Significance
UvrD helicase plays essential roles in multiple DNA metabolic processes, including methyl-directed mismatch repair. UvrD monomers can translocate along single-stranded DNA, but self-assembly or interaction with an accessory factor is required to activate processive DNA unwinding in vitro. A MutL protein dimer can activate the monomeric UvrD helicase, but the mechanism of activation is not known. The 2B subdomain of UvrD is regulatory and is rotationally flexible. Here we show that binding of MutL to a UvrD–DNA substrate complex is accompanied by rotation of the 2B subdomain to form an active helicase. The results show the important role of the 2B subdomain in regulating helicase activity.
Keywords: helicase, single molecule fluorescence, mismatch repair, conformational selection, activation
Abstract
Escherichia coli UvrD is a superfamily 1 helicase/translocase that functions in DNA repair, replication, and recombination. Although a UvrD monomer can translocate along single-stranded DNA, self-assembly or interaction with an accessory protein is needed to activate its helicase activity in vitro. Our previous studies have shown that an Escherichia coli MutL dimer can activate the UvrD monomer helicase in vitro, but the mechanism is not known. The UvrD 2B subdomain is regulatory and can exist in extreme rotational conformational states. By using single-molecule FRET approaches, we show that the 2B subdomain of a UvrD monomer bound to DNA exists in equilibrium between open and closed states, but predominantly in an open conformation. However, upon MutL binding to a UvrD monomer–DNA complex, a rotational conformational state is favored that is intermediate between the open and closed states. Parallel kinetic studies of MutL activation of the UvrD helicase and of MutL-dependent changes in the UvrD 2B subdomain show that the transition from an open to an intermediate 2B subdomain state is on the pathway to helicase activation. We further show that MutL is unable to activate the helicase activity of a chimeric UvrD containing the 2B subdomain of the structurally similar Rep helicase. Hence, MutL activation of the monomeric UvrD helicase is regulated specifically by its 2B subdomain.
Escherichia coli UvrD is an SF1A DNA helicase/translocase involved in methyl-directed mismatch DNA repair (1), nucleotide excision repair (2), replication restart (3, 4), recombination (5, 6), and transcriptional control (7). E. coli UvrD and the structurally similar SF1A helicases, E. coli Rep and Bacillus stearothermophilus PcrA, share 2 core ATPase subdomains, 1A and 2A, and 2 less conserved auxiliary subdomains, 1B and 2B (Fig. 1A) (8). The monomeric forms of these helicases are processive single-stranded DNA translocases (9–15) but have little to no helicase activity by themselves in vitro (11, 16–21). In the absence of accessory proteins, UvrD, Rep, and PcrA must assemble to form at least a dimer to activate helicase activity (11, 16–20). Structural and solution studies show that the 2B subdomains of UvrD (22–24), Rep (25, 26), and PcrA (27–30) are flexible and can rotate 130 to 160°. The 2B subdomain of the Rep monomer is autoinhibitory, as its removal activates Rep monomer helicase activity (16, 31, 32), demonstrating that a Rep monomer possesses all that is needed for both translocase and helicase activities. These findings led to the hypothesis that the 2B subdomain is regulatory and that its rotational conformational state can modulate helicase activity (16, 21, 31). Indeed, single-molecule studies of UvrD showed that DNA unwinding activity correlates with a relatively closed conformation of the 2B subdomain (33), although it is not known whether this state resembles the “closed” state observed in crystal structures (22). Cross-linking of the 2B subdomain of Rep into a closed configuration also activates Rep monomer helicase activity (29). Finally, upon formation of a UvrD dimer the 2B subdomain of the lead UvrD subunit is shifted to a more closed state (24).
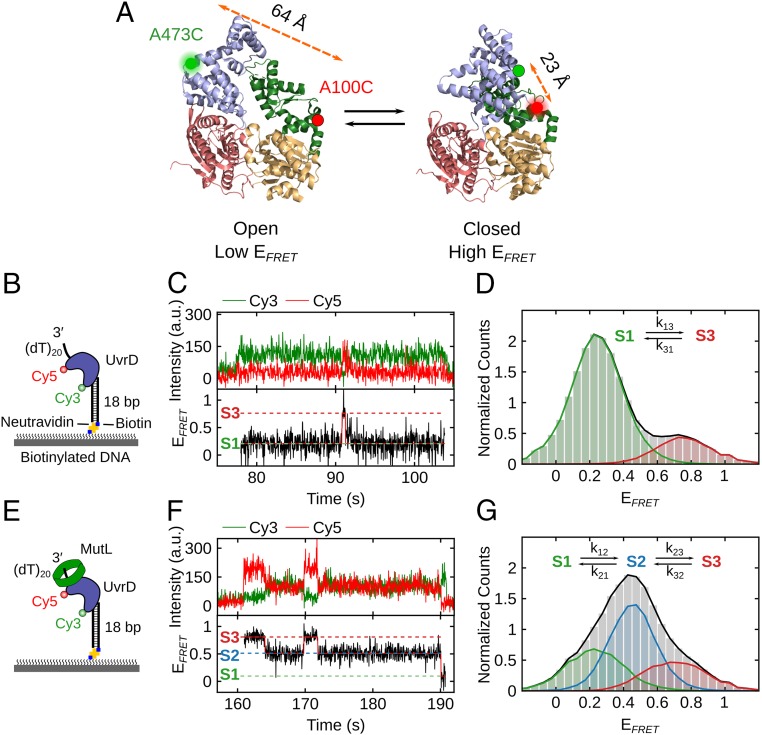
Fig. 1.
The 2B subdomain of UvrD in complex with a DNA unwinding substrate shifts to an intermediate conformation upon MutL binding. (A) The open and closed structures of UvrD are shown with subdomains 2B (blue), 1B (green), 1A (beige), and 2A (red). Rotation of the 2B subdomain results in a change in FRET of Cy3/Cy5-labeled UvrD-DM-1B/2B. The labeling positions (A100C and A473C) and the distances between them are indicated. (B) Cartoon of Cy3/Cy5-labeled UvrD binding to a 3′-(dT)20-ds18-biotin DNA tethered on a PEG surface via biotin-Neutravidin linkage. (C) Single-molecule time trajectory showing transitions of the 2B subdomain between open (S1) and closed (S3) states for Cy3/Cy5-labeled UvrD bound to DNA. (D) FRET histogram (from 346 traces) showing UvrD monomer bound to DNA transitions between primarily 2 states: S1 (open) with EFRET = 0.26 ± 0.08 (82% of population) and S3 (closed) with EFRET = 0.75 ± 0.08 (18% of population). (E) Cartoon of MutL bound to a complex of Cy3/Cy5–UvrD bound to a 3′-(dT)20-ds18-biotin DNA on the surface. (F) Single-molecule time trajectory showing transitions of the 2B subdomain for Cy3/Cy5-UvrD (250 pM) bound to the immobilized DNA in the presence of MutL (250 nM dimer). (G) FRET histogram (from 70 traces) showing UvrD monomer bound to DNA in 3 states in the presence of MutL: S1 (open), EFRET = 0.22 ± 0.11 (29% of the population); S2 (intermediate), EFRET = 0.45 ± 0.08 (49%); and S3 (closed), EFRET = 0.76 ± 0.12 (22%).
UvrD helicase activity can be activated through interactions with the MutL protein (34, 35) that is required for methyl-directed mismatch repair (36). We have shown that a single MutL dimer is sufficient to activate the UvrD monomer helicase and increase its processivity as well as stimulate the UvrD dimer helicase (37). However, the molecular basis for this activation is not known. Here, we use single-molecule and ensemble fluorescence resonance energy transfer (FRET) experiments to demonstrate that MutL binding to a UvrD–DNA complex is accompanied by formation of a rotational conformational state of the UvrD 2B subdomain that is intermediate between the open and closed states observed in crystal structures (22, 23) and this intermediate state is on the pathway for activation of the MutL–UvrD monomer helicase.
Results
The 2B Subdomain of UvrD Adopts a More Open Conformation upon Binding to a Partial Duplex DNA.
A crystal structure of a UvrD monomer complexed with a 3′-(dN)7 partial duplex DNA (18 to 28 bp) shows the 2B subdomain in a closed state and in direct contact with duplex DNA (22). However, ensemble and single-molecule FRET solution studies (23, 24) show that UvrD monomers bound to a 3′-(dT)20-duplex DNA substrate of 18 bp display a distribution of 2B subdomain rotational conformational states that center on a more open state. We investigated the distribution of 2B subdomain states for UvrD bound to a partial ss-ds DNA by using single-molecule FRET. The rotational state of the 2B subdomain was probed by using a previously characterized double-cysteine UvrD mutant, UvrD∆Cys-(A100C, A473C), referred to as UvrD-DM-1B/2B, with cysteines in the 1B (A100C) and 2B subdomains (A473C) (23, 24) (Fig. 1A). The 2 Cys residues were labeled stochastically with a mixture of Cy3 (donor) and Cy5 (acceptor) fluorophores as described previously (23, 24). As predicted from the distances between residues A100 and A473 in the crystal structures of apo UvrD (23) and UvrD in complex with partial duplex DNA (22), and as shown previously in solution (23, 24), the Cy3/Cy5-labeled UvrD-DM-1B/2B construct, hereafter referred to as Cy3/Cy5–UvrD, yields a high FRET efficiency signal, EFRET, when the 2B subdomain is in a closed state and a low EFRET signal when it is in an open state (Fig. 1A). Hence, rotations of the 2B subdomain can be monitored as a change in FRET efficiency.
To selectively observe only DNA-bound Cy3/Cy5–UvrD molecules, we immobilized an 18-bp duplex DNA with a flanking 3′-(dT)20 tail, referred to as 3′-(dT)20-ds18-biotin, to a coverslip through a biotin-Neutravidin tag at the blunt end of the duplex DNA (Fig. 1B). Cy3/Cy5–UvrD was added at low concentration (250 pM) in imaging buffer at 25 °C. Binding and the 2B conformational state of Cy3/Cy5–UvrD was monitored by exciting Cy3 donor fluorescence with a 532-nm laser and detecting Cy3 and Cy5 fluorescence emission signals using an objective-based TIRF microscope as described previously (38). Total fluorescence intensity and 1-step photobleaching/dissociation behavior indicate that Cy3/Cy5–UvrD binds to DNA as a monomer under these conditions. Fig. 1C shows an example smFRET trajectory of Cy3/Cy5–UvrD bound to DNA in which the 2B subdomain undergoes reversible transitions accompanied by anticorrelated changes in Cy3 and Cy5 fluorescence between open (S1; EFRET = 0.26 ± 0.08) and closed (S3; EFRET = 0.75 ± 0.08) states. Hidden Markov analysis was used to extract FRET states and transition rates between states as described in SI Appendix, yielding k13 = 0.159 ± 0.006 s−1 and k31 = 0.716 ± 0.025 s−1. Total EFRET distributions of 346 trajectories (Fig. 1D) and a transition density plot constructed (SI Appendix, Fig. S1A) show the existence of 2 primary states (S1 and S3), with the S1 (open) state being the most populated (82%). The observation of 2 dominant states suggests that the rates of interconversion to or from any intermediate states are faster than the rate of data acquisition of 32 ms.
Stopped-flow studies (SI Appendix, Fig. S2) show that binding of Cy3/Cy5–UvrD to an excess of the same partial duplex DNA results in an anticorrelated increase in Cy3 fluorescence and decrease in Cy5 fluorescence (decrease in FRET). This indicates that the 2B subdomain moves to a more open state upon DNA binding, in agreement with previous studies (23, 24), but not in agreement with the closed state observed in crystal structures (22).
MutL Shifts the UvrD 2B Subdomain to an Intermediate Rotational State.
Upon addition of excess MutL (250 nM dimer) and Cy3/Cy5–UvrD (250 pM) to the surface-immobilized 3′-(dT)20-ds18-biotin DNA (Fig. 1E), we observe UvrD monomers with 3 discrete EFRET states (Fig. 1 F and G); the same S1 (open; EFRET = 0.22 ± 0.11) and S3 (closed; EFRET = 0.76 ± 0.12) states that exist in the absence of MutL, but also a new intermediate state, S2, with FRET value (EFRET[S2] = 0.45 ± 0.08; Fig. 1 F and G). The FRET histogram (from 70 traces; Fig. 1G) shows that the intermediate S2 state (49%) is the most populated, followed by the open S1 state (29%) and the closed S3 state (22%). A transition density plot (SI Appendix, Fig. S1B) constructed from the time trajectories shows transitions primarily between the S1 and S2 states and the S2 and S3 states, but also less frequent transitions between the S1 and S3 states. Hidden Markov analysis yields the transition rates k12 = 0.475 ± 0.031 s−1, k21 = 0.284 ± 0.019 s−1, k23 = 0.281 ± 0.019 s−1, k32 = 0.628 ± 0.041 s−1, k13 = 0.069 ± 0.013 s−1, and k31 = 0.097 ± 0.016 s−1. The primary effect of MutL binding to the UvrD–DNA complex is to form the new intermediate S2 state (49%) at the expense of the most open S1 state decreasing from 82% to 29%. MutL has little effect on the population of the closed S3 state (18% vs. 22%).
Kinetics of Formation of the Active MutL–UvrD–DNA Complex.
We showed previously that binding of 1 MutL dimer to a UvrD monomer–DNA complex activates the UvrD monomer helicase (37). Here we performed 2 sets of stopped-flow experiments to examine the kinetics of UvrD activation by MutL and determine whether this correlates with 2B subdomain movement. In the first set of experiments, we examined the kinetics of MutL binding and formation of an active MutL–UvrD–DNA complex by monitoring the unwinding of a fluorescently labeled DNA [3′-(dT)20-ds18-BHQ2/Cy5 depicted in Fig. 2A]. In the duplex DNA, the fluorescence of Cy5 on one DNA strand is quenched by the black hole quencher, BHQ2, on the other strand; hence, the Cy5 fluorescence increases upon DNA unwinding and strand separation (37). In a second set of independent but otherwise identical experiments, we monitored the conformational changes in the monomeric UvrD 2B subdomain that accompany MutL binding by using the Cy3/Cy5–UvrD (Fig. 1A).
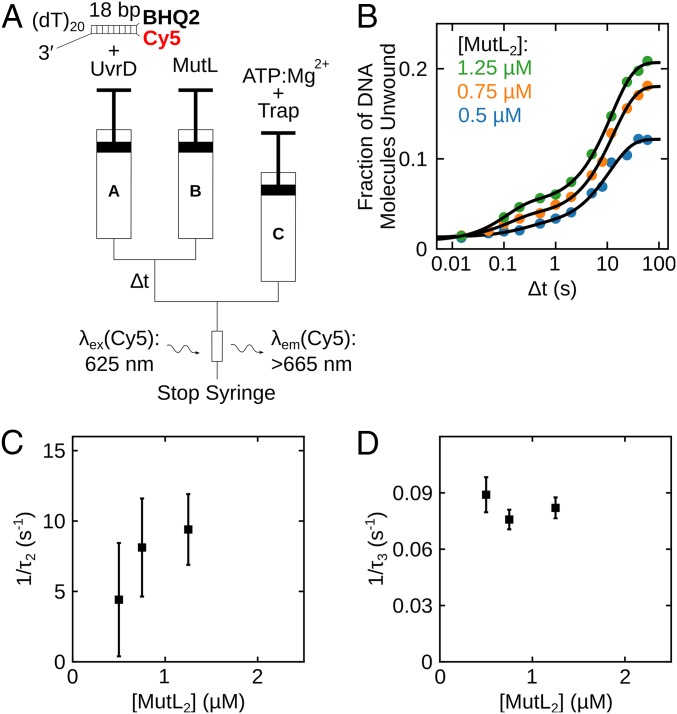
Fig. 2.
Kinetics of formation of the active MutL–UvrD–DNA helicase. (A) Schematic representation of the sequential-mixing stopped-flow fluorescence experiment. Experiments were performed in buffer T at 25 °C. (B) Each data point represents the fraction of DNA molecules unwound in a series of experiments performed with 100 nM UvrD, 250 nM 3′-(dT)20-ds18-BHQ2/Cy5 DNA substrate, and the indicated MutL concentration plotted as a function of ∆t on a log time scale. Continuous lines are simulations based on the best-fit values using SI Appendix, Eq. 2. (C and D) Reciprocal relaxation times (1/τ2 and 1/τ3) obtained from nonlinear least-squares fitting of time courses in B using SI Appendix, Eq. 2.
We first performed sequential-mixing stopped-flow experiments to monitor the kinetics of formation of an active monomeric UvrD–DNA helicase using fluorescently labeled DNA to monitor DNA unwinding upon addition of MutL (Fig. 2A) in buffer T at 25 °C. Syringe A contained UvrD (100 nM) and 3′-(dT)20-ds18-BHQ2/Cy5 (250 nM), syringe B contained excess MutL, and syringe C contained 1 mM ATP, 2 mM MgCl2, 2 µM protein trap [10-bp DNA hairpin possessing 3′-(dT)40 tail]. Under these conditions, a UvrD monomer is bound to the DNA in syringe A (19, 37). Syringes A and B were rapidly mixed in the first step and allowed to incubate for a time (∆t), after which this mixture (A + B) was rapidly mixed with syringe C to initiate DNA unwinding by any active (MutL)2–UvrD–DNA complex that assembled during the incubation period (∆t). The DNA hairpin in syringe C serves as a trap to prevent rebinding of free UvrD to the DNA substrate. As the rate of formation of the active (MutL)2–UvrD–DNA complex is much slower than the rate of DNA unwinding, the final amplitude of the Cy5 fluorescence increase observed after mixing with syringe C (SI Appendix, Fig. S3) monitors formation of active (MutL)2–UvrD helicase. From a series of experiments varying ∆t, we obtain a time course for formation of active (MutL)2–UvrD helicase (SI Appendix, Fig. S3).
Fig. 2B shows the time dependence of the fraction of DNA molecules unwound for experiments at 3 MutL concentrations (0.5, 0.75, 1.25 µM dimer; SI Appendix, Fig. S3 A–C). The time courses are biphasic, and the reciprocal relaxation times, 1/τ2 and 1/τ3, determined from a 2-exponential fit (SI Appendix, Eq. 2), are plotted in Fig. 2 C and D. The biphasic time courses suggest the presence of 2 populations of UvrD monomers bound to DNA that can both be activated by MutL. The first population shows 1/τ2 increasing with increasing MutL concentration from ∼3 s−1 to ∼9 s−1, suggesting activation by MutL binding to a UvrD–DNA complex. The second population shows 1/τ3 of ∼0.08 to 0.09 s−1 that changes little with [MutL2].
Kinetics of MutL Binding and Accompanying UvrD 2B Subdomain Conformational Changes.
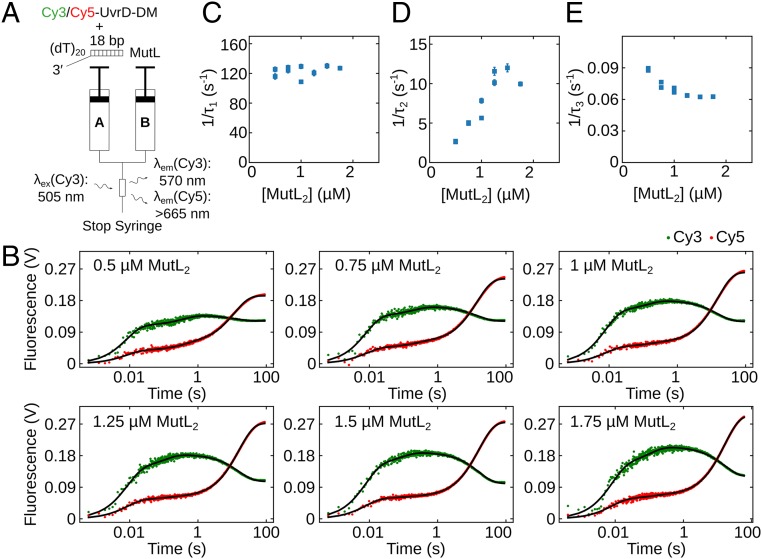
In an independent set of otherwise identical stopped-flow experiments, we also monitored the time course of conformational changes in the 2B subdomain of the monomeric UvrD–DNA complex upon MutL binding (Fig. 3A). Cy3/Cy5–UvrD (100 nM; Fig. 1A) was preequilibrated with excess of 3′-(dT)20-ds18 (250 nM) in syringe A for 5 min and then rapidly mixed with MutL at a series of concentrations (0.5, 0.75, 1, 1.25, 1.5, and 1.75 µM MutL dimer) in syringe B. The Cy3 fluorescence was excited, and both Cy3 and Cy5 fluorescence emissions were monitored. The time courses in Fig. 3B show that the changes in both Cy3 donor and Cy5 acceptor fluorescence signals occur in 3 exponential phases. The 3 reciprocal relaxation times, 1/τ1, 1/τ2, and 1/τ3, determined from a 3-exponential fit of the time courses in Fig. 3B (SI Appendix, Eq. 17), are plotted vs. the total MutL dimer concentration in Fig. 3 C–E. The observation of 3 relaxation times indicates the presence of at least 3 independent kinetic steps involving the labeled UvrD. Both 1/τ1 and 1/τ2 increase linearly with [MutL2], indicating that the first 2 phases involve MutL binding to the UvrD–DNA. However, 1/τ3 decreases with increasing [MutL2]. A decreasing reciprocal relaxation time with increasing ligand (MutL) concentration is the hallmark for a conformational selection step in the kinetic pathway (39, 40).
Fig. 3.
Kinetics of conformational changes in the UvrD 2B subdomain upon MutL binding. (A) Schematic representation of the stopped-flow experiment monitoring conformational changes in the 2B subdomain upon binding of MutL to Cy3/Cy5-UvrD-DM-1B/2B monomer–DNA complex. Experiments were performed in buffer T at 25 °C. (B) Cy3 and Cy5 fluorescence time courses from experiments performed with 100 nM Cy3/Cy5-UvrD-DM-1B/2B preequilibrated with 250 nM 3′-(dT)20-ds18 for 5 min and then rapidly mixed with MutL at the indicated concentration. Continuous lines are simulations based on the best-fit values using SI Appendix, Eq. 17. (C–E) The dependence of the reciprocal relaxation times (C, 1/τ1; D, 1/τ2; and E, 1/τ3) on the total [MutL2]. The error bars are SDs from the NLLS fitting.
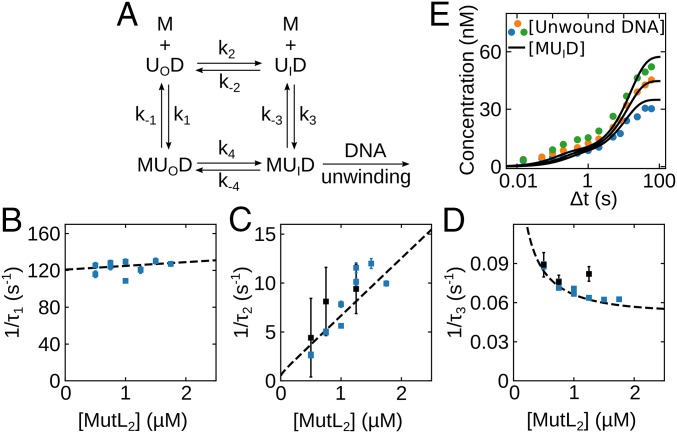
Based on the single-molecule and stopped-flow kinetics, we first considered a 6-state mechanism (SI Appendix, Fig. S4). In this scheme, UvrD–DNA complexes exist in equilibrium between open, intermediate, and closed 2B conformations (UOD ↔ UID ↔ UCD) and MutL dimer can bind to form MUOD, MUID, and MUCD, which are also in equilibrium. Although the smFRET data for the UvrD–DNA complex alone show no populated intermediate state (Fig. 1D and SI Appendix, Fig. S1), we infer that it must exist, at least transiently, along the transition pathway between the UOD (S1) and UCD (S3) states. We can simplify this 6-state scheme by eliminating the 2 closed states, UCD and MUCD. We do not consider the UCD and MUCD states to be important for activation because the population of the closed (S3) state is unaffected by MutL. It is possible that MutL does not even bind to the UCD state. Therefore, we consider the simpler mechanism in Fig. 4A that includes only the 4 states UOD, UID, MUOD, and MUID. This mechanism is also supported by the observation of only 3 relaxation times in the stopped-flow experiments. Although the 4-state mechanism has 4 kinetic steps, only 3 steps are independent, consistent with the observation of 3 relaxation times.
Fig. 4.
Kinetic mechanism for MutL binding and activation of the UvrD–DNA helicase. (A) Four states are defined by the 2B subdomain conformational state of UvrD and MutL (M) binding UOD (open 2B), and UID (intermediate 2B). (B–D) Dashed lines show the dependence of the reciprocal relaxation times on the MutL concentration simulated from the scheme in A and the rate constants in Table 1 overlaid on the experimental values. (E) Simulations of the time course for formation of the active MutL–UvrD helicase (MUID) overlaid on the experimental concentrations determined from the experiments in Fig. 2B.
We analyzed the fluorescence time courses by global nonlinear least-squares analysis (SI Appendix, Fig. S5) according to Fig. 4A using SI Appendix, Eq. 8. The resulting best-fit values of all rate constants (Table 1) and relative molar fluorescences (SI Appendix, Table S2) were well constrained. A well constrained set of rate constants could only be obtained by including the full fluorescence intensity time courses. However, because the UvrD in these stopped-flow studies is not uniformly labeled with both Cy3 and Cy5, but exists as a mixture of labeled species, one cannot assign much significance to the molar fluorescence intensities. The equilibrium constants for the steps in Fig. 4A are given in SI Appendix, Table S2. These results indicate that, before MutL binding, the UvrD monomer 2B conformation predominantly exists in an open conformation in complex with DNA, but shifts to an intermediate conformation upon MutL binding, consistent with the results of the single-molecule experiments.
Table 1.
Kinetic parameters from global NLLS analysis
| Parameter | Value |
| k1 (M−1 s−1) | (4.01 ± 0.01) ×106 |
| k−1 (s−1) | 121.3 ± 0.1 |
| k2 (s−1) | 0.0380 ± 0.0001 |
| k−2 (s−1) | 0.4076 ± 0.0005 |
| k3 (M−1 s−1) | (5.82 ± 0.01) ×106 |
| k−3 (s−1) | 0.433 ± 0.001 |
| k4 (s−1) | (9.54 ± 0.01) ×10−2 |
| k-4* (s−1) | (2.49 ± 0.01) ×10−3 |
These kinetic parameters correspond to the scheme in Fig. 4A.
During the fit, k−4 was constrained (k−4 = [k−2k−3k1k4]/[k2k3k−1]) to satisfy detailed balance.
In Fig. 4 B–D, we compare the experimental reciprocal relaxation times obtained from the fluorescence time courses (Fig. 3 C–E) and the helicase activity time courses (Fig. 2 C and D) with the relaxation times computed from the best-fit values of the rate constants in Table 1 (SI Appendix). The excellent agreement indicates that the simplified 4-state mechanism and the computed rate constants provide a good description of the data. Last, in Fig. 4E, we compare the experimental time courses for production of active MutL–UvrD helicase (Fig. 2B) with the simulated concentration time courses for MUID production (Fig. 4E, continuous lines). The good agreement supports the conclusion that the (MutL)2–UvrD–DNA complex with the 2B subdomain in an intermediate conformation between open and closed is the active form of the helicase.
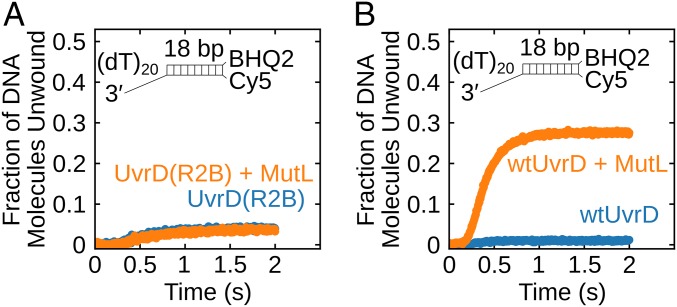
Activation of UvrD by MutL Is Specific for the UvrD 2B Subdomain.
We showed previously that MutL does not activate Rep monomer helicase activity (37), indicating that activation by MutL is specific to UvrD. Given the regulatory role of the 2B subdomain and its low-sequence conservation among UvrD-like helicases, we hypothesized that the 2B subdomain might be involved in UvrD–MutL interactions. To test this idea, we designed a chimera, UvrD(Rep2B), in which the UvrD 2B subdomain was replaced with the Rep 2B subdomain. The UvrD(Rep2B) chimera retains both ssDNA translocase activity and helicase activity under conditions of excess protein that is comparable to wtUvrD (SI Appendix, Fig. S6 A and B). The 3′ to 5′ macroscopic translocation rate is 152 ± 6 nt/s, only slightly slower than for wtUvrD monomer (191 ± 3 nt/s) (12, 14) under the same conditions. The macroscopic rate of DNA unwinding is 88 ± 7 bp/s, the same as for wtUvrD (80 ± 30 bp/s) (37) under identical conditions.
We next examined if the helicase activity of the chimeric UvrD(Rep2B) monomer can be stimulated by MutL. Single-round unwinding experiments were performed with 3′-(dT)20-ds18-BHQ2/Cy5 (50 nM) and UvrD(Rep2B) (25 nM) alone or with 250 nM MutL dimer in buffer T at 25 °C. No stimulation of monomeric UvrD(Rep2B) by MutL is observed under these conditions (Fig. 5A), in contrast to the stimulation of monomeric wtUvrD helicase by MutL under the same conditions (Fig. 5B). This suggests that MutL activation of the UvrD monomer helicase is specific for the UvrD 2B subdomain.
Fig. 5.
MutL stimulation of UvrD helicase activity is specific for the UvrD 2B subdomain. (A) Monomeric UvrD(Rep2B) shows little DNA unwinding activity and is not stimulated by MutL. DNA (50 nM) was preincubated with 25 nM UvrD(Rep2B) alone (blue) or 25 nM UvrD(Rep2B) plus 500 nM MutL dimer (orange). (B) wtUvrD monomer shows helicase activity in the presence of MutL. DNA (50 nM) was preincubated with 25 nM UvrD alone (blue) or 25 nM UvrD plus 500 nM MutL dimer (orange).
Discussion
The monomeric forms of UvrD-like helicases can rapidly and processively translocate along ssDNA with 3′ to 5′ directionality (9, 10, 12–14, 16, 21). However, significant DNA unwinding is observed in vitro only in the presence of an excess of enzyme over DNA or in the presence of accessory proteins, indicating that the monomeric form is inactive as a helicase and requires activation through self-assembly or interaction with an accessory protein (16–19, 41). Structural and functional studies show that UvrD, Rep, and PcrA possess a rotationally flexible 2B subdomain that can assume a wide range of conformational states depending on solution conditions, DNA binding, and assembly state, and it has been suggested that the rotational conformational state of this subdomain plays a regulatory role (21–25, 28).
We have shown that the helicase activity of a UvrD monomer can be activated upon binding a single MutL dimer (37). Here, we show that activation of the UvrD monomer helicase by MutL is associated with formation of an intermediate rotational conformational state of the UvrD 2B subdomain. A single-molecule experiment with FRET-labeled UvrD shows that the 2B subdomain of a UvrD monomer bound to a 3′-ssDNA duplex displays dynamic transitions between an open and closed state. However, upon binding MutL, a new intermediate state becomes populated. Stopped-flow experiments show that, under these same conditions, apo UvrD has a relatively closed state that transitions to an open state upon binding the 3′-ssDNA duplex. Upon binding of a MutL dimer, the 2B subdomain assumes an intermediate state between open and closed conformations, and this state is on the pathway to forming the active helicase. This intermediate conformation is most similar to the open state observed in the Rep-ssDNA crystal structure (23, 25) and not the fully closed state observed in a UvrD–DNA crystal structure (22).
Activation of the helicase activity of other UvrD-like enzymes has also been correlated with movement of the 2B subdomain. Rep monomer can be activated by covalent cross-linking of the 2B subdomain in a closed form (29), UvrD dimerization shifts the 2B subdomain of the lead UvrD monomer to a more closed state (24), PcrA transitions to a more closed state upon RepD binding (29), and when a pulling force is applied to the DNA UvrD monomer in a more closed state can unwind DNA (33). Whether these states are all equivalent is not clear. In fact, some of the states may actually resemble the intermediate state reported here. In this context, a molecular dynamics study has suggested that a “tilted” conformation of the 2B subdomain of UvrD that is intermediate between the open and closed states may be important for its activity. This tilted conformation may resemble the intermediate state of UvrD stabilized by MutL.
The simple mechanism in Fig. 4A provides a good description of the relaxation times and kinetic time courses. The major evidence in support of this scheme is the observation of 3 relaxation times and a decrease in 1 reciprocal relaxation time, 1/τ3, with increasing MutL concentration, which is an unequivocal indication that part of the mechanism occurs by conformational selection. Simpler 3-state mechanisms are not consistent with our data.
In Fig. 4A, the 2B subdomain of a UvrD monomer bound to DNA exists in equilibrium between 2 conformations, open and intermediate. Both the single-molecule data as well as the stopped-flow fluorescence data provide support for a preexisting equilibrium between an open form, UOD, and a closed form, UCD. Although a stable intermediate state is not observed in the absence of MutL, we infer that it must exist at some low level, at least transiently, during the transition of UvrD between the open and closed states. A MutL dimer can bind to either conformation, UOD or UID, and proceed to form the active MutL–UvrD helicase, MUID, via 2 pathways, 1 being an induced fit (IF) via step 4 and the other a conformational selection (CS) via step 3. Analysis of the complete fluorescence time courses enabled us to obtain a set of well-constrained rate constants for the scheme in Fig. 4A. The kinetics of MutL binding and subsequent effects on the 2B subdomain rotational conformation display 3 relaxation times, with the 2 slowest relaxation times being the same as the 2 relaxation times observed for formation of the active MutL–UvrD helicase. Hence, the fastest relaxation time, τ1, which is dominated by the rate constants k1 and k−1, reflecting MutL binding to the open UvrD–DNA complex, is not a dominant contributor to the formation of the active helicase mainly because k−1 is so large (∼120 s−1). The second and third relaxation times contribute to active helicase formation and reflect the steps k2, k−2, k3, and k−3, all of which involve the intermediate state, MUID. We note that, under the solution conditions used in our experiments, the MutL protein has a tendency to form higher-order assemblies beyond a dimer (42); hence, the bimolecular rate constants estimated here for MutL dimer binding to the UvrD–DNA complex, k1 = (4.01 ± 0.01) × 106 M−1 s−1 and k3 = (5.82 ± 0.01) × 106 M−1 s−1, are likely underestimates.
Although there has been much discussion in the literature as to whether 2-step binding processes occur via a CS (steps 2 and 3) or IF pathway (39, 40), the answer is generally both (43, 44), with the relative flux through each pathway depending on the concentration of the binding ligand, in this case MutL. By using the rate constants in Table 1, we calculated the time courses for formation of all UvrD species in Fig. 4A. As shown in SI Appendix, Fig. S6B (SI Appendix), before addition of 1 μM MutL dimer, the dominant species is the open UvrD bound to DNA, UOD. Although MutL can bind to both UOD and UID, most of the MUID formed goes through the CS pathway (steps 2 and 3). SI Appendix, Fig. S6C shows a plot of the fractional equilibrium flux for the CS pathway (43) as a function of MutL concentration (SI Appendix). Under the conditions and concentrations used in our experiments, >90% of the active MutL–UvrD helicase forms through the CS pathway. The IF pathway will ultimately dominate only at higher [MutL].
Although we show that an intermediate UvrD 2B subdomain state is on the pathway to formation of an active MutL–UvrD helicase, we do not know why this state is associated with activation. It has been suggested based on crystal structures of UvrD monomers bound to a 3′-(dN)7-duplex DNA that interaction of duplex DNA with the 2B subdomain in a closed state is involved in DNA unwinding (22). However, a UvrD monomer cannot unwind such DNA (11, 12, 19), suggesting that those structures do not represent an active helicase. Furthermore, our experiments indicate that the 2B subdomain of the active MutL–UvrD helicase is not in a fully closed state. In addition, in contrast to models that invoke a functional role of the 2B subdomain in DNA unwinding (22, 28), deletion of the 2B subdomain in Rep does not eliminate helicase activity, as would be predicted by those models, but rather activates Rep monomer helicase activity (16, 31, 32). Cross-linking of the 2B subdomain of Rep into a closed form also activates the Rep monomer, making it a very processive monomeric helicase (29). In that case, it may be that the closed 2B subdomain surrounds the DNA, preventing dissociation.
Materials and Methods
All buffers, proteins, and DNA used are described in the SI Appendix. Single-molecule FRET experiments were carried out by using an Olympus IX71 microscope as described (24, 37) (SI Appendix). Stopped-flow experiments were performed as described in SI Appendix using an SX.18MV stopped-flow spectrofluorometer (Applied Photophysics).
Supplementary Material
Acknowledgments
We thank Drs. R. Galletto and E. Galburt for insightful comments; members of the T.M.L. laboratory, especially L. Hao and M. Shinn, for discussions; and Thang Ho for oligodeoxynucleotides. This work was supported in part by NIH (GM045948 to T.M.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1905513116/-/DCSupplemental.
References
- 1.Iyer R. R., Pluciennik A., Burdett V., Modrich P. L., DNA mismatch repair: Functions and mechanisms. Chem. Rev. 106, 302–323 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Sancar A., DNA excision repair. Annu. Rev. Biochem. 65, 43–81 (1996). [DOI] [PubMed] [Google Scholar]
- 3.Atkinson J., McGlynn P., Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 37, 3475–3492 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heller R. C., Marians K. J., Non-replicative helicases at the replication fork. DNA Repair (Amst.) 6, 945–952 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Veaute X., et al. , UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J. 24, 180–189 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petrova V., et al. , Active displacement of RecA filaments by UvrD translocase activity. Nucleic Acids Res. 43, 4133–4149 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epshtein V., et al. , UvrD facilitates DNA repair by pulling RNA polymerase backwards. Nature 505, 372–377 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singleton M. R., Dillingham M. S., Wigley D. B., Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76, 23–50 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Dillingham M. S., Wigley D. B., Webb M. R., Demonstration of unidirectional single-stranded DNA translocation by PcrA helicase: Measurement of step size and translocation speed. Biochemistry 39, 205–212 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Dillingham M. S., Wigley D. B., Webb M. R., Direct measurement of single-stranded DNA translocation by PcrA helicase using the fluorescent base analogue 2-aminopurine. Biochemistry 41, 643–651 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Niedziela-Majka A., Chesnik M. A., Tomko E. J., Lohman T. M., Bacillus stearothermophilus PcrA monomer is a single-stranded DNA translocase but not a processive helicase in vitro. J. Biol. Chem. 282, 27076–27085 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Fischer C. J., Maluf N. K., Lohman T. M., Mechanism of ATP-dependent translocation of E.coli UvrD monomers along single-stranded DNA. J. Mol. Biol. 344, 1287–1309 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Tomko E. J., Fischer C. J., Lohman T. M., Single-stranded DNA translocation of E. coli UvrD monomer is tightly coupled to ATP hydrolysis. J. Mol. Biol. 418, 32–46 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomko E. J., Fischer C. J., Niedziela-Majka A., Lohman T. M., A nonuniform stepping mechanism for E. coli UvrD monomer translocation along single-stranded DNA. Mol. Cell 26, 335–347 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomko E. J., Lohman T. M., Modulation of Escherichia coli UvrD single-stranded DNA translocation by DNA base composition. Biophys. J. 113, 1405–1415 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brendza K. M., et al. , Autoinhibition of Escherichia coli Rep monomer helicase activity by its 2B subdomain. Proc. Natl. Acad. Sci. U.S.A. 102, 10076–10081 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng W., Hsieh J., Brendza K. M., Lohman T. M., E. coli Rep oligomers are required to initiate DNA unwinding in vitro. J. Mol. Biol. 310, 327–350 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Maluf N. K., Ali J. A., Lohman T. M., Kinetic mechanism for formation of the active, dimeric UvrD helicase-DNA complex. J. Biol. Chem. 278, 31930–31940 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Maluf N. K., Fischer C. J., Lohman T. M., A Dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J. Mol. Biol. 325, 913–935 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Lee K. S., Balci H., Jia H., Lohman T. M., Ha T., Direct imaging of single UvrD helicase dynamics on long single-stranded DNA. Nat. Commun. 4, 1878 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohman T. M., Tomko E. J., Wu C. G., Non-hexameric DNA helicases and translocases: Mechanisms and regulation. Nat. Rev. Mol. Cell Biol. 9, 391–401 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Lee J. Y., Yang W., UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell 127, 1349–1360 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jia H., et al. , Rotations of the 2B sub-domain of E. coli UvrD helicase/translocase coupled to nucleotide and DNA binding. J. Mol. Biol. 411, 633–648 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen B., Ordabayev Y., Sokoloski J. E., Weiland E., Lohman T. M., Large domain movements upon UvrD dimerization and helicase activation. Proc. Natl. Acad. Sci. U.S.A. 114, 12178–12183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korolev S., Hsieh J., Gauss G. H., Lohman T. M., Waksman G., Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell 90, 635–647 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Myong S., Rasnik I., Joo C., Lohman T. M., Ha T., Repetitive shuttling of a motor protein on DNA. Nature 437, 1321–1325 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Subramanya H. S., Bird L. E., Brannigan J. A., Wigley D. B., Crystal structure of a DExx box DNA helicase. Nature 384, 379–383 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Velankar S. S., Soultanas P., Dillingham M. S., Subramanya H. S., Wigley D. B., Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell 97, 75–84 (1999). [DOI] [PubMed] [Google Scholar]
- 29.Arslan S., Khafizov R., Thomas C. D., Chemla Y. R., Ha T., Protein structure. Engineering of a superhelicase through conformational control. Science 348, 344–347 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park J., et al. , PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell 142, 544–555 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng W., et al. , The 2B domain of the Escherichia coli Rep protein is not required for DNA helicase activity. Proc. Natl. Acad. Sci. U.S.A. 99, 16006–16011 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makurath M. A., Whitley K. D., Nguyen B., Lohman T. M., Chemla Y. R., Regulation of Rep helicase unwinding by an auto-inhibitory subdomain. Nucleic Acids Res. 47, 2523–2532 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Comstock M. J., et al. , Protein structure. Direct observation of structure-function relationship in a nucleic acid-processing enzyme. Science 348, 352–354 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamaguchi M., Dao V., Modrich P., MutS and MutL activate DNA helicase II in a mismatch-dependent manner. J. Biol. Chem. 273, 9197–9201 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Mechanic L. E., Frankel B. A., Matson S. W., Escherichia coli MutL loads DNA helicase II onto DNA. J. Biol. Chem. 275, 38337–38346 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Lahue R. S., Au K. G., Modrich P., DNA mismatch correction in a defined system. Science 245, 160–164 (1989). [DOI] [PubMed] [Google Scholar]
- 37.Ordabayev Y. A., Nguyen B., Niedziela-Majka A., Lohman T. M., Regulation of UvrD helicase activity by MutL. J. Mol. Biol. 430, 4260–4274 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen B., et al. , Diffusion of human replication protein A along single-stranded DNA. J. Mol. Biol. 426, 3246–3261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galletto R., Jezewska M. J., Bujalowski W., Kinetics of allosteric conformational transition of a macromolecule prior to ligand binding: Analysis of stopped-flow kinetic experiments. Cell Biochem. Biophys. 42, 121–144 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Vogt A. D., Di Cera E., Conformational selection or induced fit? A critical appraisal of the kinetic mechanism. Biochemistry 51, 5894–5902 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ali J. A., Maluf N. K., Lohman T. M., An oligomeric form of E. coli UvrD is required for optimal helicase activity. J. Mol. Biol. 293, 815–834 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Niedziela-Majka A., Maluf N. K., Antony E., Lohman T. M., Self-assembly of Escherichia coli MutL and its complexes with DNA. Biochemistry 50, 7868–7880 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hammes G. G., Chang Y. C., Oas T. G., Conformational selection or induced fit: A flux description of reaction mechanism. Proc. Natl. Acad. Sci. U.S.A. 106, 13737–13741 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galburt E. A., Rammohan J., A kinetic signature for parallel pathways: Conformational selection and induced fit. Links and disconnects between observed relaxation rates and fractional equilibrium flux under pseudo-first-order conditions. Biochemistry 55, 7014–7022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.