Summary
In eukaryotes with multiple small RNA pathways the mechanisms that channel RNAs within specific pathways are unclear. Here, we reveal the reactions that account for channeling in the siRNA biogenesis phase of the Arabidopsis RNA-directed DNA methylation pathway. The process begins with template DNA transcription by NUCLEAR RNA POLYMERASE IV (Pol IV) whose atypical termination mechanism, induced by nontemplate DNA basepairing, channels transcripts to the associated RNA-dependent RNA polymerase, RDR2. RDR2 converts Pol IV transcripts into double-stranded RNAs then typically adds an extra untemplated 3' terminal nucleotide to the second strands. The dicer endonuclease, DCL3 cuts resulting duplexes to generate 24 and 23nt siRNAs. The 23nt RNAs bear the untemplated terminal nucleotide of the RDR2 strand and are underrepresented among ARGONAUTE4-associated siRNAs. Collectively, our results provide mechanistic insights into Pol IV termination, Pol IV-RDR2 coupling and RNA channeling, from template DNA transcription to siRNA strand discrimination.
Keywords: Nuclear RNA Polymerase IV, noncoding RNA, RNA silencing, RNA-directed DNA methylation, transcription termination, dicing, ncRNA processing
Graphical Abstract
eTOC blurb
siRNA-directed DNA methylation is important for genome defense against transposons and viruses. Singh et al. recapitulate siRNA biogenesis in vitro using NUCLEAR RNA POLYMERASE IV, RNA-DEPENDENT RNA POLYMERASE 2 and DICER-LIKE 3, providing mechanistic insights into Pol IV termination, RDR2 activation, siRNA precursor strand specification and DCL3 dicing.
Introduction
In eukaryotes, RNA-based surveillance systems control viruses and transposable elements to minimize the deleterious consequences of genetic invasion, transposition, mutation and chromosome instability (Slotkin and Martienssen, 2007). Surveillance involves small RNAs that associate with Argonaute family proteins and basepair with target RNAs to bring about their cleavage or translational inhibition, or to guide chromatin modifications that repress gene transcription (Borges and Martienssen, 2015; Ghildiyal and Zamore, 2009; Holoch and Moazed, 2015).
In plants, the dominant process for transcriptional silencing is RNA-directed DNA methylation (RdDM; Figure 1A) guided by 24 nt siRNAs whose synthesis requires NUCLEAR RNA POLYMERASE IV (Pol IV), RNA-DEPENDENT RNA POLYMERASE 2 (RDR2) and DICER-LIKE 3 (DCL3) (Herr et al., 2005; Onodera et al., 2005; Xie et al., 2004). Once synthesized, 24 nt siRNAs associate with ARGONAUTE4 (AGO 4) (Zilberman et al., 2003), or a related AGO family member, and guide the complexes to target sites transcribed by NUCLEAR RNA POLYMERASE V (Pol V) (Wierzbicki et al., 2008; Wierzbicki et al., 2009). Through basepairing with Pol V transcripts and AGO-Pol V interactions (El-Shami et al., 2007; Wierzbicki et al., 2009), siRNA-AGO silencing complexes mediate recruitment of chromatin modifiers that include the de novo DNA methyltransferase, DRM2 (Bohmdorfer et al., 2014; Cao and Jacobsen, 2002; Wierzbicki et al., 2009). Histone post-translational modifications occur in cross-talk with DNA methylation, generating chromatin environments that suppress promoter-dependent transcription by DNA-dependent RNA Polymerases I, II or III (Du et al., 2015); for recent reviews see (Matzke et al., 2015)(Wendte and Pikaard, 2017). In the germline of mammals, an analogous pathway involves piRNAs, so-named for their association with proteins of the PIWI subfamily of Argonaute proteins (Ozata et al., 2018), which guide transposon methylation by DNMT3a and DNMT3b, the orthologs of plant DRM2 (Chedin, 2011; Skvortsova et al., 2018).
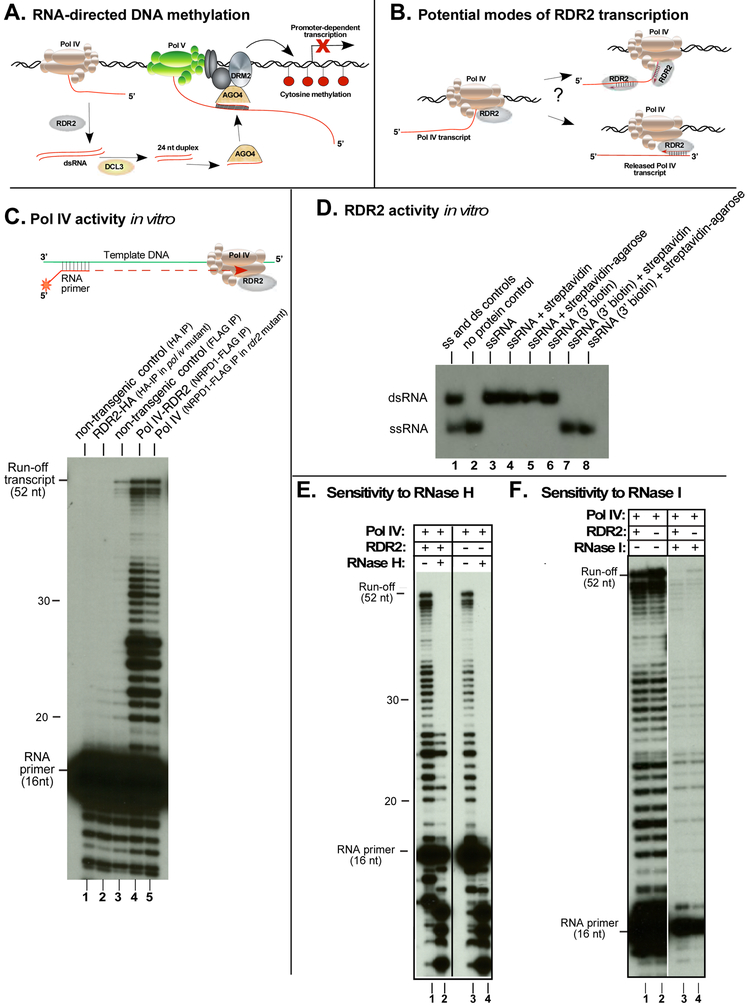
Figure 1. RNA polymerase activities of Pol IV and RDR2.
A. A simplified model for RNA-directed DNA methylation.
B. Potential modes of Pol IV-RDR2 cooperation in dsRNA synthesis.
C. Pol IV transcription is independent of RDR2. Affinity purified Pol IV and/or RDR2 were tested for the ability to extend a 5’ end-labeled 16 nt RNA primer hybridized to a DNA oligonucleotide template (see Figure S1A). Transcripts resolved by denaturing polyacrylamide gel electrophoresis were visualized by autoradiography. HA-tagged RDR2 purified from a Pol IV mutant background (nrpd1) was tested in lane 2. FLAG-tagged Pol IV purified with associated RDR2 or from a rdr2 null mutant was tested in lanes 4 and 5, respectively. Lanes 1 and 3 are controls testing non-transgenic plant lysates subjected to anti-HA or anti-FLAG affinity purification.
D. RDR2 can convert ssRNA into dsRNA in vitro. Purified, recombinant RDR2 was assayed for activity using a 37 nt 5' end-labeled ssRNA template. Reaction products resolved by non-denaturing PAGE were visualized by autoradiography. Lane 1 shows 37 nt ssRNA and 37 bp dsRNA controls. In lanes 2-5, the ssRNA template tested had a 3’OH group. RDR2 was included in all reactions except for lane 2. Reactions of lanes 4 and 5 included free streptavidin or streptavidin-agarose resin. Reactions of lanes 6-8 tested ssRNAs with a biotin group on the 3' terminal nucleotide. Streptavidin or streptavidin-agarose were included in lanes 7 and 8.
E. Pol IV transcripts generated from ssDNA templates are sensitive to RNase H. Pol IV-RDR2 was tested in lanes 1 and 2; Pol IV isolated in the rdr2 mutant background was tested in lanes 3 and 4. Transcripts in lanes 2 and 4 were subjected to RNase H treatment. Positions of run-off transcripts and the labeled RNA primer are shown.
F. Pol IV transcripts generated from ssDNA templates are sensitive to RNase I. Pol IV-RDR2 was tested in lanes 1 and 3; Pol IV isolated from rdr2 mutant plants was tested in lanes 2 and 4. Transcripts in lanes 3 and 4 were subjected to RNase I treatment.
Transcription by multisubunit DNA-dependent RNA Polymerase II (Pol II) is the first step in piRNA or siRNA biogenesis in most eukaryotes (Holoch and Moazed, 2015). However, in plants, 24 nt siRNA biogenesis requires Pol IV (Herr et al., 2005; Onodera et al., 2005), whose 12-subunits (Ream et al., 2009) reveal its origin as a specialized form of Pol II. Pol IV acts in close partnership with RDR2. The enzymes copurify (Haag et al., 2012; Law et al., 2011) and genetic and genomic evidence indicate that both are needed to produce double-stranded (ds) precursors of siRNAs (Blevins et al., 2015; Li et al., 2015; Zhai et al., 2015). These dsRNA precursors, averaging only ~32 bp (Blevins et al., 2015; Zhai et al., 2015), accumulate in dcl3 mutants but can be cut into 24 nt RNAs by purified DCL3 in vitro (Blevins et al., 2015).
Interestingly, neither strand of siRNA precursor duplexes is detected in pol IV or rdr2 single mutants indicating an enzymatic codependence whose molecular basis is unknown. Precursor RNA strands tend to begin with a purine (A or G) (Blevins et al., 2015; Zhai et al., 2015), as is common for DNA-dependent RNA polymerases (Basu et al., 2014). However, precursor strands also tend to have pyrimidines at their 3' ends, such that complementary strands are also expected to have 5’ purines and 3’ pyrimidines. The resulting inability to definitively identify Pol IV or RDR2 transcripts has led us to refer to siRNA-precursors as P4R2 RNAs (Blevins et al., 2015). An intriguing feature of P4R2 RNAs is that their 3' terminal nucleotides often do not match the corresponding DNA template (Blevins et al., 2015; Wang et al., 2016; Zhai et al., 2015). One hypothesis has suggested that nucleotide misincorporation by Pol IV, especially at methylated cytosines, induces Pol IV termination, explaining both the 3’-terminal DNA-mismatched nucleotides and the short size of Pol IV transcripts (Zhai et al., 2015). However, RDR2 has terminal transferase activity that can add untemplated nucleotides to RNA 3' ends, suggesting an alternative hypothesis for the mismatched nucleotides (Blevins et al., 2015).
Pol IV transcribes single-stranded (ss) DNA but lacks significant activity using sheared double-stranded (ds) DNA in vitro (Haag et al., 2012; Onodera et al., 2005). Our current study provides an explanation, showing that Pol IV engaged in transcription of a ssDNA strand terminates within 12-18 nt after encountering dsDNA. Importantly, Pol IV termination induced in this manner is needed to channel the transcript to RDR2, which converts the Pol IV transcript into dsRNA. We show that single-stranded M13 bacteriophage DNA can template siRNA biogenesis in vitro, with Pol IV synthesizing first strand transcripts, RDR2 synthesizing second strands and DCL3 dicing the duplexes into 24 and 23 nt siRNAs, as in vivo. DNA-mismatched nucleotides are present at precursor and siRNA 3' ends, as in vivo, and sequencing shows them to be hallmarks of RDR2 transcripts, not Pol IV transcripts. Collectively, the reactions of Pol IV, RDR2 and DCL3 are sufficient for siRNA biogenesis and account for the short length of P4R2 RNAs, the origin of untemplated 3’ nucleotides, the mechanism of Pol IV-RDR2 coupling and the channeling of RNAs, from initial DNA transcription to siRNA strand discrimination.
Results
Pol IV and RDR2 functions are separable in vitro
Pol IV has long been assumed to transcribe DNA into RNA transcripts that are then made double-stranded by RDR2 (Figure 1A), but definitive evidence is lacking (Haag et al., 2012; Matzke et al., 2015; Pikaard et al., 2012). RDR2 could potentially act co-transcriptionally with Pol IV (Figure 1B, model at upper right). Alternatively, RDR2 might only engage terminated and released Pol IV transcripts, transcribing them end-to-end (Figure 1B, model at lower right).
To investigate the order of Pol IV and RDR2 action, we purified the enzymes from transgenic plants expressing FLAG-tagged NRPD1 (the largest subunit of Pol IV) or HA-tagged RDR2 from transgenes rescuing homozygous nrpd1 or rdr2 null mutations (Haag et al., 2012). Although Pol IV and RDR2 copurify, affinity purification of Pol IV from a rdr2 mutant background, or RDR2 from a nrpd1 mutant, allows each enzyme to be isolated free of the other (Haag et al., 2012).
Using a single-stranded DNA template hybridized to an RNA primer, Pol IV extends the primer in a templated manner when supplied with ribonucleotide triphosphates (Haag et al., 2012). 5' end-labeling of the primer with 32P allows resulting transcripts, resolved by denaturing polyacrylamide gel electrophoresis (PAGE), to be visualized by autoradiography (Figure 1C; see Figure S1A for template and primer sequences). Purified RDR2-HA displays no significant DNA-dependent RNA polymerase activity in this assay (Figure 1C, lane 2). By contrast, Pol IV-RDR2 (purified Pol IV with associated RDR2) generates full-length 52 nt transcripts as well as shorter RNAs (lane 4). Pol IV isolated from a rdr2 mutant background displays the same activity as Pol IV-RDR2 (lane 5), indicating that RDR2 is not required for Pol IV activity.
RDR2 can likewise function independently of Pol IV (Figure 1D). Recombinant RDR2, expressed in insect cells (Blevins et al., 2015) transcribes a 37 nt ssRNA template to generate 37bp dsRNA (Figure 1D, lane 3). To test predictions of the models of Figure 1B, the 3' end of the ssRNA template was biotinylated and tested as a template in the presence of soluble streptavidin or streptavidin immobilized on agarose beads (Figure 1D). RNAs with unmodified 3' hydroxyl groups, or 3' biotin moieties, were both converted into dsRNA by RDR2 (Figure 1D, lanes 3 and 6). Neither streptavidin nor streptavidin-agarose beads inhibit RDR2 transcription If the RNA is not biotinylated, (lanes 4 and 5). However, if the RNA is 3' biotinylated, both free streptavidin and streptavidin-agarose (lanes 7 and 8) block RDR2 activity. These experiments suggest that RDR2 requires RNA templates with free 3' ends, as in the model at the lower right of Figure 1B.
Because Pol IV and RDR2 can function independently, one might expect that double-stranded RNAs would be produced from a single-stranded DNA template if Pol IV and RDR2 are both present. However, ribonuclease sensitivity tests indicate that this is not the case (Figure 1E, F). Ribonuclease H, which specifically degrades RNA strands of RNA-DNA hybrids, digests full-length transcription products of Pol IV (Figure 1E, lanes 2 and 4), as well as most shorter primer extension products, indicating that most Pol IV transcripts form RNA-DNA hybrids, not dsRNAs. A subset of short Pol IV transcripts that are 21 or 24-26 nt in size are resistant to RNase H when made by Pol IV-RDR2, but not by Pol IV isolated in the rdr2 mutant background (compare lanes 2 and 4), suggesting that they might be dsRNAs. However, RNase I, which digests ssRNA, but not dsRNA, digests these bands (Figure 1F). We conclude that Pol IV-RDR2 transcription of single-stranded DNA does not produce appreciable levels of dsRNA.
Pol IV termination is induced by basepaired nontemplate DNA.
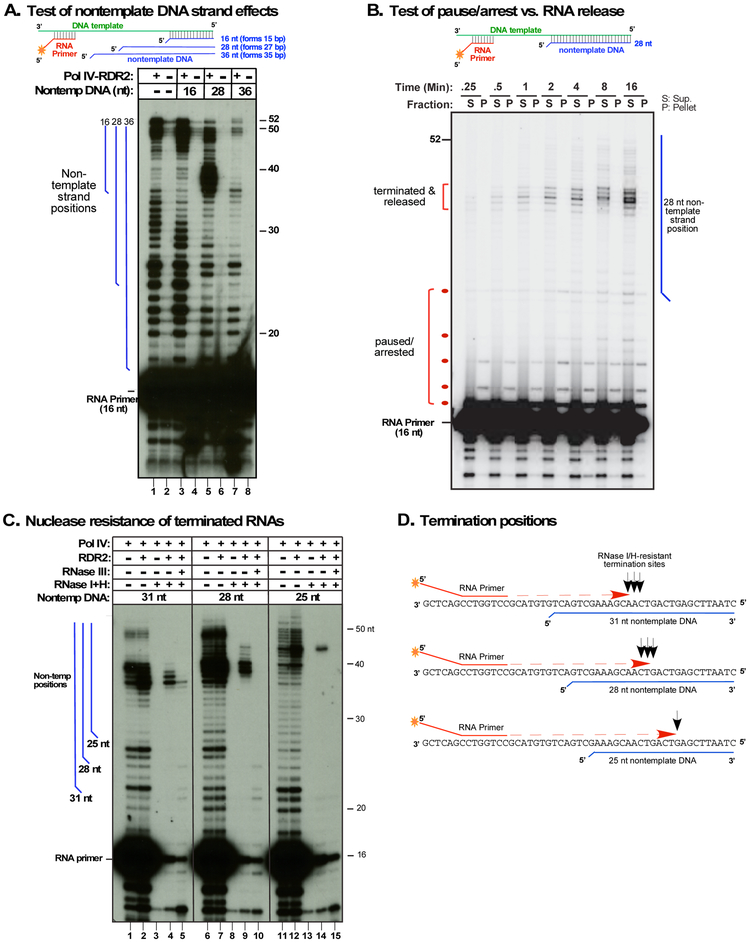
Pol IV has negligible activity using double-stranded (ds) template DNA (Haag et al., 2012; Onodera et al., 2005), thus we asked what happens when Pol IV engaged in transcription of a template DNA strand encounters a basepaired nontemplate strand (Figure 2). In the absence of nontemplate DNA, transcripts up to full-length (52 nt) are produced (Figure 2A, lane 1; see also Figure S1B). Upon annealing a 16 nt nontemplate DNA strand, which forms 15 bp with the template (and has a 1 nt 5' flap), long transcripts are still obtained, but 44-47 nt transcripts, slightly shorter than full-length (52 nt) become more abundant (Figure 2A, lane 3). A nontemplate strand of 28 nt, forming 27 bp with the template, induced abundant transcripts of 36-41 nt (Figure 2A, lane 5) whose 3' ends map to positions 12-15 nt beyond the point where the dsDNA region began. A 36 nt nontemplate strand, forming a dsDNA region that extends to within 2 nt of the RNA primer, allowed elongation of the primer for ~ 10 nt, but longer transcripts were substantially reduced. Whether full-length transcripts result from nontemplate strand displacement or a failure of template-nontemplate strand annealing is unclear.
Figure 2. Nontemplate DNA induces Pol IV termination and dsRNA synthesis.
A. Test of non-template DNA strand effects on Pol IV transcription. Primer-initiated transcription was conducted as in Figure 1C but with nontemplate DNA strands of 16, 28 or 36 nt annealed to the template DNA (see also Figure S1B). Nontemplate DNA strands have an unpaired nucleotide at their 5' ends. Lanes 1, 3, 5, and 7 show reaction products of Pol IV-RDR2 complexes purified by virtue of the FLAG-tag on NRPD1. Lanes 2, 4, 6, and 8 are controls using non-transgenic plant extracts subjected to anti-FLAG affinity purification. Blue vertical lines to the left of the autoradiogram depict the positions of nontemplate strands relative to the transcripts.
B. Pol IV termination is induced by nontemplate DNA. Primer initiated transcription, using the template - 28 nt nontemplate strand pair, was conducted using Pol IV-RDR2 complexes immobilized on anti-FLAG resin. At the indicated timepoints, transcription reactions were subjected to centrifugation and RNAs of supernatant (S) and pellet (P) fractions subjected to denaturing PAGE and autoradiography.
C. Nontemplate DNA affects termination position and induces production of RNAs with the nuclease sensitivities of dsRNA. Pol IV affinity purified from RDR2 wild-type (+) or rdr2 mutant (−) backgrounds was tested for primer-initiated transcription of the DNA template annealed to 31nt, 28nt or 25nt nontemplate DNAs. Transcription products were incubated with RNAses I and H or RNase III, as indicated. Blue vertical lines represent positions of nontemplate strands.
D. Pol IV termination positions are not sequence-specific. The cartoon shows the positions (black arrows) of 3' ends of RNase I/H-resistant Pol IV transcripts relative to DNA template, RNA primer, and nontemplate DNA strands.
In addition to inducing shortened transcripts, nontemplate DNA strands suppress some transcripts produced in their absence. For instance, RNAs of 32-37 nt observed with the template strand alone are suppressed by the 16 nt nontemplate strand (Figure 2A, compare lanes 1 and 3). The 3’ end of a 37 nt transcript corresponds to the position where the double-stranded DNA region begins, but 32-36 nt transcripts end prior to reaching the dsDNA region. Likewise, the 28 nt nontemplate strand suppresses production of transcripts whose 3' ends occur prior to reaching the dsDNA region (Figure 2A, compare lanes 1 and 5). These observations suggest that template-nontemplate strand basepairing may prevent cis-basepairing within the template strand in a way that affects Pol IV pausing or termination.
To determine if transcripts induced by nontemplate DNA result from Pol IV termination, pausing, or arrest, we tested whether transcripts are released from, or remain associated with, Pol IV. Using Pol IV-RDR2 complexes immobilized on anti-FLAG resin (by virtue of FLAG-tagged NRPD1), transcription was conducted using end-labeled primer RNA and the DNA template annealed to the 28 nt nontemplate strand. At varying times, reactions were subjected to brief centrifugation, then resin-associated (pellet, P) and supernatant (S) fractions were subjected to denaturing PAGE and autoradiography (Figure 2B). Nontemplate strand-induced 36-41 RNAs were detected within 15-30 seconds, accumulated throughout the time-course, and were found exclusively in supernatant fractions, indicative of terminated, released transcripts. Shorter RNAs detected in the pellets are interpreted to be paused or arrested transcripts still associated with Pol IV.
To further test how Pol IV termination sites relate to dsDNA encounter positions, we conducted transcription assays using the template strand annealed to 31, 28 or 25 nt nontemplate DNAs (Figure 2C). The 31 nt nontemplate strand, extending nearest the RNA primer, induced terminated Pol IV transcripts of 35-38 nt (lanes 1 and 2), the 28 nt nontemplate strand induced 36-41 nt transcripts (lanes 6 and 7) and the 25 nt nontemplate strand induced 40-44 nt transcripts (lanes 11 and 12). In each case, Pol IV termination occurs 12-16 nt beyond the point where the dsDNA region begins, at sites sharing no obvious sequence similarity (Figure 2D; see also Figure S1B).
Nuclease sensitivity tests provided the first indication that nontemplate DNA induces Pol IV-RDR2 complexes to synthesize dsRNA. In the absence of RDR2, Pol IV transcripts induced by 25, 28 or 31nt nontemplate strands are sensitive to a mixture of RNase I and RNase H (Figure 2C, lanes 3,8,13), consistent with the transcripts being ssRNAs or ssRNA-DNA hybrids. However, Pol IV transcripts generated in the presence of RDR2 are resistant to the nucleases, consistent with being strands of dsRNA (lanes 4, 9, 14). Moreover, Pol IV-RDR2 transcripts resistant to RNases I and H are digested by E. coli RNase III, which specifically degrades dsRNA (lanes, 5, 10 and 15). Importantly, the labeled Pol IV products released into the supernatant in the experiment of Figure 2B correspond to the nuclease resistant RNAs of Figure 2C (lanes 9 and 10), indicating that dsRNAs are the released products of Pol IV-RDR2 transcription complexes.
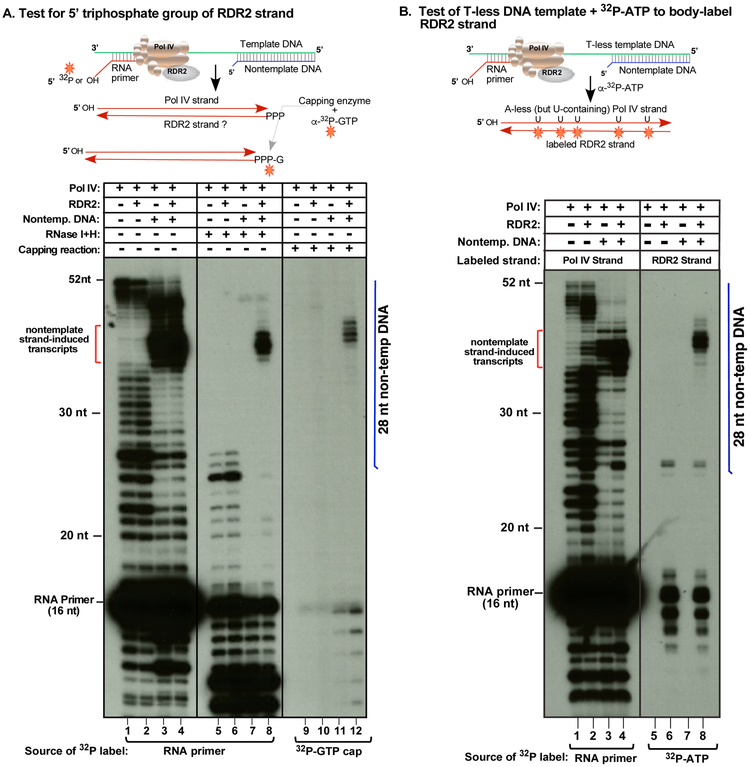
Detection of second strand synthesis by direct labeling of RDR2 transcripts
In Figures 1 and 2, the first RNA strand, synthesized by Pol IV, has the labeled 5' monophosphate of the primer. If a complementary strand is synthesized by RDR2, it should have a 5' triphosphate, making it a substrate for GTP addition by capping enzyme. To test this hypothesis, RNAs labeled by virtue of the 32P end-labeled primer (Figure 3A, lanes 1-8) were compared to RNAs initiated using unlabeled primer but then incubated with capping enzyme and α-32P-GTP (Figure 3A, lanes 9-12). As in Figure 2, labeled primer extension products are synthesized by Pol IV, independent of RDR2 (Fig. 3, compare lanes 1 and 2), and terminate early in the presence of the 28 nt nontemplate DNA strand (lanes 3 and 4). Only if RDR2 is present are RNAs induced by the nontemplate strand resistant to RNAses I and H (Figure 3A, compare lanes 7 and 8), indicative of dsRNA. Moreover, the RDR2-dependent transcripts can be labeled by 32P-GTP capping (lane 12) and are resistant to Terminator Exonuclease (Lucigen Corporation), which degrades RNAs with 5’ monophosphate but not triphosphate groups (Figure S2).
Figure 3. Tests of second strand synthesis by RDR2.
A. Capping assay to test for a 5’ triphosphate on RDR2-dependent transcripts. First strands, synthesized by Pol IV associated with RDR2 (+) or purified from a rdr2 mutant (−), were initiated using an RNA primer with a labeled 5' monophosphate (lanes 1-8) or 5' hydroxyl group (lanes 9-12). In half of the reactions, the template was annealed to the 28 nt nontemplate DNA strand to induce Pol IV termination; remaining reactions lacked the nontemplate strand. Lanes 1-4 differ from lanes 5-8 with respect to RNase I/H treatment. In lanes 9-12, vaccinia virus capping enzyme catalyzed labeling with α-32-P-GTP. For related experiments, see Figure S2.
B. Use of a T-less DNA template and α-32P-ATP incorporation to label second strands. All reactions contain Pol IV, associated with RDR2 (+) or purified from an rdr2 mutant (−) background. In the reactions of lanes 3, 4, 7 and 8, the T-less template was annealed to a 28nt nontemplate DNA strand whose relative position is depicted by the vertical blue line (see Figure S1C for details). Transcripts in lanes 1-4 were initiated using a 5' end-labeled RNA primer to label Pol IV transcripts. In lanes 5-8, RDR2 transcripts were body-labeled using α-32P-ATP. Related information is provided in Figure S3.
As an independent test of second-strand synthesis by RDR2, a DNA template that lacks thymidines was used to generate transcripts in the presence of α-32P-ATP (Figure 3B; see also Figure S1C). With no T's in the template, A's are not incorporated into the initial Pol IV strand. However, U's in the first strand template A incorporation into second strands (see Figure 3B diagram). Using T-less template DNA only, Pol IV generates full length as well as shorter RNAs (lanes 1 and 2). Annealing of a 28 nt nontemplate strand induces early Pol IV termination, generating prominent 37-40 nt transcripts (lanes 3 and 4). If RDR2 is present, these 37-40 nt transcripts are converted into dsRNAs, such that 32P-ATP is incorporated into the RDR2 strands (compare lanes 5-8). The slower mobility of RDR2 strands compared to Pol IV strands is explained, in part, by their different sequences and molecular masses (see Figure S3).
Collectively, the results of Figure 3 indicate that Pol IV termination induced by nontemplate DNA is coupled to second strand RNA synthesis by RDR2.
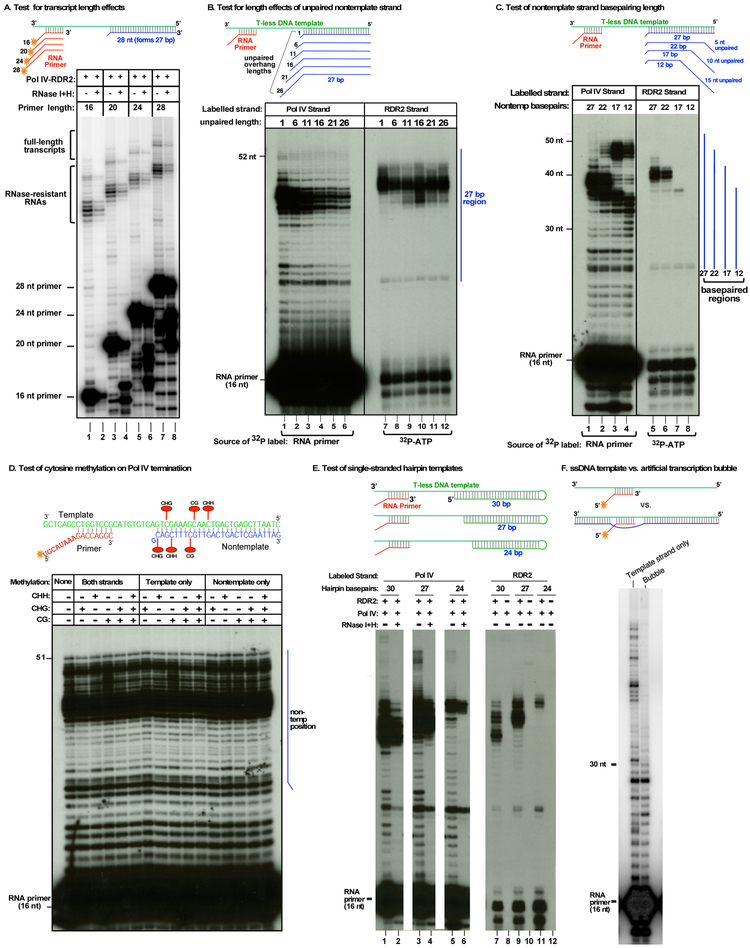
The extent of template-nontemplate basepairing is critical for Pol IV termination-RDR2 coupling.
Multiple variables might affect Pol IV-RDR2 coupling. To test whether Pol IV transcript length matters, RNA primers with 5' tails ranging from 8 nt to 20 nt were tested (Figure 4A). Each primer yielded transcripts that terminated at the same template positions, generating dsRNAs resistant to RNAses I and H (lanes 2,4,6,8). These results indicate that transcript length does not dictate Pol IV termination.
Figure 4. Tests of RNA and DNA parameters for effects on Pol IV termination and RDR2 coupling.
A. Test of transcript length on Pol IV termination/RDR2 coupling sites. End-labelled RNA primers of 16-28 nt, varying at their 5' ends (see diagram), were used to initiate transcription by Pol IV-RDR2. Duplicate reactions were differentially subjected to RNase I/H digestion.
B. Test of displaced nontemplate DNA length on Pol IV termination/RDR2 coupling. Nontemplate DNA strands of 28-53 nt, with variable unpaired 5' end lengths (1-26 nt), were annealed to the T-less template (see diagram) and tested in Pol IV-RDR2 transcription reactions initiated using an RNA primer. In the reactions of lanes 1-6, the primer was 5' end-labelled, allowing Pol IV transcript detection. In the reactions of lanes 7-12 the primer was unlabeled and α-32P-ATP was used to body-label second strand RDR2 transcripts. The vertical blue line denotes the 27 basepaired region common to all template-nontemplate combinations.
C. Test of the number of basepaired nontemplate nucleotides distal to the Pol IV encounter position. 28 nt nontemplate DNA strands forming 12-27 bp with the T-less template, but varying at their 3 ' ends, were tested in Pol IV-RDR2 transcription reactions initiated using an RNA primer. In the reactions of lanes 1-4, the primer was 5' end-labelled, allowing Pol IV transcripts to be visualized. In the reactions of lanes 5-8 the primer was unlabelled and α-32P-ATP was used to body-label RDR2 transcripts.
D. Test for effects of template and/or nontemplate DNA cytosine methylation on Pol IV termination. Template and nontemplate strands that were unmethylated (far left lane), or methylated at CG, CHG or CHH (in 5' to 3' orientation) positions indicated in the cartoon at top, were tested in Pol IV-RDR2 transcription reactions initiated using a 5' end-labelled primer to visualize Pol IV transcripts.
E. Test of single-stranded hairpin templates for Pol IV termination/RDR2 coupling. The T-less template was lengthened to allow formation of hairpins with 24-30 bp stems, as indicated in the cartoon. Pol IV-RDR2 transcription reactions were initiated using a 5' end-labeled primer to visualize Pol IV transcripts (lanes 1-6) or using an unlabeled primer and α-32P-ATP to body-label RDR2 transcripts (lanes 7-12). Pol IV purified from rdr2 null mutant plants was tested in lanes 8, 10 and 12. Reaction products of lanes 2, 4 and 6 were treated with RNases I and H. Images shown are lanes from the same gel and autoradiogram exposure.
F. Test for Pol IV transcription from an initiation bubble. Primer-initiated Pol IV-RDR2 transcription reactions were conducted using single-stranded template DNA or template DNA annealed to a partially non-complementary DNA strand, as depicted in the cartoon. The primer was 5' end-labelled to visualize Pol IV transcripts.
We next asked whether the length of displaced non-template strand DNA affects Pol IV termination and/or RDR2 coupling. The T-less template was used (as in Fig. 3B), annealed to nontemplate strands that each form 27 bp with the template but have unpaired 5' ends ranging from 1-26 nt in length (Figure 4B). An end-labeled RNA primer was used to detect Pol IV transcripts (lanes 1-6) and α-32P-ATP incorporation was used to label second strands made by RDR2 (lanes 7-12). No effect was observed on the positions of Pol IV termination nor on Pol IV-RDR2 coupling.
To test whether the length of the double-stranded region matters, we tested 28 nt nontemplate strands that form 27, 22, 17 or 12 bp with the T-less template (Figure 4C). Nontemplate strands forming 27 or 22 bp strongly induced Pol IV termination, generating 35-40 nt transcripts that terminated 12-17 nt prior to the end of the template (lanes 1 and 2). Reducing basepairing to 17 or 12 bp resulted in longer readthrough transcripts of 46-50 nt, with 35-40 nt transcripts greatly reduced in abundance (lanes 3 and 4). The transcript profile for the reaction using the nontemplate strand forming only 12 bp with the template resembles that obtained using template DNA alone, in the absence of nontemplate DNA (compare lane 4 to Figure 3B, lane 2).
Using α-32P-ATP to label second strands synthesized by RDR2 revealed that abundant RDR2 transcripts are only detected for template-nontemplate pairs that formed 27 or 22 bp (Figure 4C, lanes 5 and 6). Collectively, the results of Figure 4C indicate that the double-stranded region needs to be longer than 17 bp to induce Pol IV termination and Pol IV-RDR2 coupling.
To test whether DNA cytosine methylation affects Pol IV termination, we tested template and/or nontemplate strands that were unmethylated, methylated at individual CG, CHG or CHH motifs, or methylated in all three sequence contexts (Figure 4D; see diagram for methylcytosine positions). Cytosine methylation had no effect on Pol IV transcription or termination (Figure 4D).
We next asked whether Pol IV termination, and its coupling to RDR2 transcription, requires separable template and nontemplate strands of DNA, or merely basepaired DNA. For this test, we extended the T-less template to allow stem-loop formation, generating basepaired stems of 30, 27 or 24 bp, thus recapitulating the experiment of Figure 2C but using a single DNA molecule (Figure 4E). The results essentially mirror those of Figure 2C, with the length of Pol IV transcripts dictated by the position where dsDNA begins.
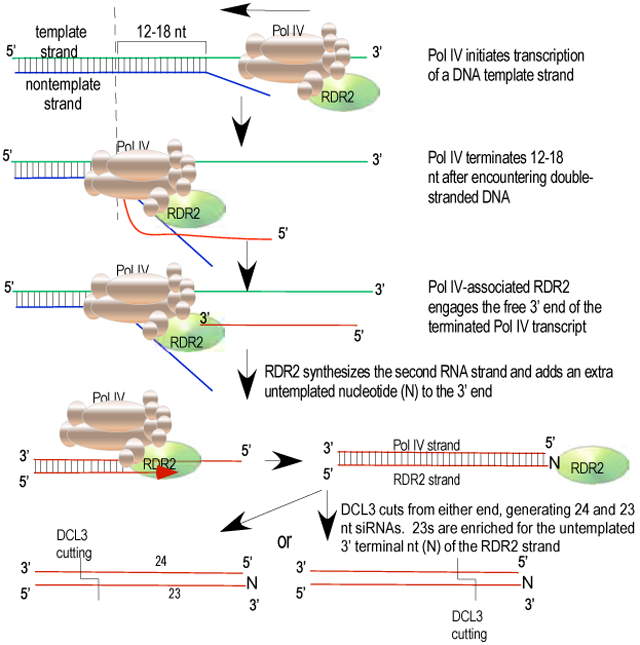
In the context of double-stranded chromosomal DNA, Pol IV presumably initiates within a locally melted region of duplex DNA, like other multisubunit RNA polymerases (Bae et al., 2015; Barnes et al., 2015; Holstege et al., 1997; Kahl et al., 2000). To test Pol IV’s ability to carry out transcription in the context of a DNA bubble (Figure 4F), we annealed template and non-template strands that have 12 nt of internal non-complementarity, and hybridized an RNA primer to one strand within the resulting bubble. Using the bubble template, Pol IV extends the primer only 14 -16 nt whereas using the template strand alone, long transcripts are produced (Figure 4F). These results are consistent with all prior results using non-bubble templates.
Pol IV, RDR2 and DCL3 are sufficient to reconstitute 24 nt siRNA biogenesis in vitro
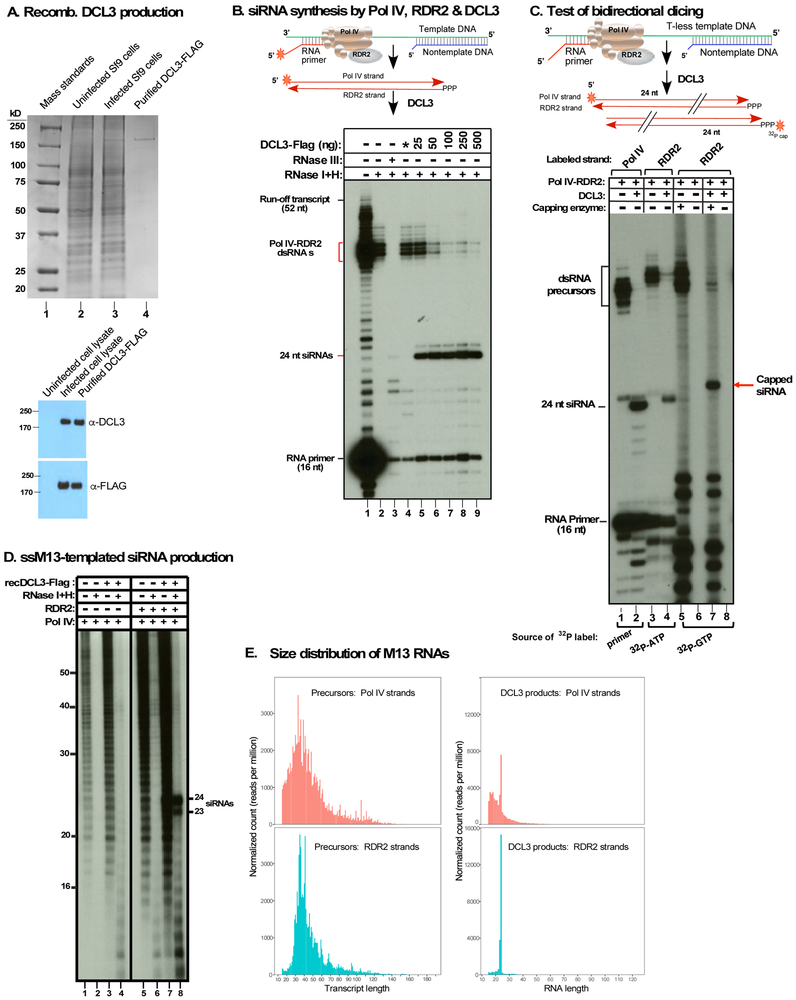
Using recombinant FLAG-tagged DCL3 produced in insect cells (Figure 5A), we tested whether Pol IV-RDR2 transcripts induced by the 28 nt nontemplate DNA strand are substrates for DCL3 (Figure 5B). Pol IV-RDR2 transcripts, whose Pol IV strands are 5’ end-labeled, are resistant to RNases I/ H (Figure 5B, lane 2) but degraded by RNase III (lane 3), indicative of double-strandedness. Addition of DCL3 converted the Pol IV-RDR2 transcripts into 24 nt RNA products (lanes 5-9).
Figure 5. Pol IV, RDR2 and DCL3 are sufficient for siRNA biosynthesis in vitro.
A. Baculovirus-mediated production of recombinant DCL3. Top panel: Stained SDS-PAGE gel showing protein standards (lane 1), uninfected Sf9 cell lysate (lane 2), DCL3-baculovirus infected cell lysate (lane 3) or affinity purified DCL3-FLAG. Bottom panel: immunoblots using anti-DCL3 or anti-FLAG antibodies.
B. siRNA synthesis by Pol IV, RDR2 and DCL3. Template DNA was annealed to a 28 bp nontemplate strand and Pol IV-RDR2 transcription was initiated using an end-labeled RNA primer. The autoradiogram shows untreated (lane 1), RNase I/H (lane 2), RNase III (lane 3) or DCL3-FLAG digested (25-500 ng; lanes 5-9) transcription products. Uninfected Sf9 cell lysate subjected to anti-FLAG affinity purification serves as a control in lane 4.
C. Pol IV-RDR2 dsRNA products are diced from either end. Using the T-less template, primer-initiated Pol IV-RDR2 reactions were labeled using end-labeled primer to detect Pol IV strands, using α-32P-ATP incorporation to body-label RDR2 strands, or using capping enzyme and α-32P- GTP to label RDR2 strand 5’ ends. For each labeling scheme, total transcription products and DCL-diced products are compared.
D. Pol IV, RDR2 and DCL3 produce siRNAs from single-stranded M13 bacteriophage DNA. Transcription reactions using α-32P-ATP to body-label transcripts were conducted using Pol IV-RDR2 (lanes 5-8) or Pol IV purified from rdr2 mutant plants (lanes 1-4). Transcripts in even lanes were subjected to digestion with RNases I and H. Transcripts in lanes 3, 4, 7 and 8 were incubated with DCL3. Related information is provided in Figure S4.
E. Size distribution and frequency of Pol IV (pink) and RDR2 (blue) strands of precursors and DCL3-diced siRNAs synthesized from M13 template DNA. Related information is provided in Figure S5.
To examine the directionality of DCL3 dicing, we used the T-less template to generate dsRNAs that were 5' end-labeled on the Pol IV strand, body-labeled throughout the RDR2 strand, or 5' end-labeled on the RDR2 strand by 32P-GTP capping following dicing (Figure 5C). Labeled 24 nt DCL3 products were observed in each case, showing that DCL3 can process precursor duplexes from either end.
In vitro biosynthesis of siRNAs from single-stranded M13 DNA
Pol IV transcription initiated using an RNA primer generates transcripts with defined 5' ends, but Pol IV does not require a primer and will initiate de novo on single-stranded DNA(Haag et al., 2012), including bacteriophage M13 (+) strands (Blevins et al., 2015). This prompted us to test whether Pol IV, RDR2 and DCL3 are sufficient to generate siRNAs from M13mp18 (+) DNA (Figure 5D), whose ~7.3 kb circular genome can fold into extensive secondary and tertiary structures that might facilitate Pol IV-RDR2 coupling (Figure S4). Pol IV transcription of M13 DNA, monitored by α-32P-ATP incorporation, yields a ladder of transcripts (Figure 5D, lane 1) that are RNAse I/ H-sensitive (lane 2) and are not substrates for DCL3 (lanes 3 and 4). However, if RDR2 is present, a portion of the labeled Pol IV transcripts become resistant to RNases I and H (compare lanes 5 and 6) and can be diced by DCL3 into 24 and 23 nt products (lane 8) in a ratio resembling their relative abundance in vivo (Kasschau et al., 2007). How DCL3 produces both 24 and 23 nt siRNAs is currently unknown.
Because M13 is single-stranded, yet is transcribed into dsRNAs by Pol IV and RDR2, the polymerases responsible for sense and antisense transcripts can be ascertained. Deep sequencing of RNAs made by Pol IV alone (isolated from a rdr2 mutant) or of RNAse I/H-resistant RNAs made by Pol IV-RDR2, before and after DCL3 dicing, shows that Pol IV makes first-strand transcripts whose polarity is opposite that of the DNA template (Figure 5E, see also Figure S5). Only if RDR2 is present are second-strand transcripts produced, which match the polarity of the ssDNA template and thus cannot be DNA-templated; instead, they can only be generated by transcription of first-strand RNAs made by Pol IV. More than 75,000 unique RNAse I/H-resistant RNAs generated by Pol IV-RDR2 were sequenced. The size distribution of Pol IV and RDR2 transcripts is similar, with most being 30-50 nt in length (Figure 5F, left plots). However, more transcripts shorter than 30 nt were detect for Pol IV than for RDR2, suggesting that RDR2 coupling is inefficient if Pol IV transcripts are shorter than ~30 nt.
Sequencing of DCL3-diced products revealed a major peak at 24 nt and a shoulder at 23 nt for both Pol IV and RDR2 strands (Figure 5E, right plots), consistent with the gel image of Figure 5D (lane 8). RNAs shorter than 23 nt are presumably transcripts too short to be diced.
Untemplated 3' terminal nucleotides are characteristic of RDR2 strands
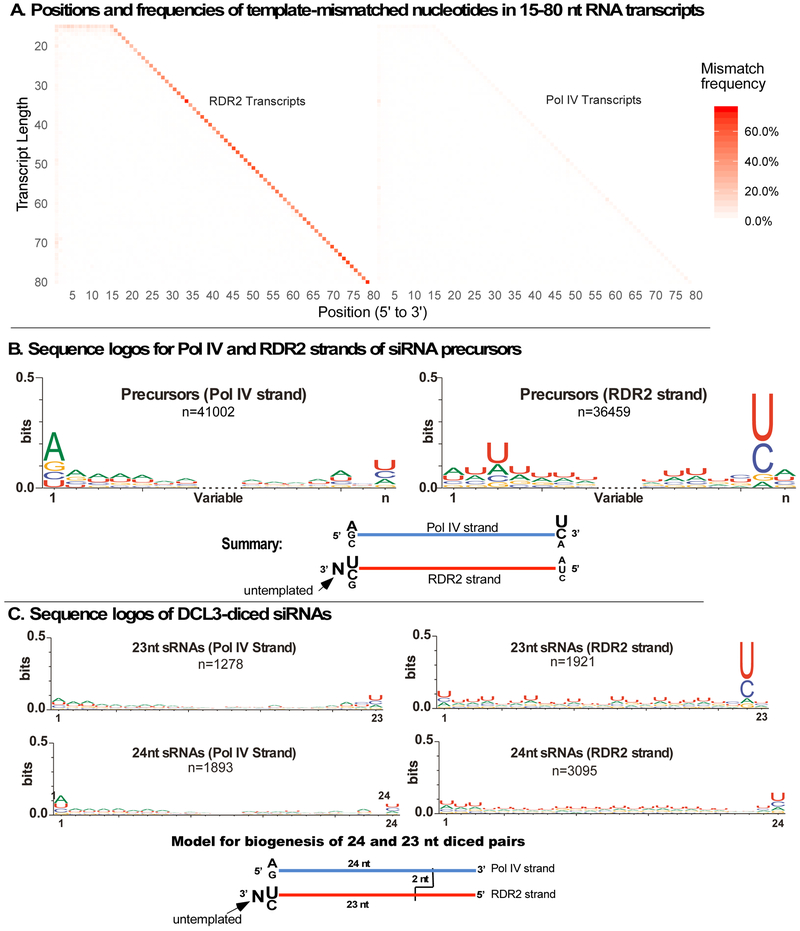
In vivo, P4R2 RNAs and siRNAs frequently have 3' nucleotides that are mismatched to the DNA template (Blevins et al., 2015; Zhai et al., 2015). Analysis of M13 transcripts revealed that mismatched nucleotides are characteristic of RDR2 transcript 3' ends, regardless of transcript length (Figure 6A). Among Pol IV transcripts, 3’ mismatched nucleotides occur at the same frequency as 5’ mismatched nucleotides and represent background levels typical of RNA-seq reads.
Figure 6. RDR2 transcripts have untemplated 3’ terminal nucleotides.
A. Positions and frequencies of template-mismatched nucleotides within Pol IV and RDR2 transcripts generated using M13 template DNA. Transcripts of 15-80 nt were examined. The frequency of template-mismatched nucleotides at each position is denoted in shades of orange/red. Related information is provided in Figure S6.
B. Consensus sequences for unique Pol IV and RDR2 transcripts longer than 26 nt. The 7 nt nearest the 5’- and 3’ ends are shown. Redundant sequences were condensed into unique sequences prior to analysis. Sequence logos were generated using WebLogo 3.6. In the summary cartoon, N denotes the untemplated nucleotide ascribed to RDR2 terminal transferase activity.
C. Sequence logos for Pol IV and RDR2 strands of 23 and 24 nt DCL3-diced siRNAs. The summary cartoon proposes a model to account for 23 nt siRNAs with 3’ terminal untemplated nucleotides.
Among RNAs longer than 26 nt, Pol IV transcripts generated from M13 DNA tend to initiate with a purine, in agreement with prior studies (Blevins et al., 2015; Zhai et al., 2015), with A>G (Figure 6B). Pol IV strand 3' ends show some enrichment for U>C (Figure 6B). RDR2 strands have a weak preference for A at their 5' ends, consistent with the weak preference for U at the 3' end of complementary Pol IV strands (Figure 6B). However, the most striking feature of RDR2 strands is the strong U/C consensus one nucleotide prior to the 3’ end (Figure 6B). We deduce that this strong U/C signature is the complement of the strong A/G signature of Pol IV strand 5' ends, with the 3' terminal nucleotide of RDR2 strands resulting from untemplated nucleotide addition after dsRNA synthesis is complete. We further deduce that RDR2’s terminal nucleotidyl transferase activity (Blevins et al., 2015) adds the untemplated nucleotide. Sequence data indicate that addition of more than one untemplated nucleotide is rare (Figure S6).
Examination of diced Pol IV-RDR2 precursors generated from M13 template DNA shows that untemplated 3’-terminal nucleotides are greatly enriched among 23 nt siRNAs derived from the RDR2 strand (Figure 6C), adjacent to the strong U/C consensus at the penultimate position. This suggests a model whereby DCL3 cuts the Pol IV strand 24 nt from its 5' end, creating a 2 nt 3' overhang with respect to the RDR2 strand. Because the paired RDR2 strand is extended 1 nt at the 3’ end by RDR2’s terminal transferase activity, the resulting RDR2 strand siRNA is 23 nt (Figure 6C, bottom left). Duplexes can also be cut from the other end, generating 24 nt siRNAs that correspond to the 5’ end of RDR2 strands (see Figure 5C). The alternative ways in which precursor duplexes can be diced presumably dilutes the A/G consensus signature of Pol IV transcript 5’ ends, making this signature less prominent among diced siRNAs.
Discussion
Pol IV and RDR2 represent a unique partnership between DNA- and RNA-dependent RNA polymerases whose concerted reactions convert the information of single-stranded DNA into double-stranded precursors of 24 nt siRNAs. In vivo, it has been unclear which enzyme synthesizes which strand of dsRNA precursors (Blevins et al., 2015; Zhai et al., 2015). Our results show definitively that Pol IV synthesizes first-strand RNAs and that Pol IV’s unusual sequence-independent mode of termination, induced by template-nontemplate strand basepairing, is needed to channel Pol IV transcripts to RDR2 for second strand synthesis.
Multisubunit RNA polymerases generally terminate in response to RNA signals (Porrua et al., 2016). In E. coli, RNA stem-loops followed by a string of uracils, or RNA-encoded Rho protein recruitment sites, mediate most termination events (Ray-Soni et al., 2016). In eukaryotes, sequence-specific cleavage of nascent transcripts enables RNA binding by termination factors that induce Pol I and Pol II termination (Birse et al., 1997; Connelly and Manley, 1988; El Hage et al., 2008; Goodfellow and Zomerdijk, 2013; Kuehner et al., 2011). Pol III termination is induced by strings of uridines in nascent transcripts, reminiscent of rho-independent termination in E. coli (Arimbasseri and Maraia, 2015; Nielsen et al., 2013), with thymidines of the nontemplate DNA strand, complementary to the uridines of the nascent transcript, also playing a role (Arimbasseri and Maraia, 2015). Pol IV termination appears to be distinct in that it is sequence-independent and specified by the extent of template-nontemplate strand basepairing.
The precise mechanisms that halt Pol IV elongation and induce transcript release are unclear, but weak motor activity associated with deletions affecting the trigger loop (Landick, 2009) may limit Pol IV’s ability to displace nontemplate DNA. Pol IV also has a ten amino acid deletion in the vicinity of the “rudder” loop, first observed in bacterial RNA polymerase (Zhang et al., 1999) and subsequently in yeast Pol II (Cramer et al., 2001). The rudder is thought to help separate template and nontemplate DNA strands downstream of the catalytic site, suggesting that Pol IV’s ability to propagate a transcription bubble may be impaired (Figure S7). Interestingly, 12 amino acids are also deleted in the vicinity of the “zipper” loop thought to facilitate template - nontemplate strand reannealing upstream of the catalytic center, suggesting that transcription bubble closure may also be impaired (Figure S7). However, amino acids corresponding to the “lid” loop, located at the upstream edge of the DNA-RNA hybrid, near the base of the RNA exit channel, are present in NRPD1 (Figure S7). Collectively, the trigger, lid, rudder and zipper loops affect multiple steps of the transcription cycle (Lee and Borukhov, 2016; Naji et al., 2008; Toulokhonov and Landick, 2006), such that deletions and amino acid changes within these elements may contribute to Pol IV’s atypical mode of termination.
An intriguing hypothesis suggested that Pol IV misincorporation may induce Pol IV termination, especially at methylcytosines (Zhai et al., 2015), thus accounting for template-mismatched nucleotides at the 3’ ends of P4R2 RNAs in vivo. Pol IV transcription is error-prone (Marasco et al., 2017), but our results show that untemplated nucleotides are found almost entirely at the 3' ends of RDR2 strands, not Pol IV strands. Moreover, methylated cytosines have no discernable effect on Pol IV termination (Figure 4 D) or Pol IV misincorporation in vitro (Marasco et al., 2017). These results argue against the Pol IV misincorporation hypothesis and point to RDR2’s terminal transferase activity as the likely explanation for the 3’ untemplated nucleotides of P4R2 RNAs and siRNAs.
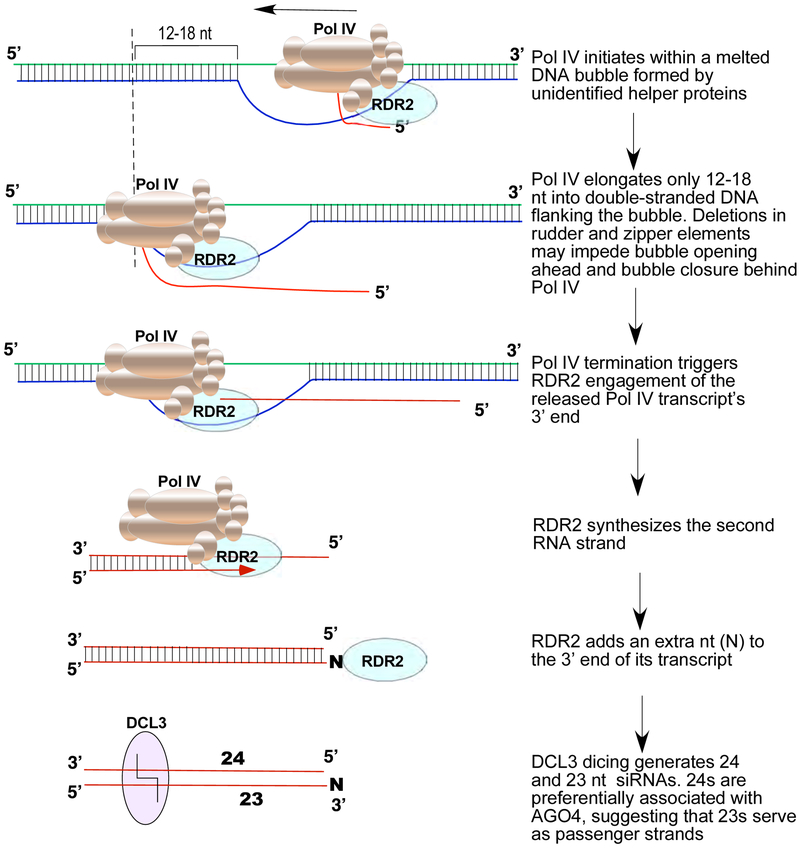
Our results also argue against the hypothesis that Pol IV transcripts are short because the high methylcytosine density of Pol IV-transcribed regions induces Pol IV misincorporation and termination. Instead, our data are consistent with a simple model (Figure 7) in which Pol IV initiates within the context of a transcription bubble, like other multisubunit RNA polymerases (Bae et al., 2015; Barnes et al., 2015; Holstege et al., 1997; Kahl et al., 2000). Pol IV then transcribes one strand to the edge of the bubble, encounters basepaired DNA and extends only 12-18 nt more before terminating, generating short transcripts that average only ~32 nt in vivo. However, the size of the initial bubble, and the factors that generate it, are unknown at this time.
Figure 7.
Model for the sequential reactions of Pol IV, RDR2 and DCL3, collectively accounting for siRNA biogenesis and RNA channeling in the RNA-directed DNA methylation pathway. Related information pertaining to Pol IV sequences that may influence activity is provided in Figure S7.
Pol IV termination induced by nontemplate DNA is critical for RDR2 to engage the free 3’ end of the released Pol IV transcript and use it as a template to synthesize the complementary strand and then add an untemplated nucleotide to the 3’ end (see Figure 7). This untemplated terminal nucleotide is enriched among 23 nt siRNAs in the M13 system (Figure 6), and in vivo (Wang et al., 2016) and is present at the same end of precursor duplexes as Pol IV strand 5’ ends (see Figure 7). Pol IV strands labeled at their 5’ ends give rise to labeled 24 nt siRNAs following dicing (Figures 5B,C). This suggests a model in which DCL3 measure 24 nt from the 5’ end of the Pol IV strand and cuts to leave a 2 nt 3' overhang, like other Dicers (Park et al.,2011). Due to the extra untemplated nucleotide at the 3' end of the RDR2 strand, the resulting DCL3-cleaved RDR2 strand is 23 nt. Importantly, siRNAs found in association with AGO4 are almost exclusively 24 nt (Havecker et al., 2010; Qi et al., 2006). This leads us to propose that 23 nt RNAs typically serve as the passenger strands for the 24 nt guide siRNAs that become stably associated with AGO4. If so, the terminal transferase activity of RDR2 has a purpose, enabling passenger and guide strands to be discriminated (Figure 7). Collectively, the enzymatic properties of Pol IV, RDR2 and DCL3 can account for RNA channeling from initial transcription to Argonaute loading.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Craig S. Pikaard (cpikaard@indiana.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Arabidopsis thaliana plants used in the study were all of the Col-0 genetic background. Plants used in this study were grown under long day conditions (16-hours of light followed by 8-hours of dark) in a growth room illuminated with fluorescent lights. Transgenic line genotypes used for Pol IV and/or RDR2 affinity purification were: NRPD1-FLAG nrpd1-3, NRPD1-FLAG nrpd1-3 rdr2-1 and RDR2-HA rdr2-1 nrpd1-3 (Haag et al., 2012). Leaves of 3-4 week old plants were flash frozen in liquid nitrogen and stored at −80°C. A Sf9 cell line (Thermo Fisher) was used to express recombinant RDR2 and DCL3 proteins cloned into baculovirus vectors. Cells were grown at 27°C in supplemented Grace’s insect cell media (Thermo Fisher) containing 10% v/v fetal bovine serum (Thermo Fisher), as either monolayers or suspension cultures.
METHOD DETAILS
Affinity purification of proteins from transgenic plants
Four grams of frozen leaf tissue was ground in liquid-nitrogen using a mortar and pestle, resuspended in 14 mL of extraction buffer (20 mM Tris-HCl pH-7.6, 150 mM sodium sulfate, 5 mM magnesium sulfate, 20 μM zinc sulfate, 1 mM PMSF, 5 mM DTT, and 1X plant protease inhibitor cocktail (Sigma), passed through two layers of Miracloth and centrifuged at 18,000 × g for 15 min to pellet debris. The supernatant was incubated with 25 μL of anti-FLAG M2 or anti-HA resin (Sigma) for 2.5 hours. The resin was pelleted by centrifugation at 200 × g for 2 min and washed twice with 14 mL of extraction buffer minus protease inhibitors. The resin, with associated affinity-captured proteins, was then suspended in 50 μL of 20 mM HEPES-KOH pH7.6, 100 mM potassium acetate, 5 mM magnesium sulfate, 10% v/v glycerol, 20 μM zinc sulfate, 0.1 mM PMSF, 1 mM DTT.
Synthetic nucleic acids used in transcription assays
DNA and RNA oligonucleotides used in the study were purchased from Integrated DNA Technologies, Inc. and are listed in Table S1.
DNA template-RNA primer hybridization
Equimolar amounts of template DNA and RNA primer oligos were mixed in annealing buffer (30 mM HEPES-KOH pH 7.6, 100 mM potassium acetate), brought to a boil in a water bath and slowly cooled to room temperature. For reactions involving nontemplate DNA, a 10% excess of nontemplate oligonucleotide was included in annealing reactions. RNA primer end-labeling was accomplished using T4 polynucleotide kinase (T4 PNK, NEB) and 25 μCi of ATP, [γ32P]- 6000 Ci/mmol (Perkin Elmer).
In-vitro transcription
Transcription reactions used 50 μL of affinity resin slurry with associated Pol IV, RDR2 or Pol IV-RDR2 (or non-specifically associated proteins of non-transgenic plant controls) mixed with transcription reaction buffer to bring the final volume to 100 μL. Final concentrations of reaction components were 20 mM HEPES-KOH pH 7.6, 100 mM potassium acetate, 60 mM ammonium sulfate, 10 mM magnesium sulfate, 10% v/v glycerol, 20 μM zinc sulfate, 0.1mM PMSF, 1mM DTT, 0.8U/μL RibolockTM (Thermo Fisher). Reactions involving end-labelled primer RNA included 25nM each of the DNA template and primer and 1 mM each of ATP, GTP, CTP and UTP. For body labelling of RDR2 strands, 250 nM T-less template DNA and 1 mM each of GTP, CTP and UTP, 40 μM ATP, 10 μCi of ATP, [α32P]-3000 Ci/mmol (Perkin Elmer) were used. Transcription reactions were incubated 1h at room temperature on a rotating mixer, then stopped by addition of 25 mM EDTA and incubation at 75° C for 10 min. Transcription reactions were then passed though PERFORMA spin columns (Edge Bio) according to the manufacturer’s protocol, and adjusted to 0.3 M sodium acetate (pH 5.2). 15 μg GlycoblueTM (Thermo Fisher) was added and RNAs precipitated with 3 volumes of isopropanol at −20°C overnight. Following centrifugation, pellets were washed with 70% ethanol, resuspended in 5 μL of 2X RNA loading dye (New England Biolabs) and heated 5 min at 75° C. RNAs were resolved on 15% polyacrylamide 7M Urea gels. Gels were transferred to filter paper, vacuum dried and subjected to autoradiography or phosphorimaging using a TyphoonTM scanner (GE Healthcare).
Transcript release assay
In-vitro transcription assays were carried out as described above. At various timepoints, transcription was stopped by EDTA addition. Resin-immobilized Pol IV-RDR2 was pelleted by centrifugation at 200 × g for 1 min and the supernatant collected. The pellet was washed once and then resuspended in 100 μL of transcription buffer. Following heat treatment at 75°C, supernatant and pellet fractions were precipitated and subjected to PAGE as described above.
RNase sensitivity assays
RNase-treated transcription reactions involved incubation with 2.5 units of RNase H (New England Biolabs), 2.5 units of RNase I (Promega), or both enzymes, at 37° C for 30 min. For RNase III tests, 1 unit of enzyme (Epicentre) was added 10 min after addition of RNases I and H. Reactions were stopped by adding SDS to a final concentration of 0.15% (w/v). For RNase H-only tests, reactions were stopped with EDTA and incubated at 75° C for 20 min.
Capping reactions
Transcripts were alcohol precipitated and pellets washed twice with 70% ethanol. RNA was resuspended and incubated 1 hr, 37°C with vaccinia virus capping enzyme (New England Biolabs), as per the manufacturer’s protocol, in the presence of 100 μM unlabeled GTP and 10 μCi of [α32P]- GTP, 3000 Ci/mmol (Perkin Elmer).
Cloning, expression, and purification of recombinant DCL3
A DCL3 cDNA, codon optimized for protein expression in insect cells and including a N-terminal 10X His-tag and C-terminal FLAG and Strep tags, was synthesized by GenScript® and cloned into pUC57. The cDNA was subcloned into a pFastBacTM HT B vector (Thermo Fisher Scientific) and used to make recombinant bacmid DNA in E coli DH10Bac cells, according to the supplier’s protocol (Bac-to-Bac® Baculovirus expression system; Thermo Fisher Scientific). DCL3-expressing virus was obtained by celfectin-mediated transfection of bacmid DNA into the sf9 cells, using a multiplicity of infection (MOI) of 2. Cells were grown for 72 hr at 27 °C, collected by centrifugation at 350 × g, 10 min, and lysed in hypertonic lysis buffer (50 mM HEPES-KOH pH 7.5, 400 mM NaCl, 10% glycerol, 1mM PMSF, 1% protease inhibitor cocktail). The lysate was centrifuged at 39,200 × g at 4°C, 30 min. The supernatant was incubated with ANTI-FLAG® M2 affinity beads (Sigma-Aldrich) on a rotating mixer for 2 hrs at 4°C. Beads were collected by centrifugation for 5 min at 200 × g, and washed 3X with 20 volumes of lysis buffer and 1X with 20 volumes of elution buffer: 50 mM HEPES-KOH pH 7.5, 150 mM NaCl and 10% glycerol, 250 μg/mL 3x FLAG peptide (APExBIO). The eluted fraction was concentrated using a centrifugal filter unit (EMD Millipore) with a 30 KDa cutoff size and analyzed by electrophoresis on a 4-20% gradient SDS-PAGE gel and immunoblotting using anti- DCL3 or anti-FLAG® M2 (Sigma Aldrich) antibodies. Recombinant DCL3 was stored at −20°C in storage buffer (HEPES-KOH pH7.5, 150 mM NaCl and 50% glycerol).
Over-expression and purification of recombinant RDR2
Recombinant RDR2 was over-expressed and purified as described previously (Blevins et al., 2015). Briefly, Sf9 cells were used to produce RDR2 using baculovirus mediated protein expression. The baculovirus infected sf9 cells were lysed in a buffer containing 50 mM HEPES-KOH (pH 7.5), 400 mM KCl, 1 mM PMSF and 10% glycerol. The cell lysate was clarified by centrifugation at 39,000 × g for 30 mins at 4 °C and subjected to affinity chromatography using anti-V5 antibody agarose beads. RDR2 was eluted using V5 peptide in an elution buffer containing 50 mM HEPES-KOH (pH 7.5), 150 mM NaCl, 0.01% NP-40 and 10% glycerol. The eluted protein was concentrated using a Centricon filter.
RDR2 activity assays
Recombinant RDR2 transcription was carried out in 100 μl reactions containing ~200 ng RDR2, 25 nM end-labeled 37 nt template RNA, 25 mM HEPES-KOH pH 7.5, 2 mM MgCl2, 0.1 mM EDTA, 0.1% Triton X100, 20 mM ammonium acetate, 3% PEG 8000, 0.1 mM of rATP, rGTP, rCTP and rUTP, respectively and 0.8 U/ÛL RiboLock (Thermo Fisher Scientific). Biotinylated template RNA was incubated with 250 ng streptavidin or 50 μl streptavidin-agarose resin (Thermo Fisher Scientific). Reactions were incubated at room temperature, 60 min and stopped with 10 mM EDTA. RNAs were then purified by TRIzol extraction and ethanol precipitation. Reaction products were resolved by electrophoresis through 15% native PAGE gels and visualized by autoradiography.
DCL3 dicing
RNAs of transcription reactions were diced in 50 μL reactions containing 100 ng DCL3, 50 mM HEPES-KOH (pH 7.6), 150 mM sodium chloride, 5 mM magnesium chloride, 5 mM ATP, 0.5 mM GTP and 10% glycerol, for 60 min at room temperature. Reactions were stopped by 10 mM EDTA addition and incubation at 75° C, 10 min. To detect diced body-labeled RNAs, transcription involved the T-less template and inclusion of 10 μCi α32P- UTP (3000 Ci/mmol) UTP (Perkin Elmer) in the reactions.
Transcription of single-stranded M13 DNA template
Pol IV purified with or without RDR2 (as described above) was incubated 1μg of single-stranded M13mp18 (+) template DNA in transcription buffer. The transcripts were body labeled using 10 μCi of α32P-ATP, (3000 Ci/mmol; Perkin Elmer). For deep sequencing library preparation, reactions were performed using unlabeled ribonucleotides. Transcription reactions were then subjected to RNase H treatment to eliminate RNA-DNA hybrids followed by treatment with Turbo-DNase (Thermo Fisher) to eliminate template DNA. The reactions were then treated with RNA pyrophosphohydrolase (RppH, NEB) to convert any 5’ triphosphate groups of RNAs into 5’monophosphates and allow adapter ligation for RNA library preparation. The libraries were generated using a TruSeq Small RNA Library Prep Kit (Illumina) according to the manufacturer’s protocol.
Deep Sequencing and analysis of M13-templated RNAs
RNA transcript and DCL3 cleavage product libraries were subjected to 80 cycles of paired-end sequencing using a NextSeq 500 instrument. Raw data were processed using 3’ adapter trimming script ‘PE_trimadapter.py’. In brief, if 5’ ends of forward and reverse reads are complementary, and remaining 3’ ends are adapter sequences, reads were merged and treated as single-end sequences. Merged sequences <15 nt were discarded. Remaining paired reads lacking adapter sequences were kept in paired format. For Pol IV and RDR2 transcripts, processed reads were first aligned to the A. thalianaTAIR10 genome sequence using Bowtie version 1.2.2 (Langmead et al., 2009), allowing 0 mismatches, to remove any contaminating RNAs. For recombinant DCL3 digestion products, reads mapping to a Spodoptera frugiperda draft genome assembly (WGS Project: NJHR01) were similarly removed. Bowtie options -a -v 0 were used for single-end sequences and options -a -v 0 --allow-contain used for paired-end reads. Filtered reads were then aligned to M13mp18 (Bayou Labs) allowing up to 3 mismatches, with option -a -v 3 for single-end sequences and -a -v 0 --allow-contain for paired-end reads. Alignment outputs from single-end sequences and paired-end reads were then merged into one file. Sequences present in both diced and un-diced Pol IV-RDR2 transcription samples were removed to enrich for DCL3 cleavage products.
RNAs analyzed in supplemental figures were single-end sequenced for 73 cycles, and 3’ adapter trimming performed using Cutadapt version 1.9.1 (Martin, 2011), with options -a TGGAATTC --discard-untrimmed -e 0 -m 15 -O 8. Processed reads were then analyzed as described above. The strandedness of aligned sequences was determined by the Flag values in the SAM output. The ‘MD:Z’ field from the alignment output was used to determine positions and numbers of mismatches between reads and the reference. Fractions of reads containing mismatched nucleotides at all positions were calculated. To prepare sequence logos for Pol IV and RDR2 transcripts, the sequences for the 5’-most and 3’-most 7 nt were prepared by WebLogo 3.6 (Crooks et al., 2004) with options --format eps --size large --colorscheme classic -- errorbars NO. The sequence logos representing 5’ and 3’ sequence features were then merged into one figure. Sequence logos for 23 nt and 24 nt DCL3 cleavage products were prepared similarly.
QUANTIFICATION AND STATISTICAL ANALYSIS
To determine the frequencies of Pol IV and RDR2 transcripts of different length, RNA-seq reads corresponding to Pol IV or RDR2 transcripts were first separated into different bins according to length. The sum of number of reads in each bin was then normalized relative to the total number of reads obtained from the library, allowing the relative frequencies of each read length to be displayed in Figure5E as the number of reads per million total reads.
Frequencies of template-mismatched nucleotides at each position within Pol IV and RDR2 transcripts varying in length from 15-80 nucleotides (Figure 6E) were calculated by dividing the number of mismatches detected at a given position by the total number of reads at that position. This was done using the custom script, Mismatch: https://github.com/wangfeng3392/AtPol-IV-RDR2-in-intro-transcription.
In total, 112689 RDR2 transcripts and 157961 Pol IV transcripts were analyzed
For sequence logo analyses to determine small RNA consensus sequences, n represents the number of unique RNA sequences analyzed, after removal of duplicate reads, using the WebLogo 3 algorithm (http://weblogo.threeplusone.com/).
DATA AND SOFTWARE AVAILABILITY
The RNA-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE126086 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126086).
Raw gel images used to generate figures in the paper are available as a Mendeley Dataset at http://dx.doi.org/10.17632/v2bbfkvymy.1
Supplementary Material
KEY RESOURCES
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-FLAG M2 affinity agarose gel | Sigma Aldrich | Cat# A2220 |
| Anti-FLAG M2 HRP Conjugate | Sigma Aldrich | Cat# A8592 |
| Anti-V5 Tag Monoclonal Antibody, HRP | Invitrogen | Cat# R961-25 |
| Anti-AtDCL3 rabbit polyclonal | (Blevins et al., 2015) | N/A |
| Anti-AtRDR2 rabbit polyclonal | (Haag et al., 2012) | N/A |
| Secondary goat anti-rabbit-HRP conjugate | Santa Cruz | Cat# SC-2004 |
| Bacterial and Virus Strains | ||
| Mix & Go Competent Cells - Strain DH5 Alpha | Zymo Research | Cat# T3007 |
| DH10Bac | Invitrogen | Cat# 10361012 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Plant protease inhibitor cocktail 100X (in DMSO) | Sigma Aldrich | Cat# P9599 |
| PMSF | Sigma Aldrich | Cat# P7626 |
| Glycoblue | Thermo Fisher | Cat# AM9515 |
| Ribolock RNase Inhibitor | Thermo Fisher | Cat# EO0384 |
| RNase H | NEB | Cat# M0297S |
| RNase I | Promega | Cat# M4261 |
| RNase III | Epicentre | Cat# RN02950 |
| Terminator Exonuclease | Lucigen | Cat# TER51020 |
| Vaccinia capping system | NEB | Cat# M2080S |
| RNA 5’ pyrophosphohydrolase (RppH) | NEB | Cat# M0356S |
| T4 Polynucleotide Kinase | NEB | Cat# M0201S |
| T4 RNA Ligase | NEB | Cat# M0204S |
| T4 RNA Ligase 2, truncated KQ | NEB | Cat# M0373L |
| Finale herbicide | Bayer | CAS# 77182822 |
| Adenosine 5′-triphosphate disodium salt hydrate | Sigma Aldrich | Cat# A2383 |
| Set of rATP, rUTP, rCTP, rGTP | Promega | Cat# E6000 |
| Turbo DNA-Free Kit | Thermo Fisher | Cat# AM1907 |
| TriZol reagent | Thermo Fisher | Cat# 15596026 |
| Grace’s insect cell media, supplemented | Thermo Fisher | Cat# 11605-102 |
| Fetal Bovine Serum, certified, United States | Thermo Fisher | Cat# 16000069 |
| Critical Commercial Assays | ||
| TruSeq small RNA library prep kit | Illumina | Cat# RS-200-0012 |
| High pure viral RNA kit | Roche | Cat# 11858882001 |
| Deposited Data | ||
| Raw and analyzed data | This paper | GEO: GSE126086 |
| Experimental Models: Cell Lines | ||
| Sf9 in Grace’s | Thermo Fisher | Cat# B82501 |
| Experimental Models: Organisms/Strains | ||
| Col-0 | N/A | N/A |
| NRPD1-FLAG in Col-0 | (Haag et al., 2012) | N/A |
| NRPD1-FLAG, rdr2-1 in Col-0 | (Haag et al., 2012) | N/A |
| RDR2-HA, nrpd1-11 in Col-0 | (Haag et al., 2012) | N/A |
| Oligonucleotides | ||
| Please see Table S1 | ||
| Recombinant DNA | ||
| pUC57-DCL3 (Codon optimized for insect cells) | This Paper (Genscript) | N/A |
| pFastBacTM HT B-DCL3 | This Paper | N/A |
| M13 mp18 (+) DNA | Bayou Biolabs | P-107 |
| Software and Algorithms | ||
| Samtools v1.5 | Li et al., 2009 | http://samtools.sourceforge.net/ |
| Bowtie v1.2.2 | Langmead et al., 2009 | https://sourceforge.net/projects/bowtie-bio/files/bowtie/1.2.2/ |
| WebLogo 3 | Crooks et al. 2004 | http://weblogo.threeplusone.com/ |
| Read me | This Paper | https://github.com/wangfeng3392/AtPol-IV-RDR2-in-intro-transcription |
| PE Trim adapter | This Paper | https://github.com/wangfeng3392/AtPol-IV-RDR2-in-intro-transcription |
| Transcript seq | This Paper | https://github.com/wangfeng3392/AtPol-IV-RDR2-in-intro-transcription |
| Mismatch | This Paper | https://github.com/wangfeng3392/AtPol-IV-RDR2-in-intro-transcription |
| Deposited data | This Paper | Mendeley raw data images: http://dx.doi.org/10.17632/v2bbfkvymy.1 GEO Series accession number GSE126086: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126086 |
Highlights.
RNA Polymerase IV, RDR2 and DCL3 are sufficient for siRNA synthesis in vitro
Non-template strand-induced Pol IV termination triggers RDR2 synthesis of dsRNA
RDR2 adds an untemplated terminal nucleotide (nt) to its transcripts’ 3’ ends
DCL3 generates 24 and 23 nt siRNAs, with untemplated terminal nt’s enriched in 23s
Acknowledgments
JS dedicates this work in loving memory of his father, S. Tejinder Pal Singh. We thank Jeremy Haag for valuable contributions at the onset of the work, Michele Marasco for pioneering the M13 system, and the Center for Genomics and Bioinformatics and the Drosophila Genome Resource Center at Indiana University for providing sequencing and cell culture facilities. This research was supported by NIH grant GM077590 and funds to CSP as an Investigator of the Howard Hughes Medical Institute. JS was supported, in part, by a Carlos O. Miller fellowship (Indiana University). HYH was supported by NIH NRSA award F32GM125334.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arimbasseri AG, and Maraia RJ (2015). Mechanism of Transcription Termination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element. Molecular cell 58, 1124–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae B, Feklistov A, Lass-Napiorkowska A, Landick R, and Darst SA (2015). Structure of a bacterial RNA polymerase holoenzyme open promoter complex. eLife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes CO, Calero M, Malik I, Graham BW, Spahr H, Lin G, Cohen AE, Brown IS, Zhang Q, Pullara Fv et al. (2015). Crystal Structure of a Transcribing RNA Polymerase II Complex Reveals a Complete Transcription Bubble. Molecular cell 59, 258–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu RS, Warner BA, Molodtsov V, Pupov D, Esyunina D, Fernandez-Tornero C, Kulbachinskiy A, and Murakami KS (2014). Structural basis of transcription initiation by bacterial RNA polymerase holoenzyme. J Biol Chem 289, 24549–24559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse CE, Lee BA, Hansen K, and Proudfoot NJ (1997). Transcriptional termination signals for RNA polymerase II in fission yeast. The EMBO journal 16, 3633–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blevins T, Podicheti R, Mishra V, Marasco M, Wang J, Rusch D, Tang H, and Pikaard CS (2015). Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. Elife 4, e09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmdorfer G, Rowley MJ, Kucinski J, Zhu Y, Amies I, and Wierzbicki AT (2014). RNA-directed DNA methylation requires stepwise binding of silencing factors to long non-coding RNA. Plant J 79, 181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, and Martienssen RA (2015). The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol 16, 727–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X, and Jacobsen SE (2002). Role of the Arabidopsis DRM Methyltransferases in De Novo DNA Methylation and Gene Silencing. Curr Biol 12, 1138–1144. [DOI] [PubMed] [Google Scholar]
- Chedin F (2011). The DNMT3 family of mammalian de novo DNA methyltransferases. Prog Mol Biol Transl Sci 101, 255–285. [DOI] [PubMed] [Google Scholar]
- Connelly S, and Manley JL (1988). A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes & development 2, 440–452. [DOI] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, and Kornberg RD (2001). Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science 292, 1863–1876. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, and Brenner SE (2004). WebLogo: a sequence logo generator. Genome Res 14, 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Johnson LM, Jacobsen SE, and Patel DJ (2015). DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol 16, 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Hage A, Koper M, Kufel J, and Tollervey D (2008). Efficient termination of transcription by RNA polymerase I requires the 5' exonuclease Rat1 in yeast. Genes & development 22, 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shami M, Pontier D, Lahmy S, Braun L, Picart C, Vega D, Hakimi MA, Jacobsen SE, Cooke R, and Lagrange T (2007). Reiterated WG/GW motifs form functionally and evolutionarily conserved ARGONAUTE-binding platforms in RNAi-related components. Genes Dev 21, 2539–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, and Zamore PD (2009). Small silencing RNAs: an expanding universe. Nat Rev Genet 10, 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow SJ, and Zomerdijk JC (2013). Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes. Subcell Biochem 61, 211–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag JR, Ream TS, Marasco M, Nicora CD, Norbeck AD, Pasa-Tolic L, and Pikaard CS (2012). In vitro transcription activities of Pol IV, Pol V, and RDR2 reveal coupling of Pol IV and RDR2 for dsRNA synthesis in plant RNA silencing. Mol Cell 48, 811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, and Baulcombe DC (2010). The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22, 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, and Baulcombe DC (2005). RNA polymerase IV directs silencing of endogenous DNA. Science 308, 118–120. [DOI] [PubMed] [Google Scholar]
- Holoch D, and Moazed D (2015). RNA-mediated epigenetic regulation of gene expression. Nat Rev Genet 16, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Fiedler U, and Timmers HT (1997). Three transitions in the RNA polymerase II transcription complex during initiation. The EMBO journal 16, 7468–7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl BF, Li H, and Paule MR (2000). DNA melting and promoter clearance by eukaryotic RNA polymerase I. Journal of molecular biology 299, 75–89. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, and Carrington JC (2007). Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 5, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehner JN, Pearson EL, and Moore C (2011). Unravelling the means to an end: RNA polymerase II transcription termination. Nature reviews Molecular cell biology 12, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landick R (2009). Functional divergence in the growing family of RNA polymerases. Structure 17, 323–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Vashisht AA, Wohlschlegel JA, and Jacobsen SE (2011). SHH1, a homeodomain protein required for DNA methylation, as well as RDR2, RDM4, and chromatin remodeling factors, associate with RNA polymerase IV. PLoS Genet 7, e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, and Borukhov S (2016). Bacterial RNA Polymerase-DNA Interaction-The Driving Force of Gene Expression and the Target for Drug Action. Front Mol Biosci 3, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Vandivier LE, Tu B, Gao L, Won SY, Li S, Zheng B, Gregory BD, and Chen X (2015). Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res 25, 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco M, Li W, Lynch M, and Pikaard CS (2017). Catalytic properties of RNA polymerases IV and V: accuracy, nucleotide incorporation and rNTP/dNTP discrimination. Nucleic Acids Res 45, 11315–11326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17, 10–12. [Google Scholar]
- Matzke MA, Kanno T, and Matzke AJ (2015). RNA-Directed DNA Methylation: The Evolution of a Complex Epigenetic Pathway in Flowering Plants. Annu Rev Plant Biol 66, 243–267. [DOI] [PubMed] [Google Scholar]
- Naji S, Bertero MG, Spitalny P, Cramer P, and Thomm M (2008). Structure-function analysis of the RNA polymerase cleft loops elucidates initial transcription, DNA unwinding and RNA displacement. Nucleic Acids Res 36, 676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S, Yuzenkova Y, and Zenkin N (2013). Mechanism of eukaryotic RNA polymerase III transcription termination. Science (New York, NY) 340, 1577–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, and Pikaard CS (2005). Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120, 613–622. [DOI] [PubMed] [Google Scholar]
- Ozata DM, Gainetdinov I, Zoch A, O'Carroll D, and Zamore PD (2018). PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet. [DOI] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, and Kim VN (2011). Dicer recognizes the 5' end of RNA for efficient and accurate processing. Nature 475, 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard CS, Haag JR, Pontes OM, Blevins T, and Cocklin R (2012). A transcription fork model for Pol IV and Pol V-dependent RNA-directed DNA methylation. Cold Spring Harb Symp Quant Biol 77, 205–212. [DOI] [PubMed] [Google Scholar]
- Porrua O, Boudvillain M, and Libri D (2016). Transcription Termination: Variations on Common Themes. Trends in genetics : TIG 32, 508–522. [DOI] [PubMed] [Google Scholar]
- Qi Y, He X, Wang XJ, Kohany O, Jurka J, and Hannon GJ (2006). Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443, 1008–1012. [DOI] [PubMed] [Google Scholar]
- Ray-Soni A, Bellecourt MJ, and Landick R (2016). Mechanisms of Bacterial Transcription Termination: All Good Things Must End. Annual review of biochemistry 85, 319–347. [DOI] [PubMed] [Google Scholar]
- Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, Zhu JK, Hagen G, Guilfoyle TJ, Pasa-Tolic L, and Pikaard CS (2009). Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell 33, 192–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skvortsova K, Iovino N, and Bogdanovic O (2018). Functions and mechanisms of epigenetic inheritance in animals. Nat Rev Mol Cell Biol 19, 774–790. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, and Martienssen R (2007). Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8, 272–285. [DOI] [PubMed] [Google Scholar]
- Toulokhonov I, and Landick R (2006). The role of the lid element in transcription by E. coli RNA polymerase. J Mol Biol 361, 644–658. [DOI] [PubMed] [Google Scholar]
- Wang F, Johnson NR, Coruh C, and Axtell MJ (2016). Genome-wide analysis of single non-templated nucleotides in plant endogenous siRNAs and miRNAs. Nucleic Acids Res 44, 7395–7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendte JM, and Pikaard CS (2017). The RNAs of RNA-directed DNA methylation. Biochim Biophys Acta Gene Regul Mech 1860, 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, and Pikaard CS (2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135, 635–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzbicki AT, Ream TS, Haag JR, and Pikaard CS (2009). RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet 41, 630–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, and Carrington JC (2004). Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2, E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J, Bischof S, Wang H, Feng S, Lee TF, Teng C, Chen X, Park SY, Liu L, Gallego-Bartolome J, et al. (2015). A One Precursor One siRNA Model for Pol IV-Dependent siRNA Biogenesis. Cell 163, 445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, and Darst SA (1999). Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 A resolution. Cell 98, 811–824. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, and Jacobsen SE (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299, 716–719. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE126086 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE126086).
Raw gel images used to generate figures in the paper are available as a Mendeley Dataset at http://dx.doi.org/10.17632/v2bbfkvymy.1