Significance
Integrating evolutionary and ecological factors is an essential step toward reaching a more complete understanding of the mechanisms underlying biodiversity patterns. While oceanic islands are often used as natural laboratories, typical analyses lack information about species abundances and distributions within islands that is necessary to disentangle the effects of biogeographical and ecological factors on biodiversity. We analyze forests across the Hawaiian archipelago and show that the effects of island age on diversity patterns of native species percolate across spatial scales via the distribution of rare species. However, we found that biological invasions are eroding the signal of island age on biodiversity. Overall, we show that diversity patterns across a biodiversity hotspot reflect the joint influence of contemporary and historical drivers.
Keywords: Hawaii, biodiversity, forests, island biogeography, habitat heterogeneity
Abstract
Biodiversity patterns emerge as a consequence of evolutionary and ecological processes. Their relative importance is frequently tested on model ecosystems such as oceanic islands that vary in both. However, the coarse-scale data typically used in biogeographic studies have limited inferential power to separate the effects of historical biogeographic factors (e.g., island age) from the effects of ecological ones (e.g., island area and habitat heterogeneity). Here, we describe local-scale biodiversity patterns of woody plants using a database of more than 500 forest plots from across the Hawaiian archipelago, where these volcanic islands differ in age by several million years. We show that, after controlling for factors such as island area and heterogeneity, the oldest islands (Kaua’i and O’ahu) have greater native species diversity per unit area than younger islands (Maui and Hawai’i), indicating an important role for macroevolutionary processes in driving not just whole-island differences in species diversity, but also local community assembly. Further, we find that older islands have a greater number of rare species that are more spatially clumped (i.e., higher within-island β-diversity) than younger islands. When we included alien species in our analyses, we found that the signal of macroevolutionary processes via island age was diluted. Our approach allows a more explicit test of the question of how macroevolutionary factors shape not just regional-scale biodiversity, but also local-scale community assembly patterns and processes in a model archipelago ecosystem, and it can be applied to disentangle biodiversity drivers in other systems.
Biodiversity is unequally distributed across the globe. For example, the number of co-occurring tree species ranges from boreal forests of only a few species that spread across thousands of hectares (1) to tropical forests with upwards of 1,000 species co-occurring within only tens to hundreds of hectares (2). This variation in biodiversity has emerged over evolutionary time (3) and also reflects current patterns of available habitat area, climate, and disturbance (including anthropogenic disturbances) (4–6), as well as historical patterns of climate, interactions with other species, and biogeography (7–10).
Research on oceanic islands has generated some of the most influential theories on biodiversity dynamics via evolutionary (11, 12) and ecological processes (13). For example, historical biogeographic factors can play an important role in driving diversity patterns, such as differences in island age resulting from volcanic processes or differences in historical connections to mainland areas (14–16). At the same time, ecological factors such as island area and heterogeneity, as well as climatic factors, also explain a large amount of the variation in biodiversity across islands (17, 18). While it is impossible to fully separate evolutionary effects from contemporary ecological effects in driving patterns of biodiversity on islands (10, 19, 20), islands have served as a model system in which to evaluate their relative importance, to understand how biodiversity is maintained, and to predict how it may be altered by anthropogenic factors.
The Hawaiian archipelago, and similar hotspot archipelagos, provides an ideal testing ground for studying the interplay between ecological and evolutionary processes generating and maintaining island-level diversity (e.g., refs. 21–23). This is because such archipelagos form over volcanic hotspots that create a temporal sequence of islands of different ages. Among the main Hawaiian islands, Kaua’i is ∼5 million years old, O’ahu is ∼3 million years old, Maui Nui (the combination of Maui, Moloka’I, and Lana’i, which until ∼20,000 years ago, was a single island) is 1 to 2 million years old, and the island of Hawai’i is less than 0.5 million years old and still growing (24). While this age gradient has served as a natural experiment for examining the influence of time for diversification on contemporary diversity patterns (25, 26), hotspot archipelagos such as Hawai’i do not allow for an unconfounded test of island age because there are at least 2 ecological factors that covary with island age: island area and island heterogeneity. The youngest island (Hawai’i) is the largest and the most heterogeneous (e.g., largest elevational gradient), while the oldest island (Kaua’i) is smaller and has lost much of its heterogeneity due to erosion (24). These covarying ecological and evolutionary factors are repeated on hotspot archipelagos around the world and, as a result, evolutionary radiations and species extinctions appear to track ontogenetic changes in island characteristics (27); clades rapidly accumulate species as islands grow and subsequently lose species as islands decay. More recently, these remote islands have become hotspots for the human-mediated establishment of alien species (28), which can influence patterns of biodiversity both positively (via increases in the size of the species pool) and negatively (via alterations of dominance patterns and extinctions of native species) (29, 30).
While the conceptual framework for the interplay between evolutionary and ecological factors in driving patterns of biodiversity on islands is compelling (31), the data used to test these hypotheses are often limited. Macroecological and biogeographic data used to examine species richness and endemism patterns are typically available at very coarse scales (e.g., from island checklist data) and thus do not allow for robust inference regarding confounded within-island variables, such as island heterogeneity. Therefore, more detailed information on the abundances and small-scale distributions of species is required to disentangle these ecological and evolutionary hypotheses. With such data, one can assess whether island-level differences in species richness reflect differences in local scale richness (i.e., higher coexistence) or within-island β-diversity, as well as how that diversity is distributed among species in terms of their relative commonness and rarity. While a few studies have begun to apply smaller-scale data to larger-scale ecoevolutionary questions on islands (22, 32–34), these studies have been greatly limited in scope (e.g., few sampling locations across gradients of island age), and thus have not been able to fully disentangle the effects of island age, area, and heterogeneity on patterns of biodiversity.
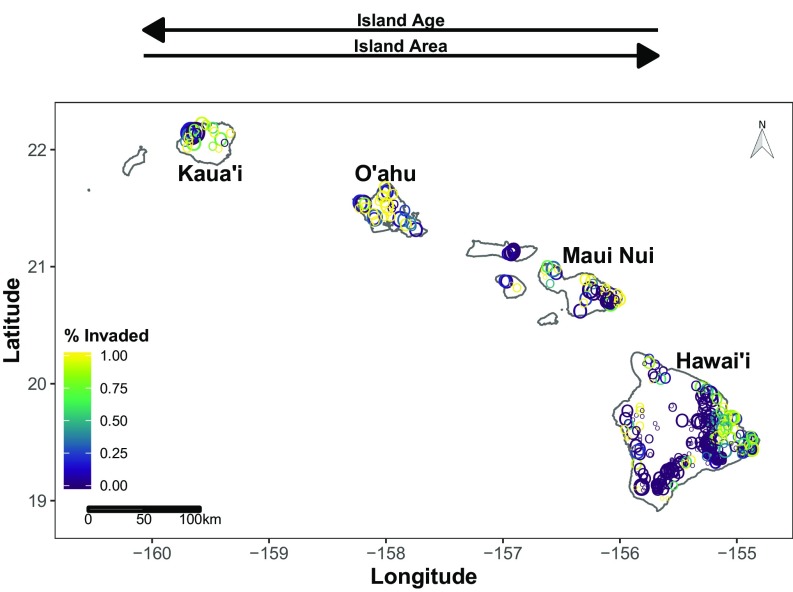
Our primary objectives are to describe patterns of plant biodiversity in the Hawaiian archipelago across spatial scales and to distinguish the roles that island age and habitat heterogeneity likely play in determining these observed patterns. We compared patterns of native species only, which reflect macroevolutionary and biogeographic processes of the islands, with patterns including alien species that have been brought to the islands over the past few hundred years. First, we used island-level data that are typically available for macroscale biodiversity research. Specifically, we used data on the total number of native and alien woody species, as well as single-island endemic woody species (i.e., the number of species endemic to an island) (31), from the flora of the Hawaiian archipelago (35). Second, we used a database of forest plots collected using similar methods from across the Hawaiian archipelago (36). This database consisted of 517 sampling plots containing 28,844 individuals of 104 native species and 62 alien species (Fig. 1), including the numbers of individuals of each species and the spatial location of plots. We performed an array of complementary analyses using rarefaction curves and relative abundance distributions, as well as diversity parameters derived from them, to disentangle factors that correlate with biodiversity patterns (37–40). If macroevolutionary patterns drive both whole-island and local patterns of coexistence and species diversity, we would expect that measures of local biodiversity (e.g., native species per unit area) would be higher on older islands. Alternatively, if macroevolutionary patterns only affect whole-island diversity but not local coexistence (e.g., via heterogeneity and/or allopatric speciation), we would expect local biodiversity to be the same among younger and older islands. Finally, we evaluated whether biological invasions influence these local patterns of diversity by combining native and alien species into the same analyses to determine whether biological invasions attenuate the influence of macroevolutionary factors on local patterns of diversity.
Fig. 1.
Species diversity patterns of plants across Hawaiian islands at local spatial scales. Each point represents a forest plot (n = 517) that is scaled by rarefied species richness (n = 100 individuals). Color corresponds to the relative abundance of alien species, expressed as the proportion of individuals, in each plot.
Results and Discussion
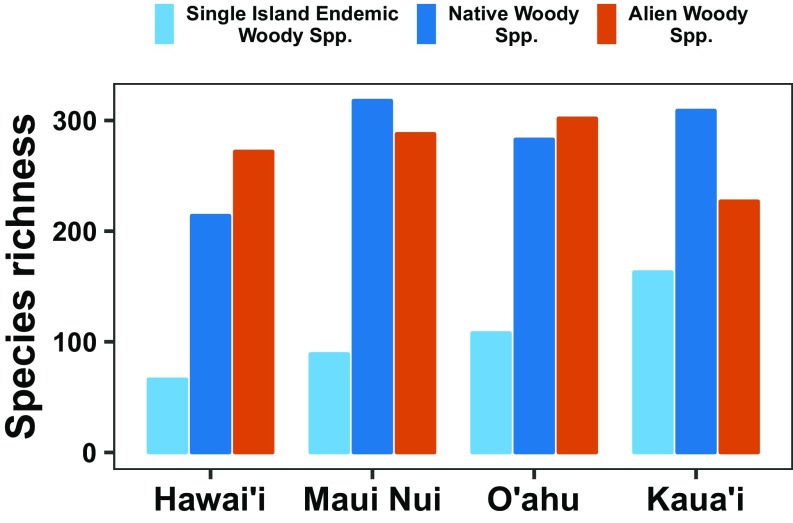
At the island level, our results are consistent with the idea that island age plays an important role in generating native biodiversity patterns (Fig. 2). The youngest island, Hawai’i, has the fewest native woody species and the fewest single island endemic woody species. Kaua’i, the oldest island, has the highest number of single island endemic woody species. However, these island-level patterns do not fully disentangle island age from ecological effects. For example, Hawai’i is considerably larger, yet has lower species diversity than the other islands. Controlling for island area, therefore, may accentuate differences in species diversity between it and the other islands in the archipelago. Likewise, Maui Nui (the second youngest island) has more native woody species than any of the other islands. This may be due, in part, to its relatively large area and high habitat heterogeneity compared with the older islands. When including alien woody species, the general pattern of total species diversity remained similar because most islands had similar numbers of alien woody species. The exception is the oldest island (Kaua’i), which has considerably fewer alien species than the other islands. However, these macroscale patterns across entire islands do not allow us to further disentangle the impact of island age (and time for diversification), relative to island area and habitat heterogeneity, on patterns of biodiversity.
Fig. 2.
Species diversity patterns of native woody plants across the Hawaiian archipelago at the island scale. Species diversity is estimated for native woody species, single-island endemic woody species, and alien woody species using an island-level checklist of the Hawaiian flora. Islands are ordered by age from youngest to oldest (Left to Right).
To discern whether island age (and time for diversification) may influence local biodiversity patterns, we first present results from rarefaction analyses of native species. For rarefaction analyses, it is critical to establish appropriate sampling scenarios before analysis to avoid misinterpretations of diversity patterns. For example, if we simply used the entire database, we would have concluded that there were more native species per unit area on the younger, larger islands (Hawai’i and Maui Nui) and fewer species per unit area on the smaller, older islands (Kaua’i and O’ahu) (SI Appendix, Fig. S1). However, this result is driven by there being more sample plots on the larger, younger islands (Fig. 1 and SI Appendix, Table S1). When we rarefied species richness to a seemingly standardized sampling effort of 10,000 individuals, estimated species richness remained higher for Hawai’i compared with Kaua’i (SI Appendix, Fig. S1). This pattern is similarly misleading because the plots on the larger islands are located along broader environmental gradients than the plots on the smaller islands (SI Appendix, Table S1). Consequently, species richness estimated via rarefaction, which randomizes individuals from across the entire extent of plots sampled on each island, is confounded by habitat heterogeneity among plots.
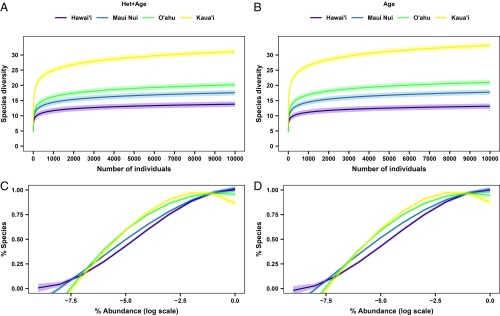
To unveil the influence of island age on diversity patterns relative to other factors, such as island area, sampling effort, and habitat heterogeneity, a more controlled sampling process is necessary. First, we controlled for differences in sampling effort, but not habitat heterogeneity (“Het”) or island age, by randomly selecting a fixed number of sampling plots (10) with a standardized area (0.72 to 0.79 ha) (“Het+Age”; SI Appendix, Table S2). Standardizing sampling at this scale, we found that the older, smaller islands have significantly steeper rarefaction curves with higher asymptotes than the younger, larger islands (P < 0.001; Fig. 3A and SI Appendix, Table S3). This indicates that, after controlling for differences in sampling effort, the older islands—especially Kaua’i—have more native species co-occurring together than younger islands. Examination of the relative abundance distributions (RADs) indicate that the 2 older islands (Kaua’i and O’ahu) have proportionately more rare species (41) than the 2 younger islands (Maui and Hawai’i) (P < 0.001; Fig. 3C and SI Appendix, Table S4). While results from this sampling scenario are suggestive of an island age effect on local patterns of native species diversity and relative abundance, they do not account for the potentially confounding effects of habitat heterogeneity on diversity patterns (31, 42).
Fig. 3.
Rarefaction curves and relative abundance distributions of native forest communities across the Hawaiian archipelago, estimated for 2 sampling scenarios that influence biodiversity. In the first scenario (“Het+Age”; A and C) sampling effort was controlled for and in the second scenario, both sampling effort and habitat heterogeneity were controlled for (“Age”; B and D). Individual-based rarefaction curves (A and B) were estimated with interpolation and extrapolation of up to 10,000 individuals. Relative abundance distributions (C and D) are presented as rescaled empirical cumulative distribution functions. Individual-based rarefaction curves and relative abundance distributions were estimated for the “Het+Age” and “Age” scenarios by randomly selecting 10 plots per island 100 times to control for sampled area. For the “Age” scenario, habitat heterogeneity was controlled for by randomly selecting 10 plots per island with a restricted range in aridity. Colored bands are 95% confidence intervals and solid lines are fitted using linear mixed-effects models.
Next, we controlled for sampling effort and habitat heterogeneity simultaneously to isolate the effects of island age (“Age”). We did this by randomly selecting 10 plots with a standardized area and a lower range in aridity (SI Appendix, Table S2). We inferred habitat heterogeneity using range in aridity because water availability is one of the most limiting resources for plant communities (43, 44) and is strongly associated with the distribution of habitat types across Hawai’i (45). Here, we found patterns similar to the sampling scenario above for both the rarefaction curves (Fig. 3B and SI Appendix, Table S3) and the shape of the RADs (Fig. 3D and SI Appendix, Table S4). Native diversity patterns estimated while controlling for both sampling effort and habitat heterogeneity indicate that there is a strong effect of island age. Specifically, there are more native species per unit area on the older islands and this is because older islands have more rare species, as depicted by changes in the shape of their RADs. This suggests that it is likely that not only the total number of native species on these islands, but also local-scale patterns of species richness and RADs are influenced by island age via differences in time for diversification. We performed sensitivity analyses to test whether our results differed if we controlled for habitat heterogeneity using elevation and found similar results to those of the “Age” sampling scenario (SI Appendix, Fig. S2 and Tables S5–S7).
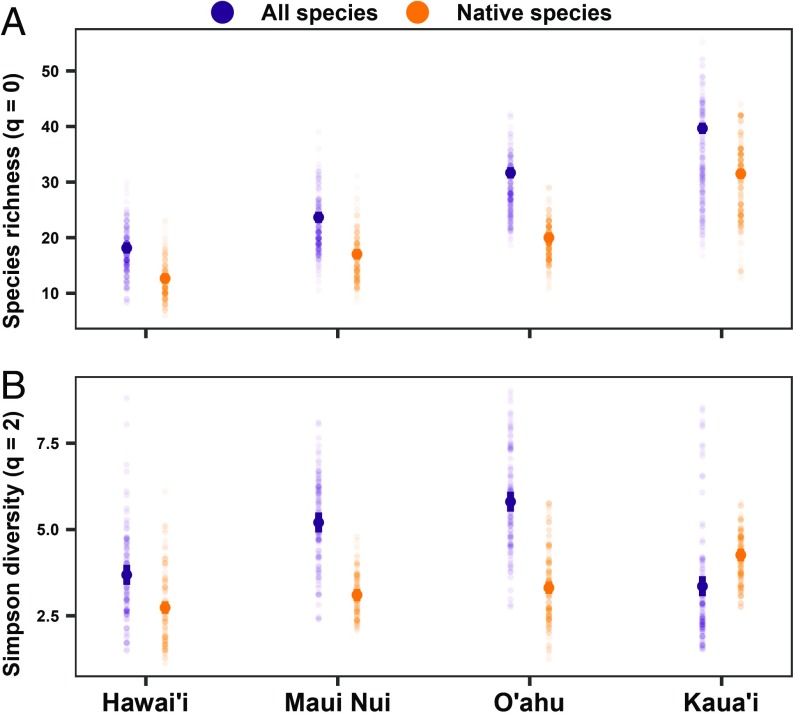
The rarefaction analyses above (Fig. 3) imply that the older islands have higher native species richness locally (per unit area), but also change in species relative abundances (i.e., more rare species). We illustrate this by comparing diversity measures that differentially incorporate species relative abundances and evenness. First, we compared patterns of native species richness (rarefied to 10,000 individuals) across the islands when both habitat heterogeneity and sampling effort were controlled (“Age”). As expected from our rarefaction analyses, we observe a systematic increase in species richness along the age gradient (Fig. 4A and SI Appendix, Table S8). Likewise, when we included alien species in the analysis, this pattern of increasing local richness from the youngest (Hawai’i) to oldest (Kaua’i) island remained, despite there being fewer overall alien species on Kaua’i than the other islands (Fig. 2). We found similar results when only sampling effort, but not habitat heterogeneity, was controlled (SI Appendix, Fig. S3). These results show that after controlling for sampling effort, island area, and habitat heterogeneity, more native species coexist locally on older islands. This result is consistent with the idea that macroevolutionary processes play an important role not just in the generation of regional patterns of biodiversity, but also in patterns of local coexistence (6, 46).
Fig. 4.
Species diversity patterns of forest communities across the Hawaiian archipelago for all species (native and alien) and only native species. Species diversity was estimated as species richness (A) and Simpson diversity (B) using sample-based interpolation for 10,000 individuals; both diversity indices are expressed in terms of effective species numbers. Species diversity was estimated using a sampling scenario (“Age”) that controlled for sampling effort and habitat heterogeneity. Means and 95% confidence intervals are estimated with 1-way ANOVAs (SI Appendix, Table S8).
Next, we calculated a diversity measure that includes aspects of the evenness of species: the effective number of species estimated from Simpson diversity (see Materials and Methods). With this measure, we found that the oldest island (Kaua’i) had the highest native species diversity, but we did not observe differences among the other islands (Fig. 4B). This suggests that the sharp increase in rarefied species richness from the youngest (Hawai’i) to the second oldest (O’ahu) island is largely due to relatively rare species. Together, these results show that the increase in local co-occurrence with increasing island age is primarily mediated by rare species. Species diversity patterns, however, changed markedly when we included alien species in our analyses: local-scale Simpson diversity was highest on the 2 intermediate-aged islands (Maui and O’ahu; Fig. 4B), mirroring the island-level patterns of total species richness (Fig. 2), but not the local-level patterns of species richness (Fig. 4A). This pattern appears to be driven primarily by alien species, which are more diverse at the local level on Maui and O’ahu (SI Appendix, Fig. S4), possibly because they have exploited the large amounts of anthropogenically disturbed habitat on these islands (47). This suggests that while biological invasions have not yet altered patterns of local species richness, they are dramatically changing the local neighborhoods experienced by native species. Our understanding of the biodiversity impacts of biological invasions is incomplete, as our analysis does not consider the potential impacts of nonnative animals, which are widespread across the Hawaiian islands and likely reduce the abundance of native plant species while facilitating the establishment of alien plant species (48). These observed patterns could represent an extinction debt (49), whereby global change drivers such as biological invasions progressively dilute the macroevolutionary signature on local species coexistence and diversity across the island-age gradient.
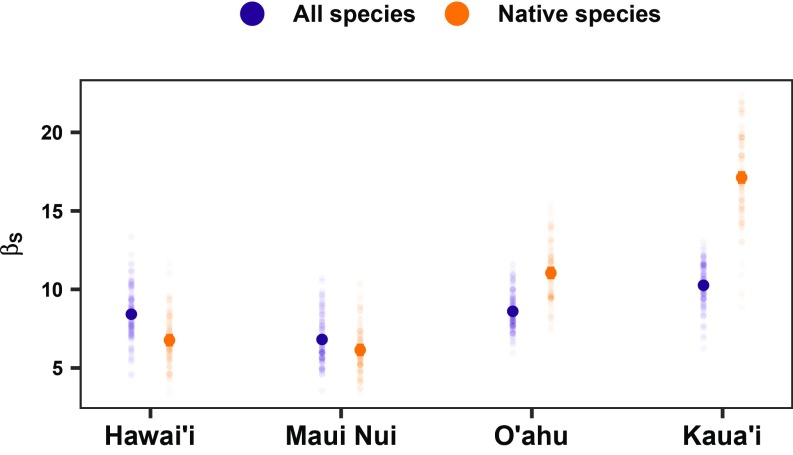
Finally, we examined whether there were differences in the spatial variation in species relative abundances (i.e., within-island β-diversity) among islands. We compared β-diversity of native species among islands (50), and found higher levels of β-diversity on the older islands (Kaua’i and O’ahu) for both sampling scenarios (Fig. 5 and SI Appendix, Fig. S5). Moreover, to ensure that β-diversity patterns did not emerge simply due to spatial distances among plots, but rather inherent compositional differences among the islands, we used variation partitioning and found that environmental factors, but not geographical distance, played a predominant role (SI Appendix, Fig. S6). This suggests that there is a strong macroevolutionary signal on both the local coexistence and spatial distribution of native species on the older islands, which have higher levels of within-site and between-site diversity than the younger islands. This result—greater β-diversity on older islands—could have emerged for a number of reasons. For example, higher levels of β-diversity of rare species could reflect that longer time for divergence on the older islands has allowed for higher rates of allopatric speciation with little secondary contact (25, 26). Higher levels of β-diversity also could result if ecoevolutionary and frequency-dependent feedbacks (e.g., interspecific interactions or interactions with enemies) have created higher levels of spatial dispersion among otherwise similar species (46, 51). When including alien species in our analyses, we found that the signal of island age on β-diversity weakened. While the older islands continued to have higher β-diversity than the younger islands, the differences are less pronounced than those observed for native species only (Fig. 5). This is consistent with the idea that the introduction of alien species frequently leads to biotic homogenization, i.e., reduced β-diversity, among local communities (52).
Fig. 5.
Beta-diversity patterns of forest communities across the Hawaiian archipelago estimated for all species (native and alien) and only native species. Beta-diversity (βs) was calculated multiplicatively using species richness, which highlights contributions of rare species. Beta-diversity was estimated under a sampling scenario (“Age”) that controlled for sampling effort and habitat heterogeneity. Means and 95% confidence intervals are estimated with 1-way ANOVAs (all species: R2 = 42.1%, F = 96.1, P < 0.001; native species: R2 = 82.5%, F = 622.2, P < 0.001).
Conclusion
Integrating evolutionary and ecological factors has been considered an essential step (10, 19, 53) toward deepening current understanding of biodiversity patterns and the mechanisms that underpin them. Here, we leverage a comprehensive database of forest plots distributed across the Hawaiian archipelago and use it to show that the effects of island age on patterns of native diversity percolate across scales, from macroecological to local, via the differences in the presence of rare species, as well as in their spatial distribution. In doing so, we provide evidence that species diversity patterns across islands bear the imprint of both ecological and historical evolutionary factors. However, when we included alien species in our analyses, these patterns weakened, suggesting that anthropogenic processes are eroding macroevolutionary signals across the island-age gradient. Our general approach can be readily applied to other systems for which there is local scale sampling of communities across relevant ecological and macroevolutionary gradients.
Materials and Methods
Data Acquisition and Description.
We estimated species richness of native woody species and single-island endemic woody species (i.e., the number of species endemic to an island; ref. 31), for each island across the Hawaiian archipelago. To do so, we classified all plant species in the flora of the Hawaiian Islands (35) as native or alien, and whether they were woody, herbaceous, or variable (based on plant growth form) using the Global Woodiness Database (SI Appendix and refs. 54 and 55). Species not found in the Global Woodiness Database were classified using the same methodology by consulting electronic sources (35, 56). We then removed nonwoody species (e.g., herbaceous or variable) before analysis.
We assembled a database containing 517 plots and 104 native and 62 alien species, including trees, shrubs, and tree ferns from publicly available sources and published studies with forest plot data in Hawaii where species identity and size of individuals ≥5-cm diameter at 1.3 m were reported (36). Forest plots range in area from 100 to 1,018 m2 (median = 1,000 m2) and are unevenly distributed across the main islands of the archipelago (71.4% of all plots are located on Hawai’i Island; Fig. 1). Consequently, sampled area per island also varies strongly, from ∼1.5 ha on Kaua’i Island to 27.6 ha on Hawai’i Island (SI Appendix, Table S1). We extracted mean annual temperature, precipitation, and potential evapotranspiration (PET Penman-Monteith, calculated using temperature, solar radiation, air pressure, and wind speed) for each plot from local interpolated climate data (57, 58). We calculated aridity as the ratio of mean annual precipitation to mean PET following (59), which is widely used to examine the impacts of water availability on plant communities in terms of diversity and ecosystem functioning (43). This index is useful because it is continuous and captures gradual transitions between habitat types, while also reflecting how habitat types have been classified in Hawai’i (60). We also extracted elevation (61) and soil substrate age (62) for each plot (SI Appendix, Table S1). Species names were standardized using The Plant List v 1.1 (63) and native status was obtained from the flora of the Hawaiian Islands (35). Species abundances were calculated on a per hectare basis in each plot to facilitate data aggregation. Throughout our analysis, we treat islands in the Maui Nui complex, i.e., Maui, Moloka’i, Lana’i, and Kaho’olawe, as one island because they have formed a single landmass during most of their history (45).
Sampling Scenarios for Dissecting Species Diversity Patterns.
To examine the impacts of island age and habitat heterogeneity on patterns of woody plant co-occurrence across the Hawaiian Islands, we used data generated from 2 nested sampling scenarios. Each sampling scenario eliminates the influence of a hypothesized driver of biodiversity patterns across islands. In the first sampling scenario (“Het+Age”), we controlled for sampling effort (i.e., differences in sampled area across islands), by randomly selecting 10 plots per island 100 times but did not control for habitat heterogeneity or island age (SI Appendix, Table S2). To further ensure that sampled area is similar across islands, we excluded iterations whose sampled area was less than or greater than the 90th and 10th quantiles of the sampled area of Kaua’i, the island with the fewest number of plots in our database (SI Appendix, Table S1); across the 100 samples, mean sampled area per island ranged from 0.72 to 0.79 ha and the mean minimum distance among plots (per sample) was 2.25 km. In the second sampling scenario (“Age”), we controlled for sampling effort and habitat heterogeneity simultaneously by randomly selecting 10 plots per island (as described above) where the range in aridity (aridityrange = ariditymax − ariditymin) among selected plots was less than the 50th quantile for each island in the “Het+Age” scenario (SI Appendix, Table S2). On average, aridityrange was 32.7% lower in the “Age” scenario than in the “Het+Age” scenario across islands. We generated both sampling scenarios for native species only (SI Appendix, Table S2) and native and alien species together (SI Appendix, Table S9).
We performed sensitivity analyses to test whether our results would differ if we used a different proxy for habitat heterogeneity by generating an additional sampling scenario (for native species only). We controlled for habitat heterogeneity by randomly selecting 10 plots per island (as described above) where the range in elevation among selected plots was less than the 50th quantile for each island in the “Het+Age” scenario (SI Appendix, Table S5). On average, range in elevation was 13.3% lower in the “Age” scenario than in the “Het+Age” scenario across islands.
Data Analysis.
For all analyses, we pooled plot data per island to ensure sufficiently large sample sizes for sample completeness and estimation of species diversity (64). We first performed all analyses for native species only and then performed the same analyses for native and alien species together. We first estimated species richness for native plants of each island using sample-based rarefaction for 100, 1,000, 10,000, and all individuals (drawn from all plots) with “vegan” (65). For each island and iteration of both sampling scenarios, we estimated species accumulation curves (SACs) up to 10,000 individuals using coverage-based interpolation and extrapolation with “iNEXT” (38, 66). We calculated RADs, which depict proportional changes in species abundances as a function of their relative rank within a community because they facilitate comparison among communities by correcting for different species diversities (41). We fit SACs and rarefaction curves across all iterations of each sampling scenario for each island using linear mixed effects with a Gaussian distribution where iteration was the random group effect nested within island using “lme4” (67). To account for the nonlinear relationship between species’ rank and relative abundance, we included relative abundance (natural-log transformed to meet normality assumptions) and 2 orthogonal polynomial terms. For SACs, we natural-log transformed the number of individuals based on the expectation that species would accumulate fast initially, followed by constant growth. We checked model assumptions visually, inspecting residual plots for homogeneity and quantile-quantile plots for normality. We used Satterthwaite’s method to calculate denominator degrees of freedom and F statistics. For all models, we calculated 95% confidence intervals with “ggeffects” (68).
Species richness and Simpson diversity (equivalent to Hill numbers where q = 0 and q = 2; ref. 69) were estimated using sample-based interpolation for 10,000 individuals for the “Het+Age” and “Age” sampling scenarios to facilitate comparisons of diversities across communities. Species richness gives equal weight to the contribution of rare species, while Simpson diversity gives more weight to abundant species and thus is influenced by the evenness of species abundance distributions. Diversity estimates were made using iNEXT (66). Beta-diversity was estimated using species richness, which gives equal weight to the contribution of common and rare species (40). Beta-diversity was calculated multiplicatively as the turnover between local () and island () scales using “mobr” (40). As β-diversity was calculated for each plot, we calculated mean β-diversity for each island per sample. Species and β-diversity estimates are expressed in terms of effective numbers of species (69). We tested for variation in species and beta-diversities across islands were tested using 1-way ANOVAs and calculated 95% confidence intervals with “ggeffects” (68).
We used partial regression with variation partitioning to examine the relative influence of environmental and spatial factors (alone and combined) on β-diversity for both sampling scenarios (per sample) using the rda function in “vegan” (65). Spatial factors were calculated using multiscale principal coordinates of neighbor matrices (PCNMs) from the geographic distance matrix among plots using the pcnm function in “vegan” (65). Environmental factors were estimated by using the first 2 axes of a principal components analysis (PCA) of mean annual precipitation, mean annual temperature, aridity, elevation, and soil substrate age with the PCA function in “FactoMineR” (70). We used corrected estimates of explained variation, which were R2 values adjusted for the number of sites and explanatory variables for each variation component (71). All data manipulation and analyses were performed using R 3.6.0 (72).
Data and Code Availability.
Data supporting the findings of this study are available for download from ref. 36 and the code used for all analyses and figures is available via GitHub (https://github.com/dylancraven/Hawaii_diversity) and mirrored at Zenodo (https://zenodo.org/record/3250638).
Supplementary Material
Acknowledgments
D.C., T.M.K., and J.M.C. acknowledge funding by the German Centre for Integrative Biodiversity Research (iDiv) Halle-Jena-Leipzig, funded by the German Research Foundation (FZT 118). T.M.K. also acknowledges support from the Alexander von Humboldt Foundation. T.M.K. and K.E.B. also acknowledge support from the National Geographic Society and the Pacific Island Climate Science Center. We thank Valentin Stefan and Elena Motivans for assistance preparing data, and data providers for collecting the field data.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Our code and data are available via GitHub (https://github.com/dylancraven/Hawaii_diversity) and mirrored on Zenodo (https://zenodo.org/record/3250638).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901954116/-/DCSupplemental.
References
- 1.Gauthier S., Bernier P., Kuuluvainen T., Shvidenko A. Z., Schepaschenko D. G., Boreal forest health and global change. Science 349, 819–822 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Anderson-Teixeira K. J., et al. , CTFS-ForestGEO: A worldwide network monitoring forests in an era of global change. Glob. Change Biol. 21, 528–549 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Schluter D., Pennell M. W., Speciation gradients and the distribution of biodiversity. Nature 546, 48–55 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Rosenzweig M. L., Species Diversity in Space and Time (Cambridge University Press, 1995). [Google Scholar]
- 5.Gaston K. J., Global patterns in biodiversity. Nature 405, 220–227 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Ricklefs R. E., A comprehensive framework for global patterns in biodiversity. Ecol. Lett. 7, 1–15 (2004). [Google Scholar]
- 7.Kier G., et al. , A global assessment of endemism and species richness across island and mainland regions. Proc. Natl. Acad. Sci. U.S.A. 106, 9322–9327 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandel B., et al. , The influence of Late Quaternary climate-change velocity on species endemism. Science 334, 660–664 (2011). [DOI] [PubMed] [Google Scholar]
- 9.LaManna J. A., et al. , Plant diversity increases with the strength of negative density dependence at the global scale. Science 356, 1389–1392 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Whittaker R. J., Fernández-Palacios J. M., Matthews T. J., Borregaard M. K., Triantis K. A., Island biogeography: Taking the long view of nature’s laboratories. Science 357, eaam8326 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Darwin C., On the Origins of Species by Means of Natural Selection (John Murray, United Kingdom, 1859). [Google Scholar]
- 12.Lack D., Darwin’s Finches (CUP Archive, 1947). [Google Scholar]
- 13.MacArthur R. H., Wilson E. O., The Theory of Island Biogeography (Princeton University Press, 1967). [Google Scholar]
- 14.Emerson B. C., Evolution on oceanic islands: Molecular phylogenetic approaches to understanding pattern and process. Mol. Ecol. 11, 951–966 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Gillespie R. G., Island time and the interplay between ecology and evolution in species diversification. Evol. Appl. 9, 53–73 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weigelt P., Steinbauer M. J., Cabral J. S., Kreft H., Late Quaternary climate change shapes island biodiversity. Nature 532, 99–102 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Kreft H., Jetz W., Mutke J., Kier G., Barthlott W., Global diversity of island floras from a macroecological perspective. Ecol. Lett. 11, 116–127 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Economo E. P., Ricklefs R. E., Triantis K. A., Guilhaumon F., Diversity regulation at macro‐scales: Species richness on oceanic archipelagos. Glob. Ecol. Biogeogr. 24, 594–605 (2015). [Google Scholar]
- 19.Losos J. B., Ricklefs R. E., Adaptation and diversification on islands. Nature 457, 830–836 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Warren B. H., et al. , Islands as model systems in ecology and evolution: Prospects fifty years after MacArthur-Wilson. Ecol. Lett. 18, 200–217 (2015). [DOI] [PubMed] [Google Scholar]
- 21.Gillespie R., Community assembly through adaptive radiation in Hawaiian spiders. Science 303, 356–359 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Gruner D. S., Geological age, ecosystem development, and local resource constraints on arthropod community structure in the Hawaiian Islands. Biol. J. Linn. Soc. Lond. 90, 551–570 (2007). [Google Scholar]
- 23.Price J. P., Wagner W. L., A phylogenetic basis for species-area relationships among three Pacific Island floras. Am. J. Bot. 98, 449–459 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Clague D. A., Sherrod D. R., “Growth and degradation of Hawaiian volcanoes” in Characteristics of Hawaiian Volcanoes, (US Geological Survey, Reston, VA, 2014), pp. 97–146. [Google Scholar]
- 25.Baldwin B. G., Sanderson M. J., Age and rate of diversification of the Hawaiian silversword alliance (Compositae). Proc. Natl. Acad. Sci. U.S.A. 95, 9402–9406 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Givnish T. J., et al. , Origin, adaptive radiation and diversification of the Hawaiian lobeliads (Asterales: Campanulaceae). Proc. Biol. Sci. 276, 407–416 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim J. Y., Marshall C. R., The true tempo of evolutionary radiation and decline revealed on the Hawaiian archipelago. Nature 543, 710–713 (2017). [DOI] [PubMed] [Google Scholar]
- 28.Moser D., et al. , Remoteness promotes biological invasions on islands worldwide. Proc. Natl. Acad. Sci. U.S.A. 115, 9270–9275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sax D. F., Gaines S. D., Colloquium paper: Species invasions and extinction: The future of native biodiversity on islands. Proc. Natl. Acad. Sci. U.S.A. 105 (suppl. 1), 11490–11497 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vilà M., et al. , Ecological impacts of invasive alien plants: A meta-analysis of their effects on species, communities and ecosystems. Ecol. Lett. 14, 702–708 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Whittaker R. J., Triantis K. A., Ladle R. J., A general dynamic theory of oceanic island biogeography. J. Biogeogr. 35, 977–994 (2008). [Google Scholar]
- 32.Rominger A. J., et al. , Community assembly on isolated islands: Macroecology meets evolution. Glob. Ecol. Biogeogr. 25, 769–780 (2016). [Google Scholar]
- 33.Borges P. A. V., Hortal J., Time, area and isolation: Factors driving the diversification of Azorean arthropods. J. Biogeogr. 36, 178–191 (2009). [Google Scholar]
- 34.Ibanez T., et al. , Regional forcing explains local species diversity and turnover on tropical islands. Glob. Ecol. Biogeogr. 27, 474–486 (2018). [Google Scholar]
- 35.Wagner W. L., Herbst D. L., Lorence D. H., Flora of the Hawaiian islands website (2005). https://naturalhistory2.si.edu/botany/hawaiianflora/. Accessed 1 May 2017.
- 36.Craven D., et al. , OpenNahele: The open Hawaiian forest plot database. Biodivers. Data J. 6, e28406 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gotelli N. J., Colwell R. K., Quantifying biodiversity: Procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 4, 379–391 (2001). [Google Scholar]
- 38.Chao A., Jost L., Coverage-based rarefaction and extrapolation: Standardizing samples by completeness rather than size. Ecology 93, 2533–2547 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Chase J. M., Knight T. M., Scale-dependent effect sizes of ecological drivers on biodiversity: Why standardised sampling is not enough. Ecol. Lett. 16 (suppl. 1), 17–26 (2013). [DOI] [PubMed] [Google Scholar]
- 40.McGlinn D. J., et al. , Measurement of Biodiversity (MoB): A method to separate the scale-dependent effects of species abundance distribution, density, and aggregation on diversity change. Methods Ecol. Evol. 10, 258–269 (2019). [Google Scholar]
- 41.Magurran A. E., McGill B. J., Biological Diversity: Frontiers in Measurement and Assessment (Oxford University Press, 2011). [Google Scholar]
- 42.Borregaard M. K., et al. , Oceanic island biogeography through the lens of the general dynamic model: Assessment and prospect. Biol. Rev. Camb. Philos. Soc. 92, 830–853 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Maestre F. T., et al. , Plant species richness and ecosystem multifunctionality in global drylands. Science 335, 214–218 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rey P. J., Alcántara J. M., Manzaneda A. J., Sánchez-Lafuente A. M., Facilitation contributes to Mediterranean woody plant diversity but does not shape the diversity-productivity relationship along aridity gradients. New Phytol. 211, 464–476 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Price J. P., Floristic biogeography of the Hawaiian islands: Influences of area, environment and paleogeography. J. Biogeogr. 31, 487–500 (2004). [Google Scholar]
- 46.Ricklefs R. E., Intrinsic dynamics of the regional community. Ecol. Lett. 18, 497–503 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Jacobi J. D., Price J. P., Fortini L. B., Gon S. M., Berkowitz P., “Baseline land cover. Baseline and projected future carbon storage and carbon fluxes in ecosystems of Hawai’i” (US Geological Survey Professional Paper 1834, US Department of the Interior, US Geological Survey, Reston, VA, 2017), pp. 9–20.
- 48.Nogueira-Filho S. L. G., Nogueira S. S. C., Fragoso J. M. V., Ecological impacts of feral pigs in the Hawaiian Islands. Biodivers. Conserv. 18, 3677 (2009). [Google Scholar]
- 49.Triantis K. A., et al. , Extinction debt on oceanic islands. Ecography 33, 285–294 (2010). [Google Scholar]
- 50.Jost L., Partitioning diversity into independent alpha and beta components. Ecology 88, 2427–2439 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Ricklefs R. E., Host-pathogen coevolution, secondary sympatry and species diversification. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 1139–1147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dornelas M., et al. , Assemblage time series reveal biodiversity change but not systematic loss. Science 344, 296–299 (2014). [DOI] [PubMed] [Google Scholar]
- 53.Donoghue M. J., Colloquium paper: A phylogenetic perspective on the distribution of plant diversity. Proc. Natl. Acad. Sci. U.S.A. 105 (suppl. 1), 11549–11555 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zanne A., et al. , Data from “Three keys to the radiation of angiosperms into freezing environments.” Dryad Digital Repository. 10.5061/dryad.63q27.2. Accessed 15 May 2019. [DOI] [Google Scholar]
- 55.Zanne A. E., et al. , Three keys to the radiation of angiosperms into freezing environments. Nature 506, 89–92 (2014). [DOI] [PubMed] [Google Scholar]
- 56.USDA NRCS , The PLANTS database (2018). https://plants.sc.egov.usda.gov/java/.
- 57.Giambelluca T. W., et al. , Online rainfall atlas of Hawai ‘i. Bull. Am. Meteorol. Soc. 94, 313–316 (2013). [Google Scholar]
- 58.Giambelluca T., et al. , Evapotranspiration of Hawai‘i. Final report submitted to the US Army Corps of Engineers—Honolulu District, and the Commission on Water Resource Management, State of Hawai‘i (2014, http://climate.geography.hawaii.edu/).
- 59.Zomer R. J., Trabucco A., Bossio D. A., Verchot L. V., Climate change mitigation: A spatial analysis of global land suitability for clean development mechanism afforestation and reforestation. Agric. Ecosyst. Environ. 126, 67–80 (2008). [Google Scholar]
- 60.Gagne W., Cuddihy L., “Vegetation” Manual of the Flowering Plants of Hawaii (University of Hawaii Press, Honolulu, 1990), p. 1952. [Google Scholar]
- 61.Jarvis A., Reuter H. I., Nelson A., Guevara E., Hole-filled SRTM for the Globe (Version 4). http://srtm.csi.cgiar.org. Accessed 15 May 2019.
- 62.Sherrod D. R., Sinton J. M., Watkins S. E., Brunt K. M., Geologic map of the State of Hawaii (Geological Survey (US), 2007).
- 63.Cayuela L., Stein A., Oksanen J., Taxonstand: Taxonomic standardization of plant species names (2017). https://cran.r-project.org/web/packages/Taxonstand/index.html. Accessed 1 May 2018.
- 64.Chao A., Chiu C., Jost L., Statistical challenges of evaluating diversity patterns across environmental gradients in mega‐diverse communities. J. Veg. Sci. 27, 437–438 (2016). [Google Scholar]
- 65.Oksanen J., et al. , vegan: Community Ecology Package (R package Version 2.5-5) https://CRAN.R-project.org/package=vegan. Accessed 15 May 2019.
- 66.Hsieh T. C., Ma K. H., Chao A., iNEXT: An R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol. 7, 1451–1456 (2016). [Google Scholar]
- 67.Bates D., Mächler M., Bolker B., Walker S., Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48 (2015). [Google Scholar]
- 68.Lüdecke D., ggeffects: Tidy data frames of marginal effects from regression models. J. Open Source Softw. 3, 772 (2018). [Google Scholar]
- 69.Jost L., Entropy and diversity. Oikos 113, 363–375 (2006). [Google Scholar]
- 70.Lê S, Josse J, Husson F, others, FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 25, 1–18 (2008). [Google Scholar]
- 71.Peres-Neto P. R., Legendre P., Dray S., Borcard D., Variation partitioning of species data matrices: Estimation and comparison of fractions. Ecology 87, 2614–2625 (2006). [DOI] [PubMed] [Google Scholar]
- 72.R Core Team , R: A Language and Environment for Statistical Computing (Version 3.6.0, R Foundation for Statistical Computing, Vienna, Austria, 2018). https://www.R-project.org/. Accessed 15 May 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available for download from ref. 36 and the code used for all analyses and figures is available via GitHub (https://github.com/dylancraven/Hawaii_diversity) and mirrored at Zenodo (https://zenodo.org/record/3250638).