Significance
Heterozygous in-frame mutations in human STAT3 coding regions underlie the only known autosomal dominant form of hyper IgE syndrome (AD HIES). About 5% of familial cases remain unexplained. We report a deep intronic heterozygous STAT3 mutation, c.1282-89C>T, in 7 relatives with AD HIES. This mutation creates a new exon, encoding a new mRNA (D427ins17) and a mutant loss-of-function, dominant-negative STAT3 protein. This mutant protein was not detected in heterozygous cells from the patient. We show that the D427ins17 mutant allele is dominant-negative despite the production of significantly smaller amounts of mutant than of wild-type protein in heterozygous cells. These findings highlight the importance of searching for deep intronic mutations in STAT3 before considering alternative genetic etiologies of HIES.
Keywords: hyper IgE syndrome, immunodeficiency, STAT3, infectious diseases, dominant negative
Abstract
Heterozygous in-frame mutations in coding regions of human STAT3 underlie the only known autosomal dominant form of hyper IgE syndrome (AD HIES). About 5% of familial cases remain unexplained. The mutant proteins are loss-of-function and dominant-negative when tested following overproduction in recipient cells. However, the production of mutant proteins has not been detected and quantified in the cells of heterozygous patients. We report a deep intronic heterozygous STAT3 mutation, c.1282-89C>T, in 7 relatives with AD HIES. This mutation creates a new exon in the STAT3 complementary DNA, which, when overexpressed, generates a mutant STAT3 protein (D427ins17) that is loss-of-function and dominant-negative in terms of tyrosine phosphorylation, DNA binding, and transcriptional activity. In immortalized B cells from these patients, the D427ins17 protein was 2 kDa larger and 4-fold less abundant than wild-type STAT3, on mass spectrometry. The patients’ primary B and T lymphocytes responded poorly to STAT3-dependent cytokines. These findings are reminiscent of the impaired responses of leukocytes from other patients with AD HIES due to typical STAT3 coding mutations, providing further evidence for the dominance of the mutant intronic allele. These findings highlight the importance of sequencing STAT3 introns in patients with HIES without candidate variants in coding regions and essential splice sites. They also show that AD HIES-causing STAT3 mutant alleles can be dominant-negative even if the encoded protein is produced in significantly smaller amounts than wild-type STAT3.
Hyper IgE syndrome (HIES) is a primary immunodeficiency (Online Mendelian Inheritance in Man #147060), first described in 1966 by Wedgwood and coworkers as Job’s Syndrome (1–4). In 1972, Buckley and coworkers reported additional features of this condition, including high serum IgE levels (5). Further studies documented the autosomal dominant (AD) inheritance of this disorder and gradually delineated various clinical phenotypes (6). AD HIES confers chronic selective susceptibility to infection with certain bacteria, including various staphylococci infecting the skin and lungs, and certain fungi, causing chronic mucocutaneous candidiasis (CMC) in particular (7). One of the hallmarks of these infections is that the associated inflammation is mild or delayed, corresponding to the “cold abscesses” originally reported by Davis et al. (4). Patients also display cutaneous and systemic allergic manifestations and extrahematopoietic features, including facial dysmorphism, the retention of deciduous teeth, osteopenia, hyperextensibility, and vascular abnormalities (6, 8). Clinical outcome is very poor, due largely to the immunodeficiency and infectious diseases of these patients, and patient management is difficult. Hematopoietic stem cell transplantation (HSCT) has been reported for 5 patients, and was apparently successful in 3, with a normalization of STAT3 signaling in hematopoietic cells and a restoration of the corresponding immune responses (9, 10). The other 2 patients had poorer outcomes: One died from posttransplantation complications (11), whereas the other displayed no immunological or clinical improvement despite full donor chimerism post-HSCT (12).
In 2007, heterozygous mutations of STAT3 underlying AD HIES were discovered by Minegishi et al. (13) and confirmed by Holland et al. (14). The STAT3 gene encodes 2 isoforms: STAT3α and STAT3β (15–17). STAT3α is the major isoform; it consists of 770 amino acids and has a molecular weight (MW) of 92 kDa. In mice, STAT3α is essential for embryonic survival (18, 19). STAT3β (85 kDa) is a truncated and less abundant isoform; it is generated by the alternative splicing of exon 23 of STAT3α, leading to deletion of the first 50 nucleotides encoded by this exon. This deletion results in a frameshift, with the addition of 7 alternative amino acids followed by a stop codon. STAT3β therefore lacks the last 55 C-terminal amino acids of STAT3α, including serine 727, which must be phosphorylated for transactivation (TA). Tyrosine 705, which must also be phosphorylated for STAT3 activation, is present in the TA domains of both isoforms (18, 20, 21). Human STAT3 is almost ubiquitously expressed and can be activated by various cytokines in different tissues (22–24). A mouse model of AD HIES has been established by the transgenic expression of a recurrent pathogenic dominant-negative (DN) STAT3 mutation in the germline (25). Moreover, an autosomal recessive (AR) form of HIES was recently attributed to loss-of-function (LOF) mutations of ZNF341, encoding a transcription factor required for STAT3 transcription, expression, activation, and activity (26–28). The disruption of STAT3 activity is, therefore, the unifying feature of AD HIES.
Since 2007, 114 causal mutations in STAT3 have been reported in 477 patients from Africa (Algeria, Comoros, and Morocco), Asia (China, Israel, Korea, Iran, Lebanon, Pakistan, Russia, Saudi Arabia, and Turkey), Australia, and Europe (Belgium, France, Germany, Hungary, Italy, the Netherlands, Poland, Portugal, Sweden, and the United Kingdom). The reported cases include 71 patients from 29 multiplex kindreds (13, 14, 29–53). All of the variants identified, including 5 recurrent mutations (H332Y, R382W, R382Q, V637M, and del463V), are private to the AD HIES group. They are located in the coding exons or the essential splice sites of their flanking introns (8, 13, 33). They can affect the N-terminal domain, DNA-binding domain (DBD), the linker domain, the Src homology 2 (SH2) domain, or the TA domain of STAT3. Most are missense mutations (78%), but some are small (<9 amino acids) in-frame deletions (9%), small (<3 amino acids) in-frame insertions or duplication (2%), essential splice-site mutations resulting in small (<10 amino acids) in-frame deletions or exon skipping (exon 11; 20 amino acids, exon 12; 10 amino acids) (11%) (8, 13, 14, 33, 42, 53, 54), or a large in-frame deletion of 2 exons (38). Remarkably, all of the mutations identified to date are in-frame mutations. However, potential exceptions to this apparent rule were recently reported, in the form of out-of-frame mutations due to nonsense (S381*) (31) or frameshift variants (R13Vfs*23, F493Lfs*508, Y657*) (29). However, no causal link between these out-of-frame mutations and HIES was established, because dominance was not experimentally investigated at the cellular level. The potential mechanism of dominance thus remains elusive.
Only 19 and 5 of the 114 in-frame mutant alleles have been shown experimentally to be LOF (37, 39, 54–57) and DN (13), respectively (SI Appendix, Table S1). In overexpression experiments, 5 mutant STAT3 proteins were found to be produced and phosphorylated in normal amounts, but to be LOF in terms of DNA-binding and transcriptional activity, resulting in a DN effect (13). Given that all of the other mutations identified were also in frame, by inference, these other STAT3 mutations have also been widely assumed to be DN (8, 14). It has been suggested, but not proved, that the recently reported out-of-frame STAT3 mutations are dominant, and pathogenic via haploinsufficiency (31). Moreover, the production of in-frame mutant STAT3 proteins in heterozygous cells has not been documented, making it impossible to define a threshold level above which mutants exert DN effects. Finally, although STAT3 mutations are the only genetic etiology of AD HIES identified to date (13, 14), a small percentage (<5%) of multiplex kindreds meeting the clinical criteria for AD HIES do not have identifiable pathogenic variants of STAT3 (58). In this context, we studied a multiplex family with AD HIES whose STAT3 exons and surrounding intronic regions carried no mutations. We hypothesized that the patients carried a deep intronic mutation, as recently reported for a growing number of inborn errors of immunity (59–61), and we reasoned that the study of this unique family might cast light on the mechanisms of dominance underlying AD HIES.
Results
A Family with AD HIES and a Deep STAT3 Intron Mutation.
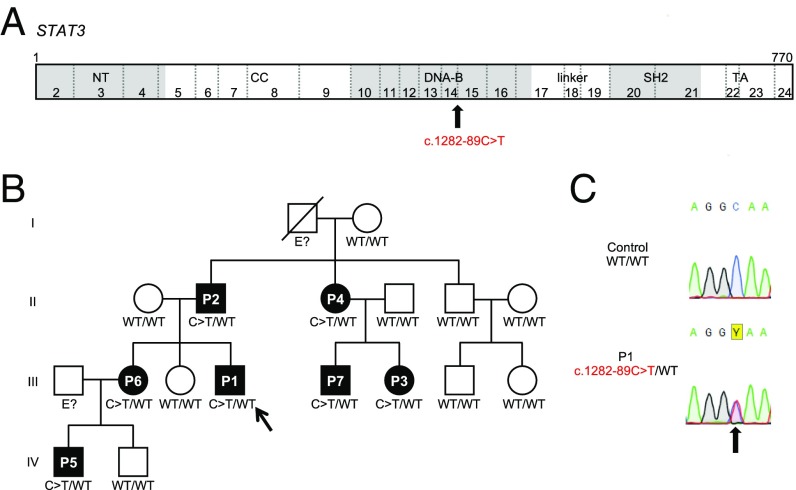
We studied a large kindred originating from and living in France. Over 3 generations, 7 individuals from this kindred (P1 to P7), all born to nonconsanguineous parents, had both bacterial and fungal infections, beginning in childhood, and high serum IgE levels (from 858 IU/mL [P5] to 22,331 IU/mL [P2]). Some of the patients had hypereosinophilia, facial dysmorphism, impaired shedding of deciduous teeth, and hyperextensibility. Their National Institutes of Health HIES scores ranged from 17 to 52 (with a score of 40 typically considered the most appropriate threshold for HIES diagnosis) (6) (Table 1). Sanger sequencing of the exons and flanking intron regions of STAT3 in P1, P2, and P3 identified no rare synonymous or nonsynonymous variants. We performed whole-exome sequencing (WES) on these 3 patients (P1, P2, and P3). Biallelic mutations of DOCK8 (62–65), PGM3 (66), and ZNF341 (26–28) underlie AR disorders that resemble HIES (65, 67, 68); our patients carried no nonsynonymous or essential splice site variations in any of these genes. We performed a genome-wide linkage study on this multiplex family (69). Fourteen regions were linked to HIES in an AD model with complete penetrance: One was on chromosome 17 (29,421,944 to 67,144,147) and contained the STAT3 locus; the other regions contained no candidate genes or variants. WES analysis confirmed the absence of rare nucleotide variants in the exons and flanking intron regions of STAT3. There were no heterozygous copy number variants at the STAT3 locus either. However, a heterozygous intron nucleotide substitution (g.17:40,478,306G>A, c.1282-89C>T) (Fig. 1A), only weakly covered by WES (3, 6, and 8 reads for P1, P2 and P3, respectively), was found 89 nucleotides upstream from exon 15 and 3,121 nucleotides downstream from exon 14. This heterozygous variant was confirmed by Sanger sequencing genomic DNA from the patients’ leukocytes (Fig. 1 B and C). All 7 AD HIES patients from this kindred carried the heterozygous c.1282-89C>T mutation, whereas the 8 asymptomatic relatives tested did not. This mutation had a combined annotation-dependent depletion (CADD) score of 11.9, below the mutation significance cutoff for STAT3 (15.3), suggesting that it was not deleterious. However, the c.1282-89C>T mutation was not found in the Human Gene Mutation Database of pathological variants (HGMD), or in the 1000 Genomes Project (2,504 healthy individuals), Genome Aggregation Database (gnomAD; 15,000), and the National Heart, Lung, and Blood Institute (NHLBI) Trans-Omics for Precision Medicine (TOPMed) Whole Genome Sequencing Program (62,784) database. These results suggest that the deep intronic mutation c.1282-89C>T in STAT3 is private and may be responsible for AD HIES in this kindred.
Table 1.
Infectious and clinical phenotypes of STAT3-deficient patients
| Patient | Sex | Status | Age* | HIES score | Age at first sign | Age at diagnosis | Infections | Dermatitis | Connective tissue signs | Immunological signs | ||||||||||||
| Skin | Pneumatocyst | Bacterial infections | ENT | CMC | Aspergillus | Dysmorphism | Tooth retention | Osteopenia | Fracture | Hyperextensibility | Vascular abnormalities | IgE (IU/mL) | Eosinophils (/mm3) | |||||||||
| P1 | III.3 | M | A | 29 | 52 | <1 | 23 | + | + | + | + | + | + | + | — | — | + | — | — | — | 18,881 | 900 |
| P2 | II.2 | M | A | 57 | 47 | >5 | 51 | + | nd | — | + | + | — | + | + | — | nd | + | — | nd | 22,331 | 2,800 |
| P3 | III.5 | F | A | 34 | 46 | >5 | 28 | + | + | + | + | — | + | + | + | + | nd | — | + | + | 2,747 | — |
| P4 | II.3 | F | A | 59 | 41 | >5 | 55 | + | nd | + | — | + | — | + | + | + | nd | — | + | nd | 4,622 | — |
| P5 | IV.1 | M | A | 5 | 22 | <1 | 2 | + | nd | — | + | — | — | + | — | na | nd | — | — | nd | 858 | 1,690 |
| P6 | III.1 | F | A | 36 | 17 | >5 | 32 | + | — | — | — | + | — | + | — | — | + | — | — | nd | 4,264 | 600 |
| P7 | III.4 | M | A | 36 | 19 | >5 | 32 | — | nd | — | — | — | — | — | + | — | nd | — | + | nd | 1,697 | — |
A, alive; na, not applicable; nd, not done; ENT, ear, nose and throat. Abnormal biological values are shown in bold. +, present; —, absent.
In June 2019. Normal values: IgE < 150 IU/mL, polymorphonuclear eosinophils < 600/mm3.
Fig. 1.
Familial segregation of the STAT3 mutation. (A) Schematic representations of the STAT3 coding sequence annotated with its exons and its various functional domains: N-terminal domain (NT), coiled-coil domain (CC), DBD (DNA-B), linker, SH2 domain, and TA domain. The exons are numbered with Arabic numerals (2 through 24), and exon 1 is a noncoding exon. The position of the STAT3 mutation reported in patients is indicated by a black arrow. (B) Pedigree of the STAT3-deficient kindred. Each generation is designated by a Roman numeral (I through IV), and each patient is designated by an Arabic numeral. Solid black shapes indicate HIES patients. Individuals whose genetic status could not be determined are indicated by “E?” The proband P1 is indicated by an arrow. “C > T” indicates the mutated allele. (C) Sequence chromatograms for STAT3, showing the presence of the heterozygous mutation (black arrow) in the patient’s genomic DNA: c.1282-89C>T (red).
Abnormal Splicing of the Mutant STAT3 Messenger RNA.
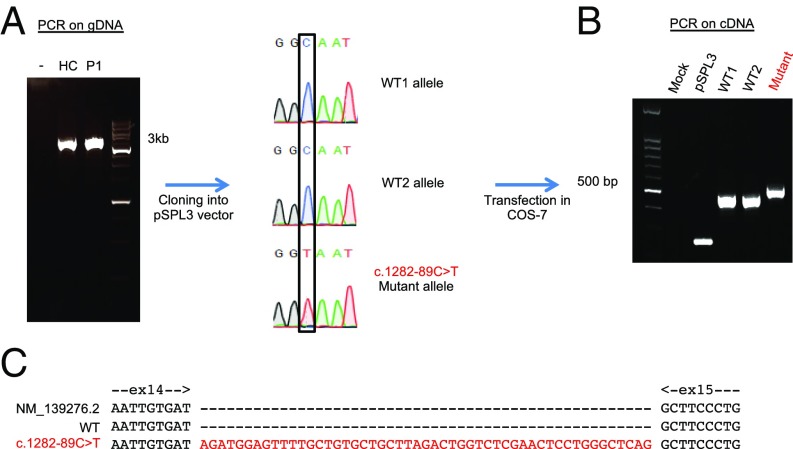
We assessed the potential impact of this deep intronic mutation, by analyzing its surrounding region with a splice site predictor (70–72). The c.1282-89C>T substitution was predicted to create a new donor splicing site in intron 14, which, in combination with a cryptic acceptor site at c.1282-141, may create a new exon of 51 nucleotides. We therefore studied STAT3 messenger RNAs (mRNAs) extracted from a healthy control and from the patients’ Epstein−Barr virus-transformed B cells (EBV-B cells). Amplification of the full-length STAT3 mRNA from the cells of a healthy control, P1, P2, and P3 yielded only one fragment (SI Appendix, Fig. S1A). We then amplified the region of the complementary DNA (cDNA) lying between exons 13 and 15. A single STAT3 amplicon was generated in healthy control cells, whereas 2 different amplicons were generated from P1, P2, and P3 cells (SI Appendix, Fig. S1B). Cloning and sequencing of the cDNA amplified from the cells of P1 revealed an insertion, in about 50% of the clones, of 51 nucleotides between the sequences corresponding to exons 14 and 15 (SI Appendix, Fig. S1C). This 51-nucleotide insertion was predicted to encode an in-frame stretch of 17 amino acids (RWSFAVLLRLVSNSWAQ), between the aspartate residue in position 427 and the alanine residue in position 428, in the DBD of the STAT3 protein. We then inserted a 3.6-kb genomic region from STAT3, encompassing exons 13, 14, and 15, from a healthy control and from P1 into the exon-trapping pSPL3 plasmid (Fig. 2A). We transfected COS-7 cells with the pSPL3 mock vector, or with pSPL3 containing the STAT3 exons 13 to 15 sequence, with or without the c.1282-89C>T mutation (Fig. 2B). All of the transcripts from the DNA carrying the c.1282-89C>T mutation contained the 51 nucleotides inserted between exons 14 and 15, whereas this insertion was not detectable in cells transfected with wild-type (WT) DNA (Fig. 2C). Thus, the c.1282-89C>T mutation creates a new donor site for splicing in intron 14, which, by association with an acceptor site in position c.1282-141, creates a new exon (exon 14b) between exons 14 and 15 of STAT3. Splicing was not leaky, as all of the mRNA products of the mutant allele contained this insertion, which was absent from all of the mRNA products generated from the WT allele. The mutant allele is therefore referred to as D427ins17 in this study.
Fig. 2.
Abnormal mRNA splicing resulting from the c.1282-89C>T mutation. (A) DNA from a healthy donor (HC) or P1 was amplified from nucleotide chr17:40481719 to chr17:40478099 (GRCh37 reference) and inserted into the pSPL3 plasmid. Sanger sequencing was used to check the sequence of the insert. (B) COS-7 cells were transfected with no vector (mock), pSPL3 empty vector (pSPL3), 2 pSPL3 vectors containing the WT genomic STAT3 insert (WT1; WT2), and a pSPL3 vector containing the c.1282-89C>T-mutated genomic STAT3 insert (Mutant). RT-PCR was performed with dUSD2 and dUSA4 primers, to amplify the splicing products 24 h after transfection. (C) The 2 mRNA splicing products from the WT genomic sequence (exons 14 and 15) and from the c.1282-89C>T-mutated genomic sequence (Mutant: exons 14 and 15 plus 51 bp from intron 14 of the STAT3 gene).
Impaired Production and Activation of the D427ins17 STAT3 Protein.
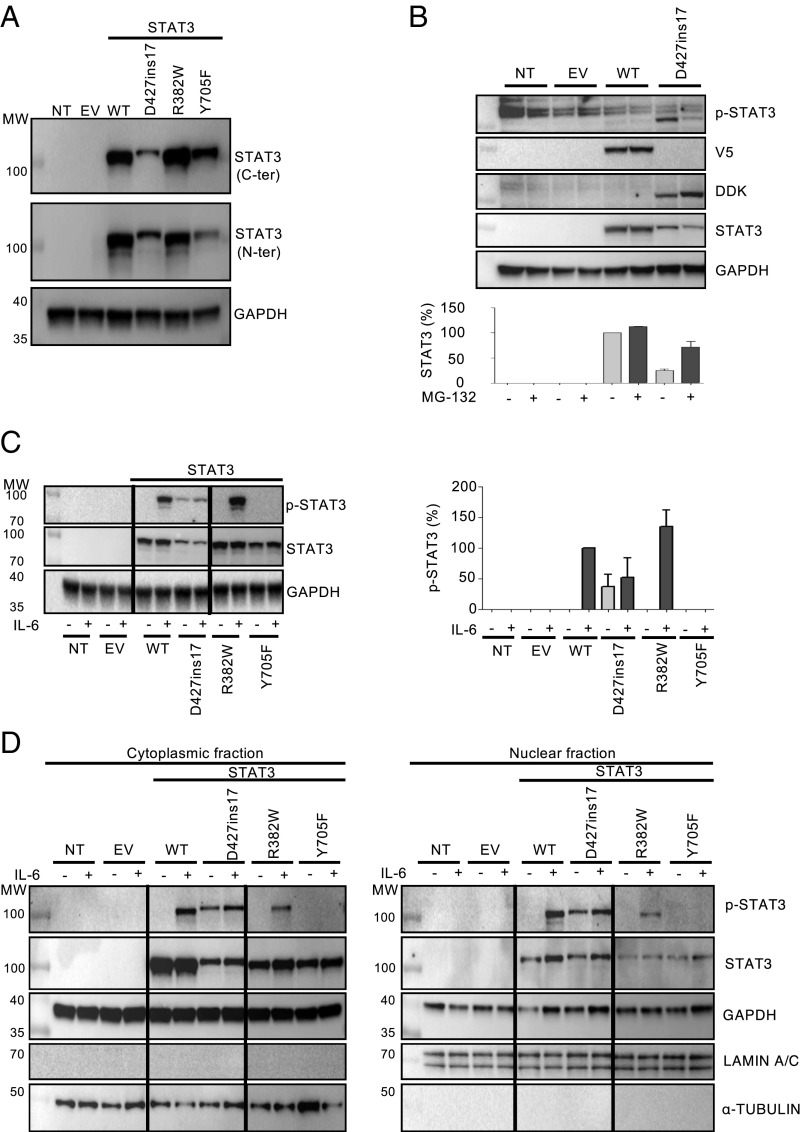
We expressed the cDNA corresponding to this new allele, D427ins17, in the STAT3-deficient colon cancer cell line A4 (STAT3−/− A4). We compared it with 2 known DN mutant alleles: R382W, a missense mutation affecting the DBD in an AD HIES patient demonstrated to be DN through effects on STAT3 binding to the response element in the nucleus and thus on the transcriptional activity of STAT3 (13), and Y705F, a DN missense mutation that abolishes phosphorylation and was generated artificially for the biochemical characterization of STAT3 (73). We measured protein levels with monoclonal antibodies (mAbs) directed against the C- or N-terminal part of STAT3 (Fig. 3A). As expected, neither mAb detected STAT3 protein in untransfected STAT3−/− A4 cells, or STAT3−/− A4 cells transfected with an empty vector (STAT3−/−-EV). With the C-terminal mAb, total STAT3 levels were found to be similar in cells transfected with WT, R382W, and Y705F STAT3, whereas total STAT3 levels were 50% lower following transfection with D427ins17 (Fig. 3A). With the N-terminal mAb, total STAT3 levels were 30% lower in cells transfected with the STAT3 D427ins17 allele (Fig. 3A). We hypothesized that the STAT3 mutant protein was misfolded and degraded by the ubiquitin-dependent proteasome. We measured STAT3 protein levels by Western blots on STAT3−/− A4 cells reconstituted with WT or mutant cDNA, with or without pretreatment with MG-132, a proteasome inhibitor. STAT3 protein levels were similar in cells transfected with WT STAT3 before and after MG-132 treatment, whereas STAT3 protein levels were 3 times higher in cells transfected with STAT3 D427ins17 (Fig. 3B). We then analyzed the phosphorylation of the Y705 residue of STAT3 following IL-6 stimulation. D427ins17 phosphorylation levels were 80% and 90% lower than those of the WT and the R382W mutant, respectively (Fig. 3C). As expected, no STAT3 phosphorylation was detected in Y705F-transfected STAT3−/− A4 cells. Intriguingly, some baseline STAT3 phosphorylation was observed for the D427ins17 mutant, but not for any of the other STAT3 constructs (Fig. 3C). STAT3−/− A4 and STAT3−/−-EV cells served as negative controls. We then investigated the subcellular distribution and nuclear accumulation of the WT and mutant STAT3 proteins in transfected STAT3−/− A4 cells, with and without IL-6 treatment, by Western blotting (Fig. 3D). The WT and all mutant STAT3 proteins were present in the cytoplasm, but D427ins17 protein levels were very low. D427ins17 accumulated similarly to the WT protein in the nucleus following IL-6 stimulation, whereas less R382W was translocated to the nucleus (Fig. 3D), as previously reported (57). Thus, the D427ins17 mutant STAT3 protein is unstable, produced in small amounts, and poorly phosphorylated upon cell stimulation, but such stimulation nevertheless results in its translocation to the nucleus.
Fig. 3.
Characterization of the new STAT3 mutant alleles. Western blot of extracts from nontransfected STAT3−/− A4 cells (NT), A4 cells transfected with Myc-DDK-PCMV6 EV, the STAT3 WT allele, or the STAT3 mutant allele of interest (D427ins17). (A) STAT3 levels in total extracts from STAT3−/− A4 NT or transfected cells. All extracts were probed with antibodies against the C-terminal and the N-terminal parts of the STAT3 protein. (B) Total protein extracts from NT or transfected STAT3−/− A4 cells, after treatment (+) with 20 µM MG-132 for 3 h. All extracts were probed with mAbs specific for the N-terminal part of the STAT3 protein. Values indicate the level of STAT3 protein normalized as a percentage (percent) relative to GAPDH. (C) (Left) Total extracts from NT or transfected STAT3−/− A4 cells, after treatment (+) with 50 ng/mL IL-6 for 20 min. All extracts were probed with mAbs specific for p-STAT3, or the N-terminal part of the STAT3 protein. (Right) Quantification of STAT3 Y705 amino acid phosphorylation in 2 independent experiments. Values indicate the percentage (percent) STAT3 phosphorylation. Values are normalized relative to GAPDH. (D) Cytoplasmic and nuclear extracts of STAT3−/- A4 cells, after treatment (+) with 50 ng/mL IL-6 for 20 min. All extracts were probed with an Ab specific for p-STAT3, and the N-terminal part of the STAT3 protein. The lanes were run on the same gel but were noncontiguous.
The D427ins17 STAT3 Allele Is LOF.
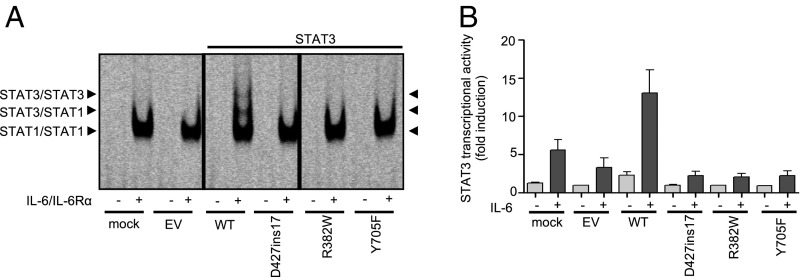
We studied the capacity of the D427ins17 mutant protein to bind cis-regulatory elements in response to IL-6/IL-6Rα, in electrophoretic mobility shift assays (EMSAs) with the m67SIE probe (74). We compared the results obtained with those for the LOF and DN mutants R382W (13) and Y705F (73), respectively, which cannot bind DNA (Fig. 4A). The different DNA−protein complexes observed on EMSA for cells transfected with the WT STAT3 allele were confirmed to be specific in competition experiments including specific unlabeled probe (data not shown). In supershift experiments with specific mAb probes, we found that these complexes contained STAT1 homodimers (lower band), STAT1/STAT3 heterodimers (middle band), and STAT3 homodimers (upper band) (SI Appendix, Fig. S2). The DNA-binding capacity of STAT3 was strongly impaired in cells transfected with D427ins17, R382W, and Y705F, whereas STAT1-binding capacity appeared to be unaffected (Fig. 4A). Only STAT1 DNA-binding complexes were observed in STAT3−/− A4 cells left untransfected or transfected with EV. Finally, we analyzed the transcriptional activity of the mutants in luciferase assays with an IL6-inducible reporter vector. STAT3−/− A4 cells, untransfected or transfected with EV, WT, or mutant STAT3 (D427ins17, R382W, Y705F) alone or together with the luciferase reporter gene, were left untreated or were treated with IL-6. Transfection with the WT STAT3 construct led to an approximately 5-fold increase in IL-6−dependent luciferase activity, whereas no such increase was observed for any of the STAT3 mutants (Fig. 4B). The novel STAT3 mutant allele D427ins17 is LOF in terms of its ability to bind cis-regulatory elements and its transcriptional activity.
Fig. 4.
LOF of the STAT3 mutant alleles in terms of cytokine signaling. (A) EMSA with the m67SIE probe on STAT3−/− A4 cells (NT), and A4 cells transfected with no vector (mock), EV, WT, or STAT3 mutants (D427ins17, R382W, Y705F), without (−) and with (+) stimulation with 100 ng/mL IL-6/IL-6Rα for 30 min. The arrows indicate the STAT3/STAT3 homodimers, STAT3/STAT1 heterodimers, and STAT1/STAT1 homodimers. The lanes were run on the same gel but were noncontiguous. (B) Luciferase assay on STAT3−/− A4 cells transfected with no vector (mock), EV, WT, or STAT3 mutants (D427ins17, R382W, Y705F), together with the luciferase reporter gene, with and without treatment with 100 ng/mL IL-6 for 24 h. The transcriptional activity of the STAT3 promoter normalized against unstimulated activity in EV-transformed cells is plotted on the y axis, and the alleles used for transfection are shown on the x axis.
Detection of the Mutant STAT3 Protein in the Cells of Heterozygous Patients.
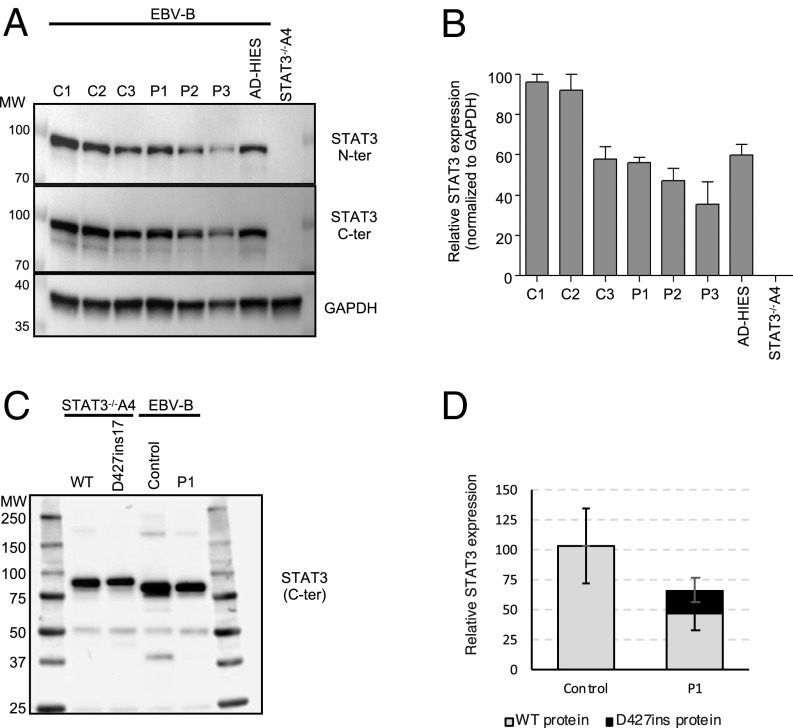
We analyzed STAT3 protein levels by Western blotting in EBV-B cell lines from healthy controls and patients. We studied 3 healthy controls (C1, C2, C3), 3 patients heterozygous for the STAT3 D427ins17 mutation (P1, P2, and P3), and an AD HIES patient heterozygous for the R382W mutation (13). Nontransfected STAT3−/− A4 cells were used as negative controls. We assessed protein levels by probing the blots with mAbs directed against the N-terminal or C-terminal part of STAT3. Similar levels of STAT3 were detected in EBV-B cells from healthy controls and the AD HIES patient with the R382W mutation (16, 18, 75). By contrast, STAT3 levels were about 60 to 70% lower in EBV-B cells from P1 to P3 (Fig. 5 A and B). Furthermore, higher MW proteins were observed when D427ins17 was overexpressed in STAT3−/− A4 cells, but not in cells from the patients (P1, P2, and P3). Given the difference in predicted MW between the WT and D427ins17 STAT3 isoforms (∼2 kDa), we attempted to detect the mutant protein by mass spectrometry (MS). We used an anti-STAT3 mAb for immunoprecipitation (IP) with whole-cell lysates from STAT3−/−A4 cells transfected with WT or D427ins17 STAT3 cDNA, and whole-cell lysates from EBV-B cells from a healthy control and P1 (Fig. 5C). We then performed liquid chromatography−tandem MS (LC-MS/MS) to determine the nature and estimate the relative amounts of the STAT3 isoforms present. Immunoblot analysis of the immunoprecipitated fractions showed that both the WT and D427ins17 STAT3 proteins were efficiently immunoprecipitated from cell extracts (Fig. 5C). Peptide analysis by LC-MS/MS of the D427ins17 mutant protein overproduced in STAT3−/− A4 cells confirmed the presence of 2 unique peptides (WSFAVLLR and LVSNSWAQASLIVTEELHLITFETEVYHQGLK) corresponding to an insertion of 17 amino acids (SI Appendix, Fig. S3). One peptide specific for the WT isoform was detectable, together with 5 other peptides common to the 2 isoforms. Peptide analysis on EBV-B cells from P1 demonstrated the presence of the same specific peptides (SI Appendix, Fig. S3B). IP and LC-MS/MS are not strictly quantitative, but we estimated the level of the mutant isoform in heterozygous EBV-B cells from P1 to be about 5 to 20% that of the WT protein (Fig. 5D). The mutant D427ins17 protein was thus produced in the patients’ lymphocytes, albeit at lower levels than the WT protein, contributing to the lower total STAT3 protein levels in these cells.
Fig. 5.
STAT3 levels in the patient’s EBV-B cells. (A) Endogenous STAT3 levels, as assessed by Western blotting of total extracts from the EBV-B cells of healthy controls (C1, C2, C3), P1, P2, P3, and an AD HIES patient heterozygous for a STAT3 mutation (WT/R382W). STAT3−/−A4 cells were used as a control for STAT3 mAb specificity. All extracts were probed with mAbs against the C- or N-terminal part of the STAT3 protein. (B) Quantification of STAT3 levels (with N- and C-terminal mAbs) relative to GAPDH. The percentage (percent) STAT3 expression is indicated on the y axis, and the cell sample is indicated on the x axis. (C) IP of STAT3 from WT or P1 EBV-B cells and of STAT3 from STAT3−/− A4 cells expressing WT or D427ins17 STAT3. All extracts were probed with an Ab specific for the C-terminal part of the STAT3 protein. (D) Relative amounts of STAT3 D427ins17. Peptide areas were initially normalized by densitometry of the Western blot. Relative quantification was performed for each specific peptide by comparing the normalized peptide area to the areas for each of the 5 common peptides. WT or D427ins17 STAT3-expressing cells were used as a reference.
Impaired STAT3-Dependent Responses in Patients’ EBV-B Cells.
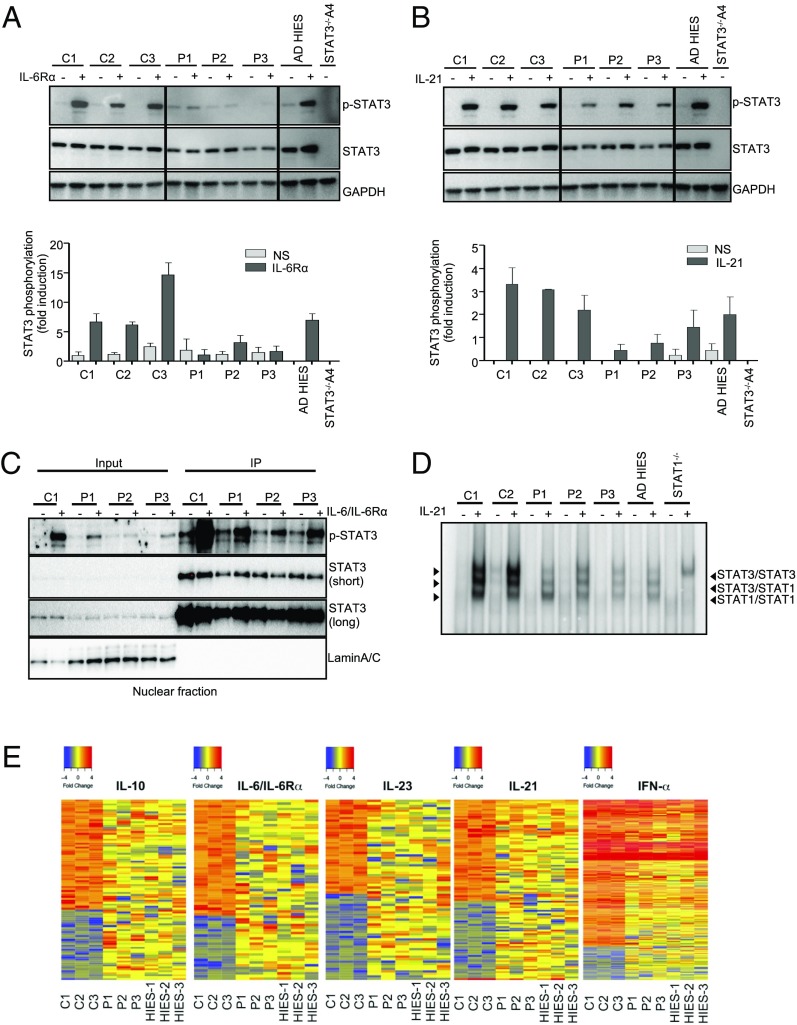
STAT3 is involved in cellular responses to at least IL-6, IL-10, IL-21, IL-27, and type I IFNs (76–78). We performed Western blots to analyze the cellular responses to IL-6 and IL-21 in EBV-B cells from P1, P2, and P3, comparing these responses with those of 3 controls (C1, C2, C3), and an AD HIES patient heterozygous for R382W. Nontransfected STAT3−/− A4 cells were used as a negative control. Following stimulation with IL-6/IL-6Rα (Fig. 6A), and, to a lesser extent, IL-21 (Fig. 6B), STAT3 phosphorylation was detected in the cells of P1, P2, and P3, but at lower levels than in the cells of healthy controls; by contrast, STAT3 phosphorylation levels were normal in the cells of heterozygous R382W patients (13). These results are consistent with the overexpression data, which revealed a phosphorylation defect for the D427ins17 allele (Fig. 3B). Upon stimulation with IL-6/IL-6Rα, we observed similar levels of weak basal phosphorylation in P1, P2, C1, and C2 EBV-B cells. As basal phosphorylation of the D427ins17 STAT3 protein was observed in both the cytoplasmic and nuclear fractions of transfected STAT3−/− A4 cells (Fig. 3D), we measured the levels of phosphorylated STAT3 (p-STAT3) in the nucleus of heterozygous cells from the patients. An analysis of the nuclear fractions of EBV-B cells from a healthy control, P1, P2, and P3, with IL-6/IL-6Rα or without stimulation, showed that p-STAT3 and total STAT3 levels were similar in cells from patients and controls (Fig. 6C). We also analyzed the capacity of the patients’ EBV-B cells to drive gene expression. We first measured the capacity of STAT3 dimers to bind cis-regulatory elements in response to IL-21 by EMSA, using the m67SIE probe in EBV-B cells from healthy controls, P1, and a WT/R382W AD HIES patient (13). Supershift experiments confirmed that the DNA-binding complexes detected by EMSA in healthy control cells were STAT1 homodimers (lower band), STAT1/STAT3 heterodimers (middle band), and STAT3 homodimers (upper band). The DNA-binding capacity of STAT3 was severely impaired in the cells of P1, P2, and P3 (Fig. 6D). Our results for the R382W mutation are consistent with previous studies showing a severe binding impairment for this mutant protein in patients’ EBV-B cells (13, 57). Transcriptome comparisons by RNA sequencing (RNA-seq) on EBV-B cells from D427ins17 patients (P1, P2, P3), AD HIES patients (heterozygous for STAT3 mutations R382W, V463del, and T708N, respectively) and healthy controls (n = 3) revealed that all AD HIES patients had similarly impaired responses to IL-6/IL-6Rα, IL-10, IL-21, and IL-23 stimulation, but a normal response to IFN-α (Fig. 6E). The B lymphocytes of patients heterozygous for D427ins17 had impaired STAT3-dependent responses, implying that heterozygosity for the mutant allele underlies a dominant cellular and clinical phenotype.
Fig. 6.
STAT3 biochemical phenotype in cells from heterozygous patients. Total extracts from the EBV-B cells of healthy controls (C1, C2, C3), P1, P2, P3, and an AD HIES patient heterozygous for a STAT3 mutation (WT/R382W) after treatment (+) with (A) 100 ng/mL IL-6/IL-6Rα or (B) 100 ng/mL IL-21 for 20 min. All extracts were probed with an Ab specific for p-STAT3, and for total STAT3 protein. The graphs show the quantification of the assay, with fold-phosphorylation on the y axis and the EBV-B cell line on the x axis. (C) IP of STAT3 from a healthy control (C1) or patients’ (P1, P2, P3) EBV-B cells. All extracts were probed with mAbs specific for p-STAT3, and the C-terminal part of the STAT3 protein. (D) EMSA with the m67SIE probe on nuclear extracts from EBV-B cells from 3 healthy controls (C1, C2, C3), P1, and an AD HIES patient heterozygous for a STAT3 mutation (WT/R382W), without (−) and with (+) stimulation with 100 ng/mL IL-21 for 30 min. The arrows indicate the STAT3/STAT3 homodimers, the STAT3/STAT1 heterodimers, and the STAT1/STAT1 homodimers. The lanes were run on the same gel but were noncontiguous. (E) RNA-seq data revealing differences in the gene expression patterns of EBV-B cells left unstimulated or stimulated with 50 ng/mL IL-10, 100 ng/mL IL-6/IL-6Rα, 100 ng/mL IL-21, 100 ng/mL IL-23, or 104 IU/mL IFNα. The heat map shows the fold change (FC) in gene expression between the values before and after stimulation on a log2 scale. Red indicates that the gene is up-regulated, and blue indicates that the gene is down-regulated. For each stimulation, the up-regulated genes were filtered according to the criterion that the FC in expression was at least 2, relative to unstimulated conditions, in all 3 healthy control subjects. Accordingly, down-regulated genes were filtered according to the criterion that the FC in expression was at least 2, in all 3 healthy control subjects.
Impaired Cytokine Production by the CD4+ T Cells of STAT3-Deficient Patients.
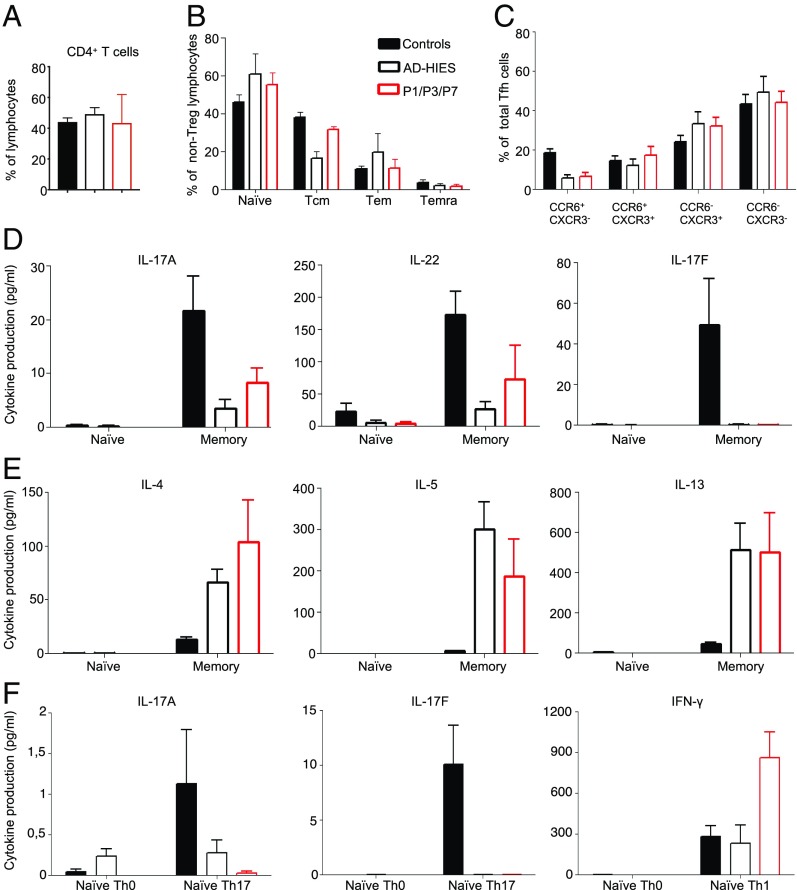
AD HIES patients have normal myeloid and lymphoid cell development, but impaired antigen-dependent differentiation and function (39, 51, 79–83). The lymphocyte subsets are severely affected, with abnormally low proportions of CD4+ and CD8+ central memory T cells (51, 80–82) and memory B cells (39, 84). In this context, we analyzed the distribution of T and B lymphocytes, by flow cytometry, in healthy controls, P1, P3, P7, and AD HIES patients with previously reported heterozygous STAT3 mutations (R382W, I568F, S668Y, T708N, K709E). P1, P3, and P7 had normal frequencies of total CD4+ (Fig. 7A) and CD8+ (not shown) T cells, but impaired differentiation in vivo resulted in low proportions of central memory, effector memory, and terminally differentiated effector memory CD4+ T cells, with a corresponding higher proportion of naive CD4+ T cells than controls (Fig. 7B). Further analysis of memory CD4+ T cells revealed that P1, P3, P7, and other AD HIES patients heterozygous for STAT3 mutations had low proportions of CXCR5−CCR6+CXCR3− CD4+ T cells (Fig. 7C), corresponding to Th17-type cells (85), whereas the proportions of CXCR5−CCR6−CXCR3− (Th2), CXCR5−CCR6+CXCR3+ (Th1*), and CXCR5−CCR6−CXCR3+ (Th1) CD4+ T cells were normal to high in the patients, relative to healthy donors. This aberrant distribution of leukocyte subsets resembles that of other patients with AD HIES (8, 80–82, 85). We then studied the in vitro differentiation of naive CD4+ T cells into effector subsets induced by specific polarizing culture conditions (Th17, Th2, and Th1) (81), and cytokine production upon stimulation with PMA/ionomycin in memory T cells from healthy controls, P1, P3, P7, and other AD HIES patients with heterozygous STAT3 mutations. Under Th0 stimulation, production of the Th17 cytokines IL-17A, IL-17F, and IL-22 by P1, P3, and P7 memory CD4+ T cells was severely impaired or abolished (Fig. 7D and SI Appendix, Fig. S4A). By contrast, P1, P3, and P7 had enhanced Th2 responses, as shown by both the higher frequencies of memory CD4+ T cells expressing the Th2 cytokines IL-4 and IL-13 (SI Appendix, Fig. S4B) and the strong increase in the secretion of IL-4, IL-5, and IL-13 cytokines following in vitro stimulation (Fig. 7E). This enhanced Th2 response is a signature of the high IgE levels typically observed in AD HIES (85). The defects of Th17 cell generation in vivo observed for P1, P3, and P7 were intrinsic to CD4+ T cells, because naïve CD4+ T cells from these patients failed to produce IL-17A and IL-17F in vitro when subjected to Th17-polarizing culture conditions (Fig. 7F). Similarly, the memory CD4+ T cells of these patients did not display the increase in IL-17A and IL-17F secretion in vitro under Th17-polarizing culture conditions typically observed for memory CD4+ T cells from healthy donors (SI Appendix, Fig. S4C). By contrast, in naïve (Fig. 7F) and memory (SI Appendix, Fig. S4C) CD4+ T cells from P1, P3, and P7, the induction and increase, respectively, in IFN-γ secretion following exposure to Th1 polarizing conditions were normal or even stronger than normal.
Fig. 7.
Immunophenotyping and function of the patients’ CD4+ T cells. (A–C) Frequencies (percent) of (A) total CD4+ T cells; (B) naïve, central memory (Tcm), effector memory (Tem), and terminal effector memory (Temra) CD4+ T cells; and (C) Th1 (CXCR3+CCR6−), Th17 (CCR6+CXCR3−), Th1* (CXCR3+CCR6+), and Th2 (CXCR3−CCR6−) type CD4+ T cells. (D and E) Memory CD4+ T cells were purified by sorting from healthy donors (n = 12), patients with STAT3 mutations and AD HIES patients (n = 7), and P1, P3, and P7. They were cultured for 5 days under Th0 conditions (anti-CD3/CD2/CD28 antibody-coated beads). The secretion (picograms per milliliter) of (D) Th17 (IL-17A, IL-22, IL-17F) and (E) Th2 (IL-4, IL-5, IL-13) cytokines was then assessed with cytometric bead arrays or by enzyme-linked immunosorbent assay. (F) Naive CD4+ T cells were purified by sorting from healthy donors (n = 12), patients with STAT3 mutations and AD HIES (n = 7), and P1, P3, and P7. The cells were cultured for 5 days under Th0, Th17, or Th1 polarizing conditions. The secretion (picograms per milliliter) of IL-17A, IL-17F, and IFNγ was then assessed. The data shown are the mean ± SEM.
Impaired Humoral Immunity due to STAT3 DN Mutations.
All patients tested had normal frequencies of CD20+ B cells (SI Appendix, Fig. S5A). However, the proportion of naïve B cells was higher than normal, whereas the proportion of memory B cells was lower than normal (SI Appendix, Fig. S5B), as previously reported for AD HIES patients (39, 83). The stimulation of human naïve B cells with IL-21 strongly induces their differentiation into Ab-secreting cells (84), but this process is almost entirely abolished by typical pathogenic STAT3 mutations (39, 83) (SI Appendix, Fig. S5C). By contrast, the residual memory B cell population of heterozygous STAT3 individuals responded almost normally to IL-21 (83) (SI Appendix, Fig. S5D). Consistent with these findings, naïve B cells from P1, P3, and P7 produced much less (10- to 50-fold less) IgM, IgG, and IgA in response to IL-21 than normal naïve B cells, whereas the memory B cells of these patients displayed responses 30 to 100% as strong as those of memory B cells from healthy donors (SI Appendix, Fig. S5 C and D). In conclusion, the defects of in vivo and in vitro T and B lymphocyte differentiation in D427ins17 heterozygous patients observed in phenotypic and functional analyses are similar to those previously described for AD HIES. Thus, heterozygosity for the D427ins17 allele underlies an AD phenotype of adaptive immunity that is typical of AD HIES.
DN Effect of the D427ins17 STAT3 Allele.
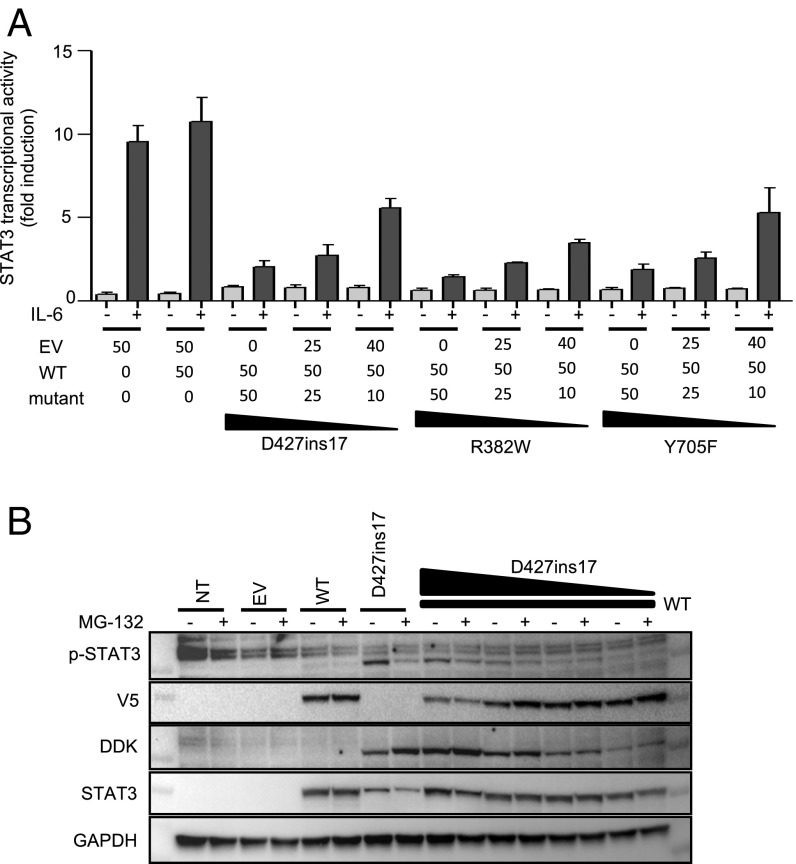
We analyzed the molecular mechanism underlying the dominance of the D427ins17 allele, by expressing its cDNA in the presence of a WT STAT3 cDNA and measuring the transcriptional activity of the STAT3 proteins produced. We used HEK293T cells, which express endogenous WT STAT3. We cotransfected these cells with various amounts of mutant cDNA (from 10 to 50 ng) and a constant amount of WT STAT3 cDNA (50 ng). The cells were then stimulated with IL-6 and their activation was measured in a luciferase reporter assay. In cells transfected with the reporter luciferase and an EV, IL-6 up-regulated luciferase activity by up to 10 times. This transcriptional activity was due to the endogenous STAT3 present in HEK293T cells (Fig. 8A). For the D427ins17 mutant, transfection with increasing amounts of mutant cDNA resulted in a dose-dependent decrease in luciferase activity (Fig. 8A), as for 2 STAT3 mutant constructs (R382W and Y705F) already demonstrated to be DN (13, 73, 86). Thus, the D427ins17 allele has a DN effect on endogenous STAT3 function in HEK293T cells (SI Appendix, Fig. S6). The mutant STAT3 protein was stabilized in presence of proteasome inhibitor in STAT3−/− A4 cells (Fig. 3B). We analyzed the coexpression of mutant and WT STAT3 proteins, by cotransfecting STAT3−/− A4 cells was various amounts of mutant STAT3 plasmid (DDK-tagged) and a constant amount of WT-STAT3 plasmid (V5-tagged) (Fig. 8B). WT protein expression stabilized the mutant protein. WT protein levels were lower in the presence of the mutant protein, and this effect was dose-dependent, suggesting that the mutant protein has a negative effect on the WT protein. Overall, our findings show that heterozygosity for the D427ins17 deep intronic variant underlies a dominant phenotype in patients’ cells through a mechanism of negative dominance, as opposed to haploinsufficiency.
Fig. 8.
DN effect of the D427ins17 allele. (A) Luciferase assay on HEK293T cells transfected with EV or with the WT STAT3 plasmid (50 ng), or cotransfected with the WT STAT3 plasmid (50 ng) and various amounts (50, 25, and 10 ng) of STAT3 mutants (D427ins17, R382W, Y705F), together with the pGL4.47 reporter construct, and an expression vector for Renilla luciferase. After 24 h, the transfected cells were stimulated with 100 ng/mL IL-6 for 24 h. The transcriptional activity of the STAT3 promoter normalized relative to unstimulated conditions in cells transfected with EV is shown on the y axis, and the alleles used for transfection are indicated on the x axis. (B) Total extracts of STAT3−/− A4 cells transfected with either the WT STAT3 (V5-tagged) allele or the D427ins17 mutant STAT3 allele (DDK-tagged) or cotransfected with various amounts of mutant STAT3 plasmid with constant amounts of WT STAT3 plasmid (50 ng). The transfected cells were treated with 20 µM MG-132 for 3 h. Extracts were probed with Abs specific for p-STAT3, the V5 tag, the DDK tag, and the N-terminal part of the STAT3 protein.
Discussion
Human STAT3 is the only known AD HIES-causing gene, with at least 114 heterozygous mutations reported (13, 14, 29–52). A small proportion (<5%) of multiplex kindreds with AD HIES, and of sporadic cases with HIES, have no identifiable pathogenic variants of STAT3, suggesting the existence of additional, as yet unidentified genetic lesions (58). Some sporadic cases have recently been shown to carry biallelic mutations of ZNF341, resulting in a close phenocopy of AD HIES (26–28). We report here a deep intronic mutation (c.1282-89C>T; p.D427ins17) of human STAT3 underlying a classic form of AD HIES. This private mutation creates a new donor splicing site that, in association with a cryptic acceptor splicing site, leads to the creation of a new in-frame 51-bp exon and the production of a new transcript, encoding the D427ins17 STAT3 protein. Only 2 of the previously reported 114 HIES-associated STAT3 alleles are in-frame insertions. Deep intronic mutations have been reported for only 11 AD inborn errors of immunity (60), none of which is dominant by a mechanism of DN. AD collagen VI-related dystrophy was recently shown to be due to a recurrent heterozygous deep intronic mutation in COL6A1, leading to a 72-bp insertion that is dominant by DN (87). This and other reports (59–61) highlight the importance of searching for deep intronic mutations before considering alternative genetic etiologies, particularly for conditions in which most patients are found to have mutations of a single gene, as for AD HIES and STAT3 (58). Based on the findings reported here, it is not impossible that most other multiplex kindreds with unexplained AD HIES, and many or most sporadic cases, carry heterozygous deep intronic mutations of STAT3. Other regulatory mutations are less likely, as they would not be expected to be DN. Whole-genome sequencing should be considered in such patients, with analyses initially focusing on the STAT3 gene, and candidate variants should be preferentially studied by analyzing STAT3 mRNA structure.
Our findings are also of interest from a purely mechanistic and biochemical perspective. The pathogenic mechanism of the first 5 heterozygous STAT3 mutations identified was shown in 2007 to involve negative dominance (13). None of the other human mutations since reported (n = 109) have been experimentally tested to determine the mechanism involved. The DN mechanism for those that have been studied involves impairments of STAT3 production, phosphorylation, translocation, DNA binding, or a combination of these processes (13, 37, 54–57, 88, 89). We show that the D427ins17 STAT3 allele acts in a DN manner by increasing WT STAT3 degradation and impairing DNA binding by mutant-containing heterodimers. Almost all AD HIES-causing STAT3 mutations are in frame and located in specific domains of the protein, strongly suggesting that negative dominance is the general rule for the mechanism of action of these mutations. Frameshift mutations of STAT3 have recently been reported, and this discovery was interpreted as evidence that dominance might also operate by haploinsufficiency (29, 31). Dominance effects based on negative dominance require the encoded mutant protein to inhibit the WT protein, by interfering with its function (90). The levels of mutant protein required to exert negative dominance differ between genes. Negative dominance is typically documented in overexpression systems. Such studies can suggest, but not prove, that the mutant protein is DN in the cells of heterozygous patients. We measured the levels of mutant STAT3 protein in the patients’ cells. We showed, by MS, that the mutant D427ins17 STAT3 protein was produced at relatively low levels, accounting for between 5% and 20% of the total STAT3 protein present. We therefore conclude that levels of LOF in-frame mutant STAT3 protein as low as 5% of the total amount of STAT3 protein present may be sufficient for DN. Low levels of DN proteins have already been reported for other inborn errors of immunity, such as AD TRAF3 (91) and CARD11 deficiencies (92), and for other types of AD inborn errors (93).
Materials and Methods
Methods are available in SI Appendix. They describe all of the genomic, molecular biology, cellular, and proteomic approaches used in this article. The institutional review board of Necker Hospital approved the study and informed consent was obtained from all patients or their families (for minors), in accordance with the Helsinki Declaration (CNIL authorization no. 908256, 14 October 2008).
Supplementary Material
Acknowledgments
We thank the patients and their families for participating in this study. We thank James E. Darnell and Claudia Mertens for providing the STAT3-deficient A4 cell line. We thank the members of the laboratory, especially David Hum for his technical assistance and Lahouari Amar, Yelena Nemirovskaya, Dominick Papandrea, Mark Woollett, Céline Desvallées, and Cécile Patissier for administrative assistance; Alicia Fernandes from the Vecteurs Viraux et Transfert de Gènes (Platform VVTG), Necker Hospital, for generating immortalized B cell lines for the patients; and members of Sidra Medicine’s genomics core facility team for their contribution to the mRNA-Seq library preparation and Illumina sequencing. This work was supported by the St. Giles Foundation, the Rockefeller University, INSERM, Paris Descartes University, HHMI, Sidra Medicine, the Job Research Foundation, and the French National Research Agency (ANR) under the “PNEUMOPID” project (Grant ANR 14-CE15-0009-01). J.K. was supported by the ANR (Grant ANR-14-CE15-0009-01) and the Imagine Institute (Imagine 4th year PhD scholarship). V.B. was supported by the ANR (Grant NKIRP-ANR-13-PDOC-0025-01). S.G.T. is supported by the National Health and Medical Research Council of Australia and the Job Research Foundation.
Footnotes
Conflict of interest statement: S.G.T., J.D.M., and M.A.C. are coauthors on a 2017 Letter to the Editor. C.P., A.P., J.-L.C., and J.D.M. are coauthors of a 2016 research article (PMID: 27114460); C.P., A.P., and J.-L.C. did not collaborate actively with J.D.M. for this article.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901409116/-/DCSupplemental.
References
- 1.Bousfiha A., et al. , The 2017 IUIS phenotypic classification for primary immunodeficiencies. J. Clin. Immunol. 38, 129–143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Picard C., et al. , International Union of Immunological Societies: 2017 Primary Immunodeficiency Diseases Committee report on inborn errors of immunity. J. Clin. Immunol. 38, 96–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Q., Boisson B., Béziat V., Puel A., Casanova J. L., Human hyper-IgE syndrome: Singular or plural? Mamm. Genome 29, 603–617 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis S. D., Schaller J., Wedgwood R. J., Job’s syndrome. Recurrent, “cold”, staphylococcal abscesses. Lancet 1, 1013–1015 (1966). [DOI] [PubMed] [Google Scholar]
- 5.Buckley R. H., Wray B. B., Belmaker E. Z., Extreme hyperimmunoglobulinemia E and undue susceptibility to infection. Pediatrics 49, 59–70 (1972). [PubMed] [Google Scholar]
- 6.Grimbacher B., et al. , Hyper-IgE syndrome with recurrent infections–An autosomal dominant multisystem disorder. N. Engl. J. Med. 340, 692–702 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Li J., Casanova J. L., Puel A., Mucocutaneous IL-17 immunity in mice and humans: Host defense vs. excessive inflammation. Mucosal Immunol. 11, 581–589 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandesris M. O., et al. , Autosomal dominant STAT3 deficiency and hyper-IgE syndrome: Molecular, cellular, and clinical features from a French national survey. Medicine (Baltimore) 91, e1–e19 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goussetis E., et al. , Successful long-term immunologic reconstitution by allogeneic hematopoietic stem cell transplantation cures patients with autosomal dominant hyper-IgE syndrome. J. Allergy Clin. Immunol. 126, 392–394 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Patel N. C., Gallagher J. L., Torgerson T. R., Gilman A. L., Successful haploidentical donor hematopoietic stem cell transplant and restoration of STAT3 function in an adolescent with autosomal dominant hyper-IgE syndrome. J. Clin. Immunol. 35, 479–485 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Nester T. A., et al. , Effects of allogeneic peripheral stem cell transplantation in a patient with job syndrome of hyperimmunoglobulinemia E and recurrent infections. Am. J. Med. 105, 162–164 (1998). [DOI] [PubMed] [Google Scholar]
- 12.Gennery A. R., Flood T. J., Abinun M., Cant A. J., Bone marrow transplantation does not correct the hyper IgE syndrome. Bone Marrow Transplant. 25, 1303–1305 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Minegishi Y., et al. , Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature 448, 1058–1062 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Holland S. M., et al. , STAT3 mutations in the hyper-IgE syndrome. N. Engl. J. Med. 357, 1608–1619 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Caldenhoven E., et al. , STAT3β, a splice variant of transcription factor STAT3, is a dominant negative regulator of transcription. J. Biol. Chem. 271, 13221–13227 (1996). [DOI] [PubMed] [Google Scholar]
- 16.Schaefer T. S., Sanders L. K., Nathans D., Cooperative transcriptional activity of Jun and Stat3 beta, a short form of Stat3. Proc. Natl. Acad. Sci. U.S.A. 92, 9097–9101 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong Z., Wen Z., Darnell J. E. Jr, Stat3 and Stat4: Members of the family of signal transducers and activators of transcription. Proc. Natl. Acad. Sci. U.S.A. 91, 4806–4810 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maritano D., et al. , The STAT3 isoforms alpha and beta have unique and specific functions. Nat. Immunol. 5, 401–409 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Takeda K., et al. , Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc. Natl. Acad. Sci. U.S.A. 94, 3801–3804 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker S., Groner B., Müller C. W., Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature 394, 145–151 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Marino F., et al. , STAT3β controls inflammatory responses and early tumor onset in skin and colon experimental cancer models. Am. J. Cancer Res. 4, 484–494 (2014). [PMC free article] [PubMed] [Google Scholar]
- 22.Casanova J.-L., Holland S. M., Notarangelo L. D., Inborn errors of human JAKs and STATs. Immunity 36, 515–528 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Shea J. J., et al. , The JAK-STAT pathway: Impact on human disease and therapeutic intervention. Annu. Rev. Med. 66, 311–328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kane A., et al. , STAT3 is a central regulator of lymphocyte differentiation and function. Curr. Opin. Immunol. 28, 49–57 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Steward-Tharp S. M., et al. , A mouse model of HIES reveals pro- and anti-inflammatory functions of STAT3. Blood 123, 2978–2987 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.August A., Who regulates whom: ZNF341 is an additional player in the STAT3/TH17 song. Sci. Immunol. 3, eaat9779 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Béziat V., et al. , A recessive form of hyper-IgE syndrome by disruption of ZNF341-dependent STAT3 transcription and activity. Sci. Immunol. 3, eaat4956 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frey-Jakobs S., et al. , ZNF341 controls STAT3 expression and thereby immunocompetence. Sci. Immunol. 3, eaat4941 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abolhassani H., et al. , Clinical, immunologic, and genetic spectrum of 696 patients with combined immunodeficiency. J. Allergy Clin. Immunol. 141, 1450–1458 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Jiao H., et al. , Novel and recurrent STAT3 mutations in hyper-IgE syndrome patients from different ethnic groups. Mol. Immunol. 46, 202–206 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Natarajan M., et al. , Aspergillosis, eosinophilic esophagitis, and allergic rhinitis in signal transducer and activator of transcription 3 haploinsufficiency. J. Allergy Clin. Immunol. 142, 993–997.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Renner E. D., et al. , STAT3 mutation in the original patient with Job’s syndrome. N. Engl. J. Med. 357, 1667–1668 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Woellner C., et al. , Mutations in STAT3 and diagnostic guidelines for hyper-IgE syndrome. J. Allergy Clin. Immunol. 125, 424–432 e8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Freeman A. F., et al. , Lung parenchyma surgery in autosomal dominant hyper-IgE syndrome. J. Clin. Immunol. 33, 896–902 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Heimall J., et al. , Paucity of genotype-phenotype correlations in STAT3 mutation positive Hyper IgE Syndrome (HIES). Clin. Immunol. 139, 75–84 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma C. S., et al. , Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J. Exp. Med. 205, 1551–1557 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Papanastasiou A. D., Mantagos S., Papanastasiou D. A., Zarkadis I. K., A novel mutation in the signal transducer and activator of transcription 3 (STAT3) gene, in hyper-IgE syndrome. Mol. Immunol. 47, 1629–1634 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Schimke L. F., et al. , Diagnostic approach to the hyper-IgE syndromes: Immunologic and clinical key findings to differentiate hyper-IgE syndromes from atopic dermatitis. J. Allergy Clin. Immunol. 126, 611–617.e1 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Avery D. T., et al. , B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J. Exp. Med. 207, 155–171 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim H. J., Kim J. H., Shin Y. K., Lee S. I., Ahn K. M., A novel mutation in the linker domain of the signal transducer and activator of transcription 3 gene, p.Lys531Glu, in hyper-IgE syndrome. J. Allergy Clin. Immunol. 123, 956–958 (2009). [DOI] [PubMed] [Google Scholar]
- 41.Powers A. E., et al. , Coccidioides immitis meningitis in a patient with hyperimmunoglobulin E syndrome due to a novel mutation in signal transducer and activator of transcription. Pediatr. Infect. Dis. J. 28, 664–666 (2009). [DOI] [PubMed] [Google Scholar]
- 42.van de Veerdonk F. L., et al. , Milder clinical hyperimmunoglobulin E syndrome phenotype is associated with partial interleukin-17 deficiency. Clin. Exp. Immunol. 159, 57–64 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Felgentreff K., et al. , Severe eczema and Hyper-IgE in Loeys-Dietz-syndrome—Contribution to new findings of immune dysregulation in connective tissue disorders. Clin. Immunol. 150, 43–50 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Giacomelli M., et al. , SH2-domain mutations in STAT3 in hyper-IgE syndrome patients result in impairment of IL-10 function. Eur. J. Immunol. 41, 3075–3084 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Kumánovics A., et al. , Rapid molecular analysis of the STAT3 gene in Job syndrome of hyper-IgE and recurrent infectious diseases. J. Mol. Diagn. 12, 213–219 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma C. S., et al. , Functional STAT3 deficiency compromises the generation of human T follicular helper cells. Blood 119, 3997–4008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Merli P., et al. , Hyper IgE syndrome: Anaphylaxis in a patient carrying the N567D STAT3 mutation. Pediatr. Allergy Immunol. 25, 503–505 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Mogensen T. H., Jakobsen M. A., Larsen C. S., Identification of a novel STAT3 mutation in a patient with hyper-IgE syndrome. Scand. J. Infect. Dis. 45, 235–238 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Sundin M., et al. , Novel STAT3 mutation causing hyper-IgE syndrome: Studies of the clinical course and immunopathology. J. Clin. Immunol. 34, 469–477 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Wolach O., et al. , Variable clinical expressivity of STAT3 mutation in hyperimmunoglobulin E syndrome: Genetic and clinical studies of six patients. J. Clin. Immunol. 34, 163–170 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Zhang L. Y., et al. , Clinical features, STAT3 gene mutations and Th17 cell analysis in nine children with hyper-IgE syndrome in mainland China. Scand. J. Immunol. 78, 258–265 (2013). [DOI] [PubMed] [Google Scholar]
- 52.Hagl B., et al. , Key findings to expedite the diagnosis of hyper-IgE syndromes in infants and young children. Pediatr. Allergy Immunol. 27, 177–184 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Al Khatib S., et al. , Defects along the T(H)17 differentiation pathway underlie genetically distinct forms of the hyper IgE syndrome. J. Allergy Clin. Immunol. 124, 342–348.e5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Renner E. D., et al. , Novel signal transducer and activator of transcription 3 (STAT3) mutations, reduced T(H)17 cell numbers, and variably defective STAT3 phosphorylation in hyper-IgE syndrome. J. Allergy Clin. Immunol. 122, 181–187 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alcántara-Montiel J. C., et al. , Functional characterization of two new STAT3 mutations associated with hyper-IgE syndrome in a Mexican cohort. Clin. Genet. 89, 217–221 (2016). [DOI] [PubMed] [Google Scholar]
- 56.He J., et al. , STAT3 mutations correlated with hyper-IgE syndrome lead to blockage of IL-6/STAT3 signalling pathway. J. Biosci. 37, 243–257 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Pelham S. J., Lenthall H. C., Deenick E. K., Tangye S. G., Elucidating the effects of disease-causing mutations on STAT3 function in autosomal-dominant hyper-IgE syndrome. J. Allergy Clin. Immunol. 138, 1210–1213.e5 (2016). [DOI] [PubMed] [Google Scholar]
- 58.Hsu A. P., Davis J., Puck J. M., Holland S. M., Freeman A. F., “Autosomal dominant hyper IgE syndrome” in GeneReviews, Adam M. P. et al. , Eds. (University of Washington, Seattle, 2010). [Google Scholar]
- 59.Boisson B., et al. , Rescue of recurrent deep intronic mutation underlying cell type-dependent quantitative NEMO deficiency. J. Clin. Invest. 129 587–597 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vaz-Drago R., Custódio N., Carmo-Fonseca M., Deep intronic mutations and human disease. Hum. Genet. 136, 1093–1111 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Hagl B., et al. , Somatic alterations compromised molecular diagnosis of DOCK8 hyper-IgE syndrome caused by a novel intronic splice site mutation. Sci. Rep. 8, 16719 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Engelhardt K. R., et al. , Large deletions and point mutations involving the dedicator of cytokinesis 8 (DOCK8) in the autosomal-recessive form of hyper-IgE syndrome. J. Allergy Clin. Immunol. 124, 1289–1302.e4 (2009). Erratum in: J. Allergy Clin. Immunol. 125, 743 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Su H. C., Jing H., Zhang Q., DOCK8 deficiency. Ann. N. Y. Acad. Sci. 1246, 26–33 (2011). [DOI] [PubMed] [Google Scholar]
- 64.Zhang Q., Davis J. C., Dove C. G., Su H. C., Genetic, clinical, and laboratory markers for DOCK8 immunodeficiency syndrome. Dis. Markers 29, 131–139 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Q., et al. , Combined immunodeficiency associated with DOCK8 mutations. N. Engl. J. Med. 361, 2046–2055 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y., et al. , Autosomal recessive phosphoglucomutase 3 (PGM3) mutations link glycosylation defects to atopy, immune deficiency, autoimmunity, and neurocognitive impairment. J. Allergy Clin. Immunol. 133, 1400–1409.e5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sassi A., et al. , Hypomorphic homozygous mutations in phosphoglucomutase 3 (PGM3) impair immunity and increase serum IgE levels. J. Allergy Clin. Immunol. 133, 1410–1419.e13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watford W. T., O’Shea J. J., Human tyk2 kinase deficiency: Another primary immunodeficiency syndrome. Immunity 25, 695–697 (2006). [DOI] [PubMed] [Google Scholar]
- 69.Belkadi A., et al. ; Exome/Array Consortium , Whole-exome sequencing to analyze population structure, parental inbreeding, and familial linkage. Proc. Natl. Acad. Sci. U.S.A. 113, 6713–6718 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cartegni L., Wang J., Zhu Z., Zhang M. Q., Krainer A. R., ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31, 3568–3571 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Desmet F. O., et al. , Human splicing finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 37, e67 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pertea M., Lin X., Salzberg S. L., GeneSplicer: A new computational method for splice site prediction. Nucleic Acids Res. 29, 1185–1190 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaptein A., Paillard V., Saunders M., Dominant negative stat3 mutant inhibits interleukin-6-induced Jak-STAT signal transduction. J. Biol. Chem. 271, 5961–5964 (1996). [DOI] [PubMed] [Google Scholar]
- 74.Mertens C., Haripal B., Klinge S., Darnell J. E., Mutations in the linker domain affect phospho-STAT3 function and suggest targets for interrupting STAT3 activity. Proc. Natl. Acad. Sci. U.S.A. 112, 14811–14816 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang Y., et al. , Stat3 isoforms, alpha and beta, demonstrate distinct intracellular dynamics with prolonged nuclear retention of Stat3beta mapping to its unique C-terminal end. J. Biol. Chem. 282, 34958–34967 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Minegishi Y., Hyper-IgE syndrome. Curr. Opin. Immunol. 21, 487–492 (2009). [DOI] [PubMed] [Google Scholar]
- 77.Minegishi Y., Karasuyama H., Defects in Jak-STAT-mediated cytokine signals cause hyper-IgE syndrome: Lessons from a primary immunodeficiency. Int. Immunol. 21, 105–112 (2009). [DOI] [PubMed] [Google Scholar]
- 78.Schindler C., Levy D. E., Decker T., JAK-STAT signaling: From interferons to cytokines. J. Biol. Chem. 282, 20059–20063 (2007). [DOI] [PubMed] [Google Scholar]
- 79.Saito M., et al. , Defective IL-10 signaling in hyper-IgE syndrome results in impaired generation of tolerogenic dendritic cells and induced regulatory T cells. J. Exp. Med. 208, 235–249 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ives M. L., et al. , Signal transducer and activator of transcription 3 (STAT3) mutations underlying autosomal dominant hyper-IgE syndrome impair human CD8(+) T-cell memory formation and function. J. Allergy. Clin. Immunol. 132, 400–411.e9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ma C. S., et al. , Unique and shared signaling pathways cooperate to regulate the differentiation of human CD4+ T cells into distinct effector subsets. J. Exp. Med. 213, 1589–1608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Siegel A. M., et al. , A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity 35, 806–818 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Deenick E. K., et al. , Naive and memory human B cells have distinct requirements for STAT3 activation to differentiate into antibody-secreting plasma cells. J. Exp. Med. 210, 2739–2753 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bryant V. L., et al. , Cytokine-mediated regulation of human B cell differentiation into Ig-secreting cells: Predominant role of IL-21 produced by CXCR5+ T follicular helper cells. J. Immunol. 179, 8180–8190 (2007). [DOI] [PubMed] [Google Scholar]
- 85.Ma C. S., et al. , Monogenic mutations differentially affect the quantity and quality of T follicular helper cells in patients with human primary immunodeficiencies. J. Allergy Clin. Immunol. 136, 993–1006.e1 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mohr A., Fahrenkamp D., Rinis N., Müller-Newen G., Dominant-negative activity of the STAT3-Y705F mutant depends on the N-terminal domain. Cell Commun. Signal. 11, 83 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bolduc V., et al. ; COL6A1 Intron 11 Study Group , A recurrent COL6A1 pseudoexon insertion causes muscular dystrophy and is effectively targeted by splice-correction therapies. JCI Insight 4, 124403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bocchini C. E., et al. , Protein stabilization improves STAT3 function in autosomal dominant hyper-IgE syndrome. Blood 128, 3061–3072 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kreins A. Y., et al. , Human TYK2 deficiency: Mycobacterial and viral infections without hyper-IgE syndrome. J. Exp. Med. 212, 1641–1662 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Herskowitz I., Functional inactivation of genes by dominant negative mutations. Nature 329, 219–222 (1987). [DOI] [PubMed] [Google Scholar]
- 91.Pérez de Diego R., et al. , Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity 33, 400–411 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dorjbal B., et al. , Hypomorphic caspase activation and recruitment domain 11 (CARD11) mutations associated with diverse immunologic phenotypes with or without atopic disease. J. Allergy Clin. Immunol. 143, 1482–1495 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Khajavi M., Inoue K., Lupski J. R., Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur. J. Hum. Genet. 14, 1074–1081 (2006). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.