Abstract
Objective
This research aimed to investigate the role and mechanism of long noncoding RNA (lncRNA) HCP5 in skin cutaneous melanoma (SKCM).
Materials and methods
Survival analysis was performed using The Cancer Genome Atlas (TCGA)-SKCM data and SKCM patients’ clinical data. Primary SKCM cells were derived from patients’ pathologic tissue specimens. HCP5 overexpression was achieved by lentiviral transduction. Malignancy of SKCM cells was evaluated in vitro by cell proliferation, colony formation, apoptosis and transwell invasion assays. RARRES3 knockdown was achieved by siRNA transfection. DIANA microT-CDS algorithm was used to predict miRNAs that might interact with HCP5 and 3ʹ untranslated region of RARRES3 mRNA. microRNA target luciferase reporter assay and AGO2-RNA immunoprecipitation were used to verify the interaction between HCP5, 3ʹ UTR of RARRES3 mRNA and miR-1286.
Results
HCP5 level was decreased in SKCM tissue specimens compared to noncancerous counterparts. Low expression of HCP5 was associated with SKCM patients’ poor overall survival and disease progression. HCP5 overexpression significantly reduced the malignancy of primary SKCM cells in vitro. RARRES3 was found as a HCP5-co-expressing gene in SKCM cells. HCP5 overexpression significantly increased RARRES3 expression in SKCM cells. RARRES3 knockdown partially abolished the anti-SKCM effect of HCP5 overexpression. MiR-1286 was found interacting with both HCP5 and 3ʹ UTR of RARRES3 mRNA.
Conclusion
HCP5 is a cancer-suppressive lncRNA in SKCM. HCP5 overexpression decreased SKCM cell malignancy in vitro by upregulating RARRES3, possibly via sponging miR-1286.
Keywords: skin cutaneous melanoma, long noncoding RNA, HCP5, RARRES3, miR-1286
Introduction
Cutaneous melanoma (or skin cutaneous melanoma, SKCM) is the least common but most aggressive skin cancer.1,2 Hyperactivation of PI3K-Akt and MEK-ERK signaling pathways are fundamental to the malignant transformation and progression of SKCM.3,4 Target therapies aiming to dampen these two signaling pathways have achieved significant progress, but the resistance of SKCM to these therapies has been increasingly reported.5,6 A more in-depth understanding of the pathological mechanism of SKCM might help overcoming this obstacle in SKCM management and improving patients’ welfare.
Long noncoding RNAs (lncRNAs) are single-stranded RNA over 200 nt in length without protein-coding potential.7 One of the major functions of lncRNAs is to regulate gene expression by interacting with microRNAs, thus preventing the latter from inducing translational repression and mRNA degradation mediated by the RNA-induced silencing complex (RISC) via interacting with mRNA 5ʹ or 3ʹ UTRs.8 By regulating the expression of oncogenes or cancer suppressive genes, lncRNA also participates in oncogenesis and cancer progression,9,10 making them not only as potential biomarkers for diagnosis or prognosis but also as possible therapeutic targets for treatment.10,11 Although several cancer-promotive or -suppressive lncRNAs have been reported in SKCM, research on the lncRNAs in SKCM is still at its dawn.12,13
The initial aim of this research was to find lncRNA(s) with prognostic value in SKCM. After querying some transcriptomic studies, we found that high expression of lncRNA HCP5 been proposed as a risk factor of SKCM progression.14,15 Research on the role and mechanism of this lncRNA in cancer development is lacking, and previous publications have described this lncRNA as either cancer promotive or suppressive in different cancer types.16–22 In this study, we investigated the clinical relevance of HCP5 expression level to SKCM progression and patients’ survival. Using patient-derived SKCM cells, we preliminarily investigated the role of HCP5 in SKCM cells. To our surprise, we found that low expression of HCP5 was associated with SKCM stage progression, lymph node metastasis, distal metastasis and SKCM patients’ shortened overall survival. Overexpression of HCP5 significantly inhibited SKCM cell proliferation, survival, colony formation and transwell invasion but promoted serum deprivation-induced apoptosis in vitro. By querying the TCGA-SKCM data, we found RARRES3 as a potential HCP5-co-expressing gene. We further confirmed that HCP5 regulated the malignancy of SKCM cells partially through RARRES3, and HCP5 overexpression increased RARRES3 gene expression in SKCM cells by preventing the degradation from RISC-mediated degradation, possibly via sponging miR-1286.
Materials and methods
Bioinformatic analysis
We obtained the HCP5 relative expression level and patients’ overall survival data in the TCGA-SKCM data from the OncoLnc website.23 To predict the HCP5 and 3ʹ UTR of RARRES3 mRNA-interacting miRNAs, we used the DIANA microT-CDS algorithm24 and DIANA microT25 websites, respectively. The predicted miRNAs with an overall score >0.6 were subject to cross-referencing (Figure 5C).
Figure 5.
HCP5-regulated RARRES3 expression in SKCM cells by sponging miR-1286. (A) HCP5 overexpression increased the expression of Gaussia luciferase (Gluc) governed by the 3ʹ UTR of RARRES3 mRNA. (B) HCP5 overexpression reduced the association of RARRES3 mRNA with AGO2 protein. (C) Putative HCP5 and RARRES3 mRNA interacting miRNAs, predicted by the DIANA microT-CDS algorithm with prediction score >0.6. (D) HCP5 overexpression increased the association of miR-1286 as well as other miRNAs with AGO2 protein in SKCM cells. (E) Putative miR-1286-binding site on the wild type 3ʹ UTR of RARRES3 (WT), which was mutated (MUT). (F) miR-1286 mimic transfection reduced the-expression of Gluc governed by wild-type 3ʹ UTR of RARRES3. (G and H) RARRES3 mRNA and protein level in SKCM was reduced by miR-1286 mimic transfection. *p<0.05. Abbreviations: SKCM, skin cutaneous melanoma; NC, normal control.
Analysis of SKCM patients’ tissue specimens
This research was approved by the ethical review committee of the Third Hospital of Ji’nan and all patients informed consent in written. This study was performed in accordance with the Declaration of Helsinki. The 54 patients included 33 males at age of 56.18±9.76 and 21 females at age of 51.81±9.62. All patients received anti-SKCM surgery and other treatment in our facility during 2007–2017. Each patient’s SKCM pathologic tissue specimen and normal tissue specimens obtained from the contralateral limb of the same patient were stored in liquid nitrogen immediately after sampling. Patients’ survival data were recorded during the postoperative follow-ups. Patients’ clinical features, including their age, gender, cancer stage, lymph node metastasis and distal metastasis, were recorded during their diagnosis and analyzed in Table 1.
Table 1.
Clinical relevance of HCP5 expression level in SKCM patients
| Patients’ features | High | Low | Fisher’s exact p-value | |
|---|---|---|---|---|
| Gender | Male | 15 | 18 | 0.5772 |
| Female | 12 | 9 | ||
| Age (years) | ≤55 | 15 | 12 | 0.5867 |
| >55 | 12 | 15 | ||
| Stage | I–II | 19 | 10 | 0.0281 |
| III–IV | 8 | 17 | ||
| Distal metastasis | No | 25 | 18 | 0.0394 |
| Yes | 2 | 9 | ||
| Lymph node metastasis | No | 24 | 15 | 0.0135 |
| Yes | 3 | 12 | ||
Abbreviations: HCP5, human leucocyte antigen complex P5; SKCM, skin cutaneous melanoma.
Establishment of primary SKCM cell lines
Two SKCM cell lines were established using SKCM pathologic tissue specimens from two patients (Chinese male, at age of 46 and 55) who received treatment in our facility in 2017. The excised SKCM tissue from each patient was minced and transplanted subcutaneously on two NU/NU nude mice (Vital River laboratory, Beijing, China). The animal experiments were approved by the Institutional Animal Care and Use Committee of the Third Hospital of Ji’nan and performed in accordance with the guidelines of the National Animal Care and Ethics Institution. The xenografts were excised from the mice when the volume reached 2,000 mm3 and grounded through a 100 μm cell strainer (F613463, Sangon Biotech, Shanghai, China). The cells were cultured using a complete cell culture media (GlutaMAX-supplemented RPMI-1640 media, supplemented with 10% FBS and 1% of penicillin‒streptomycin solution, all from Thermo Fisher Scientific, Waltham, MA, USA) in a cell incubator with a humidified atmosphere at 37°C with 5% CO2. The cells were subcultured every 2–3 days for 20 passages before further analysis.
HCP5 overexpression, RARRES3 knockdown and miR-1286 mimic transfection
We overexpressed HCP5 in the primary SKCM cells by lentiviral transduction. The HCP5-overexpressing (OE) (LPP-Y1990-Lv105-400) and empty lentiviral particles were purchased from Genecopoeia (Rockville, MD, USA) and transduced into SKCM cells at log phase at the MOI of 5 TU/cell using EndoFectin Lenti transfection reagent (EF002, Genecopoeia) following the manufacturer’s instructions. Cells with positive transduction were enriched by puromycin selection. For RARRES3 knockdown, the SKCM cells were transfected with a siRNA pool (GenePharma, Shanghai, China) composed of 4 different siRNA oligos that target RARRES3 mRNA. The cells were transfected with siRNA pool at the overall concentration of 50 nM using Lipofectamine 2000 (Thermo Fisher Scientific). To simulate miR-1286 overexpression, SKCM cells were transfected with single-stranded miR-1286 mimic (GenePharma) at 30 nM using Lipofectamine 2000. Transfection efficacy was assayed at 24 hrs after transfection by qRT-PCR or western blotting. A miRNA mimic negative control with scrambled sequence was used as negative control for miRNA mimic transfection.
qRT-PCR
Total RNA from cells cultured in vitro or from tissue specimens was extracted using Trizol reagent (R0016, Beyotime, Shanghai, China). HCP5 or RARRES3 mRNA in the total RNA was detected by qRT-PCR with 2−ΔΔCt method using a one-step SYBR green RT-qPCR Kit (QP084, Genecopoeia) and the following primers (Genecopoeia): RARRES3 (HQP108527), GAPDH (HQP006940), HCP (Fw, CAGCCTGAGAGAAGTAGGGC; Rev, TCAGTCGCATTTCCAGGTAATTT; sequences from PrimerBank).26 GAPDH was used as an internal reference when applicable. Detection of miRNAs levels in the total RNA was performed by qRT-PCR with 2−ΔΔCt method using All-in-One miRNA qRT-PCR detection kit (QP016, Genecopoeia) and the following primers (5ʹ to 3ʹ, Genecopoeia): hsa-miR-1286 (TGCAGGACCAAGATGAGCCCTAA); hsa-miR-6783-3p (TTCCTGGGCTTCTCCTCTGTAGAA); hsa-miR-6866-5p (TTAGAGGCTGGAATAGAGATTCTAA); hsa-miR-6858-3p (CAGCCAGCCCCTGCTCACCCCTAA); hsa-miR-504-5p (AGACCCTGGTCTGCACTCTATCAA); hsa-miR-4725-5p (AGACCCTGCAGCCTTCCCACCAA) and hsa-miR-6874-5p (ATGGAGCTGGAACCAGATCAGGCAA).
Western blotting
Western blotting detecting RARRES3 protein level in SKCM cells in vitro was performed by using RIPA lysis buffer (P0013B, Beyotime) and separating the cellular proteins in the cell lysate by SDS-PAGE in reducing condition, followed by transferring the separated proteins onto nitrocellulose membrane and probing with primary and secondary antibodies. Protein bands were then developed using ECL substrate (32,106, Thermo Fisher Scientific) and X-ray films (34,090, Thermo Fisher Scientific). The following antibodies purchased from Abcam (Cambridge, MA, USA) were used for western blotting: rabbit polyclonal anti-TIG3 antibody (ab96468), rabbit polyclonal anti-GAPDH antibody (ab9485) and goat anti-rabbit IgG H&L antibody (HRP-conjugated, ab205718).
Cell proliferation, colony formation, apoptosis and transwell invasion assays
SKCM cell proliferation was evaluated by CCK-8 cell viability assay. SKCM cells after transfection were inoculated on 96-well-plate at 5,000 cells/well. Cell viability was assessed using CCK-8 (CK04, Dojindo, Kumamoto, Japan) at 2, 24, 48, or 72 hrs postinoculation using a microplate reader. Clonogenicity of SKCM cells was evaluated by colony formation assays. SKCM cells after transfection were inoculated on 24-well-plate at 100 cells/well. Cells were cultured for 2 weeks before fixation and staining with crystal violet. Colonies with more than 50 cells were counted under a microscope. SKCM cell survival was evaluated by apoptosis assay. Cells after transfection were seeded on 24-well-plate at 2×105 cells/well and cultured for 2 hrs, followed by serum deprivation for 8 hrs. Cell apoptosis was assessed by detecting the percentage of Annexin V-positive and PI-positive or -negative staining cells using Annexin V-FITC/PI Apoptosis detection kit (640,914, Biolegend, San Diego, CA, USA) and flow cytometry. Invasiveness of SKCM cells was evaluated by transwell invasion assay. 5×104 SKCM cells after transfection were inoculated in a matrigel-coated transwell chamber (08-774-122, Thermo Fisher Scientific) in serum-free culture media supplemented with 2% BSA. The transwell chamber was then incubated in complete culture media for 16 hrs, and invasive cells were fixed and stained for counting under a microscope (Olympus, Tokyo, Japan) with five random fields (x200 magnification).
microRNA target luciferase reporter assay
Three different reporter plasmids were constructed by Genecopoeia for the miRNA target luciferase reporter assay: for the wild-type (WT) plasmids, the cDNA of RARRES3 mRNA 3ʹ UTR was inserted at the 3ʹ flank of Gaussia luciferase (Gluc) gene on the reporter plasmids; for the Scrambled plasmids, the cDNA sequence of RARRES3 mRNA 3ʹ UTR was scrambled; for mutant (MUT) plasmids, mutations were induced into the cDNA of RARRES3 mRNA 3ʹ UTR (as demonstrated in Figure 5E). All plasmids carried the secreted alkaline phosphatase (SEAP) gene for normalization. The plasmids were transfected into SKCM cells in vitro using EndofectinMAX (Genecopoeia). At 48 hrs after transfection, activity of Gluc and SEAP in the cell culture media was assessed using Secrete-Pair Gaussia luciferase assay kit (LF032, Genecopoeia) and a microplate reader.
AGO-RNA immunoprecipitation (AGO-RIP)
AGO-RIP was performed using Imprint RNA immunoprecipitation kit (RIP-12RXN) with rat anti-AGO2 antibody and rabbit anti-rat IgG antibody. SKCM cells after lentiviral transduction were cultured on 6-well plate until ~80% confluence. The cells were lysed on the plate with a mild lysis buffer (P0013, Beyotime), and the cell lysate was centrifuged at 15,000× g for 10 min at 4°C. The supernatant was then incubated with Protein A magnetic beads preloaded with rabbit anti-rat IgG antibody and rat anti-AGO2 antibody at 4°C for 16 hrs. The beads were then enriched with a magnetic stand, and RNAs associated with the beads were purified following the product manual. RARRES3 mRNA and miRNAs in the purified RNA were detected by qRT-PCR as described above.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 7 software. Each data group in Figures 2, 4 and 5 represent 6 replicates from 2 primary SKCM cell lines (2 biological replicates ×3 technical replicates). Data were presented as mean ± SD fashion, unless otherwise indicated. Fisher’s exact test was performed for significance test in Table 1. Log-rank test was performed for survival analysis. Mann‒Whitney test was performed for significance test in Figures 1B and 3A. Pearson and Spearman correlation analysis was performed for significance test in Figure 3B. Student’s t-test or ANOVA was performed for significance test in the rest of the figures. A difference was considered statistically significant when p<0.05.
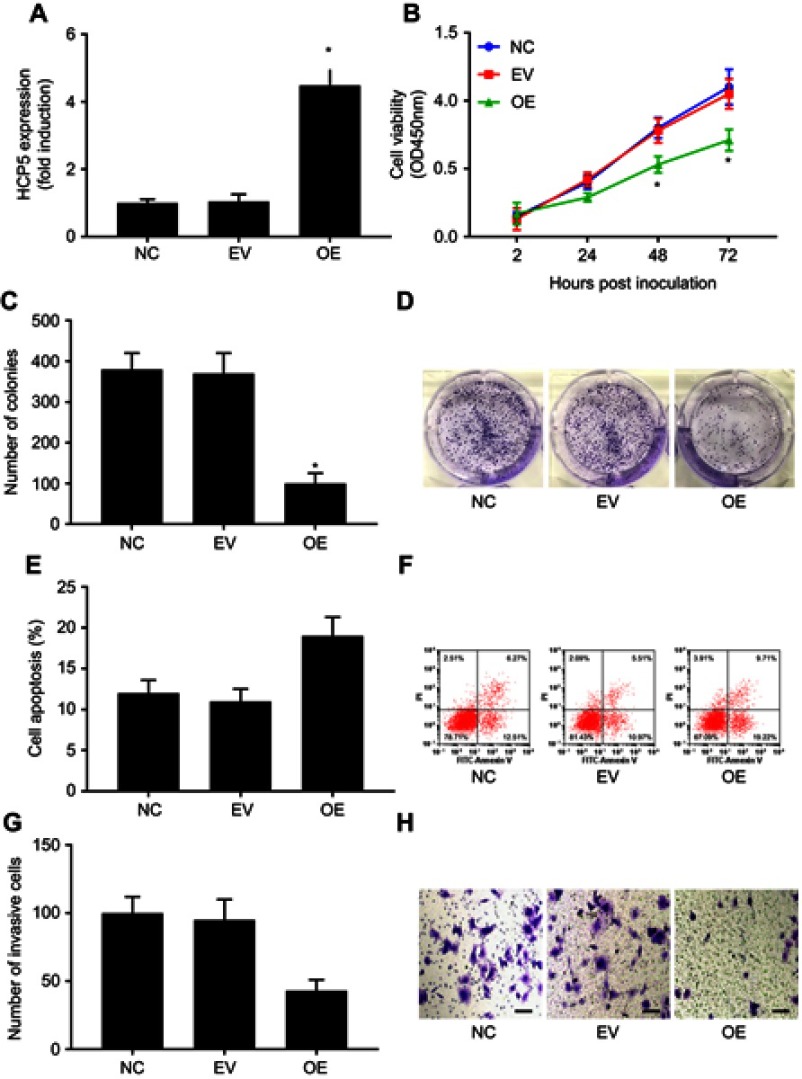
Figure 2.
HCP5 overexpression inhibited primary SKCM cell proliferation, survival, clonogenicity and invasiveness in vitro. SKCM cells were transduced with empty (EV) or HCP5 (overexpressing; OE) lentiviral vectors for 24 hrs before assay. (A) HCP5 levels in SKCM cells with or without HCP5 overexpression were compared by qRT-PCR. (B) SKCM cell proliferation was assessed by cell proliferation assay. (C and D) Clonogenicity of SKCM cells was assessed by colony formation assay. (E and F) Survival of SKCM cells was assessed by apoptosis assay. Cell apoptosis was induced by serum deprivation for 8 hrs. G and H, invasiveness of SKCM cells was assessed by transwell invasion assays. Bar indicates 100 μM. *p<0.05. Abbreviations: SKCM, skin cutaneous melanoma; NC, normal control.
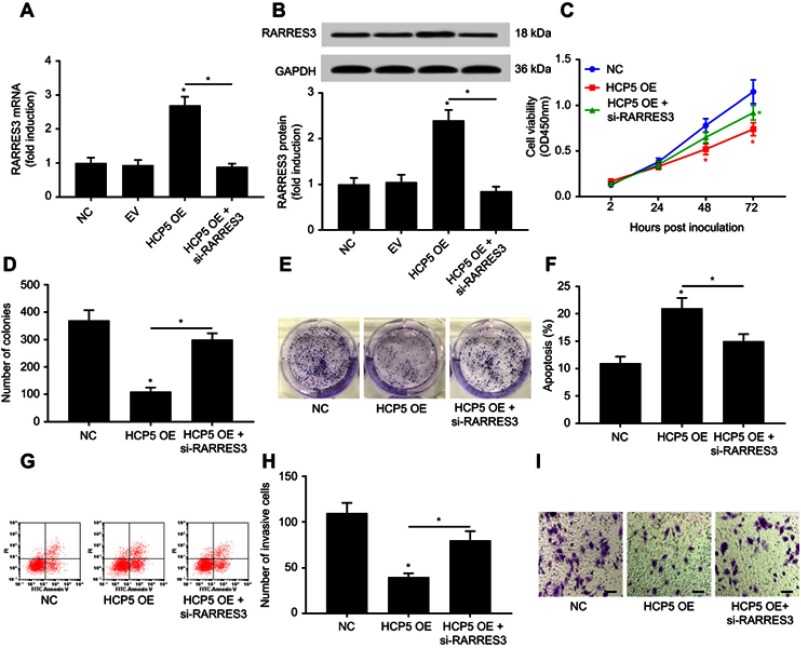
Figure 4.
HCP5 overexpression reduced SKCM cell malignancy through RARRES3. After transducing with HCP5 overexpressing lentiviral vector (HCP5 OE), SKCM cells were transfected with siRNA for RARRES3 knockdown (si-RARRES3) for 24 hrs. (A and B) Upregulation of RARRES3 mRNA and protein level by HCP5 overexpression was abolished by RARRES3 knockdown. (C–I) SKCM cell malignancy was evaluated by cell proliferation, colony formation, survival and transwell invasion assays, as described in Figure 2. In panel 4C, red asterisks labeled the comparison to NC group, while the green asterisks labeled the comparison to HCP5 OE group. Bar indicates 100 μM. *p<0.05. Abbreviations: SKCM, skin cutaneous melanoma; NC, normal control.
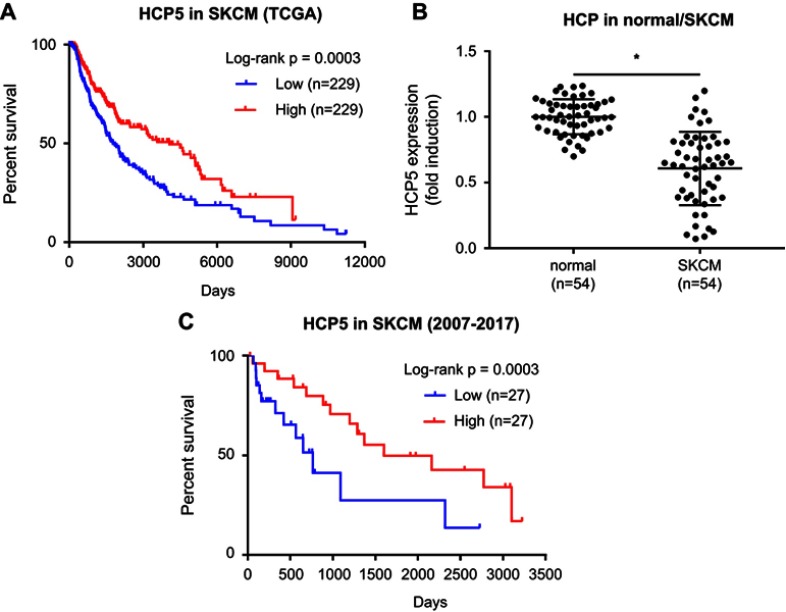
Figure 1.
Clinical significance of HCP5 expression in SKCM. (A) Survival analysis of HCP5 using TCGA-SKCM data. (B) HCP5 levels in 54 pairs of SKCM tissue specimens and normal skin tissue specimens were compared by qRT-PCR. Each dot represents one tissue specimen. (C) Survival analysis of HCP5 using the 54 SKCM patients’ clinical data. *p<0.05. Abbreviation: TCGA-SKCM, The Cancer Genome Atlas-skin cutaneous melanoma.
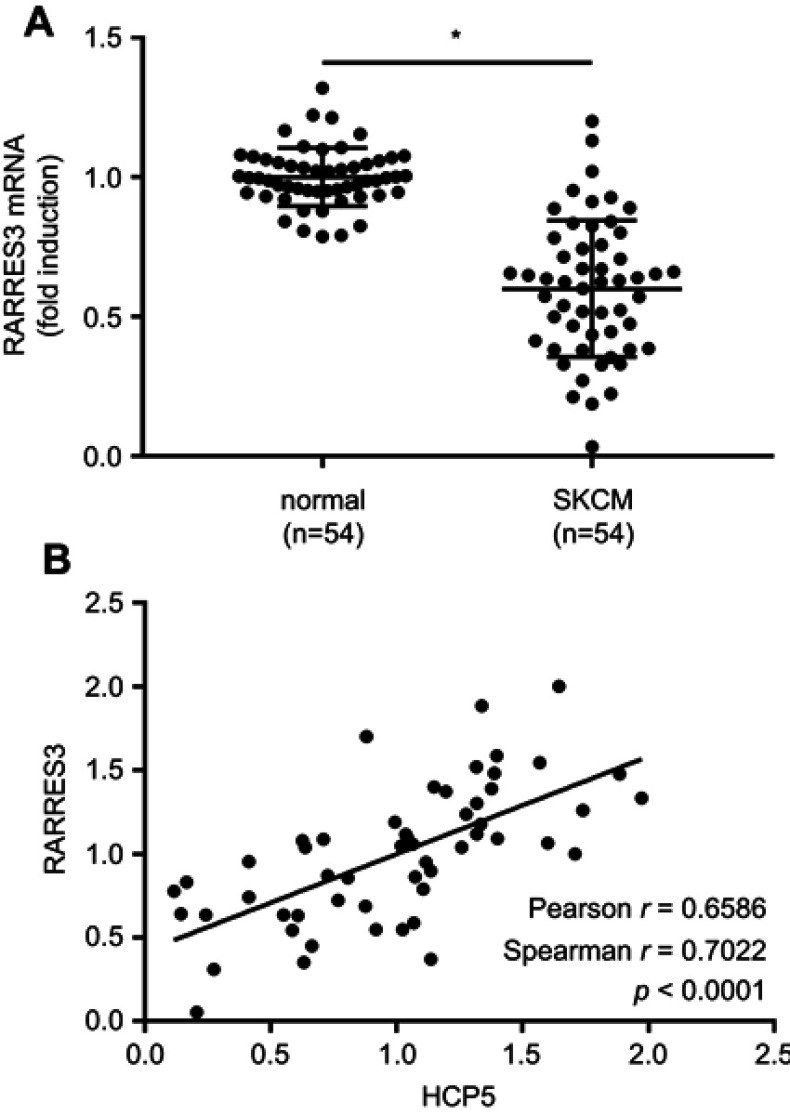
Figure 3.
RARRES3 mRNA expression correlated with that of HCP5 in SKCM. (A) RARRES3 mRNA levels in 54 pairs of SKCM tissue specimens and normal skin tissue specimens were compared by qRT-PCR. (B) Correlation analysis of RARRES3 mRNA and HCP5 expression levels. Each dot represents one tissue specimen. *p<0.05. Abbreviation: SKCM, skin cutaneous melanoma.
Ethical Statement
This research was approved by the ethical review committee of the Third Hospital of Ji’nan and all patients provided written informed consent. This study was performed in accordance with the Declaration of Helsinki. The animal experiments were approved by the Institutional Animal Care and Use Committee of the Third Hospital of Ji’nan. All experiments were performed following the guidelines and the regulations of the Third Hospital of Ji’nan.
Results
Downregulation of HCP5 in SKCM was correlated with patients’ unfavored prognosis
Previous researches have implicated dysregulation of HCP5 in SKCM progression. We thought to verify whether the increased expression of HCP5 would correlate with SKCM patients’ worsened outcome. After querying the TCGA-SKCM data, to our surprise, we found that high expression of HCP5 actually associated with increased survival of SKCM patients (Figure 1A), implying that HCP5 might limit SKCM progression. To investigate the role of HCP5 in SKCM progression, we analyzed the expression level of HCP5 in 54 SKCM patients’ tissue specimens preserved during 2007–2017, and found that HCP5 expression level in the SKCM pathological tissue specimens was significantly lower than that in the normal skin tissue specimens obtained from the contralateral limbs of the same patients (Figure 1B). Moreover, we also found that low expression of HCP5 was associated with disease progression, lymph node and distal metastasis of SKCM (Table 1) as well as the worsened outcome of these 54 SKCM patients (Figure 1C). These data implied that HCP5 might seve as a tumor suppressor in SKCM.
HCP5 overexpression significantly reduced primary SKCM cell proliferation, survival, colony formation and transwell invasion but promoted apoptosis in vitro
To investigate whether HCP5 would regulate the malignancy of SKCM cells, we established 2 patient-derived SKCM cell lines and transduced these cells with either HCP5-OE or empty (EV) lentiviral vectors for 24 hrs. Transduction with HCP5-OE vectors significantly increased HCP5 expression level in the primary SKCM cells in vitro (Figure 2A). Moreover, HCP5 overexpression significantly reduced cell proliferation, survival, clonogenicity and transwell invasion capacity but increased serum deprivation-induced apoptosis of these cells (Figures 2B–H and S1). These results suggested that HCP5 addition inhibited SKCM progression in vitro.
HCP5 overexpression reduced SKCM cell malignancy by upregulating RARRES3 gene expression
lncRNAs are known to regulate gene expression via miRNA sponging. To investigate whether HCP5 would regulate the malignancy of SKCM cells through similar mechanisms, we queried the TCGA-SKCM data in an attempt to find HCP5 co-expressing genes. The top 20 genes whose mRNA levels showed the highest correlation with that of HCP5 in TCGA-SKCM data are listed in Table 2. Among these genes, we found RARRES3, a recently proposed cancer-suppressive gene, in the top 3 genes. Thus, we investigated whether HCP5 would regulate RARRES3 gene expression in SKCM cells. We found that RARRES3 gene expression was also significantly reduced in the 54 SKCM tissue specimens comparing to their normal counterparts, significantly correlating with that of HCP5 (Figure 3A and B). Overexpression of HCP5 also significantly increased RARRES3 mRNA and protein levels in the primary SKCM cells in vitro (Figure 4A and B). To investigate whether HCP5 would regulate SKCM cell malignancy through RARRES3, we transfected siRNA targeting RARRES3 (si-RARRES3) for 24 hrs to reduce its gene expression (Figure 4A and B). RARRES3 knockdown partially attenuated HCP5-mediated inhibition of cell proliferation, survival, colony formation and transwell invasion and promotion of apoptosis in SKCM cells (Figures 4C–I and S2), suggesting that HCP5 would regulate the malignancy of SKCM cells in vitro partially through RARRES3.
Table 2.
Top 20 HCP5 co-expressing genes in the TCGA-SKCM data
| Gene | Description | Spearman’s r | p-value | q-value |
|---|---|---|---|---|
| HCG26 | HLA complex group 26 | 0.9258 | 1.14E−200 | 2.31E−196 |
| HLA-B | Major histocompatibility complex, class I, B | 0.7836 | 3.22E−99 | 3.25E−95 |
| RARRES3 | Retinoic acid receptor responder 3 | 0.7644 | 1.27E−91 | 8.53E−88 |
| BTN3A3 | Butyrophilin subfamily 3 member A3 | 0.7572 | 5.87E−89 | 2.96E−85 |
| APOL3 | Apolipoprotein L3 | 0.7503 | 1.66E−86 | 6.67E−83 |
| B2M | Beta-2-microglobulin | 0.7471 | 2.18E−85 | 7.34E−82 |
| CD2 | CD2 molecule | 0.7448 | 1.31E−84 | 3.79E−81 |
| NLRC5 | NLR family CARD domain-containing 5 | 0.7422 | 1.03E−83 | 2.61E−80 |
| GPR171 | G protein-coupled receptor 171 | 0.7419 | 1.30E−83 | 2.91E−80 |
| SLA2 | Src-like adaptor 2 | 0.7393 | 9.44E−83 | 1.90E−79 |
| CD3D | CD3d molecule | 0.739 | 1.20E−82 | 2.21E−79 |
| PSMB9 | Proteasome subunit beta 9 | 0.7372 | 4.88E−82 | 8.20E−79 |
| HLA-C | Major histocompatibility complex, class I, C | 0.7329 | 1.18E−80 | 1.83E−77 |
| TIGIT | T-cell immunoreceptor with Ig and ITIM domains | 0.7326 | 1.46E−80 | 2.11E−77 |
| IRF1 | Interferon-regulatory factor 1 | 0.7309 | 5.48E−80 | 7.37E−77 |
| APOL1 | Apolipoprotein L1 | 0.7297 | 1.28E−79 | 1.62E−76 |
| SLAMF6 | SLAM family member 6 | 0.7278 | 5.17E−79 | 6.13E−76 |
| HLA-F | Major histocompatibility complex, class I, F | 0.7259 | 2.01E−78 | 2.25E−75 |
| GBP5 | Guanylate-binding protein 5 | 0.7242 | 7.18E−78 | 7.62E−75 |
| CXCL9 | C-X-C motif chemokine ligand 9 | 0.7237 | 9.98E−78 | 1.01E−74 |
HCP5 protected RARRES3 mRNA from RISC-mediated degradation via sponging miR-1286
We performed the miRNA target luciferase reporter assay and AGO2-RIP assay to investigate whether HCP5 would regulate RARRES3 gene expression through interacting with miRNAs. Our data showed that OE HCP5 significantly increased the activity of Gluc, whose expression was governed by the WT 3ʹ UTR of RARRES3 in SKCM cells, and scrambling the sequence of this 3ʹ UTR of RARRES3 abolished the regulation of HCP5 on Gluc expression (Figure 5A). HCP5 overexpression also significantly reduced the association of RARRES3 mRNA with AGO2 protein in the RISC complex in SKCM cells (Figure 5B). These data suggested that HCP5 would protect RARRES3 mRNA from miRNA and the RISC complex-mediated degradation. To identify the miRNA(s) regulated by HCP5 in maintaining RARRES3 gene expression, we employed the DIANA microT-CDS algorithm to predict miRNAs that would interact with HCP5 or mRNA of RARRES3. By cross-referencing the miRNA prediction results, we found 7 miRNAs putatively interacting with both HCP5 and 3ʹ UTR of RARRES3 mRNA (Figure 5C). However, by analyzing the enrichment of these miRNAs by HCP5 overexpression in the AGO2-RIP products, we found that miR-1286 was the one miRNA that most significantly associated with HCP5, and some other miRNAs were not significantly enriched (Figure 5D). Further analysis using microRNA target luciferase assay confirmed the binding site of miR-1286 on the 3ʹ UTR of RARRES3 mRNA (Figure 5E and F), and miR-1286 overexpression significantly reduced RARRES3 mRNA and protein levels in SKCM cells after transfection with miR-1286 mimic for 24 hrs (Figure 5G and H). These data suggested that HCP5 would regulate RARRES3 gene expression in SKCM cells by sponging miR-1286.
Discussion
The present research aimed to investigate the role of HCP5 in SKCM development. Previous studies described the potential association between high expression of HCP5 and SKCM patients’ decreased survival.14,15 In the present research, however, we found that high expression of HCP5 might favor SKCM patients’ good prognosis, and that this lncRNA was significantly decreased in SKCM pathologic tissue specimens comparing to normal tissue specimens. These results implied HCP5 as a cancer suppressor in SKCM.
Previous studies have described HCP5 as a cancer promoter in follicular thyroid carcinoma,16 cervical cancer17 and glioma,21 while the expression level of this lncRNA was also found to be downregulated in lung adenocarcinoma,19 nasopharyngeal carcinoma20 and ovarian cancer22 tissue specimens comparing to nonmalignant counterparts. The mechanism of HCP5 is largely underexplored. In this research, we used patient-derived SKCM cells to demonstrate that HCP5 overexpression could significantly reduce the malignancy of these cells, marked by the significant decrease of cell proliferation, survival, colony formation and transwell invasion and increase of apoptosis caused by serum deprivation.
To further investigate the mechanism underlying the SKCM-suppressive role of HCP5, we explored HCP co-expressing genes in TCGA-SKCM data. We found that the expression of a novel cancer-suppressive gene, RARRES3, was significantly correlated with that of HCP5 in SKCM that were sampled in TCGA. Our qRT-PCR results also suggested the downregulation of RARRES3 gene expression in SKCM and its correlation with HCP5, and our in vitro data further showed that overexpression of HCP5 could significantly increase RARRES3 mRNA and protein level in primary SKCM cells. We transfected the HCP5-OE SKCM cells with siRNA for RARRES3 knockdown at a series of continuously diluted concentrations to downregulate their RARRES3 expression to a “near wild-type” level. RARRES3 knockdown partially attenuated the SKCM-suppressive effect of HCP5 overexpression, suggesting that HCP5 might suppress SKCM development through RARRES3.
The cancer-suppressive role of RARRES3 was first described in 1998, when DiSepio et al reported that overexpression of this gene would decrease the proliferation of various types of malignant or nonmalignant cells in vitro.27 Since then, RARRES3 has been found as a cancer-suppressive gene in liver cancer,28 breast cancer,29,30 B cell leukemia31 and skin cancer.32 RARRES3 gene was also found to promote cancer cell differentiation30,33–36 and suppress cancer metastasis.28,30,37,38 Mechanistically, Hsu et al reported that RARRES3 protein would suppress WNT/β-catenin signaling by mediating the acylation of Wnt proteins as well as LRP6, thus reducing the epithelial-to-mesenchymal transition (EMT) and cancer stem cell property of breast cancer cells.39 An early report from Ou et al showed that RARRES3 protein transcriptionally repressed the expression of HER2, resulting in the inactivation of PI3K/Akt/mTOR signaling.40 These findings suggested that RARRES3 as a cancer suppressor is multifunctional.
However, how HCP5 would regulate RARRES3 gene expression remains unclear. We hypothesized that this lncRNA might sustain RARRES3 mRNA level via miRNA sponging. The miRNA target luciferase reporter assay and AGO2-RIP assay results confirmed that HCP5 overexpression would protect the RARRES3 mRNA from miRNA-RISC-mediated degradation. By bioinformatic analysis and AGO2-RIP assay, we identified miR-1286 as a candidate miRNA. Through miRNA target luciferase reporter assay and transfecting SKCM cells with miR-1286 mimic, we for the first time confirmed RARRES3 mRNA as a target of miR-1286 in SKCM cells.
Previous research has demonstrated that RARRES3 might suppress EMT in cancer cells,37,39 and the EMT of SKCM cells was closely related to their proliferation, survival and metastasis. Here we also detected the protein levels of N-cadherin, Vimentin and E-cadherin in SKCM cells with or without HCP5 overexpression after the transfection for 48 or 72 hrs. However, the western blotting results suggested that overexpression of HCP5 seemed to have no statistically significant impact on the protein level of these markers in SKCM in vitro (data not shown). The EMT markers might be affected at other time points because their expressions follow a time course which maybe has been skipped in their unique western blot. Although these observations need to be verified, we speculate that, as a lncRNA, HCP5 might regulate the expressions or functions of other genes and proteins, and the overall effect of HCP5 overexpression on SKCM cells might be different from that of RARRES3 overexpression. Besides, previous studies indicated that HCP5 might be involved in T cell- or NC cell-mediated cytolysis of cancer cells.20,41 By querying the TCGA-SKCM data, we also found that HCP5 expressions might be significantly associated with that of some MHC I genes in SKCM. Whether HCP5 would regulate the immunogenicity of cancer cells will be investigated in our future research.
Conclusion
In the present research, we found HCP5 as a cancer-suppressive lncRNA in SKCM with high clinical relevance. Overexpression of HCP5 significantly reduced the proliferation, survival, clonogenicity and invasiveness of SKCM cells and contributed to serum deprivation-induced apoptosis in vitro through RARRES3. We found that HCP5 overexpression significantly increased RARRES3 gene expression in SKCM cells in vitro by sponging miR-1286. Our data suggested HPC5 as a possible prognostic factor and therapeutic target in clinical practice.
Supplementary materials
Materials and methods
Cell viability
SKCM cells after transfection were seeded into 96-well plates at a density of 5x103/well. After the incubation for 2, 24, 48 or 72 hrs, cells were interacted with 0.5 mg/mL MTT solution (Sigma-Aldrich, St. Louis, MO, USA) for another 4 hrs. Subsequently, the formazan was solubilized by DMSO. The absorbance at 490 nm was measured using a microplate reader (Bio-Rad, Hercules, CA, USA).
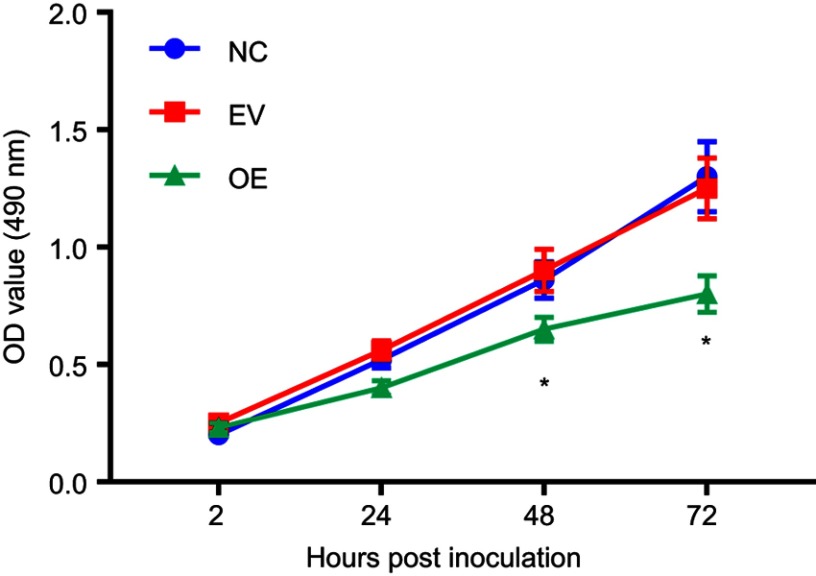
HCP5 overexpression inhibited SKCM cell proliferation. SKCM cells were transduced with empty (EV) or HCP5 (overexpressing; OE) lentiviral vectors before assay. Cell proliferation was measured by MTT assay. *p<0.05. Abbreviations: HCP5, human leucocyte antigen complex P5; SKCM, skin cutaneous melanoma; NC, negative control; EV, empty; OE, HCP-ovreexpressing.
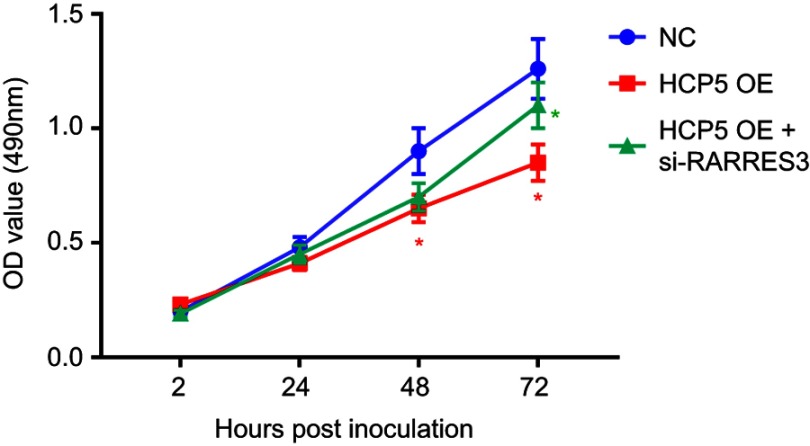
HCP5 overexpression suppressed SKCM cell proliferation by RARRES3. After transducing with HCP5 overexpressing lentiviral vector (HCP5 OE), SKCM cells were transfected with siRNA for RARRES3 knockdown (si-RARRES3). Cell proliferation was measured by MTT assay. *p<0.05. Red asterisks labeled the comparison to NC group, while the green asterisks labeled the comparison to HCP5 OE group.Abbreviation: SKCM, skin cutaneous melanoma.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lai V, Cranwell W, Sinclair R. Epidemiology of skin cancer in the mature patient. Clin Dermatol. 2018;36(2):167–176. doi: 10.1016/j.clindermatol.2017.10.008 [DOI] [PubMed] [Google Scholar]
- 2.Gandhi SA, Kampp J. Skin cancer epidemiology, detection, and management. Med Clin North Am. 2015;99(6):1323–1335. doi: 10.1016/j.mcna.2015.06.002 [DOI] [PubMed] [Google Scholar]
- 3.Schadendorf D, van Akkooi ACJ, Berking C, et al. Melanoma. Lancet. 2018;392(10151):971–984. doi: 10.1016/S0140-6736(18)31559-9 [DOI] [PubMed] [Google Scholar]
- 4.Schadendorf D, Fisher DE, Garbe C, et al. Melanoma. Nat Rev Dis Primers. 2015;1:15003. doi: 10.1038/nrdp.2015.3 [DOI] [PubMed] [Google Scholar]
- 5.Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat Rev Clin Oncol. 2017;14(8):463–482. doi: 10.1038/nrclinonc.2017.43 [DOI] [PubMed] [Google Scholar]
- 6.Johnson DB, Sosman JA. Therapeutic advances and treatment options in metastatic melanoma. JAMA Oncol. 2015;1(3):380–386. doi: 10.1001/jamaoncol.2015.0565 [DOI] [PubMed] [Google Scholar]
- 7.Kurokawa R, Rosenfeld MG, Glass CK. Transcriptional regulation through noncoding RNAs and epigenetic modifications. RNA Biol. 2009;6(3):233–236. [DOI] [PubMed] [Google Scholar]
- 8.Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi: 10.1038/nrg.2016.20 [DOI] [PubMed] [Google Scholar]
- 9.Adams BD, Parsons C, Walker L, Zhang WC, Slack FJ. Targeting noncoding RNAs in disease. J Clin Invest. 2017;127(3):761–771. doi: 10.1172/JCI84424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin C, Yang L. Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol. 2018;28(4):287–301. doi: 10.1016/j.tcb.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi: 10.1158/0008-5472.CAN-16-2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aftab MN, Dinger ME, Perera RJ. The role of microRNAs and long non-coding RNAs in the pathology, diagnosis, and management of melanoma. Arch Biochem Biophys. 2014;563:60–70. doi: 10.1016/j.abb.2014.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leucci E, Coe EA, Marine JC, Vance KW. The emerging role of long non-coding RNAs in cutaneous melanoma. Pigment Cell Melanoma Res. 2016;29(6):619–626. doi: 10.1111/pcmr.12537 [DOI] [PubMed] [Google Scholar]
- 14.Chen X, Guo W, Xu XJ, et al. Melanoma long non-coding RNA signature predicts prognostic survival and directs clinical risk-specific treatments. J Dermatol Sci. 2017;85(3):226–234. doi: 10.1016/j.jdermsci.2016.12.006 [DOI] [PubMed] [Google Scholar]
- 15.Ma X, He Z, Li L, Yang D, Liu G. Expression profiles analysis of long non-coding RNAs identified novel lncRNA biomarkers with predictive value in outcome of cutaneous melanoma. Oncotarget. 2017;8(44):77761–77770. doi: 10.18632/oncotarget.20780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang L, Xu J, Wang M, et al. LncRNA HCP5 promotes follicular thyroid carcinoma progression via miRNAs sponge. Cell Death Dis. 2018;9(3):372. doi: 10.1038/s41419-018-1111-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Y, Shen HM, Fang DM, Meng QJ, Xin YH. LncRNA HCP5 promotes the development of cervical cancer by regulating MACC1 via suppression of microRNA-15a. Eur Rev Med Pharmacol Sci. 2018;22(15):4812–4819. doi: 10.26355/eurrev_201808_15616 [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, Lu B, Sun L, Yan X, Xu J. Identification of candidate genes or microRNAs associated with the lymph node metastasis of SCLC. Cancer Cell Int. 2018;18:161. doi: 10.1186/s12935-018-0653-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu TG, Xiao X, Wei Q, Yue M, Zhang LX. Revealing potential long non-coding RNA biomarkers in lung adenocarcinoma using long non-coding RNA-mediated competitive endogenous RNA network. Braz J Med Biol Res. 2017;50(9):e6297. doi: 10.1590/1414-431X20176071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Low JS, Chin YM, Mushiroda T, et al. A genome wide study of copy number variation associated with nasopharyngeal carcinoma in Malaysian Chinese identifies CNVs at 11q14.3 and 6p21.3 as candidate loci. PLoS One. 2016;11(1):e0145774. doi: 10.1371/journal.pone.0145774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teng H, Wang P, Xue Y, et al. Role of HCP5-miR-139-RUNX1 feedback loop in regulating malignant behavior of glioma cells. Mol Ther. 2016;24(10):1806–1822. doi: 10.1038/mt.2016.103 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Liu N, Zhang R, Zhao X, et al. A potential diagnostic marker for ovarian cancer: involvement of the histone acetyltransferase, human males absent on the first. Oncol Lett. 2013;6(2):393–400. doi: 10.3892/ol.2013.1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anaya JJPCS. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput Sci. 2016;2:e67. [Google Scholar]
- 24.Paraskevopoulou MD, Vlachos IS, Karagkouni D, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. 2015;44(D1):D231–D238. doi: 10.1093/nar/gkv1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paraskevopoulou MD, Georgakilas G, Kostoulas N, et al. DIANA-microT web server v5. 0: service integration into miRNA functional analysis workflows. 2013;41(W1):W169–W173. doi: 10.1093/nar/gkt393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. 2011;40(D1):D1144–D1149. doi: 10.1093/nar/gkr1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiSepio D, Ghosn C, Eckert RL, et al. Identification and characterization of a retinoid-induced class II tumor suppressor/growth regulatory gene. Proc Natl Acad Sci U S A. 1998;95(25):14811–14815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei L, Chiu DK, Tsang FH, et al. Histone methyltransferase G9a promotes liver cancer development by epigenetic silencing of tumor suppressor gene RARRES3. J Hepatol. 2017;67(4):758–769. doi: 10.1016/j.jhep.2017.05.015 [DOI] [PubMed] [Google Scholar]
- 29.Errico A. Breast cancer: RARRES3-suppressing metastases to the lung in breast cancer. Nat Rev Clin Oncol. 2014;11(7):378. doi: 10.1038/nrclinonc.2014.103 [DOI] [PubMed] [Google Scholar]
- 30.Morales M, Arenas EJ, Urosevic J, et al. RARRES3 suppresses breast cancer lung metastasis by regulating adhesion and differentiation. EMBO Mol Med. 2014;6(7):865–881. doi: 10.15252/emmm.201303675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Casanova B, de la Fuente MT, Garcia-Gila M, et al. The class II tumor-suppressor gene RARRES3 is expressed in B cell lymphocytic leukemias and down-regulated with disease progression. Leukemia. 2001;15(10):1521–1526. [DOI] [PubMed] [Google Scholar]
- 32.Duvic M, Helekar B, Schulz C, et al. Expression of a retinoid-inducible tumor suppressor, Tazarotene-inducible gene-3, is decreased in psoriasis and skin cancer. Clin Cancer Res. 2000;6(8):3249–3259. [PubMed] [Google Scholar]
- 33.Wu CC, Shyu RY, Chou JM, et al. RARRES1 expression is significantly related to tumour differentiation and staging in colorectal adenocarcinoma. Eur J Cancer. 2006;42(4):557–565. doi: 10.1016/j.ejca.2005.11.015 [DOI] [PubMed] [Google Scholar]
- 34.Lotz K, Kellner T, Heitmann M, et al. Suppression of the TIG3 tumor suppressor gene in human ovarian carcinomas is mediated via mitogen-activated kinase-dependent and -independent mechanisms. Int J Cancer. 2005;116(6):894–902. doi: 10.1002/ijc.21127 [DOI] [PubMed] [Google Scholar]
- 35.Jiang SY, Chou JM, Leu FJ, et al. Decreased expression of type II tumor suppressor gene RARRES3 in tissues of hepatocellular carcinoma and cholangiocarcinoma. World J Gastroenterol. 2005;11(7):948–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shyu RY, Jiang SY, Chou JM, et al. RARRES3 expression positively correlated to tumour differentiation in tissues of colorectal adenocarcinoma. Br J Cancer. 2003;89(1):146–151. doi: 10.1038/sj.bjc.6601049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Wang L, Hu J, et al. RARRES3 suppressed metastasis through suppression of MTDH to regulate epithelial-mesenchymal transition in colorectal cancer. Am J Cancer Res. 2015;5(6):1988–1999. [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson AM, Kalimutho M, Harten S, Nanayakkara DM, Khanna KK, Ragan MA. The metastasis suppressor RARRES3 as an endogenous inhibitor of the immunoproteasome expression in breast cancer cells. Sci Rep. 2017;7:39873. doi: 10.1038/srep39873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu TH, Jiang SY, Chang WL, Eckert RL, Scharadin TM, Chang TC. Involvement of RARRES3 in the regulation of Wnt proteins acylation and signaling activities in human breast cancer cells. Cell Death Differ. 2015;22(5):801–814. doi: 10.1038/cdd.2014.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ou CC, Hsu SC, Hsieh YH, et al. Downregulation of HER2 by RIG1 involves the PI3K/Akt pathway in ovarian cancer cells. Carcinogenesis. 2008;29(2):299–306. doi: 10.1093/carcin/bgm263 [DOI] [PubMed] [Google Scholar]
- 41.Bauer S, Groh V, Wu J, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285(5428):727–729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HCP5 overexpression inhibited SKCM cell proliferation. SKCM cells were transduced with empty (EV) or HCP5 (overexpressing; OE) lentiviral vectors before assay. Cell proliferation was measured by MTT assay. *p<0.05. Abbreviations: HCP5, human leucocyte antigen complex P5; SKCM, skin cutaneous melanoma; NC, negative control; EV, empty; OE, HCP-ovreexpressing.
HCP5 overexpression suppressed SKCM cell proliferation by RARRES3. After transducing with HCP5 overexpressing lentiviral vector (HCP5 OE), SKCM cells were transfected with siRNA for RARRES3 knockdown (si-RARRES3). Cell proliferation was measured by MTT assay. *p<0.05. Red asterisks labeled the comparison to NC group, while the green asterisks labeled the comparison to HCP5 OE group.Abbreviation: SKCM, skin cutaneous melanoma.