Significance
Sepsis is a multifactorial syndrome with increasing incidence and significant mortality. While previous work implicated tumor necrosis factor (TNF)-induced cell death in sepsis, a role for interferon-gamma (IFNγ), which synergizes with TNF to activate macrophages, is incompletely understood. We demonstrate using genome-wide CRISPR/Cas9 screening that genes regulating the cytosolic degradative pathway of autophagy protect against IFNγ-induced cell death. This cell death requires TNF and its receptor and depends on the downstream cell death mediators caspase-8 and RIPK1. Moreover, mice with myeloid cell autophagy gene deficiency are hypersusceptible to fatal TNF-induced shock, which also depends on IFNγ signaling and RIPK1. These findings identify autophagy genes as important regulators of IFNγ- and TNF-mediated cell death with implications for fatal systemic inflammatory responses.
Keywords: autophagy, sepsis, TNF, interferon, cell death
Abstract
Host inflammatory responses must be tightly regulated to ensure effective immunity while limiting tissue injury. IFN gamma (IFNγ) primes macrophages to mount robust inflammatory responses. However, IFNγ also induces cell death, and the pathways that regulate IFNγ-induced cell death are incompletely understood. Using genome-wide CRISPR/Cas9 screening, we identified autophagy genes as central mediators of myeloid cell survival during the IFNγ response. Hypersensitivity of autophagy gene-deficient cells to IFNγ was mediated by tumor necrosis factor (TNF) signaling via receptor interacting protein kinase 1 (RIPK1)- and caspase 8-mediated cell death. Mice with myeloid cell-specific autophagy gene deficiency exhibited marked hypersensitivity to fatal systemic TNF administration. This increased mortality in myeloid autophagy gene-deficient mice required the IFNγ receptor, and mortality was completely reversed by pharmacologic inhibition of RIPK1 kinase activity. These findings provide insight into the mechanism of IFNγ-induced cell death via TNF, demonstrate a critical function of autophagy genes in promoting cell viability in the presence of inflammatory cytokines, and implicate this cell survival function in protection against mortality during the systemic inflammatory response.
IFN gamma (IFNγ) and tumor necrosis factor (TNF) mediate critical immune responses to intracellular and extracellular pathogens by synergistically activating macrophages (1). The pleiotropic functions of this synergy include priming of cell-intrinsic antimicrobial responses to intracellular pathogens, enhancement of antigen presentation, and antitumor effects (1). Since IFNγ and TNF also induce cell death, pathways that ensure cell viability must exist to maintain robust effector functions driven by these cytokines. Widespread efforts have detailed at least 3 distinct forms of TNF-induced cell death: 1) TNF-receptor interacting protein kinase 1 (RIPK1)-independent, caspase 8 (CASP8)-mediated apoptosis; 2) RIPK1- and CASP8-mediated apoptosis; and 3) RIPK1-mediated necroptosis (2). Additionally, numerous studies have illuminated a multilayered system that limits TNF-induced cell death (3–12). However, mediators of IFNγ-induced cell death, and factors that regulate these pathways, are incompletely understood.
Induction of cell death by IFNγ and synergistic antiproliferative and cytotoxic antitumor effects with TNF have been recognized for over 3 decades (13–16). Initial studies identified a role for Death Associated Proteins in mediating IFNγ-induced death in epithelial cells, although the precise mechanisms remain incompletely defined (17). Nuclear factor κB (NFκB) promotes cell viability during IFNγ or TNF responses, as mouse embryonic fibroblasts (MEFs) deficient for RelA or IκB kinase β (IKKβ) undergo RIPK1-dependent cell death upon exposure to these cytokines (18). Additionally, the dsRNA-sensing protein kinase R (PKR) promotes IFNγ-induced cell death in the absence of the Fas-associated via death domain (FADD) TNF-signaling adapter, but PKR is dispensable for TNF-induced death (19). Similarly, MEFs deficient in RIPK1 undergo cell death triggered by IFNγ that is RIPK3 and PKR dependent (20). Therefore, IFNγ can induce both apoptotic and necroptotic forms of cell death, although the role of TNF in IFNγ-induced death remains unclear. Moreover, precise mechanisms of IFNγ-induced death in macrophages—a cell type that mounts robust responses to IFNγ and TNF—have not been described.
In this study we investigated the genetic determinants of IFNγ-induced cell death in myeloid cells. Using genome-wide CRISPR/Cas9 screening in a mouse microglial cell line, we found that autophagy genes played an integral role in limiting IFNγ-induced cell death. Suppressor screening on an Atg5-deficient background identified TNF as an essential mediator of IFNγ-induced cell death. Moreover, autophagy genes in myeloid cells were required to protect mice against fatal shock triggered by TNF. RIPK1 inhibition blocked the increased susceptibility of autophagy gene-deficient cells to IFNγ-induced cell death in vitro and protected myeloid autophagy gene-deficient mice from increased TNF-induced mortality. Together, these findings provide important insights into the regulation of IFNγ-induced cell death and define a role for autophagy genes in countering this response to modulate TNF-induced shock.
Results
Identification of Autophagy Genes in Protection Against IFNγ-Induced Cell Death.
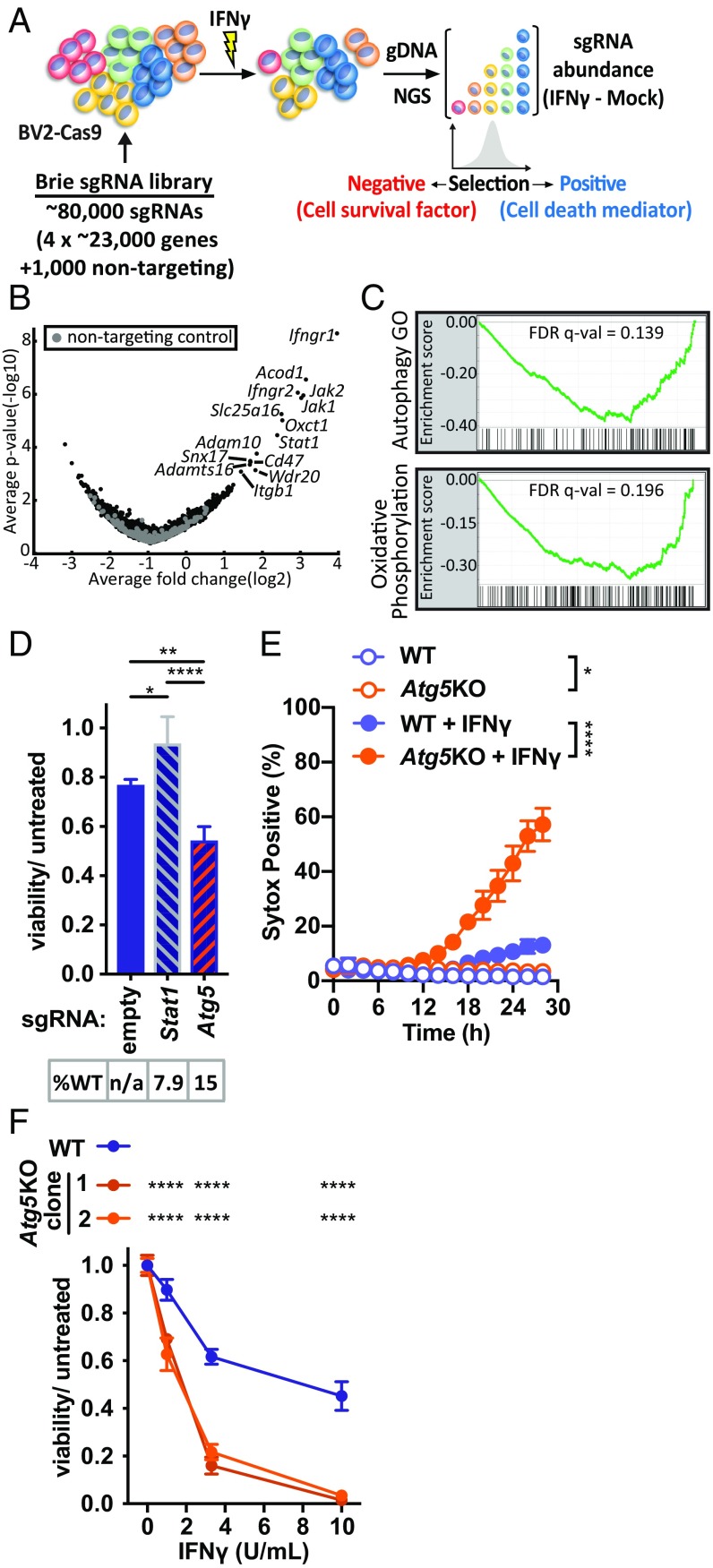
To identify regulators of IFNγ-induced cell death, we performed a genome-wide screen in the murine microglial BV2 cell line for genes that, when knocked out by CRISPR/Cas9 (CRISPRko), resulted in sensitization (negatively selected) or resistance (positively selected) to the effects of IFNγ on cell viability (Fig. 1A). Cells expressing sgRNAs targeting genes known to mediate IFNγ signaling (e.g., genes encoding heterodimeric IFNγ receptor subunits Ifngr1 and Ifngr2, intracellular signaling adapters Jak1 and Jak2, or transcriptional activator Stat1) would be expected to resist effects of IFNγ. Indeed, these factors were among the most strongly positively selected hits, confirming the robustness of the screen (Fig. 1B and SI Appendix, Table S1). In contrast, using this library with moderate genome coverage (500 cells per sgRNA), our screen did not identify any statistically significant individual negatively selected sgRNAs for genes which have prosurvival functions. To identify signatures of pathways that might exert cytoprotective effects, we performed Gene Set Enrichment Analysis (GSEA) of the genome-wide rank-ordered list for Hallmark pathways (which does not include an autophagy-related gene set), and also included a custom generated Autophagy GO functional annotation-based gene set given previously identified links between immune effects of IFNγ and autophagy. GSEA demonstrated that the most significant negatively selected pathway in our screen was the Autophagy GO pathway (Fig. 1C). These data indicated that genes in the autophagy pathway might promote viability of cells treated with IFNγ.
Fig. 1.
Pooled genome-wide CRISPRko screening for genes that regulate IFNγ-induced cell death. (A) Overview of CRISPRko screening strategy. (B) Volcano plot for Brie genome-wide library in BV2-Cas9 cells treated with IFNγ. Log2 fold changes represent average of all sgRNAs for each gene (IFNγ treated–mock). Labeled genes are those with a positive STARS score FDR < 0.1. (C) GSEA for Hallmark pathways and the Autophagy GO pathway, showing only pathways with FDR P value < 0.2. (D) Viability of BV2-Cas9 cells stably expressing indicated sgRNA and treated with IFNγ (10 U/mL) proportional to untreated condition for each stable line. %WT indicates percent alleles with wild-type sequence based on next generation sequencing (NGS) of amplicon that encompasses the indicated sgRNA; n/a indicates not applicable as no sgRNA present. (E) Cell death in WT or Atg5KO clone treated with IFNγ (10 U/mL). (F) Viability of BV2 cells of genotype indicated treated with increasing dose of IFNγ indicated. *P < 0.05; **P < 0.01; ****P < 0.0001; 1-way (D and E) or 2-way (F) ANOVA with Tukey’s (D and F) or Sidak’s (E) posttest; comparisons as indicated, or area under curve of KO vs. WT for each treatment (E), or each clone vs. WT at each dose (F). Data in D–F represent mean with SD of 3 to 4 technical replicates, and similar results were observed in at least 3 independent experiments.
Autophagy Genes Encoding Distinct Regulatory Complexes Promote Viability of IFNγ-Treated Cells.
We assessed the roles of specific genes in IFNγ-induced cell death with CRISPRko using 2 approaches: 1) polyclonal cell populations stably expressing Cas9 and a sgRNA to a gene of interest were generated and allelic disruption assessed by deep sequencing (polyclonal cells hereafter) and 2) clonal cell lines were generated by transient introduction of Cas9/sgRNAs to induce mutations disrupting all alleles for the protein coding sequence of a gene (clonal knockout [KO] cell lines) (21). We first confirmed our positively selected genome-wide screen results with polyclonal Stat1 cells, which exhibited resistance to IFNγ-induced death (Fig. 1D). Among the genes that, when deleted, exhibited the greatest viability defect after IFNγ treatment was Atg5, an autophagy gene that encodes an essential component of the LC3 conjugation machinery involved in autophagosome biogenesis (22, 23) (SI Appendix, Table S1). ATG5 also performs autophagy-independent functions in innate immunity, including mediating IFNγ-induced restriction of pathogens (24–30). Loss of Atg5 in polyclonal Atg5 cells (Fig. 1D), and in Atg5KO cells (Fig. 1 E and F), resulted in sensitization to cell death after IFNγ treatment, confirming the negative selection GSEA results in our screen. Further, increased IFNγ-induced cytotoxicity accounted for the loss of viability in Atg5-deficient cells, as measured by loss of membrane integrity (Fig. 1E). Additionally, 2 independent Atg5KO cell line clones demonstrated hypersensitivity to IFNγ-induced cell death (Fig. 1F).
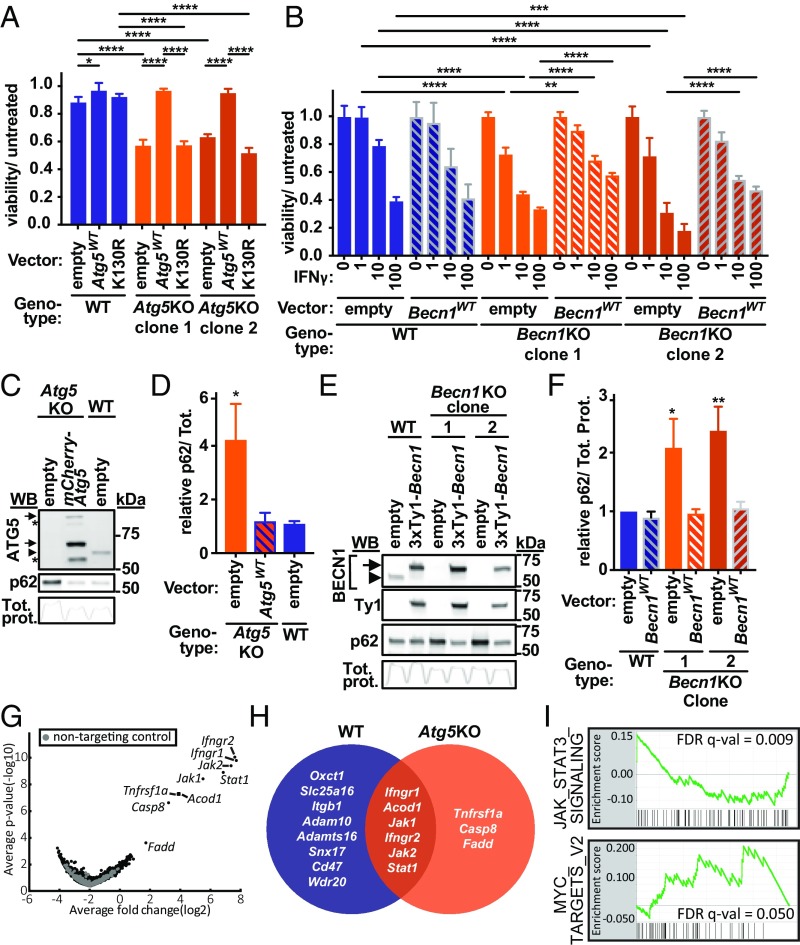
The hypersensitivity of Atg5KO cells to IFNγ was complemented by reintroducing wild-type Atg5 fused to mCherry (Atg5WT) but not by a mutant form of this protein encoding a K130R substitution that prevents the covalent linkage of ATG5 and ATG12 required for the activity of the ATG5/ATG12/ATG16L1 complex in stimulating LC3 conjugation to phosphatidylethanolamine (Fig. 2A) (31). As ATG5 has been implicated in LC3 conjugation complex-independent phenotypes (32), we evaluated a role for ATG16L1. Atg16l1KO cells were hypersensitive to IFNγ-induced cell death, which could be partially complemented by wild-type Atg16l1 but not by a mutant construct encoding a form lacking its ATG5-interacting motif (SI Appendix, Fig. S1A) (33). We next evaluated Beclin-1 (encoded by Becn1) in regulation of IFNγ-induced cell death as Beclin-1 is required both for nucleation of nascent autophagosomal membranes and for lysosomal fusion with autophagosomes (34–38). Becn1KO cells exhibited increased sensitivity to IFNγ-induced cell death, which could be partially complemented by retroviral-mediated expression of Becn1 (Fig. 2B). As Beclin-1 forms at least 2 mutually exclusive complexes with class III PI(3)K and ATG14 or with UVRAG involved in the regulation of autophagy or endocytosis/phagocytosis, respectively (36–39), we evaluated a role for ATG14 in IFNγ-induced cell death. Atg14KO cells were hypersensitive to IFNγ-induced cell death, which could be partially complemented by wild-type Atg14 but not by a mutant construct encoding a form lacking the coiled–coiled domain required for ATG14 to interact with Beclin-1 and regulate autophagy (SI Appendix, Fig. S1B) (40, 41).
Fig. 2.
Genome-wide suppressor CRISPRko screen reveals essential role for TNF pathway in IFNγ-induced death of Atg5KO cells. (A) Viability of BV2 cells of WT or Atg5KO cells stably expressing construct indicated and treated with IFNγ (3 U/mL) proportional to untreated condition for each stable line. (B) Viability of BV2 cells of genotype indicated stably expressing construct indicated and treated with indicated dose of IFNγ proportional to untreated condition for each stable line. (C and D) Western blot for endogenous ATG5 or p62 in cell line indicated (C), with p62 quantification of 3 independent experiments in D. (E and F) Western blot for endogenous BECN1, Ty1 tag, or p62 in cell line indicated (E), with p62 quantification of 3 independent experiments in F. Arrowheads indicate endogenous protein, and arrows indicate fusion proteins in C and E, as detected by antibodies indicated. Upper arrow in C is consistent with mCherry-ATG5-ATG12 conjugate, and lower arrow is unconjugated; conjugated endogenous ATG5 is observed and shown. Asterisk in C refers to unknown bands; Tot. prot. in C and E reflects intensity profile of total protein in each lane on membrane for p62 blot shown, which was used for loading control and the area under curve used for normalization in quantitation. (G) Volcano plot for Brie genome-wide screen in Atg5KO-Cas9 BV2 cells treated with IFNγ. Log2 fold changes represent average of all sgRNAs for each gene (IFNγ treated–mock). Labeled genes are those with a positive STARS score FDR < 0.1. (H) Overlap of positively selected hits from WT and Atg5KO genome-wide screens with STARS score FDR < 0.1. (I) GSEA for Hallmark pathways and the Autophagy GO pathway. Shown are pathways with FDR P value < 0.2. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; 2-way ANOVA (A and B, comparing all lines at each dose; significance for each vector or within each clone shown) or 1-way ANOVA (in D and F, vs. “WT; empty”), with Tukey’s (A and B) or Dunnett’s (D and F) posttest for multiple comparisons. Data in A and B represent mean with SD of 3 to 4 technical replicates, and similar results were observed in at least 3 independent experiments; Data in D and F represent mean with SD of 3 independent experiments.
Compared with WT cells, both Atg5KO and Becn1KO cells lacked detectable endogenous protein expression from the mutated gene, as confirmed by immunoblot (Fig. 2 C and E). Cells from these autophagy gene-deficient lines also markedly accumulated p62, which is a substrate for autophagic degradation (Fig. 2 C–F), and reintroducing the respective wild-type gene restored p62 to levels observed in WT cells (Fig. 2 C–F). While previous studies indicate that IFNγ induces autophagy (42–45), we observed constitutive conversion of LC3-II in BV2 cells as measured by LC3-II accumulation upon treatment with the vacuolar ATPase inhibitor bafilomycin A (Baf), which blocks LC3 degradation by inhibiting autophagic flux (SI Appendix, Fig. S2A) (46). IFNγ did not further increase autophagic flux (SI Appendix, Fig. S2A). We observed the expected lack of LC3-II formation in Atg5KO cells (SI Appendix, Fig. S2B). Further, cotreatment of cells with IFNγ and Baf resulted in more cytotoxicity than either treatment alone (SI Appendix, Fig. S2C). The increased sensitivity of Atg5KO and Baf-treated cells to IFNγ-induced cell death suggested that autophagic activity promoted cell survival during IFNγ responses. The increased sensitivity of independent clones to IFNγ-induced cell death and the ability to complement abnormal autophagy-related functions in autophagy gene-deficient cells support the observation that hypersensitivity was not a result of off-target effects of sgRNAs. Together these data indicate that genes encoding members of distinct regulatory complexes involved in initiation and completion of autophagy (and more specifically the interacting domains in the factors required to form these complexes) are critical to prevent the death of cells responding to IFNγ. We subsequently focused on Atg5 as a key autophagy gene for further studies on IFNγ-induced death.
The TNF Pathway Is Essential for Atg5KO Hypersensitivity to IFNγ-Induced Cell Death.
To identify genes required for the hypersensitivity of Atg5KO cells to IFNγ-induced cell death we performed a genome-wide suppressor CRISPRko screen on the Atg5KO background. We detected positive enrichment of sgRNAs targeting Tnfrsf1a (Tnfr1, TNF receptor 1), Fadd, and Casp8 in surviving cells (Fig. 2G). These genes are critical components of the TNF-induced cell death pathway (47), and they were not significantly enriched in our initial screen in WT cells that express endogenous Atg5 (Fig. 1A). Fig. 2H illustrates the overlap in positively selected hits between our WT screen and Atg5KO suppressor screen. GSEA of the suppressor screen results demonstrated positive enrichment for Myc and IL-6-JAK-STAT3 pathways (Fig. 2I). Consistent with a role for the autophagy pathway in protecting against IFNγ-induced cell death, we did not observe negative selection of Autophagy GO pathway genes by GSEA in Atg5KO cells (which are already defective in autophagy) (FDR q-val = 1).
TNF and TNFR1 Are Necessary for IFNγ-Induced Death of Atg5KO Cells.
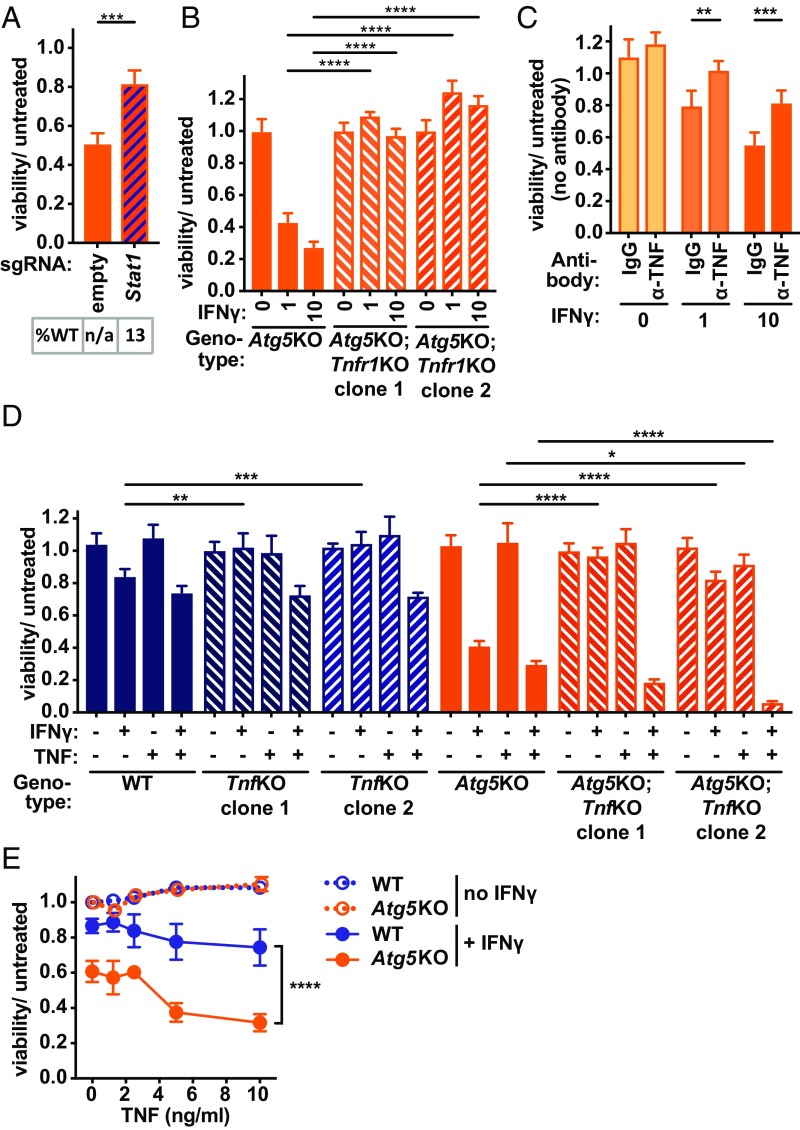
To validate results from our suppressor screen, we first analyzed Atg5KO cells with polyclonal Stat1 deletion, which protected against IFNγ-induced cell death and provided confirmation of our positively selected screen results (Fig. 3A). Next, we generated Tnfr1KO cells, which resulted in reversal of the hypersensitivity of Atg5KO cells (Fig. 3B). Although we did not identify the TNF cytokine gene in our CRISPRko screen, we reasoned this could be due to complementation in trans by the majority of cells with intact TNF production. Consistent with this possibility, antibody neutralization of endogenous TNF partially reversed IFNγ-induced death of Atg5KO cells (Fig. 3C), and a requirement for the Tnf gene was confirmed in TnfKO cells (Fig. 3D). Importantly, addition of TNF cytokine resulted in functional complementation of the loss of activity observed in TnfKO and Atg5KO;TnfKO cells treated with IFNγ alone (Fig. 3D). Therefore, IFNγ-induced death of Atg5KO cells requires TNF and its receptor TNFR1.
Fig. 3.
TNF is necessary, but not sufficient, for IFNγ-induced cell death. (A) Viability of Atg5KO-Cas9 cells stably expressing indicated sgRNA vector and treated with IFNγ (10 U/mL) proportional to untreated condition for each stable line. %WT in A indicates percent alleles with wild-type sequence based on NGS of amplicon that encompasses the indicated sgRNA; n/a indicates not applicable as no sgRNA present. (B) Viability of BV2 cells of genotype indicated treated with dose of IFNγ indicated (U/mL). (C) Viability of Atg5KO cells treated with isotype control antibody (IgG) or anti-TNF antibody (α-TNF) (100 μg/mL) just before treatment with dose of IFNγ indicated (U/mL). (D) Viability of BV2 cells of genotype indicated treated with IFNγ (1 U/mL) and/or TNF (5 ng/mL). (E) Viability of BV2 cells of genotype indicated treated with dose of TNF indicated with or without IFNγ (1 U/mL). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; significant differences for comparisons shown in (A) Stat1 vs. empty; (B) each Tnfr1KO clone vs. parental Atg5KO line for each dose; (C) α-TNF vs. IgG for each dose of IFNγ; (D) for TnfKO clones vs. parental lines for each treatment; (E) Atg5KO + IFNγ vs. WT + IFNγ at each dose of TNF, P < 0.0001; in 0 ng TNF vs. 10 ng TNF comparison for WT + IFNγ, P = 0.18, for Atg5KO + IFNγ, P < 0.0001; via unpaired t test in A, 2-way ANOVA with Dunnett’s (B), Sidak’s (C), or Tukey’s (D and E) posttests. Data represent mean with SD of 3 to 4 technical replicates, and similar results were observed in at least 3 independent experiments.
When TNF was supplied to TnfKO cells treated with IFNγ, the hypersensitivity associated with Atg5-deficiency persisted compared with WT cells (Fig. 3D). These results indicated that IFNγ-induced death of Atg5KO cells was due to hypersensitivity to events downstream of TNF production. Further, addition of increasing amounts of exogenous TNF alone was insufficient to trigger cell death of either WT or Atg5KO cells, while combined treatment with IFNγ triggered significantly more death of Atg5KO cells (Fig. 3E). Therefore, TNF and TNFR1 are essential for IFNγ-induced cell death, but TNF alone is insufficient to explain the hypersensitivity of Atg5KO cells to the combined activity of IFNγ and TNF.
Effect of ATG5 on IFNγ-Induced Transcriptomic Responses.
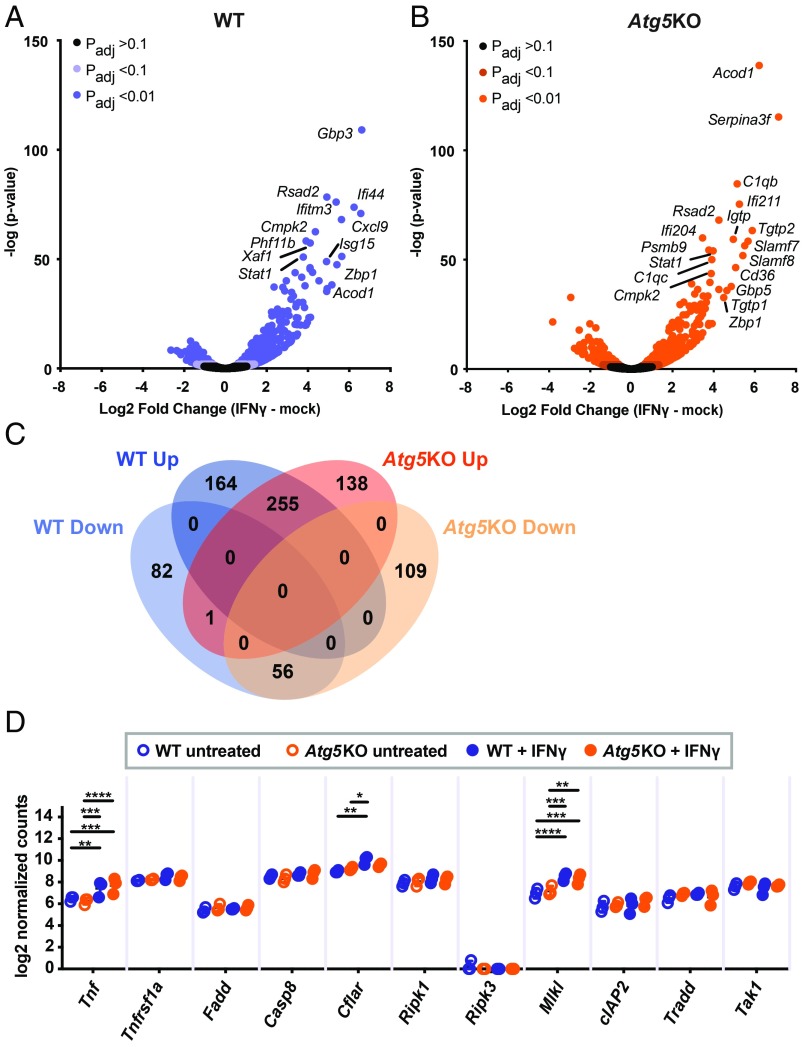
We next considered the possibility that decreased viability of Atg5-deficient cells could be due to global disruption of transcriptional responses induced by IFNγ and TNF. For example, increased IFNγ-pathway responses in Atg5KO cells could explain the hypersensitivity to cell death, whereas decreased NFκB activity downstream of TNF would be expected to sensitize cells to TNF-induced death. To test this, we performed RNA-seq on WT or Atg5KO cells with or without IFNγ treatment (Fig. 4 and SI Appendix, Table S2). IFNγ induced widespread transcriptional responses in cells of both genotypes (Fig. 4 A and B). While differences in expression of genes expressed at low levels were observed, we observed a high overall concordance of genes whose expression was altered >2-fold by IFNγ (SI Appendix, Fig. S3A). Similar responses were also reflected in GSEA of genes regulated by IFNγ in both cell lines (SI Appendix, Fig. S3B), including the IFNγ pathway itself (SI Appendix, Fig. S3 B and D). These data are consistent with a previous study of Atg5-deficient bone marrow-derived macrophages treated with IFNγ (27). We conclude that IFNγ-dependent regulation of gene expression is intact in cells lacking ATG5, leading us to examine the TNF pathway as a potential cause for the hypersensitivity of ATG5-deficient cells to IFNγ-induced cell death.
Fig. 4.
IFNγ-induced and NFκB-regulated gene expression in Atg5KO cells. (A and B) RNA-seq of WT (A) or Atg5KO (B) BV2 cells treated with IFNγ (1 U/mL). Most highly induced genes labeled for illustration purposes. (C) Overlap of RNA-seq in A and B with adjusted P < 0.01 and direction of change after IFNγ treatment. (D) Log2 normalized reads for each gene indicated from RNA-seq. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001 in D; 2-way ANOVA with Tukey’s posttest.
Evaluation of TNF Pathway in Atg5-Deficient Cells.
We detected basal Tnfrsf1a expression at similar levels in WT and Atg5KO cells, without notable induction after IFNγ (Fig. 4D). Atg5-dependent changes in IFNγ regulation of mRNAs for multiple previously described prodeath or prosurvival factors in TNF death signaling were not observed (Fig. 4D). Expression patterns for genes in the annotated “genes regulated by NF-κB in response to TNF” pathway, which exerts a prosurvival role in response to TNF, were not significantly different between WT and Atg5KO cells (SI Appendix, Fig. S3 B and C). These results indicated that ATG5 deficiency does not globally affect IFNγ regulation of the levels of factors involved in TNF-induced NFkB signaling.
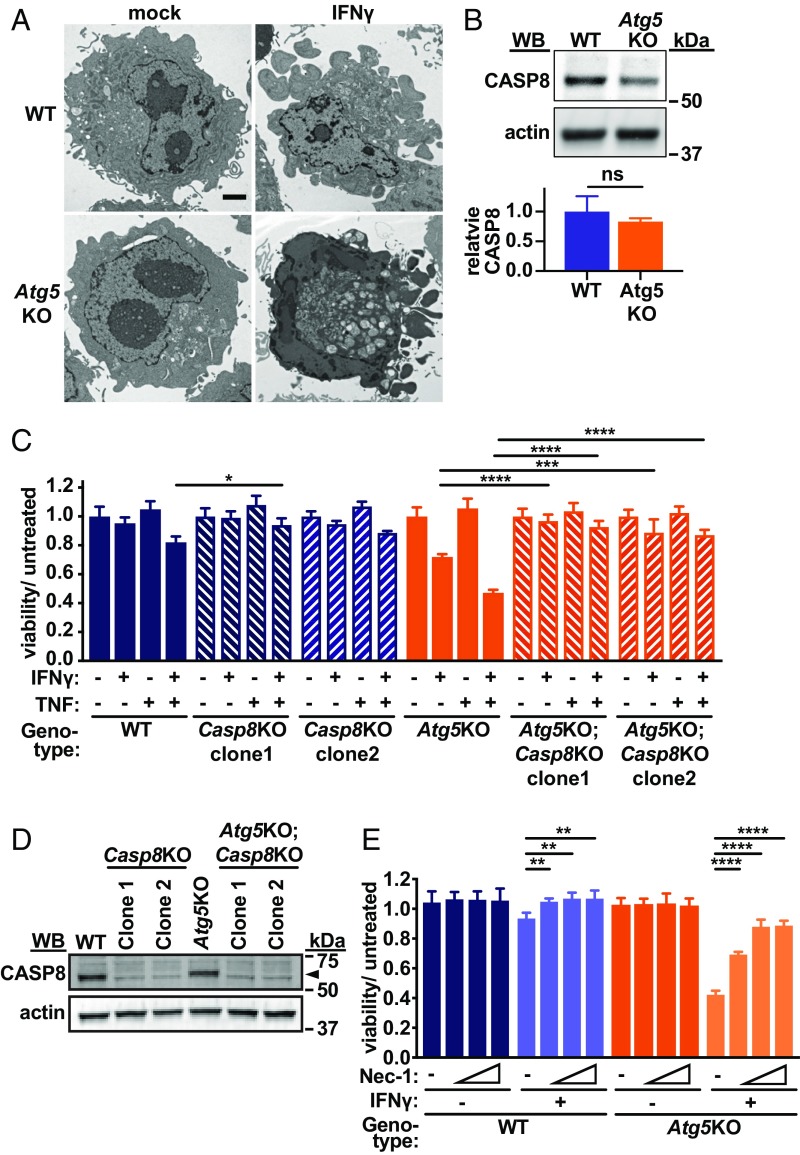
We next focused on posttranscriptional events downstream of TNF signaling. Cell death induction by TNF involves both CASP8-dependent apoptosis and CASP8-independent necroptosis (2). Apoptosis is defined in part by morphological changes such as chromatin condensation with cytoplasmic shrinkage and membrane blebbing, while necrotic cells exhibit loss of membrane integrity (48). Transmission electron microscopy revealed increased frequency of cell corpses in Atg5KO cells treated with IFNγ (22 dead/500 total cells) compared with WT cells (8/500) (P < 0.05, Fisher’s exact test). The predominant morphology in both cell lines was apoptotic (Fig. 5A), and only rare necrotic cells were observed. Further, IFNγ resulted in a dose-dependent activation of caspase 3 as measured by a fluorescent reporter, which was blocked by a caspase 3/7 inhibitor (SI Appendix, Fig. S4 A and B). We also detected cleaved caspase 3 biochemically after IFNγ treatment (SI Appendix, Fig. S4C) and markedly increased caspase 3 activity in Atg5KO cells (SI Appendix, Fig. S4D).
Fig. 5.
ATG5 protects against IFNγ-induced CASP8- and RIPK1-mediated cell death. (A) Representative transmission electron microscopy images of BV2 cells of genotype and treatment indicated. (Scale bar, 2 μm.) (B) CASP8 immunoblot of BV2 cells of genotype indicated with fluorescent quantification of triplicate experiments normalized to actin in arbitrary units. (C) Viability of BV2 cells of genotype indicated treated with IFNγ (1 U/mL) and/or TNF (10 ng/mL). (D) Immunoblot of CASP8 in cells of genotype indicated and assayed in C. Arrowhead in D indicates CASP8 band. (E) Viability of cells of genotype indicated treated with increasing doses of necrostatin-1 (Nec-1, 0.6, 2.5, 10 μM) or DMSO (−) with or without IFNγ (1 U/mL). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant; in C, for Casp8KO clones vs. parental lines for each treatment; in E, comparison with DMSO for each IFNγ condition; by unpaired t test (B) or 2-way ANOVA with Tukey’s (C) or Dunnett’s (E) posttest. Data in B represent mean with SD of 3 biological replicates, and data in C and E represent mean with SD of 3 to 4 technical replicates, with similar results observed in at least 3 independent experiments.
Our suppressor CRISPRko screen indicated a role for CASP8, which has been reported as a target of autophagic degradation in various cell types (49), although we observed no significant difference in CASP8 protein levels in Atg5KO cells (Fig. 5B). However, CRISPRko of Casp8 reversed the hypersensitivity to IFNγ-induced death in Atg5KO cells (Fig. 5 C and D). CASP8-induced cell death can be RIPK1 mediated or RIPK1 independent (2), and RIPK1 kinase activity can be inhibited by small-molecule necrostatins (50). Cotreatment of cells with necrostatin-1 (Fig. 5E), as well as the more selective and potent analog necrostatin-1s (SI Appendix, Fig. S5A), reversed the hypersensitivity of Atg5KO cells to the effects of IFNγ. We also considered whether cell death induced by IFNγ occurred via multiple TNF-induced pathways such as RIPK3- and MLKL-mediated necroptosis (2). However, polyclonal Ripk3 or Mlkl deletion in Atg5KO cells demonstrated no effect on IFNγ-induced cell death (SI Appendix, Fig. S5B). Additionally, we did not detect robust levels of Ripk3 transcripts at basal levels or after IFNγ treatment (Fig. 4D). Four distinct sgRNAs to Ripk1 resulted in limited allelic mutation (12–25% mutated alleles in polyclonal cells), and we were unable to isolate any Ripk1KO clones on the Atg5KO background, consistent with the previously described kinase-independent cell survival scaffolding function of RIPK1 (20, 51–54). In sum, we conclude that Atg5-deficient cells exhibit hypersensitivity to IFNγ-induced cell death that is dependent on TNF, TNFR1, CASP8, and RIPK1-kinase activity, with some hallmarks of apoptosis, and without a demonstrable role for the necroptotic machinery.
Autophagy Genes in Myeloid Cells Protect Mice against Fatal TNF-Induced Shock That Requires IFNγ and RIPK1 Activity.
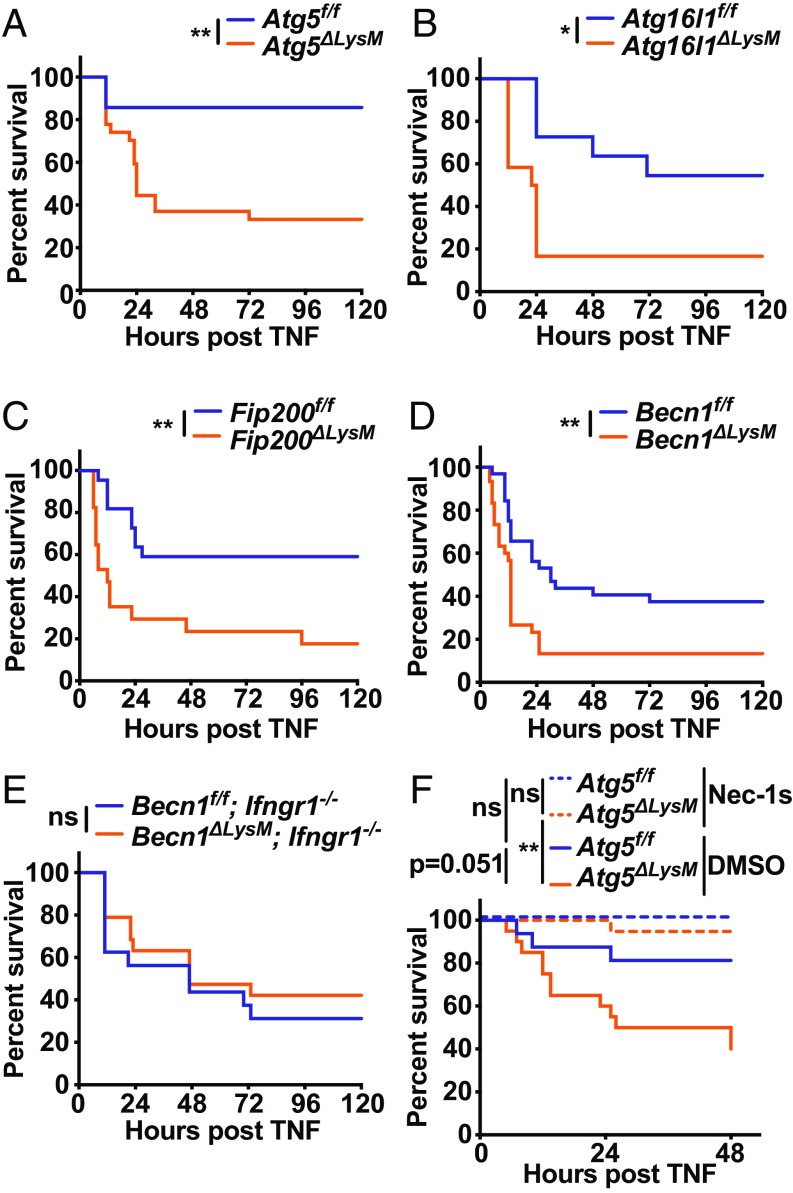
Having demonstrated a protective role for autophagy genes in controlling TNF-induced cell death after IFNγ treatment, we hypothesized that a similar process might occur in a mouse model of fatal TNF- induced shock (50, 55). We were encouraged in this hypothesis by the fact that myeloid cell responses are important for TNF-mediated fatal shock (56) and that administering individually sublethal IFNγ and TNF doses in combination leads to synergistic mortality in mice (55). First, we sought to determine if primary bone marrow-derived macrophages recapitulated the hypersensitivity to IFNγ-induced cell death observed in autophagy gene-deficient BV2 cells. IFNγ on its own, and TNF particularly when pretreated with IFNγ, resulted in significantly more cell death in cells isolated from mice lacking Atg5 in myeloid cells (ΔLysM) compared with their littermate controls (f/f) (SI Appendix, Fig. S6). To evaluate a role for myeloid cell autophagy genes in TNF-induced shock, we compared ΔLysM mice to their littermate controls after i.v. TNF injection (Fig. 6). Mice lacking Atg5, Atg16l1, Fip200, or Becn1 each demonstrated striking hypersensitivity to TNF, as demonstrated by decreased overall survival and earlier onset of illness (Fig. 6 A–D). Importantly, mice with myeloid deficiency of Becn1 bred to lack the receptor for IFNγ (Ifngr1−/−) did not exhibit increased sensitivity to TNF-induced shock (Fig. 6E). To test for the role of RIPK1-mediated cell death in the hypersensitivity of autophagy gene-deficient mice, we injected necrostatin-1s just before injection of TNF, which has been shown to reverse TNF-induced shock in wild-type mice (50, 57). Necrostatin-1s pretreatment resulted in complete protection of Atg5ΔLysM mice to fatal TNF-induced shock (Fig. 6F). These results demonstrate that autophagy genes in myeloid cells are critical to promote survival of mice in a model of TNF-induced shock and that RIPK1 kinase activity and IFNγ-signaling mediate the hypersensitive phenotype in autophagy gene-deficient animals.
Fig. 6.
Autophagy genes in myeloid cells protect against fatal TNF-induced shock. Survival of mice of genotype indicated after i.v. TNF injection. Graphs represent combined data from (A) Atg5f/f (n = 14; 1F, 13M) or Atg5ΔLysM (n = 27; 5F, 22M) mice from 3 independent experiments; (B) Atg16l1f/f (n = 11; 4F, 7M) or Atg16l1ΔLysM (n = 12; 4F, 8M) mice from 2 independent experiments; (C) Fip200f/f (n = 22; 9F, 13M) or Fip200ΔLysM (n = 17; 8F, 9M) mice from 3 independent experiments; (D) Becn1f/f (n = 32; 7F, 25M) or Becn1ΔLysM (n = 30; 7F, 23M) mice from 4 independent experiments; and (E) Becn1f/f; Ifngr1−/− (n = 16; 3F, 13M) or Becn1ΔLysM; Ifngr1−/− (n = 19; 4F, 15M) mice from 3 independent experiments. (F) Mice of genotype and treatment indicated. Graphs represent combined data from Atg5f/f + DMSO (n = 16; 8F, 8M), Atg5f/f + Nec-1s (n = 15; 7F, 8M), Atg5ΔLysM + DMSO (n = 20; 7F, 13M), and Atg5ΔLysM + Nec-1s (n = 19; 7F, 12M) mice from 3 independent experiments. Dose of TNF was 10 μg in A or 7.5 μg in B–F. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant; by log-rank (Mantel–Cox) test.
Discussion
We report an essential role for autophagy genes in protecting against IFNγ-induced macrophage cell death and identify TNF as a primary mediator of this phenotype. Furthermore, we identify RIPK1 kinase activity as the primary mediator of both hypersensitivity to IFNγ-induced cell death in vitro and the TNF-induced mortality in Atg5ΔLysM mice. Hypersensitivity of mice with Becn1ΔLysM to TNF-induced shock required the IFNγ-receptor subunit Ifngr1, providing an additional in vivo link to the IFNγ and TNF responses that we identified in vitro. Together, these findings implicate cell death as a significant mediator of the pathogenesis of systemic TNF-induced inflammatory responses, demonstrate an essential role for TNF in IFNγ-induced cell death, and identify autophagy as an important regulator of these processes.
Previous studies have evaluated autophagy genes in regulation of IFNγ-induced cell death (17, 58–65). Studies evaluating IFNγ and TNF in combination have suggested a cytoprotective role for autophagy-related genes in epithelial cells, yet the relative contribution of each cytokine was not delineated (58, 59, 64). However, additional studies have suggested that autophagy, and ATG5 specifically, performs a pro-cell death function during IFNγ and/or TNF-related responses (60–63, 65). In latently HIV-infected T cells (but not uninfected T cells), autophagosome-related structures scaffold an apoptosis-inducing complex containing RIPK1, RIPK3, FADD, and CASP8 triggered by small-molecule antagonists of inhibitor of apoptosis (IAPs) proteins (SMAC mimetics) (65). However, this form of cell death was independent of TNF (65), while the processes defined herein are TNF dependent. In mouse prostate epithelial cells lacking TAK1, treatment with the TNF-related apoptosis inducing ligand (TRAIL) leads to necroptotic or apoptotic cell death depending on the presence or absence, respectively, of the autophagy adaptor p62 (62). Since the latter 2 studies did not evaluate a role for IFNγ and given our present findings that autophagy genes limit IFNγ-induced and TNF-, CASP8-, and RIPK1-dependent death, the effect of IFNγ on the prodeath scaffolding function of autophagy components will be an important area of future study. As we investigated this pathway in cells of myeloid origin, it is likely that the context, timing, cell type, and/or specific inflammatory cues may determine whether autophagy genes perform a cytoprotective or prodeath function.
Our study further delineates the role of autophagy genes in fatal systemic responses to TNF. Previous studies have demonstrated a protective role for genes regulating multiple steps of the autophagy pathway in different tissues of mice challenged with lipopolysaccharide (LPS)/endotoxin, including Atg7 in liver and kidney (66–68) and Becn1 and Rubicon in heart (69, 70). A protective role for Atg7 and Atg16l1 in the myeloid compartment during endotoxin shock has also been demonstrated (71, 72). Mortality from LPS in mice is mediated in part by TNF (73, 74), although a specific role for TNF in systemic hyperresponsiveness to LPS in autophagy-deficient mice remains unclear. However, a role for Atg7 in protecting liver cells against TNF cytotoxicity was previously demonstrated (66, 67). We extended these findings to show multiple autophagy genes in myeloid cells protect against systemic TNF-induced fatal shock. While our study combined with previous studies implicates a set of genes regulating degradative autophagy in protection against shock, the full extent to which autophagy genes perform functions solely through degradative autophagy is not completely understood. Consistent with a degradative role for autophagy in protecting against endotoxin responses, a role for Atg16l1 in preventing Tax1BP1 accumulation and type I IFN responses in myeloid cells was recently demonstrated (72). Since Atg7 and Atg16l1 both contribute to the E3-like conjugation cascade to generate lipidated LC3 during autophagosome biogenesis, an LC3 conjugation-specific role of these genes in controlling responses of myeloid cells to endotoxin cannot be excluded.
We demonstrated here the importance of LC3 conjugation machinery (Atg5 and Atg16l1) in regulating systemic responses to TNF and also demonstrated a requirement for genes involved in upstream nucleation of autophagosome biogenesis (Becn1 and Fip200) (34, 35, 75, 76) and fusion of autophagosomes with lysosomes (Becn1) (36–38). Importantly, we also demonstrated a role for the LC3 conjugation machinery (Atg5 and Atg16l1), and the complex required for nascent autophagosome biogenesis (Becn1 and Atg14), in regulating IFNγ-induced death of BV2 cells. ATG14 has also been indicated in fusion of autophagosomes with lysosomes, which also requires its CCD (77, 78). While the most parsimonious explanation for the involvement of this set of genes is their function in degradative autophagy, we cannot rule out overlapping additional roles of these genes. For example, a role for Fip200 in countering TNF-induced lethality during embryogenesis was suggested previously (79). However, mice with mutations that abrogate the autophagy function of Fip200 have extended embryonic development but still do not survive to birth (75). In that study, double knockout of Fip200 and Tnfr1 also resulted in extended embryonic viability but also did not result in viable pups (75). Therefore, future studies should be performed to dissect degradative and alternative functions of autophagy genes in models of systemic shock.
Our findings may have important therapeutic implications. Sepsis incidence is increasing, and this syndrome has few available targeted therapies (80). Although a causative role remains to be proven, patients with promoter polymorphisms that result in diminished ATG5 expression have significantly decreased survival in sepsis (81). A recent study showed that treatment of mice with a therapeutic autophagy-inducing peptide was cardioprotective in an LPS model of sepsis, exemplifying the potential utility of therapeutically manipulating autophagy during systemic inflammatory responses (70). Accordingly, drugs that systemically inhibit autophagy may sensitize the host to sepsis. Further preclinical studies will be required to elucidate the precise role of autophagy in cell survival during different models of sepsis and infection, with augmentation of autophagy or more precise strategies targeting cell survival emerging as an attractive therapeutic target in treating human sepsis.
Materials and Methods
Mammalian Cells and Treatments.
BV2 microglial cells have been described previously (82, 83). Primary bone marrow-derived macrophages were generated as described (27). CRISPRko editing efficiency and clone isolation in BV2 cells were performed as described (21). Recombinant cytokines, chemicals, and antibodies are detailed in SI Appendix.
Retroviruses and Lentiviruses.
pMXs retrovirus encoding mCherry-Atg5 and K130R mutant have been described (84). Generation of additional vectors and lentiviruses is described in detail in SI Appendix. Transduced cells were selected and maintained in media containing puromycin (Puro, 4 μg/mL; Thermo Fisher Scientific) and/or blasticidin (4 μg/mL; Thermo Fisher Scientific).
CRISPRko Screening.
Primary screen.
Lentiviral libraries were synthesized, cloned, and produced as previously described (85). Additional details on library generation are in SI Appendix.
Atg5KO suppressor screen.
Suppressor screening with the Brie library on Atg5KO background was performed essentially as for the WT screen. Atg5KO-Cas9 cells were confirmed to exhibit equivalent IFNγ hypersensitivity as the parental Atg5KO line. Additional details on Atg5KO suppressor screen are in SI Appendix.
Next generation sequencing and data analysis.
Genomic DNA was isolated from cell pellets thawed on ice (Blood and Tissue kit; Qiagen). Sequencing libraries were prepared from 10-μg aliquots of gDNA by amplifying sgRNA regions by barcoded PCR, then sequenced, normalized, and analyzed as described (85). Additional details on screen analysis are in SI Appendix.
Viability and Cytotoxicity Analyses.
Cell viability was assessed using CellTiter-Glo assay (Promega). Cytotoxicity and Caspase 3 activity were assessed by live cell imaging on a Cytation 5 (Biotek) instrument. Additional details on cell death analyses are in SI Appendix.
Western Blot.
Samples were prepared in RIPA buffer and targets detected with the following primary antibodies: ATG5 (Sigma, A2859), p62 (Sigma, P0067), Ty1 Tag (Thermo Fisher Scientific, MA5-23513), Beclin 1 (Transduction Laboratories, 612113), Caspase 3 (Cell Signaling Technologies, 9665), and actin (Sigma, A2228). Secondary fluorescent antibodies were hFAB actin-rhodamine (BioRad), StarBright Blue 700 (BioRad), and IRDye 800-CW (Li-Cor). Additional details on Western Blot analyses are in SI Appendix.
RNA-Seq.
WT or Atg5KO clone 2 cells were treated with media or IFNγ (1 U/mL), and 24 h later, cells were harvested and RNA purified for library production. Sequencing was performed on an Illumina HiSeq 2500 instrument with a 2 × 101 run. Sample preparation, library production of polyadenylated RNA, and analysis are detailed in SI Appendix.
Gene Set Enrichment Analysis:
For GSEA of CRISPR screen results, average log2 fold change values from volcano plots output file were used. For GSEA of RNA-seq results, Stat values from DeSeq2 results were used. Gene Set Enrichment Analysis software (v3.0, Broad Institute) was run on these data, searching the Hallmark database (h.all.v5.2.symbols.gmt) (for RNA-seq results), with the addition of a custom gene set generated from GO annotations for mouse autophagy genes with data quality at Experimental level, which were appended as human orthologs to Hallmark set for CRISPRko screen GSEA analysis. GSEA was performed with default settings and 1,000 iterations, and pathways with FDR q value < 0.2 were considered significant.
Transmission Electron Microscopy.
Ultrastructural analyses were performed as described (86), except cells were fixed in 2% paraformaldehyde/2.5% glutaraldehyde (Polysciences Inc.) in 100 mM sodium cacodylate buffer (pH 7.2) and washed in cacodylate buffer instead of phosphate buffer. Samples were imaged with an AMT 8-megapixel digital camera and AMT Image Capture Engine V602 software (Advanced Microscopy Techniques).
Mice.
Atg5f/f (87), Atg16l1f/f (27), Fip200f/f (79), Becn1f/f (88), LysMcre (ΔLysM herein; Jax #004781) (89), and Ifngr−/− (Jax #003288) (90) mice have been described. Mice were housed in a temperature-controlled specific pathogen-free barrier vivarium with an alternating 12 h:12 h light:dark cycle. TNF injections were performed at doses indicated on 8- to 12-wk-old mice in a final volume of 200 μL in PBS/0.1% BSA. Nec-1s (6 mg/kg) or vehicle (6% DMSO in PBS/0.1% BSA) injections were performed 17 to 20 min before TNF injection. Mice were monitored every 6 to 12 h for clinical signs of morbidity and euthanized if unable to ambulate to hydrogel food or unable to maintain upright posture or if the core temperature nadir was <26 °C using rectal thermometer for rodents (Bioseb). All experimental protocols were reviewed and approved by the Washington University Institutional Animal Care and Use Committee.
Software and Statistical Analyses.
Software utilized was as indicated for each application. Statistical analyses (except as noted for CRISPRko screens and RNA-seq experiments) were performed in Prism with posttest analyses indicated for each figure (v8.0.0.0, GraphPad Software, Inc.).
Supplementary Material
Acknowledgments
We appreciate critical review of the manuscript from Melanie Yarbrough and David Hunstad. We thank the following individuals and Cores at Washington University School of Medicine: Darren Kreamalmeyer for expert mouse husbandry, Tim Schaiff and Anne Paredes for laboratory managerial assistance, Monica Sentmanat in the Genome Engineering and iPSC Center (GEiC) at Washington University School of Medicine for assistance with CRISPR cell line generation, and Wandy Beatty in the Molecular Microbiology Imaging Facility for assistance with electron microscopy. We thank Olivia Bare at the Broad Institute for assistance with CRISPR libraries and next generation sequencing. A.O. was supported by the Pediatric Infectious Diseases Society–St. Jude Children’s Research Hospital Fellowship Program in Basic Research and NIH award T32AI106688, R.C.O. was supported by NIH award K99-DK116666, G.A.S. was supported by NIH awards R01DK104946 and R01DK114047, and H.W.V. was supported by NIH award U19AI109725.
Footnotes
Conflict of interest statement: H.W.V. is a founder of Casma Therapeutics and PierianDx, neither of which funded this research. H.W.V. is an employee of and holds stock options in Vir Biotechnology, which did not fund this research. This work was performed at Washington University School of Medicine. Adi Kimchi is a coauthor with H.W.V. on a 2018 nomenclature paper and with G.A.S. on a 2016 guidelines paper.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1822157116/-/DCSupplemental.
References
- 1.Paludan S. R., Synergistic action of pro-inflammatory agents: Cellular and molecular aspects. J. Leukoc. Biol. 67, 18–25 (2000). [DOI] [PubMed] [Google Scholar]
- 2.Weinlich R., Green D. R., The two faces of receptor interacting protein kinase-1. Mol. Cell 56, 469–480 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Opipari A. W. Jr, Hu H. M., Yabkowitz R., Dixit V. M., The A20 zinc finger protein protects cells from tumor necrosis factor cytotoxicity. J. Biol. Chem. 267, 12424–12427 (1992). [PubMed] [Google Scholar]
- 4.He K.-L., Ting A. T., A20 inhibits tumor necrosis factor (TNF) alpha-induced apoptosis by disrupting recruitment of TRADD and RIP to the TNF receptor 1 complex in Jurkat T cells. Mol. Cell. Biol. 22, 6034–6045 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin Z., et al. , Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 137, 721–735 (2009). [DOI] [PubMed] [Google Scholar]
- 6.Won M., et al. , Novel anti-apoptotic mechanism of A20 through targeting ASK1 to suppress TNF-induced JNK activation. Cell Death Differ. 17, 1830–1841 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heger K., et al. , OTULIN limits cell death and inflammation by deubiquitinating LUBAC. Nature 559, 120–124 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Haas T. L., et al. , Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell 36, 831–844 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Tokunaga F., et al. , SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature 471, 633–636 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Varfolomeev E., et al. , IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 131, 669–681 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Vince J. E., et al. , IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell 131, 682–693 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Micheau O., Lens S., Gaide O., Alevizopoulos K., Tschopp J., NF-kappaB signals induce the expression of c-FLIP. Mol. Cell. Biol. 21, 5299–5305 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williamson B. D., Carswell E. A., Rubin B. Y., Prendergast J. S., Old L. J., Human tumor necrosis factor produced by human B-cell lines: Synergistic cytotoxic interaction with human interferon. Proc. Natl. Acad. Sci. U.S.A. 80, 5397–5401 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fransen L., Van der Heyden J., Ruysschaert R., Fiers W., Recombinant tumor necrosis factor: Its effect and its synergism with interferon-gamma on a variety of normal and transformed human cell lines. Eur. J. Cancer Clin. Oncol. 22, 419–426 (1986). [DOI] [PubMed] [Google Scholar]
- 15.Lee S. H., Aggarwal B. B., Rinderknecht E., Assisi F., Chiu H., The synergistic anti-proliferative effect of gamma-interferon and human lymphotoxin. J. Immunol. 133, 1083–1086 (1984). [PubMed] [Google Scholar]
- 16.Talmadge J. E., et al. , Toxicity of tumor necrosis factor is synergistic with gamma-interferon and can be reduced with cyclooxygenase inhibitors. Am. J. Pathol. 128, 410–425 (1987). [PMC free article] [PubMed] [Google Scholar]
- 17.Deiss L. P., Feinstein E., Berissi H., Cohen O., Kimchi A., Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 9, 15–30 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Thapa R. J., et al. , NF-kappaB protects cells from gamma interferon-induced RIP1-dependent necroptosis. Mol. Cell. Biol. 31, 2934–2946 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thapa R. J., et al. , Interferon-induced RIP1/RIP3-mediated necrosis requires PKR and is licensed by FADD and caspases. Proc. Natl. Acad. Sci. U.S.A. 110, E3109–E3118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dillon C. P., et al. , RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell 157, 1189–1202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sentmanat M. F., Peters S. T., Florian C. P., Connelly J. P., Pruett-Miller S. M., A survey of validation strategies for CRISPR-Cas9 editing. Sci. Rep. 8, 888 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuma A., et al. , The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 (2004). [DOI] [PubMed] [Google Scholar]
- 23.Mizushima N., et al. , A protein conjugation system essential for autophagy. Nature 395, 395–398 (1998). [DOI] [PubMed] [Google Scholar]
- 24.Biering S. B., et al. , Viral replication complexes are targeted by LC3-guided interferon-inducible GTPases. Cell Host Microbe 22, 74–85.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jounai N., et al. , The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. U.S.A. 104, 14050–14055 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choi J., et al. , The parasitophorous vacuole membrane of Toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity 40, 924–935 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hwang S., et al. , Nondegradative role of Atg5-Atg12/Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe 11, 397–409 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Selleck E. M., et al. , Guanylate-binding protein 1 (Gbp1) contributes to cell-autonomous immunity against Toxoplasma gondii. PLoS Pathog. 9, e1003320 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selleck E. M., et al. , A noncanonical autophagy pathway restricts Toxoplasma gondii growth in a strain-specific manner in IFN-γ-activated human cells. MBio 6, e01157-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Z., et al. , Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4, 458–469 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mizushima N., et al. , Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol. 152, 657–668 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimmey J. M., et al. , Unique role for ATG5 in neutrophil-mediated immunopathology during M. tuberculosis infection. Nature 528, 565–569 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J. H., et al. , Insights into autophagosome maturation revealed by the structures of ATG5 with its interacting partners. Autophagy 11, 75–87 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kihara A., Kabeya Y., Ohsumi Y., Yoshimori T., Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep. 2, 330–335 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang X. H., et al. , Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 402, 672–676 (1999). [DOI] [PubMed] [Google Scholar]
- 36.Zhong Y., et al. , Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat. Cell Biol. 11, 468–476 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Itakura E., Kishi C., Inoue K., Mizushima N., Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol. Biol. Cell 19, 5360–5372 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsunaga K., et al. , Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat. Cell Biol. 11, 385–396 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Martinez J., et al. , Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins. Nat. Cell Biol. 17, 893–906 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Matsunaga K., et al. , Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J. Cell Biol. 190, 511–521 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mei Y., et al. , Identification of BECN1 and ATG14 coiled-coil interface residues that are important for starvation-induced autophagy. Biochemistry 55, 4239–4253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gutierrez M. G., et al. , Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766 (2004). [DOI] [PubMed] [Google Scholar]
- 43.Matsuzawa T., et al. , IFN-γ elicits macrophage autophagy via the p38 MAPK signaling pathway. J. Immunol. 189, 813–818 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen Y.-D., et al. , S100A10 regulates ULK1 localization to ER-mitochondria contact sites in IFN-γ-triggered autophagy. J. Mol. Biol. 429, 142–157 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Dang A. T., et al. , Autophagy links antimicrobial activity with antigen presentation in Langerhans cells. JCI Insight 4, 126955 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamamoto A., et al. , Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 23, 33–42 (1998). [DOI] [PubMed] [Google Scholar]
- 47.Micheau O., Tschopp J., Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114, 181–190 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Galluzzi L., et al. , Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 25, 486–541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hou W., Han J., Lu C., Goldstein L. A., Rabinowich H., Autophagic degradation of active caspase-8: A crosstalk mechanism between autophagy and apoptosis. Autophagy 6, 891–900 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi N., et al. , Necrostatin-1 analogues: Critical issues on the specificity, activity and in vivo use in experimental disease models. Cell Death Dis. 3, e437 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roderick J. E., et al. , Hematopoietic RIPK1 deficiency results in bone marrow failure caused by apoptosis and RIPK3-mediated necroptosis. Proc. Natl. Acad. Sci. U.S.A. 111, 14436–14441 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi N., et al. , RIPK1 ensures intestinal homeostasis by protecting the epithelium against apoptosis. Nature 513, 95–99 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Berger S. B., et al. , Cutting edge: RIP1 kinase activity is dispensable for normal development but is a key regulator of inflammation in SHARPIN-deficient mice. J. Immunol. 192, 5476–5480 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Polykratis A., et al. , Cutting edge: RIPK1 kinase inactive mice are viable and protected from TNF-induced necroptosis in vivo. J. Immunol. 193, 1539–1543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doherty G. M., et al. , Evidence for IFN-gamma as a mediator of the lethality of endotoxin and tumor necrosis factor-alpha. J. Immunol. 149, 1666–1670 (1992). [PubMed] [Google Scholar]
- 56.Grivennikov S. I., et al. , Distinct and nonredundant in vivo functions of TNF produced by t cells and macrophages/neutrophils: Protective and deleterious effects. Immunity 22, 93–104 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Duprez L., et al. , RIP kinase-dependent necrosis drives lethal systemic inflammatory response syndrome. Immunity 35, 908–918 (2011). [DOI] [PubMed] [Google Scholar]
- 58.Sumpter R., Jr, et al. , Fanconi anemia proteins function in mitophagy and immunity. Cell 165, 867–881 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye Y.-C., et al. , TNFα-induced necroptosis and autophagy via supression of the p38-NF-κB survival pathway in L929 cells. J. Pharmacol. Sci. 117, 160–169 (2011). [DOI] [PubMed] [Google Scholar]
- 60.Bell B. D., et al. , FADD and caspase-8 control the outcome of autophagic signaling in proliferating T cells. Proc. Natl. Acad. Sci. U.S.A. 105, 16677–16682 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pyo J.-O., et al. , Essential roles of Atg5 and FADD in autophagic cell death: Dissection of autophagic cell death into vacuole formation and cell death. J. Biol. Chem. 280, 20722–20729 (2005). [DOI] [PubMed] [Google Scholar]
- 62.Goodall M. L., et al. , The autophagy machinery controls cell death switching between apoptosis and necroptosis. Dev. Cell 37, 337–349 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Young M. M., et al. , Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J. Biol. Chem. 287, 12455–12468 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pott J., Kabat A. M., Maloy K. J., Intestinal epithelial cell autophagy is required to protect against TNF-induced apoptosis during chronic colitis in mice. Cell Host Microbe 23, 191–202.e4 (2018). [DOI] [PubMed] [Google Scholar]
- 65.Campbell G. R., Bruckman R. S., Chu Y.-L., Trout R. N., Spector S. A., SMAC mimetics induce autophagy-dependent apoptosis of HIV-1-infected resting memory CD4+ T cells. Cell Host Microbe 24, 689–702.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lalazar G., et al. , Autophagy confers resistance to lipopolysaccharide-induced mouse hepatocyte injury. Am. J. Physiol. Gastrointest. Liver Physiol. 311, G377–G386 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Amir M., et al. , Inhibition of hepatocyte autophagy increases tumor necrosis factor-dependent liver injury by promoting caspase-8 activation. Cell Death Differ. 20, 878–887 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mei S., et al. , Autophagy is activated to protect against endotoxic acute kidney injury. Sci. Rep. 6, 22171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun Y., et al. , Beclin-1-dependent autophagy protects the heart during sepsis. Circulation 138, 2247–2262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zi Z., et al. , Rubicon deficiency enhances cardiac autophagy and protects mice from lipopolysaccharide-induced lethality and reduction in stroke volume. J. Cardiovasc. Pharmacol. 65, 252–261 (2015). [DOI] [PubMed] [Google Scholar]
- 71.Abdel Fattah E., Bhattacharya A., Herron A., Safdar Z., Eissa N. T., Critical role for IL-18 in spontaneous lung inflammation caused by autophagy deficiency. J. Immunol. 194, 5407–5416 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Samie M., et al. , Selective autophagy of the adaptor TRIF regulates innate inflammatory signaling. Nat. Immunol. 19, 246–254 (2018). [DOI] [PubMed] [Google Scholar]
- 73.Marino M. W., et al. , Characterization of tumor necrosis factor-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 94, 8093–8098 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amiot F., Fitting C., Tracey K. J., Cavaillon J. M., Dautry F., Lipopolysaccharide-induced cytokine cascade and lethality in LT alpha/TNF alpha-deficient mice. Mol. Med. 3, 864–875 (1997). [PMC free article] [PubMed] [Google Scholar]
- 75.Chen S., et al. , Distinct roles of autophagy-dependent and -independent functions of FIP200 revealed by generation and analysis of a mutant knock-in mouse model. Genes Dev. 30, 856–869 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hara T., et al. , FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181, 497–510 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Diao J., et al. , ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature 520, 563–566 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu R., Zhi X., Zhong Q., ATG14 controls SNARE-mediated autophagosome fusion with a lysosome. Autophagy 11, 847–849 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gan B., et al. , Role of FIP200 in cardiac and liver development and its regulation of TNFalpha and TSC-mTOR signaling pathways. J. Cell Biol. 175, 121–133 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singer M., et al. , The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 315, 801–810 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shao Y., et al. , Association between genetic polymorphisms in the autophagy-related 5 gene promoter and the risk of sepsis. Sci. Rep. 7, 9399 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blasi E., Radzioch D., Durum S. K., Varesio L., A murine macrophage cell line, immortalized by v-raf and v-myc oncogenes, exhibits normal macrophage functions. Eur. J. Immunol. 17, 1491–1498 (1987). [DOI] [PubMed] [Google Scholar]
- 83.Stansley B., Post J., Hensley K., A comparative review of cell culture systems for the study of microglial biology in Alzheimer’s disease. J. Neuroinflammation 9, 115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.DeSelm C. J., et al. , Autophagy proteins regulate the secretory component of osteoclastic bone resorption. Dev. Cell 21, 966–974 (2011). Erratum in: Dev. Cell.21, 1179 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Doench J. G., et al. , Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Presti R. M., et al. , Quaranfil, Johnston Atoll, and Lake Chad viruses are novel members of the family Orthomyxoviridae. J. Virol. 83, 11599–11606 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hara T., et al. , Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 (2006). [DOI] [PubMed] [Google Scholar]
- 88.Gawriluk T. R., et al. , Autophagy is a cell survival program for female germ cells in the murine ovary. Reproduction 141, 759–765 (2011). [DOI] [PubMed] [Google Scholar]
- 89.Clausen B. E., Burkhardt C., Reith W., Renkawitz R., Förster I., Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 8, 265–277 (1999). [DOI] [PubMed] [Google Scholar]
- 90.Huang S., et al. , Immune response in mice that lack the interferon-gamma receptor. Science 259, 1742–1745 (1993). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.