Significance
Spermatogenesis originates from spermatogonial stem cells (SSCs). Although aging testes lose SSCs, SSCs are considered immortal because they proliferate for 3 y by serial transplantation. However, analysis of aging is difficult because of their small number and lack of specific markers. Here, we used cultured SSCs as a model to study the aging process. We found that 5-y-old SSCs proliferated more actively with short telomeres. Despite maintaining SSC activity, they gradually lost sperm-forming potential. Hyperproliferation was associated with decreased Polycomb-repressive complex 2 activity, which activates the WNT7B-JNK pathway and alters SSC metabolism. These aged phenotypes were confirmed in mouse and rat aging models. Thus SSCs are virtually immortal but gradually lose sperm-forming potential, which is accompanied by abnormal proliferation, metabolism, and epigenetic alterations.
Keywords: aging, glycolysis, spermatogenesis
Abstract
Because spermatogonial stem cells (SSCs) are immortal by serial transplantation, SSC aging in intact testes is considered to be caused by a deteriorated microenvironment. Here, we report a cell-intrinsic mode of SSC aging by glycolysis activation. Using cultured SSCs, we found that aged SSCs proliferated more actively than young SSCs and showed enhanced glycolytic activity. Moreover, they remained euploid and exhibited stable androgenetic imprinting patterns with robust SSC activity despite having shortened telomeres. Aged SSCs showed increased Wnt7b expression, which was associated with decreased Polycomb complex 2 activity. Our results suggest that aberrant Wnt7b expression activated c-jun N-terminal kinase (JNK), which down-regulated mitochondria numbers by suppressing Ppargc1a. Down-regulation of Ppargc1a probably decreased reactive oxygen species and enhanced glycolysis. Analyses of the Klotho-deficient aging mouse model and 2-y-old aged rats confirmed JNK hyperactivation and increased glycolysis. Therefore, not only microenvironment but also intrinsic activation of JNK-mediated glycolysis contributes to SSC aging.
Because stem cells produce a large number of progenitors, they are often considered to be immortal. However, aged stem cells generally have impaired function, but the mechanism, particularly in terms of cell metabolism, is not well known. For example, although glycolysis promotes survival of hematopoietic stem cells (HSCs) in vitro (1), its impact on self-renewal division or aging has not been established. Metabolism in neural stem cells (NSCs) depends on glycolysis, but metabolic change during aging has not been determined (2). Unlike HSCs and NSCs, which are largely quiescent, spermatogonial stem cells (SSCs) continuously divide throughout the life span (3, 4), suggesting a distinct mode of aging.
SSC self-renewal depends on glycolysis (5). Despite the continuous self-renewal of SSCs, their number gradually declines during aging (6). However, SSC self-renewal continues long past the normal life span of the animal because SSCs proliferate for more than 3 y after being serially transplanted 9 times into immature testes (7). It was concluded that repeated transplantation into the young environment rejuvenated SSCs. Likewise, cultured SSCs, designated as germline stem (GS) cells, proliferated and produced offspring after 2 y of culture (1085-fold expansion) (8, 9).
Although these results suggest that the fate of aging SSCs depends greatly on their microenvironment, cell intrinsic factors might also contribute to SSC aging because aging of many stem cell types is often accompanied by intrinsic cell changes. However, direct analysis of deteriorating SSCs in aged testes or serially transplanted testis cells is technically challenging due to their small number and lack of specific markers. To overcome these problems, we used 5-y-old GS cell cultures and analyzed their phenotypic changes. We reasoned that long-term GS cell cultures would mimic continuously dividing SSCs in vivo and hoped to uncover features specific to aged SSCs.
Results
Lack of Senescence and Defective Spermatogenesis of Aged GS Cells.
Two GS cell lines with the Egfp transgene were cultured for 60 mo (60M-GS cells). Although we expected that telomere loss would occur at ∼34 mo after culture initiation (9), the cells proliferated after this period without apparent changes (374–379 passages) (Fig. 1A). In contrast, 60M-GS cells proliferated more actively than 5M-GS cells (Fig. 1B), and responsiveness to GDNF or FGF2, both of which are SSC self-renewal factors, was enhanced (SI Appendix, Fig. S1A). Neither line was positive for the senescence-associated (SA) β-galactosidase stain (SI Appendix, Fig. S1B).
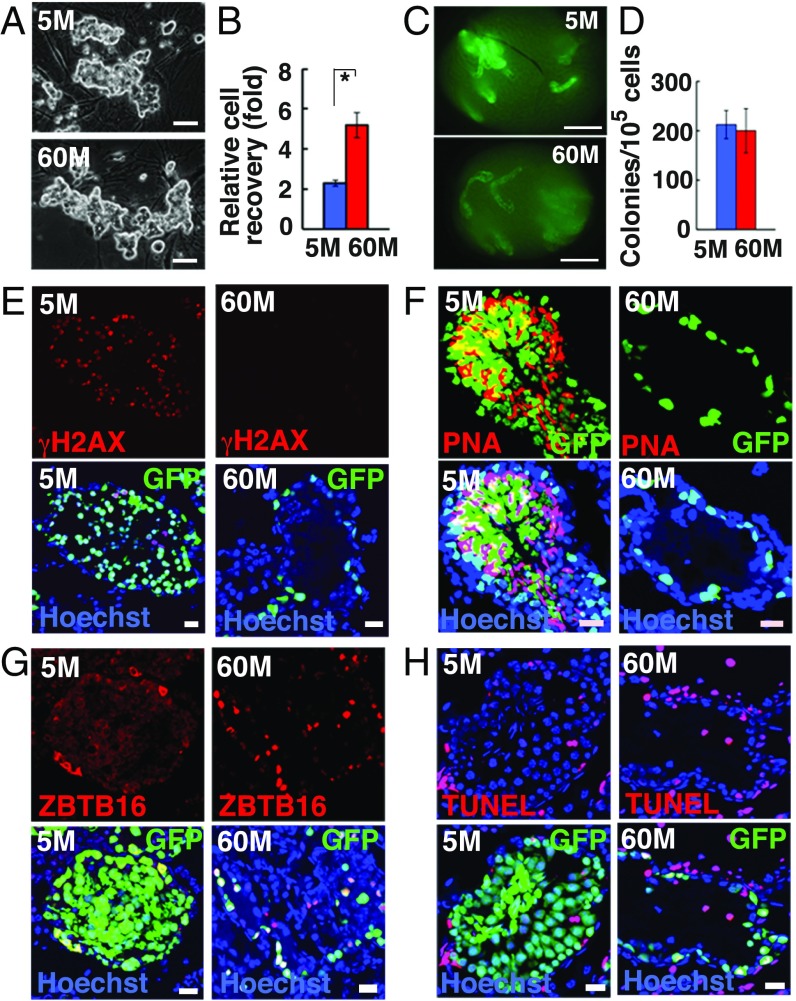
Fig. 1.
Long-term culture of GS cells. (A) Appearance of 5M-GS and 60M-GS cells. (B) Cell recovery after 5 d (n = 3). (C) Appearances of W recipient testes. (D) Colony counts (n = 8–12). (E–G) Immuno- and lectin staining of recipient testes with anti-γH2AX (E), PNA (F), or ZBTB16 (G) antibodies. (H) TUNEL staining of recipient testes 3 mo after transplantation. (Scale bars: A, 50 μm; C, 1 mm; E–H, 20 μm.) Asterisk indicates statistical significance.
Flow cytometric analyses showed increased expression of ITGA6, but the remainder of the SSC markers did not change significantly (SI Appendix, Fig. S1C). RNA sequencing (RNA-seq) analyses showed that 60M-GS cells up-regulated several differentiation-associated genes, including Kit, while self-renewal–associated genes, such as Bcl6b, were more strongly expressed in 5M-GS cells (SI Appendix, Fig. S1D and Dataset S1). To test whether 60M-GS cells possessed SSC activity, cells were transplanted into infertile WBB6F1-W/Wv (W) mice (10). Analyses of the recipient testes revealed weaker fluorescence of 60M-GS cell transplants (Fig. 1C), and only a single layer of cells were found on the basement membrane (BM) (SI Appendix, Fig. S1E). Although SSC activity remained constant (Fig. 1D and SI Appendix, Table S1), no sperm developed when 30M-GS cells were transplanted (10105 to 10106-fold expansion) (SI Appendix, Fig. S1 E and F). Interestingly, when we examined the recipient testes at 2 and 10 mo after transplantation, proliferation of CDH1+ spermatogonia from 30M-GS cells was significantly enhanced compared with that from 20M-GS cells (SI Appendix, Fig. S2). Because they were exposed to the same testis environment, this result also suggested that GS cells proliferate more actively in an autonomous manner.
Reverse transcription PCR (RT-PCR) analysis and immunostaining confirmed the lack of meiotic or haploid cells in testes with 60M-GS cells (Fig. 1 E and F and SI Appendix, Fig. S1 G–I). However, there were more ZBTB16+ spermatogonia in colonies of 60M-GS cell recipients (Fig. 1G and SI Appendix, Fig. S1J), and apoptosis was significantly enhanced (Fig. 1H and SI Appendix, Fig. S1K). Thus, GS cells maintained SSC activity, but could not produce sperm after 30 mo.
Telomere Shortening in GS Cells.
Karyotype analysis revealed that ∼64 to 68% of 5M-GS and 60M-GS cells contained 40 chromosomes (Fig. 2A). However, one of the lines showed a partial deletion of chromosome 17 (Fig. 2B). Because both lines had spermatogenic defects, it was unlikely that a partial deletion of chromosome 17 was responsible for the distinct phenotype of the 60M-GS cells. Next, WGBS was carried out (SI Appendix, Fig. S3A). The imprinting control regions showed no statistical differences in their DNA methylation levels (SI Appendix, Fig. S3B). The only exception was Snrpn, a maternally methylated gene, the methylation levels of which were significantly increased in the 60M-GS cells. However, real-time PCR showed no significant changes (SI Appendix, Fig. S3C). These results suggest that 60M-GS cells maintained androgenetic imprinting patterns.
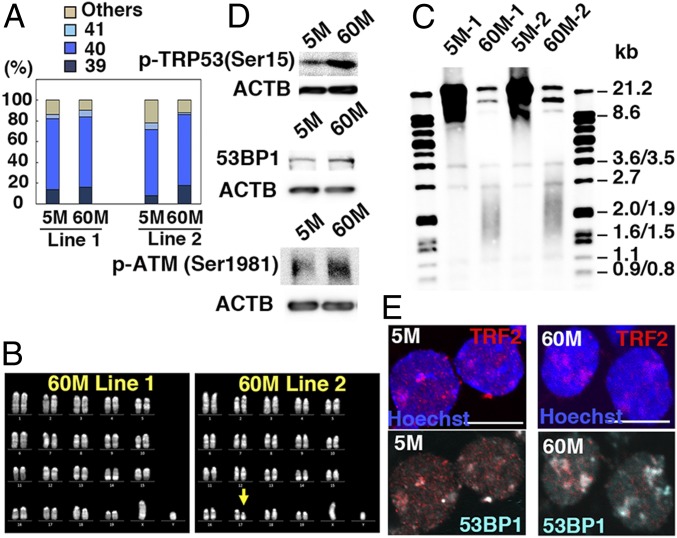
Fig. 2.
Telomere shortening in GS cells. (A) Karyotypic analyses (n = 50). (B) Quinacrine mustard and Hoechst 33258 staining of 60M-GS cells. (C) Telomere length assay. (D) Western blot analyses of p-TRP53(Ser15) (Top), 53BP1 (Middle), and p-ATM (Bottom). (E) Immunostaining of GS cells showing colocalization of 53BP1 and TRF2. (Scale bar: E, 10 μm.)
However, a comparison in 500-kb windows revealed that genome-wide methylated CpG residue (mCpG) levels slightly increased in 60M-GS cells (74.5 vs. 75.7%). Because these values were somewhat higher than that of fresh KIT− undifferentiated spermatogonia (73.9%) (11), DNA methylation appeared to increase during culture. When we compared the methylation levels in these cells strictly, 13,556 differentially methylated regions (DMRs) showed increased DNA methylation in 60M-GS cells, but 5M-GS cells contained only 858 DMRs that showed increased DNA methylation (Dataset S2). The increase in DNA methylation occurred in all chromosomes, but DMRs (5M > 60M) were predominantly found in introns, while DMRs (60M > 5M) were more frequently located in intergenic regions (SI Appendix, Fig. S3D). This difference depended on the GC content. While mCpG levels were similar (∼70–75%) in GC-rich regions (GC content > 44%), increased methylation levels were found in GC-poor regions (GC content < 39%), which often corresponded to the constitutive nuclear lamina-associated domains (cLADs) (SI Appendix, Fig. S3E). cLADs help to organize chromosomes inside the nucleus, are associated with gene repression (12), and contain fewer genes compared with GC-rich regions. Because similar methylation patterns around cLADs have been found in fresh spermatogonia (11), 60M-GS cells appear to maintain basic nuclear architecture.
We examined telomere length because late generation (∼G6) telomerase knockout (KO) mice have defective spermatogenesis (13). Telomeres were significantly shorter in 60M-GS cells (Fig. 2C). Because their lengths did not change between 54 and 60 mo, 60M-GS cells were thawed to confirm that the telomere length did not change even at 72 mo (SI Appendix, Fig. S4A), suggesting that GS cells proliferate for 18 mo even with very short telomeres. To examine telomerase expression level, we carried out real-time PCR analysis and found that 60M-GS cells express significantly lower levels of Tert and Terc, both of which are important components of telomerase (SI Appendix, Fig. S4B).
Because chromosomal abnormalities cause DNA damage, we performed Western blotting analyses and found that markers of DNA damage (53BP1, phosphorylated TRP53, and ATM) are more strongly expressed in 60M-GS cells (Fig. 2D). This was likely due to telomere damage because TRF2 (a telomere marker) and 53BP1 signals often colocalized (Fig. 2E). Therefore, 60M-GS cells proliferated despite DNA damages.
Increased c-Jun N-Terminal Kinase (JNK) Activity Promotes GS Cell Proliferation.
To understand the mechanism of enhanced proliferation of 60M-GS cells, we performed Western blotting and examined the activation levels of major signaling pathways and cell cycle regulators. The expression levels of p-JNK1-3 (MAPK8-10), p-MAPK14, and nonphosphorylated CTNNB1, an activated form of CTNNB1, were enhanced in 60M-GS cells (Fig. 3A). Regarding cell cycle-related molecules, 60M-GS cells up-regulated CDKN2A (p19ARF) (Fig. 3A), although its high expression did not interfere with spermatogonia proliferation (14).
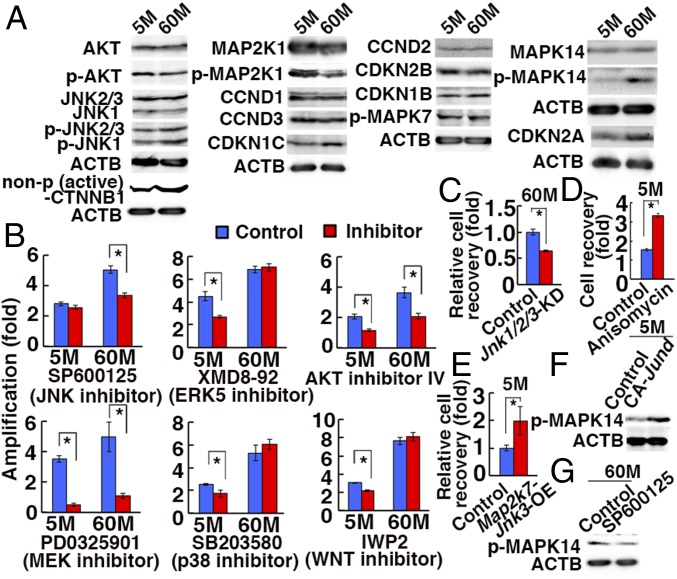
Fig. 3.
Enhanced JNK signaling in 60M-GS cells. (A) Western blot analyses. (B) Cell recovery after treatment with indicated inhibitors for 4 d (n = 3). (C) Reduction of 60M-GS cell recovery 4 d after cotransduction with shRNA against Jnk1, Jnk2, and Jnk3 (n = 3). (D) Enhanced proliferation of 5M-GS cells 2 d after anisomycin treatment (n = 3). (E) Enhanced proliferation of 5M-GS cells 4 d after Map2k7-Jnk3 OE (n = 3). (F) Western blot analyses of 5M-GS cells 15 h after transfection of CA Jund. (G) Western blot analyses of 60M-GS cells 6 h after SP6000125 treatment. Asterisks indicate statistical difference.
Chemical inhibitor studies revealed that SP600125 abrogated the proliferation of 60M-GS cells without influencing 5M-GS cells (Fig. 3B). By contrast, SB203580 and XMD8-92 inhibited the growth of 5M-GS cells, but not 60M-GS cells, suggesting that 5M-GS cells depended more on the p38 mitogen-activated protein kinase (MAPK) and MAPK7 (ERK5) pathways. Because these results suggest the involvement of the JNK pathway in enhanced proliferation of 60M-GS cells, we confirmed this by triple depletion of Jnk1/2/3 by short hairpin RNA (shRNA) in 60M-GS cells, which significantly decreased cell recovery (Fig. 3C and SI Appendix, Fig. S5A). Moreover, anisomycin, an activator of the JNK and p38 MAPK pathways, enhanced proliferation of 5M-GS cells (Fig. 3D). We also transfected Map2k7-Jnk3 into 5M-GS cells to activate the JNK pathway. Recovery of cells after transfection was significantly enhanced after Map2k7-Jnk3 overexpression (OE) (Fig. 3E). When we specifically expressed constitutively active (CA) Jund, which is the most strongly expressed Jun family gene in GS cells, the cells showed increased MAPK14 phosphorylation (Fig. 3F). Because MAPK14 phosphorylation was inhibited by SP600125 (Fig. 3G), JNK activation was necessary and sufficed for MAPK14 phosphorylation. However, MAPK14 activation per se did not appear to be responsible for hyperproliferation because SB203580 inhibited the proliferation of 5M-GS, but not 60M-GS cells (Fig. 3B). These results raised a possibility that JNK activation is responsible for enhanced proliferation of 60M-GS cells.
Decreased Polycomb Repressive Complex 2 (PRC2) Activity and Increased WNT7B in 60M-GS Cells.
Because the culture environment remained the same during the entire period, the enhancement of JNK signaling was likely caused by an autonomous cell mechanism. Although RNA-seq data did not show apparent genes involved in JNK signaling, Chromatin Immunoprecipitation Sequencing (ChIP-seq) using H3K4me3 and H3K27me3 antibodies showed that H3K27me3 peaks were significantly decreased in the promoter regions of Wnt7b and Wnt9b (Fig. 4A). While Wnt9b expression was not detectable in either type of GS cells by RT-PCR, real-time PCR analyses showed that Wnt7b was significantly up-regulated in 60M-GS cells (Fig. 4B). The enrichment of H3K27me3 in the promoter region of Wnt7b was confirmed by ChIP analysis (Fig. 4C). Although we noted similar H3K27me3 enrichment in the Tert and Terc promoters, H3K4me3 enrichment was also found in the Tert promoter, which suggested that telomerase is regulated in a different manner from Wnt7b (SI Appendix, Fig. S4C).
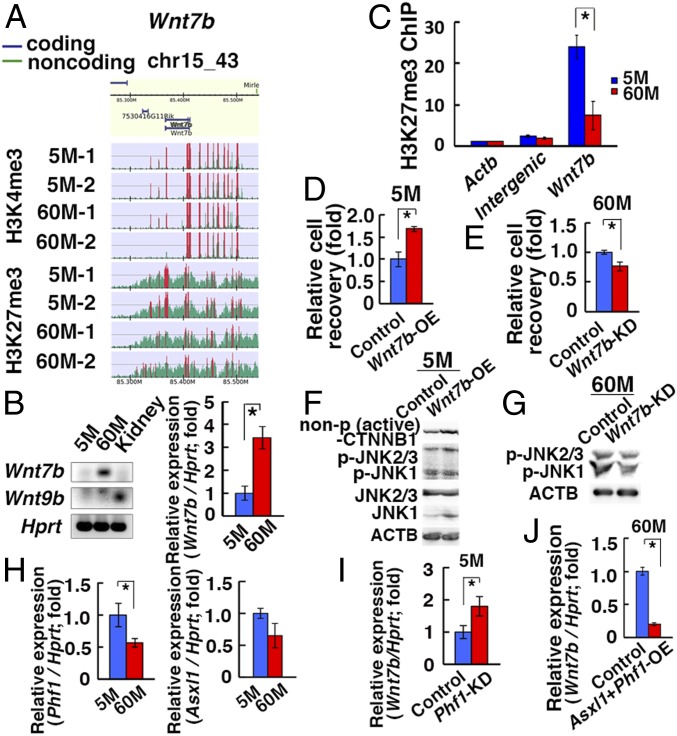
Fig. 4.
Induction of Wnt7b after long-term culture. (A) ChIP-Seq analyses of the Wnt7b promoter region. The green histogram indicates the ChIP-read line, and the region above the threshold is highlighted in red. The red regions in ChIP-read line indicate peak regions. (B) RT- and real-time PCR analyses of Wnt7b and Wnt9b (n = 5–9). (C) ChIP analyses of the Wnt7b promoter region. (D) Enhanced cell recovery of 5M-GS cells 7 d after Wnt7b OE (n = 3). (E) Reduced cell recovery of 60M-GS cells 7 d after Wnt7b KD (n = 3). (F) Western blot analyses of 5M-GS cells 7 d after Wnt7b OE. (G) Western blot analyses of 60M-GS cells 4 d after Wnt7b KD. (H) Real-time PCR analyses of Phf1 and Asxl1 (n = 8, for Phf1; n = 4, for Asxl1). (I) Real-time PCR analyses of Wnt7b 3 d after Phf1 KD in 5M-GS cells (n = 8). (J) Real-time PCR analyses of Wnt7b 3 d after cotransfection of Phf1 and Asxl1 in 60M-GS cells (n = 8). Asterisks indicate statistical difference.
Hence, we investigated the involvement of Wnt7b in GS cell proliferation by Wnt7b OE. Cell recovery was significantly enhanced by Wnt7b OE (Fig. 4D). By contrast, Wnt7b knockdown (KD) reduced 60M-GS cell proliferation (Fig. 4E and SI Appendix, Fig. S5B). Because Wnt7b activates both canonical and noncanonical WNT signaling, we transfected Wnt7b in 5M-GS cells and found that Wnt7b OE induced JNK phosphorylation (Fig. 4F and SI Appendix, Fig. S6A). Likewise, Wnt7b KD in 60M-GS cells reduced JNK phosphorylation (Fig. 4G and SI Appendix, Fig. S6B). Therefore, Wnt7b expression level was closely associated with JNK phosphorylation in GS cells.
Genome-wide analyses of ChIP sequence data revealed ∼10-fold more H3K4me3 peaks than H3K27me3 peaks in both 5M-GS and 60M-GS cells. The number of H3K27me3 peaks was slightly decreased in 60M-GS cells (SI Appendix, Fig. S7A). Although previous studies using fresh spermatogonia showed bivalent loci with underlying DNA hypomethylation (15, 16), genomic regions with decreased DNA methylation levels in 5M-GS and 60M-GS cells had high H3K4me3 levels regardless of H3K27me3 levels (SI Appendix, Fig. S7B).
Interestingly, the widths and heights of the H3K4me3 and H3K27me3 peaks changed in an opposite manner: H3K4me3 peaks became wider and higher in 60M-GS cells, and the ratio of height to width decreased (average 95.4%). By contrast, H3K27me3 peaks became narrower and lower in 60M-GS cells, and the ratio of height to width increased (average 106.2%) (SI Appendix, Fig. S7C). Consequently, while there were more genes associated with H3K4me3 in 5M-GS cells, 60M-GS cells had relatively fewer genes associated with H3K27me3. This change in histone modification pattern resulted in a smaller number of genes with bivalent modification in 60M-GS cells than in 5M-GS cells (1,216 vs. 958) (Dataset S3). Gene Ontology (GO) analyses indicated that genes enriched in H3K27me3 only and bivalent promoters were commonly involved in positive regulation of the metabolic process, animal organ development, and the cellular metabolic process (SI Appendix, Fig. S7D). We hypothesized that the globally decreased activity of the PRC2 complex was responsible for the aging of SSCs.
The PRC2 complex consists of several components, including EZH1/2 and SUZ12 (17). We examined the expression of PRC2 components using real-time PCR and found that Phf1 was significantly decreased in 60M-GS cells (Fig. 4H). Therefore, we focused on Phf1. PHF1 modulates the enzymatic activity of PRC2 or recruits PRC2 to specific genomic loci (17). To test the impact of Phf1, we depleted Phf1 in 5M-GS cells and noted a significant increase in Wnt7b expression (Fig. 4I and SI Appendix, Fig. S5C). However, Phf1 OE alone in 60M-GS cells did not change Wnt7b expression significantly. Because Asxl1, another PRC2 component, was down-regulated in 60M-GS cells (Fig. 4H), we cotransfected Phf1 and Asxl1, which resulted in significant reduction of Wnt7b expression (Fig. 4J). Therefore, a decrease in PRC2 activity may partly contribute to the increased Wnt7b expression in 60M-GS cells.
Reduced Reactive Oxygen Species (ROS) and Mitochondria Dysfunction in 60M-GS Cells.
Because ROS accumulate in many aged cells (18), ROS levels were measured in GS cells. Contrary to our expectations, ROS levels were significantly lower in 60M-GS cells than in 5M-GS cells (n = 3) (SI Appendix, Fig. S8A), as were H2O2 levels (n = 3) (SI Appendix, Fig. S8B). To study this mechanism, several ROS inhibitors were used. We found that mitochondrial respiratory chain inhibitors, such as antimycin C and rotenone, and the NAD(P)H oxidase (NOX) inhibitors diphenyleneiodonium (DPI) and apocynin, significantly suppressed proliferation of both cell types (SI Appendix, Fig. S8C).
Although Nox1 deficiency impairs SSC self-renewal (19), Nox1 significantly increased in 60M-GS cells (SI Appendix, Fig. S8D). Next we analyzed the status of the mitochondria, which are another major source of ROS. As expected, Mitotracker staining showed a significant reduction of mitochondria in 60M-GS cells (SI Appendix, Fig. S8 E and F). This decrease was also suggested by decreased mitochondrial DNA (0.6-fold) (SI Appendix, Fig. S8G). Therefore, the reduction in mitochondrial activity is likely responsible for the decreased ROS formation in 60M-GS cells.
60M-GS Cells Have Reduced Mitochondrial Function.
Because 60M-GS cells showed significant loss of mitochondria and GO analyses suggested metabolic changes, we reasoned that 60M-GS cells had altered metabolic activity. As expected, maximal respiration and spare capacity were significantly reduced in 60M-GS cells (SI Appendix, Fig. S9A). An increased glycolytic pathway appeared to compensate for this defect because 2-deoxy-d-glucose (2DG), a glucose analog that inhibits glycolysis, suppressed 60M-GS cell proliferation more significantly, suggesting that 60M-GS cells depended more heavily on glycolysis than 5M-GS cells (SI Appendix, Fig. S9B). Indeed, glycolysis and glycolytic capacity were significantly increased in 60M-GS cells (SI Appendix, Fig. S9C). As expected, Map2k7-Jnk3 OE increased the extracellular acidification rate (ECAR) in 5M-GS cells (SI Appendix, Fig. S9D).
To examine the mechanism underlying mitochondrial defects, we searched for the genes responsible for this phenomenon by real-time PCR. Through screening of genes involved in mitochondria-related genes, we found that Ppargc1a was significantly down-regulated in 60M-GS cells (SI Appendix, Fig. S9E). Western blot analyses confirmed this result (SI Appendix, Fig. S9F). Because Ppargc1a positively regulates the number and activity of mitochondria (20), decreased Ppargc1a expression is likely responsible for mitochondrial deficiency. Indeed, while Ppargc1a KD significantly reduced mitochondria in 5M-GS cells (SI Appendix, Figs. S5D and S9G), Ppargc1a OE increased their number in 60M-GS cells (SI Appendix, Fig. S9H). Ppargc1a expression levels were also closely related with ROS levels; Ppargc1a KD decreased ROS (SI Appendix, Fig. S9I), while Ppargc1a OE increased ROS (SI Appendix, Fig. S9J).
To examine whether enhanced JNK signaling was involved in ROS regulation and mitochondria activity, we transfected CA Jund in 5M-GS cells and found that the transfected cells decreased Ppargc1a expression (SI Appendix, Fig. S9K). In contrast, when SP600125 was added to inhibit JNK in 60M-GS cells, Ppargc1a significantly increased (SI Appendix, Fig. S9L). Consistent with these observations, anisomycin decreased mitochondrial staining and ROS generation in 5M-GS cells (SI Appendix, Fig. S9 M and N), suggesting that JNK was necessary and sufficient for Ppargc1a expression. These results provided evidence that down-regulation of mitochondrial activity in 60M-GS cells was caused by JNK activation.
JNK Hyperactivation and Enhanced Glycolysis in Spermatogonia of Rodent Aging Models.
To confirm the in vitro findings, we analyzed rodent aging models. Klotho KO mice show many features of aged mice, including infertility (21). Klotho KO testis was significantly smaller by 7 wk after birth (SI Appendix, Fig. S10A). These testes contained significantly fewer tubules with PNA+ haploid cells (SI Appendix, Fig. S10 B and C). However, immunostaining indicated increased proliferation of ZBTB16+ cells (SI Appendix, Fig. S10D). Although the number of mitotic KIT+ cells was comparable (SI Appendix, Fig. S10E), 53BP1 expression was significantly elevated in Klotho KO mice (SI Appendix, Fig. S10F), suggesting more DNA damage. Germ cell apoptosis was elevated in spermatocytes without apparent changes in Sertoli cells (SI Appendix, Fig. S10 G–K).
We also analyzed Brown Norway (BN) rats, the best representative rodent model of human testicular aging (22). In this model, the frequency of atrophied testes increases with age; 5 of 10 animals had bilateral atrophied testes when testes were collected from 24-mo-old animals (SI Appendix, Fig. S11A). The atrophied testes showed significantly lower weights than those from young (8 wk) or aged animals (SI Appendix, Fig. S11B). Histological analysis showed very few germ cells (SI Appendix, Fig. S11C). However, they still contained a small number of mitotic ZBTB16+ spermatogonia that proliferate more actively than those in young testes (SI Appendix, Fig. S11D).
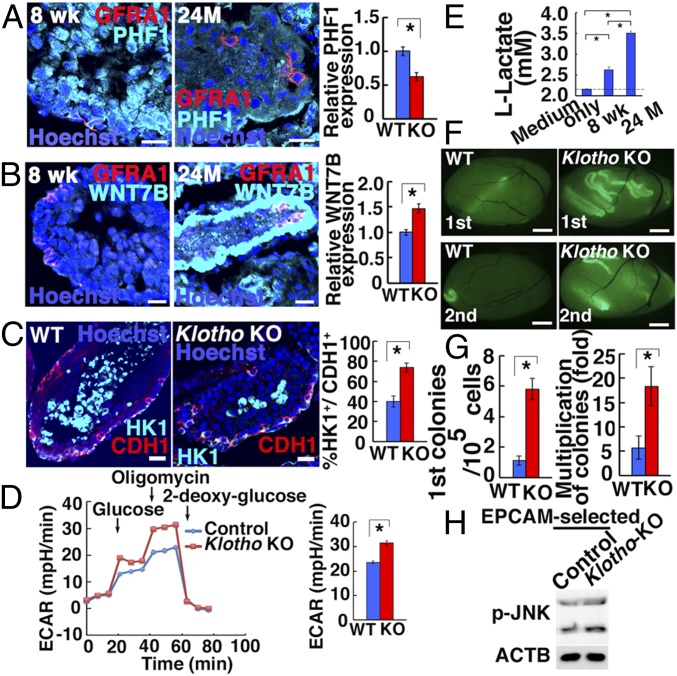
Because of the 60M-GS cell phenotype, the expression of molecules involved in GS cell aging was evaluated. Although Phf1 and Wnt7b expression did not show statistical significance in Klotho KO mice, we noted decreased PHF1 expression and increased WNT7B expression in BN rats (Fig. 5 A and B). Consistent with the increased glycolysis in aged GS cells, HK1 expression increased in undifferentiated spermatogonia of both models (Fig. 5C and SI Appendix, Fig. S11D). Because HK1 mediates an irreversible step of glycolysis, this result suggested that aged spermatogonia depend more heavily on glycolysis. Indeed, spermatogonia from Klotho KO testes showed enhanced glycolytic capacity (Fig. 5D). Moreover, lactate production was increased in BN rat spermatogonia from atrophied testes (Fig. 5E). Because maximal respiratory capacity did not change significantly in newborn Klotho KO mice (SI Appendix, Fig. S10L), we carried out flow cytometric analyses and found that Klotho KO germ cells had significantly less mitochondrial staining (SI Appendix, Fig. S10M), suggesting that Klotho KO mitochondria at this stage can still accommodate metabolic changes before they exhibit defective spermatogenesis.
Fig. 5.
Functional analyses of SSCs in Klotho KO mice and BN rats. (A and B) Immunostaining of BN rat testes by GFRA1 and PHF1 (A: n = 22) or WNT7B (B: n = 20–21). (C) Immunostaining of Klotho KO mouse testes by HK1 and CDH1 (n = 25 to 29). (D) ECAR of Klotho KO spermatogonia (n = 21 to 22). (E) Lactate production of BN rat spermatogonia (n = 6). (F) Appearance of primary (Top) and secondary (Bottom) recipient testes. (G) Colony counts (n = 16, for first recipients; n = 12, for second recipients). (H) Western blot analyses of phosphorylated JNK in EPCAM-selected Klotho KO testis cells (n = 7). (Scale bars: A–C, 20 μm; F, 1 mm.) Asterisks indicate statistical difference.
To evaluate the SSC activity in a functional manner, we performed spermatogonial transplantation. Cell recovery from Klotho KO testes significantly decreased (SI Appendix, Fig. S10N). Analysis of recipients revealed significant enrichment of SSCs in Klotho KO testes (Fig. 5 F and G). Because it is possible that SSC enrichment was caused by a loss of differentiating germ cells, we performed serial transplantation to confirm the enhanced self-renewal activity of Klotho KO SSCs. Assuming 10% colonization efficiency (23), the multiplication of colony numbers (total regenerated colony number × 10/primary colony number) significantly increased (Fig. 5G). Consistent with increased proliferation, Klotho KO spermatogonia showed increased expression of phosphorylated JNK (Fig. 5H and SI Appendix, Fig. S6C). We also confirmed increased JNK phosphorylation in GFRA1+ spermatogonia in 2-y-old atrophied BN testes (SI Appendix, Fig. S11D). Thus, JNK hyperactivation and increased glycolysis appear to be the common features of aged rodent SSCs.
Discussion
An interesting feature of 60M-GS cells was the lack of apparent senescence. Both lines proliferated even with very short telomeres but maintained SSC activity. However, both lines gradually lost sperm-forming potential. Although we currently do not know whether lack of senescence and loss of spermatogenic potential are related to each other, at least 2 possibilities exist to explain the loss of sperm-forming potential. First, it is possible that failure to complete spermatogenesis was caused by telomere shortening because spermatogenic arrest was also found in late generation (∼G6) telomerase KO mice the spermatogenesis of which was interrupted at the onset of meiosis (13). DNA damages found around the telomeres in 60M-GS cells may similarly inhibit meiosis. However, indirect influences on spermatogenesis in the KO mice cannot be excluded completely, and transplantation of telomerase KO mice into normal recipients is necessary to define the impact of telomerase in meiosis. The second possibility is the increased Wnt7b expression. Because Wnt family genes promote proliferation of spermatogonia (24), increased Wnt7b may have promoted self-renewal and inhibited meiosis. Thus, SSCs are virtually immortal but the mechanism underlying the loss of haploid cell-forming capacity needs to be determined in future.
Another notable feature of aged GS cells was their enhanced proliferation. Our analysis suggested that reduced PRC2 activity caused by Phf1 down-regulation is responsible for Wnt7b up-regulation and JNK hyperactivation. Although age-induced changes in H3K27 methylation levels have previously been reported for HSCs and muscle stem cells (25), the peaks become broader and more intense upon aging. By contrast, H3K27me3 peaks become narrower and lower in 60M-GS cells. Although we noted Phf1 down-regulation in 60M-GS cells, the reason for this difference from other stem cell types and why Phf1 and Asxl1 were down-regulated remain to be determined. Because PHF1 and ASXL1/BAP1 are involved in DNA repair (26, 27), increased DNA damage during aging may influence their expression levels. In the germline, PRC2 is important for regulating SSC number (28). Our results now suggest that PRC2 may play additional roles during aging.
Activation of the WNT-JNK pathway caused abnormal metabolism in GS cells. Although AKT activation promotes glycolysis in GS cells (5), we found that JNK activation enhances glycolysis in 60M-GS cells. On the other hand, 60M-GS cells had lower levels of ROS. We initially thought that decreased NOX1 activity was responsible for this change because NOX1 is important for SSCs (19). However, Nox1 was expressed more strongly in 60M-GS cells. In addition, ROS reduction was caused by augmentation of WNT-JNK signaling via mitochondrial defect. When this signaling pathway was activated in 5M-GS cells, the number of mitochondria decreased significantly. Down-regulation of Ppargc1a was ultimately responsible for the decrease in mitochondria activity. Considering that 60M-GS cells proliferate more actively despite the down-regulation of mitochondria-derived ROS, NOX-derived ROS probably play more important roles in SSC self-renewal. How the difference in the origin of ROS influences GS cell proliferation is a next important question.
Undifferentiated spermatogonia in Klotho KO mice and aged BN rats also show JNK phosphorylation and enhanced glycolysis. Although we could not carry out transplantation experiments with BN rats due to lack of donor cell markers, serial transplantation of Klotho KO testes showed more frequent self-renewing divisions. Moreover, the increased generation of secondary colonies of Klotho KO SSCs also suggests that the aging phenotype was not rescued by transplantation into the wild-type environment (primary recipients). These defects are probably caused by intrinsic defects in the germ cells because only germ cells express KLOTHO (29). On the other hand, while down-regulation of PHF1 and increased WNT7B expression in BN rats were confirmed, Wnt7b did not change significantly in Klotho KO mice. We speculate that Klotho mutations might have occurred downstream of Wnt7b signaling.
Our study suggests that cell-autonomous factors also contribute to SSC aging. Although we cannot completely exclude the possible involvement of other factors, we think that down-regulation of PRC2 activity likely contributed to the aged phenotype of 60M-GS cells by activating WNT7B-JNK signaling. One of the next important unsolved questions is how the telomeres are maintained in aged SSCs. Since there were no apparent karyotype abnormalities, SSCs must have a unique machinery to protect telomere fusions. Because the Klotho KO mouse phenotype is caused by hyperphosphatemia caused by abnormal calcium metabolism (21), further investigation of the link between calcium metabolism and glycolysis may also reveal the mechanisms of SSC aging. Studies of these issues are warranted in the future, which opens up new possibilities for controlling germ cell aging and infertility.
Materials and Methods
Cell Culture.
GS cells were derived from C57BL/6 Tg14(act-EGFP)OsbY01 on a DBA/2 background (green) (M. Okabe, Osaka University, Osaka, Japan) (8). GS cell culture conditions were previously described (8). The cells were maintained on mouse embryonic fibroblasts treated with mitomycin C (Sigma). SA β-galactosidase activity was examined as described previously (30).
To determine the effects of chemicals, GS cells were treated with allopurinol (50 μM; Sigma), antimycin A (5 μM; Santa Cruz), rotenone (1 μM; Sigma), DPI (1 μM; Sigma), apocynin (1 mM; TCI), L-NAME (100 μM; Santa Cruz), anisomycin (300 ng/mL Santa Cruz), H2O2 (30 μM; Wako). Dimethyl sulfoxide (Sigma) or medium was used as a control. For analyses of the signal transduction pathway, GS cells were treated with SP600125 (13.3 μM; Selleck), XMD8-92 (1.67 μM; Tocris), AKT inhibitor IV (26.7 μM; Calbiochem), PD0325901 (1 μM; Stemgent), SB203580 (10 μM; Selleck), or IWP2 (0.5 μM; Stemgent). In some experiments, 2DG (5 mM; Wako) was used to study the impact of glycolysis.
Statistical Analyses.
Results are presented as means ± SEMs. Significant differences between means for single comparisons were determined using the Student’s t test. Multiple comparison analyses were performed using ANOVA followed by Tukey’s HSD test.
Accession Numbers.
All of the sequencing data generated in this study have been deposited in the GEO and DDBJ databases (GSE120778 for WGBS and ChIP-seq and DRA007373 for RNA-seq data, respectively).
Supplementary Material
Acknowledgments
We thank Ms. S. Ikeda, M. Kataba, and S. Sakurai for technical assistance. Financial support was provided by the Ministry of Education, Culture, Sports, Science, and Technology, Japan (18H05281, 19H05750, and 17H05639). This research is also supported by the Platform Project for Supporting in Drug Discovery and Life Science Research (Platform for Drug Discovery, Informatics, and Structural Life Science) from the Japan Agency for Medical Research and AMED-CREST (JP18gm1110008).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo [accession no. GSE120778 for Whole-Genome Bisulfite Sequencing (WGBS) and Chromatin Immunoprecipitation Sequencing (ChIP-seq) data] and in the DNA Data Bank of Japan, https://www.ddbj.nig.ac.jp/index-e.html (accession no. DRA007373 for RNA-seq data).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904980116/-/DCSupplemental.
References
- 1.Ito K., Suda T., Metabolic requirements for the maintenance of self-renewing stem cells. Nat. Rev. Mol. Cell Biol. 15, 243–256 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stoll E. A., et al. , Aging neural progenitor cells have decreased mitochondrial content and lower oxidative metabolism. J. Biol. Chem. 286, 38592–38601 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Rooij D. G., Russell L. D., All you wanted to know about spermatogonia but were afraid to ask. J. Androl. 21, 776–798 (2000). [PubMed] [Google Scholar]
- 4.Meistrich M. L., van Beek M. E. A. B., “Spermatogonial stem cells” in Cell and Molecular Biology of the Testis, Desjardins C. C., Ewing L. L., Eds. (Oxford University Press, 1993), pp. 266–295. [Google Scholar]
- 5.Kanatsu-Shinohara M., et al. , Myc/Mycn-mediated glycolysis enhances mouse spermatogonial stem cell self-renewal. Genes Dev. 30, 2637–2648 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang X., Ebata K. T., Robaire B., Nagano M. C., Aging of male germ line stem cells in mice. Biol. Reprod. 74, 119–124 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Ryu B. Y., Orwig K. E., Oatley J. M., Avarbock M. R., Brinster R. L., Effects of aging and niche microenvironment on spermatogonial stem cell self-renewal. Stem Cells 24, 1505–1511 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanatsu-Shinohara M., et al. , Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol. Reprod. 69, 612–616 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Kanatsu-Shinohara M., Shinohara T., Spermatogonial stem cell self-renewal and development. Annu. Rev. Cell Dev. Biol. 29, 163–187 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Brinster R. L., Zimmermann J. W., Spermatogenesis following male germ-cell transplantation. Proc. Natl. Acad. Sci. U.S.A. 91, 11298–11302 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubo N., et al. , DNA methylation and gene expression dynamics during spermatogonial stem cell differentiation in the early postnatal mouse testis. BMC Genomics 16, 624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Steensel B., Belmont A. S., Lamina-associated domains: Links with chromosome architecture, heterochromatin, and gene repression. Cell 169, 780–791 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemann M. T., et al. , Telomere dysfunction triggers developmentally regulated germ cell apoptosis. Mol. Biol. Cell 12, 2023–2030 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Churchman M. L., Roig I., Jasin M., Keeney S., Sherr C. J., Expression of arf tumor suppressor in spermatogonia facilitates meiotic progression in male germ cells. PLoS Genet. 7, e1002157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammoud S. S., et al. , Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 15, 239–253 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Hammoud S. S., et al. , Transcription and imprinting dynamics in developing postnatal male germline stem cells. Genes Dev. 29, 2312–2324 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Margueron R., Reinberg D., The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finkel T., The metabolic regulation of aging. Nat. Med. 21, 1416–1423 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Morimoto H., et al. , ROS are required for mouse spermatogonial stem cell self-renewal. Cell Stem Cell 12, 774–786 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Austin S., St-Pierre J., PGC1α and mitochondrial metabolism: Emerging concepts and relevance in ageing and neurodegenerative disorders. J. Cell Sci. 125, 4963–4971 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Nabeshima Y., Discovery of alpha-Klotho unveiled new insights into calcium and phosphate homeostasis. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85, 125–141 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C., Leung A., Sinha-Hikim A. P., Reproductive aging in the male brown-Norway rat: A model for the human. Endocrinology 133, 2773–2781 (1993). [DOI] [PubMed] [Google Scholar]
- 23.Nagano M., Avarbock M. R., Brinster R. L., Pattern and kinetics of mouse donor spermatogonial stem cell colonization in recipient testes. Biol. Reprod. 60, 1429–1436 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lombardi A. P. G., et al. , Physiopathological aspects of the Wnt/β-catenin signaling pathway in the male reproductive system. Spermatogenesis 3, e23181 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beerman I., Rossi D. J., Epigenetic control of stem cell potential during homeostasis, aging, and disease. Cell Stem Cell 16, 613–625 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong Z., et al. , A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell. Nucleic Acids Res. 36, 2939–2947 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abdel-Wahab O., et al. , Deletion of Asxl1 results in myelodysplasia and severe developmental defects in vivo. J. Exp. Med. 210, 2641–2659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mu W., Starmer J., Fedoriw A. M., Yee D., Magnuson T., Repression of the soma-specific transcriptome by Polycomb-repressive complex 2 promotes male germ cell development. Genes Dev. 28, 2056–2069 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S. A., et al. , Immunohistochemical localization of Klotho protein in brain, kidney, and reproductive organs of mice. Cell Struct. Funct. 29, 91–99 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Debacq-Chainiaux F., Erusalimsky J. D., Campisi J., Toussaint O., Protocols to detect senescence-associated beta-galactosidase (SA-betagal) activity, a biomarker of senescent cells in culture and in vivo. Nat. Protoc. 4, 1798–1806 (2009). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.