Significance
Controlling vascular inflammation is critical for limiting the progression of chronic vascular diseases such as atherosclerosis. Although poorly studied in the context of human vascular inflammation, long noncoding RNAs (lncRNAs) have the potential to regulate their neighboring genes. However, what constitutes a neighboring lncRNA is currently not well defined. In this study, we took an innovative approach to define IL-1β−regulated neighboring mRNA−lncRNA pairs based on colocalization within the same chromatin neighborhood and divergent transcriptional orientation. This approach led to the discovery of lncRNA-CCL2, which positively regulates its neighboring gene, CCL2, an important player in atherogenesis. Furthermore, lncRNA-CCL2 is relevant to human disease, as it is elevated in human atherosclerotic plaques, and, given its regulatory role, it may contribute to atherogenesis.
Keywords: long noncoding RNA, chromatin, atherosclerosis, endothelium, epigenetics
Abstract
Atherosclerosis is a chronic inflammatory disease that is driven, in part, by activation of vascular endothelial cells (ECs). In response to inflammatory stimuli, the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling pathway orchestrates the expression of a network of EC genes that contribute to monocyte recruitment and diapedesis across the endothelium. Although many long noncoding RNAs (lncRNAs) are dysregulated in atherosclerosis, they remain poorly characterized, especially in the context of human vascular inflammation. Prior studies have illustrated that lncRNAs can regulate their neighboring protein-coding genes via interaction with protein complexes. We therefore identified and characterized neighboring interleukin-1β (IL-1β)−regulated messenger RNA (mRNA)−lncRNA pairs in ECs. We found these pairs to be highly correlated in expression, especially when located within the same chromatin territory. Additionally, these pairs were predominantly divergently transcribed and shared common gene regulatory elements, characterized by active histone marks and NF-κB binding. Further analysis was performed on lncRNA-CCL2, which is transcribed divergently to the gene, CCL2, encoding a proatherosclerotic chemokine. LncRNA-CCL2 and CCL2 showed coordinate up-regulation in response to inflammatory stimuli, and their expression was correlated in unstable symptomatic human atherosclerotic plaques. Knock-down experiments revealed that lncRNA-CCL2 positively regulated CCL2 mRNA levels in multiple primary ECs and EC cell lines. This regulation appeared to involve the interaction of lncRNA-CCL2 with RNA binding proteins, including HNRNPU and IGF2BP2. Hence, our approach has uncovered a network of neighboring mRNA−lncRNA pairs in the setting of inflammation and identified the function of an lncRNA, lncRNA-CCL2, which may contribute to atherogenesis in humans.
Atherosclerosis is a chronic inflammatory vascular disease characterized by fatty plaque build-up within the arterial wall, and culminates in the development of coronary artery disease. Vascular inflammation plays a pivotal role in the initiation and progression of atherosclerosis (1). In response to inflammatory stimuli, vascular endothelial cells (ECs) become activated via the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling pathway. This NF-κB−dependent transcriptional program leads to the increased production of a network of proinflammatory cytokines, chemokines, and adhesion molecules that facilitate monocyte recruitment and adhesion (1). Following extravasation into the vessel wall, monocytes differentiate into macrophages, which contribute to the growth and subsequent destabilization of plaques. Understanding the molecular mechanisms that control vascular inflammation is imperative for developing new methodologies to limit the progression of atherosclerosis.
Accumulating evidence has shown that long noncoding RNAs (lncRNAs) are dysregulated in atherosclerosis (2, 3); however, only a handful of studies have assessed their functional involvement in regulating inflammatory pathways (4–12), and relatively little is known about their contribution to human vascular EC inflammation. LncRNAs are defined as a heterogeneous class of noncoding transcripts >200 base pairs in length (13, 14). They can be found within the nucleus or the cytoplasm or in both cellular compartments (14). A proportion of lncRNAs have been reported to act in cis, meaning that they act near their site of transcription to regulate the expression of neighboring messenger RNA (mRNA) transcripts (15–19), while others have been shown to act in trans in areas far from their site of transcription (20, 21). The mechanisms of action of lncRNAs are varied and stem from their ability to interact with RNA, DNA, and proteins (13, 14). For example, lncRNAs can act as guides or decoys to either help recruit or repel a wide range of proteins, including chromatin remodeling complexes, transcription factors, and RNA-binding proteins (12, 22–25). The functionality of the lncRNA, however, is not limited to the transcript itself, as others have shown that lncRNA transcription can be functionally important, as is the case with Airn and Blustr (16, 26); meanwhile, others have found lncRNA-associated regulatory DNA elements to be important, as is the case with Haunt, Lockd, and Pantr1 (27–29).
Given the potential for lncRNAs to regulate their neighboring genes, we aimed to discover cis-regulatory lncRNAs involved in vascular inflammation. Features defining cis-regulatory lncRNAs, such as the genomic distance between the lncRNA and its regulated mRNA, however, are not well established in the literature (15, 30, 31). Therefore, we set out to systematically define what constitutes a neighboring mRNA−lncRNA pair. We identified neighboring mRNA−lncRNA pairs differentially expressed in ECs stimulated with the proinflammatory cytokine, interleukin-1β (IL-1β). A strong positive correlation in IL-1β responsiveness of the mRNA−lncRNA pairs was observed, but it significantly weakened once the pairs were farther than 100 kilobase pairs (kbp) away from each other. Interestingly, chromatin organization played a more important role than physical distance along the chromosome, as mRNA−lncRNA pairs localized within the same topologically associated domain (TAD) showed the highest correlation, irrespective of the distance between them (32). Additionally, we noted that IL-1β−responsive mRNA−lncRNA pairs were predominantly transcribed in a divergent orientation to each other, and shared regulatory elements, including NF-κB−bound enhancers and promoters. Follow-up functional experiments were performed on lncRNA-CCL2, an lncRNA transcribed divergently to the proatherosclerotic chemokine gene, CCL2, which encodes monocyte chemoattractant protein 1 (MCP1) (33). Due to its role in monocyte recruitment, MCP1 has been implicated in multiple human diseases, including atherosclerosis (33). We validated lncRNA-CCL2 to be a cis-regulatory lncRNA, as it positively regulated CCL2 levels. Pull-down experiments suggested that lncRNA-CCL2 may regulate CCL2 levels via interaction with RNA-binding proteins. In particular, insulin growth factor 2 binding protein (IGF2BP2) and HNRNPU bound to lncRNA-CCL2, and modulation of their abundance affected CCL2 levels during IL-1β stimulation. Apart from being up-regulated during acute inflammation in vitro, lncRNA-CCL2 was found to be elevated and correlated with CCL2 expression in unstable symptomatic human atherosclerotic plaques, which suggests its potential regulatory role during disease development. Overall, our work has taken a unique approach to identify IL-1β−regulated neighboring mRNA−lncRNA pairs and has led to the discovery of a functional lncRNA that is dysregulated in human disease.
Results
Identification of IL-1β−Regulated mRNA−lncRNA Neighboring Pairs.
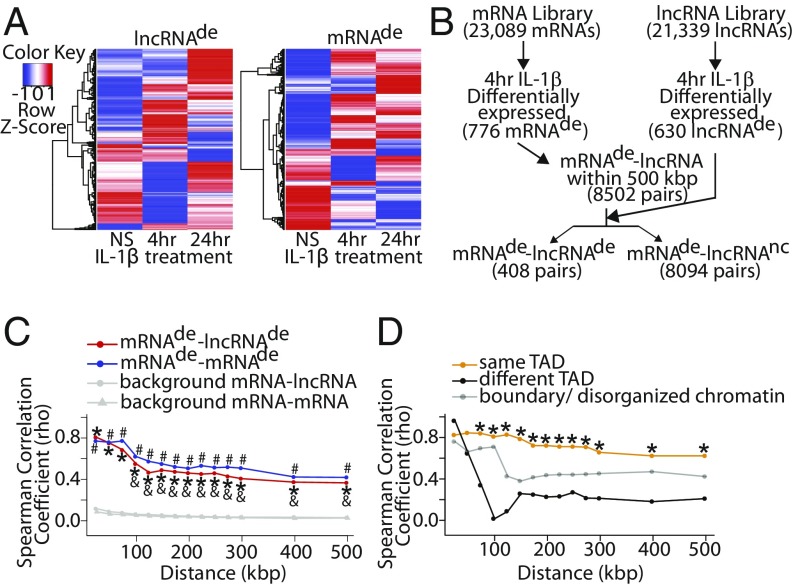
To catalog expression changes in neighboring mRNA−lncRNA pairs during the EC response to inflammatory stimuli, human umbilical vein ECs (HUVECs) were treated with the proinflammatory cytokine, IL-1β, for either 4 or 24 h. RNA was isolated and converted to complementary DNA (cDNA), which was then hybridized to the Arraystar Human lncRNA Expression Microarray V3.0 (containing unique probes for 23,089 mRNAs and 21,339 lncRNAs; Dataset S1) (Gene Expression Omnibus [GEO] ID code GSE127990). In total, 630 lncRNAs and 776 mRNAs were differentially expressed (>2-fold change, P < 0.05) upon 4-h IL-1β treatment (Fig. 1 A and B and Dataset S2). Meanwhile, 583 lncRNAs and 873 mRNAs were differentially expressed (>2-fold change, P < 0.05) upon 24-h IL-1β treatment (Fig. 1A and Dataset S2). For simplicity, we chose to focus our subsequent analyses on the earlier 4-h IL-1β treatment time point, as changes at this time point are more likely to be primary responses to IL-1β treatment. To identify all IL-1β−regulated neighboring mRNA−lncRNA pairs, any lncRNA whose gene body was located up to 500 kbp from the transcriptional start site (TSS) of a differentially expressed (de) mRNA (mRNAde) after 4 h of IL-1β treatment was selected, yielding a total of 8,502 mRNAde−lncRNA pairs (Fig. 1B). Neighboring mRNAde−lncRNA pairs were then segregated depending on whether the lncRNA was IL-1β−regulated (lncRNAde) or whether there was no change (nc) in lncRNA expression (lncRNAnc), resulting in 8,094 mRNAde−lncRNAnc pairs and 408 mRNAde−lncRNAde pairs. For comparison, an identical pipeline was used to catalog neighboring IL-1β−regulated mRNA−mRNA pairs (mRNAde−mRNAnc or mRNAde−mRNAde) (SI Appendix, Fig. S1A). This revealed that mRNAde−mRNAnc pairs were more common (11,763 pairs) than mRNAde−mRNAde pairs (450 pairs). Next, the distribution of genomic distances from the mRNAde to its neighboring transcript(s) was tabulated (SI Appendix, Fig. S1B). The mRNAde were found to be physically closer to neighboring lncRNAde than lncRNAnc (median 142.6 kbp vs. 205.3 kbp, P = 3.11 × 10−8). A similar observation was made with mRNAde vs. mRNAnc (median 176.0 kbp vs. 217.9 kbp, P = 1.09 × 10−3). Subsequent analyses used genomic distances representative of the first quartile (50 kbp), median (150 kbp), and third quartile (300 kbp) of mRNAde−lncRNAde distances (Dataset S3). Across these genomic distances, there were more mRNAde that had a neighboring lncRNAnc than a neighboring lncRNAde (426 vs. 82, 568 vs. 107, and 732 vs. 200, respectively) (SI Appendix, Fig. S1C and Dataset S3). Additionally, there were fewer lncRNAde neighbors than lncRNAnc neighbors (median 1 vs. 2 to 6, P < 1 × 10−15) per individual mRNAde (SI Appendix, Fig. S1D). Altogether, these results suggest that, although almost all IL-1β−regulated mRNAs have an lncRNA located within 500 kbp, very few of these lncRNAs are also IL-1β−regulated.
Fig. 1.
Identification of IL-1β−regulated mRNA−lncRNA neighboring pairs. (A) Microarray heat maps of differentially expressed (de) (>2-fold change, P < 0.05) lncRNAs and mRNAs upon 4 or 24 h IL-1β treatment. NS, not stimulated. (B) Schematic of the computational pipeline used to identify neighboring mRNAde−lncRNAde pairs (4 h IL-1β vs. NS); nc, no change. (C) Spearman correlation coefficients for mRNAde−lncRNAde and mRNAde−mRNAde pairs as a function of genomic distance between them. Background correlation was established using all mRNA−lncRNA and all mRNA−mRNA neighboring pairs possible given the genomic distance between them. Decreasing correlation was observed with increasing distance between mRNAde−lncRNAde pairs. *P < 0.05, Fisher z transformation with Bonferroni correction, mRNAde−lncRNAde vs. all mRNA−lncRNA; #P < 0.05, Fisher z transformation with Bonferroni correction, mRNAde−mRNAde vs. all mRNA−mRNA; &P < 0.05, Fisher z transformation with Bonferroni correction, mRNAde−lncRNAde at specified distance vs. mRNAde−lncRNAde 25 kbp. (D) Spearman correlation coefficients of transcript pairs found either on the same TAD, different TAD, or on boundary/disorganized chromatin. *P < 0.05, Fisher z transformation with Bonferroni correction, same TAD vs. different TAD at the specified distance.
mRNAde−lncRNAde Pairs Demonstrate a Coordinated Response to IL-1β, Especially When Localized on the Same Topologically Associated Domain.
Next, we explored features pertaining to the mRNAde−lncRNAde pairs identified in our study. First, we noted that mRNAde and their neighboring lncRNAde had a coordinate response to IL-1β stimulation, meaning that the neighboring transcript pairs were either both up- or down-regulated by IL-1β. Particularly, we observed a moderate to strong positive correlation in expression fold change upon IL-1β stimulation of mRNAde−lncRNAde pairs which weakened with increasing genomic distance between the mRNAde−lncRNAde pair (Spearman ρ = 0.805 for 25 kbp vs. Spearman ρ = 0.549 for 100 kbp, P = 0.023) (Fig. 1C, SI Appendix, Fig. S2A, and Dataset S4). This coordinate response to IL-1β was not unique to mRNAde−lncRNAde pairs, as the correlation was not significantly different from that of mRNAde−mRNAde pairs (Spearman ρ = 0.488 vs. 0.548 for 150 kbp, P = 1.0). These findings are not unexpected and are consistent with those reported by other groups (14, 34–37). We next assessed the natural background correlation in responsiveness to IL-1β of neighboring genes by measuring the correlation of all mRNA−lncRNA or mRNA−mRNA neighboring pairs possible given the microarray probes available within a given distance of each other (Fig. 1C, SI Appendix, Fig. S2B, and Dataset S4). The correlation observed in mRNAde−lncRNAde and mRNAde−mRNAde pairs was significantly higher than that of the natural background (Spearman ρ = 0.805–0.366 for mRNAde-lncRNAde vs. spearman ρ = 0.086–0.026 for all mRNA-lncRNA, P < 1 × 10−8). This result is in line with our previous observation that, although most IL-1β−regulated transcripts will have a neighboring transcript, very few of those will also be IL-1β−regulated (SI Appendix, Fig. S1 C and D). Lastly, permutation tests were performed, and no correlation was found between random mRNAde−lncRNAde and random mRNAde−mRNAde pairs that are not neighboring each other (SI Appendix, Fig. S2C).
It is becoming increasingly apparent that the genome is divided into chromatin neighborhoods, which are composed of TADs, where there is a high degree of chromatin interaction within, but not between, adjacent TADs (38). Gene coregulation is typically observed within a TAD (32, 39), but has not been explored in the setting of mRNA−lncRNA pairs. We found that, when the mRNAde−lncRNAde pair was located on the same TAD, this was associated with stronger correlation in expression of mRNAde−lncRNAde pairs compared with being on different TADs (Spearman ρ = 0.79 vs. 0.26 for 150 kbp, P = 9.56 × 10−5) (Fig. 1D and Dataset S4) (GEO ID code for endothelial TADs: GSE63525) (40). Strikingly, the correlation of mRNAde−lncRNAde pairs located on the same TAD did not significantly weaken with increasing genomic distance (Spearman ρ = 0.825 for 25 kbp vs. Spearman ρ = 0.656 for 300 kbp, P = 1.0) (Fig. 1D and Dataset S4). Hence, genomic distance alone may be an overly simplistic approach for defining neighboring mRNAde−lncRNAde pairs. Instead, our data suggest that localization within the same chromatin neighborhood (i.e., TAD) should be considered when seeking to identify neighboring genes.
Divergent Transcription Is a Common Feature of mRNAde−lncRNAde Pairs.
To gain further mechanistic insight into the regulation of mRNAde−lncRNAde pairs, the presence of genomic regulatory elements in their vicinity was explored. Previously published RELA (NF-κB subunit) and H3K27ac (histone mark characteristic of active enhancers and promoters) chromatin immunoprecipitation sequence (ChIP-seq) data (GEO ID code GSE89970) from 4-h IL-1β−stimulated human aortic ECs was utilized for analysis (41). Intriguingly, mRNAde−lncRNAde pairs, on average, had stronger intensities of RELA and H3K27ac ChIP-seq signals between the 2 transcripts compared with 50 kbp upstream or downstream of the transcript pair, or compared with background (random sampling within the chromosomes containing neighboring transcript pairs of the same size range as the neighboring transcript pair). This observation held true for mRNAde−lncRNAde pairs 50 and 150 kbp apart and was evident, to a lesser extent, for those 300 kbp apart (SI Appendix, Fig. S3 A–C). In contrast, mRNAde−mRNAde pairs did not follow the same trend, which may suggest differences in how they are organized in the genome relative to mRNAde−lncRNAde pairs (SI Appendix, Fig. S3 A–C).
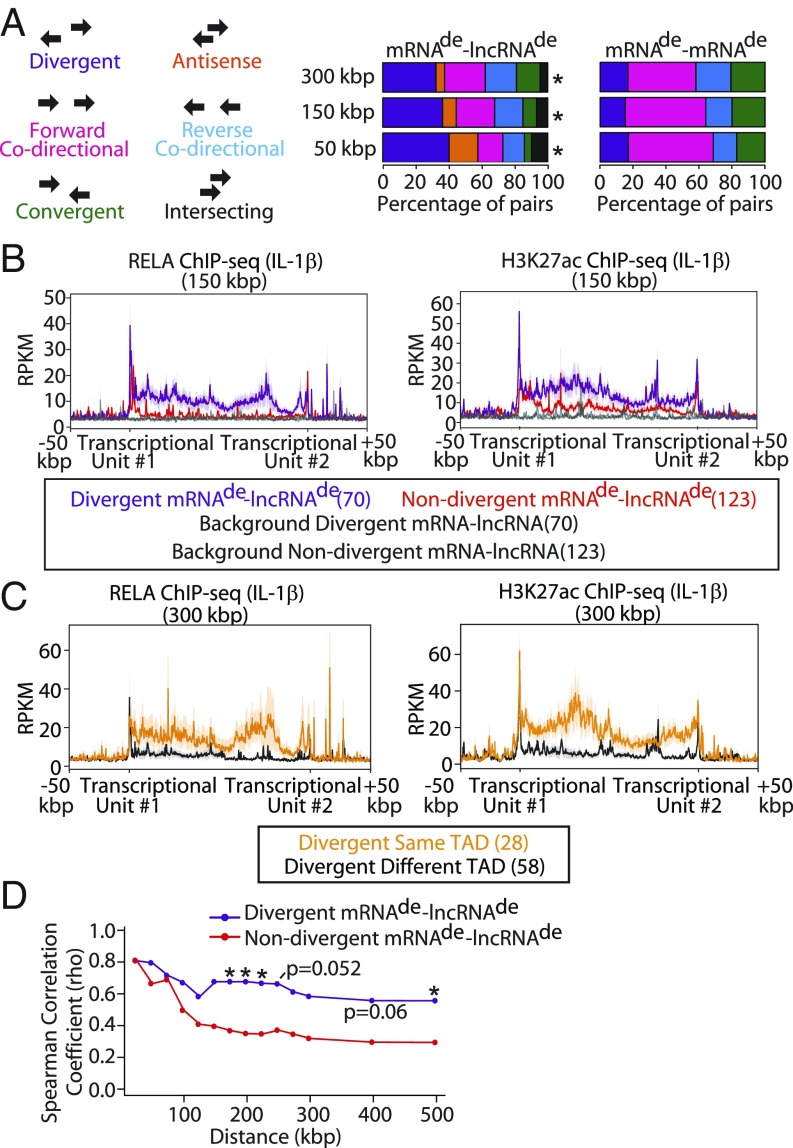
The presence of regulatory elements between mRNAde−lncRNAde pairs prompted us to examine the transcriptional orientation of mRNAde−lncRNAde pairs with respect to each other and the shared regulatory element(s). Specifically, mRNAde−lncRNAde pairs were characterized as being divergent, antisense, forward codirectional, reverse codirectional, convergent, or intersecting with respect to each other. The proportion of different transcriptional orientation types significantly differed between mRNAde−lncRNAde and mRNAde−mRNAde pairs that were 50, 150, and 300 kbp apart (Fig. 2A and Dataset S5). While mRNAde−lncRNAde pairs were most frequently divergent (40.2/36.3/32.2% vs. 17.1/15.3/16.9% of total, 50/150/300 kbp mRNAde−lncRNAde vs. mRNAde−mRNAde, respectively, P < 0.004), mRNAde−mRNAde pairs were most commonly forward codirectional (15.2/23.3/24.8% vs. 51.4/48.8/41.3% of total, 50/150/300 kbp mRNAde−lncRNAde vs. mRNAde−mRNAde, respectively, P < 0.004). However, we did observe that all possible mRNA−lncRNA pairs in our dataset also enriched for divergent transcription—suggesting inherent organization of mRNA−lncRNA pairs into divergent orientations across the genome (40.2/36.3/32.2% vs. 32.0/28.4/28.3% of total, 50/150/300 kbp mRNAde−lncRNAde vs. all mRNA−lncRNA, respectively, P > 0.25) (Fig. 2A and Dataset S5). We then assessed whether divergent mRNAde−lncRNAde pairs were more likely to have shared regulatory elements between them compared with nondivergent pairs. Indeed, divergent mRNAde−lncRNAde pairs that were 50, 150, and 300 kbp apart, on average, had stronger intensities of RELA and H3K27ac ChIP-seq signals between them compared with nondivergent mRNAde−lncRNAde pairs (Fig. 2B and SI Appendix, Fig. S3 D and E). This observation was especially pronounced for divergent mRNAde−lncRNAde pairs that were on the same TAD (Fig. 2C). Furthermore, divergent mRNAde−lncRNAde pairs exhibited stronger correlation in IL-1β−induced expression fold change than mRNAde−lncRNAde pairs transcribed in nondivergent orientations (i.e., Spearman ρ = 0.68 vs. ρ = 0.35 at 200 kbp, P = 0.028; Spearman ρ = 0.56 vs. ρ = 0.29 at 500 kbp, P = 0.045) (Fig. 2D, SI Appendix, Fig. S4A, and Dataset S5). Hence, we interpret these data to indicate that these divergent mRNAde−lncRNAde pairs are coregulated through shared regulatory elements located between their transcriptional units.
Fig. 2.
Divergent transcription is a common feature of mRNAde−lncRNAde pairs. (A) Percentage distribution of orientations of mRNAde−lncRNAde vs. mRNAde−mRNAde transcript pairs that are <50, <150, and <300 kbp apart. *P < 0.05, χ2 test with Bonferroni correction, mRNAde−lncRNAde vs. mRNAde−mRNAde. (B) Profile plot of RELA and H3K27ac ChIP-seq signals in IL-1β−treated ECs that fall within the region between divergent and nondivergent mRNAde−lncRNAde that are <150 kbp apart. Background signal was determined based on a random sampling of the same-size regions within the chromosomes containing the neighboring transcript pairs. (C) Profile plot of RELA and H3K27ac ChIP-seq signals with IL-1β treatment for regions between divergently transcribed mRNAde−lncRNAde pairs that are on either the same or a different TAD within a 300-kbp window. The number of pairs is indicated in brackets. (D) Spearman correlation coefficients for divergent and nondivergent mRNAde−lncRNAde and mRNAde−mRNAde pairs that are of increasing genomic distances apart. *P < 0.05, Fisher z transformation with Bonferroni correction, divergent vs. nondivergent at the specified distance.
Next, the functional significance of divergently transcribed lncRNAs was pursued. Given that cis-acting lncRNAs have been reported to regulate their neighboring mRNAs (15, 36), we tested whether the presence of a neighboring lncRNAde influences mRNAde responsiveness to IL-1β. Taken as a whole, having a neighboring lncRNAde had only a modest influence on mRNAde responsiveness to IL-1β (SI Appendix, Fig. S4B). However, upon subdividing mRNAde−lncRNAde based on transcriptional orientation, we found that up-regulated mRNAde neighboring a divergent lncRNAde trended toward a higher response to IL-1β than those near a nondivergent lncRNAde (median fold change for 150-kbp distance, 6.07 for divergent, 2.97 for nondivergent, P = 0.19) (SI Appendix, Fig. S4 C and D). A possible explanation for this phenomenon is that divergent mRNAde−lncRNAde pairs share stronger regulatory elements, as a recent study utilizing massively parallel reporter assays has shown that promoters of divergent lncRNAs tend to be stronger at driving transcription (42). However, it is plausible that the trend in greater responsiveness to IL-1β could also be attributed to the divergent lncRNA positively regulating its neighboring gene, as another group showed that chromatin-tethered lncRNAs can drive the expression of their neighboring mRNAs. Hence, we further assessed divergent mRNAde−lncRNAde pairs to discover functional lncRNAs.
Identification and Characterization of Divergent mRNAde−lncRNAde Pairs.
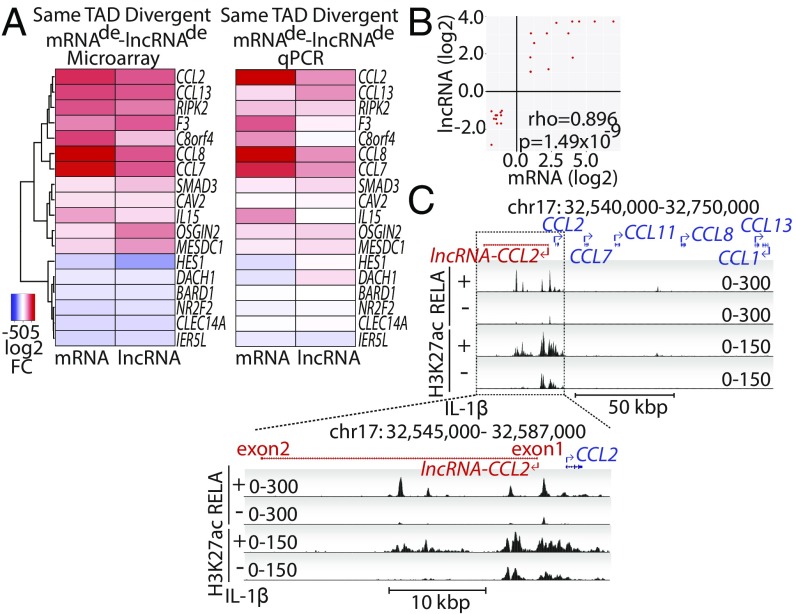
To further investigate the regulation of mRNAde−lncRNAde pairs, we identified all divergent pairs that were localized on the same TAD. In total, there were 18 such divergent mRNAde−lncRNAde pairs after removing splice variants, which correspond to 18 unique mRNAs and 15 unique lncRNAs (Fig. 3A and Dataset S6). These divergent mRNAde−lncRNAde pairs had a coordinated response to IL-1β demonstrated by strong correlation (Spearman ρ = 0.896, P = 1.49 × 10−9) in IL-1β−induced fold change (Fig. 3 A and B). As expected, the mRNAde from the list were enriched for inflammation-related gene ontology biological processes such as “response to wounding,” “inflammatory response,” “defense response,” and “regulation of cell proliferation” (SI Appendix, Fig. S5A). The responsiveness of mRNAde−lncRNAde pairs to 4-h IL-1β treatment was successfully validated by qRT-PCR for 10/15 (69%) lncRNAs and 17/18 (94.4%) mRNAs (Fig. 3A). Examples of genomic loci of a few successfully validated mRNAde−lncRNAde pairs are shown, namely those of lncRNA-F3, lncRNA-BARD1, and lncRNA-CCL2 (Fig. 3C and SI Appendix, Fig. S5B). Importantly, RELA (NF-κB) and H3K27ac ChIP-seq signals were found to be enriched between the mRNAde and lncRNAde transcriptional units.
Fig. 3.
Identification and characterization of divergent mRNAde−lncRNAde pairs. (A) Divergent mRNAde−lncRNAde pairs found within the same TAD are displayed. Heatmap showing the log2 fold-change (FC) in response to 4 h of IL-1β treatment based on microarray (Left) and qPCR validation (Right) are shown. (B) Scatterplot showing the log2 FCs of mRNAde on the y axis and the log2 FCs of their neighboring divergent lncRNAde on the x axis. Spearman rho correlation coefficient and its associated P value are indicated. (C) University of California, Santa Cruz genome browser tracks depicting lncRNA-CCL2. ChIP-seq tracks are shown for RELA and H3K27ac under either basal conditions or upon IL-1β treatment. Zoom-in of lncRNA-CCL2 shows that it is a 2-exon transcript spanning 32.8 kbp of DNA.
CCL2 and lncRNA-CCL2 Are Coordinately Regulated during Acute Inflammation.
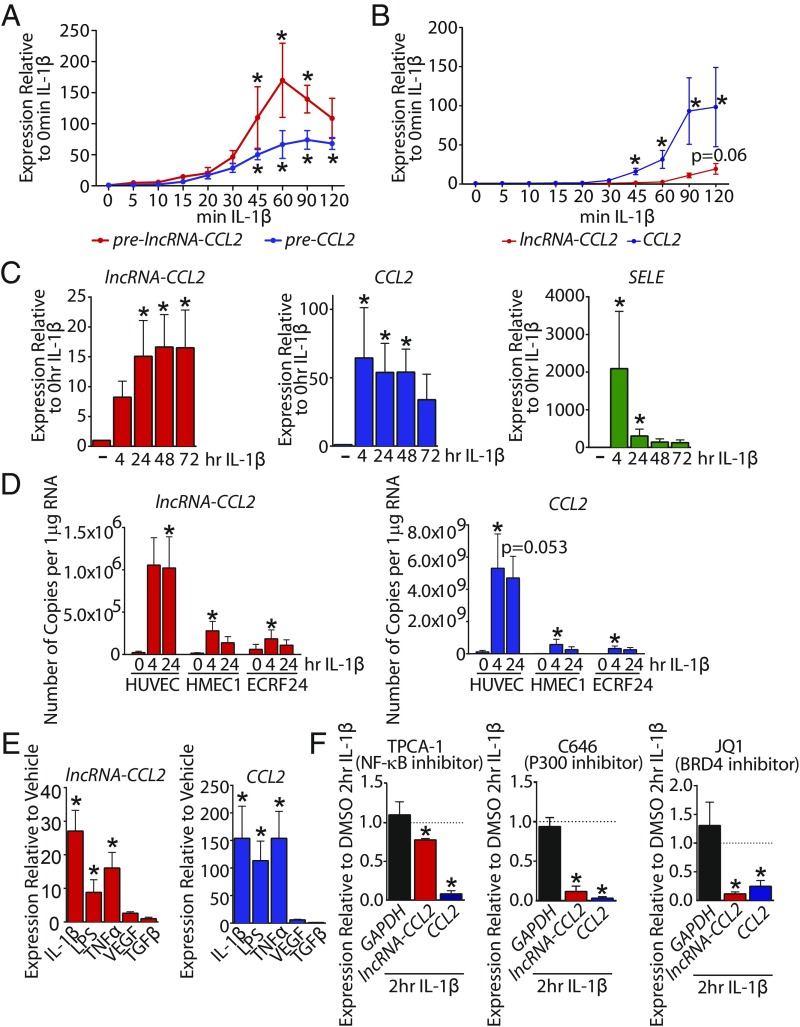
For follow-up experiments, we chose to assess the function of lncRNA-CCL2 (a previously uncharacterized lncRNA annotated as TCONS_00025610), due to the functional relevance of CCL2/MCP1 to inflammatory diseases, including atherosclerosis (43). The lncRNA-CCL2 is a 2-exon transcript spanning ∼33 kbp of DNA (Fig. 3C and SI Appendix, Table S2). We confirmed its existence by amplification of the full-length spliced transcript from HUVEC cDNA (SI Appendix, Table S2). Not only does lncRNA-CCL2 neighbor CCL2, but a number of other CCL genes are located in the genomic vicinity, including CCL7, CCL11, CCL13, CCL8, and CCL1 (Fig. 3C). The lncRNA-CCL2 is transcribed through a superenhancer (44) that spans ∼20 kbp and contains multiple RELA (NF-κB subunit) binding peaks (Fig. 3C). In primary HUVEC, the transcription of lncRNA-CCL2 and CCL2 (as assessed by measuring the unspliced primary transcripts, pre-lncRNA-CCL2 and pre-CCL2) is induced around the same time (i.e., 45 min after IL-1β treatment) (Fig. 4A). However, there is a delay in the processing of lncRNA-CCL2, as the spliced transcript does not increase in expression until 2 h of IL-1β treatment (Fig. 4B). This is consistent with the large intron in lncRNA-CCL2 that must be spliced out to generate the mature transcript (SI Appendix, Table S2). The expression of lncRNA-CCL2 and CCL2 is also sustained during long-term treatment with IL-1β, something atypical of other IL-1β−responsive genes, such as SELE (Fig. 4C). Moreover, the expression of lncRNA-CCL2 and CCL2 is maintained after removal of IL-1β, and they do not exhibit desensitization to IL-1β exposure, which is in contrast to SELE (SI Appendix, Fig. S6). The induction of lncRNA-CCL2 and CCL2 in response to IL-1β was also evident in EC lines, namely human microvascular ECs (HMEC1) and immortalized umbilical vein ECs (EC-RF24) (Fig. 4D). However, the absolute copy number of lncRNA-CCL2 and CCL2 differed between EC types. In particular, the EC-RF24 and HMEC1 cell lines had lower copy numbers of CCL2 and lncRNA-CCL2 than HUVEC. Across all cell lines, CCL2 transcript copy number was ∼5,300-fold higher than that of lncRNA-CCL2 (CCL2, ∼3.2 × 108 vs. 5.3 × 109 copies per 1 µg of RNA; lncRNA-CCL2, ∼1.8 × 105 vs. 1 × 106 copies per 1 µg of RNA; EC-RF24/HMEC1 vs. HUVEC). Aside from IL-1β stimulation, lncRNA-CCL2 and CCL2 were also coordinately induced by other proinflammatory stimuli such as lipopolysaccharide (LPS) and tumor necrosis factor alpha (TNF-α) (Fig. 4E). Neither transcript was inducible by vascular endothelial cell growth factor (VEGF) or transforming growth factor beta (TGF-β) stimulation. Lastly, inhibitor experiments demonstrated that lncRNA-CCL2 and CCL2 were regulated by NF-κB, p300, and BRD4, as seen using inhibitors of IκB kinase 2 (i.e., TPCA-1), p300/CBP (i.e., C646), and BRD4 (i.e., JQ1) (Fig. 4F). Thus, in ECs, lncRNA-CCL2 and CCL2 are a divergently transcribed transcript pair that shows synchronized responses to proinflammatory stimuli. Interestingly, a number of other IL-1β−regulated chemokine genes are clustered near the CCL2 superenhancer, including CCL7, CCL11, CCL8, CCL13, and CCL1 (Fig. 3C). While lncRNA-CCL2, CCL2, and CCL7 followed similar kinetics, the other CCL genes in this locus responded to IL-1β with differing kinetics, suggesting potential differences in their regulation (SI Appendix, Fig. S7).
Fig. 4.
CCL2 and lncRNA-CCL2 are coordinately regulated during acute inflammation. (A) The qRT-PCR data of unspliced pre-lncRNA-CCL2 and pre-CCL2 upon treatment with IL-1β for various lengths of time. *P < 0.05, Friedman test with Dunn’s multiple comparisons test, relative to basal, n = 3 to 4. (B) The qRT-PCR data of spliced lncRNA-CCL2 and CCL2 upon treatment with IL-1β for various lengths of time. *P < 0.05, Friedman test with Dunn’s multiple comparisons test, relative to basal, n = 5. (C) The qRT-PCR data showing the kinetic response of lncRNA-CCL2, CCL2, and SELE to 4 to 72 h of IL-1β in HUVEC. *P < 0.05, Friedman test with Dunn’s multiple comparisons test, relative to basal, n = 6. (D) Copy number of lncRNA-CCL2 and CCL2 transcripts per 1 µg of RNA measured using qRT-PCR in HUVEC, HMEC1, and EC-RF24 at multiple time points of IL-1β stimulation. *P < 0.05, Friedman test with Dunn’s multiple comparisons test, relative to basal, n = 4 to 7. (E) The qRT-PCR data showing the responsiveness of lncRNA-CCL2 and CCL2 to 2 h of IL-1β, LPS, TNFα, VEGF, and TGF-β stimulation in HUVEC. *P < 0.05, Friedman test with Dunn’s multiple comparisons test, n = 3 to 7. (F) The qRT-PCR data of GAPDH, lncRNA-CCL2, and CCL2 expression upon 1-h pretreatment with chemical inhibitors of NF-κB signaling (TPCA-1), p300 activity (C646), and BRD4 (JQ1) followed by 2 h of IL-1β stimulation in HUVEC. All data are relative to expression values upon DMSO pretreatment followed by 2 h of IL-1β stimulation. *P < 0.05, paired Student’s t test, n = 3.
LncRNA-CCL2 Positively Regulates Levels of Its Neighboring CCL2 Gene.
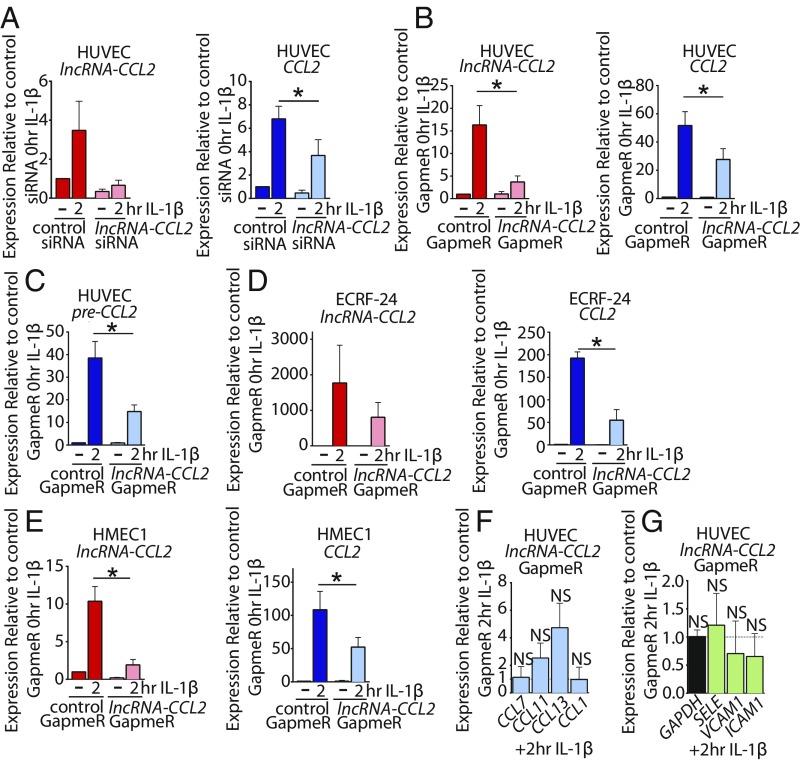
The functional role of lncRNA-CCL2 was explored using 2 different knockdown approaches. Small interfering RNA (siRNA) and antisense locked nucleic acid (LNA) GapmeR were designed to target exon 2 of lncRNA-CCL2. Exon 2 was targeted as it is 15 kbp distal to the superenhancer, and therefore siRNAs and GapmeRs targeting this region should not directly interfere with the activity of the superenhancer (Fig. 3C). We were able to achieve sufficient knockdown of the spliced lncRNA-CCL2 transcript (Fig. 5 A and B), but not the pre-lncRNA-CCL2 transcript (SI Appendix, Fig. S8A). Knockdown of lncRNA-CCL2 using either siRNA or GapmeR led to a ∼50% decrease in CCL2 expression upon IL-1β stimulation of HUVEC (Fig. 5 A and B). Levels of the unspliced pre-CCL2 were likewise decreased upon lncRNA-CCL2 knockdown (Fig. 5C and SI Appendix, Fig. S8B). Furthermore, we were able to recapitulate the effect of lncRNA-CCL2 knockdown on CCL2 levels in the EC lines, EC-RF24 and HMEC1 (Fig. 5 D and E). A strong positive correlation between the degree of lncRNA-CCL2 knockdown and the decrease in CCL2 induction was observed (r = 0.8040, P = 0.0634) (SI Appendix, Fig. S8C). However, we did not find significant changes in basal CCL2 levels, which may be due to the low basal levels of lncRNA-CCL2 (Fig. 5 A–E). Next, we assessed whether lncRNA-CCL2 can regulate any of the other genes in the CCL gene cluster. No significant changes were observed in the expression of other CCL genes upon IL-1β stimulation (Fig. 5F), suggesting that lncRNA-CCL2 only regulates the most proximal CCL gene of the cluster, namely CCL2. Additionally, no consistent change in expression of the NF-κB responsive genes SELE, ICAM1, and VCAM1 was observed (Fig. 5G and SI Appendix, Fig. S8D). This finding suggests that lncRNA-CCL2 is unlikely to be involved in directly regulating NF-κB signaling. Furthermore, ectopic overexpression of spliced lncRNA-CCL2 in HUVEC had no effect on CCL2 expression (SI Appendix, Fig. S9). This finding is in line with previous studies in which ectopic expression of cis-acting lncRNAs had no effect on their target mRNA, potentially due to the inability of the ectopically expressed lncRNA to properly localize to its target locus (18, 45).
Fig. 5.
The lncRNA-CCL2 positively regulates levels of its neighboring CCL2 gene. (A and B) The qRT-PCR data of lncRNA-CCL2 and CCL2 gene expression upon knockdown of lncRNA-CCL2 using (A) siRNA or (B) GapmeR under basal condition and upon stimulation with IL-1β for 2 h in HUVEC. *P < 0.05, paired Student’s t test, (A) n = 3, (B) n = 8. (C) The qRT-PCR data of unspliced pre-CCL2 gene expression upon knockdown of lncRNA-CCL2 using GapmeR under basal condition and upon stimulation with IL-1β for 2 h in HUVEC. *P < 0.05, paired Student’s t test, n = 5. (D and E) qRT-PCR data of lncRNA-CCL2 and CCL2 expression upon knockdown of lncRNA-CCL2 using GapmeR under basal conditions or upon stimulation with IL-1β for 2 h in (D) EC-RF24 and (E) HMEC1. *P < 0.05, paired Student’s t test, (D) n = 3, (E) n = 5. (F) The qRT-PCR data of CCL genes upon knockdown of lncRNA-CCL2 using GapmeR and stimulation with IL-1β for 2 h in HUVEC. *P < 0.05, paired Student’s t test, n = 4; NS, nonsignificant. (G) The qRT-PCR data of inflammatory genes upon knockdown of lncRNA-CCL2 using GapmeR and stimulation with IL-1β for 2 h in HUVEC. *P < 0.05, paired Student’s t test, n = 4.
LncRNA-CCL2 Regulation of CCL2 Levels May, in Part, Be Mediated through Interaction with RNA-Binding Proteins.
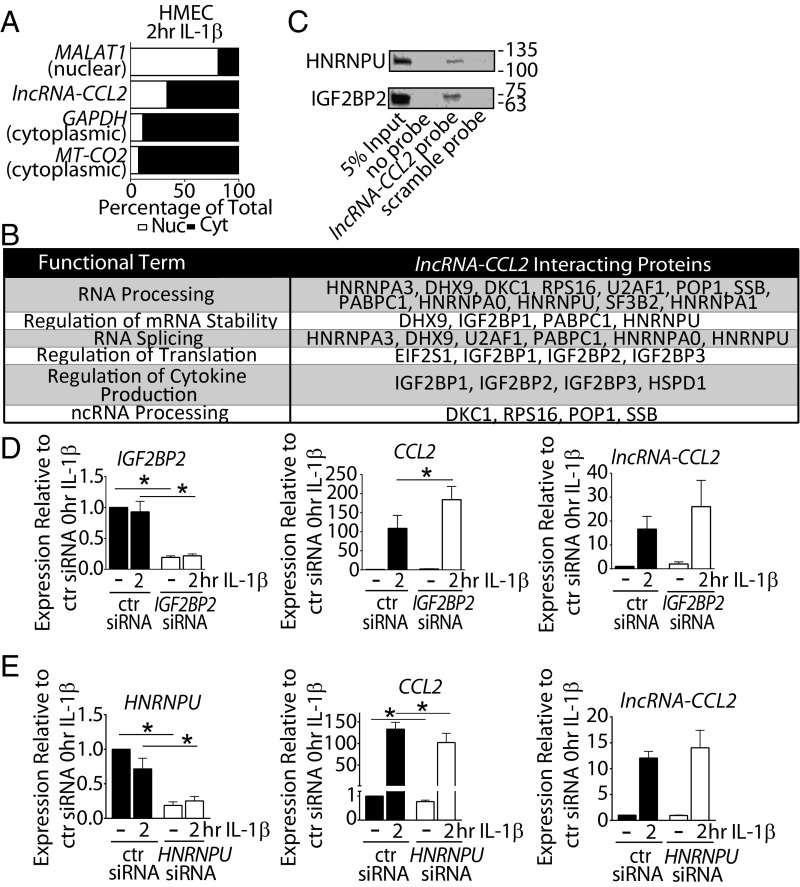
We next sought to explore the mechanism(s) through which lncRNA-CCL2 regulates CCL2 levels. Determining the subcellular localization of an lncRNA can give insight into its potential mechanism of action. LncRNA-CCL2 was detectable in both the nuclear and cytoplasmic compartments, with a modest enrichment in the cytoplasm (Fig. 6A). Given the presence of lncRNA-CCL2 in the cytoplasm, we first verified that it does not encode any proteins. The coding potential of lncRNA-CCL2 is very low (0.002, according to the Coding-Potential Assessment Tool, with a coding probability of <0.364 indicating a noncoding sequence; SI Appendix, Fig. S10A) (46). Although it has a potential open reading frame of 29 amino acids, overexpression of lncRNA-CCL2 did not produce any observable peptides (SI Appendix, Fig. S10B). Moreover, cytoplasmic lncRNAs typically act in trans (31), but we found no evidence of lncRNA-CCL2 globally regulating inflammatory genes (i.e., SELE, VCAM1, and ICAM1). Thus, it is unlikely that lncRNA-CCL2 acts in trans to indirectly regulate CCL2 via modulating NF-κB signaling. For this reason, we next queried whether the nuclear fraction of lncRNA-CCL2 is responsible for regulating CCL2 in cis.
Fig. 6.
The regulation of CCL2 levels by lncRNA-CCL2 may, in part, be mediated through interaction with RNA-binding proteins. (A) The qRT-PCR data showing the distribution of MT-CO2 (cytoplasmic control), GAPDH (cytoplasmic control), lncRNA-CCL2, and MALAT1 (nuclear control) in the nuclear and cytoplasmic compartments of 2-h IL-1β−stimulated HMEC1, n = 3. (B) Table showing the functional terms associated with the proteins uniquely pulled down or enriched in pull-downs using a biotinylated lncRNA-CCL2 probe in nuclear extracts of IL-1β−stimulated HMEC1. (C) Validation of IGF2BP2 and HNRNPU pull-down with the biotinylated lncRNA-CCL2 probe by Western blot. (D) The qRT-PCR data of IGF2BP2, lncRNA-CCL2, and CCL2 expression upon knockdown of IGF2BP2 using siRNA under basal and 2-h IL-1β−stimulated conditions in HMEC1. *P < 0.05, paired Student’s t test, n = 4. (E) The qRT-PCR data of HNRNPU, lncRNA-CCL2, and CCL2 expression upon knockdown of HNRNPU using siRNA under basal and 2-h IL-1β−stimulated conditions in HMEC1. *P < 0.05, paired Student’s t test, n = 4.
Cis-acting lncRNAs have previously been shown to interact with transcription factors and chromatin remodeling complexes (15). Hence, we probed for the interaction of lncRNA-CCL2 with CCL2 superenhancer-associated chromatin machinery (44). However, we found no reproducible binding of lncRNA-CCL2 to RELA, p300, or BRD4 (SI Appendix, Fig. S11A). An unbiased approach was next used to identify nuclear interacting protein partners of lncRNA-CCL2 by performing an RNA pull-down experiment. To accomplish this, a biotinylated lncRNA-CCL2 probe or a scramble probe (SI Appendix, Table S2) was incubated with nuclear lysates of IL-1β−stimulated HMECs. Mass spectrometry of the isolated proteins revealed 49 proteins that uniquely bound and 9 proteins that enriched for binding to the lncRNA-CCL2 probe (Dataset S7). Interaction of CHD4 with lncRNA-CCL2 was confirmed by CHD4 immunoprecipitation (SI Appendix, Fig. S11B). However, CHD4 knockdown experiments revealed that CHD4 played no role in modulating CCL2 levels during inflammation (SI Appendix, Fig. S11C).
Functional annotation of the proteins pulled down with lncRNA-CCL2 revealed that they were predominantly enriched for RNA-binding proteins such as members of the IGF2BP and the heterogeneous ribonucleoprotein (HNRNP) families (Fig. 6B). This finding was intriguing, as we found differences in the transcript stability of lncRNA-CCL2 and CCL2 (SI Appendix, Fig. S12), implying that RNA-binding proteins may participate in their regulation. In particular, we noted lncRNA-CCL2 to have a longer half-life than CCL2 in actinomycin D experiments (>240 min for lncRNA-CCL2 vs. <60 min for CCL2). Hence, we focused on RNA-binding proteins, IGF2BP2, IGF2BP3, and HNRNPU, as they have previously been shown to interact with lncRNAs (47–51) and have been shown to modulate mRNA stability (52–54). We confirmed that HNRNPU and IGF2BP2 bind specifically to an lncRNA-CCL2 probe, but not to a scramble probe (Fig. 6C). We then explored whether these proteins could modulate CCL2 levels by performing siRNA-mediated knock down of IGF2BP2, IGF2BP3, and HNRNPU in HMECs. Varying effects on CCL2 levels were observed with each of these 3 proteins. In the case of IGF2BP3, its knockdown resulted in no change in CCL2 or lncRNA-CCL2 levels during IL-1β stimulation (SI Appendix, Fig. S13). Meanwhile, knockdown of IGF2BP2 significantly increased the level of CCL2, while there was no significant change in lncRNA-CCL2 during IL-1β stimulation (Fig. 6D). Lastly, knockdown of HNRNPU resulted in a significant decrease in CCL2 but not lncRNA-CCL2 during IL-1β stimulation (Fig. 6E). These results imply that IGF2BP2 and HNRNPU may, in part, contribute to the regulation of CCL2 by lncRNA-CCL2. Taken together, these findings suggest that lncRNA-CCL2 may act as a scaffold for RNA-binding proteins that both positively and negatively regulate CCL2 levels.
LncRNA-CCL2 Is Elevated in Unstable Symptomatic Human Atherosclerotic Plaques Where It Correlates with CCL2 Expression.
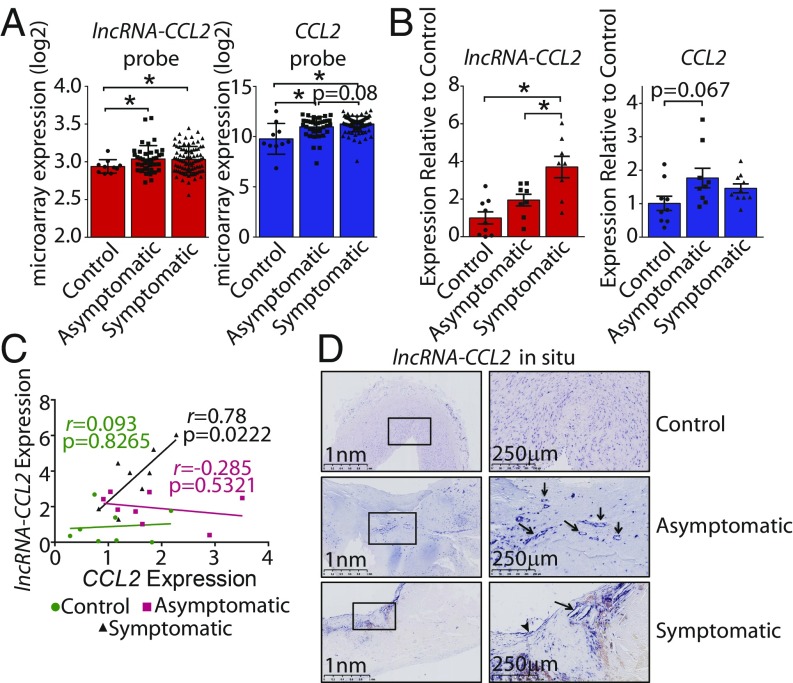
While we have provided evidence that lncRNA-CCL2 is regulated in vitro by inflammatory stimuli, we sought to determine whether the expression of lncRNA-CCL2 is relevant to chronic inflammatory pathologies such as atherosclerosis. To do so, levels of lncRNA-CCL2 and CCL2 were assessed in human atherosclerotic plaques isolated from patients undergoing carotid endarterectomy (CEA, n = 127) and belonging to the Biobank of Karolinska Endarterctomies (BiKE), Karolinska Institute. Patients were characterized as either stable asymptomatic (n = 40) or unstable symptomatic (n = 87), where “symptomatic” indicates that the patient has experienced a mild embolic event, particularly, either a transient ischemic attack, a minor stroke, or temporary loss of vision (i.e., amaurosis fugax) (55). As a control, healthy arteries (n = 10) were used from organ donors with no previous history of cardiovascular disease. Based on the microarray, lncRNA-CCL2 and CCL2 were significantly elevated in both unstable symptomatic and stable asymptomatic plaques (Fig. 7A). However, the probe capturing lncRNA-CCL2 (236469_at) was not uniquely specific to lncRNA-CCL2 and aligned to other mRNAs in the human genome. Thus, to validate the microarray and to more accurately assess lncRNA-CCL2 expression, real-time qPCR (RT-qPCR) was performed with primers that spanned exon 1 and exon 2 (as used in our in vitro experiments) on a smaller set of CEA and control samples. RT-qPCR revealed that lncRNA-CCL2 was significantly elevated in unstable symptomatic plaques (Fig. 7B). Furthermore, we observed a positive correlation between the expression of lncRNA-CCL2 and CCL2 in unstable symptomatic plaque samples (r = 0.78, P = 0.0222) (Fig. 7C). This observation in unstable symptomatic plaques is in line with our previous in vitro findings that lncRNA-CCL2 and CCL2 are correlated in expression in response to IL-1β treatment and that lncRNA-CCL2 positively regulates CCL2 expression in ECs. However, atherosclerotic plaques comprise more than just ECs, and changes in the cell type composition of plaques could also affect the relative levels of lncRNA-CCL2 and CCL2 (56, 57). Next, using RNA in situ hybridization, it was observed that lncRNA-CCL2 was expressed in the highly inflamed shoulder regions and fibrous caps of stable asymptomatic plaques (Fig. 7D). In unstable symptomatic plaques, lncRNA-CCL2 was particularly highly expressed in the ECs overlying the carotid plaque (Fig. 7D, arrowhead). Strikingly, lncRNA-CCL2 appeared to also be expressed in the lining of neovessels within stable asymptomatic and unstable symptomatic plaques (Fig. 7D, arrows). Unlike advanced atherosclerotic plaques, only low levels of diffuse signal corresponding to lncRNA-CCL2 could be detected in undiseased healthy arteries. Altogether, these data suggest that lncRNA-CCL2 is dysregulated during human atherosclerosis.
Fig. 7.
LncRNA-CCL2 is elevated in unstable symptomatic human atherosclerotic plaques where it correlates with CCL2 expression. (A) Normalized log2 expression of lncRNA-CCL2 and CCL2 probes in microarrays performed on human unstable symptomatic or stable asymptomatic atherosclerotic plaques or healthy artery controls. *P < 0.05, ordinary 1-way ANOVA with Sidak correction. (B) The qRT-PCR expression of lncRNA-CCL2 and CCL2 in human unstable symptomatic or stable asymptomatic atherosclerotic plaques or healthy artery controls. *P < 0.05, ordinary 1-way ANOVA with Sidak correction. (C) Scatter plot showing the correlation in qRT-PCR expression of lncRNA-CCL2 and CCL2 in samples from human unstable symptomatic or stable asymptomatic atherosclerotic plaques or healthy artery controls. The Pearson correlation coefficient (r) and its associated P values are shown. (D) RNA in situ hybridization showing the presence and increased expression of lncRNA-CCL2 in unstable symptomatic human atherosclerotic plaques compared with healthy artery controls from the BiKE biobank. LncRNA-CCL2 is expressed in neovessels (arrows) and in the EC lining of the atheroma (arrowhead). Images from 2 different magnifications, 2.5× and 10×, are shown. A higher-quality Fig. 7 is available at Figshare (DOI: 10.6084/m9.figshare.8956868.v1).
Discussion
The discovery that thousands of lncRNAs are encoded in the human genome adds an extra layer to the regulation of gene expression, yet the vast majority of lncRNAs remain uncharacterized. Our study aimed to uncover functional lncRNAs involved in human vascular inflammation with the hopes of finding ways to fine-tune the inflammatory response. A common approach to identifying functional lncRNAs has been to focus on lncRNAs that act in cis to regulate their neighboring mRNAs. However, there is currently no systematic way to identify cis-acting lncRNAs, as there are no evidence-based guidelines as to what constitutes a “neighboring” mRNA (15, 30). Thus, we sought to precisely define the concept of a neighboring mRNA−lncRNA pair by identifying and characterizing all IL-1β−regulated mRNA−lncRNA pairs separated by varying genome distances. We found that IL-1β−regulated mRNA−lncRNA pairs were correlated in their responsiveness to IL-1β and that this correlation generally decreased with increasing genomic distance. Similar observations have been reported by others who observed coexpression of lncRNAs with their neighboring protein coding genes (14, 58–60), but this had not been previously assessed in the setting of EC activation. Importantly, genomic distance alone does not reflect the actual physical interactions that occur between genomic regions, since the genome is compartmentalized into chromatin neighborhoods that have been referred to as TADs (32, 38, 40). Indeed, our analysis revealed that IL-1β−regulated mRNA−lncRNA pairs on the same TAD were highly correlated regardless of the genomic distance between them. Hence, we argue that defining neighboring transcript pairs based on a genomic distance cutoff alone may be superficial, as it does not take into consideration the genomic neighborhoods in which the transcripts reside.
Currently, “lncRNA” is an umbrella term for a broad class of noncoding transcripts. Subgroups of lncRNAs include competing endogenous RNAs, enhancer RNAs, and antisense RNAs, yet the majority of lncRNAs have not been categorized into functional subgroups. A further distinction is in regards to the transcriptional orientation of the lncRNA to its neighboring mRNA. Our work noted that IL-1β−regulated mRNA−lncRNA pairs are disproportionally divergently transcribed relative to IL-1β−regulated mRNA−mRNA pairs. These divergent mRNA−lncRNA pairs showed enrichment of RELA binding and H3K27ac marks in the intervening regions between them, suggesting that they share regulatory elements. Moreover, mRNAs near an IL-1β−regulated divergent lncRNA trended toward enhanced up-regulated response to IL-1β compared with those that were near a nondivergent lncRNA. These data are in line with evidence showing that promoters of divergent lncRNAs have higher activity than promoters of other classes of lncRNAs (42). However, in addition to sharing regulatory elements, we found that lncRNA-CCL2 positively regulates its neighboring mRNA, CCL2. Other groups have also noted the high frequency of divergently transcribed neighboring mRNA−lncRNA pairs (18, 58). One of these groups successfully knocked down 16 divergent and antisense lncRNAs and found 10 of them to positively regulate the expression of their neighboring genes; meanwhile the other 6 divergent lncRNAs had no effect on their neighboring genes (18). It is intriguing that functional divergent lncRNAs, including lncRNA-CCL2, positively but never negatively regulated their neighboring genes. Thus, there may be something inherently unique to the function of divergent lncRNAs, and, in the future, there may be enough evidence for them to be considered as their own subclass of lncRNAs.
Teasing apart the mechanism of action of lncRNAs remains an arduous task due to the vast number of proteins that lncRNAs have been shown to interact with (30, 31). Adding to the complexity, some lncRNAs are found in both the nucleus and cytoplasm and have a distinct mechanism of action within each subcellular compartment (61). A systematic RNA fluorescence in situ hybridization screen of 61 lncRNAs revealed a wide range of distributions of lncRNAs throughout the cell. The majority of lncRNAs were found to be nuclear-enriched, but the amount present in the cytoplasmic fraction was highly variable (62). In the case of lncRNA-CCL2, it is detectable in both cellular compartments, with an enrichment in the cytoplasm, but our work has focused solely on the nuclear role of lncRNA-CCL2 in regulating CCL2 expression. We found that RNA-binding proteins that bind to nuclear lncRNA-CCL2, namely HNRNPU and IGF2BP2, can enhance or suppress CCL2 transcript levels, respectively, without altering the abundance of lncRNA-CCL2. This implicates lncRNA-CCL2 as a scaffold for RNA-binding proteins that may modulate CCL2 expression in cis. An lncRNA expressed in adipocytes, Linc-ADAL, is also present in both the nuclear and cytoplasmic compartments, and, like lncRNA-CCL2, it has been shown to interact with HNRNPU and IGF2BP2. In the case of Linc-ADAL, it regulates levels of multiple mRNAs involved in adipocyte differentiation (49). Thus, it remains possible that lncRNA-CCL2 may also act in trans to regulate other mRNAs via interaction with HNRNPU and IGF2BP2, and this should be investigated in future studies.
An additional layer of complexity to lncRNA function is discerning between the role of the lncRNA transcript itself vs. the contribution of transcription through the locus from which it is transcribed. The act of transcribing an lncRNA has previously been shown to mediate chromatin remodeling (63). In particular, halting lncRNA transcription through insertion of a polyA termination signal has been shown to influence neighboring gene expression via a decrease in enhancer marks (64). Given that lncRNA-CCL2 is transcribed through a superenhancer, it is possible that its transcription could also have a functional consequence on enhancer activity. We attempted to use CRISPR/Cas9 to insert a polyA termination signal immediately after the first exon of lncRNA-CCL2. Despite the successful insertion of the polyA termination signal, we were unable to halt the transcription of lncRNA-CCL2. Perhaps the polyA signal was not strong enough to block the recruitment of abundant elongation factors and transcription machinery to the CCL2 superenhancer (44). Another explanation could be that other TSSs are present within the CCL2 superenhancer, which could give rise to an alternative lncRNA-CCL2 transcript. It is therefore not possible, at the present time, to comment on the possible contribution of lncRNA-CCL2 transcription to CCL2 regulation. However, transcription alone cannot be solely responsible for its function, as targeted knock-down using siRNAs or GapmeRs directed to exon 2 was able to decrease CCL2 mRNA levels without apparently impacting lncRNA-CCL2 transcription.
Taken together, our studies have identified principals of dynamic lncRNA−mRNA networks in the setting of EC activation. We demonstrate that divergently transcribed lncRNAs located in the same TAD can positively regulate the expression of neighboring mRNAs during the inflammatory response. The applicability of lncRNAs for therapeutic intervention in vascular inflammatory diseases is exciting, but many questions remain. Their high cell type specificity makes them an attractive drug target due to fewer cell type off-target effects (59, 65). In our study, we found lncRNA-CCL2 to be dysregulated in human atherosclerotic plaques. Therefore, exploring the therapeutic potential of lncRNA-CCL2 may be of interest. A potential approach to target lncRNAs could be to use inhibitors to block the binding of lncRNAs to their interacting protein complexes. This approach has already been tested using “mixmer” oligomers, a mixture of LNA monomers combined with DNA monomers, in a spinal muscular atrophy model (66). Hence, using mixmers to block the interaction of lncRNA-CCL2 with its interacting RNA-binding proteins, such as HNRNPU and IGF2BP2, could be of future interest to suppress vascular inflammation.
Methods
Cell Culture and Reagents.
ECs utilized in this study included primary HUVEC (ScienCell 8000), immortalized human dermal microvascular ECs (HMEC1, ATCC CRL-3243), and immortalized HUVECs (EC-RF24, ABM T0003). HUVECs were cultured in Endothelial Cell Medium (ECM) with 5% FBS and Endothelial Cell Growth Supplement (ScienCell); meanwhile, HMEC1 and EC-RF24 were cultured in Endothelial Cell Media MV2 (PromoCell). Cells were treated with the following stimulants: IL-1β (10 ng/mL, Gibco), LPS (100 ng/mL, InvivoGen), TNFα (10 ng/mL, Gibco), VEGF (50 ng/mL, Gibco), TGFβ (10 ng/mL, Cell Signaling) for specified times. Cells were pretreated with the following inhibitors 1 h before IL-1β stimulation: TPCA-1 (IKK inhibitor, 3 µM, Sigma-Aldrich), C646 (p300 inhibitor, 10 µM, Sigma-Aldrich), JQ1 (BRD4 inhibitor, 500 nM, Sigma-Aldrich), and actinomycin D (transcriptional inhibitor, 5 µM, BioShop). All drugs were prepared as 1,000× stocks in dimethyl sulfoxide (DMSO). Human material analysis was performed as described in SI Appendix.
Cloning, Transfection, and Nucleoporation.
Cells were transfected at 40% confluency with siRNAs and GapmeRs listed in SI Appendix, Table S1 using Lipofectamine RNAiMAX Reagent (1 µL/20 nM siRNA or GapmeR, Thermo Fisher Scientific) in Opti-MEM I Reduced Serum Media (100 µL, Thermo Fisher Scientific). Media was changed back to regular Endothelial Cell Media after 4 to 6 h, and cells were harvested after 48 h. For lncRNA-CCL2 overexpression studies and in vitro transcription, lncRNA-CCL2 was cloned into pCS2+ (Addgene #2295) (SI Appendix, Table S2). Alternatively, a scramble sequence of the same length as lncRNA-CCL2 was cloned into pGEM-T Easy (Promega) for in vitro transcription (SI Appendix, Table S2). Overexpression experiments in HUVEC were performed using the P5 Primary Cell 4D-Nucleofector X Kit (Lonza) and the Amaxa 4D-Nucleofector System (Lonza) as per manufacturer’s instructions for HUVEC using 1 µg of plasmid.
RNA Isolation, Reverse Transcription, and Quantitative PCR.
RNA was isolated from cells using TRIzol Reagent (Thermo Fisher Scientific) as per the manufacturer’s instruction. Nuclear fractionation was performed as described in SI Appendix. Reverse transcription was performed with between 500 ng to 1 µg RNA using the high-capacity cDNA reverse transcription kit (Applied Biosystems). The qRT-PCR was performed in triplicate using the LC480 SYBR Green I Master (Roche) and the Roche Lightcycler 480 (Roche). Data were analyzed using the delta−delta Ct method and were normalized to TATA-binding protein. Copy number quantification was performed using Roche Lightcycler 480 (Roche) using 107 to 103 copy numbers of qPCR products for the creation of a standard curve. Primers used are listed in SI Appendix, Tables S3 and S4.
Microarray Analysis.
Expression changes of lncRNAs and mRNAs were assessed by the manufacturer using the Human LncRNA Expression Microarray V3.0 (Arraystar). The expression data were normalized using the Robust Multi-Array Average method. The lncRNAs and mRNAs were considered differentially expressed if they had a >2-fold change and were statistically significant (paired t test P < 0.05) for any of the time points of IL-1β treatment. Heatmaps were constructed using the “heatmap.2” function from the “gplots” Bioconductor package (67). Additionally, the “ggplot2,” “sm,” and “violinplot” Bioconductor packages were used for graph visualization. Neighboring transcript pairs were identified based on genomic distance (20 to 500 kbp) between them with the help of the “GenomicRanges” Bioconductor package (68). Spearman correlation coefficient (rho) was calculated using base R, and statistical significance between 2 rho values was determined using Fisher Z-transformation. ChIP followed by sequencing analysis was performed as described in SI Appendix.
RNA Pull-Down.
RNA pull-down was performed as described elsewhere (69). To generate in vitro transcribed probes, lncRNA-CCL2 was cloned downstream of the SP6 promoter in pCS2+ plasmid, and a scramble sequence of same length as lncRNA-CCL2 was cloned downstream of the T7 promoter in pGEM-T Easy plasmid (SI Appendix, Table S2). In vitro transcription was performed using SP6 RNA Polymerase (P1085, Promega) and T7 RNA Polymerase (P2075, Promega) with the Biotin RNA Labeling Mix (Roche) using 1 µg of linearized plasmid. To pull down interacting partners, nuclear extracts from IL-1β−stimulated HMEC1 cells (5 × 106 cells per condition) were used. Precleared nuclear extracts were incubated with 1 µg of lncRNA-CCL2 probe, scramble probe, or no probe. After pull-down, samples were either sent for mass spectrometry analysis (as described in SI Appendix) or run on Western blot (as described in SI Appendix). RNA immunoprecipitation was performed as described in SI Appendix.
Data Availability.
Microarray data generated in this study have been deposited in the GEO database under the accession number GSE127990. Code used in the analysis can be accessed at https://github.com/nadiyakhyzha/lncRNA_mRNA_neighboring_analysis.
Statistical Analyses.
Experiments were performed at least 3 times unless stated otherwise. Statistical analyses performed for each experiment are specified in the figure legends. P < 0.05 was considered to be statistically significant. Plotted data represent the mean ± SEM.
Supplementary Material
Acknowledgments
We wish to thank Jonathan Krieger of SPARC BioCentre, The Hospital for Sick Children, Toronto, Canada, for assistance with mass spectrometry. This work was supported by Project Grants from the Canadian Institutes of Health Research (CIHR) (PJT 148487 [to J.E.F.] and 364832 [to J.E.F. and M.D.W.]). J.E.F. and M.D.W. were funded by CIHR Tier 2 Canada Research Chairs and Early Researcher Awards from the Ontario Ministry of Research and Innovation. J.E.F. was supported by a CIHR-funded Transnational Team Grant. Infrastructure in the J.E.F. laboratory was funded by the John R. Evans Leaders Opportunity Fund from the Canada Foundation for Innovation and the Ontario Research Fund. N.K. received a Canada Graduate Scholarship from the Natural Sciences and Engineering Research Council of Canada (NSERC). M.K. received an Undergraduate Student Research Award from NSERC. M.D.W. was supported by an NSERC grant (436194-2013). The BiKE project was financed by the Swedish Heart and Lung Foundation, Swedish Research Council, and Stockholm County Council. L. Maegdefessel is the recipient of fellowships from the Swedish Society for Medical Research and Heart and Lung Foundation, and is further supported by the European Research Council (Starting Grant NORVAS) and the German Center for Cardiovascular Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Microarray data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE127990). Code used in the analysis can be accessed at https://github.com/nadiyakhyzha/lncRNA_mRNA_neighboring_analysis. A higher-quality Fig. 7 is available at Figshare (DOI: 10.6084/m9.figshare.8956868.v1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1904108116/-/DCSupplemental.
References
- 1.Libby P., Inflammation in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 32, 2045–2051 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arslan S., et al. ; Cardiolinc™ network , Long non-coding RNAs in the atherosclerotic plaque. Atherosclerosis 266, 176–181 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Cremer S., et al. , Hematopoietic deficiency of the long non-coding RNA MALAT1 promotes atherosclerosis and plaque inflammation. Circulation 139, 1320–1334 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Helgadottir A., et al. , A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 316, 1491–1493 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Sallam T., et al. , Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat. Med. 24, 304–312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jarinova O., et al. , Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler. Thromb. Vasc. Biol. 29, 1671–1677 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Atianand M. K., et al. , A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 165, 1672–1685 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan J., et al. , Cutting edge: A natural antisense transcript, AS-IL1α, controls inducible transcription of the proinflammatory cytokine IL-1α. J. Immunol. 195, 1359–1363 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenter S., et al. , A long noncoding RNA mediates both activation and repression of immune response genes. Science 341, 789–792 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou X., et al. , Long non-coding RNA ANRIL regulates inflammatory responses as a novel component of NF-κB pathway. RNA Biol. 13, 98–108 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.IIott N. E., et al. , Long non-coding RNAs and enhancer RNAs regulate the lipopolysaccharide-induced inflammatory response in human monocytes. Nat. Commun. 5, 3979 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rapicavoli N. A., et al. , A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife 2, e00762 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hon C.-C., et al. , An atlas of human long non-coding RNAs with accurate 5′ ends. Nature 543, 199–204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabili M. N., et al. , Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 25, 1915–1927 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guil S., Esteller M., Cis-acting noncoding RNAs: Friends and foes. Nat. Struct. Mol. Biol. 19, 1068–1075 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Engreitz J. M., et al. , Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452–455 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joung J., et al. , Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 548, 343–346 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo S., et al. , Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 18, 637–652 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Quinodoz S., Guttman M., Long noncoding RNAs: An emerging link between gene regulation and nuclear organization. Trends Cell Biol. 24, 651–663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kung J. T. Y., Colognori D., Lee J. T., Long noncoding RNAs: Past, present, and future. Genetics 193, 651–669 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchese F. P., Raimondi I., Huarte M., The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 18, 206 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaneko S., Son J., Shen S. S., Reinberg D., Bonasio R., PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat. Struct. Mol. Biol. 20, 1258–1264 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cifuentes-Rojas C., Hernandez A. J., Sarma K., Lee J. T., Regulatory interactions between RNA and polycomb repressive complex 2. Mol. Cell 55, 171–185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sigova A. A., et al. , Transcription factor trapping by RNA in gene regulatory elements. Science 350, 978–981 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun X., Haider Ali M. S. S., Moran M., The role of interactions of long non-coding RNAs and heterogeneous nuclear ribonucleoproteins in regulating cellular functions. Biochem. J. 474, 2925–2935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Latos P. A., et al. , Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338, 1469–1472 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Yin Y., et al. , Opposing roles for the lncRNA Haunt and its genomic locus in regulating HOXA gene activation during embryonic stem cell differentiation. Cell Stem Cell 16, 504–516 (2015). [DOI] [PubMed] [Google Scholar]
- 28.Paralkar V. R., et al. , Unlinking an lncRNA from its associated cis element. Mol. Cell 62, 104–110 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goff L. A., et al. , Spatiotemporal expression and transcriptional perturbations by long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. U.S.A. 112, 6855–6862 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Signal B., Gloss B. S., Dinger M. E., Computational approaches for functional prediction and characterisation of long noncoding RNAs. Trends Genet. 32, 620–637 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Kopp F., Mendell J. T., Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393–407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixon J. R., Gorkin D. U., Ren B., Chromatin domains: The unit of chromosome organization. Mol. Cell 62, 668–680 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Melgarejo E., Medina M. A., Sánchez-Jiménez F., Urdiales J. L., Monocyte chemoattractant protein-1: A key mediator in inflammatory processes. Int. J. Biochem. Cell Biol. 41, 998–1001 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Frank S., et al. , yylncT defines a class of divergently transcribed lncRNAs and safeguards the T-mediated mesodermal commitment of human PSCs. Cell Stem Cell 24, 318–327.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Spurlock C. F., 3rd, et al. , Expression and functions of long noncoding RNAs during human T helper cell differentiation. Nat. Commun. 6, 6932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Werner M. S., et al. , Chromatin-enriched lncRNAs can act as cell-type specific activators of proximal gene transcription. Nat. Struct. Mol. Biol. 24, 596–603 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trimarchi T., et al. , Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell 158, 593–606 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dixon J. R., et al. , Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nora E. P., et al. , Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao S. S., et al. , A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hogan N. T., et al. , Transcriptional networks specifying homeostatic and inflammatory programs of gene expression in human aortic endothelial cells. eLife 6, e22536 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattioli K., et al. , High-throughput functional analysis of lncRNA core promoters elucidates rules governing tissue specificity. Genome Res. 29, 344–355 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aiello R. J., et al. , Monocyte chemoattractant protein-1 accelerates atherosclerosis in apolipoprotein E-deficient mice. Arterioscler. Thromb. Vasc. Biol. 19, 1518–1525 (1999). [DOI] [PubMed] [Google Scholar]
- 44.Brown J. D., et al. , NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell 56, 219–231 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang K. C., et al. , A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472, 120–124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang L., et al. , CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 41, e74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu Y., et al. , The NF-κB-responsive long noncoding RNA FIRRE regulates posttranscriptional regulation of inflammatory gene expression through interacting with hnRNPU. J. Immunol. 199, 3571–3582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishitsuji H., et al. , Long noncoding RNA #32 contributes to antiviral responses by controlling interferon-stimulated gene expression. Proc. Natl. Acad. Sci. U.S.A. 113, 10388–10393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X., et al. , Interrogation of nonconserved human adipose lincRNAs identifies a regulatory role of linc-ADAL in adipocyte metabolism. Sci. Transl. Med. 10, eaar5987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mineo M., et al. , The long non-coding RNA HIF1A-AS2 facilitates the maintenance of mesenchymal glioblastoma stem-like cells in hypoxic niches. Cell Rep. 15, 2500–2509 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hosono Y., et al. , Oncogenic role of THOR, a conserved cancer/testis long non-coding RNA. Cell 171, 1559–1572.e20 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yugami M., Kabe Y., Yamaguchi Y., Wada T., Handa H., hnRNP-U enhances the expression of specific genes by stabilizing mRNA. FEBS Lett. 581, 1–7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Degrauwe N., Suvà M. L., Janiszewska M., Riggi N., Stamenkovic I., IMPs: An RNA-binding protein family that provides a link between stem cell maintenance in normal development and cancer. Genes Dev. 30, 2459–2474 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bell J. L., et al. , Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): Post-transcriptional drivers of cancer progression? Cell. Mol. Life Sci. 70, 2657–2675 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perisic L., et al. , Gene expression signatures, pathways and networks in carotid atherosclerosis. J. Intern. Med. 279, 293–308 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Finn A. V., Nakano M., Narula J., Kolodgie F. D., Virmani R., Concept of vulnerable/unstable plaque. Arterioscler. Thromb. Vasc. Biol. 30, 1282–1292 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Tabas I., García-Cardeña G., Owens G. K., Recent insights into the cellular biology of atherosclerosis. J. Cell Biol. 209, 13–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sigova A. A., et al. , Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 110, 2876–2881 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brazão T. F., et al. , Long noncoding RNAs in B-cell development and activation. Blood 128, e10–e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Purmann A., et al. , Genomic organization of transcriptomes in mammals: Coregulation and cofunctionality. Genomics 89, 580–587 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Chen L.-L., Linking long noncoding RNA localization and function. Trends Biochem. Sci. 41, 761–772 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Cabili M. N., et al. , Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 16, 20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hirota K., et al. , Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature 456, 130–134 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Anderson K. M., et al. , Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 539, 433–436 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wahlestedt C., Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat. Rev. Drug Discov. 12, 433–446 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Woo C. J., et al. , Gene activation of SMN by selective disruption of lncRNA-mediated recruitment of PRC2 for the treatment of spinal muscular atrophy. Proc. Natl. Acad. Sci. U.S.A. 114, E1509–E1518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wickham H., Wickham H., ggplot2, Elegant Graphics for Data Analysis (Springer; ), pp. 33–74 (2016). [Google Scholar]
- 68.Lawrence M., et al. , Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marín-Béjar O., Huarte M., RNA pulldown protocol for in vitro detection and identification of RNA-associated proteins. Methods Mol. Biol. 1206, 87–95 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray data generated in this study have been deposited in the GEO database under the accession number GSE127990. Code used in the analysis can be accessed at https://github.com/nadiyakhyzha/lncRNA_mRNA_neighboring_analysis.