Significance
In the present study, we identify and describe an important cross-talk between leptin signaling and macrophage functions in the context of Salmonella Typhimurium infection. Genetic ablation of leptin receptor or pharmacological antagonization of leptin augmented lysosomal functions in macrophages, reduced S. Typhimurium burden, and diminished inflammation both in vitro and in vivo. Leptin signaling activates mTORC2/Akt pathway through the down-regulation of Phlpp1 phosphatase, thus impairs lysosome-mediated pathogen clearance.
Keywords: macrophages, leptin, Salmonella, AKT, lysosomes
Abstract
The dynamic interplay between metabolism and immune responses in health and disease, by which different immune cells impact on metabolic processes, are being increasingly appreciated. However, the potential of master regulators of metabolism to control innate immunity are less understood. Here, we studied the cross-talk between leptin signaling and macrophage function in the context of bacterial infections. We found that upon infection with Gram-negative pathogens, such as Salmonella Typhimurium, leptin receptor (Lepr) expression increased in both mouse and human macrophages. Unexpectedly, both genetic Lepr ablation in macrophages and global pharmacologic leptin antagonization augmented lysosomal functions, reduced S. Typhimurium burden, and diminished inflammation in vitro and in vivo. Mechanistically, we show that leptin induction activates the mTORC2/Akt pathway and subsequently down-regulates Phlpp1 phosphatase, allowing for phosphorylated Akt to impair lysosomal-mediated pathogen clearance. These data highlight a link between leptin signaling, the mTORC2/Phlpp1/Akt axis, and lysosomal activity in macrophages and have important therapeutic implications for modulating innate immunity to combat Gram-negative bacterial infections.
Antibiotic-resistant Gram-negative bacteria are on the rise globally and narrows antibiotic options to alarming levels in the clinic (1). Thus, the World Health Organization has released a global action plan and a pathogen priority list highlighting the development of new treatment options against infections specifically caused by Gram-negative bacteria (2). Gram-negative pathogens, including Salmonella Typhimurium (S. Typhimurium), cause fatal infections due to their ability to evade host immune defense mechanisms (3). To develop new treatment strategies, the investigation into the molecular mechanisms of infections due to Gram-negative bacteria is essential.
Macrophages are sentinel immune cells at the forefront of the innate immune response. These cells detect invading pathogens via pattern recognition receptors and engulf pathogens into phagosomes that are destined for destruction via lysosomes fusion (4). Lysosomes not only function in cellular degradation processes, but also integrate signals from growth factors and regulate cellular metabolism (5). Several pathogens, including S. Typhimurium, have evolved mechanisms to evade the hostile milieu of lysosomes and induce necroptosis of macrophages (6, 7). In the case of S. Typhimurium, Salmonella pathogenicity island (SPI)-I and SPI-II–encoded virulence effectors are injected into host cells via the type III secretion system (6). We recently showed that S. Typhimurium also targets metabolic sensors, namely AMPK and SIRT1, for lysosomal degradation in a SPI-II–dependent manner (8).
Metabolism and immunity are closely linked at both the cellular and organismal levels. At the organismal level, epidemiological studies have shown that patients with diabetes or obesity are predisposed to infections, indicating a clinically evident dysfunction of the immune response in metabolic disease (9, 10). Metabolic diseases also alter the composition and functional states of various immune cells in metabolic tissues (11). At the cellular level, various metabolic pathways and intracellular metabolites can modulate the innate and adaptive immune responses (12, 13). In light of these developments, a new research area known as “immunometabolism” has emerged that concerns the interplay between metabolic pathways and immune system function (14).
Circulating adipokines and hormones, such as leptin and insulin, have key roles in regulating metabolic homeostasis in the body, much in the same way that cytokines and chemokines mediate intercellular communication in the immune system. Insulin is produced by pancreatic β-cells to lower glucose levels in the blood stream by allowing glucose to migrate into cells while leptin release through the adipose tissue controls food intake by reducing the appetite. Notably, insulin and leptin can also regulate immune responses in various ways. For instance, insulin regulates macrophage-mediated inflammatory responses in the context of obesity (15), while leptin enhances proinflammatory cytokine secretion in macrophages and chemotaxis in granulocytes, and also inhibits specific immune responses such as natural killer cell cytotoxicity (16–19). Despite the close relationship between metabolic hormones and the immune response, the roles of insulin and leptin signaling in macrophage functions remain elusive. In particular, it is unclear whether there is functional cross-talk between macrophages and leptin signaling in the context of acute inflammatory conditions such as infections caused by Gram-negative bacteria. In this study, we focused on the role of leptin in macrophage defenses toward bacterial pathogens. To address this question, we used mouse bone marrow-derived macrophages (BMDMs) to investigate the roles of leptin signaling in the context of bacterial infections. We found that leptin receptor (Lepr) expression is highly up-regulated in mouse and human macrophages infected with S. Typhimurium and other Gram-negative bacteria. Genetically ablating Lepr in macrophages or pharmacologically antagonizing leptin augmented the clearance of Gram-negative bacteria due to elevated lysosomal function. In sum, we found that leptin signaling impinges on the lysosomal mTORC2/Phlpp1/Akt axis involved in chaperone-mediated autophagy (20), thus critically reorienting this pathway in the innate immune response.
Results
S. Typhimurium Infection Induces Leptin Signaling in Macrophages.
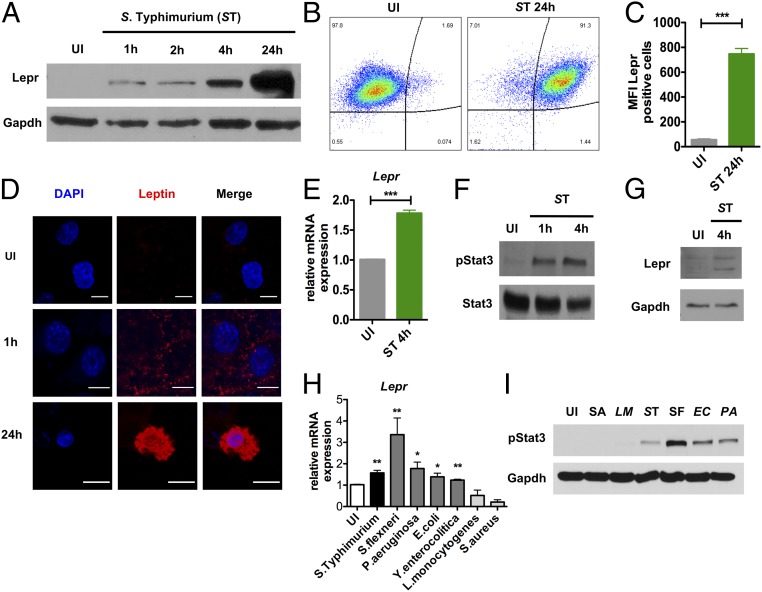
To study the role of leptin signaling in macrophages, we first infected a mouse macrophage cell line (RAW-264.7) with S. Typhimurium and analyzed Lepr expression at 1, 2, 4, and 24 h after infection. Here, we observed strong Lepr protein induction over time (Fig. 1A). Consistently, flow cytometry indicated increased binding of fluorescently labeled leptin to its receptor (Fig. 1 B and C), and confocal microscopy imaging confirmed leptin uptake into RAW macrophages (Fig. 1D). This induction of Lepr expression at both the mRNA and protein levels was also evident in BMDMs from WT C57BL/6 mice at 1 and 4 h after S. Typhimurium infection (Fig. 1E) (SI Appendix, Fig. S1A). These time points were chosen for experiments with BMDMs due to increased cell death induced by S. Typhimurium. We also found an associated increase in signal transducer and activator of transcription 3 (Stat3) phosphorylation levels at 1 and 4 h, which lies downstream of leptin signaling (Fig. 1F). Finally, we confirmed that Lepr expression and activation of Stat3 are also induced in human macrophages isolated from peripheral blood mononuclear cells (PBMCs) upon S. Typhimurium infection (Fig. 1G). We also assessed whether Lepr up-regulation was virulence-dependent during S. Typhimurium infection. Here, we infected BMDMs with S. Typhimurium SPI-I (invA) or SPI-II (ssaV) mutants or heat-killed (HK) S. Typhimurium and found that Lepr and Stat3 activation was reduced upon infection with ssaV mutant strain of S. Typhimurium (SI Appendix, Fig. S1 B and C). Moreover, increased Lepr and Stat3 activation were specific for Gram-negative pathogens, including Pseudomonas aeruginosa, Shigella flexneri, Escherichia coli, and Yersinia enterocolitica, but not with Gram-positive pathogens, such as Staphylococcus aureus and Listeria monocytogenes (Fig. 1 H and I).
Fig. 1.
S. Typhimurium infection induces leptin signaling in macrophages. (A) Western blot analysis of LepR expression in uninfected (UI) and 1, 2, 4, and 24 h after S. Typhimurium infection in RAW-264.7 macrophages (n = 4). (B) Uptake of fluorescent leptin in UI and 24-h infected S. Typhimurium RAW macrophages measured by FACS (n = 2). (C) MFIs of fluorescent leptin-positive macrophages. (D) Confocal microscopy of fluorescent leptin uptake in UI and 1 and 24 h in RAW macrophages. (E) Relative Lepr mRNA expression in UI and 4-h S. Typhimurium-infected WT BMDM (n = 4). (F) Protein expression of phospho-Stat3 and Gapdh in WT BMDMs in UI and 1 and 4 h after S. Typhimurium infection (n = 6). (G) Western blot analysis of LepR and Gapdh expression in human macrophages of UI and 4 h after S. Typhimurium infection (n = 2). (H) Relative Lepr expression in WT BMDMs 4 h after S. Typhimurium (ST), S. flexneri (SF), P. aeruginosa (PA), E. coli (EC), Y.enterocolitica (YE), L. monocytogenes (LM), and S. aureus (SA). (I) Immunoblot analysis of phospho-Stat3. Data are shown as mean ± SEM and statistical significance calculated using Student t test and represented as *P < 0.05; **P < 0.01; ***P < 0.001. (Scale bars: 10 μm.)
We next sought to understand the mechanisms leading to elevated Lepr expression. We found that Lepr induction was independent of LPS (SI Appendix, Fig. S1D) and canonical TLR pathways (SI Appendix, Fig. S1E). Since Lepr expression has been shown to be regulated by Stat3 (21, 22) and having found that Stat3 activation is specifically induced upon Gram-negative bacteria infection, we examined whether Lepr expression is regulated by Stat3. Stat3 knockdown in WT BMDMs by siRNA (SI Appendix, Fig. S1F) resulted in reduced Lepr transcription 1 h after S. Typhimurium infection (SI Appendix, Fig. S1G). These data suggest that Lepr expression is Stat3-dependent. Taken together, we have found that Gram-negative pathogens specifically induce leptin signaling in macrophages upon infection.
Leptin Signaling Inhibits Bacterial Clearance and Promotes Inflammation.
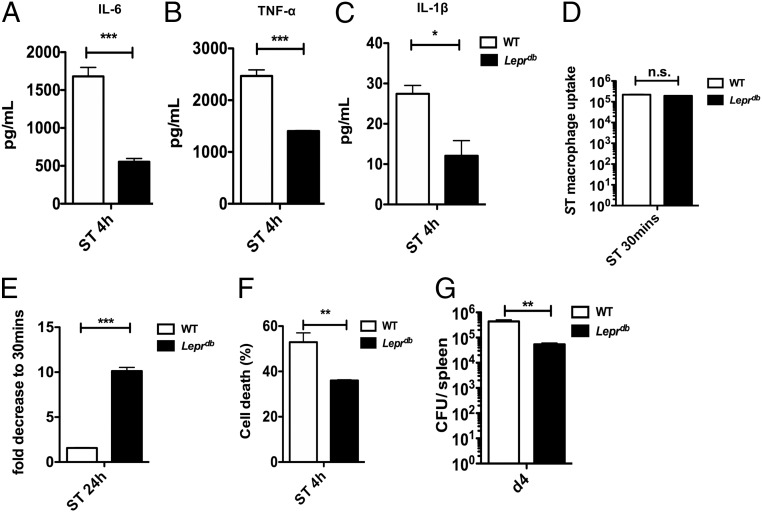
We next investigated how genetic ablation of leptin signaling impacts macrophages defense mechanisms against S. Typhimurium challenge. We infected BMDMs isolated from obese, Lepr-deficient B6.BKS (D)-Leprdb/J (Leprdb) mice infected with S. Typhimurium and observed a significant reduction in the levels of IL-6, TNF-α, and IL-1β proinflammatory cytokines compared with WT C57BL/6 controls (Fig. 2 A–C); however, these cytokines were not differentially expressed in uninfected BMDMs from WT and Leprdb mice (SI Appendix, Fig. S2A). We also did not observe any differences in the initial phagocytosis of bacteria in BMDMs from WT and Leprdb mice at 30 min (Fig. 2D). Interestingly, we found that S. Typhimurium burden (Fig. 2E) and cell death (Fig. 2F) were significantly diminished in Leprdb-deficient BMDMs. Conversely, supplementing WT BMDMs from C57BL/6 mice in vitro with leptin markedly increased the cellular bacterial burden (SI Appendix, Fig. S2B). We further confirmed a significant reduction in S. Typhimurium burden in the spleens of Leprdb mice on day 4 (Fig. 2G), but no significant difference in bacterial burden was observed at day 2 after infection (SI Appendix, Fig. S2 C and D), supporting the notion that leptin signaling is detrimental for the host immune defense mechanisms against S. Typhimurium in vivo.
Fig. 2.
Leptin signaling negatively regulates bacterial clearance and inflammation. (A) IL-6, TNF-α (B), and IL-1β (C) in supernatants of WT and Leprdb BMDMs 4 h after S. Typhimurium infection (n = 3). (D) In vitro bacterial burden expressed as CFU after 30 min of S. Typhimurium infection in WT BMDMs compared with Leprdb (n = 3). (E) In vitro bacterial burden expressed as fold decrease (CFU) after 24 h of S. Typhimurium infection in WT BMDMs compared with Leprdb (n = 3). (F) Cell viability of 4-h S. Typhimurium-infected WT vs. Leprdb BMDMs (n = 2). (G) In vivo bacterial burden expressed as CFU in the spleen from Leprdb mice 4 d after i.v. S. Typhimurium infection compared with WT. Data are shown as mean ± SEM and statistical significance calculated using Student t test and represented as *P < 0.05; **P < 0.01; ***P < 0.001.
Leptin Signaling Regulates Macrophage-Mediated S. Typhimurium Elimination.
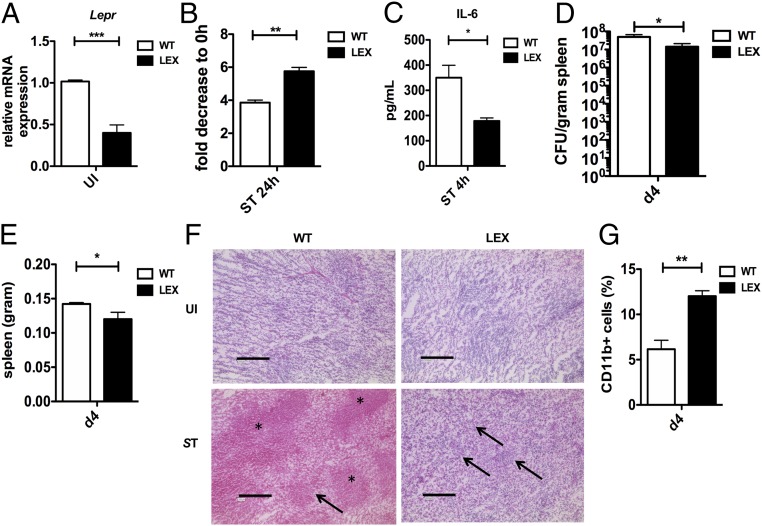
The macrophage defense mechanisms toward S. Typhimurium in Leprdb obese mice could be secondary to global chronic inflammation and metabolic abnormalities. We thus generated a mouse model lacking the leptin receptor only in myeloid cells by crossing Leprfl/fl mice with transgenic mice expressing LysMCre+/+ (hereafter termed as LEX), to create myeloid-specific knockdowns (23, 24). Consistent with a cell intrinsic role for leptin-signaling LEX mice were not obese and Lepr expression in LEX BMDMs was significantly reduced upon infection (Fig. 3A). Notably, and similar to our findings using Leprdb macrophages, bacterial burden (Fig. 3B) and IL-6 (Fig. 3C) and TNF-α (SI Appendix, Fig. S3A) levels were significantly reduced upon S. Typhimurium infection. Similarly, LEX BMDMs also showed enhanced elimination of another Gram-negative pathogen, P. aeruginosa (SI Appendix, Fig. S3B), while bactericidal activity against the Gram-positive bacteria L. monocytogenes did not differ between WT and LEX macrophages (SI Appendix, Fig. S3C).
Fig. 3.
Leptin signaling intrinsically regulates the ability of macrophages to eliminate S. Typhimurium. (A) Relative Lepr mRNA expression in Leprfl/fl LysMCre+/+ (LEX) compared with WT (n = 3). (B) In vitro bacterial burden after 24 h of S. Typhimurium infection in WT compared with LEX BMDMs (n = 3). (C) IL-6 expression in supernatants of WT and LEX BMDMs 4 h infected with S. Typhimurium (n = 3). (D) In vivo bacterial burden expressed as CFU in spleens from LEX versus WT mice per gram of tissue (n = 3). (E) Weight of spleens taken from LEX and WT mice 4 d after S. Typhimurium infection. (F) HE-staining of spleen tissue sections from UI and 4 d after i.v. S. Typhimurium infected WT and LEX mice. In F, *, necrosis; →, lymphoid cells. (G) Percentage of CD11b-positive cells from WT and LEX spleens 4 d after infection. Data are shown as mean ± SEM and statistical significance calculated using Student t test and represented as *P < 0.05; **P < 0.01; ***P < 0.001. (Scale bars: 200 μm.)
Consistent with our in vitro results, we also observed reduced S. Typhimurium burden in the spleen (Fig. 3D) and liver of LEX mice (SI Appendix, Fig. S3D) compared with WT controls. Accordingly, the weight of these organs after infection was also reduced in LEX mice (Fig. 3E and SI Appendix, Fig. S3E). Histological examination of spleen sections from infected LEX mice strikingly revealed reduced inflammation-mediated tissue damage and an enhanced presence of lymphoid cells compared with WT mice (Fig. 3F). Consistently, flow cytometric analysis of the spleen cells showed increased recruitment of CD11b+ myeloid cells in LEX mice (Fig. 3G), which are essential for pathogen clearance. Finally, we detected significantly reduced IL-1β levels in the spleen and liver of S. Typhimurium-infected LEX mice (SI Appendix, Fig. S3 F and G). Taken together, these data suggest that macrophage-specific leptin signaling inhibits bactericidal functions in macrophages.
Leptin Antagonism Reduces Bacterial Burden and Inflammation in Vivo.
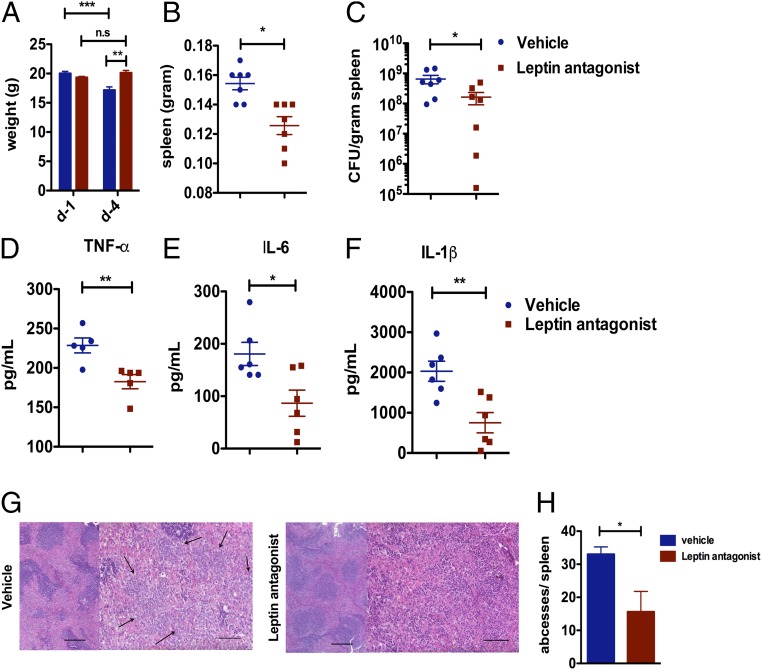
We next asked whether a leptin antagonist could also reduce bacterial burden in WT C57BL/6 mice infected with S. Typhimurium. Here, we tested the efficiency of pegylated leptin antagonists, previously shown to block leptin signaling (25), in controlling bacterial burden and inflammation. WT mice were administered 6.25 mg/kg leptin antagonist once daily from 1 d before infection until 4 d after infection when mice were killed, and the spleens were collected. Over the course of infection, we observed that the vehicle-treated mice lost weight as expected, but leptin antagonist-treated mice maintained their starting weight (Fig. 4A and SI Appendix, Fig. S4A). Spleens from leptin antagonist-treated mice also weighed less compared with vehicle-treated mice (Fig. 4B and SI Appendix, Fig. S4B) and showed a reduced bacterial burden (Fig. 4C) similar to that of the Leprdb and LEX mice. Consistently, TNF-α (Fig. 4D), IL-6 (Fig. 4E), and IL-1β (Fig. 4F) levels were reduced in the spleens of leptin antagonist-treated mice compared with vehicle-treated mice. Finally, histopathological analysis revealed a significantly reduced number of abscesses in the spleens of the leptin antagonist-treated mice compared with vehicle-treated mice upon S. Typhimurium infection (Fig. 4 G and H). Summing up, we could successfully demonstrate that pharmacological inhibition of leptin signaling significantly improves bacterial elimination, which is consistent with our data from mouse models lacking leptin signaling.
Fig. 4.
Leptin antagonist reduces bacterial burden and inflammation in vivo. (A) Leptin antagonist and vehicle-treated WT mice were infected with S. Typhimurium and the weights of the mice were monitored on day 1 (d-1) and day 4 (d-4) after infection. (B) On day 4, spleens were isolated and weighed. (C) Bacterial burden in the spleen was estimated, and the inflammatory cytokines TNF-α (D), IL-6 (E), and IL-1β (F) were measured by ELISA. (G) HE staining of spleen tissue sections from 4 d after i.p. S. Typhimurium-infected vehicle and leptin antagonist-treated mice. →, abscesses. (H) Bar graph showing the number of abscesses per spleen from S. Typhimurium-infected vehicle and leptin antagonist-treated mice. Data are shown as mean ± SEM and statistical significance calculated using Student t test and represented as *P < 0.05; **P < 0.01; ***P < 0.001. (Scale bars: 200 μm.)
Enhanced Phagolysosomal Processing in Lepr-Deficient Macrophages Reduces Bacterial Burden.
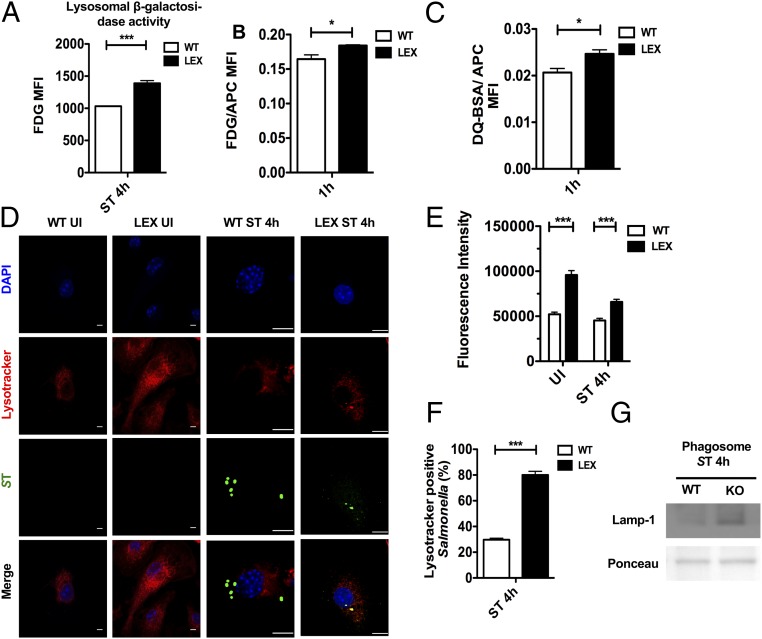
Phagolysosomal processing is a major mechanism by which macrophages eliminate invading pathogens. Because the loss of leptin signaling led to reduced bacterial burden, we investigated the effects of phagosome maturation in BMDMs isolated from LEX and WT mice. We infected LEX and WT BMDMs with C12FDG-coated S. Typhimurium or pulsed with C12FDG-coated fluorescent, inert beads, and then analyzed β-galactosidase and proteolytic activities in phagolysosomes by flow cytometry, as described previously (26). As beads are taken up much more rapidly by macrophages than S. Typhimurium infection, the 1-h time point was chosen to show the effects whenever beads were used for experiments. Here, we found that β-galactosidase activity was enhanced in LEX BMDMs upon S. Typhimurium infection (Fig. 5A), indicative of increased lysosomal activity. Moreover, this effect was independent of bacterial virulence, because lysosomal β-galactosidase and proteases activity were also up-regulated in bead-containing phagolysosomes of LEX macrophages (Fig. 5 B and C). Interestingly, we also detected increased intensity of LysoTracker—a red fluorescent dye that tracks acidic organelles in cells—in uninfected and S. Typhimurium-infected LEX BMDMs (Fig. 5 D and E). Furthermore, we observed increased colocalization between S. Typhimurium and LysoTracker (Fig. 5F) and increased lysosomal-associated membrane protein 1 (Lamp-1) expression in S. Typhimurium-containing phagosomes isolated from LEX BMDMs compared with WT BMDMs (Fig. 5G). Moreover, LEX BMDMs exhibited an increased number of bacteria in Lamp-1–positive phagosomes compared with that of WT BMDMs (SI Appendix, Fig. S5 A and B). These results suggest that lysosomal functions are elevated in LEX macrophages.
Fig. 5.
Reduced S. Typhimurium burden is mediated through enhanced phagolysosomal processing in Lepr-deficient macrophages. (A) Flow cytometric analysis of WT versus LEX BMDMs infected with C12FDG-coated S. Typhimurium 4 h after infection (n = 3). Flowcytometric analysis of WT and LEX BMDMs pulsed with C12FDG-coated beads (n = 3) (B) and DQ-BSA–coated beads (n = 3) (C). Bar graphs represent MFIs of C12FDG and DQ-BSA normalized to MFIs of red fluorescence. (D) Confocal microscopy analysis of UI and 4-h infected WT and LEX BMDMs pulsed with Lysotracker and stained for S. Typhimurium LPS. (E) Bar graph shows the fluorescent intensity of Lysotracker measured with ImageJ. (F) S. Typhimurium-LysoTracker colocalization in WT and LEX BMDMs 4 h after infection. (G) Lamp-1 expression on S. Typhimurium containing phagosomes 4 h after infection. Data are shown as mean ± SEM and statistical significance calculated using Student t test and represented as *P < 0.05; **P < 0.01; ***P < 0.001. (Scale bars: 20 μm.)
Leptin-Dependent mTORC2 Signaling Regulates Enhanced Lysosomal Activity.
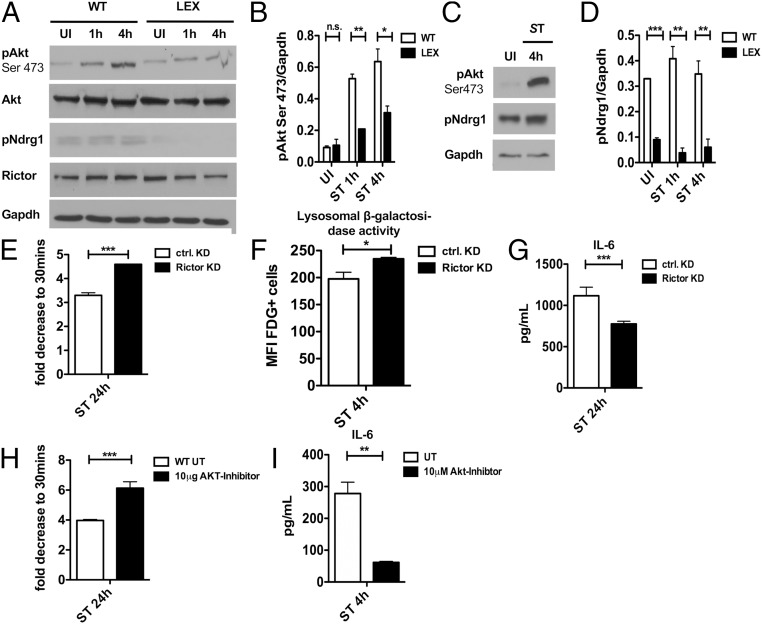
Leptin regulates the activity of mTOR (27), which has a critical role in regulating lysosomal functions. We hypothesized, therefore, that mTOR complexes (mTORC) may be differentially regulated in Lepr-deficient BMDMs compared with WT BMDMs upon S. Typhimurium infection. We previously reported that S. Typhimurium infection led to increased phosphorylation of mTORC1 targets (8), namely S6kinase (pS6K) and Akt (pAkt-Thr308). Here, we found that these phosphorylation events had similar kinetics in both WT and LEX BMDMs upon S. Typhimurium infection (SI Appendix, Fig. S6A). Conversely, Akt phosphorylation at serine 473 (pAkt-Ser473), which is mediated by mTORC2 (28), was induced upon S. Typhimurium infection in both WT and LEX BMDMs (Fig. 6 A and B) and human monocyte-derived macrophages (HMDMs) (Fig. 6C). Despite this induction, total pAkt-Ser473 levels were reduced between WT and LEX BMDMs at all time points (Fig. 6 A and B).
Fig. 6.
Enhanced lysosomal activity is mediated through leptin-dependent mTORC2 signaling. (A) Immunoblot analysis of mTORC2 pathway in WT versus LEX BMDMs infected with S. Typhimurium for 1 h and 4 h compared with UI (n = 3). (B) Densitometric analysis of phosphorylated Akt to Gapdh (n = 3). (C) Immunoblot analysis of mTORC2 pathway in human macrophages (n = 1). (D) Densitometric analysis of phosphorylated Ndrg1 to Gapdh (n = 3). (E) Bacterial load in Rictor siRNA transfected WT BMDMs compared with control siRNA-transfected cells infected with S. Typhimurium for 24 h. (F) Flow cytometric analysis in Rictor siRNA transfected WT BMDMs compared with control siRNA transfected cells with C12FDG-coated S. Typhimurium 4 h after infection (n = 3). (G) IL-6 expression in supernatants from Rictor siRNA-transfected WT BMDMs compared with control siRNA transfected cells infected with for 24 h. (H) Bacterial load 24 h after infection in WT BMDMs treated with 10 μM Akt-Inhibitor compared with untreated. (I) IL-6 expression in supernatants in 24-h infected WT BMDMs treated with 10 μM Akt-Inhibitor compared with untreated. Data are shown as mean ± SEM and statistical significance calculated using Student t test and represented as *P < 0.05; **P < 0.01; ***P < 0.001.
We also found that phosphorylation of Ndrg1, another target of mTORC2 complex, was induced upon S. Typhimurium infection in WT BMDMs (Fig. 6 A and C) and HMDMs (Fig. 6D), but not in LEX BMDMs. Moreover, Ndrg1 was already reduced in uninfected BMDMs. Finally, Rictor, a specific component of mTORC2, was also modestly reduced in LEX BMDMs upon S. Typhimurium infection. These data suggest that leptin signaling specifically regulates mTORC2 activity upon S. Typhimurium infection.
We further investigated whether enhanced bacterial clearance in Lepr-deficient (LEX) BMDMs is dependent on mTORC2 activity. We first knocked down Rictor by siRNA in WT BMDMs, which resulted in reduced Akt phosphorylation at Ser473 (SI Appendix, Fig. S5B). Rictor depletion enhanced S. Typhimurium elimination (Fig. 6E) and phagolysosomal activity to a similar level as seen in LEX BMDMs (Fig. 6F). IL-6 secretion was also dampened in Rictor-depleted BMDMs (Fig. 6G).
Akt is a downstream target of mTORC2 and has a reported role in lysosomal function (29). During infection, S. Typhimurium activates Akt, thereby inhibiting AMPK activity and autophagy and promoting pathogenesis (8). We asked, therefore, whether reducing Akt function can restore the pathogen response in WT BMDMs. Here, we found reduced pAkt-Ser473 in infected BMDMs treated with an Akt VIII-Inhibitor (SI Appendix, Fig. S6C), as well as reduced S. Typhimurium burden and inflammatory-cytokine levels (Fig. 6 H and I). Taken together, these data suggest that pathogen-induced leptin signaling activates mTORC2-dependent Akt phosphorylation at Ser473, leading to reduced lysosomal function and clearance.
Loss of Leptin Signaling Enhances Phlpp1-Dependent Akt Dephosphorylation and Subsequent Lysosomal Function.
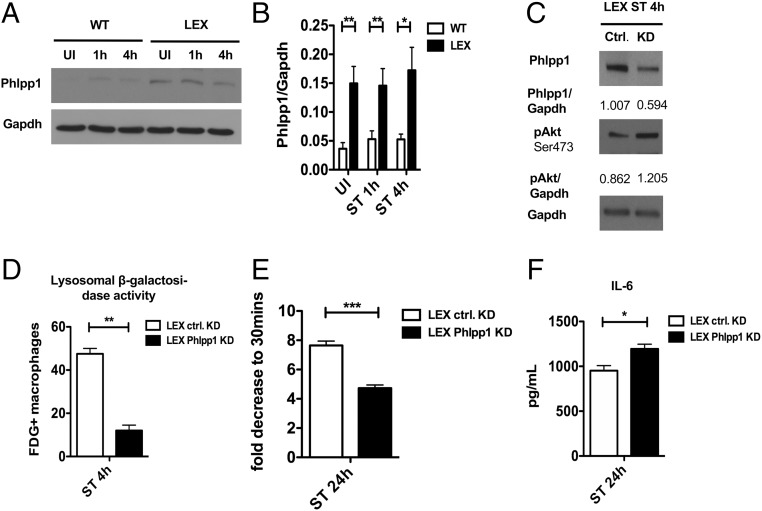
The Pleckstrin homology (PH) domain and leucine-rich repeat protein phosphatase 1 (Phlpp1) dephosphorylates phospho-Akt at serine 473 and thus enhances lysosome-dependent clearance of protein aggregates (20). Therefore, we asked whether leptin signaling regulates Phlpp1 function in the context of S. Typhimurium infection. We found that Phlpp1 expression was markedly increased in LEX BMDMs (Fig. 7 A and B), which correlated with decreased pAkt-Ser473 levels (Fig. 7 A and B). siRNA-mediated Phlpp1 knockdown enhanced the Akt phosphorylation at serine 473, supporting this critical function of Phlpp1 (Fig. 7C).
Fig. 7.
Loss of leptin signaling enhances Phlpp-1-dependent dephosphorylation of Akt to enhance lysosomal function. (A) Immunoblot analysis of Phlpp1 expression in WT versus LEX BMDMs at 1 and 4 h after infection compared with UI (n = 3). (B) Densitometric analysis of Phlpp1 to Gapdh (n = 3). (C) Expression of Phlpp1 and phospho-AKT Ser473 in Phlpp1-depleted LEX BMDMs compared with control cells (n = 3). (D) Bar graph represents flow cytometric measurement of FDG-positive cells in Phlpp1-depleted LEX BMDMs compared with control cells. (E) Bacterial burden in Phlpp1-deficient LEX BMDMs compared with control cells 24 h after infection. (F) IL-6 expression in supernatants of Phlpp1-depleted LEX BMDMs compared with control cells 24 h after infection. Data are shown as mean ± SEM and statistical significance calculated using Student t test and represented as *P < 0.05; **P < 0.01; ***P < 0.001.
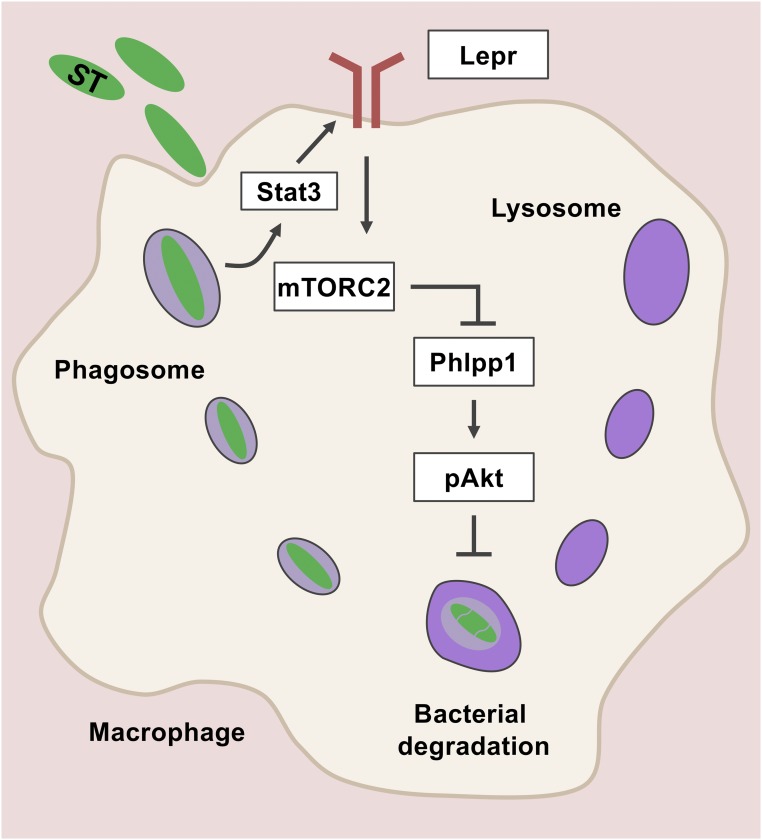
Because Akt inhibition reduced S. Typhimurium burden in macrophages (Fig. 6G), we asked whether Phlpp1 depletion could reverse this phenotype. Indeed, siRNA-mediated Phlpp1 knockdown in LEX BMDMs resulted in a significant decrease in lysosomal activity and increased S. Typhimurium burden and IL-6 secretion (Fig. 7 D–F). Overall, these results suggest a cross-talk between leptin signaling and lysosome-mediated clearance of Gram-negative bacteria through an mTORC2/Phlpp1/Akt axis in macrophages (Fig. 8).
Fig. 8.
Schematic diagram represents that leptin signaling-dependent activation of mTORC2/Phllp1/Akt pathway abrogates lysosomal function and negatively regulates macrophage defense during S. Typhimurium infection.
Discussion
Metabolic regulation of the innate and adaptive immune responses is an active and expanding area of investigation. In particular, the roles of metabolic hormones and adipokines in regulating the functions of different immune cells are increasingly recognized. Leptin is a pleiotropic hormone that regulates appetite and modulates immunity (30). Distinct roles of leptin signaling describing protective or harmful effects for the host during infection have also been proposed, through the utility of various mouse models (19). In addition, a cohort study found that children from Bangladesh with at least one copy of the leptin receptor 223R mutation, who thus suffer from obesity, are more susceptible to amebic colitis (31). While leptin treatment enhances proinflammatory cytokine secretion in a human macrophage cell line (16), the role of leptin signaling on macrophage function is elusive. In this study, we delineated the functional interplay between leptin signaling and bacterial clearance mechanisms in BMDMs and in vivo using various mouse models.
We first observed marked Lepr induction in murine BMDMs, RAW-264.7 macrophages, and human macrophages upon S. Typhimurium infection. Lepr induction was also noted in response to infection with other Gram-negative bacteria, including S. flexneri, P. aeruginosa, E. coli, and Y. enterocolitica, but not Gram-positive bacteria, such as L. monocytogenes or S. aureus. This finding suggests a specific role for Lepr activation during Gram-negative bacterial infection.
Importantly, we found that Lepr was neither induced by heat-killed S. Typhimurium nor S. Typhimurium-derived LPS; however, it was dependent on Stat3 activation. Consistently, Stat3 is activated only upon Gram-negative pathogens. Together, these data suggest that either Gram-negative bacteria target the leptin signaling pathway to enhance pathogenesis or that activating leptin signaling is a countermeasure employed by macrophages to adapt to the metabolic alterations specific to Gram-negative bacteria.
Previous studies and our own experience using db/db mice have supported the idea that leptin signaling positively regulates immune defense mechanisms against microbial infections (32–34). These db/db mice harbor an additional mutation in the misty gene, now renamed as dock7m, which affects growth (35). Dock7m also regulates the actin cytoskeleton, which is critical for phagosome maturation (36, 37). Mancuso et al. (38) recently showed using leptin-receptor-deficient alveolar macrophages that leptin positively regulates Streptococcus pneumoniae clearance. This finding is consistent with the differential leptin response we observed between Gram-positive and Gram-negative bacteria.
Mancuso et al. (39) earlier showed that leptin-deficient mice (ob/ob) have a compromised ability to defend against the Gram-negative K. pneumoniae, which they attributed to impaired phagocytic activity of alveolar macrophages. By contrast, we show here, using three different mouse models, that inhibiting leptin signaling enhances the ability of macrophages to eliminate S. Typhimurium. Our models included: (i) B6.BKS-Leprdb mice carrying the WT dock allele (40), (ii) myeloid cell-specific Lepr knockout (LEX) mice, and (iii) leptin antagonist-treated WT mice. It is unclear whether leptin has a variant function in alveolar macrophages or if other Gram-negative bacteria, which we have not investigated, differentially regulate leptin signaling similar to Gram-positive bacteria. Discrepancies could also be due to the different mouse models used and altered metabolism associated with the obese phenotype.
We also found an increased number of CD11b+ cells and thus reduced inflammation-mediated tissue damage and cytokine release in the spleens of LEX mice upon infection, which could be attributed to increased survival of LEX macrophages. We previously reported a similar increase in the number of macrophages and reduced bacterial burden in type I IFN receptor-deficient mice infected with S. Typhimurium as they endure pathogen-induced cell death (7). An important finding from our present study is that Lepr-deficient BMDMs have enhanced lysosomal functions, resulting in increased bacterial clearance in vitro and in vivo and reduced inflammation.
Many have highlighted the additional role of lysosomes in sensing metabolic changes and regulating metabolic homeostasis (5). Lysosomes communicate metabolic alterations to metabolic hubs, such as mTORC1, which are assembled and activated on lysosomes (41, 42). Although leptin activates mTORC1 in macrophages (43) and Tregs (44), here we found that mTORC1 was highly active during S. Typhimurium infection despite loss of leptin signaling. Rather, we observed that mTORC2, which also localizes on endosomal membranes, was instead down-regulated in LEX BMDMs. The discrepancy could be due to an infection model tested in this study because S. Typhimurium virulence factors such as sopB are known to activate mTORC1 (8). The partitioning of mTORC2 to endosomes is critical for Akt phosphorylation as it counteracts phosphatases (45). Consistently, Lepr-deficient macrophages showed increased Phlpp1 expression, reduced pAkt-Ser473 levels, and consequently decreased mTORC2-dependent Akt activation. Consistent with previous reports (46), we found that bacterial clearance was dependent on Akt activity, as its inhibition in WT BMDMs led to improved bacterial clearance. Reduction in Akt-activation could also lead to increased autophagy (47), thereby increasing the capture and elimination of S. Typhimurium (48, 49). However, we found that bacterial clearance was dependent on Akt activity, as its inhibition in WT BMDMs led to improved bacterial clearance. These findings corroborate with a study showing lysosomal mTORC2/Phlpp1/Akt signaling regulates chaperone-mediated autophagy in response to cellular stress (20) and a recent report that mTORC2 disrupts lysosomal acidification in macrophages (50). Furthermore, S. Typhimurium can evade cell-autonomous defenses and autophagy by activating Akt (8). Early Akt activation is dependent on SopB, a SPI-I–encoded virulence factor of the pathogen (51), but its constitutive phosphorylation is dependent on focal adhesion kinase (FAK) possibly regulated by a SPI-II–encoded virulence factor (46). Here, we show that the Lepr expression is reduced when macrophages are infected with the SPI-II mutant ssaV. Whether focal adhesion kinase is regulated by leptin during S. Typhimurium infection remains unknown. Additionally, we have shown an important function of Phlpp1 governed by leptin signaling in the context of macrophage defense against S. Typhimurium infection.
Our study has shown that leptin signaling is detrimental to S. Typhimurium infection. We have comprehensively studied leptin signaling applying in vitro conditions and various mouse models upon S. Typhimurium infection. Moreover, we could detect a role for other Gram-negative pathogens in vitro, which needs to be further evaluated in vivo. Although we could show that S. Typhimurium infection also induces leptin signaling in human macrophages, further studies testing the whole idea and its therapeutic potential in a clinical setting are needed.
In conclusion, our study delineates a mechanistic link between leptin signaling and macrophage functions in the context of bacterial infections (Fig. 8). Distorted lysosomal function is a common pathology in obesity, which impairs lipid degradation leading to lipid accumulation in adipose tissue macrophages. Our finding that a leptin antagonist reduces bacterial burden and inflammation support that leptin signaling may be a suitable therapeutic target to treat lysosomal dysfunction disorders. Our study extends the current understanding of leptin signaling in macrophages upon infection and suggests that targeting the leptin signaling pathway could provide a host-targeted strategy to treat infections caused by Gram-negative pathogens.
Methods
Study Approval.
All animal procedures were conducted in accordance with the institutional guidelines on animal welfare and were approved by the North Rhine-Westphalian State Agency for Nature, Environment, and Consumer Protection (Landesamt für Natur, Umwelt and Verbraucherschutz Nordrhein-Westfalen; File nos.: 84-02.05.40.14.082 and 84-02.04.2015.A443) and the University of Cologne, Germany. The study involving human blood samples from healthy donors was approved by the Local Ethics Commission of the University Hospital of Cologne, Germany (file no.: 12-164). Written consent of participants was received from participants before inclusion in the study. All clinical investigations were conducted according to the Declaration of Helsinki principles.
Human Subjects.
After obtaining written consent from healthy donors, blood samples were obtained from two female donor in the age of 20–30. PBMCs were isolated for cell culture ex vivo experiments.
Animals.
For the in vivo study, Lepr-deficient B6.BKS (D)-Leprdb/J (Leprdb) and WT C57BL/6 female mice at the age of 8 to 12 wk were commercially obtained from Charles River Laboratories. Furthermore, a mouse model lacking the leptin receptor in myeloid cells by crossing Leprfl/fl mice with transgenic mice expressing LysMCre+/+ (termed as LEX) and used littermate controls, which did not carry the LysMCre+/+ as previously described, was generated. For the experiments, female animals at the age of 8 to 12 wk (10th generation) were used. Mice were killed with cervical dislocation, and organs were taken for experiment analysis or bone marrow isolation.
Cell Line.
RAW-264.7 mouse macrophage cell line was cultured in RPMI supplemented with 10% fetal bovine serum (FBS) and tested for contamination due to Mycoplasma on a regular basis.
Pathogens.
The S. Typhimurium strain (SL1344) was commercially purchased. The S. Typhimurium ΔssaV and ΔinvA mutants were kindly provided by Ivan Dikic, Institute for Biochemistry II, Goethe University, Frankfurt, Germany. Other pathogens used as controls in this study S. flexneri (American Type Culture Collection [ATCC] 12022), P. aeruginosa (ATCC 27853), Y. enterocolitica (ATCC 9610), E. coli (ATCC 25922), S. aureus (ATCC 29213) or L. monocytogenes (EGD-e) were kindly provided by Georg Plum, Institute for Medical Microbiology, Immunology and Hygiene, University of Cologne, Cologne, Germany, and are used as control strains in the routine patient diagnostics at the Institute of Medical Microbiology, Immunology and Hygiene of the University Hospital Cologne. These strains are registered in ATCC.
Bacteria Preparation.
Bacteria from a single colony was inoculated into 5 mL of brain heart infusion (BHI) medium and incubated overnight at 37 °C with constant agitation. Next, 1 mL of bacterial suspension was transferred into 50 mL of BHI and grown until the OD600 reached 1. The bacteria concentration was estimated by plating serial dilutions on BHI agar plates. To prepare HK, bacteria were inactivated for 10 min at 95 °C.
Cell Culture and Bacterial Infection.
Bone marrow was differentiated into macrophages for 7 to 10 d in RPMI medium supplemented with 20% L929 supernatant and 10% FBS with or without leptin. All experiments used FBS from the same lot containing the same amount of leptin to feed cultured macrophages. BMDMs were infected with specified bacteria at a multiplicity of infection (MOI) of 10. After infection, BMDMs were incubated with the bacteria for 10 min at room temperature (RT) and then 30 min at 37 °C. This incubation time was sufficient for bacteria to be internalized by macrophages. Cells were then washed with RPMI medium and incubated in medium supplemented with 50 µg/mL gentamicin. After 2 h, the gentamicin concentration was reduced to 10 µg/mL BMDMs were treated with 100 ng/mL recombinant mouse leptin (R&D), 10 µM AKT-Inhibitor VIII (Enzo Lifesciences), or 100 ng/mL S. Typhimurium LPS (Sigma) dissolved in medium for 2 h before infection. Human PBMCs were isolated from 30 mL of ethylenediaminetetraacetic acid blood using Lymphoprep (Stem Cells), and the monocytes were differentiated in RPMI containing 5% autologous serum.
Leptin Antagonist Treatment.
Pegylated leptin antagonist (Protein Laboratories Rehovot) was dissolved in sterile H2O and administered (i.p.) or the vehicle (sterile H2O) to C57B6/J WT mice at 6.25 mg/kg once daily from 1 d before infection until day 4 after infection when mice were killed and the spleens were collected. The control mice were administered the same volume of vehicle at the same time points.
Uptake of Fluorescently Labeled Leptin.
RAW macrophages (106) were plated in a six-well dish on the previous day. Next-day infection was performed according the protocol described above. At the desired time points, the cells were washed with sterile phosphate buffered saline (PBS) and treated with 1 nM/mL Cy5-labeled, recombinant leptin (FC5-003-13, Phoenix Pharmaceuticals, Inc.) for 15 min at 37 °C. After washing with PBS, the cells were fixed with 1% formaldehyde for 15 min. Cells were harvested in 1% bovine serum albumin (BSA) for fluorescence-activated cell sorting (FACS) acquisition in a FACSCanto flow cytometer. Analysis was carried out using FlowJo software.
Phagosomal β-Galactosidase Activity Assay.
To assess β-galactosidase activity in phagolysosomes, either S. Typhimurium (SL1344) or red fluorescent beads (Bangs Laboratories) were coated with 5-dodecanoylaminofluorescein Di-β-d-Galactopyranoside (C12FDG, Life Technologies) for 60 min at 37 °C in NaHCO3 buffer (pH 9.6). Infection was carried out at MOI of 10, as described above, or 100 beads per cell were added to BMDMs and incubated for 10 min at RT and then 10 min at 37 °C, followed by washes with RPMI to remove extracellular beads. After 10 min or 0.5, 1, 2, or 4 h, the cells were washed with cold PBS and fixed with 1% formaldehyde for 10 min. After intensive washing, the cells were collected in 1% BSA/PBS and samples were acquired on a FACS Canto flow cytometer (26), and the data were analyzed using FlowJo software. The mean fluorescence intensities (MFIs) of C12FDG was normalized to the red MFI of the beads for every sample.
Phagosomal Proteolytic Activity Assay.
To assess the proteolytic activity in the phagolysosomes, red fluorescent beads were coated with green DQ-BSA (Life Technologies) dissolved in carbodiimide solution (25 mg/mL in PBS) for 30 min at RT. After washing, beads were resuspended in 0.1 M sodium tetraborate decahydrate solution (pH 8.0 in ddH2O) and incubated overnight at RT. Beads were then washed, resuspended in RPMI, and added to BMDMs at 100 beads per cell. After 10-min incubation at RT and 10-min incubation at 37 °C, cells were washed to remove noninternalized beads. After 10 min and 0.5, 1, and 2 h, the cells were washed with cold PBS and resuspended in 1% formaldehyde. Samples were acquired on a BD FACS Canto flow cytometer, and the data were analyzed in FlowJo. The DQ-BSA MFI was normalized to the red MFI of beads for every sample.
S. Typhimurium Phagosome Isolation.
S. Typhimurium was grown in BHI broth until the OD600 reached 1 and then biotinylated with EZ-link NHS-Biotin reagent (Thermo Fisher Scientific). After washing, biotinylated bacteria were incubated with siMAG Streptavidin ferrofluid (Chemicell) at 37 °C. Biotinylated S. Typhimurium bound to Streptavidin ferrofluid was then separated using a magnet, and the bacteria were quantified using BHI agar plates. Subsequently, BMDMs were infected with the biotinylated S. Typhimurium bound to the Streptavidin ferrofluid at MOI of 10. At each time point, S. Typhimurium-containing phagosomes were isolated using equilibration and lysis buffer, as previously described for bead phagosome isolation (52).
In Vitro Bacterial Burden and Enzyme-Linked Immunosorbent Assay (ELISA).
After 30 min and 24 h postinfection, BMDMs were lysed with 1% Triton X-100, 0.01% SDS in PBS. Several dilutions of the lysate were plated on BHI plates and incubated overnight at 37 °C. The next day, S. Typhimurium colony forming units (CFU) were enumerated. Supernatants were collected and analyzed for IL-6, TNF-α, and IL-1β secretion using ELISA kits (R&D, DY406-05, DY410-05, DY2786-05) or Proteom Profiler Mouse Chemokine Array (R&D, ARY020) according to the manufacturer’s instructions.
Estimation of Bacterial Burden In Vivo.
Mice were infected (i.v.) with 100 CFU S. Typhimurium. After 4 d of infection, mice were euthanized. The spleen and liver were isolated, weighted, and homogenized using a gentleMACS Dissociator (Miltenyi Biotec) in sterile PBS. Extracts of the homogenized livers were plated on BHI Agar plates. After 24-h incubation at 37 °C, the bacterial colonies were enumerated. The number of colonies was normalized to per gram of tissue. Spleen and liver extracts were also used for ELISA (described above) and stained for CD11b, and subsequently analyzed by FACS.
Tissue Histology.
Spleen and liver samples were obtained from in vivo experiments and cross-sectioned in equal-sized slices 2 mm in width. The respective slices were snap-frozen in liquid nitrogen covered with Tissue-Tek and stored at −80 °C until further use. Cryostat sections were cut to 3 µm and attached to adhesive glass slides (Dako), fixed with 4% phosphate-buffered formalin for 30 s, and stained with hematoxylin and eosin. After rinsing in water, the slides were dehydrated via an ascending ethanol gradient, immersed in xylene, and mounted on glass coverslips. The tissue sections were imaged using a Hitachi HV-F202 UXGA 3CCD camera attached to Leica DM5500B microscope. Pictures were generated using the DISKUS program (Hilgers Technisches Büro).
Western Blotting.
BMDMs were lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with a 1× protease/phosphatase inhibitor mixture (Thermo Fisher Scientific). Protein concentrations were estimated using Pierce BCA Protein Assay Kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Equal amounts of protein were separated on 10% or 12% SDS/PAGE gels. The proteins were then transferred onto a polyvinylidene difluoride membrane and probed with the following antibodies: Lepr (ab5593, Abcam), phospho-Stat3 (9138, Cell Signaling), Stat3 (9139, Cell Signaling) Lamp-1 (sc-17768, Santa Cruz Biotechnology), phospho-Akt Ser473 (4060, Cell Signaling), phospho-Akt Thr-308 (2965, Cell Signaling), Akt (4691, Cell Signaling), phospho-Ndrg1 Thr-346 (3271, Cell Signaling), Rictor (2140, Cell Signaling), Phllp1 (07-1341, Merck Millipore), phospho-mTOR Ser-2448 (2971, Cell Signaling), phospho-s6K (9205, Cell Signaling) and Gapdh (2118S, Cell Signaling). Gapdh was used as a loading control. For phagosome samples, Ponceau staining was used as loading control (sc-301558, Santa Cruz Biotechnology). After incubation with secondary horseradish peroxidase-conjugated antibodies (R&D), the blots were developed using enhanced chemiluminescence reagent (GE Healthcare).
Immunofluorescence Staining and Confocal Microscopy.
BMDMs grown on glass coverslips were infected with S. Typhimurium and fixed with 4% formaldehyde in PBS for 15 min at RT. When indicated, the cells were treated with LysoTracker (L12492, Life Technologies) or Cy5-labeled, recombinant leptin (FC5-003-13, Phoenix Pharmaceuticals) before fixation at the indicated time points. The samples were then permeabilized with 0.3% Triton X-100 in PBS for 5 min and then incubated with Image-iT R FX (I36933, Invitrogen) and blocked with 5% (wt/vol) normal goat serum (Life Technologies). The cells were then incubated overnight with specific primary antibodies against Ob-R (Ab5593 Abcam) and S. Typhimurium LPS (MA1-83451, Thermo Fisher Scientific) at 4 °C. After washing with 0.03% Triton in PBS, the cells were incubated with either Alexa Fluor 594-conjugated goat anti-rabbit or anti-mouse secondary antibody or Alexa Fluor 488-conjugated goat anti-rabbit or anti-mouse (Life Technologies) for 1 h at RT protected from light. The coverslips were then mounted using ProLong Gold antifade DAPI (P36953, Life Technologies) to stain the nuclei. The cells were imaged under a confocal microscope (Olympus IX81) with an UPlanSApo 60× N.A. 1.40 oil objective lens or 60× oil PlanApo objective, numerical aperture 1.4 at RT on an Olympus IX81 inverted confocal microscope equipped with photomultiplier tube detectors for imaging. Olympus Fluoview FV10-ASW 4.2 software was used for acquisition.
Rictor, Phlpp1, and Stat3 siRNA Knockdown.
BMDMs were transfected with either 50 nM nontargeting siRNA (SR-CL000-005, Eurogentec) or siRNA for Rictor (L-064598-01-0005, Dharmacon), Phlpp1 (L-058853-00-0005, Dharmacon) or Stat3 (L-040794-01-0005, Dharmacon) using the transfection reagent Lipofectamine 3000 (Life Technologies), according to the manufacturer’s instructions.
qRT-PCR.
Total RNA from BMDMs (106 cells per well) was isolated using an RNeasy Mini Kit (74106, Qiagen) and cDNA (500 ng) was synthesized with random hexamers using reverse transcriptase (SuperScript III; 18080-044, Invitrogen). Primers (Invitrogen) were designed using Primer3 software and Basic Local Alignment Search Tool (National Center for Biotechnology Information). PCRs (20 µL) contained 10 ng of cDNA, 0.4 µmol/L of each forward and reverse primer, and master mix (SsoFast EvaGreen Supermix; 1725201, Bio-Rad). Real-time PCR was performed on a Bio-Rad CFX Touch Real-time PCR System with the following conditions: initial denaturation step of 95 °C for 2 min, 40 cycles of 95 °C for 5 s, and 60 °C for 15 s, followed by a denaturation step of 95 °C for 60 s and subsequent melt curve analysis to check amplification specificity. The data were analyzed using the Ct method with hypoxanthine-guanine phosphoribosyltransferase (Hprt) as the endogenous reference gene for all reactions. The relative mRNA levels of uninfected BMDMs were used as normalized controls for infected BMDMs. All assays were performed in triplicate, and a nontemplate control was included in all experiments to exclude DNA contamination. The primer sequences were as follows:
Hprt (for) 5′-GTTGGATACAGGCCAGACTTTGTTG-3′
Hprt (rev) 5′-GATTCAACTTGCGCTCATCTTAGGC-3′
Lepr (for) 5′-TGCTTTGGGAATGAGCAAGG -3′
Lepr (rev) 5′-AGCTGGCGAAAAACTGAAGC-3′
Cell Viability Assay.
BMDMs (0.1 × 106 cells per well) were plated into a 96-well plate. Cell viability was measured at the indicated time points using CytoTox 96 Lactate Dehydrogenase Assay (G1780, Promega), according to manufacturer’s instructions. Luminescence was measured on a multimode plate reader (Perkin-Elmer).
Statistical Analysis.
Statistical analyses were performed using GraphPad Prism software (Version 5.0b). A two-tailed Student’s t test was conducted for most datasets unless specified otherwise, to determine statistical significance. All data are represented as the means ± SEM. For all tests, a P value < 0.05 was considered statistically significant (*P < 0.05; **P < 0.01; ***P < 0.005).
Supplementary Material
Acknowledgments
We thank the N.R. laboratory members for helpful discussion. We thank Zahra Hejazi, Monika Keiten-Schmitz, Jennifer Klimek, Julia Cassens, and Sara Khorasani for technical support. This work was supported by funding from DZIF and Cologne Fortune Starter Grant (to J.F.) and from CECAD (funded by the Deutsche Forschungsgemeinschaft [DFG] within the Excellence Initiative by the German federal and state governments), Köln Fortune, and Maria-Pesch, University clinic, Cologne, grants from DFG (SFB 670) (to N.R.). The LepRfl/fl mice were kindly provided by Jens C. Brüning and Streamson Chua, who was funded by NIH New York Nutrition Obesity Research Center Grant 1PO1DK26687. Moreover, we thank Dr. Dominik Fischer for technical support with the graphical abstract and Dr. Jessica Tamanini (Insight Editing London) for editing the manuscript prior to submission.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.V.-T. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904885116/-/DCSupplemental.
References
- 1.Laxminarayan R., et al. , Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 13, 1057–1098 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Tacconelli E., et al. ; WHO Pathogens Priority List Working Group , Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Monack D. M., Mueller A., Falkow S., Persistent bacterial infections: The interface of the pathogen and the host immune system. Nat. Rev. Microbiol. 2, 747–765 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Ginhoux F., Guilliams M., Tissue-resident macrophage ontogeny and homeostasis. Immunity 44, 439–449 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Lim C. Y., Zoncu R., The lysosome as a command-and-control center for cellular metabolism. J. Cell Biol. 214, 653–664 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ilyas B., Tsai C. N., Coombes B. K., Evolution of salmonella-host cell interactions through a dynamic bacterial genome. Front. Cell. Infect. Microbiol. 7, 428 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson N., et al. , Type I interferon induces necroptosis in macrophages during infection with Salmonella enterica serovar Typhimurium. Nat. Immunol. 13, 954–962 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganesan R., et al. , Salmonella Typhimurium disrupts Sirt1/AMPK checkpoint control of mTOR to impair autophagy. PLoS Pathog. 13, e1006227 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaspersen K. A., et al. , Obesity and risk of infection: Results from the Danish blood donor study. Epidemiology 26, 580–589 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Maccioni L., et al. , Obesity and risk of respiratory tract infections: Results of an infection-diary based cohort study. BMC Public Health 18, 271 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cildir G., Akıncılar S. C., Tergaonkar V., Chronic adipose tissue inflammation: All immune cells on the stage. Trends Mol. Med. 19, 487–500 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Kornberg M. D., et al. , Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 360, 449–453 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mills E. L., et al. , Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113–117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O’Neill L. A., Kishton R. J., Rathmell J., A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 16, 553–565 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mauer J., et al. , Myeloid cell-restricted insulin receptor deficiency protects against obesity-induced inflammation and systemic insulin resistance. PLoS Genet. 6, e1000938 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsiotra P. C., Boutati E., Dimitriadis G., Raptis S. A., High insulin and leptin increase resistin and inflammatory cytokine production from human mononuclear cells. BioMed Res. Int. 2013, 487081 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naylor C., et al. , Leptin receptor mutation results in defective neutrophil recruitment to the colon during Entamoeba histolytica infection. MBio 5, e02046-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamas B., et al. , Leptin modulates dose-dependently the metabolic and cytolytic activities of NK-92 cells. J. Cell. Physiol. 228, 1202–1209 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Naylor C., Petri W. A. Jr, Leptin regulation of immune responses. Trends Mol. Med. 22, 88–98 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Arias E., et al. , Lysosomal mTORC2/PHLPP1/Akt regulate chaperone-mediated autophagy. Mol. Cell 59, 270–284 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inagaki-Ohara K., et al. , Enhancement of leptin receptor signaling by SOCS3 deficiency induces development of gastric tumors in mice. Oncogene 33, 74–84 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Cascio S., et al. , Expression of angiogenic regulators, VEGF and leptin, is regulated by the EGF/PI3K/STAT3 pathway in colorectal cancer cells. J. Cell. Physiol. 221, 189–194 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Abram C. L., Roberge G. L., Hu Y., Lowell C. A., Comparative analysis of the efficiency and specificity of myeloid-Cre deleting strains using ROSA-EYFP reporter mice. J. Immunol. Methods 408, 89–100 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radigan K. A., et al. , Impaired clearance of influenza A virus in obese, leptin receptor deficient mice is independent of leptin signaling in the lung epithelium and macrophages. PLoS One 9, e108138 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elinav E., et al. , Pegylated leptin antagonist is a potent orexigenic agent: Preparation and mechanism of activity. Endocrinology 150, 3083–3091 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson N., et al. , Mycobacterial phenolic glycolipid inhibits phagosome maturation and subverts the pro-inflammatory cytokine response. Traffic 9, 1936–1947 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Procaccini C., et al. , Leptin-induced mTOR activation defines a specific molecular and transcriptional signature controlling CD4+ effector T cell responses. J. Immunol. 189, 2941–2953 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M., Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Saric A., et al. , mTOR controls lysosome tubulation and antigen presentation in macrophages and dendritic cells. Mol. Biol. Cell 27, 321–333 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pérez-Pérez A., et al. , Role of leptin as a link between metabolism and the immune system. Cytokine Growth Factor Rev. 35, 71–84 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Duggal P., et al. , A mutation in the leptin receptor is associated with Entamoeba histolytica infection in children. J. Clin. Invest. 121, 1191–1198 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park S., Rich J., Hanses F., Lee J. C., Defects in innate immunity predispose C57BL/6J-Leprdb/Leprdb mice to infection by Staphylococcus aureus. Infect. Immun. 77, 1008–1014 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodgson K. A., Morris J. L., Feterl M. L., Govan B. L., Ketheesan N., Altered macrophage function is associated with severe Burkholderia pseudomallei infection in a murine model of type 2 diabetes. Microbes Infect. 13, 1177–1184 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Lemos M. P., Rhee K. Y., McKinney J. D., Expression of the leptin receptor outside of bone marrow-derived cells regulates tuberculosis control and lung macrophage MHC expression. J. Immunol. 187, 3776–3784 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Truett G. E., Tempelman R. J., Walker J. A., Wilson J. K., Misty (m) affects growth traits. Am. J. Physiol. 275, R29–R32 (1998). [DOI] [PubMed] [Google Scholar]
- 36.Garrity P. A., et al. , Drosophila photoreceptor axon guidance and targeting requires the dreadlocks SH2/SH3 adapter protein. Cell 85, 639–650 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Jahraus A., et al. , ATP-dependent membrane assembly of F-actin facilitates membrane fusion. Mol. Biol. Cell 12, 155–170 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mancuso P., et al. , Ablation of the leptin receptor in myeloid cells impairs pulmonary clearance of Streptococcus pneumoniae and alveolar macrophage bactericidal function. Am. J. Physiol. Lung Cell. Mol. Physiol. 315, L78–L86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mancuso P., et al. , Leptin-deficient mice exhibit impaired host defense in Gram-negative pneumonia. J. Immunol. 168, 4018–4024 (2002). [DOI] [PubMed] [Google Scholar]
- 40.The Jackson Laboratory , Mouse Strain Datasheet - 00697. https://www.jax.org/strain/000697. Accessed 7 November 2019 (2018).
- 41.Sancak Y., et al. , The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sancak Y., et al. , Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maya-Monteiro C. M., et al. , Leptin induces macrophage lipid body formation by a phosphatidylinositol 3-kinase- and mammalian target of rapamycin-dependent mechanism. J. Biol. Chem. 283, 2203–2210 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Procaccini C., et al. , An oscillatory switch in mTOR kinase activity sets regulatory T cell responsiveness. Immunity 33, 929–941 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ebner M., Sinkovics B., Szczygieł M., Ribeiro D. W., Yudushkin I., Localization of mTORC2 activity inside cells. J. Cell Biol. 216, 343–353 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Owen K. A., Meyer C. B., Bouton A. H., Casanova J. E., Activation of focal adhesion kinase by Salmonella suppresses autophagy via an Akt/mTOR signaling pathway and promotes bacterial survival in macrophages. PLoS Pathog. 10, e1004159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmieri M., et al. , mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat. Commun. 8, 14338 (2017). Erratum in: Nat. Commun.8, 15793 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia K., et al. , Autophagy genes protect against Salmonella typhimurium infection and mediate insulin signaling-regulated pathogen resistance. Proc. Natl. Acad. Sci. U.S.A. 106, 14564–14569 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Birmingham C. L., Smith A. C., Bakowski M. A., Yoshimori T., Brumell J. H., Autophagy controls Salmonella infection in response to damage to the Salmonella-containing vacuole. J. Biol. Chem. 281, 11374–11383 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Monteith A. J., et al. , mTORC2 activity disrupts lysosome acidification in systemic lupus erythematosus by impairing caspase-1 cleavage of Rab39a. J. Immunol. 201, 371–382 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tattoli I., et al. , Amino acid starvation induced by invasive bacterial pathogens triggers an innate host defense program. Cell Host Microbe 11, 563–575 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Gutiérrez S., Wolke M., Plum G., Robinson N., Isolation of Salmonella typhimurium-containing phagosomes from macrophages. J. Vis. Exp. e56514 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.