Significance
A hypothesis for the paradoxical evolution of programmed cell death posits that suicide mechanisms were co-opted from processes that were originally pro-survival. Here we show that meiotically programmed mitochondrial release of the Endonuclease G (EndoG) homolog Nuc1 occurs during sporulation in budding yeast. While EndoG release is implicated in animal cell apoptotic death, we find that Nuc1 release is pro-survival, protecting spores from the detrimental consequences of unrestrained viral expression.
Keywords: yeast, gametogenesis, Nuc1, mitochondria, viruses
Abstract
The programmed release of apoptogenic proteins from mitochondria is a core event of apoptosis, although ancestral roles of this phenomenon are not known. In mammals, one such apoptogenic protein is Endonuclease G (EndoG), a conserved mitochondrial nuclease that fragments the DNA of dying cells. In this work, we show that budding yeast executes meiotically programmed mitochondrial release of an EndoG homolog, Nuc1, during sporulation. In contrast to EndoG’s ostensible pro-death function during apoptosis, Nuc1 mitochondrial release is pro-survival, attenuating the cytosolic L-A and Killer double-stranded RNA mycoviruses and protecting meiotic progeny from the catastrophic consequences of their derepression. The protective viral attenuation role of this pathway illuminates a primordial role for mitochondrial release of EndoG, and perhaps of apoptosis itself.
An outstanding question of evolution concerns how programmed cell death (PCD) arose given its maladaptive consequences for the executing cell. Paradoxically, PCD is widely found in unicellular organisms, although it is not well understood in them. Mathematical modeling approaches suggest that host–virus interactions drove the evolution of PCD in unicellular eukaryotes concurrent with an emerging capacity for multicellularity (1, 2). Unicellular yeast species harbor numerous cytosolic viruses that have no extracellular phase and are only vertically transmitted through cytoplasmic inheritance (3). The most comprehensively studied of these is Killer, a paired system of the L-A and M double-stranded RNA viruses in Saccharomyces cerevisiae. L-A produces a viral particle that houses and propagates M, which itself encodes a secreted toxin that kills neighboring uninfected cells and also confers toxin immunity to the host (3). Differing Killer toxins that act through distinct mechanisms are encoded by varied M genomes, but each requires the L-A virus (4). Genetic studies reveal that M1, which encodes the K1 Killer toxin, exists in clear conflict with its host, although how and if this relates to PCD or other developmental occurrences in yeast is not known (5).
Yeast sporulation employs internal meiotic divisions and culminates in the development of spore progeny within the remnant of the mother cell (6). PCD of this remnant cell occurs as an intrinsic aspect of sporulation and is executed through vacuolar rupture, leading to mother cell autolysis (7, 8). Spores survive this process, called meiotic PCD, through coordinated development of their protective spore coats (7). Curiously, discarded meiotic nuclei are frequently swept up in meiotic PCD, where their DNA is fragmented into nucleosomal ladders in a manner requiring NUC1, the yeast homolog of the Endonuclease G (EndoG) family of mitochondrial nucleases (8). This finding is reminiscent of EndoG promoting DNA fragmentation in apoptotic cells following its mitochondrial release, although EndoG is not required for apoptosis per se and its roles in this process are debated (9–13). NUC1 function in yeast is similarly puzzling, as meiotic PCD is unperturbed in nuc1∆/∆ mutants beyond the absence of fragmented DNA (8). Indeed, although activated genome fragmentation pathways accompany diverse forms of PCD in plants, animals, protists, and fungi, the adaptive purposes of this association are unclear (14–16).
In this work, we show that Nuc1 enters the cytosol through meiotically programmed release from mitochondria. Mitochondrial release of Nuc1 requires the yeast voltage-dependent anion channel (VDAC), a mitochondrial outer membrane protein that is implicated in the mitochondrial release of apoptogenic proteins in humans (17, 18). We hypothesized that cytosolic Nuc1 might have additional targets beyond discarded meiotic nuclei and demonstrate that Nuc1 attenuates the L-A and Killer viruses that reside in the cytosol. When developing nuc1∆ spores also lack the function of SKI3, an essential component of the most well-characterized yeast antiviral pathway, they exhibit Killer-dependent lethality. Genetic and biochemical analyses indicate that mitochondrially released Nuc1 collaborates with SKI3 to prevent the lethal accumulation of the Killer toxin within the spores. We also investigated Nuc1 in the context of the L-A virus, as Killer viruses are relatively rare in wild isolates (19, 20). In strains harboring L-A but lacking M, our studies reveal that synergistically acting meiotic attenuation of L-A by Nuc1 and Ski3 prevents a massive accumulation of L-A. Independent of M, L-A causes respiratory growth defects in nuc1∆ ski3∆ meiotic segregants that are suggestive of mitochondrial dysfunction, revealing a fitness consequence of infection with the L-A virus. Similarly, sporulation of a strain lacking both copies of NUC1 is associated with a dramatic accumulation of L-A and spores that exhibit respiratory deficiency, confirming NUC1’s meiotic role in viral attenuation.
Results and Discussion
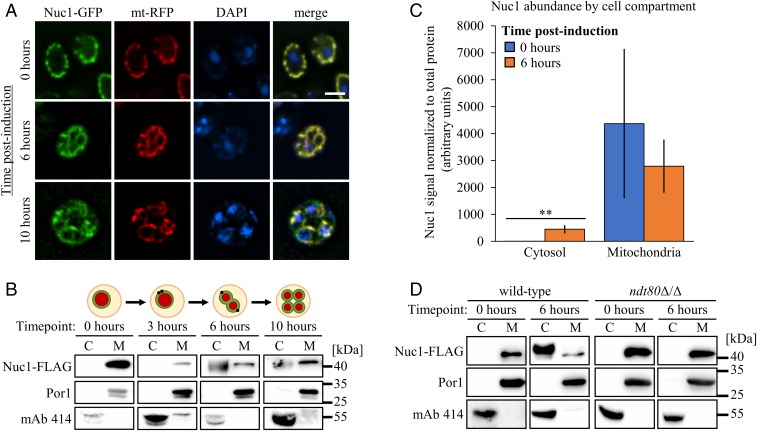
The observation of NUC1-dependent DNA ladders that can accompany meiotic PCD suggests that Nuc1 is released from mitochondria during sporulation (8). We first confirmed Nuc1 mitochondrial localization during meiosis using fluorescence microscopy of chromosomally integrated Nuc1-GFP (green fluorescent protein) together with a plasmid encoding a mitochondrial targeted red fluorescent protein (RFP) in the efficiently sporulating SK1 strain background, which accomplishes meiotic nuclear divisions in 8 to 10 h in batch culture (8) (Fig. 1A). With one exception noted below, all experiments described here were performed in isogeneic SK1 strains. Interestingly, some Nuc1-GFP signal detected at the midmeiotic 6-h time point did not appear to overlap with mitochondria, although this was difficult to quantify (Fig. 1A).
Fig. 1.
Developmental regulation of Nuc1 location during meiosis. (A) Nuc1 localization assessed at 0, 6, and 10 h postinduction of sporulation in fixed cells expressing Nuc1-GFP and mito-RFP (mt-RFP). Cells were stained with DAPI to visualize nuclei and mitochondrial nucleoids. (Scale bar, 4 μm.) (B) Anti–Nuc1-FLAG Western blots of mitochondrial (M) and concentrated cytosolic (C) cell fractions sampled at the indicated time points from a synchronized sporulation timecourse of a strain expressing Nuc1-FLAG. A schematic depiction of meiotic progression at each time point is shown above the corresponding Western blots. Fractions were probed with antibodies against Por1 and mAb 414 to validate successful enrichment of mitochondrial and cytosolic contents, respectively. Molecular weight markers are given on the far right. (C) Quantification of Nuc1 abundance in cytosolic and mitochondrial cell fractions at 0 and 6 h postinduction . Data points represent mean of biological replicates (n = 4 per time point) ± SD. Statistical significance was determined by 2-tailed Student’s t test (**P < 0.01). (D) Anti–Nuc1-FLAG Western blots of cell fractions sampled at meiotic time points from wild-type and a mutant ndt80Δ/Δ strain expressing Nuc1-FLAG (fractionation controls included). Strains used: MEY322, MEY783, MMY7415, MMY7599, and SCY53.
To more sensitively assess the subcellular localization of Nuc1, we measured the abundance of a FLAG epitope-tagged allele of Nuc1 in biochemically prepared cytosolic and mitochondrial fractions from cells undergoing sporulation. The integrity of these cell compartment fractions was first confirmed using Western blotting to detect proteins known to localize to the mitochondria or the cytosol (SI Appendix, Fig. S1). As expected, Nuc1-FLAG fractionated exclusively with mitochondria, as marked by the Por1 mitochondrial protein, at the onset of sporulation (Fig. 1B). Beginning at the 6-h time point and persisting through the postmeiotic 10-h time point, we detected robust Nuc1-FLAG accumulation in concentrated cytosolic fractions (Fig. 1B). We quantified this in 4 biological replicates and determined that a small but significant fraction of total Nuc1 (∼13% of the total signal) accumulated in the cytosol during meiosis, suggesting that a portion of Nuc1 had been released from mitochondria (Fig. 1C). Furthermore, Nuc1-FLAG mitochondrial release required NDT80, a transcription factor that commits cells to sporulation and directs meiotic PCD (6, 7, 21), confirming this phenomenon as an intrinsic and NDT80-programmed aspect of sporulation (Fig. 1D).
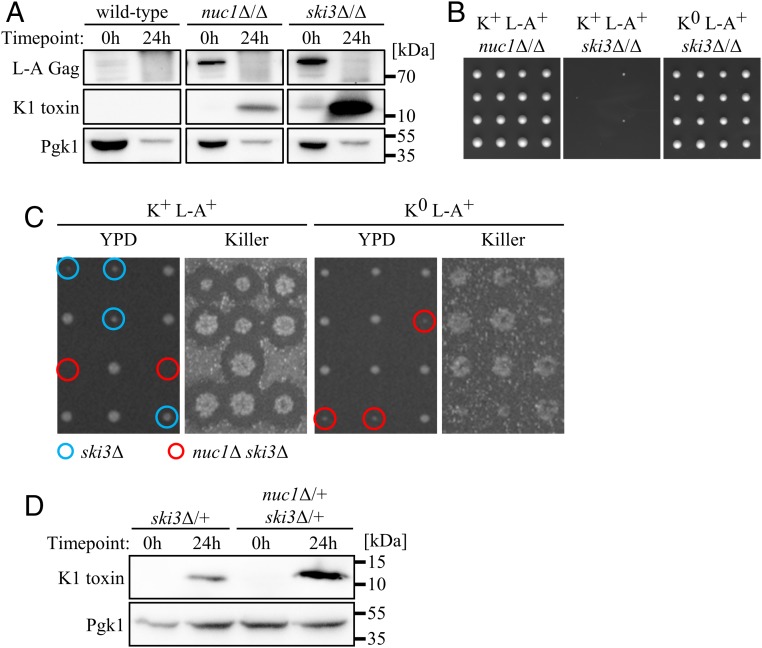
Our fractionation experiments were performed using standard sporulation conditions that do not exhibit NUC1-dependent DNA ladders from uncellularized meiotic nuclei (8). We therefore considered other roles for cytosolic Nuc1, which, like EndoG, can digest RNA as well as DNA (22, 23). The SK1 strain background expresses the K1 form of Killer toxin, encoded by the M1 genome (24, 25). Consistent with an early study (26), we found that an M1-infected and Killer-positive (K+) nuc1∆/∆ diploid strain exhibited increased abundance of L-A Gag, the major coat protein of the L-A viral particle (Fig. 2A). Curiously, L-A Gag levels plummeted following sporulation in both wild-type and nuc1∆/∆ strains (Fig. 2A). We hypothesized that this decrease in L-A Gag was a consequence of M1, which has a known parasitic relationship toward L-A (SI Appendix, Fig. S2A) (27). Using an antibody with specificity for the 11-kDa alpha subunit of K1, we were unable to detect Killer toxin within wild-type cells before or following sporulation, consistent with its nature as a secreted protein (Fig. 2A) (28, 29). In contrast, we found that a sporulated nuc1∆/∆ strain accumulated substantial amounts of Killer toxin (Fig. 2A). We compared the mobility of the secreted form of mature K1 toxin with the intracellular accumulating form we detected during sporulation and found that they were identical (SI Appendix, Fig. S2B).
Fig. 2.
Nuc1 mediates viral repression in meiotic yeast. (A) Anti-K1 toxin and anti–L-A Gag Western blots of whole-cell lysates sampled from synchronized sporulations of homozygous K+ strains at 0 and 24 h postinduction of sporulation with Pgk1 loading control. (B) Subsection of homozygous mutant dissections with 4 tetrads per strain, imaged 3 d postdissection on YPD at 30 °C. (C) Tetrad dissections of K+ L-A+ (Left) and K0 L-A0 (Right) nuc1Δ/+ ski3Δ/+ mutants. Viral genotypes were confirmed by RT-PCR. (Left) Two days postdissection on YPD at 30 °C; Right, Killer plates showing killing activity. Known or inferred haploid genotypes of interest are circled in blue (ski3Δ, K+ dissection only) and red (nuc1Δ ski3Δ). (D) Anti-K1 toxin Western blots of whole-cell lysates sampled from synchronized sporulations of heterozygous K+ L-A+ strains at 0 and 24 h postinduction of sporulation. Pgk1 as loading control. Strains used: MMY5914, MMY5977, MMY6117, MMY6170, MMY6461, MMY7464, MMY7974, MMY7977, and MMY7978.
Although the observation of intracellular mature K1 toxin accumulation is unusual, perturbation of the secretory pathway through mutation of SEC1 has been shown to cause the accumulation of an apparent mature form of K1 toxin (29). During sporulation, post-Golgi secretory vesicles are directed toward prospore membranes, suggesting that secretory pathway rewiring may lead to intracellular K1 accumulation (6, 30). Sporulation may thus be a developmental scenario in which Killer attenuation is particularly crucial. Indeed, using RT-qPCR, we found that bulk M1 RNA levels were repressed 5-fold during wild-type sporulation (SI Appendix, Fig. S2C). Surprisingly, nuc1∆/∆ mutants exhibited a similar degree of M1 repression during sporulation (SI Appendix, Fig. S2C). These findings suggest that, rather than targeting M1 transcripts for digestion, Nuc1 may act indirectly to reduce K1 toxin levels, perhaps by repressing translation of M1 transcripts or through some activity directed toward the L-A virus. However, the parasitic relationship between M1 and L-A complicates this line of inquiry.
Our findings illuminate a sporulation-specific role of NUC1 in Killer attenuation and we therefore considered the roles of established anti-Killer pathways during sporulation. The SKI2, 3, and 8 (superkiller) genes encode subunits of a well-characterized cytoplasmic complex that attenuates Killer expression in vegetative cells at least in part through translational repression of M1-encoded transcripts (3, 31). Consistent with a previous study, we found that deletion of both copies of SKI3 caused increased L-A Gag levels in mitotic cells (Fig. 2A) (5). Analogous to our observations in nuc1Δ/Δ mutants, L-A Gag was depleted during sporulation of a K+ ski3∆/∆ mutant strain (Fig. 2A). Sporulation of this strain caused a large accumulation of K1 toxin and an according defect in bulk M1 RNA repression (Fig. 2A and SI Appendix, Fig. S2C). While nuc1∆/∆ and ski3∆/∆ both sporulated efficiently, ski3∆/∆ produced nearly 100% inviable spores, suggesting that the high degree of Killer toxin accumulation within them caused their lethality (Fig. 2B). To confirm this, we generated a ski3∆/∆ strain lacking the M1 virus (K0 L-A+) and found that Killer loss completely suppressed the lethal phenotype (Fig. 2B). Moreover, deletion of both copies of KEX1, which encodes a protease that accomplishes the final alpha-subunit trimming steps required for K1 maturation, also suppressed the lethality exhibited, as observed following sporulation of a ski3∆/∆ kex1∆/∆ strain (SI Appendix, Fig. S2D) (3, 32). This suppression was associated with accumulation of K1 toxin with slightly reduced mobility, suggesting that the action of intracellular accumulated mature K1 toxin accounted for spore lethality (SI Appendix, Fig. S2E). These findings identify a heretofore-unknown essential role for Killer attenuation during sporulation that is dependent on the SKI complex and show that NUC1 contributes to this attenuation.
To assess the relative contributions of NUC1 and SKI3 for meiotic Killer attenuation, we dissected a doubly heterozygous K+ nuc1∆/+ ski3∆/+ diploid strain and found that nuc1∆ ski3∆ spores were inviable (Fig. 2C). Sporulation of this nuc1∆/+ ski3∆/+ strain caused an accumulation of Killer toxin that was substantially greater than that exhibited by a ski3∆/+ strain, suggesting that elevated Killer toxin levels in nuc1∆ ski3∆ spores accounted for their lethality (Fig. 2D). Indeed, eradication of M1 in a K0 L-A+ nuc1∆/+ ski3∆/+ strain suppressed nuc1∆ ski3∆ synthetic lethality, confirming that NUC1 and SKI3 attenuate lethal Killer toxin accumulation during sporulation through parallel mechanisms (Fig. 2C). To determine if the nuclease activity of Nuc1 was required for Killer attenuation, we employed a previously characterized enzymatically dead allele of Nuc1, Nuc1-H138A (33). We introduced plasmids expressing either NUC1 or NUC1(H138A) under the control of the inducible GAL1-10 promoter into a K+ nuc1∆/+ ski3∆/+ diploid strain from the W303 strain background, which exhibits a greater proficiency for galactose induction than the SK1 background. W303, like SK1, is infected with the M1 virus (34). We found that overexpression of NUC1 but not NUC1(H138A) during spore germination rescued the nuc1∆ ski3∆ synthetic lethality, indicating that the nuclease activity of Nuc1 was essential for its attenuation of Killer virus (SI Appendix, Table S1). Western blotting and cell fractionation experiments showed that Nuc1 and Nuc1(H138A) were produced at comparable levels and that some cytosolic accumulation accompanied their overexpression, consistent with attenuation of Killer by cytosolic Nuc1 (SI Appendix, Fig. S3).
Notably, ski3∆ single mutant spore clones produced by sporulation of a K+ nuc1∆/+ ski3∆/+ strain exhibited a slight degree of slow growth but were always viable (Fig. 2C). The viability of these ski3∆ spores contrasts with the Killer-dependent lethality exhibited by spores produced from a K+ ski3∆/∆ homozygous strain (Fig. 2B). Together, these genetic results highlight an essential role for SKI3 in Killer attenuation during sporulation that is conferred by the diploid genome during meiosis, preceding sporogenesis and its associated spore-autonomous gene expression. Specifically, our results show that SKI3 expressed during meiosis contributes to the health of the spore progeny, suggesting that it acts during meiosis to impact spore health and/or that inherited SKI3 gene products function within developing spores. We refer to this as the maternal stage of sporulation.
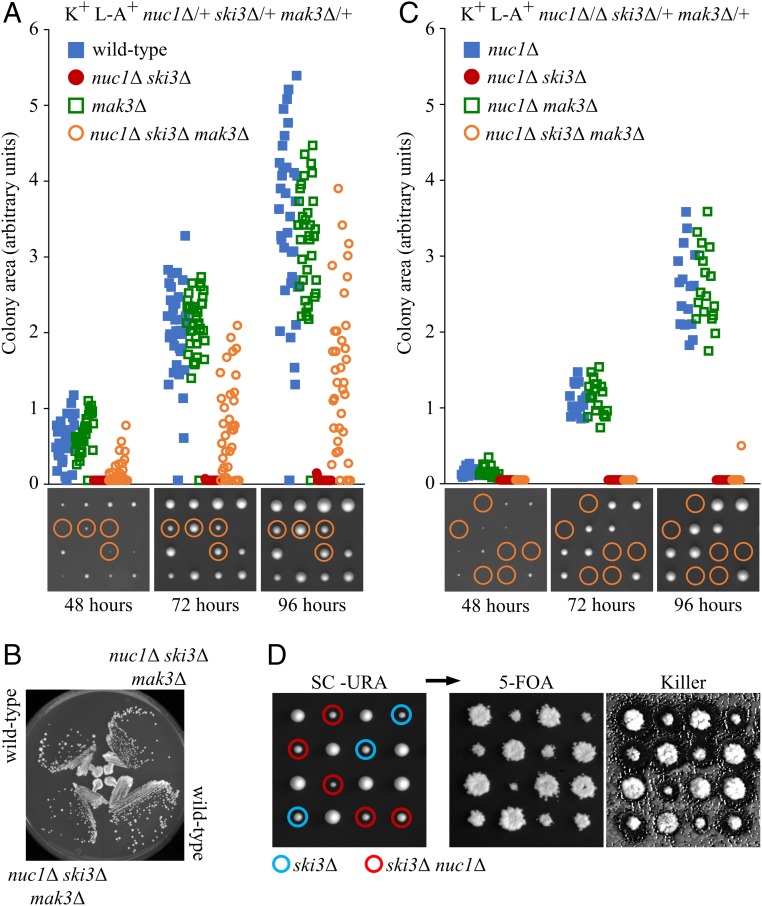
As Nuc1 mitochondrial release initiates during the maternal meiotic stage and persists into sporogenesis (Fig. 1B), we devised a genetic experiment to determine the relative contributions of maternal versus spore autonomous NUC1 function for Killer attenuation. MAK3 encodes the enzymatic subunit of the NatC N-terminal acetyltransferase complex that is essential for Killer maintenance in vegetative cells (35). Foundational studies reveal that NatC is not required for Killer maintenance during sporogenesis; rather, spore clones lacking NatC only lose Killer following germination and subsequent mitotic proliferation (36, 37). Utilizing this feature of NatC-dependent Killer maintenance, we constructed a K+ nuc1∆/+ ski3∆/+ mak3∆/+ triply heterozygous diploid strain and measured spore colony growth following its sporulation and dissection. As expected, mak3∆ always segregated with a Killer-minus phenotype. The lethality of nuc1∆ ski3∆ mutants was rescued by mak3∆ in ∼90% of the triple mutants (Fig. 3A). Although the rescued spore clones showed variably delayed postgermination growth kinetics, upon subculture, the nuc1∆ ski3∆ mak3∆ isolates exhibited normal growth (Fig. 3B). These results show that the lethality caused by loss of spore-autonomous Killer attenuation by SKI3 and NUC1 could be overcome if the nascent colony lacked MAK3 function, presumably through K1 toxin dilution during initial divisions following germination. In contrast, nuc1∆ ski3∆ mak3∆ spores produced by dissecting a K+ nuc1∆/∆ ski3∆/+ mak3∆/+ strain were inviable, revealing that the combination of the loss of maternal NUC1 and spore-autonomous SKI3 resulted in insurmountable Killer activity (Fig. 3C). These findings identify an important maternal contribution for NUC1 in Killer attenuation, corresponding with when Nuc1 mitochondrial release was initiated. Thus, both maternal and spore-autonomous NUC1 function to attenuate Killer.
Fig. 3.
The developmental context of yeast antiviral activity. (A and C) Quantifications of colony growth in dissections of nuc1Δ/+ ski3Δ/+ mak3Δ/+ and nuc1Δ/Δ ski3Δ/+ mak3Δ/+. Representative dissections shown below at the corresponding time points with nuc1Δ ski3Δ mak3Δ progeny circled. Growth of known/inferred haploid genotypes of interest over 3 d were derived by measuring colony surface area on plates imaged at each time point. Data shown in figure was assembled from multiple dissections, with (A) n = 143 colonies measured for nuc1Δ/+ ski3Δ/+ mak3Δ/+ dissections and (C) n = 60 colonies measured for nuc1Δ/Δ ski3Δ/+ mak3Δ/+. (B) Growth of wild-type and nuc1Δ ski3Δ mak3Δ haploids on YPD at 30 °C, imaged 2 d after streaking. (D) The K+ L-A+ nuc1Δ/+ ski3Δ/+ parent strain was transformed with a plasmid expressing Nuc1 from its endogenous promoter and sporulated for tetrad analysis. Plates were imaged 3 d postdissection on SC–URA at 30 °C (Left) then replica-plated to SC + 5-FOA to counter select against plasmid (Right). Genotypes of interest are denoted by blue (ski3Δ) and red (nuc1Δ ski3Δ) circles. Strains used: MMY5994, MMY7776, MMY7827, MMY7829, MMY7839, MMY7950, and MMY7951.
To confirm that the essential NUC1-mediated Killer attenuation observed in nuc1Δ ski3Δ spores occurred during sporulation, we rescued nuc1∆ ski3∆ spore synthetic lethality by dissecting a K+ nuc1∆/+ ski3∆/+ strain containing a NUC1-URA3 plasmid. Consistent with the hypothesis that NUC1’s essential function in Killer attenuation occurred during sporulation, mitotically proliferating nuc1∆ ski3∆ mutants could readily lose the NUC1 rescuing plasmid despite retaining Killer activity (Fig. 3D).
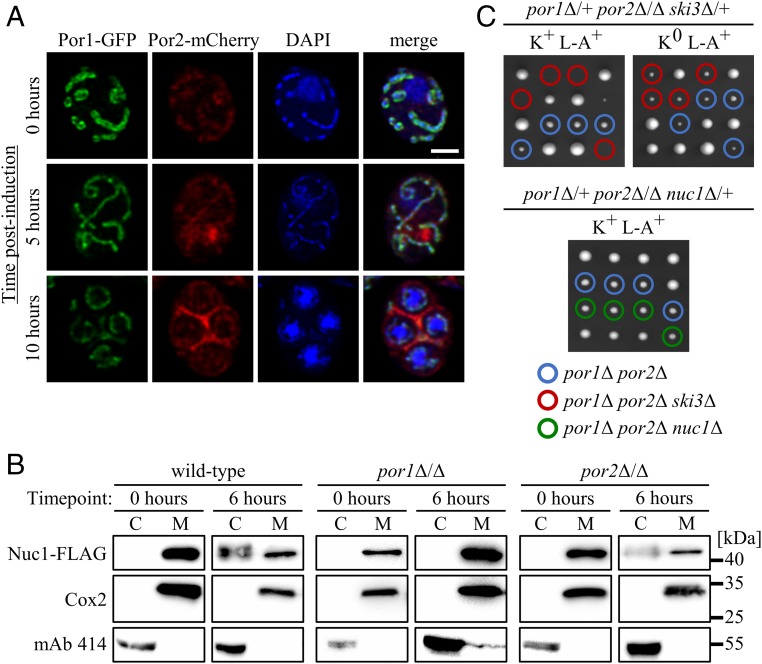
We reasoned that Nuc1 mitochondrial release during sporulation might occur through a conserved mechanism. The mitochondrial VDAC functions as a pore for small hydrophilic molecules and has been implicated in the release of apoptogenic proteins, including EndoG, through the formation of an oligomeric “megachannel” (38–41). Yeast VDACs are encoded by the paralogous POR1/2 genes. Intriguingly, ribosome profiling studies revealed that POR2 translation was dramatically up-regulated during meiosis (SI Appendix, Fig. S4) (42). To investigate the possibility of VDAC involvement in meiotic Nuc1 release, we first determined the expression and localization of Por1/2 using a Por1-GFP Por2-mCherry–expressing strain. As expected, Por1-GFP exhibited strong signal corresponding to mitochondrial networks and colocalized with mitochondrial nucleoids throughout meiosis (Fig. 4A). Although Por2-mCherry exhibited considerably less bright signal, it colocalized with Por1-GFP (Fig. 4A). As predicted by the ribosome profiling studies, we found that Por2-mCherry exhibited induced expression during meiosis. However, the induced Por2-mCherry signal did not colocalize with Por1-GFP or mitochondrial nucleoids and instead appeared as cytoplasmic foci (Fig. 4A). Although Por2-mCherry localized with Por1-GFP during sporogenesis at the 10-h time point, substantial amounts of Por2-mCherry, and not of Por1-GFP, persisted in the mother cell compartment (Fig. 4A). While we do not know the identity or significance of these Por2 structures, our findings confirm that the yeast VDAC paralogs localize to mitochondria throughout sporulation and that Por2 is meiotically induced.
Fig. 4.
Developmentally regulated mitochondrial VDACs mediate Nuc1 antiviral activity and cytosolic accumulation. (A) Meiotic imaging time course of cells expressing Por1-GFP and Por2-mCherry. Fixed cells were stained with DAPI to visualize nuclei and mitochondrial nucleoids. (Scale bar, 2 μm.) (B) Anti–Nuc1-FLAG Western blots of cell fractions sampled from a synchronized sporulation of VDAC mutants expressing Nuc1-FLAG with a wild-type strain for comparison (fractionation controls included). Note that the por2Δ/Δ and por1Δ/Δ cytosolic lanes were loaded with twice the sample volume of wild type. (C) Dissections of VDAC-NUC1/SKI3 triple mutants, imaged 3 d postdissection on YPD at 30 °C. Known/inferred haploid genotypes of interest are indicated: por1Δ por2Δ (blue), por1Δ por2Δ ski3Δ (red), and por1Δ por2Δ nuc1Δ (green). Strains used: MMY7571, MMY7572, MMY7599, SCY1, SCY28, SCY41, and SCY73.
We next assessed the roles of yeast VDAC in Nuc1 mitochondrial release using subcellular fractionations. Nuc1-FLAG was undetectable and weakly detectable respectively in cytosolic preparations of sporulating por1∆/∆ or por2∆/∆ strains at the 6-h meiotic time point (Fig. 4B). These results suggest that Nuc1 undergoes programmed mitochondrial release during meiosis through a conserved VDAC mechanism.
A model explaining our findings posits that Por1/2 VDAC-mediated release of Nuc1 into the cytosol exposes the Killer system to Nuc1, resulting in Killer attenuation. A key prediction of this model is that POR1 and POR2 should act in the NUC1 antiviral pathway. As deletion of both copies of POR1 caused a strong defect in Nuc1 mitochondrial release at the 6-h maternal meiotic stage, we first tested this prediction by sporulating and dissecting a K+ por1∆/∆ ski3∆/+ diploid strain. Double-mutant spores lacking SKI3 produced by these strains exhibited a high degree of lethality and were recovered at only one-third the expected frequency (SI Appendix, Table S2). Consistent with our model, this partially penetrant synthetic lethality was fully suppressed by loss of Killer (K0 L-A+) and not observed in K+ por1∆/∆ nuc1∆/+ dissections (SI Appendix, Table S2). We next investigated the contributions of POR2 to the putative Por1/2-Nuc1 antiviral pathway. Following sporulation of a K+ por1∆/+ por2∆/∆ ski3∆/+ strain, we observed near 100% lethality of por1∆ por2∆ ski3∆ mutant spores (Fig. 4C and SI Appendix, Table S2). Although por1∆ por2∆ spore clones were slow-growing, the synthetic lethal interaction between SKI3 and POR1/2 was Killer-dependent: por1∆ por2∆ ski3∆ synthetic lethality was fully suppressed by loss of Killer (Fig. 4C and SI Appendix, Table S2). Dissection of a K+ por1∆/+ por2∆/∆ nuc1∆/+ strain resulted in fully viable por1∆ por2∆ nuc1∆ spores, confirming that Por1/2 acts in the Nuc1 pathway to attenuate Killer during sporulation (Fig. 4C and SI Appendix, Table S2).
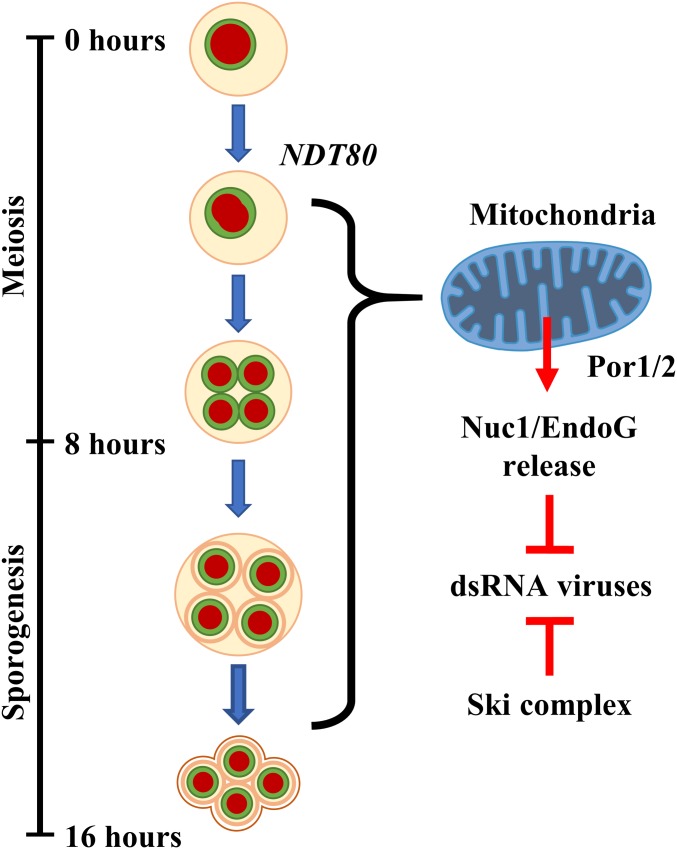
Our results reveal a meiotically programmed system of Killer attenuation involving at least 2 converging pathways. In addition to the essential maternal role played by SKI3 in this process, we find that Nuc1 mitochondrial release into the maternal cytosol also attenuates Killer during sporulation. This meiotic Nuc1 release pathway collaborates with SKI3 to prevent a lethal accumulation of the M1-encoded K1 toxin (Fig. 5). Further, our studies indicate that the role of NUC1 in viral attenuation persists into sporogenesis. Killer attenuation by NUC1 and SKI3 continues in developing spores to maintain the viability of meiotic progeny (Fig. 5).
Fig. 5.
Model of the developmentally regulated Nuc1 antiviral pathway. Diagram depicting proposed timeline of Nuc1 mitochondrial release in the context of sporulation. The timing (at hours postinduction) for each event and the phase of sporulation progression is given on the left. On the right, depiction of model of mitochondrial Nuc1 release and meiotic antiviral activity in conjunction with the Ski complex.
Could EndoG mitochondrial release in other organisms similarly target viruses and might this relate to the origins of apoptosis? While further studies are needed to investigate this in animal models, we address here the generality of Nuc1 viral attenuation in yeast. In contrast to the relative rarity of Killers (19, 20), the L-A virus is found in virtually all laboratory strains. As noted above, we found that eradicating M1 from a K+ strain to create a K0 L-A+ strain resulted in elevated L-A Gag levels, reflecting the known parasitic nature of M1 for L-A (SI Appendix, Fig. S2A) (27). Therefore, the following studies on L-A were performed in M1-free strains to circumvent complications presented by the relationship between M1 and L-A.
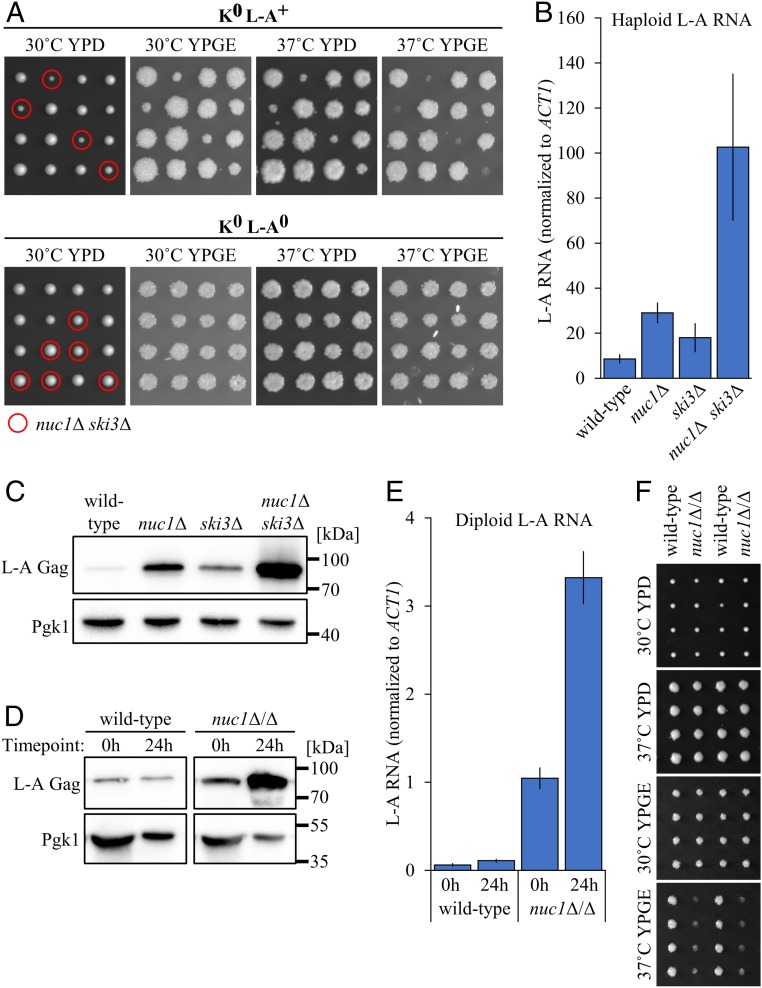
Interestingly, nuc1∆ ski3∆ mutants dissected from a K0 L-A+ strain were noticeably slow-growing (Fig. 6A). We found that this phenotype was accentuated by growth at high temperature, or on nonfermentable carbon sources that require mitochondrial respiration. Further, combining these conditions resulted in growth arrest, suggesting that mitochondrial function was compromised (Fig. 6A). These phenotypes were associated with a dramatic increase in levels of L-A RNA and its encoded Gag protein (Fig. 6 B and C). To determine if these growth phenotypes were caused by L-A overproduction, we evicted L-A to generate an isogenic K0 L-A0 nuc1∆/+ ski3∆/+ diploid strain. Following sporulation of this strain we found that the nuc1∆ ski3∆ growth phenotypes were completely suppressed (Fig. 6A). These findings demonstrate a fitness consequence of harboring the L-A virus, which has thus far been regarded as commensal to the host cell (43).
Fig. 6.
Nuc1-mediated meiotic viral attenuation targets the L-A virus. (A) Tetrad dissections of K0 L-A+ (Top) and K0 L-A0 (Bottom) nuc1Δ/+ ski3Δ/+ mutants. Leftmost panels show growth at 2 d postdissection on YPD at 30 °C. Dissections were replica-plated and incubated for 1 d in the following conditions (left to right): YPD at 37 °C, YPGE at 30 °C, and YPGE at 37 °C. Haploid genotypes of interest are circled in red (nuc1Δ ski3Δ). (B) L-A RNA in saturated cultures of haploid K0 L-A+ strains was quantified by qPCR and normalized to endogenous ACT1 RNA. Data points represent mean of biological replicates (n = 3) ± SD. (C) Anti–L-A Gag Western blots of isogenic haploid K0 L-A+ strains sampled from saturated cultures. Pgk1 used as loading control. (D) Anti–L-A Gag Western blots of isogenic K0 L-A+ wild-type and nuc1Δ/Δ cell lysates sampled from premeiotic (0 h) and terminal sporulation (24 h) cultures. Pgk1 used as loading control. (E) L-A RNA levels in K0 L-A+ strains were quantified by qPCR across a meiotic time course at premeiotic (0 h postinduction ) and terminally sporulated (24 h postinduction ) time points. Samples were normalized to endogenous ACT1 RNA. Data points represent mean of biological replicates (n = 3) ± SD. (F) Tetrad dissections of isogenic K0 L-A+ wild type and nuc1Δ/Δ. Topmost image shows growth at 2 d postdissection on YPD at 30 °C. Dissections were replica-plated and incubated for 1 d in the following conditions (top to bottom): YPD at 37 °C, YPGE at 30 °C, and YPGE at 37 °C. Strains used: MMY6469, MMY6470, MMY6471, MMY6472, MMY7506, MMY7507, MMY7960, MMY7961, MMY8226, and MMY8242.
As NUC1 functions both maternally and spore-autonomously to attenuate K1 during sporulation of K+ strains, we investigated the maternal contribution of NUC1 to L-A attenuation in a K0 L-A+ nuc1∆/∆ strain. Consistent with previous findings, mitotic K0 L-A+ nuc1∆/∆ cells exhibited increased levels of L-A Gag and RNA compared with an isogenic wild-type strain (Fig. 6 D and E) (26). Upon sporulation, we saw that nuc1∆/∆ exhibited large increases in L-A RNA and L-A Gag protein (Fig. 6 D and E). This is in contrast to a wild-type strain, where L-A viral product abundance was virtually unchanged from presporulation levels (Fig. 6 D and E). Given this sporulation-enhanced attenuation of L-A by Nuc1, we posit that the increased L-A levels in nuc1∆ meiotic progeny (Fig. 6 B and C) were in part a consequence of increased L-A copy number inherited during sporulation. We speculate that a cell’s viral load is established during sporulation through the interplay between viruses and host antiviral factors. The consequences of this rampant L-A accumulation in sporulating K0 L-A+ nuc1Δ/Δ cells were observed upon phenotypic analysis of the spore progeny. Spore clones derived from a K0 L-A+ nuc1Δ/Δ strain displayed reduced growth on nonfermentable carbon at high temperature, albeit with lower penetrance compared with the K0 L-A+ nuc1Δ ski3Δ mutant phenotype (Fig. 6F).
Like with NUC1, mutations of POR1 have been associated with increased L-A abundance in haploid cells following saturating growth and media exhaustion (26, 44). Thus, it seems possible that the Por1/2-Nuc1 pathway we have identified can function outside of meiosis, although our results show its meiotic role is much more consequential. It is important to note that undomesticated S. cerevisiae exist essentially exclusively as diploids and that the response of these diploid cells to nutritional exhaustion is to sporulate (8). Thus, vestigial applications of the Por1/2-Nuc1 meiotic pathway in artificially neutered haploid cells may be expected to occur in response to similar nutritional cues. More broadly, much of the controversial phenomenon of yeast “apoptosis” observed to occur in haploid cells following exhaustion of growth media may relate to vestigial manifestations of meiotic PCD (45).
Transferring the paradigm of altruistic PCD from the multicellular context to unicellular organisms remains dubious and may cloud our understanding of the nature and origin of microbial PCD (46–48). Our findings identify meiosis as a context where crucial host–virus interactions are mediated within the milieu of PCD in budding yeast. While the Por1/2-Nuc1 pathway we identify here is similar to VDAC-EndoG pathways that accompany canonical apoptosis, rather than death the Por1/2-Nuc1 pathway promotes survival. As cell death pathways must have arisen from nonsuicidal roles, our findings may thus illuminate transitional forms of PCD (49).
Materials and Methods
Strains and Media.
Standard S. cerevisiae genetic and strain manipulation techniques were used for strain construction and tetrad analysis (50). Refer to SI Appendix, Table S3 for strains used in this paper. For tetrad dissections, sporulated strains were incubated in 3 mg/mL 20T Zymolyase (Seikagaku Glycobiology) for 15 to 20 min at room temperature and spread on 1% yeast extract, 2% peptone, 0.004% adenine, and 2% dextrose (YPD) plates for dissecting.
Plasmids.
The p5472 NUC1 CEN/ARS URA3 MOBY plasmid was obtained from Brenda Andrews, University of Toronto, Toronto, ON, Canada (51). The pRS424 ADH-mtRFP plasmid was obtained from Janet Shaw, University of Washington, Seattle, WA (52). The pME1 NUC1::EGFP URA3 plasmid was generated from the pUG35 NUC1::EGFP URA3 plasmid (33) through replacement of the MET25 promoter with 500 bp of the NUC1 upstream sequence via subcloning by restriction digest and ligation.
Killer Activity Assay.
To determine the Killer phenotypes, strains were replica-plated onto lawns of Killer-sensitive yeast cells obtained from Reed Wickner, National Institutes of Health, Bethesda, MD, on buffered plates (0.0034% wt/vol methylene blue in YPD, adjusted to pH 4.5 with citric acid monohydrate and sodium phosphate dibasic) and grown at 20 °C for 3 d to observe killer activity.
Eviction of Viruses.
Cells were treated with 32 mM Anisomycin (BioShop ANS245) in liquid YPD for 4 d at 30 °C and plated for single colonies on YPD plates to assay for Killer activity. Non-Killer isolates were confirmed to have lost the M1 genome by RT-PCR and by Western blotting. Some of the isolates identified in this chemical screen also lost the L-A virus. These progenitor K0 L-A+ and K0 L-A0 strains were used for subsequent back-crossing strategies using mak3∆ to engineer the varied K0 L-A+ and K0 L-A0 strains described here. The presence or absence of M and/or L-A in these strains was confirmed using RT-PCR.
SK1 Synchronized Sporulation Protocol.
Methods described by Eastwood and Meneghini (7) were used to synchronize sporulation progression in SK1. Strains were streaked to YPGE (1% yeast extract, 2% peptone, 0.004% adenine, 3% glycerol, and 2% ethanol) agar plates and incubated at 30 °C overnight. Cells from the YPGE plate were streaked to a YPD plate and incubated for 2 d at 30 °C, whereupon single colonies were inoculated in liquid YPD and grown to saturation overnight at 30 °C. For presporulation growth, the YPD culture was diluted in YPA (1% yeast extract, 2% peptone, 0.004% adenine, and 2% acetate) to an optical density of 0.3 at 600 nm (OD600) and incubated for 12 to 14 h at 30 °C until the OD600 reached 1.3 to 1.7. To induce sporulation, the presporulation culture was washed twice and resuspended in sporulation medium (1% potassium acetate and 0.02% raffinose) at an OD600 of 2.2 and incubated at 30 °C. All sporulations were performed in standard (high-carbon) conditions unless otherwise specified.
Yeast Subcellular Fractionation.
The Mitochondrial Yeast Isolation kit and protocol (ab178779; Abcam) was used to fractionate yeast cells by differential centrifugation. Following pelleting of the mitochondrial fraction, the soluble cytosolic fraction was collected by precipitation with 2% trichloroacetic acid at 4 °C for 30 min and washed 3 times with chilled acetone before resuspension in SDS/PAGE sample buffer (50 mM Tris⋅HCl, pH 6.8, 2% SDS, 10% glycerol, 0.025% bromophenol blue, and 100 mM dithiothreitol) and heating at 100 °C for 10 min. Mitochondrial and whole-cell pellets were resuspended and heated in SDS/PAGE sample buffer.
Yeast Whole-Cell Protein Extraction.
Vegetative cells were permeabilized with 0.1 N NaOH for 5 min at room temperature before resuspension in SDS/PAGE buffer and heating at 100 °C for 10 min. The soluble fraction was isolated by centrifugation and used for subsequent Western blotting. Sporulated cells were disrupted by bead-beating cells suspended in SDS/PAGE buffer for 10 min before heating at 100 °C.
Western Blots.
Cell fractions and whole-cell lysates prepared as described above were electrophoresed on 8 to 10% SDS/PAGE and transferred to poly(vinylidene difluoride) membranes. Blots were incubated overnight in diluted primary antibody, probed with horseradish peroxidase (HRP)-conjugated horse anti-mouse (7075; Cell Signaling Technology) or goat anti-rabbit (7074; Cell Signaling Technology) secondary antibody and detected using Luminata Forte Western HRP Substrate (EMD Millipore). Blots were imaged with the Bio-Rad ChemiDoc XRS+ system; image processing was performed with the Image Lab software package (Bio-Rad). The primary antibodies and dilutions used were 1:1,000 anti-FLAG M2 (F1804; Sigma-Aldrich), 1:2,000 mouse anti-nuclear pore complex protein mAb 414 (obtained from Alexander Palazzo, University of Toronto, Toronto, ON, Canada), 1:1,000 anti-VDAC1/Porin (ab110326; Abcam), 1:1,000 anti-Cox2 (obtained from Tom Fox, Cornell University, Ithaca, NY), 1:2,500 anti-GAPDH (obtained from Cordula Enenkel, University of Toronto, Toronto, ON, Canada), 1:5,000 anti-Pgk1 (ab113687; Abcam), 1:2,000 anti-Killer toxin (obtained from Motomasa Tanaka, Tokyo Institute of Technology, Tokyo, Japan) (28), and 1:2,000 anti–L-A Gag (obtained from Reed Wickner).
Quantification of Nuc1 Abundance.
Western blots were analyzed with ImageJ analysis software to determine the background-subtracted pixel density of Nuc1-FLAG bands (53). To ensure accurate quantification, all blots selected for analysis had similar exposure times. The protein concentration of each fraction loaded for the Western blots was determined using the RD DC Protein Assay (Bio-Rad) and the total protein loaded per lane was derived from this value. Pixel density was normalized to total protein loaded per lane to calculate Nuc1 abundance in each cell fraction.
Fluorescent Microscopy.
Cells were fixed with 4% formaldehyde and washed 3 times with 1× phosphate-buffered saline. Fixed cells were then stained with DAPI and mounted on a microscope slide for imaging. Nuc1-GFP images were taken using the Zeiss LSM 880 with Airyscan confocal laser scanning microscope (LSM) on the Fast module and processed with ZEN lite (blue edition) software from Carl Zeiss Microscopy. Por1-GFP/Por2-mCherry images were acquired with the Leica Sp8 confocal LSM and processed with the Leica Application Suite (LAS X) software package. Deconvolution was applied to all images obtained by microscopy.
Quantification of Colony Growth Kinetics.
Dissections were imaged at 24-h intervals between 48 and 96 h postdissection and the colony sizes of all haploid progeny from tetrad dissections were quantified using ImageJ analysis software (53). The resulting growth curve dataset was plotted to follow the growth kinetics of each spore from the imaged dissections, including nonviable spores with inferable genotypes.
RNA Extraction.
Strains were sporulated and harvested by flash freezing in liquid nitrogen at time points of 0 and 24 h postinduction of sporulation. Twenty-four-hour samples were subjected to bead-beating (Mini Bead beater; Biospec Products) for 1 min in TRIzol (Invitrogen) followed by 2-min incubations on ice for 8 cycles. All samples were incubated at 65 °C for 30 min in phenol solution (P4682; Sigma-Aldrich), SDS, and buffer AE (10 mM Tris⋅Cl and 0.5 mM EDTA, pH 9.0) and the phase-separated supernatant was washed with chloroform, precipitated in isopropanol, and washed with 70% ethanol before resuspension in H2O. The crude RNA samples were purified using the RNEasy Mini Kit (Qiagen) and residual DNA was removed by DNase I digestion.
Two-Step RT-qPCR.
Five micrograms of purified RNA were reverse-transcribed using random nonamers and Maxima H Minus Reverse Transcriptase (Thermo Fisher) according to supplier protocols. The resulting cDNA product was isolated by alkaline hydrolysis and RNase A digestion of remaining RNA. Subsequently, qPCR was performed with 1/10 dilutions of cDNA product in 2× SYBR Green/PCR buffer (54) on the iQ5 platform (Bio-Rad). Refer to SI Appendix, Table S4 for qPCR primers used in this study.
Normalization and Data Analysis.
The relative starting quantities (SQs) of M1 and ACT1 in each sample were determined on the iQ5 software platform (Bio-Rad) using 6-point standard curves. M1 SQs were normalized to endogenous ACT1 to account for variation in total cDNA concentration between samples. To determine fold change in viral RNA levels, a second normalization to wild-type SQs at 0 h postinduction was performed for each set of biological replicates.
Supplementary Material
Acknowledgments
We thank Reed Wickner for strains and the L-A antibody as well as Motomasa Tanaka for the K1 toxin antibody. We thank Charlie Boone, Brenda Andrews, Janet Shaw, and Frank Madeo for providing plasmids, as well as to Tom Fox, Cordula Enenkel, and Alex Palazzo for providing us with additional antibodies. Work in the M.D.M. laboratory is supported by a Natural Sciences and Engineering Research Council of Canada Discovery grant and Canadian Institutes of Health Research Grant MOP-89996 to M.D.M.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 16167.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1900751116/-/DCSupplemental.
References
- 1.Koonin E. V., Viruses and mobile elements as drivers of evolutionary transitions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iranzo J., Lobkovsky A. E., Wolf Y. I., Koonin E. V., Virus-host arms race at the joint origin of multicellularity and programmed cell death. Cell Cycle 13, 3083–3088 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wickner R. B., Fujimura T., Esteban R., Viruses and prions of Saccharomyces cerevisiae. Adv. Virus Res. 86, 1–36 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitt M. J., Breinig F., Yeast viral killer toxins: Lethality and self-protection. Nat. Rev. Microbiol. 4, 212–221 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Ridley S. P., Sommer S. S., Wickner R. B., Superkiller mutations in Saccharomyces cerevisiae suppress exclusion of M2 double-stranded RNA by L-A-HN and confer cold sensitivity in the presence of M and L-A-HN. Mol. Cell. Biol. 4, 761–770 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neiman A. M., Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189, 737–765 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eastwood M. D., Meneghini M. D., Developmental coordination of gamete differentiation with programmed cell death in sporulating yeast. Eukaryot. Cell 14, 858–867 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eastwood M. D., Cheung S. W., Lee K. Y., Moffat J., Meneghini M. D., Developmentally programmed nuclear destruction during yeast gametogenesis. Dev. Cell 23, 35–44 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Li L. Y., Luo X., Wang X., Endonuclease G is an apoptotic DNase when released from mitochondria. Nature 412, 95–99 (2001). [DOI] [PubMed] [Google Scholar]
- 10.van Loo G., et al. , Endonuclease G: A mitochondrial protein released in apoptosis and involved in caspase-independent DNA degradation. Cell Death Differ. 8, 1136–1142 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Zhang J., et al. , Endonuclease G is required for early embryogenesis and normal apoptosis in mice. Proc. Natl. Acad. Sci. U.S.A. 100, 15782–15787 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.David K. K., Sasaki M., Yu S. W., Dawson T. M., Dawson V. L., EndoG is dispensable in embryogenesis and apoptosis. Cell Death Differ. 13, 1147–1155 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Parrish J., et al. , Mitochondrial endonuclease G is important for apoptosis in C. elegans. Nature 412, 90–94 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Deponte M., Programmed cell death in protists. Biochim. Biophys. Acta 1783, 1396–1405 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Dickman M., Williams B., Li Y., de Figueiredo P., Wolpert T., Reassessing apoptosis in plants. Nat. Plants 3, 773–779 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Shlezinger N., Goldfinger N., Sharon A., Apoptotic-like programed cell death in fungi: The benefits in filamentous species. Front. Oncol. 2, 97 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shimizu S., Matsuoka Y., Shinohara Y., Yoneda Y., Tsujimoto Y., Essential role of voltage-dependent anion channel in various forms of apoptosis in mammalian cells. J. Cell Biol. 152, 237–250 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCommis K. S., Baines C. P., The role of VDAC in cell death: Friend or foe? Biochim. Biophys. Acta 1818, 1444–1450 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Philliskirk G., Young T. W., The occurrence of killer character in yeasts of various genera. Antonie van Leeuwenhoek 41, 147–151 (1975). [DOI] [PubMed] [Google Scholar]
- 20.Nakayashiki T., Kurtzman C. P., Edskes H. K., Wickner R. B., Yeast prions [URE3] and [PSI+] are diseases. Proc. Natl. Acad. Sci. U.S.A. 102, 10575–10580 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pak J., Segall J., Regulation of the premiddle and middle phases of expression of the NDT80 gene during sporulation of Saccharomyces cerevisiae. Mol. Cell. Biol. 22, 6417–6429 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dake E., Hofmann T. J., McIntire S., Hudson A., Zassenhaus H. P., Purification and properties of the major nuclease from mitochondria of Saccharomyces cerevisiae. J. Biol. Chem. 263, 7691–7702 (1988). [PubMed] [Google Scholar]
- 23.Kalinowska M., Garncarz W., Pietrowska M., Garrard W. T., Widlak P., Regulation of the human apoptotic DNase/RNase endonuclease G: Involvement of Hsp70 and ATP. Apoptosis 10, 821–830 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Chang S. L., Leu J. Y., Chang T. H., A population study of killer viruses reveals different evolutionary histories of two closely related Saccharomyces sensu stricto yeasts. Mol. Ecol. 24, 4312–4322 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Johnson A. W., Kolodner R. D., Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role for these genes in translation control. Mol. Cell. Biol. 15, 2719–2727 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y. X., Dieckmann C. L., Overproduction of yeast viruslike particles by strains deficient in a mitochondrial nuclease. Mol. Cell. Biol. 9, 3323–3331 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ball S. G., Tirtiaux C., Wickner R. B., Genetic control of L-a and L-(Bc) dsrna copy number in killer systems of Saccharomyces cerevisiae. Genetics 107, 199–217 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki G., Weissman J. S., Tanaka M., [KIL-d] protein element confers antiviral activity via catastrophic viral mutagenesis. Mol. Cell 60, 651–660 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bussey H., Saville D., Greene D., Tipper D. J., Bostian K. A., Secretion of Saccharomyces cerevisiae killer toxin: Processing of the glycosylated precursor. Mol. Cell. Biol. 3, 1362–1370 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neiman A. M., Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J. Cell Biol. 140, 29–37 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Masison D. C., et al. , Decoying the cap- mRNA degradation system by a double-stranded RNA virus and poly(A)- mRNA surveillance by a yeast antiviral system. Mol. Cell. Biol. 15, 2763–2771 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dmochowska A., Dignard D., Henning D., Thomas D. Y., Bussey H., Yeast KEX1 gene encodes a putative protease with a carboxypeptidase B-like function involved in killer toxin and alpha-factor precursor processing. Cell 50, 573–584 (1987). [DOI] [PubMed] [Google Scholar]
- 33.Büttner S., et al. , Endonuclease G regulates budding yeast life and death. Mol. Cell 25, 233–246 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Drinnenberg I. A., Fink G. R., Bartel D. P., Compatibility with killer explains the rise of RNAi-deficient fungi. Science 333, 1592 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wickner R. B., Chromosomal and nonchromosomal mutations affecting the “killer character” of Saccharomyces cerevisiae. Genetics 76, 423–432 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Somers J. M., Bevan E. A., The inheritance of the killer character in yeast. Genet. Res. 13, 71–83 (1969). [DOI] [PubMed] [Google Scholar]
- 37.Wickner R. B., Twenty-six chromosomal genes needed to maintain the killer double-stranded RNA plasmid of Saccharomyces cerevisiae. Genetics 88, 419–425 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shoshan-Barmatz V., De S., Meir A., The mitochondrial voltage-dependent anion channel 1, Ca2+ transport, apoptosis, and their regulation. Front. Oncol. 7, 60 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X., Bian X., Kong J., The proapoptotic protein BNIP3 interacts with VDAC to induce mitochondrial release of endonuclease G. PLoS One 9, e113642 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zalk R., Israelson A., Garty E. S., Azoulay-Zohar H., Shoshan-Barmatz V., Oligomeric states of the voltage-dependent anion channel and cytochrome c release from mitochondria. Biochem. J. 386, 73–83 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keinan N., Tyomkin D., Shoshan-Barmatz V., Oligomerization of the mitochondrial protein voltage-dependent anion channel is coupled to the induction of apoptosis. Mol. Cell. Biol. 30, 5698–5709 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brar G. A., et al. , High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335, 552–557 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McBride R. C., Boucher N., Park D. S., Turner P. E., Townsend J. P., Yeast response to LA virus indicates coadapted global gene expression during mycoviral infection. FEMS Yeast Res. 13, 162–179 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Dihanich M., van Tuinen E., Lambris J. D., Marshallsay B., Accumulation of viruslike particles in a yeast mutant lacking a mitochondrial pore protein. Mol. Cell. Biol. 9, 1100–1108 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herker E., et al. , Chronological aging leads to apoptosis in yeast. J. Cell Biol. 164, 501–507 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bayles K. W., Bacterial programmed cell death: Making sense of a paradox. Nat. Rev. Microbiol. 12, 63–69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allocati N., Masulli M., Di Ilio C., De Laurenzi V., Die for the community: An overview of programmed cell death in bacteria. Cell Death Dis. 6, e1609 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nedelcu A. M., Driscoll W. W., Durand P. M., Herron M. D., Rashidi A., On the paradigm of altruistic suicide in the unicellular world. Evolution 65, 3–20 (2011). [DOI] [PubMed] [Google Scholar]
- 49.Aram L., Arama E., Sporoptosis: Sowing the seeds of nuclear destruction. Dev. Cell 23, 5–6 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Smith J. S., Burke D., Eds., Yeast Genetics: Methods and Protocols (Methods in Molecular Biology, Springer, New York, 2014). [Google Scholar]
- 51.Ho C. H., et al. , A molecular barcoded yeast ORF library enables mode-of-action analysis of bioactive compounds. Nat. Biotechnol. 27, 369–377 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gorsich S. W., Shaw J. M., Importance of mitochondrial dynamics during meiosis and sporulation. Mol. Biol. Cell 15, 4369–4381 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider C. A., Rasband W. S., Eliceiri K. W., NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aparicio O., et al. , Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Mol. Biol. Chapter 21, Unit 21.3 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.