Significance
Our study identified SLAMF9 (which belongs to the conserved lymphocytic activation molecule family) as a plasmacytoid dendritic cell receptor that can regulate the function and maintenance of these cells in health and during inflammation. We therefore believe that blocking SLAMF9 might be a promising strategy to mediate plasmacytoid dendritic cell function and frequencies during autoimmune diseases.
Keywords: SLAMF9, pDCs, EAE
Abstract
SLAMF9 belongs to the conserved lymphocytic activation molecule family (SLAMF). Unlike other SLAMs, which have been extensively studied, the role of SLAMF9 in the immune system remained mostly unexplored. By generating CRISPR/Cas9 SLAMF9 knockout mice, we analyzed the role of this receptor in plasmacytoid dendritic cells (pDCs), which preferentially express the SLAMF9 transcript and protein. These cells display a unique capacity to produce type I IFN and bridge between innate and adaptive immune response. Analysis of pDCs in SLAMF9−/− mice revealed an increase of immature pDCs in the bone marrow and enhanced accumulation of pDCs in the lymph nodes. In the periphery, SLAMF9 deficiency resulted in lower levels of the transcription factor SpiB, elevation of pDC survival, and attenuated IFN-α and TNF-α production. To define the role of SLAMF9 during inflammation, pDCs lacking SLAMF9 were followed during induced experimental autoimmune encephalomyelitis. SLAMF9−/− mice demonstrated attenuated disease and delayed onset, accompanied by a prominent increase of immature pDCs in the lymph node, with a reduced costimulatory potential and enhanced infiltration of pDCs into the central nervous system. These results suggest the crucial role of SLAMF9 in pDC differentiation, homeostasis, and function in the steady state and during experimental autoimmune encephalomyelitis.
The lymphocytic activation molecule family (SLAMF) consists of 9 family members within a shared locus, both in humans and in mice. Their expression is mainly restricted to hematopoietic cells (1), where they act as immunomodulating receptors that are occupied by homophilic and heterophilic interactions (2). Engagement of SLAMs initiates phosphorylation of the tyrosine switch-based motif (ITSM) at their intracellular domain and results in recruitment of slam-associated protein (SAP) or Ewing’s sarcoma-activated transcript-2 (EAT2) adaptor proteins, which convey downstream signals (3).
The SLAMF9 gene (CD84H, SF2001, CD2F10) resides outside of the SLAM locus (4). Its transcript is primarily present in peripheral monocytes and human monocyte-derived dendritic cells (DCs). Moreover, RT-PCR analysis revealed SLAMF9 expression in cancerous cell lines of monocytic or lymphocytic origin (5). Furthermore, sequence alignment with other SLAMF members identified a relatively short cytoplasmic tail that does not contain an ITSM motif (6, 7), raising the intriguing question of whether SLAMF9 is an active member of the SLAM family able to mediate immune functions. The role of SLAMF9 is mainly unexplored, and was described only in a double SLAMF9- and SLAMF8-deficient mouse strain (8), in which a specific effect of SLAMF8/9 deficiency in macrophages was revealed.
Plasmacytoid DCs (pDCs) are a rare subpopulation of immune cells that exhibit a remarkable capacity to secrete type I IFN in response to viral infections and are involved in the progression of autoimmune diseases (9–11). Their development begins in the bone marrow (BM) from progenitor cells derived from lymphoid or myeloid origin (12–15). Once their differentiation is completed, pDCs enter the peripheral lymphoid tissues through the blood, where they reside under steady-state conditions (16). Although much progress has been made in understanding pDC development and functionality, many questions remain regarding molecular pathways regulating their ontogeny, plasticity, and survival at the periphery (17).
Several transcription factors were shown to play an important role in pDC development, such as E2-2 (TCF-4) and SpiB (PU.1 related) (18). E2-2 is exclusively expressed in pDCs and executes a unique gene transcription program involved in the development of these cells, and their capacity to secrete IFN-α (19, 20). One of the targets of E2-2 is SpiB, which is highly abundant in pDCs and was shown to directly support pDC maturation and function (21).
Along their maturation steps in the BM, pDCs acquire the expression of receptors that can promote or restrict their IFN response. Murine pDC receptors, such as Ly49Q (22, 23) and PDC-TREM (24), support the production of IFN-α, whereas murine pDC receptor SiglecH (25) and human ILT7 and BDCA-2 (26, 27) act as inhibitory receptors mediating suppression of the type I IFN response.
In this study, we investigated the role of SLAMF9 in immune cells with a focus on pDC, which preferentially express SLAMF9 mRNA. By generating SLAMF9 knockout mice, we revealed a role for SLAMF9 in pDC differentiation and function. In its absence, pDCs accumulate at the immature stage, and secrete lower levels of IFN-α in the steady state and during inflammation.
Materials and Methods
Generation of SLAMF9−/− Mice Using CRISPR/Cas9.
C57BL/6 WT mice were purchased from Harlan Biotech Israel. SLAMF9 knockout mice were generated on a C57BL/6 background, as previously described (28). CRISPR guide sequences were as follows: upstream guide: 5′ATCATCTGACTGTTAGACGG3′; downstream guide: 5′ACTCGCTCTGCAATAAACAT3′. Guide sequences were chosen by optimizing for the lowest off-target and maximum on-target potential using programs described in ref. 29 (for off-target) and refs. 30 and 31 (for on-target). Mouse sequences were analyzed using Sequencher v5.4 (GeneCodes Corp.) and visualized in the University of California, Santa Cruz Genome Browser (32). Primers used for screening genomic screening were as follows: forward 5′GATTTCTACGGGATCTGGGC3′ and reverse 5′CAATCATCCTTTGGTGCCCC3′.
Mice were maintained under sterile pathogen-free conditions, and all experiments were approved by the Animal Research Committee at the Weizmann Institute.
RNA Isolation and qRT-PCR.
RNA was extracted from sorted cell populations using a ZR RNA microprep kit (Zymo Research) according to the manufacturer’s instructions, followed by single-strand cDNA synthesis using a qScript cDNA Synthesis Kit. Gene-expression levels were analyzed by quantitative real-time PCR using a Light-Cycler 480 instrument and SYBR Green I mix kit (Roche Diagnostics). The primers were purchased from Sigma Aldrich, and their sequences are as follows: SLAMF9 forward 5′CACAAGTCAGTGCGGTTCAC3′ and reverse 5′GTGTTTCCTGTGTTAATGCCAC3′; L32 forward 5′TTAAGCGAAACTGGCGGAAAC3′ and reverse 5′TTGTTGCTCCCATAACCGATG3′; TCF4 forward 5′-CCAGGAACCCTTTCGCCCACCAAAC-3′ and reverse 5′TGCTGGCTGCTGGCTTGGAGGAA3′; SpiB forward 5′AGAGGACTTCACCAGCCAGA3′ and reverse 5′GGGCTGTCCAGCATAATGTC3′.
Competitive BM Chimera.
WT CD45.1(C57BL/6) mice were lethally irradiated (950 Rad), and reconstituted with an intravenous injection of 2 × 106 BM cells per recipient derived from a 1:1 mixture of CD45.1 (WT) and CD45.2 (WT), or from a 1:1 mixture of CD45.1 (WT) with CD45.2 (SLAMF9 KO) cells. Mice were killed 8 wk postreconstitution.
Experimental Autoimmune Encephalomyelitis Induction and Assessment.
Experimental autoimmune encephalomyelitis (EAE) was induced in C57BL/6 mice at the age of 10 to 12 wk by subcutaneous injection with 200 µg of MOG33–55 peptide (GenScript) emulsified in complete Freund’s adjuvant (CFA) containing 3.3 mg/mL of heat-killed Mycobacterium tuberculosis (BD Bioscience), and intraperitoneal injection with 200 ng of Pertussis toxin per mouse (Sigma) in PBS on days 0 and 2. Clinical signs of EAE were assessed daily, and scored as follows: 0, normal behavior; 1, tail low tonus; 2, hind leg weakness; 3, hind leg paralysis; 4, full paralysis; 5, death.
CpG In Vivo Challenge.
Mice were injected intravenous with 6 μg of CpGA ODN1585 (Invivogen) mixed with 30 µL of DOTAP (Roche) to a final volume of 200 µL. Mice were killed after 24 h, and lymph nodes (LN) were isolated and cultured for 5 h in the presence of 1 µM of CpGA ODN1585.
Ex Vivo Stimulation.
For stimulation of pDCs, 1 × 106 spleen cells were resuspended in RPMI supplemented with 10% FCS, 1% penicillin streptomycin, 1% l-glutamine solution, and 1% sodium pyruvate solution (Beit Ha’emek), and stimulated for 16 h with 1 μM of ODN1585 (Invivogen).
For stimulation of pDCs derived from EAE mice and CpGA-induced mice, LN cultures were stimulated for 5 or 24 h, as indicated in the presence of 1 μM of ODN1585 (Invivogen).
Tissue Extraction.
Cells were extracted from the BM by flushing the tibia and femur with PBS. Brain and spinal cord samples were harvested and homogenized, and resuspended in 30% Percoll before density centrifugation at 7,800 × g for 30 min at room temperature, followed by resuspension in 50% Ficoll, and centrifugation at 2,500 rpm for 25 min at room temperature with no brake.
Flow Cytometry.
For flow cytometry analysis, the following antibodies were used: SLAMF9 (R&D Systems), CD11b (clone: M1/70), CD11C (clone: N418), B220 (clone: RA3-6B2), PDCA-1 (clone: JF05-1C2.4.1), CCR9 (clone: CW1.2), SCA-1 (clone: D7), CD19 (clone: eBio1D3), Ly49Q (clone: 2E6), Ly6C (clone AL21), CD45.1 (clone: A20), CD45.2 (clone: 104), IFN-α (clone: RMMA-1), TNF-α (clone: MP6-XT22), CCR5 (clone:HM-CCR5), SiglecH (clone: 551.3D3), CD40 (clone: FGK45.4), CD80 (clone: 16–10A1), I-A/E (clone: M5/114.15.2), SpiB (ab42436), CD3 (clone: 17A2), CD4 (clone: GK1.5), IFN-γ (clone: XMG1.2), IL-17 (clone: TC11-18H10.1), ROγT (clone: AFKJS-9), TBET (clone: eBio4B10), FOXP3 (clone: FJK-16s), and CD25 (clone: PC61.5).
For intracellular staining, cells were fixed and permeabilized using the CytoFix/Cytoperm kit (BD Bioscience) or with Fixation/permeabilization kit (eBioscience) according to the manufacturer’s instructions. Cells were acquired using FACS Canto II flow cytometer (BD Bioscience) and analyzed with FlowJo software (v10). Cell sorting was performed using FACS Aria II system (BD Bioscience).
RNA Sequencing.
A total of 1 × 105 pDCs (CD19−, CD11B−, CD11Cinter+, B220+, PDCA+) were collected in triplicates from the BM of WT and SLAMF9-deficient mice. RNA was extracted and fragmented followed by reverse-transcription and second-strand cDNA synthesis. Libraries were evaluated by Qubit (Thermo Fisher Scientific) and TapeStation (Agilent). Sequencing libraries were constructed with barcodes to allow multiplexing. Approximately 17 to 38 × 106 single-end 75-bp reads were sequenced per sample on Illumina Nextseq 500 high-output run. Gene-expression levels were quantified using htseq-count (33). Differential gene-expression analysis was performed using DESeq2 (34). Raw P values were modified for multiple testing using the Benjamini and Hochberg procedure. Identification of differentially expressed genes focused on fold-difference equal or greater to 1.5 and P ≤ 0.05. Pathway analysis was performed using ingenuity pathway analysis (Qiagen, https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis).
Statistics.
Data analysis was performed using Graphpad Prism (v7 GraphPad Software), and results are provided as means and ±SEM.
Statistical analysis was conducted using an unpaired t test, a ratio t test to correct for normalized data or 2-way ANOVA, depending on the experiment. Results were defined as significant with a P ≤ 0.05.
Results
The pDC Population Is Elevated in the LNs of SLAMF9-Deficient Mice in a Cell-Intrinsic Manner.
According to Immgene data, among nonactivated immune cells, expression of SLAMF9 is restricted to pDCs, while its expression on B and T cells is hardly detected (35).
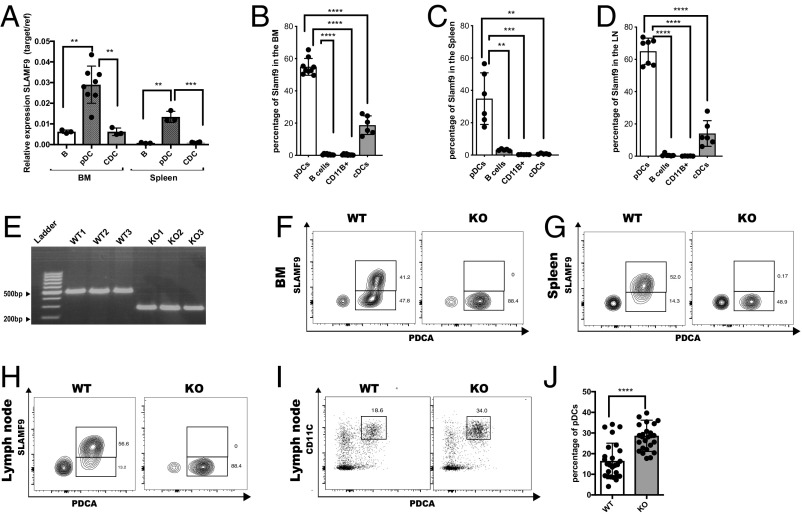
To confirm these data, we determined SLAMF9 expression on selected sorted immune populations using qRT-PCR. As shown in Fig. 1A, SLAMF9 mRNA was mainly expressed in BM and splenic pDCs (CD19−CD11B−CD11Cinter+B220+PDCA+) compared with B (CD19+B220+) and conventional DC (cDCs) (CD11ChighB220−CD19−CD11B+) cells. As shown in SI Appendix, Fig. S1A, mRNA levels of the pDC transcription factor TCF-4 (E2-2) were significantly higher in pDCs, confirming cell purity. Furthermore, SLAMF9 cells surface expression was detected mainly on pDCs derived from BM (Fig. 1B), spleen (Fig. 1C), and LN (Fig. 1D) with undetectable to low expression on cDCs. The proportion of SLAMF9+ pDCs was elevated in the LN compared with its ratio in the BM, suggesting a function for this receptor in the pDCs maturation.
Fig. 1.
SLAMF9 is expressed on pDCs and regulates their numbers. (A) Sorted pDCs (CD19−CD11CinterB220+PDCA+), B cells (CD19+ B220+), and cDCs (CD19−B220−CD11B+CD11Chigh) were analyzed for the mRNA levels of SLAMF9. Graph shows relative expression of target gene/reference gene (L32). Results are represented as mean percentage ± SD (unpaired t test 2-tailed **P < 0.005, ***P < 0.0005); n = 3–8. (B–D) SLAMF9 protein levels were analyzed under naïve state in pDCs (CD19−CD11CinterB220+PDCA+ CD11B−), B cells (CD19+ B220+), cDCs (CD19−B220−CD11B+CD11Chigh), and in macrophages (CD11B+CD11C− CD19-SSClow). Graph shows protein levels in the BM (B), spleen (C), and in the LN (D). (E) DNA agarose gel electrophoresis demonstrating a genomic deletion of about 200 bp in 3 SLAMF9−/− mice compared with 3 WT controls. (F–H) Representative FACS plots showing SLAMF9 expression in WT compared with SLAMF9−/− pDCs as a negative control. (F) SLAMF9 expression in the BM. (G) SLAMF9 expression in the spleen. (H) SLAMF9 expression in the LN. (I and J) LN pDCs (CD19−CD11CinterB220+PDCA+) were analyzed in WT and SLAMF9−/− mice. (I) Representative dot plot. (J) Graph shows the percent of pDCs in the LN. Results are a summary of 6 independent experiment; n = 26 mice (unpaired t test 2-tailed *P < 0.05, **P < 0.005, ***P < 0.0005, ****P < 0.0001).
To reveal the function of the SLAMF9 receptor, we generated mice deficient for this SLAM family member using the CRISPR/Cas9 system. Sequencing of a genomic PCR product obtained from the mutated SLAMF9 allele of the resulting mice revealed a 211-bp genomic deletion (SI Appendix, Fig. S1B and Fig. 1E), which included a part of the promoter, the first exon and intron, and most of the second exon, resulting in a nonfunctional locus. As shown in Fig. 1 F–H, no cell surface expression of SLAMF9 was detected on cells derived from the SLAMF9-deficient (SLAMF9−/−) mice.
Since SLAMF9 is highly expressed on pDCs, we focused on characterization of these cells in the SLAMF9−/− mice. The frequency of pDCs (CD19−CD11B−CD11C+B220+PDCA+) in the BM (SI Appendix, Fig. S1 C and D) and spleen (SI Appendix, Fig. S1E) in mutant animals was not significantly different compared with their levels in WT mice. Interestingly, a significant increase of about 1.9-fold was detected in the frequency of pDCs in the LN (Fig. 1 I and J) and among total live cells (SI Appendix, Fig. S1F), and in their absolute numbers (SI Appendix, Fig. S1G). This elevation was specific to pDCs, while the proportion of the B cell and cDC populations in the periphery (SI Appendix, Fig. S1 H and I) or macrophages (SI Appendix, Fig. S1J) was mostly unchanged, highlighting a possible exclusive function for SLAMF9 in pDCs.
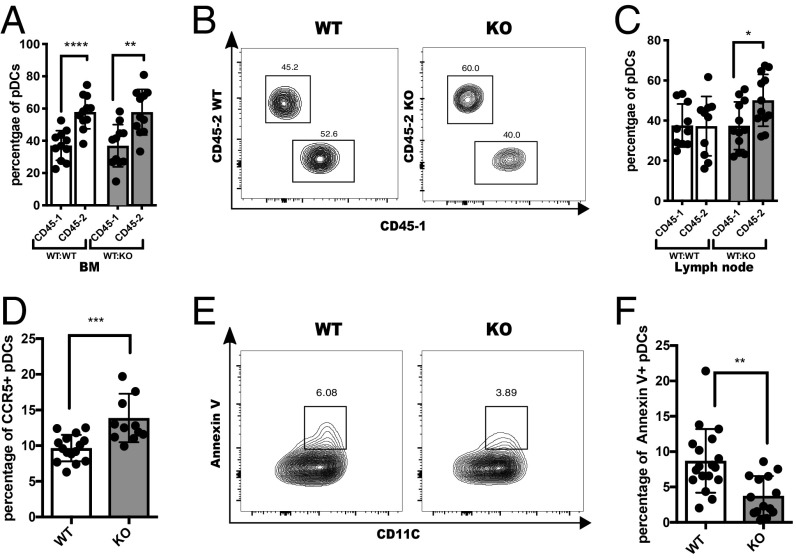
To determine whether the increase in LN pDCs was an intrinsic feature of the SLAMF9-deficient pDCs or a result of a dysregulated function of other cells in the microenvironment, mixed BM chimeras were generated. CD45.1 (WT) and CD45.2 (SLAMF9−/− or WT) BM cells were injected at a 1:1 ratio into lethally irradiated CD45.1 (WT) mice. Animals were killed after 8 wk, and pDC numbers and frequencies were analyzed in various tissues. As shown in Fig. 2A, an elevated pDC population derived from CD45.2 WT or SLAMF9−/− was observed in the BM. In addition, no changes were detected in the pDC frequencies in the spleen (SI Appendix, Fig. S2A). In contrast, in the LN (Fig. 2 B and C), only pDCs derived from CD45.2 (SLAMF9−/−) were increased compared with CD45.1 WT. The elevation was specific to pDCs, and no significant changes were detected in cDC and B cell populations in this compartment (SI Appendix, Fig. S2 B and C). These results establish that the CD45.2 background confers a developmental advantage over the CD45.1 background, as reported previously (36, 37). In contrast, the accumulation of pDCs in the LN was only observed in the mixed chimeras harboring mutant BM. This finding demonstrates that the pDC accumulation in the LN of SLAMF9−/− mice is due to the intrinsic lack of SLAMF9 in these cells.
Fig. 2.
SLAMF9−/−pDC accumulation in the LN is cell intrinsic and results from increased CCR5 expression and enhanced survival. (A) Graph shows the percent of BM pDC derived from either CD45.1 or CD45.2 in (WT CD45.1: WT CD45.2) or in (WT CD45.1: SLAMF9 KO CD45.2) chimera; ***P < 0.0001, **P = 0.0018. (B) Representative staining of CD45.1 and CD45.2 in LN pDCs in chimeric mice. (C) Graph shows the percent of LN pDC derived from either CD45.1 or CD45.2 in (WT CD45.1: WT CD45.2) or in (WT CD45.1: SLAMF9 KO CD45.2) chimera; *P < 0.0203. Results are representative of 3 independent experiments using chimera; n = 11 to 13 mice. (D–F) BM cells from WT or SLAMF9−/− mice were purified and analyzed. (D) Graph shows CCR5 expression on BM pDCs. Results are representative of 3 independent experiments; n = 12–15 mice; ***P = 0.0004. (E) Representative staining profile of Annexin V+ pDCs in WT or SLAMF9−/− mice. (F) Graphical view of Annexin V+ pDC percentages in LN pDCs derived from WT or SLAMF9−/− mice. Results are representative of 4 independent experiments; n = 14–18; **P = 0.0011.
To determine whether the elevation of LN pDCs in the in SLAMF9−/−mice results from their enhanced egress from the BM, the expression of CCR5, a chemokine receptor known to mediate the exit of pDCs to the periphery (38), was followed. As shown in Fig. 2D, CCR5 expression levels were significantly increased on SLAMF9−/− pDCs, supporting a higher potential to emerge from the BM. To further explore whether the exclusive increase of pDCs in the LN might result from enhanced cell survival at this site, Annexin V levels were analyzed by FACS. As shown in Fig. 2 E and F, a prominent decrease in Annexin V was detected in SLAMF9−/− pDCs in the LN. Collectively, these data suggest that the pDC accumulation in the LN of SLAMF9−/−mice results both from enhanced egress from the BM and enhanced pDC survival.
SLAMF9 Regulates pDC Differentiation and Activation State.
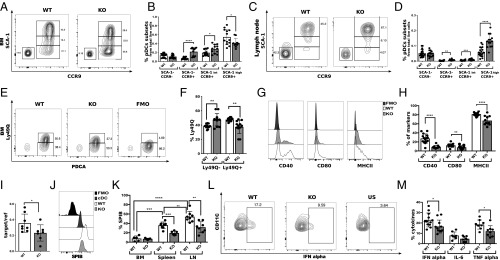
Next, we wished to explore whether SLAMF9 regulates pDC differentiation and function. We therefore compared the maturation state of WT and SLAMF9−/− pDCs by analysis of expression levels of Ly49Q, SCA-1, CCR9, and SiglecH receptors, which are involved in pDC differentiation, and can distinguish between the different stages of pDC development in the BM (23, 39–41).
Levels of SCA-1 were shown to segregate CCR9+ pDCs in the BM into 2 populations, in which higher expression levels of this molecule represent a more advanced stage of differentiation (42). A significant increase in the pDC populations expressing low to intermediate levels of SCA-1 was detected in the BM (Fig. 3 A and B), and in the LN of SLAMF9−/− mice both as a fraction of total live cells (Fig. 3 C and D) and in terms of absolute cell numbers (SI Appendix, Fig. S3 A and B). No differences were detected in frequencies or absolute numbers of the CCR9− SCA-1− precursor population in the BM (Fig. 3B and SI Appendix, Fig. S3A) or in the LN (Fig. 3D and SI Appendix, Fig. S3B). Furthermore, BM SLAMF9-deficient pDCs expressed lower levels of Ly49Q (Fig. 3 E and F), while expression of additional pDC markers, such as Ly6c, PDCA, and SiglecH, in the BM (SI Appendix, Fig. S3C) and in the LN (SI Appendix, Fig. S3 D and E) remained unchanged.
Fig. 3.
Higher numbers of immature pDCs in the BM of SLAMF9−/− mice and reduced levels of SpiB and functional pDCs in the LN. pDCs from BM and LN were isolated from WT or SLAMF9−/− mice, and stained for maturation and activation markers. (A and B) pDC subsets expressing SCA-1 and CCR9 in WT or SLAMF9−/− mice in the BM. (A) Representative dot plot of the BM populations. (B) Graph summarizes the percent CCR9− SCA-1−, CCR9+ SCA-1−, and pDCs expressing high or intermediate levels of SCA-1. Results are a summary of 3 independent experiments; n > 11; *P < 0.05, ****P < 0.0001. (C and D) pDC subsets expressing SCA-1 and CCR9 in WT or SLAMF9−/− mice in the LN. (C) Representative dot plots of pDCs subsets expressing SCA-1 and CCR9 in WT or SLAMF9−/− mice in the LN. (D) Graph summarizing the percent of LN CCR9− SCA-1−, CCR9+ SCA-1− and pDCs expressing high or intermediate levels of SCA-1. Results are a summary of 3 independent experiments; n > 14; *P < 0.005, ***P < 0.0005, ****P < 0.0001. (E and F) pDCs expressing Ly49Q in WT or SLAMF9−/− mice in the BM. (E) Representative histogram of staining for Ly49Q in WT and SLAMF9−/− pDCs compared with fluorescence-minus-one (FMO) control. (F) Graph summarizing the percent of Ly49Q− and Ly49Q+ pDCs. Results are summary of 3 independent experiments; n > 9; **P < 0.005. (G and H) pDCs were harvested from the LN of WT and SLAMF9−/− mice, and stained for the activation markers CD40, CD80, and MHC class II. (G) Representative histograms of activation markers on WT and SLAMF9−/− pDCs compared with FMO control. (H) Graph summarizes the percent of the pDC activation markers on WT and SLAMF9−/−. Results are representative of 3 independent experiments; n = 11 to 15; ****P < 0.0001, **P = 0.0094. (I) Sorted pDCs from the LN of WT and SLAMF9−/− mice were analyzed for the mRNA levels of SpiB. Graph shows relative expression of target gene/reference gene (L32). Results are representative of 3 independent experiments; n > 6 mice; *P = 0.049. (J–K) pDCs from the LN of WT and SLAMF9−/− mice were analyzed for SpiB expression. (J) Representative histograms showing SpiB expression in WT and SLAMF9−/− pDCs, compared with its expression in cDCs and FMO control. (K) Graph shows SpiB percentages in BM, spleen, and LN pDCs from WT and SLAMF9−/− mice. Results are a summary of 2 independent experiments; n = 5–8 mice; **P < 0.005, ***P < 0.0005, ****P < 0.0001. (L and M) Splenic pDCs from WT and SLAMF9−/− mice were freshly isolated and stimulated for 18 h with 1 μM of ODN1585, and analyzed for intracellular levels of IFN-α, IL-6, and TNF-α expression. (L) Representative dot plot for IFN-α staining in pDCs from WT or from SLAMF9−/− mice compared with unstimulated (US) control. (M) Graph summarizes the percent of IFN-α, TNF-α, and IL-6 expressing pDCs in WT and SLAMF9−/− following ODN1585 stimulation. Results are representative of 3 independent experiments for IFN-α (n = 12); *P = 0.0289 and 2 independent experiments (n = 9) with mice for IL-6 and TNF-α; ***P = 0.0147.
To determine the functionality of the accumulated pDCs in the LN, the expression of MHC class II and costimulatory molecules was followed. A decrease was detected in MHC class II expression and CD40 on SLAMF9−/− pDCs (Fig. 3 G and H and SI Appendix, Fig. S3F). However, the reduction in the cell surface expression of CD80 was smaller (Fig. 3 G and H). Analysis of MHC expression on pDC subsets that express high levels of SCA-1 and CCR9 revealed reduced expression of this molecule, suggesting that although these populations are present in elevated numbers, their costimulatory potential is reduced (SI Appendix, Fig. S3 G and H).
Since SpiB-deficient mice exhibit maturation defects in BM pDCs, and an increase of pDCs in the LN (21), we determined SpiB expression in SLAMF9−/− pDCs. Interestingly, SpiB mRNA levels were significantly reduced in LN SLAMF9−/− pDCs (Fig. 3I). We next determined SpiB protein expression. As shown in Fig. 3J and SI Appendix, Fig. S3I, SpiB protein was highly expressed in the pDC population. Its protein levels were significantly reduced in the SLAMF9-deficient LN pDCs (Fig. 3 J and K).
The defining feature of pDCs is their intrinsic capacity to rapidly secrete high levels of IFN-α in response to viruses and inflammatory cues (9). The magnitude of this response has been linked in part to the cell differentiation state (23, 42). Thus, to further explore the functionality of SLAMF9, pDCs were stimulated with CpG ODN1585, a synthetic ligand for Toll-like receptor 9 (TLR9) (43), and analyzed for intracellular levels of IFN-α, TNF-α, and IL-6 by FACS. Lower levels of IFN-α and TNF-α were detected in SLAMF9−/− pDCs from the spleen (Fig. 3 L and M), whereas IL-6 levels remained comparable with those in WT cells.
These results suggest that although SLAMF9 deficiency results in accumulation of pDCs, their cytokine production is aberrant.
pDCs Derived from SLAMF9−/− Mice Present a Unique Gene-Expression Profile.
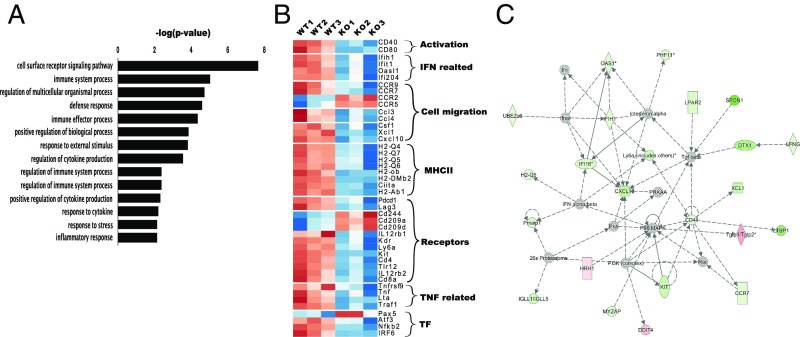
To further understand the role of SLAMF9 in regulating pDC function, we compared the transcriptional profile of SLAMF9−/− and WT BM pDCs using RNA sequencing. Principal components analysis analysis validated reproducibility of each group, and indicated 2 distinct clusters for WT and SLAMF9−/− samples (SI Appendix, Fig. S4A). Furthermore, a deletion in exon 1 was confirmed in all SLAMF9−/−samples, as illustrated by the integrative genomics viewer plots (SI Appendix, Fig. S4B).
Gene ontology (GO) analysis of significantly regulated genes in SLAMF9−/− pDCs indicated an enrichment of genes involved in immunity, inflammatory response, and cytokine secretion (Fig. 4A). Moreover, differential gene-expression analysis revealed 143 up-regulated genes and 303 down-regulated genes in pDC cells lacking SLAMF9, compared with WT cells. As shown in the heat map (Fig. 4B), genes related to the activation state and MHC class II, and genes involved in egress from the BM, such as CCR2 and CCR5, were significantly up-regulated in SLAMF9−/− pDCs, further confirming our results at the transcriptional level. Expression of the inducer of type I IFNs, IRF7, in pDCs (44) was not significantly changed. However, as illustrated in Fig. 4C, genes involved in regulating IFN levels, such as OASL1, MDA5 (IFIH1), and the DNA sensor IFI204 (45–48) were significantly down-regulated. Moreover, genes involved in TNF induction, such as TNF-α, TNF-β (LTA), and TRAF were significantly decreased, suggesting the potential dysregulation of the proinflammatory response.
Fig. 4.
Transcriptome analysis of SLAMF9 −/− pDCs reveals a unique gene-expression profile. BM pDCs (CD19−CD11B−CD11CinterB220+PDCA+) were sorted from WT and SLAMF9−/− mice. RNA was extracted and subjected to sequencing. Differentially expressed genes were in accordance with P < 0.05 and fold-change ≥1.5. (A) Differentially expressed genes in SLAMF9−/− pDCs were analyzed by GO pathway analysis for enriched biological processes with P ≤ 0.05. (B) Heat map illustrating expression of selected up-regulated and down-regulated genes with shared biological function. (C) Selected network representation obtained by ingenuity pathway analysis. Significant down-regulated genes are labeled in green, and up-regulated genes are labeled in red.
Inactive SLAMF9-Deficient pDCs Accumulate and Are Functionally Impaired in the LN In Vivo.
We next wished to determine whether the defects detected in pDC lacking SLAMF9 in the steady state could affect their immune response during inflammation. We therefore focused on EAE as an inflammatory model to study the role of SLAMF9 in vivo. pDCs have been shown to play a role during the pathogenesis of EAE (11, 49, 50); however, their function was described as either protective or pathogenic depending on the stage of the disease (51, 52).
To investigate the in vivo role of SLAMF9 during inflammation, we compared the progression of EAE (53) in WT and SLAMF9−/− mice. Interestingly, SLAMF9−/− mice showed delayed onset and milder disease compared with their WT counterparts, manifested by reduced clinical signs and weight loss from the time of disease induction to day 16 (Fig. 5A). In the BM, (Fig. 5B) and LN (Fig. 5C), pDC frequencies were higher in SLAMF9−/− mice and showed a higher fold-increase from WT during EAE compared with the elevation in steady state (SI Appendix, Fig. S5A). The number of pDCs and their frequencies among total live cells were increased in both BM and LN, but no change in pDC frequencies were detected in the spleen (SI Appendix, Fig. S5 B and C). In contrast, a small increase in macrophage frequencies was observed in the spleen, both in their proportion of total live cells and in absolute numbers (SI Appendix, Fig. S5 D and E). Furthermore, analysis of B cells and cDCs showed no changes in their frequencies (SI Appendix, Fig. S5 F and G), suggesting a specific effect of SLAMF9 on the accumulation of the pDC population in the LN during EAE progression.
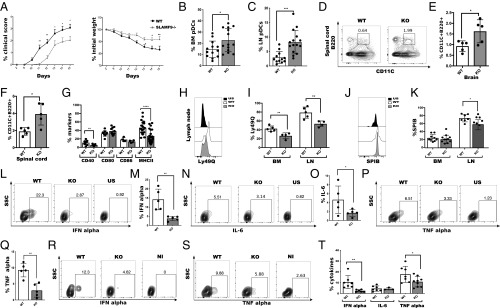
Fig. 5.
SLAMF9-deficient pDCs accumulate in the LN and CNS in vivo and are functionally impaired. EAE was induced in C57BL/6 mice WT and SLAMF9−/− mice by subcutaneous injection of MOG33–55 peptide in CFA and were further injected intraperitoneally with Pertussis toxin on days 0 and 2. EAE progression was monitored and assessed by standardized scoring until the peak of the disease on day 16. (A) Clinical scoring of EAE and weight measurements. Results are representative of 3 independent experiments; n = 11–15 mice. Data were analyzed using 2-way ANOVA for repeated measures, ***P = 0.0004, **P = 0.0013. (B and C) Graphical view of pDC percent in the BM and in the LN on day 16 after EAE. *P = 0.0272, ***P = 0.0004. (D) Representative staining profile of pDCs (CD45+CD11Cinter+, B220+CD11B) in the spinal cord. (E and F) Graphical view of pDCs in the spinal cord (E) and in the CNS (F). Results are representative of 2 independent experiments; each dot represent 2 mice; *P < 0.0308. (G) Graph summarizing the percent of CD40, CD80, CD86, and MHC class II on LN pDCs. Results are representative of 3 independent experiments; n = 9 to 15; **P = 0.0097, ****P < 0.0001. (H and I) Representative histogram of Ly49Q expression on LN pDCs from WT and SLAMF9−/− mice compared with FMO control (H). Graph shows the percent of Ly49Q on BM and LN pDCs from WT or SLAMF9−/− mice (I); n = 4 to 5 mice; **P < 0.005. (J and K) pDCs from the LN were isolated from EAE induced mice and analyzed for the expression of SpiB. Representative histogram of SpiB expression in LN pDCs from WT and SLAMF9−/− mice (J). Graph summarizing the percent of SpiB levels in BM and in LN pDCs (K). Results are a summary of 2 independent experiments; n = 7 to 10 mice; *P = 0.0101. (L–Q) Total LN cells were isolated from WT and SLAMF9−/− on day 16 following EAE induction, and stimulated for 5 h with 1 μM of ODN1585. (L and M) Representative staining and graphical view of IFN-α levels in WT and SLAMF9−/− pDCs compared with unstimulated control. (N and O) Representative staining and graphical view of IL-6 levels in WT and SLAMF9−/− compared with unstimulated control. (P and Q) Representative staining and graphical view of TNF-α levels in WT and SLAMF9−/− compared with unstimulated control. *P = 0.0173 and **P < 0.005. (R–T) WT and SLAMF9−/− mice were injected intravenously with 6 μg of CpG. After 24 h, LN cells were isolated and cultured for 5 h with 1 μM of ODN1585. Representative staining of IFN-α (R) and TNF-α (S), and graph view of cytokines levels (T) in WT and SLAMF9−/− pDCs compared with noninjected control. *P = 0.0395, **P = 0.0081.
pDCs were detected in cerebrospinal fluid both in humans and in mice during multiple sclerosis and EAE (52, 54). Therefore, the presence of SLAMF9−/− pDCs was analyzed in the CNS. The levels of SLAMF9−/− pDCs were significantly elevated in the brain and spinal cord, suggesting the enhanced infiltration of pDCs into the CNS in SLAMF9−/− mice (Fig. 5 D–F), whereas B cells and cDC frequencies remained unchanged (SI Appendix, Fig. S5H).
To investigate whether the increase in SLAMF9−/− pDCs in EAE is a result of enhanced BM exit, CCR5 levels were compared. As shown in SI Appendix, Fig. S5 I and J, increased expression of CCR5 was detected on SLAMF9−/− pDCs, which was about 1.8-fold higher than in the WT, supporting the notion of increased egress of SLAMF9−/− pDCs into the periphery. In the LN, SLAMF9−/− pDCs exhibited reduced levels of activation markers, such as MHC class II and CD40 (Fig. 5G and SI Appendix, Fig. S5K). Interestingly, while under physiological conditions, Ly49Q expression was not changed in the periphery in SLAMF9−/− pDCs compared with WT cells; in EAE, pDCs derived from SLAMF9−/− showed a significant decrease of Ly49Q expression both in the BM and in the LN (Fig. 5 H and I) and in mean fluorescence intensity values in the LN (SI Appendix, Fig. S5L). This decrease was accompanied by reduction in SpiB levels (Fig. 5 J and K), supporting their aberrant differentiation during EAE. Analysis of SLAMF9 expression in the LN of EAE induced mice showed that SLAMF9 is mainly expressed on pDCs compared with other immune cells (SI Appendix, Fig. S5M). To directly show the functionality of LN pDCs, their cytokine production was analyzed. pDCs derived from EAE-induced mice expressed lower levels of IFN-α following 18 h of CpGA stimulation (SI Appendix, Fig. S5 N and O). In addition, this down-regulation was also detected following short-term induction with CpGA, accompanied by reduced levels of IL-6 and TNF-α (Fig. 5 L–Q), further demonstrating the aberrant functionality of these cells.
Upon viral stimulation, pDCs are the principal sources of IFN-α, primarily induced by the ligation of viral nucleic acids with TLR7, TLR8, or TLR9 (55). While TLR7 and TLR8 are involved in the response to binding of single-stranded viral RNA, TLR9 recognizes unmethylated viral CpG DNA motifs (50). To demonstrate the role of SLAMF9 in pDCs in vivo, we analyzed the cytokine profile of WT pDCs compared with SLAMF9−/− in CpG-induced mice. Analysis of the cytokine profile in SLAMF9−/− pDCs derived from CpG-induced mice compared with the WT control, revealed a reduction in IFN-α levels and in proinflammatory cytokines, such as TNF-α and IL-6 (Fig. 5 R–T). Thus, in the absence of SLAMF9, pDCs present a reduced potential to propagate a type I IFN response in reaction to TLR9 activation.
Discussion
The role of SLAMF9 in the immune response has been mostly unexplored. Compared with other SLAMF members that are broadly expressed by cells of the immune system, SLAMF9 expression in the steady state is limited mainly to myeloid cells (5), with abundant expression in pDCs compared with cells of the lymphoid lineage (35). Its role was previously studied during inflammation in a double SLAMF9 and SLAMF8-deficient mice (8). Therefore, the specific effect of SLAMF9 was not directly addressed. The main population that was followed in the recent study was macrophages, while the role of SLAMF9 in pDCs was not reported.
This restricted expression of SLAMF9 in pDCs led us to investigate its function in these cells. Gene-expression datasets from previous studies indicated abundant mRNA expression of SLAMF9 on mature pDCs compared with their immediate precursors (40, 42). Therefore, we hypothesized that SLAMF9 might be essential for differentiation of these cells, and followed the maturation state of pDCs in the BM.
The nature of the downstream cascade induced by SLAMF9 in pDCs under steady-state conditions is not clear. Human pDCs do not express slam-associated protein or EAT2, but were shown to express the inhibitory phosphatases SHP-1 and SHP-2, which are involved in inhibiting SLAM signaling (56). Nevertheless, SLAMF9 does not contain a signaling motif in its cytosolic tail, and therefore binding of SHPs is unlikely. It is possible that either an adaptor molecule with a signaling motif can bind to SLAMF9, or that engagement with another pDC through a different SLAMF member can induce downstream signals on the cognate cell.
Notably, we demonstrate that SLAMF9 deficiency in mice results in an enrichment of immature pDCs in the BM, along with a significant accumulation of immature and mature pDCs in the LN. The accumulation of pDCs in the LN is even more significant during inflammation. Since SLAMF9-deficient pDCs express intact levels of other pDCs receptors, such as CCR9, SiglecH, and PDCA, we suggest that SLAMF9 is involved in a distinct maturation step of SCA-1− pDCs into the SCA-1high subset in the BM.
At both the transcriptional and protein level, pDCs lacking SLAMF9 demonstrate reduced expression of pDC activation markers and SCA-1 compared with WT cells, and exhibit reduced capacity to secrete IFN-α and TNF-α. Interestingly, it was recently shown that SLAMF9 is expressed on tumor-associated macrophages of human and murine melanomas. Overexpression of SLAMF9 in a murine macrophage cell line resulted in attenuated migration following LPS stimulation, whereas no significant change in TNF-α levels was detected. In contrast, overexpression of SLAMF9 in a human macrophage-like cell line resulted in reduced levels of TNF-α in response to LPS, supporting a proinflammatory role for SLAMF9 (57).
SpiB was shown to induce a type I IFN response in pDCs and to act synergistically with IRF7 (21). Its deficiency in mice results in much broader defects in pDC differentiation. These defects include not only reduced expression of maturation markers, but also a severe decrease in pDC markers and numbers in the BM. The reduced expression of SpiB in SLAMF9−/− pDCs likely contributes to their impaired functionality in the periphery. Interestingly, gene-expression analysis of SpiB-deficient pDCs revealed that SLAMF9 is one of the most strongly down-regulated genes, with about 10-fold decrease in its expression compared with WT pDCs (21); it therefore remains to be further investigated whether SLAMF9 could directly regulate SpiB expression in pDCs.
The involvement of SLAMs in migration during inflammation was previously demonstrated (58), while their effect under steady state has not been addressed. An increase in migratory potential of pDCs in the LN following inflammation was observed in SLAMF9−/− mice, whereas an opposite outcome was shown in the absence of SLAMF1 (58). Here, we show that SLAMF9−/− pDCs express higher levels of CCR5, which could increase their mobilization to the periphery. Nevertheless, the increase in CCR5 was relatively modest compared with the prominent increase of pDCs in the LN; therefore, other mechanisms apart from migration were considered. Interestingly, mechanisms that control the homeostasis and survival of pDCs in the periphery are still elusive. It was demonstrated that IFN-I expression mediates pDC turnover during viral infections through induction of a proapoptotic pathway (59), and that pDC survival can be enhanced by overexpression of BCL2 (60) or diminished through its inhibition (61). Our results indicate that the lack of SLAMF9 in pDCs could enhance their survival in the LN, and therefore support their accumulation in the steady state.
To follow the role of SLAMF9 during in vivo inflammation, we utilized EAE as an inflammatory model. The accumulation of pDCs in several autoimmune diseases, such as lupus and psoriasis, was described as pathogenic due to their enhanced IFN I production (49). However, the functional significance of pDC accumulation in multiple sclerosis patients and during EAE is still controversial (51, 52).
In this study, SLAMF9 deficiency in mice resulted in a protective effect during EAE, associated with a significant elevated number of immature pDCs in the LN. The exact contribution of this phenotype to disease progression might be attributable to their quantity, quality, and their cross-talk with other immune cells in their microenvironment. It was previously shown that pDCs could play a protective role during the acute phase of EAE by regulation of T cell function. Depletion of pDCs during the acute phase of EAE was accompanied by increased TH17 and TH1 responses and resulted in exacerbated disease (62), while adoptive transfer of pDCs showed an opposite and ameliorating effect (63).
IL-6 and TNF are important mediators in EAE pathogenesis and were both diminished in pDCs lacking SLAMF9 (64). Thus, it could be hypothesized that a reduced costimulatory potential of SLAMF9−/− pDCs along with their impaired IFN response and IL-6 and TNF-α production is likely to diminish the magnitude of the inflammatory response in EAE-induced SLAMF9−/− mice. Finally, the reduced functionality of pDCs lacking SLAMF9 is not restricted to an inflammatory model. Our results show that pDCS derived from CpG immunized mice exhibit aberrant cytokine secretion. Thus, this molecule regulates functionality of pDCs during different modes of stimulation of the immune response.
Collectively, our study identified SLAMF9 as a pDC receptor that can regulate the function and maintenance of these cells in health and during inflammation. We therefore believe that blocking SLAMF9 might be a promising strategy to regulate pDC function and frequency during autoimmune disease.
Supplementary Material
Acknowledgments
The authors thank Prof. Steffen Jung for his support, Dilan Köneş and Efrat Hagai for help on the project, and members of the I.S. laboratory for fruitful discussion and support. I.S. is the incumbent of the Dr. Morton and Ann Kleiman Professorial Chair. This research was supported by the German Cancer Research Center–Israeli Ministry of Science and Technology cooperation in cancer research, ERA-NET TRANSCAN-2 program JTC 2014–project FIRE-CLL, and Binational Science Foundation Grant 711979. L.S. was supported by the Azrieli fellow program funded by The Azrieli Foundation. G.F. is the Incumbent of the David and Stacey Cynamon Research fellow Chair in Genetics and Personalized Medicine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE131633).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900079116/-/DCSupplemental.
References
- 1.Schwartzberg P. L., Mueller K. L., Qi H., Cannons J. L., SLAM receptors and SAP influence lymphocyte interactions, development and function. Nat. Rev. Immunol. 9, 39–46 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Yan Q., et al. , Structure of CD84 provides insight into SLAM family function. Proc. Natl. Acad. Sci. U.S.A. 104, 10583–10588 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cannons J. L., Tangye S. G., Schwartzberg P. L., SLAM family receptors and SAP adaptors in immunity. Annu. Rev. Immunol. 29, 665–705 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Ma C. S., Deenick E. K., The role of SAP and SLAM family molecules in the humoral immune response. Ann. N. Y. Acad. Sci. 1217, 32–44 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Zhang W., et al. , Genetic approach to insight into the immunobiology of human dendritic cells and identification of CD84-H1, a novel CD84 homologue. Clin. Cancer Res. 7 (suppl. 3), 822s–829s (2001). [PubMed] [Google Scholar]
- 6.Fennelly J. A., Tiwari B., Davis S. J., Evans E. J., CD2F-10: A new member of the CD2 subset of the immunoglobulin superfamily. Immunogenetics 53, 599–602 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Fraser C. C., et al. , Identification and characterization of SF2000 and SF2001, two new members of the immune receptor SLAM/CD2 family. Immunogenetics 53, 843–850 (2002). [DOI] [PubMed] [Google Scholar]
- 8.Zeng X., et al. , Combined deficiency of SLAMF8 and SLAMF9 prevents endotoxin-induced liver inflammation by downregulating TLR4 expression on macrophages. Cell. Mol. Immunol. 10.1038/s41423-018-0191-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y. J., IPC: Professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 23, 275–306 (2005). [DOI] [PubMed] [Google Scholar]
- 10.Siegal F. P., et al. , The nature of the principal type 1 interferon-producing cells in human blood. Science 284, 1835–1837 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Swiecki M., Gilfillan S., Vermi W., Wang Y., Colonna M., Plasmacytoid dendritic cell ablation impacts early interferon responses and antiviral NK and CD8(+) T cell accrual. Immunity 33, 955–966 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naik S., Vremec D., Wu L., O’Keeffe M., Shortman K., CD8alpha+ mouse spleen dendritic cells do not originate from the CD8alpha- dendritic cell subset. Blood 102, 601–604 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Onai N., et al. , Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 8, 1207–1216 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Shigematsu H., et al. , Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity 21, 43–53 (2004). [DOI] [PubMed] [Google Scholar]
- 15.Rodrigues P. F., et al. , Distinct progenitor lineages contribute to the heterogeneity of plasmacytoid dendritic cells. Nat. Immunol. 19, 711–722 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sozzani S., Vermi W., Del Prete A., Facchetti F., Trafficking properties of plasmacytoid dendritic cells in health and disease. Trends Immunol. 31, 270–277 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Reizis B., Colonna M., Trinchieri G., Barrat F., Gilliet M., Plasmacytoid dendritic cells: One-trick ponies or workhorses of the immune system? Nat. Rev. Immunol. 11, 558–565 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swiecki M., Colonna M., The multifaceted biology of plasmacytoid dendritic cells. Nat. Rev. Immunol. 15, 471–485 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cisse B., et al. , Transcription factor E2-2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 135, 37–48 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghosh H. S., Cisse B., Bunin A., Lewis K. L., Reizis B., Continuous expression of the transcription factor e2-2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity 33, 905–916 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasaki I., et al. , Spi-B is critical for plasmacytoid dendritic cell function and development. Blood 120, 4733–4743 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Tai L. H., et al. , Positive regulation of plasmacytoid dendritic cell function via Ly49Q recognition of class I MHC. J. Exp. Med. 205, 3187–3199 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Omatsu Y., et al. , Development of murine plasmacytoid dendritic cells defined by increased expression of an inhibitory NK receptor, Ly49Q. J. Immunol. 174, 6657–6662 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Watarai H., et al. , PDC-TREM, a plasmacytoid dendritic cell-specific receptor, is responsible for augmented production of type I interferon. Proc. Natl. Acad. Sci. U.S.A. 105, 2993–2998 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borioni R., Lavanga G., Garofalo M., Garofalo R., [Recent experience in: Saving the inferior mesenteric artery during anterior resection of the rectum] [in Italian]. Minerva Chir. 44, 2123 (1989). [PubMed] [Google Scholar]
- 26.Cao W., et al. , Regulation of TLR7/9 responses in plasmacytoid dendritic cells by BST2 and ILT7 receptor interaction. J. Exp. Med. 206, 1603–1614 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jähn P. S., Zänker K. S., Schmitz J., Dzionek A., BDCA-2 signaling inhibits TLR-9-agonist-induced plasmacytoid dendritic cell activation and antigen presentation. Cell. Immunol. 265, 15–22 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Wang H., et al. , One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ran F. A., et al. , Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chari R., Mali P., Moosburner M., Church G. M., Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat. Methods 12, 823–826 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu H., et al. , Sequence determinants of improved CRISPR sgRNA design. Genome Res. 25, 1147–1157 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kent W. J., et al. , The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heng T. S., Painter M. W.; Immunological Genome Project Consortium , The immunological genome project: Networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Waterstrat A., Liang Y., Swiderski C. F., Shelton B. J., Van Zant G., Congenic interval of CD45/Ly-5 congenic mice contains multiple genes that may influence hematopoietic stem cell engraftment. Blood 115, 408–417 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mercier F. E., Sykes D. B., Scadden D. T., Single targeted exon mutation creates a true congenic mouse for competitive hematopoietic stem cell transplantation: The C57BL/6-CD45.1(STEM) mouse. Stem Cell Rep. 6, 985–992 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sawai C. M., et al. , Transcription factor Runx2 controls the development and migration of plasmacytoid dendritic cells. J. Exp. Med. 210, 2151–2159 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamogawa-Schifter Y., et al. , Ly49Q defines 2 pDC subsets in mice. Blood 105, 2787–2792 (2005). [DOI] [PubMed] [Google Scholar]
- 40.Schlitzer A., et al. , Identification of CCR9- murine plasmacytoid DC precursors with plasticity to differentiate into conventional DCs. Blood 117, 6562–6570 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Zhang J., et al. , Characterization of Siglec-H as a novel endocytic receptor expressed on murine plasmacytoid dendritic cell precursors. Blood 107, 3600–3608 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Niederquell M., et al. , Sca-1 expression defines developmental stages of mouse pDCs that show functional heterogeneity in the endosomal but not lysosomal TLR9 response. Eur. J. Immunol. 43, 2993–3005 (2013). [DOI] [PubMed] [Google Scholar]
- 43.Krug A., et al. , Identification of CpG oligonucleotide sequences with high induction of IFN-alpha/beta in plasmacytoid dendritic cells. Eur. J. Immunol. 31, 2154–2163 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Dai J., Megjugorac N. J., Amrute S. B., Fitzgerald-Bocarsly P., Regulation of IFN regulatory factor-7 and IFN-alpha production by enveloped virus and lipopolysaccharide in human plasmacytoid dendritic cells. J. Immunol. 173, 1535–1548 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Lee M. S., Kim B., Oh G. T., Kim Y. J., OASL1 inhibits translation of the type I interferon-regulating transcription factor IRF7. Nat. Immunol. 14, 346–355 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Kawai T., et al. , IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6, 981–988 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Kato H., et al. , Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Unterholzner L., et al. , IFI16 is an innate immune sensor for intracellular DNA. Nat. Immunol. 11, 997–1004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banchereau J., Pascual V., Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity 25, 383–392 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Gilliet M., Cao W., Liu Y. J., Plasmacytoid dendritic cells: Sensing nucleic acids in viral infection and autoimmune diseases. Nat. Rev. Immunol. 8, 594–606 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Lande R., et al. , Plasmacytoid dendritic cells in multiple sclerosis: Intracerebral recruitment and impaired maturation in response to interferon-beta. J. Neuropathol. Exp. Neurol. 67, 388–401 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Bailey S. L., Schreiner B., McMahon E. J., Miller S. D., CNS myeloid DCs presenting endogenous myelin peptides ‘preferentially’ polarize CD4+ T(H)-17 cells in relapsing EAE. Nat. Immunol. 8, 172–180 (2007). [DOI] [PubMed] [Google Scholar]
- 53.Miller S. D., Karpus W. J., Davidson T. S., Experimental autoimmune encephalomyelitis in the mouse. Curr. Protoc. Immunol. Chapter 15, Unit 15.1 (2010). [DOI] [PubMed] [Google Scholar]
- 54.Pashenkov M., et al. , Two subsets of dendritic cells are present in human cerebrospinal fluid. Brain 124, 480–492 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Webster B., Assil S., Dreux M., Cell-cell sensing of viral infection by plasmacytoid dendritic cells. J. Virol. 90, 10050–10053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hagberg N., et al. , Systemic lupus erythematosus immune complexes increase the expression of SLAM family members CD319 (CRACC) and CD229 (LY-9) on plasmacytoid dendritic cells and CD319 on CD56(dim) NK cells. J. Immunol. 191, 2989–2998 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Dollt C., et al. , The novel immunoglobulin super family receptor SLAMF9 identified in TAM of murine and human melanoma influences pro-inflammatory cytokine secretion and migration. Cell Death Dis. 9, 939 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang N., et al. , Negative regulation of humoral immunity due to interplay between the SLAMF1, SLAMF5, and SLAMF6 receptors. Front. Immunol. 6, 158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Swiecki M., et al. , Type I interferon negatively controls plasmacytoid dendritic cell numbers in vivo. J. Exp. Med. 208, 2367–2374 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Genaro A. M., Gonzalo J. A., Bosca L., Martinez C., CD2-CD48 interaction prevents apoptosis in murine B lymphocytes by up-regulating bcl-2 expression. Eur. J. Immunol. 24, 2515–2521 (1994). [DOI] [PubMed] [Google Scholar]
- 61.Carrington E. M., et al. , Prosurvival Bcl-2 family members reveal a distinct apoptotic identity between conventional and plasmacytoid dendritic cells. Proc. Natl. Acad. Sci. U.S.A. 112, 4044–4049 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bailey-Bucktrout S. L., et al. , Cutting edge: Central nervous system plasmacytoid dendritic cells regulate the severity of relapsing experimental autoimmune encephalomyelitis. J. Immunol. 180, 6457–6461 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duraes F. V., et al. , pDC therapy induces recovery from EAE by recruiting endogenous pDC to sites of CNS inflammation. J. Autoimmun. 67, 8–18 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murphy A. C., Lalor S. J., Lynch M. A., Mills K. H., Infiltration of Th1 and Th17 cells and activation of microglia in the CNS during the course of experimental autoimmune encephalomyelitis. Brain Behav. Immun. 24, 641–651 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.