 2008;24:139–153. doi: 10.1016/j.cvfa.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chochua, S. et al. Increased nasopharyngeal density and concurrent carriage of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis are associated with pneumonia in febrile children. PLoS One11, 10.1371/journal.pone.0167725 (2016). [DOI] [PMC free article] [PubMed]
- 52.Rodrigues F, Foster D, Nicoli E. Relationships between rhinitis symptoms, respiratory viral infections and nasopharyngeal colonization with Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in children attending daycare. J Pediatric Infect Dis Soc. 2013;32:227–232. doi: 10.1097/INF.0b013e318298e62e. [DOI] [PubMed] [Google Scholar]
- 53.Tegtmeier C, Angen O, Ahrens P. Comparison of bacterial cultivation, PCR, in situ hybridization and immunohistochemistry as tools for diagnosis of Haemophilus somnus pneumonia in cattle. Vet Microbiol. 2000;76:385–394. doi: 10.1016/s0378-1135(00)00259-5. [DOI] [PubMed] [Google Scholar]
- 54.Angen O, Ahrens P, Bisgaard M. Phenotypic and genotypic characterization of Mannheimia (Pasteurella) haemolytica-like strains isolated from diseased animals in Denmark. Vet Microbiol. 2002;84:103–114. doi: 10.1016/s0378-1135(01)00439-4. [DOI] [PubMed] [Google Scholar]
- 55.Quinn, P. J. et al. Veterinary Microbiology and Microbial Disease Second edn, 301–313 (Blackwell Publishing Ltd., 2011).
- 56.Horwood, P. F. & Mahony, T. J. Rapid detection of bovine respiratory disease pathogens. (Meat & Livestock Australia Limited, 2007).
- 57.Sweeney, M. T., Quesnell, R., Tiwari, R., Lemay, M. & Watts, J. L. In vitro activity and rodent efficacy of clinafloxacin for bovine and swine respiratory disease. Front Microbiol4, 10.3389/fmicb.2013.00154 (2013). [DOI] [PMC free article] [PubMed]
- 58.Newcombe RG. Two-sided confidence intervals for the single proportion: Comparison of seven methods. Stat Med. 1998;17:857–872. doi: 10.1002/(SICI)1097-0258(19980430)17:8<857::AID-SIM777>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 59.Newcombe RG. Interval estimation for the difference between independent proportions: Comparison of eleven methods. Stat Med. 1998;17:873–890. doi: 10.1002/(SICI)1097-0258(19980430)17:8<873::AID-SIM779>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 60.Brown LD, et al. Interval estimation for a binomial proportion. Stat. Sci. 2001;16:101–133. [Google Scholar]
- 61.Sherman M, leCessie S. A comparison between bootstrap methods and generalized estimating equations for correlated outcomes in generalized linear models. Commun Stat Simul Comput. 1997;26:901–925. doi: 10.1080/03610919708813417. [DOI] [Google Scholar]
- 62.Tueller SJ, Van Dorn RA, Bobashev GV. Visualization of categorical longitudinal and times series data. Methods Rep RTI Press. 2016;2016:MR-0033–1602. doi: 10.3768/rtipress.2016.mr.0033.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson-Bergman C. icenReg: regression models for interval censored data in R. J Stat Softw. 2017;81:1–23. doi: 10.18637/jss.v081.i12. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information: Insights into Pasteurellaceae carriage dynamics in the nasal passages of healthy beef calves
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.