Abstract
Background
RADA16I represents one of promising hydrogel forming peptides. Several implementations of RADA16I hydrogels have proven successful in the field of regenerative medicine and tissue engineering. However, RADA16I peptides used in various studies utilize synthetic peptides and so far, only two research articles have been published on RADA16I peptide recombinant production. Moreover, previous studies utilized non- or less routine expression and purification methods to produce RADA16I peptide recombinantly.
Objectives
The main goal was to produce the self-assembling peptide, RADA16I, in Escherichia coli by exploiting routine and widely used vectors and purification methods, in shake flask.
Material and Methods
RADA16I coding sequence was inserted in pET31b+, and the construct was transformed into E. coli. Purified fusion constructs were purified using Nickel Sepharose. RADA16I unimers were released using CNBr cleavage. CD and FTIR spectroscopy were used to study recombinant RADA16I’s confirmation. TEM was used to confirm fibril formation of recombinant RADA16I. Furthermore, MTT assay was implemented to assess cytocompatibility of recombinant RADA16I.
Results
The biochemical, biophysical and structural analysis proved the ability of the recombinant RADA16I to form self-assembling peptide nanofibers. Furthermore, the nanofibers exhibited no cytotoxicity and retained their cell adhesive activity.
Conclusions
We successfully produced RADA16I in acceptable levels and established a basis for future investigation for the production of RADA16I under fermentation conditions.
Keywords: Fusion Protein, Hydrogel, Nanofibers, RADA16I, Self-Assembling Peptide
1. Background
Among many challenges in tissue engineering is the development of biomaterials that support cellular functions, such as cell adhesion, growth and differentiation (1). Small biological molecules capable of self-assembly and degradation over time into predictable metabolites represent ideal candidates for scaffolds to regenerate tissues and organs. For instance, peptide base self-assembling peptide nanofibers maintain functionality within a desired period of time, as well as the byproducts resulting from their degradation introduce no toxic constituents to their surrounding environment(2).
RADA-16( peptide the well-studied self-assembling peptide nanofiber (SAPN) was first introduced by S. Zhang (3) who has inspired the design of RADA16I from the set of tandem repeats, namely EAK16 (AEAEAKAKAEAEAKAK), present in the yeast Zuotin protein (4). In RADA-16( peptide, with the (RADARADARADARADARADA) sequence, the four amino acid RADA motif, hence annotation 16; whereas the ( is used to discriminate it from RADA-16(( which has the following sequence (RARADADARARADADADA) (5). RADA-16( exhibits an intrinsic capability to form hydrogels in the presence of monovalent salts or under physiological conditions due to the ionic interactions and the tendency of the backbone to adopt β-sheet structure (4, 6). There is extensive literature regarding the biophysical and biochemical properties of RADA-16( as well as on the various conditions at which RADA-16( forms hydrogel (7–13).
Several biological implementations of RADA-16( based hydrogels have proven successful. For example, S. Zhang has proven the ability of RADA-16( to support the attachment and differentiation of a broad range of cell types among which are neuronal stem cells, hepatic stem cells and cardiac myocytes (3, 4, 14–16). S. Zhang etal further demonstrated the ability of RADA-16( to promote angiogenesis (17). Other researchers have elaborated RADA-16( ability to repair brain, spinal and neuronal tissue injuries (18–20). In another study S. Zhang etal used RADA-16( hydrogels for the sake of cartilage tissue repair (21). Moreover, there is clear evidence on RADA-16(‘s capability to stop bleeding (complete hemostasis) immediately when directly treating brain, spinal cord, femoral artery, liver, or skin wounds(18).
2. Objectives
Here we describe a novel straight forward method for the recombinant production of RADA-16I based on the method initially described by T. Walsh(22). Briefly, Keto Steroid Isomerease (KSI)/ RADA-16( /His-tag fusion expressed in E. coli was purified using affinity chromatography and then cleaved with Cyanogen Bromide (CNBr) to release RADA-16I peptides (Fig. 1); in the end fibril formation was confirmed. Furthermore, fibrils formed proved to be non-cytotoxic, promote cell proliferation and exhibited cell adhesivity.
Figure 1.
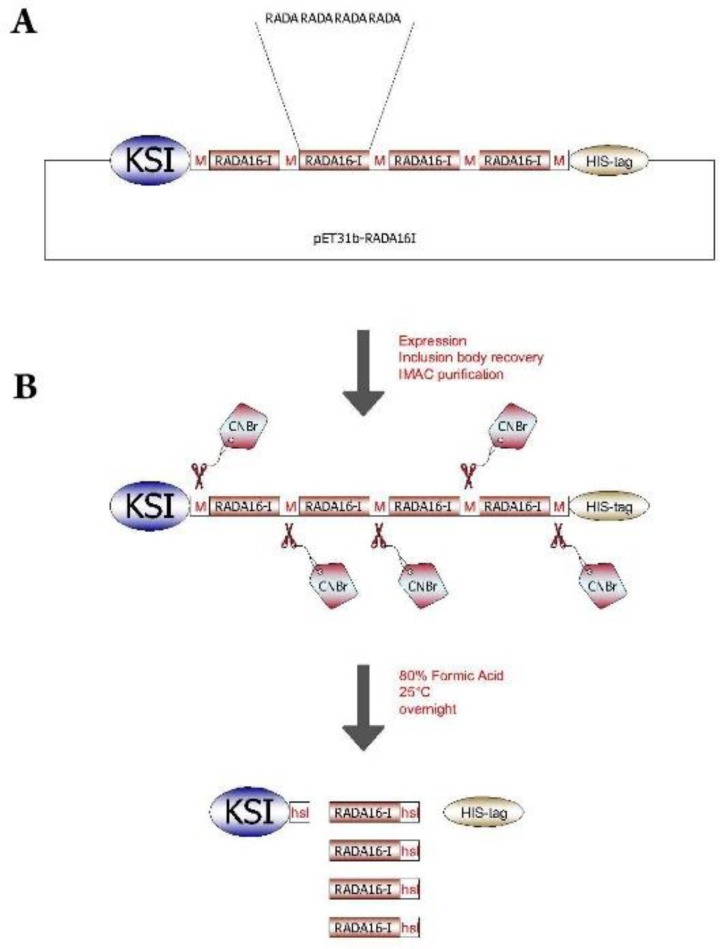
Cloning and Expression stratagies for RADA16I peptide repeats. a) Four RADA16I repeats were inserted between KSI and HIS-tag. Methionine spacers were located between the repeats, as well as between the repeats and both KSI and HIS-tag. b) Following protein expression, inclusion body recovery and IMAC purification the KSI-[RADA16I]4-HIS-tag is treated with CNBr to generate a mixture of KSI, RADA16I unimers and HIS-tag.
3. Materials and Methods
3.1. Bacterial Strains, Plasmid, and Culture Media
Escherichia coli Stbl3 strain was used for routine cloning, whilst E. coli
BL21 (DE3) pLysS strain was used for the expression of the pET31b+/ RADA-16( construct.
E. coli strains were grown in Super Optimal broth with Catabolite repression
(SOC) medium, containing 2% w.v−1 Tryptone, 0.5% w.v−1 Yeast extract, 10 mM NaCl,
2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, and 20 mM Glucose. Ampicillin was added to the media at a final concentration of 100 µg mL−1. Competent E. coli strains were prepared and transformed according to the protocol described by Inoue et al (23).
3.2. Protein Expression and Purification
In order to express the recombinant construct, pre-cultures (10 mL) from single colonies were incubated with shaking at 18°C for 18 hours. Subsequently, 25 µL of the pre-culture were added to 250 mL SOC medium in a 1 L conical flask and grown with shaking at 18°C until it reached OD600~0.8 (18 h). Isopropyl-b-D-Thiogalactopyranoside (IPTG) was added to final concentration 0.8 mM to induce protein expression. The bacteria were kept shaking at 250 rpm in an orbital incubator at 18ᵒ C for 6 hours. Afterwards, cells were harvested by centrifugation at 1792 g using a fixed angle rotor. In order to extract IBs, 10 mL Lysis Buffer (20 mM sodium phosphate, 500 mM NaCl, 8 M Urea) was added to the bacterial pellet obtained from 250 mL culture, and subsequently sonicated on ice to lyse the cells thoroughly. Lysates were then centrifuged (18928 g, 4ᵒC) and aliquots of the supernatant were stored at -20ᵒC for subsequent analysis.
A 5 ml HisTrap chelating column (GE Healthcare UK Ltd.) was used to purify KSI fusion proteins according to the manufacturer’s instructions. Before loading the supernatant, 0.45 µm syringe filter was used to remove any insoluble materials.
3.3. Cleavage of the KSI/ RADA-16I /His Protein by Cyanogen Bromide
Purified KSI/ RADA-16( /His were dialyzed against distilled water over night, and subsequently freeze-dried. KSI/ RADA-16I /His dried sample (10–20 mg.mL−1) was dissolved in 70% formic acid and then Cyanogen Bromide (CNBr) added to the protein sample at a 200-molar excess to methionine residues in a 2 mL volume. The tube was capped and incubated for 18–20 h at room temperature in the dark. Subsequently, the CNBr cleaved KSI/ RADA-16I /His mixture was dried by rotary evaporation. The gelatinous material formed was resuspended in 20 mM KH2PO4, 100 mM NaCl, and the pH adjusted to 7.4 with 1 M NaHCO3. This mixture was stirred overnight under N2 at room temperature in darkness. Afterwards, the suspension was centrifuged at 18928 g for 45 min at 4°C and the supernatant was collected.
3.4. RP-HPLC and Mass Spectrometry
RP-HPLC was performed using an analytical Zorbax SB-C18C18 column (15 cm, 3.5 μm) from Agilent, with a linear gradient of acetonitrile from 5% to 55% in a 0.1% Formic acid solution. The flow rate was 0.3 mL.min−1 for 20 min. Purified peptides were quantified via absorbance measurement at 215 nm Peak fractions obtained from RP-HPLC were freeze-dried prior to analysis by mass spectrometry. Positive Electron Spray Ionization (ESI) was carried out on Agilent G6410 Triple Quadrapole Mass spectrometer.
3.5. Circular Dichroism (CD)
Jasco Spectro-polarimeter J-715 (Tokyo, Japan) was used to perform Circular dichroism measurements. Far-UV CD spectra were recorded from 190 to 260 nm with the temperature of the cell holder controlled at 25 °C. Data obtained were smoothed with J-715 software, followed by processing and deconvoluting by ORIGIN software.
3.6. Fourier Transform Infrared Spectroscopy (FTIR)
FTIR was used to further assign the secondary structure of dissolved recombinant RADA-16(. Two RADA-16( concentrations (2.5 and 0.5 mg.mL−1) were prepared. From each sample 100 µL were deposited on Potassium Bromide (KBr) discs. The FTIR spectrum corresponding to the discs was taken at a wavenumber resolution of 4 cm−1 using Tensor 27 FT-IR spectrometer. Measurements were recorded within 500–4000 cm−1, where the amide ( band is expected to appear.
3.7. Transmission Electron Microscopy
Thirty microliters of RADA-16( samples (2.5 and 0.5 mg.mL−1 samples of RADA-16I) were applied to a 300-mesh formvar/carbon-coated nickel grid and after 2 min, stained with 3% uranyl acetate for 2 minutes, rinsed, and air-dried. Images were taken using Zeiss-EM10C-80 KV transmission electron microscope (Oberkochen, Germany).
3.8. Assessment of Purified RADA-16( Peptide Cell Toxicity
MCF-7 cells were cultured in a 96-well flask with 5x103 cells in each well. The next day, MCF-7 cells were exposed to purified recombinant RADA-16( peptide, at a final concentration 0.5% w.v−1 and 1% w.v−1, for 24 hours; control groups were treated similarly with recombinant RADA-16( free media. Later on, the medium was aspirated and a medium was added with a final concentration of MTT equal to 5 mg.mL−1. The cells were incubated for 4 h at 37° C. Then the medium was aspirated carefully without disturbing the formed formazan crystals, which were then dissolved using 100 µL of DMSO (dimethyl sulfoxide). After complete solubilization of formazan crystals, absorbance was recorded at 570 nm.
3.9. Evaluation of Purified RADA-16( Peptide Cell Adhesive Activity
For coating glass cover slips, 50 µL of purified peptides (100, 250, 500, and 1000 µM) diluted in 1X phosphate buffer saline (PBS) were spread on the top of the glass cover slips, which in turn were embedded at the bottom of 6-well plates and left to dry at 37 °C. After complete dryness, 3x103 MCF-7 cells in Dulbecco’s Modified Eagle’s medium were seeded on the glass cover slips. After Incubation for 48 h, cover slips were observed under light microscope.
4. Results
4.1. Expression and of Purification of Recombinant RADA-16I
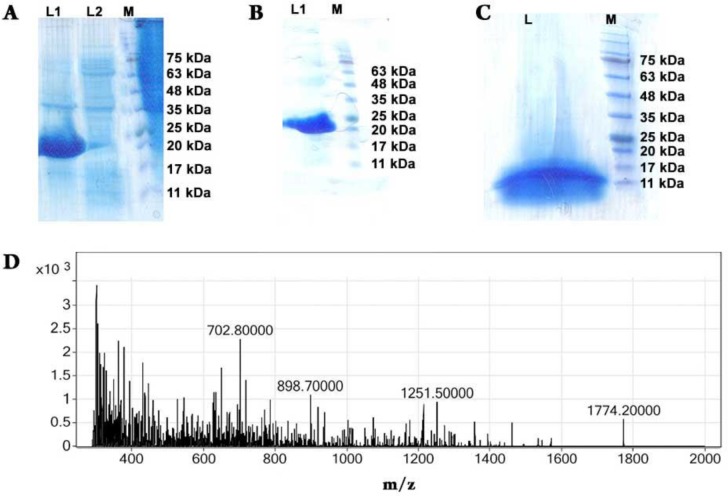
The integrity of the pET-31 KSI/ RADA-16I /His construct was confirmed by sequencing (Data not shown). The expressed recombinant protein present in the pellet of the bacterial lysate, i.e. inclusion bodies, was analyzed using SDS-PAGE (Fig. 2A). The molecular weight of the fusion protein KSI/ RADA-16I /His (21.28 kDa) was confirmed through comparison with protein ladder (Fig. 2A) were it ranged from 20 kDa to 25 kDa; both the crude lysate (Fig. 2A) and purified (Fig. 2B) fusion protein KSI/ RADA-16( /His proved to match the expected molecular weight.
Figure 2.
Expression, purification and CNBr digestion of KSI-[RADA16I]4-HIS-tag constructs. A) SDS–PAGE analysis of 0.8 mM IPTG-induced expression of KSI fusion constructs carrying RADA16I peptide repeats. L1, pelet of bacterial lysate; L2, supernatant of bacterial lysate; M, molecular lader. B) SDS–PAGE analysis of KSI-[RADA16I]4-HIS-tag constructs. L1, purified KSI-[RADA16I]4-HIS-tag constructs M, molecular lader. C) SDS–PAGE analysis of the CNBr digested KSI-[RADA16I]4-HIS-tag fusion protein. L, digestion mixture; M, molecular ladder. D) Mass spectrum of RADA16I recovered by RP-HPLC following CNBr cleavage of purified KSI-[RADA16I]4-HIS-tag constructs. The monoisotopic mass of 1772.85 Da corresponds to the theoretical mass of RADA16I, 1774.20 Da
4.2. CNBr Cleavage, Purification and Confirmation of Recombinant RADA-16I
Purified KSI/ RADA-16( /His fusion protein were dialyzed against ddH2O and subsequently freeze-dried. Dried samples were digested with CNBr which specifically cleaved the fusion protein at the Methionine junction to release insoluble KSI, the His6 tag, and recombinant RADA-16I. The digestion reaction was analyzed with SDS-PAGE (Fig. 2C). It is clear from the gel electrograph that complete digestion of the fusion protein took place since we can observe a band located between 11 kDa to 17 kDa, the molecular weight corresponding to KSI (14 kDa).
Following the incubation with CNBr, the reaction was dried using rotary evaporation in order to remove formic acid. A gelatinous material was observed at the bottom of the reaction tubes. The gelatinous material was then resuspended in 20 mM KH2PO4, 100 mM NaCl buffer. The solution was extensively centrifuged, and the resulting supernatant were RADA-16I is expected to be was subjected to RP-HPLC. The RP-HPLC purified peptides were freeze-dried and then solubilized in 65% formic acid and used for electrospray ionization mass spectrometry for further characterization. The determined molecular weight of recombinant RADA-16I obtained from CNBr cleavage is 1774.2 Da, as shown in (Fig. 2D).
4.3. Evaluation of β-Sheet Formation by RADA-16I
One of the well-recognized characteristics of RADA-16I peptide is its ability to form β-sheet structures. There are several techniques to evaluate this criterion. We examined the secondary structure of recombinant RADA-16I by CD and FTIR. Purified recombinant RADA-16I were diluted in distilled water to a final concentration of 300 mM and it exhibited a sharp minimum ellipticity at ~205 nm, slight minimum ellipticity at ~216 nm, and a maximum ellipticity at ~192 nm (Fig. 3A); which is consistent with previous reports(3, 10). J-715 software estimated the β-sheet content of the RADA-16I peptide to be ~82.9%.
Figure 3.
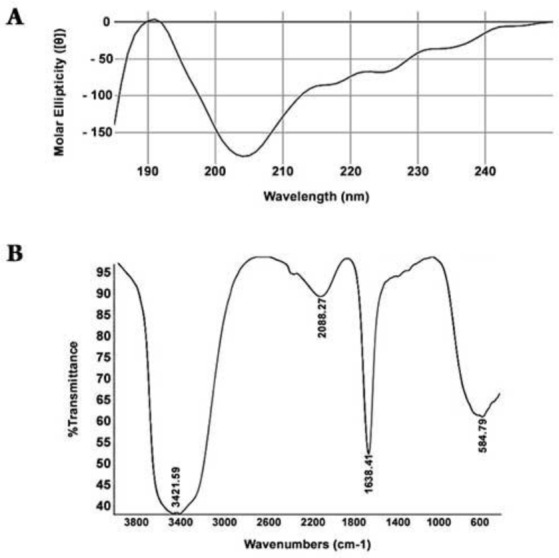
Secondary structure analysis of RADA16I. A) CD spectrum of RADA16I; B) Fourier transform IR spectra of amide I (1700-1600 cm−1) and amide II (1600-1500 cm−1) bands of RADA16I.
FTIR was also used to study the secondary structure of recombinant RADA-16I. FTIR analysis of RADA-16I further supported CD data showing that a strong peak was observed at 1638.41 which confirmed the antiparallel β-sheet configuration of RADA16I (Fig. 3B).
4.4. Fibril Observation by Transmission Electron Microscopy
After confirming the adoption of β-sheet conformation by the recombinant RADA-16I peptides, observation of fibril formation by transmission electron microscope was the next validation step. At low concentrations 300 mM, recombinant RADA-16I exhibited fibrils with ~8nm diameter (Data not shown), whilst at higher concentrations, recombinant RADA16I peptide fibrils were noted to be interconnecting. It was also evident that fibrils were extended and twisted, as well as forming a mesh-like appearance; neighboring fibrils can be seen as fibrillar bundles and entanglements (Fig. 4A).
Figure 4.
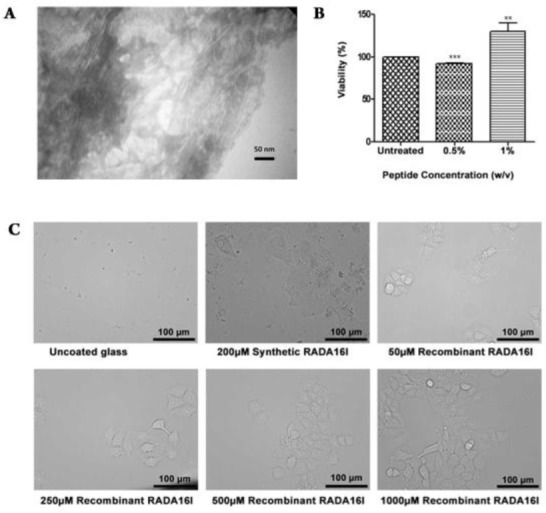
A) Transmission electron microscopy (TEM) image of RADA16I fibrils formed at a final peptide concentration of 2.5 mg.L-1. B) Evaluation of cell cytotoxicity of purified RADA16I fibrils. Blank, cells not treated with RADA16I; green, cells treated with 0.5% RADA16I; violet, cells treated with 1% RADA16I. C) Evaluation of cell adhesivity of purified RADA16I fibrils. i, uncoated glass cover slips; ii, glass cover slips coated with 200µM synthetic RADA16I; iii, glass cover slips coated with 50µM recombinant RADA16I; iv, glass cover slips coated with 250µM recombinant RADA16I; v, glass cover slips coated with 500µM recombinant RADA16I; vi, glass cover slips coated with 1000µM recombinant RADA16I.
4.5. Assessing Recombinant RADA-16I‘s Cytotoxicity and Cell Adhesion Activity
MTT assay was implemented to assess cell viability of cells exposed to recombinant RADA-16I. In this regard, MCF-7 cells were exposed to recombinant RADA-16I at final concentration 0.5% and 1% w/v. When compared to control samples, MCF-7 cells exposed to recombinant RADA-16I at final concentration 0.5% exhibited a slight decrease in cell viability 91.85 %, whereas MCF-7 cells exposed to recombinant RADA-16I at final concentration 1% exhibited an increase in cell viability 127.1 % (Fig. 4B).
The assessment of cell adhesive activity of recombinant RADA-16I followed. To this aim, glass cover slips were coated with RADA-16I, and next MCF-7 cells were seeded on these glass cover slips. FBS free DMEM was used to avoid any possible serum effects. After 48 h, applying inverted microscopy, our observations show that MCF-7 cells attached to recombinant RADA-16I coated cover slips whilst no cells were observed on the surface of cover slips treated with PBS only (Fig. 4C).
5. Discussion
RADA-16I forms nanofiber matrices which have shown great promise for tissue engineering. RADA-16I is currently produced using chemical synthesis and commercially available as PuraMatrix (BD, Franklin Lakes, NJ, USA). Besides the fact that it is an expensive process in which cost increases with peptide length, F. Albericio describes the process of synthesizing and purifying RADA-16( as a “Tough” one(24). In contrast, although there are significant challenges for the production and efficient purification of short self-assembling peptides, recombinant production of RADA-16( in a bacterial host offers a potentially more economical solution owing to its ease of use, robustness and low costs.
For various reasons E. coli represents one of the attractive choices for heterologous protein production. For instance, the genetic characteristics of E. coli are well-established and has the ability to grow rapidly to high cell density using low cost media. Frequently, the recombinant protein production in E. coli is associated with the formation of intracellular protein aggregates in the form of inclusion bodies. For most protein production systems this is undesirable since recovery of functional protein from inclusion bodies involves complex refolding. However, for peptides production this is not an obstacle and recovery from inclusion bodies represents the basis of a method introduced by Walsh et al.; in this method tandem repeats of the peptide coding region are fused to KSI, the HIS-tag for purification and intervening Met residues for consequent CNBr cleavage (Fig. 1) (22). In this study, the goal was the enhancement of the production of RADA-16( peptide as a unimers, the introduction of a rapid and simple purification strategy and finally the characterization and cytotoxicity determination of the recombinant RADA-16(. To the best of our knowledge, so far two research articles have been published regarding the recombinant production of RADA-16(. However, when compared to our strategy, both attempts implemented novel (25) or less routine (26) methods. For instance, Reed et al. (25) implemented pDR1 plasmid as well as Ralstonia eutropha for the RADA-16( peptide recombinant production. Under fermentation conditions, Reed et al. yield of production of fusion protein was 433 mg.L−1 and were able to purify 10.1 mg.L−1 of recombinant RADA-16(. Whereas in our case, the yield of production of fusion protein was ~95 mg.L−1 and were able to purify 7.8 mg.L−1 of RADA-16( (Table 1). Eiry Kobatake et al. did not report the final fusion protein or purified peptide yield in their study(26).
Table 1.
Reported levels of production and purification of RADA peptide by recombinant approaches
| Host | Fusion protein | Theoretical peptide | Peptide purified | Efficiency of peptide | Reference |
|---|---|---|---|---|---|
| yield (mg/L) | yield (mg/L) | (mg/L) | Purification (%) | ||
| Ralstonia eutropha | 433 | 116 | 10.1 | 8.7 | (25) |
| Escherichia coli | 95 | 33.86 | 7.8 | 23.03 | This work |
Circular dichroism analysis indicated that a minimum is located at ~205 nm and a maximum ellipticity at ~192 nm, characteristic of random coil. The shoulder around ~216 nm corresponds to β-sheet (Fig. 3A). This observation could be attributed to the fact that at this concentration of purified recombinant peptide RADA-16( monomers with random coil confirmation along with fibrils with β-sheet confirmation are present concomitantly in the aqueous solution(9). However, J-715 software estimated that the β-sheet content is 85%.
In the amide I, fibrils exhibit two bands; a low frequency, high intensity band around 1620–1630 cm−1 and a high frequency, low intensity band located around 1685–1695 cm−1, which correspond to anti-parallel β-sheet The concurrent appearance of these two bands correspond to anti-parallel β-sheet whilst the presence of only the low frequency band around 1630–1640 cm−1 confers to the parallel counterpart. Hence, FTIR analysis indicated the existence of parallel β-sheet conformation (Fig. 3B) since it only exhibited a single band 1638.41 cm−1 in the amide I region (27, 28). TEM observations (Fig. 4A) further confirmed that recombinant RADA-16( formed fibrils with dimensions similar to what has been reported by (26).
RADA-16( is known to exhibit two criteria useful for tissue engineering and regenerative medicine, namely cytocompatibility and cell adhesivity. Recombinant RADA-16( proved to be non-cytotoxic to MCF-7 cells grown up in contact with recombinant RADA-16( for 24 hrs. Furthermore, similar to previous reports (26) recombinant RADA-16( aided the attachment of MCF-7 cells to glass cover slips (Fig. 4B, 4C).
6. Conclusions
We have demonstrated that RADA-16I can be expressed recombinantly in E. coli using pET31b+ plasmid. Efficient purification was achieved using Nickel Sepharose, and CNBr cleavage subsequently was used to release RADA-16I unimers. Under shake flask conditions, 10.1 mg.L−1 was the final yield of the recombinant RADA-16I. Under the experimental conditions we have set, the simple culture medium SOC proved to be more suitable for the recombinant production of RADA16I peptide in comparison to the complex autoinduction medium. Spectroscopic and microscopic analysis proved that recombinant RADA-16I exhibited properties similar to the previously reported recombinant and synthetic RADA-16I peptide. RADA-16I self-assembled into fibrils, and cytotoxicity studies indicated that recombinant RADA-16I was non-cytotoxic towards MCF-7 cells. This study establishes that recombinant RADA-16I peptide can be produced in a feasible manner using routine expression and purifications methods.
Footnotes
Conflict of Interest
There is no conflict of interest with this study.
References
- 1.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260(5110):920–926. doi: 10.1126/science.8493529 pmid: 8493529 [DOI] [PubMed] [Google Scholar]
- 2.Stupp SI, Zha RH, Palmer LC, Cui H, Bitton R. Self-assembly of biomolecular soft matter. Faraday Discuss. 2013;166:9–30. doi: 10.1039/c3fd00120b pmid: 24611266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang S, Holmes TC, DiPersio CM, Hynes RO, Su X, Rich A. Self-complementary oligopeptide matrices support mammalian cell attachment. Biomaterials. 1995;16(18):1385–1393. doi: 10.1016/0142-9612(95)96874-Y pmid: 8590765 [DOI] [PubMed] [Google Scholar]
- 4.Zhang S, Lockshin C, Cook R, Rich A. Unusually stable beta-sheet formation in an ionic self-complementary oligopeptide. Biopolymers. 1994;34(5):663–672. doi: 10.1002/bip.360340508 pmid: 8003624 [DOI] [PubMed] [Google Scholar]
- 5.Zhang S, Gelain F, Zhao X. Designer self-assembling peptide nanofiber scaffolds for 3D tissue cell cultures. Semin Cancer Biol. 2005;15(5):413–420. doi: 10.1016/j.semcancer.2005.05.007 pmid: 16061392 [DOI] [PubMed] [Google Scholar]
- 6.Zhang S, Holmes T, Lockshin C, Rich A. Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. Proc Natl Acad Sci U S A. 1993;90(8):3334–3338. doi: 10.1073/pnas.90.8.3334 pmid: 7682699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aramvash A, Seyedkarimi MS. All-atom molecular dynamics study of four RADA 16-I peptides: the effects of salts on cluster formation. J Cluster Sci. 2015;26(2):631–643. doi: 10.1007/s10876-014-0836-8 [Google Scholar]
- 8.Yokoi H, Kinoshita T, Zhang S. Dynamic reassembly of peptide RADA16 nanofiber scaffold. Proc Natl Acad Sci U S A. 2005;102(24):8414–8419. doi: 10.1073/pnas.0407843102 pmid: 15939888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arosio P, Owczarz M, Wu H, Butte A, Morbidelli M. End-to-end self-assembly of RADA 16-I nanofibrils in aqueous solutions. Biophys J. 2012;102(7):1617–1626. doi: 10.1016/j.bpj.2012.03.012 pmid: 22500762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Luo H, Zhao X. Mechanistic study of self-assembling peptide rada16-i in formation of nanofibers and hydrogels J Nanotechnol Eng Med. 2010;1(1):011007 doi: 10.1115/1.4000301 [Google Scholar]
- 11.Cormier AR, Pang X, Zimmerman MI, Zhou HX, Paravastu AK. Molecular structure of RADA16-I designer self-assembling peptide nanofibers. ACS Nano. 2013;7(9):7562–7572. doi: 10.1021/nn401562f pmid: 23977885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagarkar RP, Schneider JP. Synthesis and primary characterization of self-assembled peptide-based hydrogels. Methods Mol Biol. 2008;474:61–77. doi: 10.1007/978-1-59745-480-3_5 pmid: 19031061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye Z, Zhang H, Luo H, Wang S, Zhou Q, Du X, et al. Temperature and pH effects on biophysical and morphological properties of self-assembling peptide RADA16-I. J Pept Sci. 2008;14(2):152–162. doi: 10.1002/psc.988 pmid: 18196533 [DOI] [PubMed] [Google Scholar]
- 14.Holmes TC, de Lacalle S, Su X, Liu G, Rich A, Zhang S. Extensive neurite outgrowth and active synapse formation on self-assembling peptide scaffolds. Proc Natl Acad Sci U S A. 2000;97(12):6728–6733. doi: 10.1073/pnas.97.12.6728 pmid: 10841570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Semino CE, Merok JR, Crane GG, Panagiotakos G, Zhang S. Functional differentiation of hepatocyte-like spheroid structures from putative liver progenitor cells in three-dimensional peptide scaffolds. Differentiation. 2003;71(4-5):262–270. doi: 10.1046/j.1432-0436.2003.7104503.x pmid: 12823227 [DOI] [PubMed] [Google Scholar]
- 16.Semino CE, Kasahara J, Hayashi Y, Zhang S. Entrapment of migrating hippocampal neural cells in three-dimensional peptide nanofiber scaffold. Tissue Eng. 2004;10(3-4):643–655. doi: 10.1089/107632704323061997 pmid: 15165480 [DOI] [PubMed] [Google Scholar]
- 17.Narmoneva DA, Oni O, Sieminski AL, Zhang S, Gertler JP, Kamm RD, et al. Self-assembling short oligopeptides and the promotion of angiogenesis. Biomaterials. 2005;26(23):4837–4846. doi: 10.1016/j.biomaterials.2005.01.005 pmid: 15763263 [DOI] [PubMed] [Google Scholar]
- 18.Ellis-Behnke RG, Liang YX, Tay DK, Kau PW, Schneider GE, Zhang S, et al. Nano hemostat solution: immediate hemostasis at the nanoscale. Nanomedicine. 2006;2(4):207–215. doi: 10.1016/j.nano.2006.08.001 pmid: 17292144 [DOI] [PubMed] [Google Scholar]
- 19.Guo J, Su H, Zeng Y, Liang YX, Wong WM, Ellis-Behnke RG, et al. Reknitting the injured spinal cord by self-assembling peptide nanofiber scaffold. Nanomedicine. 2007;3(4):311–321. doi: 10.1016/j.nano.2007.09.003 pmid: 17964861 [DOI] [PubMed] [Google Scholar]
- 20.Guo J, Leung KK, Su H, Yuan Q, Wang L, Chu TH, et al. Self-assembling peptide nanofiber scaffold promotes the reconstruction of acutely injured brain. Nanomedicine. 2009;5(3):345–351. doi: 10.1016/j.nano.2008.12.001 pmid: 19268273 [DOI] [PubMed] [Google Scholar]
- 21.Kisiday J, Jin M, Kurz B, Hung H, Semino C, Zhang S, et al. Self-assembling peptide hydrogel fosters chondrocyte extracellular matrix production and cell division: implications for cartilage tissue repair. Proc Natl Acad Sci U S A. 2002;99(15):9996–10001. doi: 10.1073/pnas.142309999 pmid: 12119393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuliopulos A, Walsh CT. Production, purification, and cleavage of tandem repeats of recombinant peptides. J Am Chem Soc. 1994;116(11):4599–4607. doi: 10.1021/ja00090a008 [Google Scholar]
- 23.Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96(1):23–28. pmid: 2265755 [DOI] [PubMed] [Google Scholar]
- 24.Paradís‐Bas M, Tulla‐Puche J, Zompra AA, Albericio F. RADA‐16: A Tough Peptide–Strategies for Synthesis and Purification. Eur J Organ Chem. 2013;2013(26):5871–5878. doi: 10.1002/ejoc.201300612 [Google Scholar]
- 25.Reed DC, Barnard GC, Anderson EB, Klein LT, Gerngross TU. Production and purification of self-assembling peptides in Ralstonia eutropha. Protein Expr Purif. 2006;46(2):179–188. doi: 10.1016/j.pep.2005.08.023 pmid: 16249097 [DOI] [PubMed] [Google Scholar]
- 26.Mie M, Oomuro M, Kobatake E. Hydrogel scaffolds composed of genetically synthesized self-assembling peptides for three-dimensional cell culture Polymer J. 2013;45(5):504 doi: 10.1038/pj.2012.216 [Google Scholar]
- 27.Nevskaya NA, Chirgadze YN. Infrared spectra and resonance interactions of amide-I and II vibration of alpha-helix. Biopolymers. 1976;15(4):637–648. doi: 10.1002/bip.1976.360150404 pmid: 1252599 [DOI] [PubMed] [Google Scholar]
- 28.Sarroukh R, Goormaghtigh E, Ruysschaert JM, Raussens V. ATR-FTIR: a “rejuvenated” tool to investigate amyloid proteins. Biochim Biophys Acta. 2013;1828(10):2328–2338. doi: 10.1016/j.bbamem.2013.04.012 pmid: 23746423 [DOI] [PubMed] [Google Scholar]