Abstract
Radiation-induced fibrosis (RIF) is a common delayed effect of acute ionizing radiation exposure (DEARE) affecting diverse tissues including the heart, lungs, liver and skin, leading to reduced tissue function and increased morbidity. Monocytes, which may be classified into classical (CD14++, CD16−), intermediate (CD14++, CD16+) and non-classical (CD14+/low, CD16++) subtypes in humans and non-human primates (NHPs), and monocyte-derived macrophages may play an integral role in the pathogenesis of RIF. We tested the hypothesis that moderate to high levels of total-body exposure to radiation would alter monocyte polarization and produce phenotypes that could promote multi-organ fibrosis in a well-established NHP model of DEARE. Subjects were 16 young adult male rhesus macaques, ten of which were exposed to high-energy, 4 Gy X-ray total-body irradiation (TBI) and six that received sham irradiation (control). Total monocytes assessed by complete blood counts were 89% depleted in TBI animals by day 9 postirradiation (P < 0.05), but recovered by day 30 postirradiation and did not differ from control levels thereafter. Monocytes were isolated from peripheral blood mononuclear cells (PBMCs) and sorted into classical, intermediate and non-classical subsets using fluorescence-activated cell sorting (FACS) prior to and at 6 months post-TBI. At 6 months postirradiation, monocyte polarization shifted towards lower classical (92% → 86%) and higher intermediate (7% → 12%) and non-classical monocyte subsets (0.6% → 2%) (all P < 0.05) in TBI animals compared to baseline. No change in monocyte subsets was observed in control animals. Transcriptional profiles in classical and intermediate monocyte subsets were assessed using RNAseq. Classical monocyte gene expression did not change significantly over time or differ cross-sectionally between TBI and control groups. In contrast, significant numbers of differentially expressed genes (DEGs) were detected in intermediate monocyte comparisons between the TBI animals and all animals at baseline (304 DEGs), and in the TBI versus control animals at 6 months postirradiation (67 DEGs). Intermediate monocytes also differed between baseline and 6 months in control animals (147 DEGs). Pathway analysis was used to identify genes within significant canonical pathways, yielding 52 DEGs that were specific to irradiated intermediate monocytes. These DEGs and significant canonical pathways were associated with pro-fibrotic and anti-inflammatory signaling pathways that have been noted to induce M2 macrophage polarization. These findings support the hypothesis that TBI may alter monocyte programming and polarization towards a profibrotic phenotype, providing a novel target opportunity for therapies to inhibit or prevent RIF.
INTRODUCTION
One delayed effect of acute ionizing radiation exposure (DEARE) is radiation-induced fibrosis (RIF), a debilitating condition occurring 4–12 months postirradiation in humans that has been referred to as a “wound that fails to heal” (1). Radiation-induced fibrosis has been documented in many tissues including the heart (2), lungs (3), liver (4) and skin (5, 6) and is hypothesized to involve initial injury and healing followed by a period of cellular quiescence and later progression to fibrosis (7). The initiation signals that lead to progression from quiescence to RIF are largely unknown, but may involve an aberrant healing response mediated by monocytes, macrophages, fibroblasts, endothelial cells and platelets (8–10). Ultimately, this response results in excessive deposition of extracellular matrix, which reduces tissue compliance, impairs functionality and increases morbidity. Unfortunately, there are currently no therapies that can halt or reverse RIF (1). In 2016, 3.05 million cancer survivors were estimated to have been treated with radiotherapy, and the number is expected to increase to 4.17 million by 2030 (11), representing a large at-risk population for RIF.
Monocytes are key cells of the innate immune system, constituting 5–10% of circulating white blood cells in humans and non-human primates (12). They respond to tissue injury and may transform into tissue-resident macrophages upon exiting the vasculature. In human and non-human primates, they are often classified based on cell surface expression of CD14 [a co-receptor with Toll-like receptor 4 and lymphocyte antigen 96 (MD2) that together function to detect LPS] and CD16 (FcγRIII) expression into classical (CD14++, CD16−), intermediate (CD14++, CD16+) and non-classical (CD14+/low, CD16++) monocytes (13). Classical monocytes represent the majority of monocytes (85–90%) with intermediate and non-classical monocytes comprising the remainder (5–10% each) (13). It is currently unclear whether classical monocytes differentiate over time in a linear fashion into intermediate and non-classical monocytes (12, 14, 15) or if they are programmed from macrophage dendritic cell progenitors (MDPs) in the bone marrow (16). However, it is known that these monocyte subsets have disparate phenotypes, classical monocytes being associated with inflammatory responses and anaerobic metabolism, intermediate monocytes being associated with antigen presentation and processing, and non-classical monocytes being associated with oxidative phosphorylation and protein metabolism (17, 18). Depending on the signals received from the local and systemic environment, monocytes may adopt pro-inflammatory (M1-like) or anti-inflammatory-tissue remodeling/profibrotic (M2-like) phenotypes as monocyte-derived macrophages (19).
Increased intermediate monocyte polarization and subsequent tissue infiltration has been implicated as a primary mediator of pro-fibrotic activity in pathologic fibrotic disorders including systemic sclerosis, renal fibrosis, idiopathic pulmonary fibrosis, hepatic fibrosis, cardiac fibrosis and skeletal muscle fibrosis (13, 20, 21) but their role in RIF is currently unclear. Groves et al. (22) recently provided clarification in this area, showing in a murine MCP-1 receptor (CCR2) knockout model of total-body irradiation (TBI) with bone marrow transplantation a significantly reduced influx of peripheral monocyte-derived macrophages and reduced radiation-induced fibrosis in the lung. Additionally, Gibbons et al. (23) demonstrated that early monocyte and macrophage depletion by clodronate in a bleomycin-induced pulmonary fibrosis model led to a reduction in fibrosis. In the current study, we sought to assess the effects of TBI on circulating monocyte polarization in rhesus macaques, a well-established model of acute and delayed effects of ionizing radiation exposure. We hypothesized that monocyte polarization would shift towards increased intermediate and non-classical subsets and that transcriptional changes would reflect increased profibrotic/anti-inflammatory pathways.
MATERIALS AND METHODS
Animals
Sixteen male rhesus macaques, aged 5.8 ± 0.5 years, were obtained from World Wide Primates, Inc. (Miami, FL). Animals were pre-screened for tuberculosis and simian retroviruses and upon arrival were examined by a veterinarian, and quarantined for 60 days per standard institutional protocol. After quarantine, animals were fed a Western-like diet [typical American diet (TAD); LabDiet®, St. Louis, MO] and randomized to receive 4 Gy TBI (n = 10) or sham irradiation (control; n = 6). Irradiations were designed to mimic what might occur in a nuclear event, and were not intended to replicate radiation treatments for medical procedures, although some inferences may be made in cases where TBI is employed. The 4 Gy dose was selected to be well below the LD50/60 of 7.5 Gy previously reported for male rhesus monkeys (24). Irradiations were performed on a clinical linear accelerator using two lateral fields of 6-MV X rays [174-cm source-axis-distance (SAD), 32 × 40 cm2 field size at 100 cm from the source, nominal dose rate of 0.8 Gy/min at 174 cm SAD, 2.7 min irradiation time per field]. The irradiation geometry was dosimetrically confirmed prior to animal irradiation using a calibrated dosimeter system. The same dosimetry system was used for in vivo transit dosimetry for each radiation field delivered for each animal, with real-time analysis confirming the integrated dose received from the two fields. Each animal was sedated via an intramuscular injection of 15 mg/kg ketamine and placed in the supine position between two plastic buildup screens, knees up and arms crossed at the lower abdomen, with water-bolus compensation for thinner body regions (head and lower legs). The left side was irradiated first using 2 Gy and the animal was then rotated 180° to irradiate the right side using 2 Gy, for a total dose of 4 Gy. Dose calculations were performed for 2 Gy to mid-plane per field, based on anatomical measurements (nominal depths of 8–10 cm). Sham-irradiated animals underwent all procedural steps, including sedation, positioning and observation from outside the radiation room for the elapsed time of irradiation for each field, except that ionizing radiation was not delivered.
Monkeys were monitored daily for food consumption, defecation, urination and evidence of illness. Illness was determined using methods modified from the Children’s Cancer Group Clinical Toxicity Criteria established by Uckun et al. (25). Additionally, complete blood counts and chemistry panels were conducted weekly after irradiation and intervention was guided by clinical pathologic findings as reported elsewhere by our group in DeBo et al. (26) All experimental procedures were performed in accordance with the Guide for Care and Use of Laboratory Animals and followed protocol for avoidance of pain and discomfort and conducted in compliance with the Wake Forest School of Medicine (WFSM) Institutional Animal Care and Use Committee requirements. WFSM is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International, and operates in compliance with the Animal Welfare Act.
Total Monocyte Counts
Total monocyte and other blood cell fraction counts were obtained from complete blood counts that were collected before irradiation, then weekly after irradiation for two months and then monthly thereafter. Analyses were conducted by an external commercial veterinary laboratory (IDEXX Laboratories, North Grafton, MA).
Monocyte Isolation
Classical (CD14++, CD16−), intermediate (CD14++, CD16+) and non-classical (CD14+/low, CD16++) monocytes were obtained using fluorescence-activated cell sorting (FACS) on isolated peripheral blood mononuclear cells (PBMCs) before irradiation and 6 months postirradiation. PBMCs were first isolated using 50-ml Leucosep Centrifuge Tubes (Greiner Bio-One, Monroe, NC) according to manufacturer instructions. Briefly, 15 ml of EDTA whole blood was mixed with 15 ml phosphate buffered saline (PBS) without calcium or magnesium and layered onto 15 ml of 1.077-density Ficoll-Paque® Premium (GE Healthcare Life Sciences, Marlborough, MA). Tubes were centrifuged at 800g for 20 min at room temperature and PBMCs were collected from the interphase layer. PBMCs were washed with PBS without calcium or magnesium and centrifuged at 300g for 10 min, subsequently incubated in 1:9-ratio ammonium-chloride-potassium lysing buffer for 10 min to remove residual erythrocytes, centrifuged at 300g for 10 min and re-washed with PBS. Washed PBMCs were counted and assessed for viability using ViaStain® acridine orange/propidium iodide (AOPI) staining solution (Nexcelom Bioscience LLC, Lawrence, MA) on a Cellometer Vision Cell Profiler (Nexcelom Bioscience LLC, Lawrence, MA).
After isolation, PBMCs were incubated for 20 min at 4–8°C with 80 μl/1 × 107 cells autoMACS running buffer and 20 μl/1 × 107 CD14 non-human primate MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany). PBMCs were washed with 2 ml autoMACS running buffer, centrifuged at 300g for 10 min, resuspended in 500 μl autoMACS running buffer and subsequently run over magnetic separation (MS) columns (Miltenyi Biotec, Bergisch Gladbach, Germany). Collected CD14+ monocytes were counted and assessed for viability using AOPI staining solution on the Cellometer Vision Cell Profiler, centrifuged and resuspended in 100 μl PBS per 1 × 106 cells. CD14+ positive monocytes were incubated with 10 μl/1 × 106 cells CD14-PE (Tük4 clone) conjugated and CD16-VioBlue (VEP13 clone) conjugated monoclonal antibodies and appropriate isotype controls (all from Miltenyi Biotec, Bergisch Gladbach, Germany), incubated for 1 h 30 min at 4°C during transport to the Duke Human Vaccine Institute Flow Cytometry Core (Durham, NC). Cell subset proportions and preparations were obtained by FACS through sorting into classical, intermediate and non-classical monocytes using a FACSAria™ II flow cytometer (Becton Dickinson, Franklin Lakes, NJ), then stored in 85% FBS/15% DMSO sterile freezing media within a Mr. Frosty freezing container (Thermo Fisher Scientific™, Waltham, MA) at −80°C for 24 h before transfer into a liquid nitrogen dewar.
Total RNA Isolation
Isolated classical and intermediate monocytes stored in freezing media were thawed and centrifuged at 1,000g for 5 min. The freezing media was drawn off, RLT buffer (QIAGEN®, Valencia, CA) was added and the sample was disrupted by passing over a QIAshredder column (QIAGEN). The disrupted samples were then extracted for total RNA using the AllPrep DNA/RNA Mini Kit according to manufacturer’s protocol (QIAGEN). The extracted RNA was DNase-treated and purified using the RNA Clean & Concentrator™−5 kit (Zymo Research, Irvine, CA) and assessed for RNA quality using an Agilent 2100 Bioanalyzer and the RNA 6000 Pico Kit (Agilent Technologies, Santa Clara, CA).
cDNA Library Preparation and Sequencing Methods
Total RNA was used to prepare cDNA libraries with the SMARTer® Stranded Total RNA-Seq Kit v2 - Pico Input Mammalian (TaKaRa Bio USA Inc., Mountain View, CA). Briefly, 8 ng of total RNA was subjected to fragmentation followed by reverse-transcription. Illumina sequencing adaptors were attached during PCR amplification and the double-stranded cDNA was purified using AMPure XP magnetic beads. Finally, the cDNA was subjected to rRNA depletion and the stranded libraries were pre-amplified with PCR. The library size distribution was validated and quality inspected on an Agilent 2100 Bioanalyzer using a High Sensitivity DNA chip (Agilent Technologies). The quantity of each cDNA library was measured using a Qubit® 3.0 Fluorometer (Thermo Fisher Scientific). Libraries were sequenced to a read depth of >35M reads per sample using 1 × 75 bp single-end (SE) sequencing (Illumina®, San Diego, CA) on the Illumina NextSeq 500.
Data Processing and Expression Analysis
RNA sequencing quality control, alignment, read counts and differential gene expression was facilitated using Galaxy analysis platform (27). FASTq files were quality checked using FastQC (28), aligned to the rhesus macaque (rheMac3) genome using HISAT2 (29) and count files were generated using RefSeq genes in htseq-count using union mode (30). Differential gene expression was calculated via edgeR (31, 32) using robust settings to remove outlier genes, then filtering genes with less than 1.0 counts per million, and finally normalizing gene counts between samples using the trimmed mean of M values (TMM) method. Comparisons were made to evaluate the following: 1. differences in classical monocytes between TBI and control groups at 6 months postirradiation and changes from baseline in each group; and 2. differences between intermediate monocytes between TBI and control groups at 6 months postirradiation as well as changes in each group from baseline.
Statistics
All data were analyzed for normality and are presented as mean ± SEM. All analyses, except for gene expression analyses, were performed using Statistica™ version 13.3 (TIBCO® Software Inc., Palo Alto, CA). Difference in leukocyte counts and monocyte phenotypes were assessed using analysis of variance (ANOVA) or repeated measures ANOVA with Bonferroni post hoc tests. The level of significance was set at P < 0.05. Differential gene expression was calculated using edgeR with a log2 fold change threshold of 1.0 and Benjamini and Hochberg adjusted P < 0.05. Pathway analysis thresholds for significance are presented as an enrichment score based on a Fischer’s exact test P < 0.05 in Ingenuity Pathway Analysis (IPA) software (QIAGEN).
Post-Statistical Analysis of Biomolecular Pathways Implicated in TBI Responses
For those genes found to meet the criteria of being differentially expressed in response to TBI, biomolecular pathway analysis was performed using IPA software.
RESULTS
Leukocyte Depletion and Recovery Postirradiation
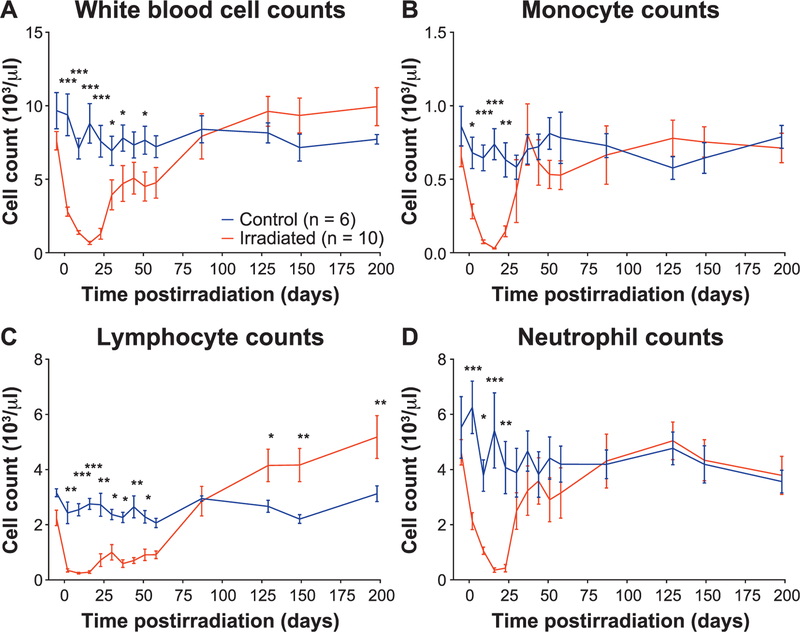
Total white blood cells, monocyte, lymphocyte and neutrophil counts were all acutely depleted in animals receiving TBI (P < 0.05). Monocytes and neutrophil counts recovered by day 30 postirradiation while total white blood cells and lymphocytes were significantly decreased until days 51 and 58 postirradiation, respectively [Fig. 1 (linear scale) or Supplementary Fig. S1; http://dx.doi.org/10.1667/RR15310.1.S1 (log scale)]. Total white blood cells, monocytes and neutrophils in TBI animals did not differ from controls post-recovery, but TBI lymphocytes increased above controls by day 129 postirradiation and remained elevated to the 200-day timepoint.
FIG. 1.
Peripheral immune cell recovery after total-body irradiation. Panels A–D: Total white blood cell, monocyte, lymphocyte and neutrophil counts, respectively, were all acutely depleted by TBI. Monocytes and neutrophils recovered by dau 30 postirradiation while total white blood cells and lymphocytes remained significantly decreased until days 51 and 58 postirradiation, respectively. After recovery, total white blood cells, monocytes and neutrophils in irradiated animals did not differ from controls, but lymphocytes increased above controls animals at day 129 postirradiation and remained elevated thereafter. *P < 0.05, **P < 0.01 and ***P < 0.001.
Monocyte Subset Alterations in Response to TBI
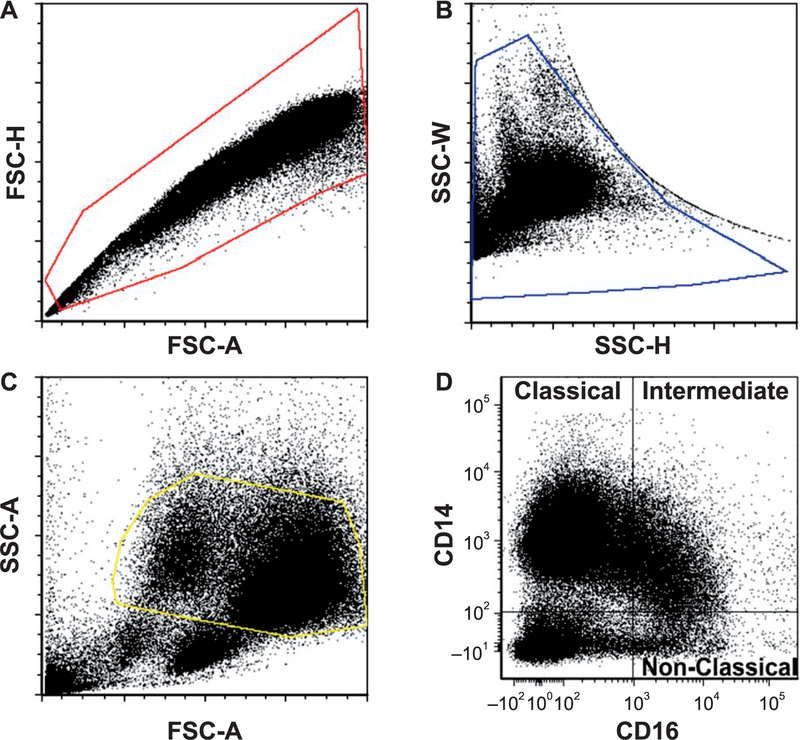
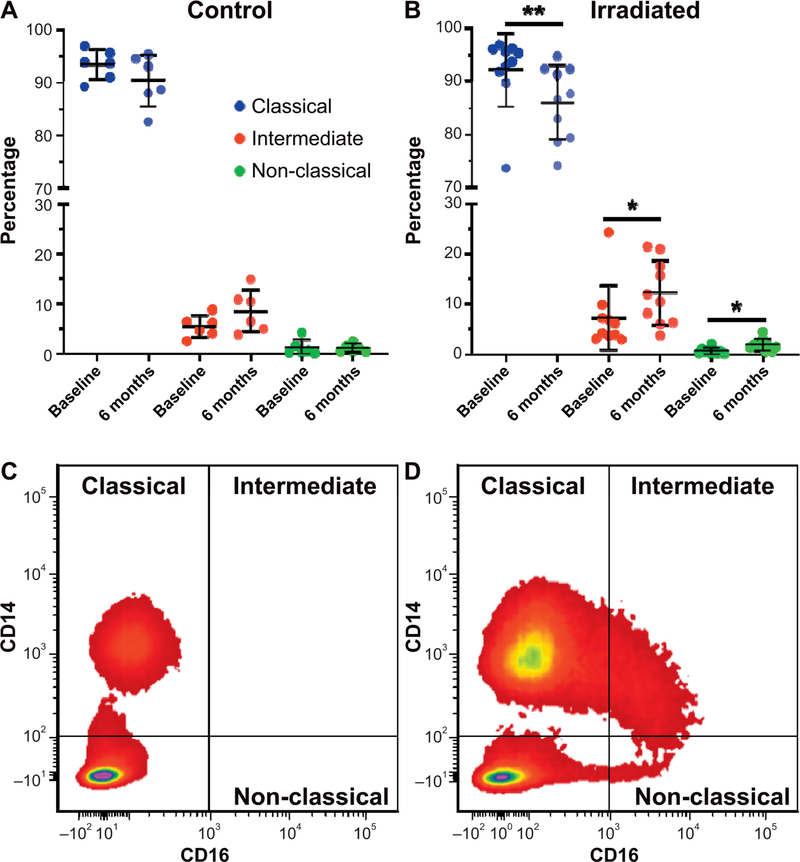
Magnetic bead-selected CD14+ monocytes were gated based on forward- and side-scatter parameters before final gating into classical (CD14++, CD16−), intermediate (CD14++, CD16+) and non-classical (CD14+/low, CD16++) subsets (Fig. 2). At 6 months postirradiation, monocyte polarization shifted towards lower classical (92% → 86%) and higher intermediate (7% → 12%) and non-classical monocyte subsets (0.6% → 2%) (all P < 0.05) in TBI animals compared to baseline. No changes in monocyte subset proportions were observed in the control animals (Fig. 3).
FIG. 2.
Gating strategy to obtain monocyte subsets. Magnetic bead selected CD14+ monocytes were gated based on forward- and side-scatter parameters before final gating into classical (CD14++, CD16−), intermediate (CD14++, CD16+) and non-classical (CD14+/low, CD16++) phenotypes. Panel A: Forward scatter plot. Panel B: Side scatter plot. Panel C: Forward scatter-side scatter. Panel D: Final representation of monocyte subsets by CD14 and CD16 labeling. Profiles from a representative sample from an irradiated animal at 6 months postirradiation are shown. FSC-H = forward scatter height; FSC-A = forward scatter area; SSC-W = side scatter width; SSC-H = height; SSC-A = side scatter area.
FIG. 3.
Alterations in distribution of monocyte subtypes after total-body irradiation. Panel A: Monocyte polarization was unaltered in control animals at 6 months after sham irradiation. Panel B: In contrast, irradiated animals had significant shifts towards lower classical and higher intermediate and non-classical monocyte subsets compared to baseline. Panels C and D: Flow cytometry density plots from a representative control and irradiated animal, respectively, are shown at 6 months postirradiation. (*P < 0.05 and **P < 0.01).
Classical Monocyte Gene Expression Responses to TBI
Differential gene expression in classical monocytes was evaluated by comparing the following: 1. TBI at 6 months to TBI baseline; 2. control at 6 months to control baseline; and 3. TBI at 6 months to control at 6 months (Table 1). The full list of genes assessed for each comparison, including log2 fold change, raw P value and false discovery rate (FDR) P value, is provided in Supplementary Table S1A (http://dx.doi.org/10.1667/RR15310.1.S2); and an abbreviated list of genes within each comparison in which the FDR is below 0.10 is provided in Supplementary Table S1B (http://dx.doi.org/10.1667/RR15310.1.S3). For the classical subset, there were three DEGs (3 upregulated) identified when TBI at 6 months was compared to TBI at baseline; and 12 DEGs (12 upregulated) were identified when control at 6 months was compared to control at baseline. No significant DEGs were identified in a comparison between TBI at 6 months and control at 6 months. For classical monocytes, no canonical pathways were identified as being significantly influenced by TBI, using IPA.
TABLE 1.
Cross-Sectional and Longitudinal Differential Gene Expression in Classical Monocytes
| Classical monocyte comparisons | Total DEGs | Upregulated DEGs | Downregulated DEGs |
|---|---|---|---|
| Irradiateda (6 months postirradiation) vs. baseline (all animals)b | 3 | 3 | 0 |
| Controlc (6 months postirradiation) vs. baseline (all animals)b | 12d | 12d | 0d |
| Irradiateda (6 months postirradiation) vs. controlc (6 months postirradiation) | 0d | 0d | 0d |
Irradiated animals (n = 10).
All animals (n = 16)
Control animals (n = 6).
Denotes potentially underpowered comparison.
Abbrevation: DEGs = differentially expressed genes.
Intermediate Monocyte Gene Expression Responses to TBI
Differences in gene expression between groups and over time in intermediate monocytes were evaluated by: 1. Comparing TBI monocytes at 6 months to baseline; 2. Comparing control monocytes at 6 months to baseline; and 3. Comparing TBI monocytes at 6 months to control at 6 months (Table 2). The full list of genes assessed for each comparison, including log2 fold change, raw P value and FDR P value are listed in Supplementary Table S2A (http://dx.doi.org/10.1667/RR15310.1.S4); and the abbreviated list of genes within each comparison, in which the FDR is below 0.10, is provided in Supplementary Table S2B(http://dx.doi.org/10.1667/RR15310.1.S5).
TABLE 2.
Cross-Sectional and Longitudinal Differential Gene Expression in Intermediate Monocytes
| Intermediate monocyte comparisons | Total DEGs | Upregulated DEGs | Downregulated DEGs |
|---|---|---|---|
| Irradiated (6 months postirradiation) vs. baseline | 304 | 145 | 159 |
| Control (6 months postirradiation) vs. baseline | 147* | 115* | 32* |
| Irradiated (6 months postirradiation) vs. control (6 months postirradiation) | 67* | 25* | 42* |
Notes. Irradiated animals (n = 10), control animals (n = 6);
Denotes potentially underpowered comparison. DEGs = differentially expressed genes.
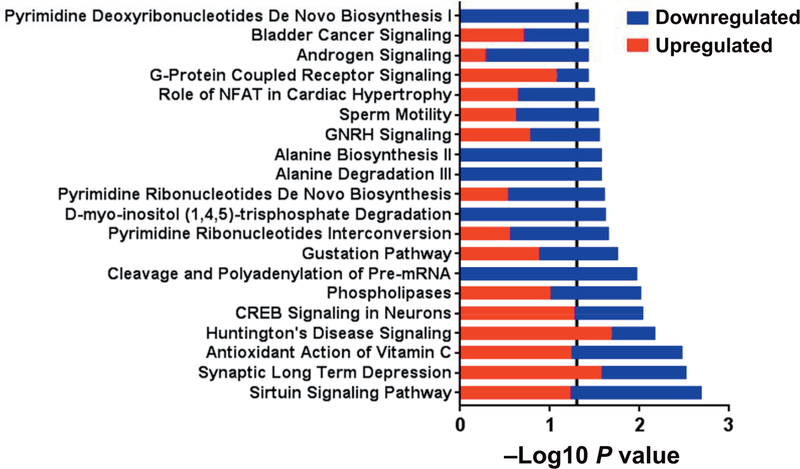
There were 304 DEGs (145 upregulated and 159 downregulated in the TBI group) identified when the TBI group at 6 months was compared to baseline. The top five diseases and disorders associated with these DEGs were endocrine system disorders, gastrointestinal disease, inflammatory disease, inflammatory response, and organismal injury and abnormalities (all P < 0.05). The top 20 significant pathways (all P < 0.05) are shown in Fig. 4. Pathways containing a majority of upregulated genes included synaptic long-term depression, Huntington’s disease signaling, cyclic AMP response element binding protein (CREB) signaling in neurons and G-protein coupled receptor signaling. Pathways containing a majority of downregulated genes included sirtuin signaling pathway, cleavage and polyadenylation of pre-mRNA, pyrimidine ribonucleotides interconversion, D-myo-inositol (1,4,5)-trisphosphate degradation, pyrimidine ribonucleotides de novo biosynthesis, alanine degradation III, alanine biosynthesis II, sperm motility, role of NFAT in cardiac hypertrophy, androgen signaling and pyrimidine deoxyribonucleotides de novo biosynthesis I. Pathways with equal number of up- and downregulated genes included antioxidant action of vitamin C, phospholipases, gustation pathway, gonadotropin-releasing hormone (GNRH) signaling and bladder cancer signaling.
FIG. 4.
Canonical pathways differentiating intermediate monocytes in irradiated subjects at 6 months (n = 10) from all subjects (n = 16) at baseline. Within each pathway red bars indicate the proportion of DEGs upregulated in irradiated subjects at 6 months, blue bars indicate the proportion of DEGs downregulated in irradiated subjects at 6 months and the overall length of the bar is the −log10 P value. The vertical line indicates the threshold for significance based on a −log10 P value of 1.3 (equal to P < 0.05).
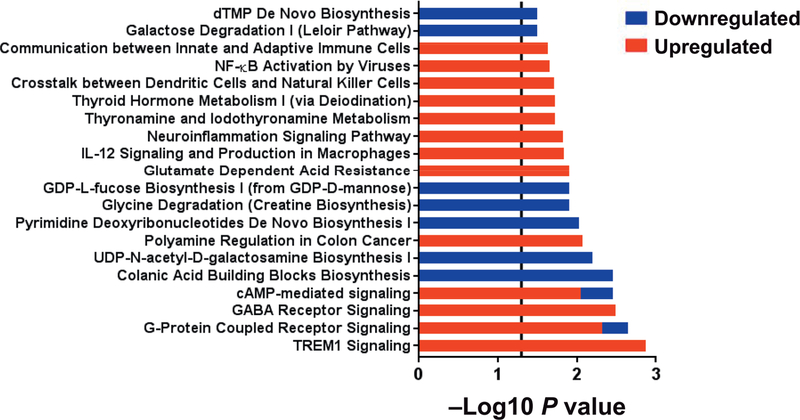
There were 147 DEGs (115 upregulated and 32 downregulated) identified when control intermediate monocytes at 6 months were compared to baseline. The top five diseases and disorders associated with these DEGs were neurological disease, psychological disorders, organismal injury and abnormalities, gastrointestinal disease and immunological disease (all P < 0.05). The top 20 significant pathways (all P < 0.05) are shown in Fig. 5. Pathways containing a majority of upregulated genes included triggering receptor expressed on myeloid cells (TREM1) signaling, G-protein coupled receptor signaling, gamma-aminobutyric acid (GABA) receptor signaling, cAMP-mediated signaling, polyamine regulation in colon cancer, glutamate dependent acid resistance, interleukin 12 (IL12) signaling and production in macrophages, neuro-inflammation signaling pathway, thyronamine and iodothyronamine metabolism, thyroid hormone metabolism I (via deiodination), crosstalk between dendritic cells and natural killer cells, nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) activation by viruses and communication between innate and adaptive immune cells. Pathways containing a majority of downregulated genes included colanic acid building blocks biosynthesis, UDP-N-acetyl-D-galactosamine biosynthesis I, pyrimidine deoxyribonucleotides de novo biosynthesis I, glycine degradation (creatine biosynthesis), GDP-L-fucose biosynthesis I (from GDP-D-mannose), galactose degradation I (Leloir pathway), and deoxythymidine monophosphate (dTMP) de novo biosynthesis.
FIG. 5.
Canonical pathways differentiating intermediate monocytes in control subjects at 6 months to all subjects (n = 16) at baseline. Within each pathway red bars indicate the proportion of DEGs upregulated in control subjects at 6 months, blue bars indicate the proportion of DEGs downregulated in control subjects at 6 months within the respective pathway and the overall length of the bar is the −log10 P value. The vertical line indicates the threshold for significance based on a −log10 P value of 1.3 (equal to P < 0.05).
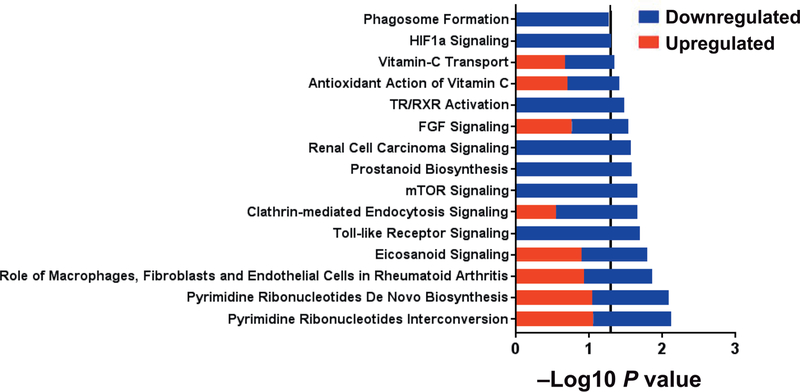
We identified 67 DEGs (25 upregulated and 42 downregulated after TBI) between TBI intermediate monocytes at 6 months and controls at 6 months. The top five diseases and disorders associated with these DEGs were inflammatory response, cancer, organismal injury and abnormalities, infectious disease and neurological disease (all P < 0.05). The significant pathways are shown in Fig. 6 (all P < 0.05). For all identified pathways, at least half of the genes were downregulated in TBI. Pathways containing a majority of TBI downregulated genes included Toll-like receptor signaling, clathrin-mediated endocytosis signaling, mTOR signaling, prostanoid biosynthesis, renal cell carcinoma signaling, fibroblast growth factor (FGF) signaling, thyroid hormone receptor (TR)/retinoid X receptor (RXR) activation, hypoxia inducible factor 1 alpha (HIF1α) signaling, and phagosome. Pathways containing equal numbers of up- and downregulated genes included pyrimidine ribonucleotides interconversion, pyrimidine ribonucleotides de novo biosynthesis, role of macrophages, fibroblasts and endothelial cells in rheumatoid arthritis, eicosanoid signaling, antioxidant action of vitamin C and vitamin-C transport.
FIG. 6.
Canonical pathways differentiating intermediate monocytes in irradiated subjects from control at 6 months. Within each pathway red bars indicate the proportion of DEGs upregulated in irradiated subjects, the blue bars indicate the proportion of DEGs downregulated in irradiated subjects within the respective pathway and the overall length of the bar is the −log10 P value. The vertical line indicates the threshold for significance based on a −log10 P value of 1.3 (equal to P < 0.05).
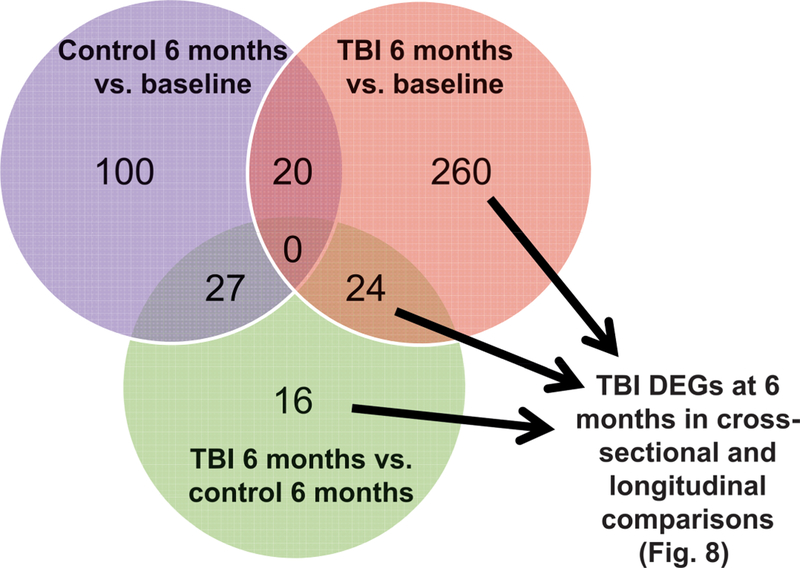
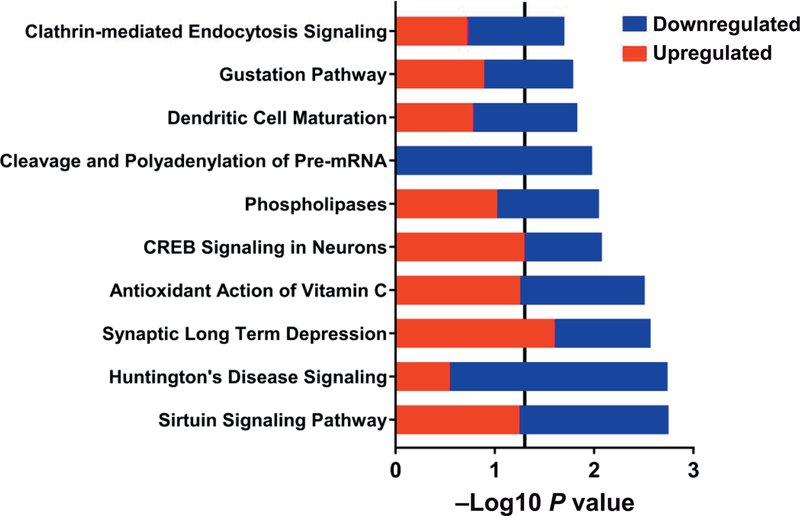
To determine DEGs that were specific to TBI intermediate monocytes at 6 months, we evaluated shared and distinct DEGs from all intermediate monocyte comparisons (TBI, 6 months vs. baseline; control, 6 months vs. baseline; and TBI, 6 months vs. control, 6 months) and removed DEGs (n = 47) that were found to be differentially expressed over the 6-month time period in the control group. (Fig. 7). Remaining DEGs were subsequently combined (n = 300) (Fig. 7) and analyzed using pathway analysis in IPA to determine canonical pathways specific to TBI, 6-month intermediate monocytes (Fig. 8). From this analysis we identified the subset of individual DEGs that were associated with significant canonical pathways. This yielded 52 DEGs (23 upregulated in TBI, 6-month monocytes compared to time-corrected control, 29 down-regulated) (Table 3). The top five diseases and disorders associated with these DEGs were cancer, organismal injury and abnormalities, neurological disease, gastrointestinal disease and skeletal and muscular disorders (all P < 0.05). The top 10 irradiated upregulated genes by log2 fold change were: Fc fragment of IgG receptor Ia (FCGR1A), collagen type III alpha 1 chain (COL3A1), gap junction protein beta 1 (GJB1), translocase of outer mitochondrial membrane 40 like (TOMM40L), glutamate metabotropic receptor 8 (GRM8), adenylate cyclase 2 (ADCY2), glutamate metabotropic receptor 1(GRM1), gap junction protein gamma 1 (GJC1), P21 (Rac1) activated kinase 6 (PAK6), and fibroblast growth factor 7 (FGF7). The top 10 downregulated genes were taste 2 receptor member 39 (TAS2R39), cyclin dependent kinase 7 (CDK7), transforming growth factor beta 1 induced transcript 1 (TGFB1I1), CD80 molecule (CD80), RNA polymerase II subunit G (POLR2G), G protein subunit beta 3 (GNB3), solute carrier family 25 member 42 (SLC25A42), H1 histone family, member X (H1FX), fibroblast growth factor 23 (FGF23), and phospholipase A2 group V (PLA2G5). The top 10 significant pathways (all P < 0.05) are shown in Fig. 8. Pathways containing a majority of upregulated genes included synaptic long-term depression and CREB signaling in neurons. Pathways containing a majority of downregulated genes included sirtuin signaling pathway, Huntington’s disease signaling, cleavage and polyadenylation of pre-mRNA, dendritic cell maturation and clathrin-mediated endocytosis. Pathways indicative of the antioxidant action of vitamin C, phospholipases and gustation had equal numbers of up- and downregulated genes identified.
FIG. 7.
Differentiating irradiated vs. control intermediate monocyte differentially expressed genes using cross-sectional and longitudinal analyses. Intermediate monocyte DEGs affected by radiation (n = 300) were determined by removing DEGs shared with longitudinal analyses of control intermediate monocytes (n = 47).
FIG. 8.
Intermediate monocyte canonical pathways differentiating irradiated vs. control subjects using cross-sectional and longitudinal differentially expressed genes. Within each pathway red bars indicate the proportion of DEGs upregulated in irradiated monocytes at 6 months, blue bars indicate the proportion of DEGs downregulated in irradiated monocytes at 6, and the overall height of the bar is the −log10 P value. The vertical line indicates the threshold for significance based on a −log10 P value of 1.3 (equal to P < 0.05).
TABLE 3.
Differentially Expressed Genes Identified in 6-Month Intermediate Monocytes from Cross Sectional and Longitudinal Comparisons
| Gene | Gene ID | Log2 fold change | FDR P |
|---|---|---|---|
| Fc fragment Of IgG receptor Iaa | FCGR1A | 5.98 | 0.0303 |
| Collagen type III alpha 1 chainb | COL3A1 | 5.29 | 0.0001 |
| Gap junction protein beta 1b | GJB1 | 4.95 | 0.0001 |
| Translocase of outer mitochondrial membrane 40 likea | TOMM40L | 4.79 | 0.0486 |
| Glutamate metabotropic receptor 8b | GRM8 | 4.25 | 0.0006 |
| Adenylate cyclase 2b | ADCY2 | 4.17 | 0.0005 |
| Glutamate metabotropic receptor 1b | GRM1 | 4.13 | 0.0012 |
| Gap junction protein gamma 1b | GJC1 | 4.13 | 0.0142 |
| P21 (Rac1) activated kinase 6b | PAK6 | 4.04 | 0.0007 |
| Fibroblast growth factor 7c | FGF7 | 3.70 | 0.0029 |
| Phospholipase C like 1b | PLCL1 | 3.69 | 0.0087 |
| Synaptosome associated protein 91b | SNAP91 | 3.65 | 0.0297 |
| Signal transducer and activator of transcription 5Ab | STAT5A | 3.14 | 0.0318 |
| Microtubule associated protein 1 light chain 3 betab | MAP1LC3B | 3.08 | 0.0297 |
| Translocase of outer mitochondrial membrane 7b | TOMM7 | 3.06 | 0.0283 |
| Matrix metallopeptidase 14b | MMP14 | 2.97 | 0.0244 |
| Translocase of outer mitochondrial membrane 40b | TOMM40 | 2.95 | 0.0370 |
| Nudix hydrolase 18c | NUDT18 | 2.90 | 0.0400 |
| Golgi snap receptor complex member 1b | GOSR1 | 2.83 | 0.0147 |
| Phospholipase A2 group XVIc | PLA2G16 | 2.80 | 0.0457 |
| Calcium voltage-gated channel auxiliary subunit beta 3b | CACNB3 | 2.63 | 0.0401 |
| Rab5A, member Ras oncogene familyb | RAB5A | 2.04 | 0.0368 |
| Phosphodiesterase 12b | PDE12 | 1.67 | 0.0022 |
| Death associated protein kinase 1b | DAPK1 | −1.22 | 0.0338 |
| Mitogen-activated protein kinase kinase kinase 1b | MAP3K11 | −1.37 | 0.0297 |
| Signal transducer and activator of transcription 2b | STAT2 | −1.41 | 0.0165 |
| Dynamin 2b | DNM2 | −1.41 | 0.0359 |
| Phospholipase C beta 2b | PLCB2 | −1.44 | 0.0057 |
| Uridine-cytidine kinase 1 like 1b | UCKL1 | −1.50 | 0.0230 |
| Autophagy related 9Ab | ATG9A | −1.55 | 0.0472 |
| Histone deacetylase 5b | HDAC5 | −1.60 | 0.0297 |
| Isocitrate dehydrogenase NADP+ 2, mitochondrialb | IDH2 | −1.63 | 0.0107 |
| GA binding protein transcription factor beta subunit 1b | GABPB1 | −1.86 | 0.0469 |
| Inositol(Myo)-1(Or 4)-monophosphatase 2b | IMPA2 | −1.97 | 0.0395 |
| Inositol polyphosphate phosphatase like 1b | INPPL1 | −2.06 | 0.0079 |
| Cleavage and polyadenylation specific factor 1b | CPSF1 | −2.07 | 0.0063 |
| Intraflagellar transport 57b | IFT57 | −2.79 | 0.0467 |
| Adenylate kinase 5c | AK5 | −3.06 | 0.0351 |
| Ribosomal protein S27Aa | RPS27A | −3.10 | 0.0450 |
| Cleavage and polyadenylation specific factor 4b | CPSF4 | −3.15 | 0.0394 |
| Insulin like growth factor 1b | IGF1 | −3.34 | 0.0351 |
| Glutamic-pyruvic transaminaseb | GPT | −3.38 | 0.0394 |
| Phospholipase A2 group Vb | PLA2G5 | −3.41 | 0.0392 |
| Fibroblast growth factor 23b | FGF23 | −3.49 | 0.0351 |
| H1 histone family, member Xb | H1FX | −3.62 | 0.0353 |
| Solute carrier family 25 member 42b | SLC25A42 | −3.66 | 0.0297 |
| G protein subunit beta 3b | GNB3 | −3.73 | 0.0277 |
| RNA polymerase II subunit Gb | POLR2G | −3.75 | 0.0297 |
| CD80 moleculeb | CD80 | −3.80 | 0.0297 |
| Transforming growth factor beta 1 induced transcript 1b | TGFB1I1 | −4.38 | 0.0109 |
| Cyclin dependent kinase 7b | CDK7 | −4.51 | 0.0097 |
| Taste 2 receptor member 39b | TAS2R39 | −4.99 | 0.0037 |
Indicates differentially expressed genes identified only in cross-sectional comparisons.
Indicates differentially expressed genes identified only in longitudinal comparisons.
Indicates differentially expressed genes identified in both cross-sectional and longitudinal comparisons.
DISCUSSION
This study provides insights into the effects of ionizing radiation on circulating populations of classical, intermediate and non-classical monocytes, as defined by CD14 and CD16 surface markers, in a well-established non-human primate model, the rhesus macaque. Monocyte populations shifted from a classical phenotype towards intermediate and non-classical phenotypes at 6 months postirradiation, gene expression changes occurred in intermediate, but not classical monocytes, and intermediate monocyte DEGs were associated with sirtuin, glutamatergic and anti-inflammatory signaling.
After radiation-induced depletion, monocyte populations returned to normal levels by approximately 30 days and remained at those levels for the study duration. A similar pattern of repopulation was observed for neutrophils, while recovery of lymphocytes was slower and actually increased above baseline in TBI compared to control animals. This apparent “overshoot” in circulating levels by the lymphocytes may reflect a “ramping up” in production after TBI and their relatively long half-life (several months) (33) relative to circulating lifespans of neutrophils and monocytes, which are only a few days (34). Despite the fact that we observed similar total monocyte numbers, monocyte phenotypic polarization was shifted 6 months postirradiation, the time frame consistent with initiation of RIF in humans, away from classical subsets towards increased intermediate and non-classical subsets. To our knowledge, this is the first time that alterations in monocyte polarization have been reported as a consequence of ionizing radiation exposure. The shift from classical to intermediate and non-classical monocyte subsets that we observed is similar to shifts observed in other pathologic fibrotic disorders (13, 20, 21).
We also found that TBI had differential effects on these two populations of monocytes. Interestingly, there were few DEGs detected in classical monocyte comparisons, whereas there were often hundreds of DEGs identified in intermediate monocyte comparisons. To our knowledge, this is the first time it has been shown that selected monocyte phenotypes have differentially altered gene expression after ionizing radiation exposure.
We identified DEGs that were specific to TBI 6-month intermediate monocytes by assessing shared and distinct DEGs from all intermediate monocyte comparisons (Table 3), and removing those that were found to be differentially expressed over time (perhaps due to effects of Western diet exposure) in the control group. This trimmed set of genes was subjected to further analysis to explore canonical pathways or groups of pathways that were more specific to TBI 6-month intermediate monocytes (Fig. 8). The pathways identified contained upregulated genes associated with neurologic signaling (synaptic long-term depression and CREB signaling in neurons), and the key genes driving these changes, GRM1 and GRM8, have also been demonstrated as pivotal mediators of inflammatory and monocyte signaling. These metabotropic glutamate receptor genes (GRM1, GRM8) are part of family C of GPCRs comprising eight total receptors (GRMs 1–8) split into three groups: group I (GRM1 and GRM5) receptors induce inositol 1,4,5, triphosphate (IP3) formation, whereas group II (GRM2 and GRM3) and group III (GRM4, GRM6, GRM7 and GRM8) receptors signal to reduce intracellular cAMP (35). Plasma glutamate has been noted to be increased in conditions associated with immunodeficiency, including HIV infection (36) and cancer (37), and decreased in autoimmune disorders like systemic lupus erythematosus (SLE) (38). The implications of these changes in glutamate concentration on monocyte/macrophage function are under-explored, but there is evidence it may lead to reduced pro-inflammatory signaling (39). When murine RAW 264.7 macrophages were transfected with the group I metabotropic glutamate receptor 5 (mGlu5), producing an environment with increased glutamate, they were found to have increased secretion of IL10, high mobility group box 1 protein (HMGB1), galectin 3 (Gal-3) and PPARγ (40). These cytokines are generally associated with anti-inflammatory signaling and IL10, HMGB1and Gal-3 specifically have been noted to induce the polarization of macrophages towards the alternatively activated-M2 macrophage state (41–43). Finally, the anti-inflammatory effects of PPARγ are accompanied by inhibition of sirtuin 1 (SIRT1) expression and activity (44). These associations may explain the identification of sirtuin signaling as the most significant (based on P value), and downregulated, pathway in our analyses of TBI intermediate monocytes 6 months postirradiation.
In monocytes and macrophages, sirtuins function as nutrient sensing proteins that respond to NAD+ production and NAD/NADH ratios to regulate the metabolic switch from glycolysis (associated with M1-like monocyte/macrophage phenotypes) to fatty acid oxidation (associated with M2-like monocyte/macrophage phenotypes) (45). Downregulation of sirtuin 3 (SIRT3) signaling has been noted to promote fibroblast-myofibroblast differentiation resulting in increased pulmonary fibrosis in murine models. This effect was found to be mediated by decreased repression of TGFβ1/mothers against decapentaplegic homolog 3 (SMAD3) signaling (46). In contrast, sirtuin 1 (SIRT1) overexpression was observed to ameliorate renal fibrosis in partially nephrectomized rats through inhibition of the TGFβ1/SMAD3 signaling pathway, while SIRT1 knockdown produced the opposite effect (47). TGFβ1 is a potential “master” regulator of fibrosis in various fibrotic conditions including those affecting the heart (48, 49), lung (50) and kidney (51) via signaling through canonical (Smad-based) or non-canonical (non-Smad-based) pathways. Downregulation of sirtuin signaling in TBI intermediate monocytes could potentiate a pro-fibrotic phenotype after radiation exposure due to upregulation and signaling of metabotropic glutamate receptors in response to increased availability of systemic glutamate.
Of the remaining top 10 pathways identified (Huntington’s disease signaling, antioxidant of vitamin C, phospholipases, cleavage and polyadenylation of pre-mRNA, dendritic cell maturation, gustation pathway, and clathrin-mediated endocytosis signaling) the majority contained predominately TBI downregulated genes. Several shared DEGs among these pathways encoded phospholipases, including phospholipase C beta 2 (PLCB2), phospholipase A2 group XVI (PLA2G16), phospholipase A2 group V (PLA2G5) and phospholipase C like 1 (PLCL1). Two DEGs, insulin like growth factor 1 (IGF1) and NFκB inhibitor alpha (NFKBIA) (both downregulated) were shared between several of these pathways. IGF-1 is a 70 amino acid hormone that is predominately produced by the liver, (52) but also produced by skeletal myocytes, fibroblasts, endothelial cells, macrophages and monocytes (53). Monocyte/macrophage-derived IGF-1 has been identified as a critical regulator of normal skeletal muscle repair in mice after injury. Conditional deletion of IGF-1 in murine macrophages resulted in increased inflammation, impaired healing and dysregulated inflammatory gene expression response consisting of increased pro-inflammatory (TNFα, NOS2, IL-1β) and anti-inflammatory (IL-10) cytokines (53). Decreased IGF1 expression in monocytes could result in excessive inflammation and aberrant wound healing. In human cirrhosis and non-alcoholic fatty liver disease, intermediate monocytes have been implicated in fibrosis progression (54) and IGF-1 deficiency may be partially responsible for progressive inflammation and matrix deposition characteristic of these diseases (52, 55). Downregulation of IGF1 combined with NFKBIA down-regulation suggests that NFκB was also downregulated, as IGF-1 has been demonstrated to activate NFκB through the PI-3 kinase pathway (56), which in turn activates NFKBIA expression, forming a self-regulating negative feedback cycle (57). Decreased IGF1 and NFKBIA expression could suggest there is reduced NFκB activation in intermediate monocytes. These findings suggest that TBI intermediate monocytes may elicit excessive and dysregulated inflammatory responses that also result in excessive fibrogenesis, which provides some support for our findings regarding glutamatergic and sirtuin related pathways.
This study utilized a well-characterized prospective cohort of male rhesus macaques in which certain limitations were inherent and unavoidable. By design, we isolated three specific monocyte subtypes based on the relative expression of CD14 and CD16, consistent with significant literature using that method, as a practical approach to begin to answer questions related to monocyte polarization responses to radiation. Newer methodologies, such as single cell sequencing of monocytes, which would provide more detail and granularity to the spectrum of monocyte subsets and phenotype variation, which might result from radiation exposure, were beyond the scope of the current project, but will be pursued in the future. The IPA software relies on human- and animal-based data from published studies, and public and third-party databases, to provide a means to identify molecular pathways and processes associated with patterns of differential gene expression and glean insights into the overall pathobiology of the system, by its nature is dependent on available data, and may be limited in its ability to identify novel pathways to explain observed outcomes. There were also some limitations in terms of statistical power. Using the power calculator provided by Hart et al. (58), we concluded that a sample size of 10 animals per group was needed to detect a log2 fold change of 0.91 with 80% power and a false discovery rate (FDR) P value < 0.05. Therefore, while comparisons between TBI and all groups are sufficiently powered, comparisons of the control group to itself and other groups are relatively underpowered. The finding that intermediate monocyte gene expression profiles changed in control animals between baseline and 6 months postirradiation could be a result of age-related changes, or the accumulating effects of consuming a Western diet over time. There was little overlap of DEGs that changed over time in the TBI vs. control groups, which suggests that the control group changed differently from the irradiated group. Control animals consuming a Western diet gained substantial weight between baseline and 6 months post-TBI (10.8 ± 0.8 to 13.3 ± 1.1 kg, ~23% increase). Increases in fat mass and obesity have been noted to lead to increased macrophage infiltration into the expanding adipose tissue and alter monocyte polarization (59, 60). Animals receiving TBI lost weight afterwards, but rebounded to baseline body weights by 6 months postirradiation (10.6 ± 0.8 vs. 11.0 ± 0.9 kg).
In summary, we show that 6 months post-TBI, monocyte phenotypes were shifted towards intermediate and non-classical phenotypes, accompanied by gene expression changes in intermediate monocytes, but not classical monocytes. The pathways identified from available algorithms are associated with pro-fibrotic and anti-inflammatory signaling pathways, which could function to push intermediate monocytes towards M2 macrophage polarization upon extravasation. In particular, the 67 DEGs (25 upregulated and 42 downregulated in TBI group) between the TBI and control intermediate monocytes at 6 months postirradiation provides support for an M2 phenotype shift, as the highest-ranked disease/disorders classification included inflammatory response and organismal injury and abnormalities (Fig. 6), in which the majority of genes were downregulated in the TBI group. These findings support the notion that selective targeting of monocyte populations and programming may present a novel target opportunity for therapies to inhibit or prevent RIF. This could prove to be an exciting new area of research, but one that needs significant further and careful examination in terms of macrophage activation and other cell phenotypes in relationship to fibrosis at the tissue level in key target (and non-target) organs. These areas will be explored in future studies in the subjects described in the current study, which includes longitudinal phenotyping of cardiac structure and function by ultrasound and cardiac fibrosis via magnetic resonance imaging approaches.
Supplementary Material
Fig. S1. Peripheral immune cell recovery after total body irradiation “log scale”.
Table S1A. Classical monocyte comparisons, full list.
Table S1B. Classical monocyte comparisons, abbreviated list.
Table S2A. Intermediate monocyte comparisons, full list.
Table S2B. Intermediate monocyte comparisons, abbreviated list.
ACKNOWLEDGMENTS
This research was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIAID/NIH award no. U19AI067773) and by NIH grant nos. U19 AI67798, R01 HL122393, T32 OD010957 and DOD W81XWH-15–1-0574, as well as the Wake Forest Baptist Comprehensive Cancer Center’s NCI Cancer Center Support Grant (no. P30CA012197–39) and Shared Instrumentation grant (no. 1S10OD023409–01). We thank the Duke Human Vaccine Institute Flow Facility for the flow cytometry work performed as well as Jean Gardin, Russell O’Donnell, Chrystal Bragg, Matt Dwyer, Michael Bennett and J. D. Bottoms for their technical assistance. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
REFERENCES
- 1.Straub JM, New J, Hamilton CD, Lominska C, Shnayder Y, Thomas SM. Radiation-induced fibrosis: mechanisms and implications for therapy. J Cancer Res Clin Oncol 2015; 141:1985–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fajardo LF, Stewart JR. Pathogenesis of radiation-induced myocardial fibrosis. Lab Invest 1973; 29:244–57. [PubMed] [Google Scholar]
- 3.Cardozo BL, Zoetelief H, van Bekkum DW, Zurcher C, Hagenbeek A. Lung damage following bone marrow transplantation: I. The contribution of irradiation. Int J Radiat Oncol Biol Phys 1985; 11:907–14. [DOI] [PubMed] [Google Scholar]
- 4.Ingold JA, Reed GB, Kaplan HS, Bagshaw MA. Radiation hepatitis. Am J Roentgenol Radium Ther Nucl Med 1965; 93:200–8. [PubMed] [Google Scholar]
- 5.Bentzen SM, Thames HD, Overgaard M. Latent-time estimation for late cutaneous and subcutaneous radiation reactions in a single-follow-up clinical study. Radiother Oncol 1989; 15:267–74. [DOI] [PubMed] [Google Scholar]
- 6.Brocheriou C, Verola O, Lefaix JL, Daburon F. Histopathology of cutaneous and subcutaneous irradiation-induced injuries. Br J Radiol Suppl 1986; 19:101–4. [PubMed] [Google Scholar]
- 7.Stewart JR, Fajardo LF. Radiation-induced heart disease: an update. Prog Cardiovasc Dis 1984; 27:173–94. [DOI] [PubMed] [Google Scholar]
- 8.Franklin TJ. Therapeutic approaches to organ fibrosis. Int J Biochem Cell Biol 1997; 29:79–89. [DOI] [PubMed] [Google Scholar]
- 9.Denham JW, Hauer-Jensen M. The radiotherapeutic injury–a complex ‘wound’. Radiother Oncol 2002; 63:129–45. [DOI] [PubMed] [Google Scholar]
- 10.Tak JK, Park JW. The use of ebselen for radioprotection in cultured cells and mice. Free Radic Biol Med 2009; 46:1177–85. [DOI] [PubMed] [Google Scholar]
- 11.Bryant AK, Banegas MP, Martinez ME, Mell LK, Murphy JD. Trends in radiation therapy among cancer survivors in the United States, 2000–2030. Cancer Epidemiol Biomarkers Prev 2017; 26:963–70. [DOI] [PubMed] [Google Scholar]
- 12.Yona S, Jung S. Monocytes: subsets, origins, fates and functions. Curr Opin Hematol 2010; 17:53–9. [DOI] [PubMed] [Google Scholar]
- 13.Wong KL, Yeap WH, Tai JJ, Ong SM, Dang TM, Wong SC. The three human monocyte subsets: implications for health and disease. Immunol Res 2012; 53:41–57. [DOI] [PubMed] [Google Scholar]
- 14.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science 2010; 327:656–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hume DA, MacDonald KP. Therapeutic applications of macrophage colony-stimulating factor-1 (CSF-1) and antagonists of CSF-1 receptor (CSF-1R) signaling. Blood 2012; 119:1810–20. [DOI] [PubMed] [Google Scholar]
- 16.Hanna RN, Carlin LM, Hubbeling HG, Nackiewicz D, Green AM, Punt JA, et al. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes. Nat Immunol 2011; 12:778–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zawada AM, Rogacev KS, Rotter B, Winter P, Marell RR, Fliser D, et al. SuperSAGE evidence for CD14++ CD16+ monocytes as a third monocyte subset. Blood 2011; 118:e50–61. [DOI] [PubMed] [Google Scholar]
- 18.Wong KL, Tai JJ-Y, Wong W-C, Han H, Sem X, Yeap W-H, et al. Gene expression profiling reveals the defining features of the classical, intermediate, and nonclassical human monocyte subsets. Blood 2011; 118:e16. [DOI] [PubMed] [Google Scholar]
- 19.Shi X, Shiao SL. The role of macrophage phenotype in regulating the response to radiation therapy. Transl Res 2018; 191:64–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Westbury CB, Yarnold JR. Radiation fibrosis – current clinical and therapeutic perspectives. Clinical Oncol 2012; 24:657–72. [DOI] [PubMed] [Google Scholar]
- 21.Liaskou E, Zimmermann HW, Li KK, Oo YH, Suresh S, Stamataki Z, et al. Monocyte subsets in human liver disease show distinct phenotypic and functional characteristics. Hepatology 2013; 57:385–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groves AM, Johnston CJ, Williams JP, Finkelstein JN. Role of infiltrating monocytes in the development of radiation-induced pulmonary fibrosis. Radiat Res 2018; 189:300–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibbons MA, MacKinnon AC, Ramachandran P, Dhaliwal K, Duffin R, Phythian-Adams AT, et al. Ly6Chi monocytes direct alternatively activated profibrotic macrophage regulation of lung fibrosis. Am J Respir Crit Care Med 2011; 184:569–81. [DOI] [PubMed] [Google Scholar]
- 24.Thrall KD, Love R, O’Donnell KC, Farese AM, Manning R, MacVittie TJ. An interlaboratory validation of the radiation dose response relationship (DRR) for H-ARS in the rhesus macaque. Health Phys 2015; 109:502–10. [DOI] [PubMed] [Google Scholar]
- 25.Uckun FM, Yanishevski Y, Tumer N, Waurzyniak B, Messinger Y, Chelstrom LM, et al. Pharmacokinetic features, immunogenicity, and toxicity of B43(anti-CD19)-pokeweed antiviral protein immunotoxin in cynomolgus monkeys. Clin Cancer Res 1997; 3:325–37. [PubMed] [Google Scholar]
- 26.DeBo RJ, Lees CJ, Dugan GO, Caudell DL, Michalson KT, Hanbury DB, et al. Late effects of total-body gamma irradiation on cardiac structure and function in male rhesus macaques. Radiat Res 2016; 186:55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Cech M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res 2016; 44:W3–W10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andrews S FastQC: a quality control tool for high throughput sequence data. 2010. (http://www.bioinformatics.babraham.ac.uk/projects/fastqc)
- 29.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015; 12:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 2015; 31:166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu R, Holik AZ, Su S, Jansz N, Chen K, Leong HS, et al. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res 2015; 43:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salvato MS, Rater M, Pauza CD. Tracking of dye-labeled lymphocytes in rhesus macaques. J Med Primatol 1996; 25:112–21. [DOI] [PubMed] [Google Scholar]
- 34.He Z, Allers C, Sugimoto C, Ahmed N, Fujioka H, Kim WK, et al. Rapid turnover and high production rate of myeloid cells in adult rhesus macaques with compensations during aging. J Immunol 2018; 200:4059–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiocchetti A, Miglio G, Mesturini R, Varsaldi F, Mocellin M, Orilieri E, et al. Group I mGlu receptor stimulation inhibits activation-induced cell death of human T lymphocytes. Br J Pharmacol 2006; 148:760–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eck HP, Frey H, Droge W. Elevated plasma glutamate concentrations in HIV-1-infected patients may contribute to loss of macrophage and lymphocyte functions. Int Immunol 1989; 1:367–72. [DOI] [PubMed] [Google Scholar]
- 37.Ollenschlager G, Karner J, Karner-Hanusch J, Jansen S, Schindler J, Roth E. Plasma glutamate–a prognostic marker of cancer and of other immunodeficiency syndromes? Scand J Clin Lab Invest 1989; 49:773–7. [DOI] [PubMed] [Google Scholar]
- 38.Poulopoulou C, Papadopoulou-Daifoti Z, Hatzimanolis A, Fragiadaki K, Polissidis A, Anderzanova E, et al. Glutamate levels and activity of the T cell voltage-gated potassium Kv1.3 channel in patients with systemic lupus erythematosus. Arthritis Rheum 2008; 58:1445–50. [DOI] [PubMed] [Google Scholar]
- 39.Eck H-P, Frey H, Droge W. Elevated plasma glutamate concentrations in HIV-1-infected patients may contribute to loss of macrophage and lymphocyte functions. Int Immunol 1989; 1:367–72. [DOI] [PubMed] [Google Scholar]
- 40.Shanshiashvili L, Tsitsilashvili E, Dabrundashvili N, Kalandadze I, Mikeladze D. Metabotropic glutamate receptor 5 may be involved in macrophage plasticity. Biol Res 2017; 50:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinha P, Clements VK, Bunt SK, Albelda SM, Ostrand-RosenbergS. Cross-talk between myeloid-derived suppressor cells and macrophages subverts tumor immunity toward a type 2 response. J Immunol 2007; 179:977–83. [DOI] [PubMed] [Google Scholar]
- 42.MacKinnon AC, Farnworth SL, Hodkinson PS, Henderson NC, Atkinson KM, Leffler H, et al. Regulation of alternative macrophage activation by galectin-3. J Immunol 2008; 180:2650–8. [DOI] [PubMed] [Google Scholar]
- 43.Novak R, Dabelic S, Dumic J. Galectin-1 and galectin-3 expression profiles in classically and alternatively activated human macrophages. Biochim Biophys Acta 2012; 1820:1383–90. [DOI] [PubMed] [Google Scholar]
- 44.Han L, Zhou R, Niu J, McNutt MA, Wang P, Tong T. SIRT1 is regulated by a PPARgamma-SIRT1 negative feedback loop associated with senescence. Nucleic Acids Res 2010; 38:7458–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vachharajani VT, Liu T, Wang X, Hoth JJ, Yoza BK, McCall CE. Sirtuins link inflammation and metabolism. J Immunol Res 2016; 2016:8167273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sosulski ML, Gongora R, Feghali-Bostwick C, Lasky JA, Sanchez CG. Sirtuin 3 deregulation promotes pulmonary fibrosis. J Gerontol A Biol Sci Med Sci 2017; 72:595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang XZ, Wen D, Zhang M, Xie Q, Ma L, Guan Y, et al. Sirt1 activation ameliorates renal fibrosis by inhibiting the TGF-beta/Smad3 pathway. J Cell Biochem 2014; 115:996–1005. [DOI] [PubMed] [Google Scholar]
- 48.Dobaczewski M, Chen W, Frangogiannis NG. Transforming growth factor (TGF)-beta signaling in cardiac remodeling. J Mol Cell Cardiol 2011; 51:600–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ahamed J, Laurence J. Role of platelet-derived transforming growth factor-beta1 and reactive oxygen species in radiation-induced organ fibrosis. Antioxid Redox Signal 2017; 27:977–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bonniaud P, Margetts PJ, Ask K, Flanders K, Gauldie J, Kolb M. TGF-beta and Smad3 Signaling Link Inflammation to Chronic Fibrogenesis. J Immunol 2005; 175:5390–5. [DOI] [PubMed] [Google Scholar]
- 51.Meng XM, Nikolic-Paterson DJ, Lan HY. TGF-beta: the master regulator of fibrosis. Nat Rev Nephrol 2016; 12:325–38. [DOI] [PubMed] [Google Scholar]
- 52.de la Garza RG, Morales-Garza LA, Martin-Estal I, Castilla-Cortazar I. Insulin-like growth factor-1 deficiency and cirrhosis establishment. J Clin Med Res 2017; 9:233–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tonkin J, Temmerman L, Sampson RD, Gallego-Colon E, Barberi L, Bilbao D, et al. Monocyte/macrophage-derived IGF-1 orchestrates murine skeletal muscle regeneration and modulates autocrine polarization. Mol Ther 2015; 23:1189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liaskou E, Zimmermann HW, Li K-K, Oo YH, Suresh S, Stamataki Z, et al. Monocyte subsets in human liver disease show distinct phenotypic and functional characteristics. Hepatology 2013; 57:385–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dichtel LE, Corey KE, Misdraji J, Bredella MA, Schorr M, Osganian SA, et al. The association between IGF-1 levels and the histologic severity of nonalcoholic fatty liver disease. Clin Transl Gastroenterol 2017; 8:e217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chetty A, Cao GJ, Nielsen HC. Insulin-like growth factor-I signaling mechanisms, type I collagen and alpha smooth muscle actin in human fetal lung fibroblasts. Pediatr Res 2006; 60:389–94. [DOI] [PubMed] [Google Scholar]
- 57.Collins EP, Mitxitorena I, Carmody JR. The ubiquitination of NF-kappaB subunits in the control of transcription. Cells 2016; 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hart SN, Therneau TM, Zhang Y, Poland GA, Kocher J-P. Calculating sample size estimates for RNA sequencing data. J Comput Biol 2013; 20:970–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kraakman MJ, Murphy AJ, Jandeleit-Dahm K, Kammoun HL. Macrophage polarization in obesity and type 2 diabetes: weighing down our understanding of macrophage function? Front Immunol 2014; 5:470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lumeng CN, Bodzin JL, Saltiel AR. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J Clin Invest 2007; 117:175–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Peripheral immune cell recovery after total body irradiation “log scale”.
Table S1A. Classical monocyte comparisons, full list.
Table S1B. Classical monocyte comparisons, abbreviated list.
Table S2A. Intermediate monocyte comparisons, full list.
Table S2B. Intermediate monocyte comparisons, abbreviated list.