Significance
Metagenomic or social genetic effects—how the genetic makeup of organisms around an individual impacts an individual’s phenotype—have been demonstrated for animals. However, in humans, where experimental manipulation is less feasible, such metagenomic effects of peers have largely been suggestive. Here, we leverage as-if-random variation in same-grade peers to document metagenomic effects on adolescent smoking outcomes. The effect of grade-mates’ genotypes is larger in magnitude than many other predictors of smoking, including ego’s own genotype, being male, and family income. Further, a minority with high genetic propensities to smoke can affect the smoking behavior of an entire grade. This approach offers a way to integrate genetic and environmental influences on human phenotypes.
Keywords: sociogenomics, adolescent smoking, social genetic effects
Abstract
Recent scholarship suggests that the genomes of those around us affect our own phenotypes. Much of the empirical evidence for such “metagenomic” effects comes from animal studies, where the socio-genetic environment can be easily manipulated. Among humans, it is more difficult to identify such effects given the nonrandom distribution of genes and environments. Here we leverage the as-if-random distribution of grade-mates’ genomes conditional on school-level variation in a nationally representative sample. Specifically, we evaluate whether one’s peers’ genetic propensity to smoke affects one’s own smoking behavior net of one’s own genotype. Results show that peer genetic propensity to smoke has a substantial effect on an individual’s smoking outcome. This is true not only when the peer group includes direct friends, and therefore where the individual plays an active role in shaping the metagenomic context but also when the peer group includes all grade-mates and thus in cases where the individual does not select the metagenomic environment. We explore these effects further and show that a small minority with high genetic risk to smoke (‘bad apples’) can greatly affect the smoking behavior of an entire grade. The methodology used in this paper offers a potential solution to many of the challenges inherent in estimating peer effects in nonexperimental settings and can be utilized to study a wide range of outcomes with a genetic basis. On a policy level, our results suggest that efforts to reduce adolescent smoking should take into account metagenomic effects, especially bad apples, within social networks.
Tobacco use habits are largely established during adolescence; indeed, 9 out of 10 smokers first try cigarettes before age 18 (1). Scholars have approached the question of which youth adopt smoking behavior from 2 perspectives. The first emphasizes individual’s biological tendencies that reward nicotine consumption (2) and in particular their genetic architecture that encodes that reward system (3). Under this framework variation in tobacco usage and likelihood of developing a nicotine dependency result from individual genetic differences shaping the biological responses to nicotine consumption.
The second approach emphasizes instead the role the social environment plays in governing the availability of cigarettes (4) and in acculturating individuals to smoke (5). For adolescents, the social environment may include many different contexts: the neighborhood (6), school (7), their grade (8), friends (9), and family (10). Across these domains, same-age peers have been shown to be particularly influential: having coeval peers who smoke is associated with increased individual smoking (11). This effect holds for many different types of peers, with the literature showing a robust association between the smoking behavior of one’s schoolmates, classmates, and close friends and one’s own smoking behavior (12, 13). Limited experimental evidence suggests that these associations are not epiphenomenal (14).
Despite the respective power of genetics and peers in accounting for variation in which adolescents smoke, little prior research has sought to integrate the two—that is, to simultaneously identify genetic influence, peer effects and the interaction between the 2 as they influence adolescent tobacco use (15). One possible avenue for integration, and the one explored in this paper, is the analysis of metagenomic effects—also referred to as “social genetic effects” or “indirect genetic effects” in animal studies (16–18)—that is, the extent to which others’ genes affect an individual’s outcomes. We borrow the metaphor of metagenomics from research on the microbiome where all organisms in the microbiome are genotyped to capture the collective genomic context which is then used to understand individual outcomes. In the case here, the metagenome, or the collective genomic context, consists of the genomes of all of one’s peers. Since smoking behavior has a genetic basis, and since others’ behaviors may matter for individual behaviors and outcomes, we should not be surprised to observe metagenomic effects. Indeed, a recent experimental study on mice found that randomly assigned cage-mates’ genotypes affect individual mice’s outcomes, accounting for as much as 29% of the phenotypic variance among the mice (19).
However, for humans in particular, extant evidence for metagenomic effects is limited. Only a few studies to date have seriously considered the question and thus for most outcomes, including smoking, there is little credible evidence that such effects exist. Specifically, in the absence of careful study design, genes and environments can act as mutual proxies, confounding the researcher’s attempts to parse their effects (20). Because of these difficulties, the few empirical studies that do consider the question of metagenomic in humans are, at best, suggestive (21, 22). Perhaps the most convincing evidence for metagenomic effects in humans comes from research that has identified the important impact of parental and sibling genotypes on an individual’s health and educational outcomes, in a dynamic called “genetic nurturance” (23, 24). Outside the family environment, however, empirical evidence remains scarce; that said, research from behavioral genetics has consistently found that nonshared environment is more salient to most phenotypes than is the shared (read: family) environment of siblings (25, 26). This may suggest that metagenomics effects are not limited to genetic nurturance in the household context.
In this study, we explore the metagenomic effects of smoking. We use data from the National Longitudinal Study of Adolescent to Adult Health (Add Health), a nationally representative cohort study of the health and behavior of US students in grades 7 through 12, first interviewed in 1994 to 1995 across a sample of 132 middle and high schools in the United States (27). In wave IV of the data collection, in 2008 and 2009, DNA specimens were collected and archived. With the advent of genotyping technology, these samples were later genotyped and linked to the survey data in 2017. This results in a one-of-a-kind dataset, which ties adolescents and their genomes to their health and behaviors within the group setting most salient to their young lives: schools.
The basic analytic approach involves predicting an individual’s smoking behavior as a function of the genetic risk for smoking in their metagenomic environment, operationalized as the mean polygenic score (PGS) for smoking of their peers, controlling for possible confounders (such as population stratification) and the individual’s own genetic risk (see Materials and Methods and SI Appendix for more information). In a second set of analyses, we relax the mean polygenic score constraint and explore potential nonlinear effects.
By using unchanging peer genotypes as our independent variable, we overcome 2 common problems in peer effects research: the reflection bias, where peer and individual behavior are observed at the same moment such that influence cannot be adjudicated, and the exclusion bias, where the fact that you cannot be friends with yourself induces a mechanical, negative correlation between peer and individual behavior (28). That said, other potential biases thwart easy estimation. Peers and individuals share the same environment and therefore may adopt similar behaviors (contextual bias) (29, 30). For example, if people with particular genotypes attend similar kinds of schools, our estimation will be biased (31, 32). To ward against context effects, we include grade and school fixed effects, leveraging random, grade-cohort fluctuations in the genetic landscape to identify metagenomic effects.
Peers may also select into relations with one another, whether in being friends or classmates, on the basis of their genetic factors even within the context of a single grade. The risk that our results will be biased by this form of genetic selection will therefore depend on how we define peers. Because adolescents are embedded in multiple social groups even within the context of a single grade, peers can be defined in a myriad of ways. We explore 3 such groups: the grade (all of the other students with whom one shares a grade, or grade-mates), the classroom (all of the other students with whom one shares classes, or classmates; see Materials and Methods for a description of how this is ascertained), and the friendship group (all of the friends one nominated or from whom one received a friendship nomination, or friends).
At the most macrolevel, we explore the relationship between the average genetic predispositions toward smoking of an adolescent’s grade-mates to the adolescent’s own smoking behavior. We posit that conditional on school-level variation, the distribution of genetic propensity to smoke is as-if-random in a given grade. Because genes are assigned at birth and being in a given grade within a school is roughly defined by the year and month of birth, it is reasonable to assume the absence of selection into the grade environment based on an individual’s smoking genes. We show evidence for this assumption in the SI Appendix. Although smoking has been shown to be correlated with lower educational performance (33) and may therefore also be correlated in being held back a grade, there is no evidence of a nation-wide trend in individuals being held back in some years but not others. We therefore posit that the grade-mate estimates are unbiased. In a second set of analyses, we evaluate the extent to which the observed metagenomic effects are social in nature and operate through the actual smoking behavior of grade-mates using an instrumental variable analysis (SI Appendix, Table S4). While this analysis is likely underpowered, it suggests a way to link genetic and social theories, which could prove fruitful as more and larger datasets incorporate social, relational and genetic data.
We then explore the relationship between the average genetic predispositions of an adolescent’s classmates and friends and the adolescent’s smoking behavior. Unlike the grade-mate analyses, these models are descriptive in nature due to the presence of genetic homophily, but they are still analytically useful for contextualizing the grade-mate results presented earlier and are substantively interesting in themselves. The grade, while perhaps not considered the most salient peer group in the sociological literature, offers us the chance to identify causal effects free from selection bias; effects of classroom peers and friends are subject to confounding due to selection; thus, we consider the grade-mates effects the purest estimate of metagenomic peer effects. The grade as an environment is also theoretically interesting because it is an ecology within which individuals in a birth cohort interact, form groups, and learn behaviors. Examining the entire grade relaxes the assumption that the diffusion of behaviors is restricted to dyadic interactions and that social contagion works like a contagion of viruses (34, 35).
Our framework allows us to explore interesting and potentially superlinear effects of both peer behaviors and peer genes on individual behavior. For instance, we might expect individuals with the highest or lowest levels of genetic risk of smoking in a given grade to have a disproportionate impact on individual smoking compared with the impact of the average grade-level genetic risk. Research on the effects of grade and classroom composition on individual outcomes has pointed to the possibility that the presence of an exceptionally excellent student may lead to positive outcomes for everyone in the class (the “shining light” model) (36). Alternatively, one or a few exceptionally disruptive students may evince negative outcomes for the entire grade/class (the “bad apple” model) (37, 38). We test these contextual dynamics empirically by examining whether the proportion of individuals within a grade who fall in the top or bottom decile of the overall smoking polygenic score distribution, has a significantly different impact on individual smoking compared with what the grade-level average PGS indicates.
Overall, our results show that adolescents’ smoking behavior is strongly predicted by the genetic propensity of their peers to smoke. This holds at multiple different levels, whether peers are conceptualized as grade-mates or friends. Further, adolescents appear to be particularly affected by peers who are in the top decile of smoking PGS, while peers in the bottom decile offer little by way of protection.
Results
Metagenomic Effects.
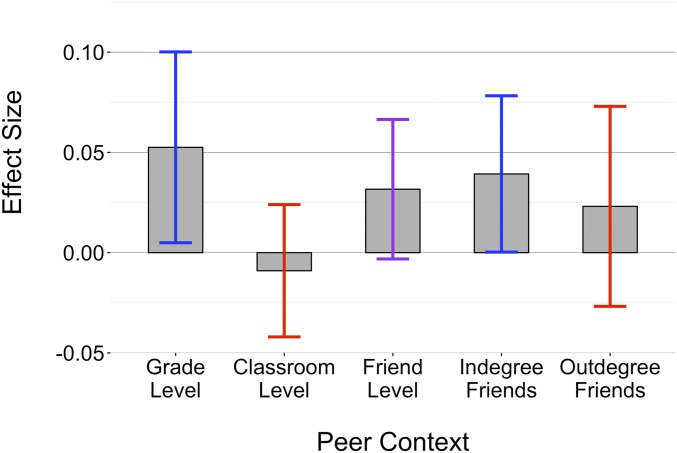
In the results presented in Fig. 1, we examine the association between the metagenomic environment, defined as grade-mates’, classmates’, and friends’ smoking polygenic scores, and individual smoking behavior. Before the analyses were run and the contextual variables constructed, polygenic scores were residualized on the first 4 principal components for the genetic data to ward against population stratification. Grade-mates include all others in an adolescent’s grade, classmates include people with whom the adolescent commonly shared classes, and friends include any student who the adolescent nominated as a friend and anyone who nominated the adolescent as a friend. Full descriptions about how grade-mates, classmates, and friends were identified and defined can be found in the Materials and Methods section. For each model, we control for race and the smoking behavior of family members, to ward against potential confounds related to population stratification, and a number of contextual variables shown to be associated with smoking behavior, both on the individual and respective peer level. That is to say, models at the grade-level control for the attributes and behaviors of the grade-mate peers, while models at the friend-level control for those very same attributes but averaged across a person’s friend group. When considering peers at a more macrolevel such as the grade, we do not exclude friend or classroom clusters because we are interested in the overall effect of the metagenomic environment and not the effect of the grade net of the friends and shared classroom contexts. The focal individual is left out of the calculation of every peer-level calculation of the mean both for the independent variable (average smoking PGS levels) and for the peer-level control variables. Thus, unlike a classical hierarchical model, the number of observations in each regression reflects the total number of students in Add Health who have membership in a grade, classroom, or friendship group, rather than the number of grades, classrooms or friendship groups in the data. That said, all models correct SEs for school-grade clusters.
Fig. 1.
This figure presents metagenomic effect sizes for smoking across different operationalizations of the peer context. Bars signify the magnitude of the coefficient for each context. CIs at the 95% are colored according to significance: blue if significant at a 0.05 level, purple if significant at a 0.10 level, and red otherwise.
For 3 of the 5 models, we find that the average levels of smoking polygenic scores among peers is positively and, depending on the model, at least marginally, significantly associated with individual smoking behavior. This association is positive and significant at the grade-level, which is the only causally identified model. It is marginally significant for friends. That the CIs of the grade- and friend-level models are overlapping suggests they do not significantly differ from one another. In the SI Appendix, we explore potential causes for the difference in magnitude between the grade-level and friend-level effects. Breaking down friendships by in-degree friends (those who nominated an individual) and out-degree friends (those whom an individual nominated) shows that in-degree friends’ average smoking PGS is positively and significantly associated with smoking behavior while out-degree smoking PGS is not. Classmates’ smoking PGS is not significantly associated with individual smoking outcomes, likely indicating that classmates are not a highly relevant social context and that classmates are inconsequential for one’s smoking behavior.
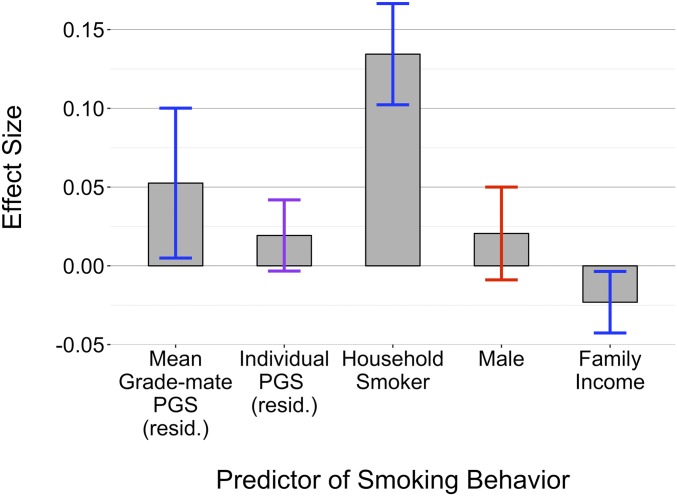
In Fig. 2, we plot the grade-level metagenomic effect alongside other coefficients included in the grade-level model, all of which have been identified as important determinates of adolescent smoking, including: individual smoking polygenic score (15), being male (39), having a household member that smokes (8), and family income (40). The sizable metagenomic effect represents a form of genetic nurturance that is “horizontal” in nature, in contrast to the vertical nurturance parental genetic nurturance documented by Kong et al. (23) and which may be driving the strong effect that we observe having a household member that smokes has on one’s smoking behavior.
Fig. 2.
This figure compares the metagenomic effect size for smoking at the grade-level to other predictors in the grade-level model. Since all of these coefficients come from the same model, the effects are conditional on each other. Polygenic scores, whether at the individual- or group-level (i.e., grade, classmates, and friends), were residualized on the first 4 genetic principal components. Bars signify the magnitude of the coefficient for each variable. CIs at the 95% are colored according to significance: blue if significant at a 0.05 level, purple if significant at a 0.10 level, and red otherwise.
Having shown a robust association between peer genes and individual smoking outcomes, we can utilize this association to answer age-old questions about peer effects. The study of peer effects, that is, the causal effect of peers on individuals’ behaviors, beliefs or outcomes, suffers from a set of difficulties that thwart its estimation: specifically, the reflection problem, contextual bias, and selection bias (see Materials and Methods for more detail). Because genes are assigned at birth and being in a given grade is roughly defined by one’s age, we argue that the distribution of genetic propensity to smoke in a given grade is as-if-random conditional on the school attended. This provides a plausible identification strategy for the estimation of the effect of peer smoking on individual smoking.
To estimate the peer effects of smoking behavior, we use the average level of genetic predisposition toward smoking within a school and grade as an instrument for the levels of smoking behavior in that grade. This instrument is then used to predict an individual student’s smoking behavior. We find a positive and significant association between peer behavior, instrumented by grade-mates’ genes, and individual smoking behavior. The coefficient is roughly the same as that of the metagenomic effects at the grade level signaling that the effect of peer genes indeed goes through peer behavior. The full table of the results can be found in the SI Appendix. Although this analytical technique provides a simple and useful framework for the study of peer effects, we may be underpowered to find robust results using two-staged least square analysis (see SI Appendix for more details).
Superlinear Metagenomic Effects.
The models above have repeatedly shown an association of higher peer smoking genetic risk and individual smoking outcomes. This effect, however, might not be strictly linear. Individuals who have the highest genetic predisposition to smoking could have a disproportionate impact on an individual’s smoking. We examine this possibility by replacing the average level of genetic risk of smoking by the proportion of the grade that has a top or bottom decile smoking PGS (see Materials and Methods for how the deciles were calculated), which we refer to as “bad apples” and “shining stars” models, respectively, following previous literature (36).
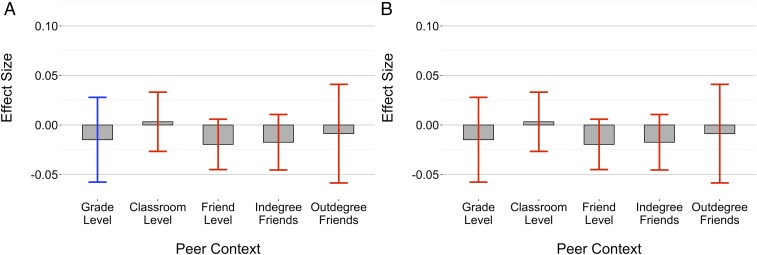
The results can be found in Fig. 3, where Fig. 3A presents a coefficient plot for the bad apple regressions and Fig. 3B presents a coefficient plot for the shining star regressions. Proportion bad apples is positively associated with one’s own smoking across all of the different operationalizations of the peer group except for the classroom, but only significantly so at the grade-level. Proportion shining stars is negatively associated with one’s own smoking across all of the different operationalizations of the peer group except for the classroom, but never significantly so. Therefore, ignoring significance levels the direction of the coefficients, the results generally paint a similar picture across all levels except for the classroom: higher proportions of bad apples are positively associated with smoking behavior, while higher proportions of shining stars do not appear to have a suppressive effect.
Fig. 3.
This figure shows the effect of bad apples and shining stars on smoking behavior across different peer contexts. A compares the effect of bad apples across different peer contexts, while B does the same for shining stars. Bars signify the magnitude of the coefficient for each context. CIs at the 95% are colored according to significance: blue if significant at a 0.05 level, purple if significant at a 0.10 level, and red otherwise.
Conclusion
Smoking behavior has long been thought to have both social and genetic etiologies. Our findings suggest that genes should be treated as an important part of the social environment and the social environment must be considered when thinking about genetic influence. In this paper, we show a robust association between the metagenomic environment and individual outcomes. Individuals’ smoking habits are associated with the smoking PGS of their peers across multiple social contexts: the grade, the friendship group and in-degree friends (those who nominated the individual as a friend). We find grade-mate smoking behavior to possibly be a stronger predictor of individual smoking than even the individual’s own genetic contributions.
Finally, we explored the contextual dynamics of metagenomic effects of smoking and find that the presence of bad apples in the peer context, that is, those in the top decile of smoking genetic risk, increases adolescents’ smoking likelihoods. More work should be done to understand under what conditions the effect of bad apples can be mitigated (or, for that matter, the effect of shining stars can be amplified), including how forces such as the grade friendship network structure, school’s administrative activities, or state’s cigarette taxation rates might moderate their effect.
Our design affords insight into the nature of peer influence and its relationship to genetic effects. It also has substantial implications for future research, even beyond the case of smoking. That an individual’s smoking behavior, and ultimately health outcomes, are affected by his or her peers’ genes is important to consider for understanding social multiplier effects. Further, our framework relies only on the assumption of random variation in the local metagenomic environment and, as a result, can be used to estimate both metagenomic effects and peer effects for any behavior that has a genetic basis. This is especially valuable given the difficulty of estimating causal effects of peers, whether they result from their behaviors or genes. Future work may seek to apply this method to other socially driven outcomes, including health outcomes, that display an element of contagion and are in part influenced by genetic disposition—be that actual communicable disease as driven by the immunological profiles of peers around us, or depression and suicide. In the meantime, public health efforts to reduce smoking update among adolescents would be wise to take account of peer environments in designing interventions.
Materials and Methods
The data for this study come from Add Health, a nationally representative cohort study on the health and behavior of adolescent school children first interviewed in 1994 to 1995 across a sample of 132 middle and high schools across the United States (41). An in-school survey was administered to every student present at each of the 132 schools asking them to self-report their social and health behaviors, friends, and family and school context. A follow-up in-home survey was administered to a random sample of the students in each school, which entailed a far more detailed questionnaire about their behaviors, attitudes, health, parents, siblings, and home life. Information on how to obtain the Add Health data files is available on the Add Health website (https://www.cpc.unc.edu/projects/addhealth).
During the fourth wave of data collection in 2008 to 2009, 96% of the respondents who participated in the Wave IV in-home survey (n = 15,159) agreed to provide biospecimen (in the form of saliva and capillary whole blood samples) to be immediately genotyped for specific single-nucleotide polymorphisms (SNPs) and candidate genes; of those individuals, 80% (n = 12,254) agreed to have their biospecimen archived for future use. With the more recent development of genotyping technology and the resulting decrease in cost, the archived samples were genotyped on about one million genetic markers, which provide genome-wide data for polygenic score construction (42). Two Illumina platforms were used for genotyping Add Health genetic data. Illumina Human Omni1-Quad BeadChip was used for roughly 80% of the sample and includes over 1.1 million genetic markers. Illumina Human Omni-2.5 Quad BeadChip was used for the remainder of the sample and includes 2.5 million markers. A series of quality control procedures were performed on the SNP and individual level. Genetic markers with call rates <90% and minor allele frequency <0.5% were excluded. SNPs with Hardy–Weinberg Equilibrium P value < 5 × 10−5 were also excluded. After the quality control measures, genotype data were available for 9,975 individuals. For more details on the Add Health genome-wide association study (GWAS) data quality, see Highland et al. (43).
Our measure of genetic propensity to smoke is a polygenic score. A polygenic score is a genome-wide score that summarizes the presence of presence and associated weights of risk alleles discovered by GWASs. In essence, a polygenic score is a weighted average or composite score that considers information across an individual’s entire genome to measure his/her genetic predisposition or risk for a particular outcome. A polygenic score for individual i is a weighted average across J SNPs of the number of reference alleles x (0, 1, or 2) at loci j multiplied by the score for that SNP β:
| [1] |
Many complex traits have been shown to be highly polygenic, that is, affected by many different genetic variants each contributing a small amount to the overall outcome (44). This is true even for clinically dichotomous outcomes, which may reflect a shift along a phenotypic continuum known as decanalization (45). In analyzing such traits, individual genes will have low penetrance, making it difficult to distinguish between the effects of genes and the effects of the environment. Polygenic scores overcome many of these difficulties. Unlike candidate-gene approaches, polygenic scores do not focus on a few prespecified genes of theoretical interest. Rather, they attempt to quantify an individual’s genetic risk to exhibit a trait by aggregating the effects of all of genes on the phenotype of interest. Polygenic scores are therefore “hypothesis-free,” in the sense the researcher does not have to understand all of the underlying biological processes giving rise to a phenotype to be able to study the effects of genes on that phenotype. As a result, they can be used to discover genetic contributions previously unknown to researchers.
Although polygenic scores are far more robust than candidate-genes, recent research has identified some of their shortcomings. Polygenic scores have been shown to pick up not only direct genetic effects, but also the effects of genetic nurturance, deep ancestry, and assortative mating, all of which can bias results when the distribution of genes to environments is nonrandom. Nevertheless, this should not impact our results as long as our assumption holds that the distribution of grade-mate polygenic scores within schools is as-if random. We provide evidence in favor of this assumption in the SI Appendix.
To identify bad apples and shining stars, respondents in the full sample were ordered according to their PGS. The top 10% of respondents in terms of PGS, those who were deemed as having the highest genetic risk of smoking, were classified as bad apples, while the bottom 10% were classified as shining stars. We performed this classificatory exercise separately for members of each race, in case racial differences in mean PGS were driving assignment to bad apple or shining star.
We use the GWAS summary statistics from the Tobacco and Genetics (TAG) Consortium on cigarettes per day (3). The SNPs genotyped in Add Health were then matched to the ones in the GWAS. A polygenic score was then constructed by aggregating the effects of the overlapping variants across the genome and weighting them by the strength of their association with the outcome or behavior. We used 529,390 to create the smoking scores. A P value threshold was not imposed in the construction of the PGS as to retain as much information as possible on genetic contributions of all SNPs, especially since SNPs with higher P values are down weighted in the composite score. As a check of robustness, we also used the most recent GWAS provided by the GWAS & Sequencing Consortium of Alcohol and Nicotine (GSCAN) (46), but found that the resulting polygenic scores were less predictive of individual smoking behavior compared with those produced with TAG (0.11 versus 0.15 correlated with cigarettes per day, respectively).
Student’s who participated in the in-school administration of Add Health were asked to identify up to 10 friends, 5 male and 5 female friends, using a roster of the other students in their school. The results from this exercise make up the friendship data used in this paper. As a consequence of the nomination design, every student is associated with as many as 10 out-degree friends, and as many in-degree friends as there are other students in the school, although the large majority of students were nominated as a friend by others zero to 4 times. Friend-level variable estimates were obtained by averaging polygenic scores over: 1) all of a student’s friends; 2) all of the people who nominated the student; or 3) all of the people who the student nominated, which we refer to as friend-level, in-degree nominations, and out-degree nominations respectively.
Identifying classmates required supplementary information, which came from the The Adolescent Health and Academic Achievement (AHAA) study. It provides the high school transcripts for Add Health Wave III sample members (original Wave I sample members who were reinterviewed at Wave III). After collecting the transcripts, the AHAA constructed academic networks using overlap in course-taking and ran clustering analyses on those networks using a p*-based algorithm fully detailed in Field et al. (47). Individuals were assigned to clusters with other students with whom they took similar sets of courses, with the idea that the resulting clusters proxy mutual exposure during course taking. Student’s who were assigned to clusters with one or no other students were reassigned to the cluster to which they had the highest probability of belonging according to their coursework. We used these clusters to identify classmates, and again, for each student, averaged over the variables of their classmates to provide estimates of their peers’ mean smoking behavior, mean smoking genetic risk, and demographic composition. We tested other specifications of classmates, including a simple binary indicator of having shared any course, all of which returned similar results as to those reported in the main text.
Individual smoking behavior measures how many cigarettes a day an individual smoked, on the days that they smoked. We include school and grade fixed effects to control for school and grade level. We further control for the sex, race, maternal education, familial smoking behavior, having an older siblings and family income of every individual in the sample as well as the average level of those variables at the grade level.
Further, following previous research on grade-level peers (48), for all analyses, we limit the sample to students in schools with a 12th grade (which results in excluding middle schools, but retaining high schools with seventh and eighth grades) and who were assigned sample weights. Then, for all grade-level analyses, we further exclude students who attended a grade with fewer than 20 total students in our sample. In all, 74 schools and 148 grades are represented in the data. This leaves us with 3,895 respondents who have genotype data and are in grades with a reasonable number of peers. For all friend analyses, we only include students who had at least one other friend in their grade, giving us 3,708 respondents.
In addition to residualizing individual smoking PGS, a number of controls were included in the analyses to again ward against various forms of population stratification. In terms of race, we include individuals of all races in the analyses, but control for the individual’s own race and the racial composition of the grade, classroom, or friend group. Because it is possible that genes are a proxy for parent or sibling smoking behavior which might directly influence other individuals in a grade, we block this pathway by including an indicator for the presence of household smokers and older sibling both on the individual and on the grade level. In addition, we control for an individual’s own smoking polygenic score in the models such that the results provide estimates of others’ polygenic scores on individual smoking net of individual’s own polygenic score.
Supplementary Material
Acknowledgments
This paper benefited from comments and discussions with Parijat Chakrabarti, Andrew McMartin, Benjamin Domingue, members of the Biosociology Lab at Princeton University, the instructors of the Russell Sage Foundation Summer Institute in Social-Science Genomics, and the reviewers who provided valuable feedback. This paper uses data from Add Health, a program project directed by K.M.H. and designed by J. Richard Udry, Peter S. Bearman, and K.M.H. at the University of North Carolina at Chapel Hill and funded by Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) Grant P01-HD31921, with cooperative funding from 23 other federal agencies and foundations. This research used Add Health GWAS data funded by NICHD Grants R01 HD073342 and R01 HD 060726.
Footnotes
The authors declare no conflict of interest.
Data deposition: The Add Health genome-wide association study (GWAS) data reported in this paper have been deposited in the database of Genotypes and Phenotypes (dbGaP), https://www.ncbi.nlm.nih.gov/gap (accession no. phs001367.v1.p1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806901116/-/DCSupplemental.
References
- 1.U.S. Department of Health and Human Services , The Health Consequences of Smoking—50 Years of Progress: A Report of the Surgeon General (U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Chronic Disease Prevention and Health Promotion, Office on Smoking and Health, Atlanta, GA, 2014). [Google Scholar]
- 2.Pidoplichko V. I., DeBiasi M., Williams J. T., Dani J. A., Nicotine activates and desensitizes midbrain dopamine neurons. Nature 390, 401–404 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Tobacco and Genetics Consortium , Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 42, 441–447 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woodruff S. I., Candelaria J. I., Laniado-Laborín R., Sallis J. F., Villaseñor A., Availability of cigarettes as a risk factor for trial smoking in adolescents. Am. J. Health Behav. 27, 84–88 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Schofield P. E., Pattison P. E., Hill D. J., Borland R., Youth culture and smoking: Integrating social group processes and individual cognitive processes in a model of health-related behaviours. J. Health Psychol. 8, 291–306 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Mathur C., Erickson D. J., Stigler M. H., Forster J. L., Finnegan J. R. Jr, Individual and neighborhood socioeconomic status effects on adolescent smoking: A multilevel cohort-sequential latent growth analysis. Am. J. Public Health 103, 543–548 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perra O., Fletcher A., Bonell C., Higgins K., McCrystal P., School-related predictors of smoking, drinking and drug use: Evidence from the Belfast Youth Development Study. J. Adolesc. 35, 315–324 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Fletcher J. M., Social interactions and smoking: Evidence using multiple student cohorts, instrumental variables, and school fixed effects. Health Econ. 19, 466–484 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Christakis N. A., Fowler J. H., The collective dynamics of smoking in a large social network. N. Engl. J. Med. 358, 2249–2258 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cambron C., Kosterman R., Catalano R. F., Guttmannova K., Hawkins J. D., Neighborhood, family, and peer factors associated with early adolescent smoking and alcohol use. J. Youth Adolesc. 47, 369–382 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nash S. G., McQueen A., Bray J. H., Pathways to adolescent alcohol use: Family environment, peer influence, and parental expectations. J. Adolesc. Health 37, 19–28 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Alexander C., Piazza M., Mekos D., Valente T., Peers, schools, and adolescent cigarette smoking. J. Adolesc. Health 29, 22–30 (2001). [DOI] [PubMed] [Google Scholar]
- 13.Ennett S. T., Bauman K. E., The contribution of influence and selection to adolescent peer group homogeneity: The case of adolescent cigarette smoking. J. Pers. Soc. Psychol. 67, 653–663 (1994). [DOI] [PubMed] [Google Scholar]
- 14.Gardner M., Steinberg L., Peer influence on risk taking, risk preference, and risky decision making in adolescence and adulthood: An experimental study. Dev. Psychol. 41, 625–635 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Domingue B. W., Belsky D., Conley D., Harris K. M., Boardman J. D., Polygenic influence on educational attainment: New evidence from The National Longitudinal Study of Adolescent to adult health. AERA Open 1, 1–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Canario L., Lundeheim N., Bijma P., The early-life environment of a pig shapes the phenotypes of its social partners in adulthood. Heredity 118, 534–541 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergsma R., Kanis E., Knol E. F., Bijma P., The contribution of social effects to heritable variation in finishing traits of domestic pigs (Sus scrofa). Genetics 178, 1559–1570 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petfield D., Chenoweth S. F., Rundle H. D., Blows M. W., Genetic variance in female condition predicts indirect genetic variance in male sexual display traits. Proc. Natl. Acad. Sci. U.S.A. 102, 6045–6050 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baud A., et al. , Genetic variation in the social environment contributes to health and disease. PLoS Genet. 13, e1006498 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domingue B. W., Belsky D. W., The social genome: Current findings and implications for the study of human genetics. PLoS Genet. 13, e1006615 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rauscher E., Conley D., Siegal M. L., Sibling genes as environment: Sibling dopamine genotypes and adolescent health support frequency dependent selection. Soc. Sci. Res. 54, 209–220 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Cawley J., Han E., Kim J. J., Norton E. C., Testing for peer effects using genetic data. 10.3386/w23719 (August 2017). [DOI]
- 23.Kong A., et al. , The nature of nurture: Effects of parental genotypes. Science 359, 424–428 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Conley D., et al. , Is the effect of parental education on offspring biased or moderated by genotype? Sociol. Sci. 2, 82–105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polderman T. J., et al. , Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat. Genet. 47, 702–709 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Polubriaginof F. C. G., et al. , Disease heritability inferred from familial relationships reported in medical records. Cell 173, 1692–1704.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harris K. M., et al. , Cohort profile: The National Longitudinal Study of Adolescent Health (Add Health). Int. J. Epidemiol., 10.1093/ije/dyz115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manski C. F., Identification of endogenous social effects: The reflection problem. Rev. Econ. Stud. 60, 531–542 (1993). [Google Scholar]
- 29.Cohen-Cole E., Fletcher J. M., Is obesity contagious? Social networks vs. environmental factors in the obesity epidemic. J. Health Econ. 27, 1382–1387 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Christakis N. A., Fowler J. H., Social contagion theory: Examining dynamic social networks and human behavior. Stat. Med. 32, 556–577 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Domingue B. W., et al. , The social genome of friends and schoolmates in the national longitudinal study of adolescent to adult health. Proc. Natl. Acad. Sci. U.S.A. 115, 702–707 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamer D., Sirota L., Beware the chopsticks gene. Mol. Psychiatry 5, 11–13 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Bryant A. L., Schulenberg J., Bachman J. G., O’Malley P. M., Johnston L. D., Understanding the links among school misbehavior, academic achievement, and cigarette use: A national panel study of adolescents. Prev. Sci. 1, 71–87 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Goldberg A., Stein S. K., Beyond social contagion: Associative diffusion and the emergence of cultural variation. Am. Sociol. Rev. 83, 897–932 (2018). [Google Scholar]
- 35.Crosnoe R., Muller C., Frank K., Peer context and the consequences of adolescent drinking. Soc. Probl. 51, 288–304 (2004). [Google Scholar]
- 36.Hoxby C. M., Weingarth G., Taking race out of the equation: School reassignment and the structure of peer effects. https://doi.org/10.1.1.75.4661(2005).
- 37.Lazear E. P., Educational production. Q. J. Econ. 116, 777–803 (2001). [Google Scholar]
- 38.Sacerdote B., “Peer effects in education: How might they work, how big are they and how much do we know thus far?” in Handbook of the Economics of Education, Hanushek E. A., Machin S., Woessmann L., Eds. (Elsevier, Amsterdam, The Netherlands, 2011), vol. 3, pp. 249–278. [Google Scholar]
- 39.Grunberg N. E., Winders S. E., Wewers M. E., Gender differences in tobacco use. Health Psychol. 10, 143–153 (1991). [PubMed] [Google Scholar]
- 40.Winkleby M. A., Jatulis D. E., Frank E., Fortmann S. P., Socioeconomic status and health: How education, income, and occupation contribute to risk factors for cardiovascular disease. Am. J. Public Health 82, 816–820 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harris K. M., An integrative approach to health. Demography 47, 1–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris K. M., et al. , Social, behavioral, and genetic linkages from adolescence into adulthood. Am. J. Public Health 103 (suppl. 1), S25–S32 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Highland H. M., Avery C. L., Duan Q., Li Y., Harris K. M., “Quality control analysis of Add Health GWAS data” (Carolina Population Center, University of North Carolina at Chapel Hill, Chapel Hill, NC, 2018).
- 44.Visscher P. M., Hill W. G., Wray N. R., Heritability in the genomics era–Concepts and misconceptions. Nat. Rev. Genet. 9, 255–266 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Gibson G., Decanalization and the origin of complex disease. Nat. Rev. Genet. 10, 134–140 (2009). [DOI] [PubMed] [Google Scholar]
- 46.Liu M., et al. ; 23andMe Research Team; HUNT All-In Psychiatry , Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 51, 237–244 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Field S., Frank K. A., Schiller K., Riegle-Crumb C., Muller C., Identifying positions from affiliation networks: Preserving the duality of people and events. Soc. Networks 28, 97–123 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.