Significance
Proteases precisely control a wide variety of physiological processes and thus are important drug targets. Compared with small-molecule inhibitors, monoclonal antibodies (mAbs) are attractive, as they provide required specificity. However, finding inhibitory mAbs is often a bottleneck, largely due to lack of a function-based selection method. We overcame this obstacle and successfully isolated mAbs that effectively inhibited 5 therapeutic targets spanning 4 basic classes of proteases. Our mAb inhibitors are highly selective and deliver desired biochemical and biological actions, including reduction of amyloid beta formation in vitro and pain relief in animal behavioral tests. Thus, the technique described here can be readily applied to many pathophysiologically important proteases for the discovery and engineering of therapeutic mAbs.
Keywords: protease inhibitor, monoclonal antibody, functional selection, MMP, BACE-1
Abstract
Critical for diverse biological processes, proteases represent one of the largest families of pharmaceutical targets. To inhibit pathogenic proteases with desired selectivity, monoclonal antibodies (mAbs) hold great promise as research tools and therapeutic agents. However, identification of mAbs with inhibitory functions is challenging because current antibody discovery methods rely on binding rather than inhibition. This study developed a highly efficient selection method for protease inhibitory mAbs by coexpressing 3 recombinant proteins in the periplasmic space of Escherichia coli—an antibody clone, a protease of interest, and a β-lactamase modified by insertion of a protease cleavable peptide sequence. During functional selection, inhibitory antibodies prevent the protease from cleaving the modified β-lactamase, thereby allowing the cell to survive in the presence of ampicillin. Using this method to select from synthetic human antibody libraries, we isolated panels of mAbs inhibiting 5 targets of 4 main protease classes: matrix metalloproteinases (MMP-14, a predominant target in metastasis; MMP-9, in neuropathic pain), β-secretase 1 (BACE-1, an aspartic protease in Alzheimer’s disease), cathepsin B (a cysteine protease in cancer), and Alp2 (a serine protease in aspergillosis). Notably, 37 of 41 identified binders were inhibitory. Isolated mAb inhibitors exhibited nanomolar potency, exclusive selectivity, excellent proteolytic stability, and desired biological functions. Particularly, anti-Alp2 Fab A4A1 had a binding affinity of 11 nM and inhibition potency of 14 nM, anti-BACE1 IgG B2B2 reduced amyloid beta (Aβ40) production by 80% in cellular assays, and IgG L13 inhibited MMP-9 but not MMP-2/-12/-14 and significantly relieved neuropathic pain development in mice.
Proteases regulate important physiological processes and are involved in essential pathogenesis of viruses, and therefore represent attractive targets for pharmaceutical development (1, 2). Human genome contains 569 proteases of 68 families (3), belonging to 5 main classes based on their catalytic mechanisms: aspartic, cysteine, threonine, serine, and metalloproteases. Existing in a delicate balance of networks with their endogenous inhibitors and substrates, proteases maintain the body’s homeostasis. Dysregulation of proteolysis, however, causes a variety of diseases ranging from cancer, inflammation, and cardiovascular disorders to osteoporosis, neuropathic pain, and degenerative diseases (4–9). Notably, it has been estimated that proteases account for 5 to 10% of all drug targets being studied for therapeutic development (10). Numerous protease inhibitors are currently on the market for the treatment of coagulation, hypertension, viral infection, cancer, and diabetes through targeting well-established proteases such as angiotensin-converting enzyme and HIV proteases (10). Nevertheless, small-molecule inhibitors are often limited by lack of specificity and/or appropriate pharmacokinetic properties required for effective and safe protease-based therapy (11). These challenges are well reflected by failed clinical trials of historical attempts to inhibit matrix metalloproteinases (MMPs) as a cancer therapy (12, 13), and more recent ones to inhibit β-secretase 1 (BACE-1) for the remediation of Alzheimer's disease (14). Consequently, despite decades of intensive efforts, conventional drug discovery strategies have only achieved a limited success by targeting a tiny fraction of all therapeutically relevant proteases.
Biologics, for example, monoclonal antibodies (mAbs), provide exquisite specificity capable of distinguishing between closely related protease family members (15–21). Their stability in serum, potential to cross blood–brain barrier, novel design as prodrugs, and improved effector functions offer significant advantages over small-molecule therapeutics (22–24). In addition to pharmaceutical applications, specific inhibitory mAbs targeting individual proteases of biomedical importance are highly valuable as research tools to characterize protease degradomes. Since the invention of mAb technology in 1975, tremendous progress has been made in mAb discovery and engineering that forms the core of the biopharmaceutical industry today. However, current mAb selection/screening methods, including hybridoma, phage panning, single-cell PCR, cell surface display coupled flow cytometry, and so forth, are primarily based on binding affinities but not inhibitory function. Potentially important inhibitory clones that have weak binding affinities can be lost during the repeated cycles of competitive binding. As a result, there is a high level of probability that few or none of the selected binders are inhibitory (25). Furthermore, generated mAbs often exhibit suboptimal inhibition properties and are vulnerable to being cleaved by the protease target (26). Aiming to facilitate the direct identification of protease inhibitory mAbs with desired pharmacological properties, we developed a functional selection method, built on our experience with periplasmic coexpression of Ab fragments and active human proteases (27). To demonstrate the feasibility and generality of our method, we selected 5 drug targets representing 4 protease classes.
Results
Design of Functional Selection for Protease Inhibitory mAbs.
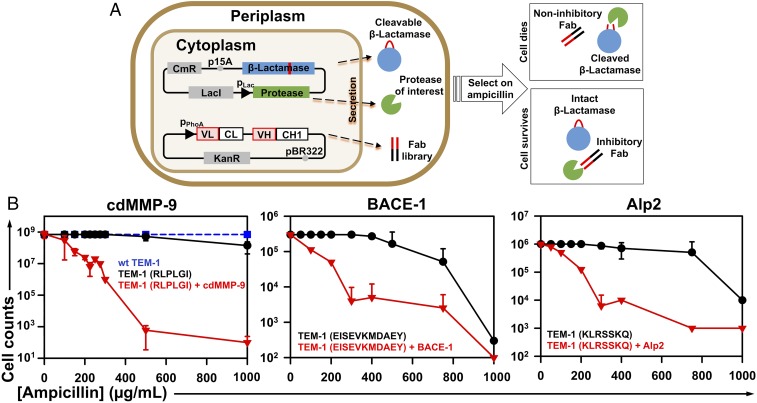
To select mAbs that inhibit proteases, 3 recombinant proteins—a clone from an antibody library, the protease of interest, and the protease substrate acting as an in vivo sensor—must be produced in the same location. We hypothesize that Escherichia coli periplasmic coexpression is ideal for this task because the oxidative environment and associated molecular chaperons facilitate disulfide formation needed to produce antibody fragments and many human proteases in their active form. In addition, large combinatorial libraries have been routinely constructed in E. coli thanks to its high transformation efficiency. The crucial aspect of this method is a cellular protease inhibition sensor; our design is to engineer β-lactamase TEM-1, a periplasmic hydrolase of β-lactam antibiotics, by inserting a protease-specific cleavable peptide sequence. When the modified TEM-1 is cleaved by the protease of interest, it loses its β-lactam hydrolytic activity, and thus the cell cannot grow in the presence of ampicillin. Conversely, when proteolytic activity of the target is blocked by a coexpressed antibody, TEM-1 is spared to confer ampicillin resistance to the host cell. Therefore, this live or die selection can identify antibody clones that specifically inhibit the activity of the targeted protease (Fig. 1A).
Fig. 1.
Functional selection design for protease inhibitory antibodies. (A) Scheme showing that 3 recombinant proteins are simultaneously coexpressed in the periplasmic space of E. coil—a clone from the Fab library, the protease of interest, and the modified β-lactamase TEM-1 with a cleavable peptide insertion. The protease extracellular/cd under a lac promoter and TEM-1 under its native promoter are cloned into a low copy number (p15A ori) plasmid of chloramphenicol resistance (CmR). The antibody Fab library under a phoA promoter is cloned into a medium copy number (pBR322 ori) plasmid carrying kanamycin resistance (KanR). If the Fab has no inhibition, the protease will cleave TEM-1 leading to cell death in the presence of ampicillin. An inhibitory Fab blocks proteolytic activity which allows TEM-1 to remain intact, resulting in cell growth on ampicillin plates. (B) Selection windows for cdMMP-9 (Left), BACE-1 (Middle), and Alp2 (Right) inhibitors. TEM-1 was modified by inserting the protease-specific cleavage peptide sequences (shown in parentheses) between Gly196 and Glu197 of TEM-1 (SI Appendix, Fig. S2). At 0 to 1,000 µg/mL ampicillin, survival curves of E. coli cells transformed with modified TEM-1s without protease genes were measured (black circles) and compared with those for cells coexpressing both modified TEM-1s and the associated proteases (red triangles). The survival curve with WT TEM-1 is shown as a blue dashed line. Experiments were repeated 3 times with 2×YT agar plates containing 0.1 mM IPTG.
To demonstrate the generality of this functional selection method, we chose 5 disease-associated targets from 4 major classes of proteases: MMP-9 (neuropathic pain) (28), MMP-14 (metastasis) (29), aspartic protease BACE1 (Alzheimer’s disease) (30), serine protease Alp2 of Aspergillus fumigatus (aspergillosis) (31), and cysteine protease cathepsin B (cancer and neurodegenerative disorders) (32). The extracellular/catalytic domains (cd) of these targets without their propeptide sequences were cloned downstream of a pLac promoter and a pelB leader for periplasmic expression. Enzymatic assays showed that produced proteases were functional with expected activities (SI Appendix, Fig. S1). Yields of 0.5 to 2.0 mg of active soluble proteases per liter of culture were typically achieved (except BACE1, which yielded 0.1 µg per liter culture), suggesting the feasibility of their inhibition by coexpressed Fabs, which are usually produced at similar level in periplasm (27).
Distinct Selection Windows for Protease Inhibitors.
To select inhibitory mAbs with high potencies, protease-specific substrates with relatively fast kinetics (kcat/Kms) were used for TEM-1 insertion sequence design. Synthetic peptide substrates RLPLGI, SGRIGFLRTA, and KLHFSKQ were chosen for cdMMP-9, cdMMP-14, and cathepsin B, respectively. For BACE1, a peptide sequence EISEVKMDAEY was derived from its physiological substrate amyloid precursor protein (APP). As the physiological substrate of Alp2 is unknown, for its TEM-1 design, we chose a generic serine protease substrate sequence KLRSSKQ, which can be efficiently cleaved by Alp2 in vitro (SI Appendix, Fig. S1). Flanked by flexible serine−glycine linkers at both ends, that is, GSG−peptide−SGG, these cleavable peptide sequences were introduced between Gly196 and Glu197 of TEM-1 (SI Appendix, Fig. S2). This site is located on an exposed surface loop away from the β-lactamase active center and has been exploited for the construction of cellular sensors (33, 34). Survival curves of E. coli cells expressing modified TEM-1s without carrying genes of associated proteases were measured on agar plates supplemented with 0 to 1,000 µg/mL ampicillin. Results showed that the minimal inhibitory concentrations (MICs) were 500 µg/mL or higher (Fig. 1B and SI Appendix, Fig. S3), suggesting that peptide insertion did not significantly disrupt TEM-1 catalytic function, although the activities of modified TEM-1s were less than that of wild-type (WT) TEM-1, especially at high ampicillin concentrations (Fig. 1B, dashed line). However, when the associated proteases were periplasmically coexpressed, the MICs dramatically decreased to 200 µg/mL or lower, indicating that proteolytic cleavages of modified TEM-1s resulted in loss of their β-lactamase activity. Overall, the significant disparity between the survival curves with (red triangles) and without (black circles) coexpressed proteases provided distinct windows for effective selection of inhibitors. To optimize selection conditions, protease expression level was also up- or down-regulated by adding isopropyl β-D-1-thiogalactopyranoside (IPTG) or glucose. With a relatively high yield and high catalytic activity, the leakage expression of cdMMP-14 in the presence of 2% glucose was sufficient to effectively cleave the associated TEM-1(SGRIGFLRTA) and resulted in an MIC of 150 µg/mL (SI Appendix, Fig. S3). As too much protease activity would result in the selection of inhibitors with high potency or high expression, and thus reduce the diversity of isolated clones, keeping cdMMP-14 expression at low levels should be beneficial. Therefore, 200 µg/mL ampicillin with 2% glucose was determined as the condition for cdMMP-14 inhibitory antibody selection. In contrast, other tested protease targets exhibited low expression levels and/or low activities, and thus overexpression is favorable to boost protease production. Therefore, 300 µg/mL ampicillin with 0.1 mM IPTG was applied for these proteases (SI Appendix, Table S1). Overall, these conditions yielded a 100% survival rate in the absence of protease and nearly complete cell death in the presence of protease, with the survival rates 2 to 3 orders of magnitudes lower.
Isolation of Multiple Potent mAb Inhibitors for Each Protease.
To promote generation of inhibitory mAbs targeting protease reaction clefts (21), a human Fab synthetic library of 1.1 × 109 diversity encoding CDR-H3s with 23 to 27 amino acids was constructed downstream of a phoA promoter and an STII leader (Fig. 1A). Obtained Fab library plasmids were transformed to E. coli competent cells bearing the reporter plasmids for each protease. Libraries of 1.5 to 8.6 × 108 diversity were generated and subjected to functional selection for each protease inhibition under predetermined conditions (SI Appendix, Table S1). Surviving colonies were then individually screened by culturing in liquid media under more stringent conditions, that is, by increasing ampicillin concentration by 100 µg/mL. Taking anti-cdMMP-14 as an example, 190 clones survived after the initial selection, and the secondary screening narrowed them down to 161. Among them, 40 were randomly picked for DNA sequencing, and 38 unique Fabs were identified. Applying similar procedures, 5 to 20 monoclonal Fabs were successfully discovered for each of the other tested proteases (SI Appendix, Table S1).
Three to 8 Fabs from each selection experiment (total 41) were produced for biochemical characterizations (Table 1). Binding affinity measurements by biolayer interferometry and ELISA confirmed that all of the tested Fabs bound to their protease targets with dissociation constants (KDs) ranging from less than 10 nM to more than 1 µM (SI Appendix, Fig. S4). Among the tested 41 Fabs, 20 of them had KD values < 250 nM. Particularly, anti-MMP9 Fab H4, anti-BACE1 Fab B3B12, anti-Alp2 Fab A4A1, and anti-cathepsin B Fab CBA3 exhibited a KD of 6.9, 10, 11, and 16 nM, respectively. Inhibitory functions of purified Fabs were assayed with associated proteases and their fluorescence resonance energy transfer (FRET) peptide substrates. Results indicated that 37 out of 41 tested Fabs were inhibitors (SI Appendix, Fig. S5), equivalent to a 90% overall success rate, that is, the ratio of inhibitors over binders. Among these noninhibitors, 2 anti-MMP9 and 2 anti-Alp2 Fabs did not prevent their target proteases from cleaving the peptide substrates. Among isolated inhibitory Fabs, 57% of them (21 Fabs) showed potent inhibition with calculated inhibition constant (KI) values < 250 nM (Table 1). Particularly, Fabs H4, B3B12, A4A1, and CBA3 had a KI of 57, 26, 14, and 91 nM, respectively. Converting 2 anti-MMP9 and 2 anti-BACE1 inhibitory Fabs of nanomolar potencies into their IgG format improved their affinities and potencies as expected; for example, B3B12 IgG exhibited KD of 7.2 nM and KI of 16 nM (Table 1). Rapid isolation of multiple potent inhibitory mAbs targeting all 5 tested proteases from >108 library clones demonstrated the effectiveness and robustness of this selection system.
Table 1.
Isolated inhibitory antibodies toward 4 classes of proteases
| Type | Target | Peptide insert | Fab* | CDR-H3 (length) | KD (nM)† | KI (nM)† |
| Metallo- | MMP-9 | RLPLGI | H4 | SSLAWAQDRVYKPVEAMTWAYGMDY (25) | 6.9 | 57 |
| (neuropathic | H3 | RFEPGLLKRNKRWISYTLCEAGYGMDY (27) | 98 (71)‡ | 71 (52)‡ | ||
| pain) | L13 | KYMVFGTRMGWVEHTDFAGQGYYAMDY (27) | 120 (53)‡ | 160 (120)‡ | ||
| H25 | CKLYTSYMIPVGSDSVNRCMSSYGMDY (27) | 450 | 300 | |||
| MMP-14 | SGRIGFLRTA | 2B4 | SDSAVYSVRRMGSSGLAAYAMDY (23) | 66 | 95 | |
| (cancer) | 2B12 | DCCSCVFSQSAGITLACVYVMDY (23) | 76 | 130 | ||
| 1A5 | LDFLMRDIYYDLGGGALGWLIKYAMDY (27) | 57 | 160 | |||
| 2B10 | QLFACWRQSILTPPLLSAMMMGYAMDY (27) | 46 | 220 | |||
| 2D9 | GVTRFTNDASVGQVWAGAYGMDY (23) | 260 | 240 | |||
| 2A6 | VVRMLPVRCIPRCIKTTLPLYGMDY (25) | 130 | 440 | |||
| Aspartic | BACE-1 | EISEVKMDAEY | B3B12 | YICGHRWRDFDMWRARTGVNYAMDY (25) | 10 (7.2)‡ | 26 (16)‡ |
| (Alzheimer’s) | B1A4 | HYYVSVGSGIDY (12) | 20 (16)‡ | 41 (37)‡ | ||
| B2B5 | WHGYPPGYSYYSSFSSSGFDY (21) | 130 | 80 | |||
| B2B2 | YWGYYAWFGSHPWAYGAFDY (20) | 52 | 85 | |||
| B2B3 | SASGIDY (7) | 100 | 110 | |||
| B2B9 | SSSSYYYGMDY (11) | 110 | 120 | |||
| B2B6 | DNSICVLTQKEVDTKFLVGQHSYVMDY (27) | 28 | 130 | |||
| B1B3 | ERSSCPVGWRDSRFGADGYGLEY (23) | 31 | 210 | |||
| Serine | Alp2 | KLRSSKQ | A4A1 | FGSWSYAIDY (10) | 11 | 14 |
| (aspergillosis) | A4A2 | KTSDQYLLVGGSFFKLRDCCYVMDY (25) | 110 | 220 | ||
| A4A7 | GRSPGPYAVCGNLFRSVSYGMDY (23) | 420 | 300 | |||
| Cysteine | cathepsin B | KLHFSKQ | CBA3 | GFAWSPGLDY (10) | 16 | 91 |
| (cancer) | CBA2 | YGYPGGYHFWGWWSSPYAFDY (21) | 200 | 120 | ||
| CBA1 | GGGSWSAMDY (10) | 300 | 210 |
Only Fabs with inhibition constants (KIs) < 500 nM are shown. Clones are ranked by their KI values.
See statistical analysis of KD and KI in SI Appendix, Table S2.
Data of associated IgGs are shown in parentheses.
Inhibitory mAbs Are Highly Selective, Functional on Physiological Substrates, and Proteolytically Stable.
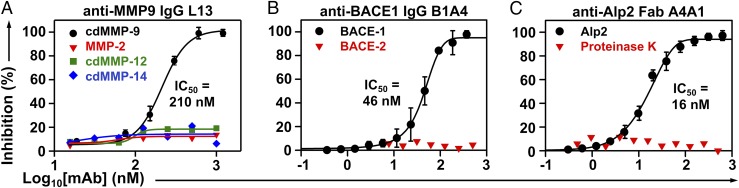
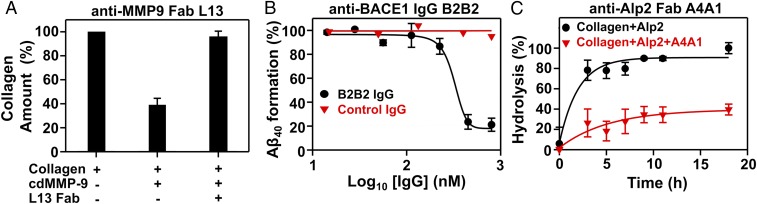
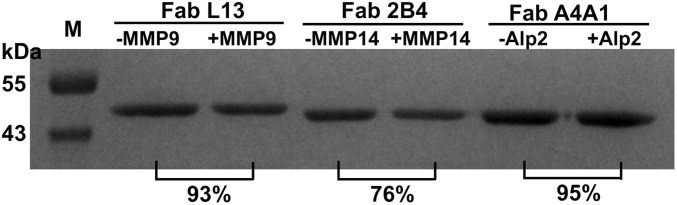
Inhibition selectivity of 11 representative mAbs was tested. As results in Fig. 2A and SI Appendix, Fig. S5 show, IgGs H3 and L13 inhibited cdMMP-9 with KIs of 52 and 120 nM, respectively, but did not inhibit structurally conserved (cd)MMP-2/-12/-14 (56 to 59% sequence similarity over cdMMP-14), even at 1 µM. Fabs 2B4, 2B12, and 1A5 inhibited cdMMP-14 with KIs of 95 to 160 nM but did not inhibit cdMMP-9 at 500 nM (SI Appendix, Fig. S5). Similarly, IgGs B3B12 and B1A4 inhibited BACE-1 with nanomolar potency (KI = 16 and 37 nM, respectively) but not BACE-2 (Fig. 2B and SI Appendix, Fig. S5) with its cd sharing 70% sequence with BACE-1; Fab A4A1 inhibited Alp2 (KI = 14 nM) but not proteinase K, a broad-spectrum serine protease (Fig. 2C); and Fabs CBA3, CBA2, and CBA1 inhibited cathepsin B completely but not cathepsin K at the same Fab concentrations (SI Appendix, Fig. S5). Next, anti-cdMMP-9, anti-Alp2, and anti-BACE1 mAbs were exanimated for their inhibitory functions on the associated physiological/macromolecular substrates. Under tested conditions, Fab L13 reduced the degradation of type I collagen by cdMMP-9 from 61 to 4% (Fig. 3A), Fab A4A1 reduced the hydrolysis of FITC-conjugated collagen by Alp2 from 90 to 38% (Fig. 3C), and IgG B1A4 reduced cleavage of APP571–696 by BACE-1 from 86 to 37% (SI Appendix, Fig. S6). Furthermore, in in vitro assays using HEK293 cells expressing APP, IgG B2B2 reduced amyloid beta (Aβ40) production 80% in a dose-dependent manner with an apparent IC50 of 330 nM (Fig. 3B). To test proteolytic stability of inhibitory antibodies with target proteases in vitro, 1 µM purified Fabs were incubated with 1 µM respective protease at 37 °C. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE) revealed that, after exposure to equal molar of the protease for 24 h, Fabs L13, 2B4, and A4A1 remained 93%, 76%, and 95% intact (Fig. 4).
Fig. 2.
Inhibition potency and selectivity. (A) Anti-MMP9 IgG L13, (B) anti-BACE1 IgG B1B4, and (C) anti-Alp2 Fab A4A1. Inhibition assays were measured using FRET peptide substrates.
Fig. 3.
Inhibitory functions of isolated mAbs on proteolysis of physiological/macromolecular substrates. (A) Fab L13 blocked MMP-9 from hydrolyzing type I collagen. (B) Inhibition of Aβ40 formation by IgG B2B2 in cellular assays. HEK293F cell cultures expressing APP571–696 were incubated with IgG for 72 h. Generated Aβ40 was measured by ELISA. (C) Fab A4A1 blocked Alp2 from hydrolyzing FITC-conjugated type I collagen.
Fig. 4.
In vitro proteolytic stability with target proteases. SDS/PAGE of 1 µM Fabs after incubation with 1 µM of the target protease for 12 h. The relative amounts of remaining Fabs were determined by densitometric analysis.
Identification of Active Site and Exosite Inhibitors.
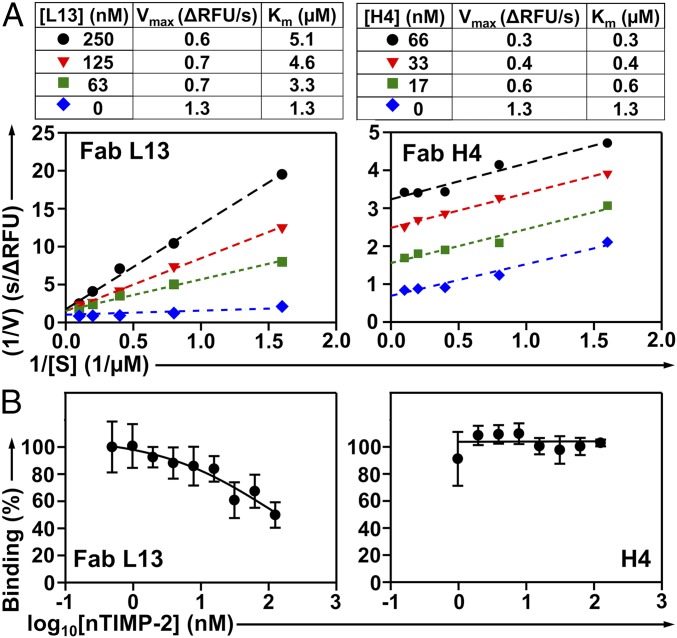
To understand the inhibition mechanism, we measured cdMMP-9 kinetics in the presence of various concentrations of its inhibitory mAbs. When concentration of Fab L13 increased from 0 to 250 nM, the Lineweaver–Burk plots of cdMMP-9 indicated an unaltered maximum velocity Vmax and increased Michaelis constant Km, suggesting that Fab L13 performed as a competitive inhibitor (Fig. 5A). In contrast, with increasing concentrations of Fab H4, the kinetics of cdMMP-9 showed decreases on both Vmax and Km values, suggesting an uncompetitive inhibition mode. In addition, ELISA of Fab L13 on immobilized cdMMP-9 with the presence of nTIMP-2, an endogenous inhibitor of MMP-9 recognizing its reaction cleft (35), revealed that high concentrations of nTIMP-2 displaced L13 on binding to cdMMP-9 (Fig. 5B). However, nTIMP-2 did not interfere with H4 on its binding to cdMMP-9 in a similar competitive ELISA. These results suggest that L13 had its epitope overlapping with that nTIMP-2, while H4 did not share any epitope with nTIMP-2. Collectively, Fab L13 is an active site competitive inhibitor, and Fab H4 is an exosite uncompetitive inhibitor presumably delivering its blockage function by an allosteric mechanism.
Fig. 5.
Inhibition mechanisms of anti-MMP9 Fabs. (A) Lineweaver–Burk plots of cdMMP-9 in the presence of 62.5, 250, and 500 nM Fab L13 (Left) or 66, 33, and 16.5 nM Fab H4 (Right). (B) Competitive ELISA of Fab L13 (Left) or Fab H4 (Right) on immobilized cdMMP-9 in the presence of 1 to 125 nM nTIMP-2.
Anti-MMP9 IgG L13 Exhibits Neuropathic Pain Attenuation Efficacy In Vivo.
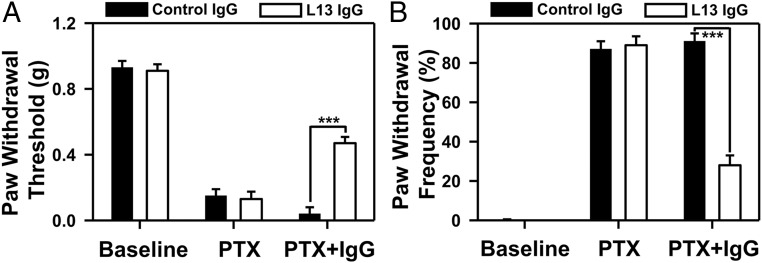
As MMP-9 is required in the early phase of neuropathic pain development after nerve injury (28), we further evaluated the pain relief efficacy of MMP-9 inhibitory IgG L13 in paclitaxel (PTX)-induced neuropathic pain in mice. PTX evoked robust mechanical allodynia, a cardinal feature of neuropathic pain, by decreasing paw withdrawal threshold (Fig. 6A) and increasing paw withdrawal frequency to a subthreshold filament (0.6 g; Fig. 6B). In contrast to polyclonal human control IgG which did not change the animals’ behavioral responses, intrathecal injection of 200 µg of IgG L13, given 15 d after the first PTX administration, significantly increased the withdrawal threshold and reduced the withdrawal frequency (P < 0.001, 2-way ANOVA) (Fig. 6).
Fig. 6.
Analgesic effects of MMP-9 inhibitor IgG L13 in neuropathic pain induced by the chemotherapy agent paclitaxel (PTX) in male mice; 200 ng IgG L13 was intravenously administered on day 15 after PTX injections. Behavioral tests of neuropathic pain symptom mechanical allodynia, evaluated by paw withdrawal threshold (A) and frequency (B), were performed in a blinded manner (n = 7 mice for control IgG, and n = 6 mice for L13 IgG). ***P < 0.001, 2-way ANOVA with Tukey’s post hoc test.
Discussion
In this study, we chose 5 disease-associated proteases representing 4 basic classes with diverse catalytic chemistries and surface topologies (SI Appendix, Fig. S7). More specifically, while MMPs and cathepsins display well-pronounced catalytic clefts and BACE-1 shows a large reaction cavity, the active site of Alp2 is relatively small and shallow. Interestingly, when 2 synthetic Fab libraries carrying CDR-H3s of 23 to 27 or 5 to 21 amino acids were separately applied for BACE-1 (SI Appendix, Table S1), we were able to isolate inhibitory mAbs from both libraries. Among 8 potent anti-BACE1 inhibitors, 5 of them carry long CDR-H3s (>20 aa), and the remaining 3 have short CDR-H3s (7 to 12 aa) (Table 1). However, when these 2 synthetic Fab libraries were combined and jointly applied for Alp2, the most potent Fab isolated (A4A1 with a KI of 14 nM) was derived from the short CDR-H3 library. These results suggest that, at least for active site inhibitors, appropriate CDR-H3 length distribution could be a considerable factor to accommodate desired paratope conformations that are compatible with targeted protease topology.
Our functional selection achieved an exceptionally high success rate of 90%, that is, 37 out of 41 tested binders were inhibitors. We further analyzed these 4 noninhibitory clones, to reason how they could survive during this live or die selection. Anti-MMP9 Fabs H44 and L6 exhibited binding affinities of 520 ± 130 nM and 230 ± 95 nM, respectively, but did not inhibit cdMMP-9 from cleaving its FRET peptide substrate. To define their epitopes, we conducted competitive ELISA on immobilized cdMMP-9 in the presence of nTIMP-2. Results showed that high concentrations of nTIMP-2 displaced Fabs on binding to cdMMP-9 (SI Appendix, Fig. S8), indicating that H44 and L6 Fabs had their epitopes at least partially overlapping with that of nTIMP-2, and thus could block the access of macromolecular substrates such as TEM-1(RLPLGI) that allowed cell survival during the selection. Unfortunately, similar competitive ELISA cannot be carried out for anti-Alp2 because Alp2 macromolecular inhibitors are not available. Interestingly, however, 2 noninhibitory anti-Alp2 Fabs were isolated with a peptide insertion KLRSSKQ which was derived from a generic substrate of serine proteases. Notably, the survival rates of E. coli cells coexpressing Alp2 and TEM-1(KLRSSKQ) gradually decreases, then plateaus when ampicillin concentration increases (Fig. 1 B, Right). This suboptimal survival curve implies the chance that noninhibitory clones are able to escape from the ampicillin selection. Therefore, the outcomes of noninhibitory clones could be potentially remedied by applying insertion peptide sequences with high cleaving efficiency and/or performing additional rounds of selection with more stringent conditions.
Other than antibody library and peptide insertion sequence designs, the selection conditions, such as concentrations of ampicillin and inducer, culture media, and temperature, can be customized for each protease target, allowing rapid downsizing of libraries. Our selection resulted in numerous clones after the secondary screening (e.g., 161 anti-MMP14 and 73 anti-BACE1), of which only small subsets were randomly picked for full characterizations, due to time constrain. Therefore, it is likely that additional inhibitory mAbs could be identified from the remaining uncharacterized pools. Among tested mAbs, more than half of identified inhibitors had a potency KI < 250 nM, while some showed a weaker potency (KI > 1 µM). Considering that all these mAbs were isolated from synthetic libraries, ranges of different affinity/potency were expected. Interestingly, we also found that highly potent anti-BACE1 B3B12 and B1A4 were produced at low yields with 0.1 mg or less purified Fabs per liter of culture, while low-potency B2B5 and B2B2 Fabs were generated at higher yield, with 0.56 and 1.3 mg per liter of culture (SI Appendix, Table S3). Presumably, these weak inhibitors were isolated because of their high titers which can compensate for their low potency. In addition, our approach of periplasmic coexpression facilitates the disulfide formation required for activities of many human proteases; for example, cds of BACE1 and cathepsin B have 3 and 6 disulfide bonds, respectively. Furthermore, proteases were produced in their propeptide-free form; thus isolated mAbs can directly inhibit the activated proteases. Certain macromolecular inhibitors of proteases, especially the canonical mechanism inhibitors including endogenous inhibitors and inhibitory mAbs, tend to be slowly cleaved by the targeted protease (36). However, the mAbs isolated in this study exhibited excellent proteolytic stability (Fig. 4). It is likely that this benefited from the nature of in vivo selection, because inhibitory function and thus the integrity of Fabs must be maintained over the entire course for cell survival.
In summary, this study described a high-throughput method for selecting protease inhibitory mAbs. Compared with recent technology developments such as epitope synthetic mimicry (37), convex paratope design (21), competitive phage elution (16), cytoplasmic genetic selection (38), and epitope-specific fluorescence-activated cell sorter (39), this method directly relies on functional inhibition and offers the following advantages: 1) an exceptionally high successful rate as the ratio of inhibitors over binders (SI Appendix, Table S1); 2) exclusive selectivity against proteases of the same family (Fig. 2 and SI Appendix, Fig. S5); 3) relatively high proteolytic resistance to the target proteases (Fig. 4); 4) both active-site and allosteric inhibitors can be identified (Fig. 5); and 5) multiple inhibitory mAbs are available for providing a rich pool for lead candidate characterization and optimization (Table 1). Considering that 2% of the human genome encode proteases and half of them are extracellular and thus targetable by mAbs, the method developed here can be readily applied for the discovery of mAbs that are able to inhibit many therapeutically important proteases. Notably, lanadelumab, a plasma kallikrein (pKal) inhibitor, was approved by the Food and Drug Administration in 2018 as the first protease inhibitory mAb for treating hereditary angioedema. As numerous BACE-1 compound inhibitors failed in clinical trials, we sincerely hope our technique can promote mAb discovery for protease inhibitors. In addition to mAbs, other biologics, especially endogenous inhibitors of proteases, can be engineered using the similar approach.
Materials and Methods
Protease cleavable peptide sequences with flanking serine−glycine linkers were assembled by using synthetic oligonucleotides and cloned between G196 and E197 of TEM-1 to generate modified TEM-1 genes. Reporter plasmids for periplasmic coexpression of proteases of interest and associated modified TEM-1s were constructed and tested for β-lactamase activities on agar plates containing 0 to 1,000 µg/mL ampicillin. Synthetic human Fab libraries were transformed to cells harboring reporter plasmids and subjected to inhibitory selection under predetermined conditions for each protease. Fabs and IgGs of isolated antibody clones were produced and biochemically characterized by biolayer interferometry, ELISA, and inhibition assays using FRET peptides and macromolecular substrates. Biological functions of anti-MMP9 IgG L13 were also tested with neuropathic pain mouse model. All animal experiments were approved by the Institutional Animal Care and Use Committees (IACUC) of Duke University. Detailed experimental procedures are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants R01GM115672 (to X.G.) and R01DE17794 (to R.-R.J.), and National Science Foundation CAREER 1453645 (to X.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1903330116/-/DCSupplemental.
References
- 1.Turk B., Turk D., Turk V., Protease signalling: The cutting edge. EMBO J. 31, 1630–1643 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deu E., Verdoes M., Bogyo M., New approaches for dissecting protease functions to improve probe development and drug discovery. Nat. Struct. Mol. Biol. 19, 9–16 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.López-Otín C., Bond J. S., Proteases: Multifunctional enzymes in life and disease. J. Biol. Chem. 283, 30433–30437 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.López-Otín C., Matrisian L. M., Emerging roles of proteases in tumour suppression. Nat. Rev. Cancer 7, 800–808 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Prassas I., Eissa A., Poda G., Diamandis E. P., Unleashing the therapeutic potential of human kallikrein-related serine proteases. Nat. Rev. Drug Discov. 14, 183–202 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Singh R. B., Dandekar S. P., Elimban V., Gupta S. K., Dhalla N. S., Role of proteases in the pathophysiology of cardiac disease. Mol. Cell. Biochem. 263, 241–256 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Troeberg L., Nagase H., Proteases involved in cartilage matrix degradation in osteoarthritis. Biochim. Biophys. Acta 1824, 133–145 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ji R. R., Xu Z. Z., Wang X., Lo E. H., Matrix metalloprotease regulation of neuropathic pain. Trends Pharmacol. Sci. 30, 336–340 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Strooper B., Proteases and proteolysis in Alzheimer disease: A multifactorial view on the disease process. Physiol. Rev. 90, 465–494 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Drag M., Salvesen G. S., Emerging principles in protease-based drug discovery. Nat. Rev. Drug Discov. 9, 690–701 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turk B., Targeting proteases: Successes, failures and future prospects. Nat. Rev. Drug Discov. 5, 785–799 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Overall C. M., Kleifeld O., Towards third generation matrix metalloproteinase inhibitors for cancer therapy. Br. J. Cancer 94, 941–946 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vandenbroucke R. E., Libert C., Is there new hope for therapeutic matrix metalloproteinase inhibition? Nat. Rev. Drug Discov. 13, 904–927 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Vassar R., BACE1 inhibitor drugs in clinical trials for Alzheimer’s disease. Alzheimers Res. Ther. 6, 89 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Y., et al. , Structural insight into distinct mechanisms of protease inhibition by antibodies. Proc. Natl. Acad. Sci. U.S.A. 104, 19784–19789 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devy L., et al. , Selective inhibition of matrix metalloproteinase-14 blocks tumor growth, invasion, and angiogenesis. Cancer Res. 69, 1517–1526 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Atwal J. K., et al. , A therapeutic antibody targeting BACE1 inhibits amyloid-β production in vivo. Sci. Transl. Med. 3, 84ra43 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Schneider E. L., et al. , A reverse binding motif that contributes to specific protease inhibition by antibodies. J. Mol. Biol. 415, 699–715 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenniston J. A., et al. , Inhibition of plasma kallikrein by a highly specific active site blocking antibody. J. Biol. Chem. 289, 23596–23608 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.David T., et al. , Factor XIa-specific IgG and a reversal agent to probe factor XI function in thrombosis and hemostasis. Sci. Transl. Med. 8, 353ra112 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Nam D. H., Rodriguez C., Remacle A. G., Strongin A. Y., Ge X., Active-site MMP-selective antibody inhibitors discovered from convex paratope synthetic libraries. Proc. Natl. Acad. Sci. U.S.A. 113, 14970–14975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu Y. J., et al. , Therapeutic bispecific antibodies cross the blood-brain barrier in nonhuman primates. Sci. Transl. Med. 6, 261ra154 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Sharma S. K., Bagshawe K. D., Antibody directed enzyme prodrug therapy (ADEPT): Trials and tribulations. Adv. Drug Deliv. Rev. 118, 2–7 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Wang X., Mathieu M., Brezski R. J., IgG Fc engineering to modulate antibody effector functions. Protein Cell 9, 63–73 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang J., et al. , Identification of inhibitory scFv antibodies targeting fibroblast activation protein utilizing phage display functional screens. FASEB J. 27, 581–589 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee K. B., Dunn Z., Ge X., Reducing proteolytic liability of a MMP-14 inhibitory antibody by site-saturation mutagenesis. Protein Sci. 28, 643–653 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nam D. H., Ge X., Direct production of functional matrix metalloproteinase–14 without refolding or activation and its application for in vitro inhibition assays. Biotechnol. Bioeng. 113, 717–723 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Kawasaki Y., et al. , Distinct roles of matrix metalloproteases in the early- and late-phase development of neuropathic pain. Nat. Med. 14, 331–336 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itoh Y., Seiki M., MT1-MMP: A potent modifier of pericellular microenvironment. J. Cell. Physiol. 206, 1–8 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Vassar R., Kovacs D. M., Yan R., Wong P. C., The β-secretase enzyme BACE in health and Alzheimer’s disease: Regulation, cell biology, function, and therapeutic potential. J. Neurosci. 29, 12787–12794 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abad A., et al. , What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev. Iberoam. Micol. 27, 155–182 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Gondi C. S., Rao J. S., Cathepsin B as a cancer target. Expert Opin. Ther. Targets 17, 281–291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galarneau A., Primeau M., Trudeau L. E., Michnick S. W., Beta-lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein protein interactions. Nat. Biotechnol. 20, 619–622 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Porter J. R., Stains C. I., Segal D. J., Ghosh I., Split β-lactamase sensor for the sequence-specific detection of DNA methylation. Anal. Chem. 79, 6702–6708 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Fernandez-Catalan C., et al. , Crystal structure of the complex formed by the membrane type 1-matrix metalloproteinase with the tissue inhibitor of metalloproteinases-2, the soluble progelatinase A receptor. EMBO J. 17, 5238–5248 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farady C. J., Craik C. S., Mechanisms of macromolecular protease inhibitors. ChemBioChem 11, 2341–2346 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sela-Passwell N., et al. , Antibodies targeting the catalytic zinc complex of activated matrix metalloproteinases show therapeutic potential. Nat. Med. 18, 143–147 (2011). [DOI] [PubMed] [Google Scholar]
- 38.Gal-Tanamy M., et al. , HCV NS3 serine protease-neutralizing single-chain antibodies isolated by a novel genetic screen. J. Mol. Biol. 347, 991–1003 (2005). [DOI] [PubMed] [Google Scholar]
- 39.Lopez T., et al. , Epitope-specific affinity maturation improved stability of potent protease inhibitory antibodies. Biotechnol. Bioeng. 115, 2673–2682 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.