Abstract
To define transcriptional dependencies of triple-negative breast cancer (TNBC), we identified transcription factors highly and specifically expressed in primary TNBCs and tested their requirement for cell growth in a panel of breast cancer cell lines. We found that EN1 (Engrailed 1) is overexpressed in TNBCs and its downregulation preferentially and significantly reduced viability and tumorigenicity in TNBC cell lines. By integrating gene expression changes after EN1 downregulation with EN1 chromatin binding patterns, we identified genes involved in WNT and Hedgehog signaling, neurogenesis, and axonal guidance as direct EN1 transcriptional targets. Quantitative proteomic analyses of EN1-bound chromatin complexes revealed association with transcriptional repressors and co-activators including TLE3, TRIM24, TRIM28, and TRIM33. High expression of EN1 correlated with short overall survival and increased risk of developing brain metastases in TNBC patients. Thus, EN1 is a prognostic marker and a potential therapeutic target in TNBC.
INTRODUCTION
Breast cancer is a heterogeneous group of diseases with different biological and clinical characteristics. Based on the presence of estrogen and progesterone receptors (ER and PR), and HER2, tumors are classified into ER+, HER2+, and ER−PR−HER2− (triple-negative) subtypes, while gene expression and epigenetic profiles divide breast tumors into luminal and basal groups (1). Knowledge of the molecular properties of luminal ER+ and HER2+ subtypes has led to the development of endocrine and HER2-targeted therapies. However, currently there is no effective targeted therapy for triple-negative breast tumors.
TNBC constitute 10–20% of breast cancer cases in the US and more commonly affects younger and African-American women (2,3). TNBCs have higher risk of developing distant metastases and in general have poor clinical outcome. TNBCs are also heterogeneous and have been grouped into luminal, basal, and mesenchymal subtypes based on gene expression patterns (2–4). However, these subtypes, besides luminal androgen receptor (AR) positive tumors, have not impacted the clinical management of TNBC patients (3) highlighting the need for additional molecular markers to guide treatment decisions. TNBC genome sequencing studies have so far failed to identify novel recurrent mutations besides TP53, PIK3CA, and PTEN (2,3), suggesting that TNBC phenotypes may be driven by non-genetic alterations such as perturbed epigenetic and transcriptional programs.
The luminal phenotype is defined by a set of lineage-specific transcription factors including ESR1, FOXA1, GATA3, and SPDEF that also represent transcriptional dependencies in luminal breast tumors (2,3). We hypothesized that transcription factors (TFs) specifically expressed in TNBCs may also exemplify such dependencies that can be exploited therapeutically and may divide TNBC into clinically relevant subsets. To test this hypothesis, first we selected TFs that are highly and specifically expressed in primary TNBCs followed by a targeted cellular viability screen for these TFs in a panel of breast cancer cell lines of different subtypes. Using this approach, we have identified several TFs specifically required for the survival of TNBCs and among these further characterized EN1, a TF with known roles in brain (5,6) and dermomyotome (7) development.
MATERIALS AND METHODS
Cell lines
Breast and colon cancer cell lines were obtained from ATCC or generously provided by Steve Ethier (SUM149 and SUM159 cell lines, University of Michigan). Cells were cultured in media recommended by the provider, except SUM149 and SUM159 cells were cultured in DMEM/F12 supplemented with 5% FBS, 10 mM HEPES pH7.4, 1 μg/ml hydrocortisone, 5 μg/ml insulin, 50 U/mL penicillin and 50 μg/ml streptomycin. MCF7 cells were cultured in 4.5mg/l glucose DMEM supplemented with 10% FBS, 10 μg/ml insulin, 50 U/mL penicillin and 50 μg/ml streptomycin. Cells were cultured at 37°C with 5% CO. The identities of the cell lines were confirmed by STR analysis; and they were regularly tested for mycoplasma.
Generation of cell line derivates
SUM149 and SUM159 cells expressing TET-inducible shRNAs targeting EN1, CTNNB1, or NLGN4X in pLKO lentiviral vector were generated by selecting with 5 ug/ml puromycin for five days after lentiviral infection. Entry cDNA ORF for EN1 in pENTR221 was obtained from human ORFeome collection v5.1. pCDNA3-CTNNB1 and pCDNA3-CTNNB1S33Y were obtained from Addgene. Lentiviral expression constructs were generated by Gateway swap into pLenti6.3/V5-Dest vector (Life Technologies) and sequence verified. MCF7-lacZ and MCF7-EN1 cells were selected with 5 ug/ml blasticidin.
Colony growth assays
500–1,500 cells expressing TET-inducible EN1- or CTNNB1-targeting shRNAs were plated into each well of a 6-well plate. Next day, regular media (no doxycycline control) or media containing 500 ng/ml doxycycline were added to induce shRNA mediated downregulation. The media was changed every 2 days and colonies stained with 0.5% crystal violet solution after 10–15 days.
Xenograft assays
Animal experiments were approved by DFCI IACUC under protocol #11–023. SUM149 or SUM159 cells (1 × 106) expressing TET-inducible Luciferase or EN1-targeting shRNAs were resuspended in 50% Matrigel (BD Biosciences) and injected orthotopically into the mammary fat pads of 6-week old female NOG mice (Taconic). When the tumors became palpable, shRNAs were induced in the treatment group by administering a doxycycline diet (625ppm). Animals were euthanized and tumors were harvested when tumors in the control group reached ~ 1.5 cm size. For histological analyses, 5μm sections of formalin-fixed paraffin-embedded (FFPE) tissue slides were stained with hematoxylin and eosin using standard protocols.
Cell cycle analyses
Thirty-six hours after induction of EN1 shRNA with doxycycline, these cells and the control cells were treated with nocodazole (200ng/ml) for 12h. Plates were tapped multiple times to detach cells arrested in G2/M. After washing twice with PBS, cells were plated in fresh medium into collagen-coated plates. Cells were collected at 0, 3, 9, 12, and 24-hour time points for FACS and immunoblot analyses. For cell cycle analysis, cells were harvested, washed in PBS, and fixed in ice-cold 70% ethanol at −20°C overnight. Fixed cells were re-suspended in a solution containing 100μg/ml RNase and incubated for 30 min. at 37°C with agitation. The cells were then resuspended in a solution containing 40μg/ml propidium iodide (Sigma), and the analysis was performed on a FACS AriaII cytometer (BD Biosciences).
METABRIC expression and survival analysis
Expression analysis was performed on METABRIC dataset. Using the mRNA EN1 expression distribution we separated EN1 expression into four clusters using a univariate Gaussian mixture model-based clustering (mclust version 5.4.2 package for R) (8–10). Using the EN1-defined clusters, Kaplan Meier survival curves were plotted and a log rank p value was computed using the function km.coxph.plot in the R package survcomp. Next, the survival in the C4 cluster was compared with the other clusters. A univariable Cox proportional hazards analysis was performed using the coxph function in R to assess the association of EN1 mRNA expression with overall survival. A multivariable Cox proportional hazard analysis was performed using the coxph function in R, and the age, the Nottingham Prognostic Index, and continuous EN1 expression were considered. The EN1 expression z-scores of 243 triple-negative tumors from the METABRIC cohort were computed and the mean levels in the primary tumor for patients that had a brain metastasis (n=17) and patients that did not (n=226) were compared using a two-sided t-test. Differential gene expression was performed using two-class unpaired significance of microarray analysis (SAM 2.0 package, R 3.2.2) in basal carcinomas in the C1, C2 and C3 cluster and the C4 cluster. Next, we performed a pre-ranked GSEA (Gene-Set Enrichment Analysis) using software provided by the Broad Institute (http://www.broadinstitute.org/gsea/msigdb/annotate.jsp) on a ranked gene list ranked (after exclusion of EN1 gene) based on the d-statistic computed from the differential expression analysis, and we assessed enrichment using the Broad Institute’s Molecular Signatures Database (mSigDB) Hallmark gene sets collection (n = 50).
Statistical Analysis
Unpaired two-tailed student’s t tests were used except otherwise stated. P<0.05 was defined as statistically significant.
ACCESSION CODES
Gene Expression Omnibus: RNA-Seq and ChIP-seq datasets have been deposited to GEO with accession number GSE120957.
RESULTS
TNBC-specific transcription factors
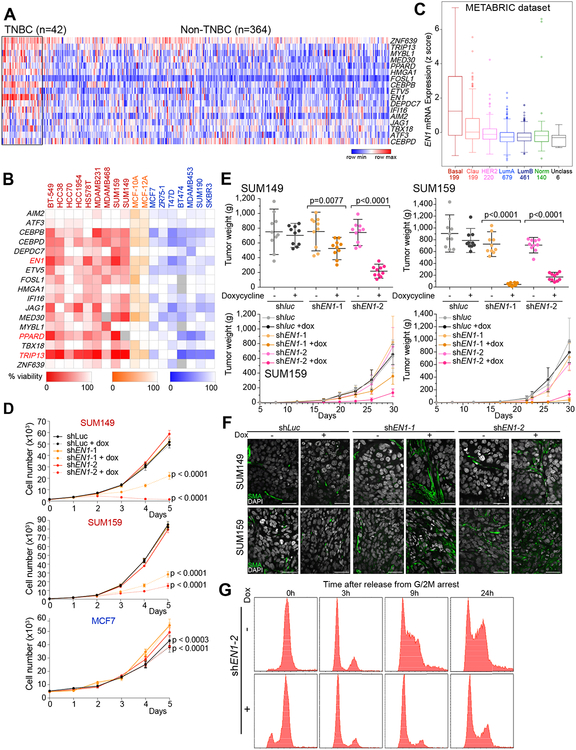
We previously analyzed the inheritance pattern of basal/mesenchymal and luminal phenotypes by generating and characterizing somatic cell fusions between luminal and basal/mesenchymal breast cancer cell lines (11). We determined that the basal/mesenchymal phenotype is dominant, and it is defined by epigenetic factors. We also identified 17 transcription factors (TFs) the expression of which strongly correlated with the inheritance of basal/mesenchymal phenotype and was also significantly higher in TNBCs compared to other subtypes in the TCGA dataset (12) (Fig. 1A) and in basal compared to other breast tumors in the GOBO dataset (13) (Supplementary Fig. S1A). To determine if these TFs are specifically required for the survival or proliferation of TNBC cells, we performed a targeted siRNA screen of all 17 TFs in 18 breast cancer cell lines of different subtypes. Downregulation of multiple factors (CEBPB, EN1, JAG1, MED30, PPARD, and TRIP13) led to a more than 50% decrease in viable cell numbers in multiple basal/mesenchymal cell lines with only modest reduction of viability in a few luminal lines (Fig. 1B and Supplementary Fig. S1B). This subtype-specificity of difference in cellular viability was statistically significant (p<0.05) for EN1, PPARD, and TRIP13. EN1 displayed the most TNBC-specific expression pattern in primary tumors and the most significant and specific effect on viability in basal/mesenchymal vs. luminal breast cancer cell lines; thus, we investigated it in further detail.
Figure 1. Transcription factors in non-luminal breast cancers.
A, Expression of selected transcription factors in TNBC and non-TNBC breast tumors in the TCGA dataset. Colors reflect z-scores normalized to depict relative values for each factor. B, Heatmap depicting cell viability siRNA screen for 17 transcription factors in 18 breast cancer cell lines; 9 non-luminal (red), 2 immortalized mammary epithelial cell lines (orange), and 7 luminal (blue). Shading indicates percentage (0–100%) of viable cells after siRNA-mediated downregulation of the selected factors compared to non-targeting siRNA control. C, Box plot depicting the expression of EN1 in breast tumor subtypes (basal, claudin-low, HER2, luminal A, luminal B, normal-like, and unclassified) in the METABRIC dataset. Numbers indicate number of tumors in each subtype. D, Proliferation of SUM149, SUM159, and MCF7 cell lines following downregulation of EN1 by TET-inducible shRNAs. shluc was used as control. E, Plots depicting tumor volume and weights at 30 days after injection of TET-inducible shluc- or shEN1-expressing SUM149 and SUM159 cells, respectively, in the presence (+Dox) and absence (-Dox) of doxycycline. F, Immunofluorescence staining of smooth muscle actin (SMA) expression in SUM149 and SUM159 xenografts expressing TET-inducible shluc or shEN1 grown in presence (+) or absence (–) of doxycycline. Scale bars correspond to 50 μm. G, Cell cycle profile of SUM159 cells expressing TET-inducible shEN1 grown in presence (+) or absence (–) of doxycycline, synchronized in G2/M and analyzed at different time points (0h, 3h, 9h, 24h) after release.
High expression and essential role of EN1 in TNBC
We further explored the expression of EN1 in primary breast tumors by analyzing the METABRIC (14), TCGA, and GOBO datasets, and confirmed its specific and high expression in basal breast tumors compared to other subtypes (Fig. 1C and Supplementary Fig. S1C). Similarly, EN1 was highly expressed in non-luminal compared to luminal breast cancer cell lines (Supplementary Fig. S1D) and there was a highly significant correlation (R2=0.85, p<0.0001) between EN1 mRNA levels and the degree of loss of cellular viability following its downregulation by siRNAs (Supplementary Fig. S1E). We could detect neither endogenous EN1 protein levels nor the effect of shEN1 on EN1 protein due to our inability to reliably detect EN1 with any of the 13 commercially available EN1 antibodies tested (see Supplementary Methods).
We validated the results of siRNA-mediated downregulation of EN1 by TET-inducible EN1-targeting shRNAs (Supplementary Fig. S1F). shRNA mediated downregulation of EN1 in SUM149 basal and SUM159 mesenchymal TNBC lines, but not in non-TNBC MCF7 luminal cells showed significant reduction in cellular viability (Fig. 1D). Overexpression of a constitutively expressed EN1 in SUM159 cells was able to partially rescue this growth inhibition in a dose-dependent manner confirming the specificity of shEN1 (Supplementary Fig. S1G). Unfortunately, we could not identify an shRNA targeting the non-coding region of EN1, thus, the overexpressed EN1 was still sensitive to the shRNA leading to partial rescue likely due to this. Downregulation of EN1 also led to significant reduction of colony forming ability of SUM149 and SUM159 cells, but not in the MDA-MB-231 TNBC cell line that has no detectable levels of EN1 mRNA further supporting that the decrease in cell viability after shEN1 expression is due to downregulation of EN1 (Supplementary Fig. S1D and S2A).
To determine if EN1 is essential for tumor growth in vivo, we performed xenograft studies using SUM149 and SUM159 cells expressing two independent TET-inducible EN1-targeting shRNAs. Xenografts were allowed to grow to palpable size before inducing shRNA expression (Supplementary Fig. S2B). Downregulation of EN1 significantly reduced tumor weight, volume, and cellularity in xenografts derived from both SUM149 and SUM159 cell lines (Fig. 1E and Supplementary Fig. S2C). Immunofluorescence analysis of smooth muscle actin (SMA), a marker of stromal myofibroblasts and myoepithelial cells, demonstrated a significant increase in SMA+ cells within tumors following EN1 downregulation (Fig. 1F). To determine whether this is due to increased recruitment of mouse stroma, we performed immuno-FISH using mouse and human-specific DNA probes combined with SMA immunofluorescence and confirmed that SMA+ cells are mouse myofibroblasts (Supplementary Fig. S2D). In line with this, immunohistochemical analysis of p63, a basal cell-specific transcription factor, showed no significant change in expression following EN1 downregulation (Supplementary Fig. S2E).
To investigate mechanisms underlying EN1-loss-induced decrease in cellular viability, we analyzed the cell cycle profile of synchronized SUM159 cells following downregulation of EN1. SUM159 cells expressing TET-inducible EN1-targeting shRNAs were synchronized in G2/M with nocodazole 36 hours after induction of shEN1. Upon release from G2/M blockade by washing off nocodazole and re-plating the cells in fresh medium, cells with downregulation of EN1 arrested in G1 whereas the majority of control cells progressed to S and G2/M phase by 9 and 24 hrs, respectively (Fig. 1G). The relatively low levels of phospho-histone H3 (phosphorylated in metaphase of mitosis) in control cells at 24 hours (Supplementary Fig. S2F) despite the high fraction of cells with 4n DNA content based on FACS could be due to the cells being more in G2/early M phase than in later phases of mitosis when histone H3 is phosphorylated. The downregulation of EN1 in synchronized cells led to a notable reduction of CCND1 and c-MYC protein levels that was the most significant at later (9 and 24 hour) time points after release when control cells entered S/G2 phase of the cell cycle (Supplementary Fig. S2F). This decrease in cyclin D1 and c-MYC could directly play a role in the EN1-loss-mediated growth suppression. CCND1 and c-MYC are targets of the WNT signaling pathway and EN1 is a known WNT target during brain development (15). Thus, the decrease in CCND1 and c-MYC levels following EN1 loss in SUM159 cells could potentially indicate downregulation of WNT signaling. At later time points following EN1 downregulation, we observed an increase in apoptosis based on an increase in sub-G1 population by FACS (Supplementary Fig. S2G,H) and increase in cleaved caspase-3 levels (Supplementary Fig. S2I). These results correlate with the anti-apoptotic function of EN1 in dopaminergic neurons and suggest similar roles for EN1 in TNBC (16).
Transcriptional and genomic targets of EN1 in TNBCs
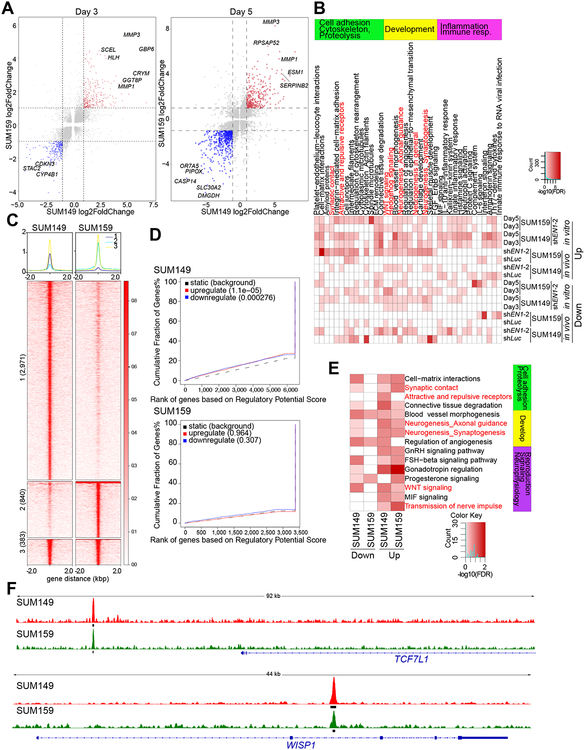
Next, we examined changes in gene expression profiles of SUM149 and SUM159 cells expressing TET-inducible shEN1 at 3 days and 5 days after doxycycline treatment. A large fraction of genes displayed similar expression changes in both cell lines analyzed (Fig. 2A and Supplementary Fig. S3A,B, and Supplementary Table S1). We also performed RNA-seq on SUM149 and SUM159 xenografts from control and doxycycline-treated mice. The number of differentially expressed genes was significantly higher in cell culture at relatively early time points after EN1 downregulation compared to the xenografts that were collected 30 days after injection into mice. Furthermore, residual tumors observed in doxycycline-treated mice apparently escaped the shRNA effect based on the lack of significant differences in EN1 mRNA levels between control and doxycycline conditions (Supplementary Table S1). Functional analysis of genes differentially expressed after EN1 knock down (KD) in vitro or in vivo using Metacore (17) revealed enrichment in development, inflammation, and cell adhesion/migration-related pathways including WNT and Hh signaling, and neurogenesis (Fig. 2B). We analyzed the expression of genes involved in WNT signaling in further detail and found that the majority showed decreased levels after shEN1 expression in both cell lines and time points highlighting the impact of EN1 expression on WNT pathway activation (Supplementary Fig. S3C). We validated the expression WISP1 and WISP2, direct transcriptional targets of β-catenin (3), by immunoblot analysis of cell lysates from cell cultures and xenografts and confirmed their decline after shEN1 expression (Supplementary Fig. S3D,E).
Figure 2. Transcriptional and genomic targets of EN1 in TNBC cells.
A, Gene expression changes in SUM149 (x-axis) and SUM159 (y-axis) cells expressing TET-inducible shEN1 grown in presence of doxycycline for 3 or 5 days. Red and blue indicates significantly up and downregulated genes, respectively, in both cell lines. B, Top process networks significantly enriched in genes differentially expressed following EN1 downregulation in SUM149 and SUM159 cell lines grown in cell culture (in vitro) or as xenografts (in vivo). C, Plot of ChIP-Seq signal of the EN1 peaks in SUM149 and SUM159 cells. Numbers indicate cell line-unique and overlapping peaks. D, BETA factor function prediction analysis to predict EN1 direct transcriptional function (activating/repressing) in SUM149 and SUM159 cells. Red and blue lines represent up and downregulated genes following EN1 knockdown. Black dashed line represents static genes. Genes are ranked by regulatory potential score, and significance relative to static genes is determined by Kolmogorov-Smirnov test. E, Top process networks significantly enriched in genes that are direct genomic targets of EN1 and are differentially expressed 3 days following EN1 downregulation in SUM149 and SUM159 cell lines. Each ChIP-Seq peak is assigned to the gene whose transcription start site (TSS) is closest to the peak, defining the factors direct genomic targets. F, EN1 ChIP-Seq signal at selected genes in SUM149 (red) and SUM159 (green) cells. Thick black lines below the signal tracks denote peaks.
To identify the direct genomic targets of EN1, we performed ChIP-seq in SUM149 and SUM159 cells for EN1 and histone H3 lysine 27 acetyl (H3K27ac) associated with transcriptionally active regions to define super-enhancers (SEs) (18). The validity of the EN1 ChIP-seq data was confirmed based on (1) the data passing all QC in the ChiLin 2.0 pipeline, (2) ChIP-seq using the antibody against endogenous EN1 and V5-tagged EN-1 showed high correlation in all three cell lines tested (Supplementary Fig. S3F), (3) the number of peaks correlated with endogenous EN1 levels; for example, SUM149 cells have higher endogenous EN1 than SUM159 cells and have more EN1 ChIP-seq peaks than SUM159 cells (3,354 vs.1,223), and (4) the peaks showed enrichment in EN1 sequence motif (Supplementary Fig. S3G). The majority of EN1 peaks were located in non-promoter regions in both cell lines (Supplementary Fig. S3H) and a significant fraction overlapped between the two cell lines with SUM149 cells having more unique peaks consistent with the higher expression level of EN1 in this cell line (Fig. 2C). EN1 and H3K27ac peaks also demonstrated significant overlap in both cell lines (Supplementary Fig. S3I). Analysis of SEs for core regulatory circuits (CRCs) (19) revealed ZNF217, POU3F3, ELF5, and KLF4 as master TFs key for the establishment of the SE landscape in SUM159 cells, while in SUM149 cells it uncovered a larger set of TFs including EN1 that formed an interconnected network centered around MYC based on STRING protein-interaction network analysis (20) (Supplementary Fig. S3I).
Next, we explored whether the up- or downregulated genes were more likely to be direct EN1 targets at different time points after EN1 downregulation. Based on the fraction of direct EN1 targets up- or downregulated (Supplementary Fig. S3K) and on BETA analysis (Fig. 2D and Supplementary Fig. S3L), EN1 appears to both positively and negatively regulate gene expression in both TNBC cell lines with more genes being up than downregulated in SUM149 cells. The top process networks enriched for direct EN1 targets that were up or downregulated after shEN1 expression, were essentially the same as observed for differentially expressed genes with WNT signaling and neurogenesis-related functions showing the most significant enrichment (Fig. 2E). Examples for EN1 targets include TCF7L1 transcription factor and WISP1 secreted growth factor mediating WNT signaling (Fig. 2F and Supplementary Table S2). Thus, similar to brain and dermomyotome, EN1 may modulate WNT signaling and neural-related functions in TNBC (15).
To investigate the EN1-WNT pathway link in more detail, we tested the effects of wild type or a constitutively active S33Y mutant β-catenin overexpression or CTNNB1 downregulation in SUM159 and SUM149 cells. Endogenous β-catenin and phospho-β-catenin levels were higher in SUM149 compared to SUM159 cells, but downregulation of EN1 had no effect on these in either cell line (Supplementary Fig. S4A). Overexpression of CTNNB1 and CTNNB1S33Y in SUM159 cells had no effect on cellular viability and did not rescue the EN1-loss induced growth arrest (Supplementary Fig. S4A–C). Similarly, downregulation of CTNNB1 in SUM159 cells did not affect cellular viability and colony growth (Supplementary Fig. S4D–F). Lastly, we tested the effect of several compounds targeting different steps of the canonical WNT signaling pathway (i.e., porcupine, tankyrase, and WNT inhibitors, and Axin2 activator) on the growth of TNBC (SUM149 and SUM159, high EN1 expression) and luminal ER+ (MCF7 and T47D, no EN1 expression) breast cancer cells, but did not find any significant subtype-specific differences or associations with the degree of growth inhibition and endogenous β-catenin or EN1 levels (Supplementary Fig. S4G,H). Thus, EN1 appears to modulate WNT signaling in TNBC cells via alternative mechanisms not directly involving CTNNB1 itself.
The effect of EN1 expression in luminal breast cancer cells
Our prior data demonstrated that EN1 was one of the few transcription factors that was able to switch MCF7 luminal breast cancer cells to a more basal/mesenchymal phenotype (11). To investigate whether this is due to the reprogramming of the epigenetic landscape by EN1, we expressed EN1 in MCF7 cells and confirmed the switch to a more mesenchymal phenotype based on morphologic changes as well as increased migration and invasion (Fig. 3A), although the cells maintained their estrogen dependence for growth (Supplementary Fig. S5A). Interestingly, EN1 expression did not have the same effect in the T47D luminal breast cancer cell line highlighting the importance of cellular context (Supplementary Fig. S5B). Similar context-dependent observations were also made for several other EMT-inducing transcription factors including ZEB1 and TWIST (21). However, genes differentially expressed after EN1 downregulation in SUM159 and SUM149 cells that are also direct EN1 targets did classify primary breast tumors into basal and non-basal subsets highlighting the role of EN1 and its targets in regulating luminal and basal subtypes (Supplementary Fig. S5C).
Figure 3. EN1-associated chromatin complexes in breast cancer cells.
A, Cell migration and invasion of MCF7 cells expressing lacZ or EN1. P-value of difference indicated (t-test). B, Plot of ChIP-Seq signal of the EN1 peaks in SUM149, SUM159, and MCF7 cells. Numbers indicate cell line-unique and overlapping peaks. C, Top process networks significantly enriched in genes that are direct genomic targets of EN1 and are differentially expressed following EN1 expression in MCF7 cells. D, Word clouds illustrating top EN1-associated proteins in MCF7 and SUM159 cells. E, Top process networks enriched in EN1-interacting proteins in MCF7 and SUM159 cells. F, Heatmap and clustering of TLE3 and EN1 peaks in SUM159 cells. G, Top process networks enriched in differentially expressed genes after EN1 knock down associated with TLE3-EN1 overlapping peaks in either control (-dox) or shEN1 (+dox) conditions. H, Top process networks enriched in β-catenin peaks in SUM149 cells associated with genes differentially expressed after EN1 knock down.
To examine if EN1 binds to and regulates the same set of genes in MCF7 cells as it does in TNBC cell lines, we performed RNA-seq and EN1 ChIP-seq comparing MCF7-lacZ and MCF7-EN1 cells. We found that a subset of EN1 peaks overlapped among the three cell lines, but there were numerous MCF7-specific EN1 peaks as well. (Fig. 3B). Integrating EN1 ChIP-seq and RNA-seq data demonstrated that EN1 targets in MCF7 cells include genes both up and downregulated after EN1 expression (Supplementary Fig. S5D,E and Supplementary Table S3). Functional analysis of differentially expressed genes that are EN1 targets showed higher number of process networks enriched in downregulated genes after EN1 overexpression including many development and neurogenesis-related pathways such as axonal guidance, WNT, and NOTCH signaling (Fig. 3C).
To determine if the luminal-to-mesenchymal switch observed in EN1-expressing MCF7 cells is due to the EN1’s modulation of chromatin binding of luminal TFs such as FOXA1, a lineage determining TF, we performed FOXA1 ChIP-seq on LacZ and EN1-expressing MCF7 cells. Overall there was a substantial overlap between FOXA1 and EN1 peaks, and while we identified some FOXA1 peaks that were only detected in MCF7-lacZ or in MCF7-EN1 cells (Supplementary Fig. S5F,G and Supplementary Table S4), the majority of FOXA1 peaks were the same in the two cells. These data imply that EN1 expression does not significantly affect FOXA1 chromatin binding, although it might still modulate FOXA1 transcriptional activity for a subset of its target genes.
EN1-interacting proteins in breast cancer cells
During brain development EN1 function is modulated by its interaction with co-activators and co-repressors such as PITX3 (22) and TLE3 (23). To identify EN1-associated factors in MCF7 and SUM159 breast cancer cells that exogenously express EN1 with a V5 tag, we performed qPLEX-RIME (quantitative Multiplexed Rapid Immunoprecipitation Mass spectrometry of Endogenous proteins) (24) using the anti-V5 antibody. We identified numerous EN1-associated proteins in both cell lines, which we also confirmed by co-immunoprecipitation (Fig. 3D and Supplementary Fig. S5H). EN1-associated proteins showed significant enrichment in chromatin modification, androgen receptor signaling, and DNA damage functional categories (Fig. 3E). Several of the EN1-interacting proteins common between the two cell lines represent transcriptional co-factors TLE3 and TRIM28 (25), and nuclear hormone receptor co-activators TRIM33 and TRIM24. Interestingly TRIM28 was recently identified as a critical regulator of TRIM24, an oncogene in prostate cancer (26) and TRIM24-TRIM28-TRIM33 complex has previously been identified as a suppressor of murine hepatocellular carcinoma growth (27). Due to prior data implying a functional role for the TLE3 - EN1 interaction during development (23), we further explored the relevance of this association in breast cancer cells.
To confirm that EN1 and TLE3 bind to the same genomic regions and to test whether downregulating EN1 changes TLE3 chromatin binding patterns, we performed TLE3 ChIP-seq in SUM149 and SUM159 cells −/+ induction of shEN1. We detected a significant overlap between TLE3 and EN1 peaks and a subset of the TLE3 peaks showed decreased (11,428) or increased (7,150) signal in shEN1-expressing cells (Fig. 3F, Supplementary Fig. S5I, and Supplementary Table S5). To investigate the functional relevance of TLE3-EN1 interaction, we integrated TLE3 ChIP-seq and differential gene expression after shEN1 induction and performed Metacore analyses. We found that the most significant network process enrichment in both SUM149 and SUM159 cells was in progesterone and GnRH signaling, angiogenesis and ECM remodeling-related pathways by genes associated with TLE3 peaks present only in shEN1 cells and upregulated after shEN1 (Fig. 3G). These data suggest that EN1 may suppress the expression of these genes by preventing TLE3 binding.
To investigate whether the EN1-mediated changes in the expression of genes involved in WNT signaling is due to direct effects on β-catenin transcriptional activity, we also performed β-catenin ChIP-seq in cells with or without EN1 expression and integrated β-catenin chromatin binding patterns with gene expression changes. We observed a significant enrichment in WNT signaling pathway among genes that are direct β-catenin targets are downregulated after shEN1, implying that EN1 loss may affect β-catenin transcriptional activity (Fig. 3H). However, EN1 expression did not significantly impact β-catenin chromatin binding, thus, EN1 may regulate the WNT signaling pathway via other indirect mechanisms.
High EN1 is associated with poor prognosis and brain metastasis in TNBC
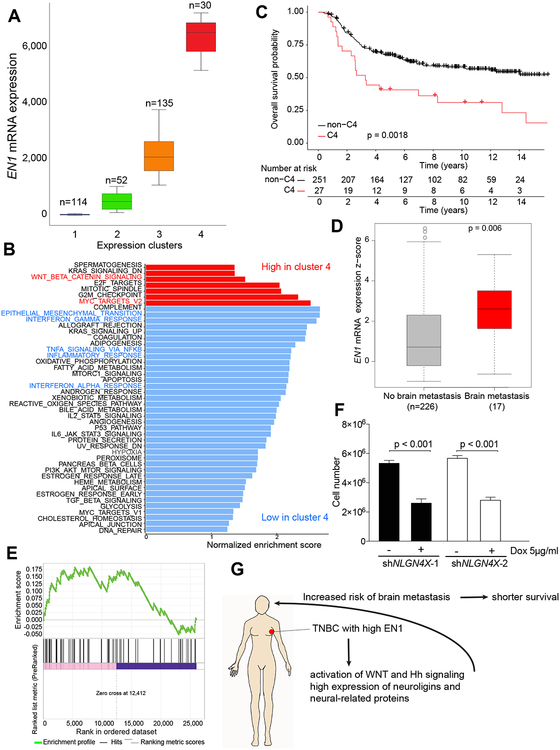
To investigate the clinical relevance of EN1 in breast cancer, we analyzed associations between EN1 expression and clinical outcome using the METABRIC and TCGA datasets. EN1 is highly expressed in TNBCs compared to non-TNBC and also in basal breast cancers compared to other subtypes (Fig. 1A, C). However, even within basal breast cancers there was a significant heterogeneity for EN1 expression and tumors could be divided into four groups based on EN1 expression distribution using finite normal mixture modelling (mclust package for R, version 5.4.2) (8,9) (Fig. 4A and Supplementary Fig. S6A). Tumors with the highest EN1 expression were enriched for WNT signaling and MYC target genes (Fig. 4B) and showed worse overall survival (Fig. 4C). A Cox multiple regression model including tumor grade, size, and numbers of positive lymph nodes demonstrated statistically significant (p=0.0085) association between metastasis-free survival and EN1 expression. Similarly, in the TCGA dataset, basal breast tumors with high EN1 expression had worse overall survival than those with low EN1 levels, but this did not reach statistical significance likely due to the relatively smaller size of this cohort and more limited numbers of basal breast cancers (Supplementary Fig. S6B). EN1 expression significantly associated with overall survival in a Cox simple regression model. A multiple regression model adjusted for the Nottingham Prognosis Index and age also showed significant association between overall survival and EN1 expression.
Figure 4. EN1 expression in breast cancer and clinical outcome.
A, Basal breast cancers in METABRIC dataset divided into four different clusters based on EN1 mRNA expression. The number of tumor samples in each cluster is indicated. B, GSEA of genes differentially expressed between tumors in cluster 4 vs. clusters 1–3 combined. C, Kaplan-Meier survival plot showing overall survival of breast cancer patients with basal breast cancer in the Metabric cohort classified into cluster 4 (C4) and all other clusters (Cluster 1–3) combined based on EN1 expression. p-value indicated the statistical significance of the observed difference. D, The expression of EN1 in primary TNBCs with and without brain metastasis in the Metabric cohort. E, GSEA analysis depicting enrichment of matched primary vs. brain metastatic TNBC samples in genes that go up after EN1 knockdown in SUM149 and SUM159 cells (p-val = 0.01). F, Cellular viability after NLGN4X downregulation in SUM159 cells. G, Schematic summary of our EN1 data.
The short survival of TNBC patients with high EN1 expression suggested that these patients might be more likely to develop brain metastases commonly associated with shorter survival in breast cancer patients. Indeed, TNBC patients who developed brain metastases had primary tumors with significantly higher EN1 levels than those who did not (Fig. 4D). The significant association with brain metastases was still observed when compared to patients with metastases in other organs (28). We also analyzed gene expression data of matched primary tumors and brain metastases and found EN1 transcriptional targets were significantly enriched in genes differentially expressed between primary breast tumors and brain metastasis (Fig. 4E). These results suggest that high expression of EN1 in primary TNBCs may increase the risk of brain metastases potentially due to its regulation of WNT signaling and neurogenesis-related pathways including the expression of several neuroligins in these breast tumors. Neural activity-induced NLGN3 was shown to promote the growth of high-grade brain tumors such as glioblastoma and DIPG (29), thus, their upregulation in primary TNBCs may select for tumors with preferential growth in the brain.
Interestingly, in our prior breast cancer somatic cell fusion studies the only SNP that was significantly associated with the inheritance of SUM159 mesenchymal-cell features was within the NLGN4X gene (11), implying that NLGN4X plays an essential role in these cells. In both SUM149 and SUM159 cell lines the expression of NLGN4X, and other neuroligins, decreased following EN1 knockdown (Supplementary Fig. S6C). To test if NLGN4X expression in SUM159 breast cancer cells may reflect a dependency for this neural growth factor, we downregulated its expression using TET-inducible shRNAs (Supplementary Fig. S6D) and analyzed changes in cell viability and proliferation. We detected a significant decrease in cell number five days following induction of two independent shNLGN4X confirming that NLGN4X positively affects SUM159 cell growth (Fig. 4F). To investigate potential mechanisms by which loss of NLGN4X leads to decreased cell growth, we analyzed gene expression changes after shNLGN4X induction. We found that downregulated genes showed significant enrichment in muscle contraction (e.g., CAPN3, CALR3, VIPR1), while upregulated genes were enriched in cell adhesion (e.g., NRXN1, TNC) and neurogenesis axonal guidance (e.g., SEMA3B and SEMA7A) proteins (Supplementary Fig. S6E and Supplementary Table S6). These results support our hypothesis that neural survival-related pathways play an important role in a subset of TNBCs with high EN1 expression.
DISCUSSION
In a targeted cellular viability screen for transcription factors selected based on their high expression in TNBCs we identified EN1, a neural-specific homeo-domain transcription factor (30) with functional and clinical relevance in TNBCs. During embryonic development EN1 plays essential roles in mid-hindbrain pattern formation, axonal guidance, and neuron specification (5). EN1 is highly expressed in dopaminergic neurons both during development and in adulthood, and it is essential for their differentiation, maintenance, and survival (16). High expression of EN2 was reported in prostrate, breast, and ovarian carcinomas (31) and EN2 has an oncogenic function in breast cancer (32). In agreement with our findings, a recent study also reported that the downregulation of EN1 reduces cellular viability in the SUM149 TNBC cell line (33).
We demonstrated an oncogenic role for EN1 in TNBCs based on the observation that its downregulation leads to G1 arrest and apoptosis, and reduced tumor growth. These findings correlate with the anti-apoptotic function of EN1 in dopaminergic neurons, where EN1 modulates mitochondrial signals and upregulates cell survival pathways (34). Functional enrichment analysis of genes differentially expressed following EN1 downregulation for process networks and pathways identified WNT and Hh signaling as top enriched networks. In the brain, EN1 is a downstream target of WNT, and EN1 and WNT pathway genes cooperate in mid-hindbrain development (15). EN1 was also shown to negatively regulate β-catenin transcriptional activity in a cell culture model (23). The identification of WNT as top enriched process network positively regulated by EN1 in TNBC suggest similar interactions between EN1 and WNT signaling in TNBCs. Indeed, our β-catenin ChIP-seq analysis showed that a subset of genes differentially expressed after EN1 downregulation are direct β-catenin targets.
We defined the genomic targets of EN1 by ChIP-seq for endogenous EN1 in SUM159 and SUM149 TNBC and for exogenously-expressed EN1 in MCF7 luminal breast cancer cells and determined that EN1 can both positively and negative affect gene transcription depending on the cell line and its association with other transcriptional regulators. Direct EN1 targets that show expression changes after EN1 knock down also showed highest enrichment for WNT and Hh signaling, and neural-related functions. Among others, neuroligins and their receptors are direct targets and regulated by EN1 in breast cancer cells. Based on qPLEX-RIME we identified TLE3, TRIM24, TRIM28, and TRIM33 as EN1 interacting proteins in breast cancer cells. ChIP-seq for TLE3 revealed that the TLE3-EN1 complex may have a negative effect on the expression of genes involved in angiogenesis and ECM remodeling, since TLE3 peaks present in shEN1-expressing cells were preferentially associated with genes showing upregulation after EN1 knock down.
Besides brain and neural development EN1 is essential for the specification of dorsal dermal fate via transmitting WNT signaling (7) and it is implicated as a regulator of dermal-derived fibroblasts involved in wound-healing and cancer-associated stroma formation (35). During early embryonic development EN1 is expressed in the dermal precursors and later on its expression is detected in dermal progenitors. Because the mammary gland is derived from the dermis, it is tempting to speculate that there could be rare EN1-expressing mammary progenitors and that a subset of TNBCs may originate from these cells.
We showed that EN1 was specifically and highly expressed in triple-negative and basal breast cancers. However, we also found significant heterogeneity in EN1 expression and patients with the highest expression of EN1 had worse overall survival in both METABRIC and TCGA datasets. Furthermore, TNBC patients with high expression of EN1 were more likely to develop brain metastases potentially due to the expression and dependency of these tumors for neural survival factors such as NLGN4X and other neuroligins. Besides neural cells, neurexins and neuroligins are also expressed in the vasculature and they promote the maturation and formation of blood vessels (36). Thus, the upregulation of neuroligins by EN1 in breast tumors could promote the growth of the primary tumor, enhance their ability to disseminate, and also promote the growth of metastatic lesions, especially in the brain.
In summary, we identified EN1 as a TNBC-specific TF the expression of which is associated with neural features, angiogenesis, and brain metastasis of breast cancer leading to poor clinical outcome (Fig. 4G). Thus, targeting of EN1 and neuroligins might be explored as a potential therapeutic strategy for the prevention and treatment of brain metastases of TNBC.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ramesh Shivdasani and members of our laboratory for their critical reading of the manuscript and for useful discussions. This work was supported by the National Cancer Institute R35CA197623 (K.P.), CDRMP W81XWH-09–1-0131 (K.P.), W81XWH-14–1-0212 (K.P.), W81XWH-09–1-0561 (A.M.), W81XWH-18–1-0027 (B.J.), the Susan G. Komen Foundation (Y.S. and F.B.), and CRUK core funding and an ERC Consolidator award (J.S.C.).
Footnotes
Disclosure of Potential Conflicts of Interest: K.P. is a Scientific Advisory Board member of Mitra Biotech and Acrivon Therapeutics. A.B. is an equity holder and employee of PathAI. J.S.C. is the founder and CSO of Azeria Therapeutics Ltd.
REFERENCES
- 1.Russnes HG, Lingjaerde OC, Borresen-Dale AL, Caldas C. Breast Cancer Molecular Stratification: From Intrinsic Subtypes to Integrative Clusters. Am J Pathol 2017;187:2152–62. [DOI] [PubMed] [Google Scholar]
- 2.Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol 2016;13:674–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garrido-Castro AC, Lin NU, Polyak K. Insights into Molecular Classifications of Triple-Negative Breast Cancer: Improving Patient Selection for Treatment. Cancer Discov 2019;9:176–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehmann BD, Jovanovic B, Chen X, Estrada MV, Johnson KN, Shyr Y, et al. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS One 2016;11:e0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchs J, Stettler O, Alvarez-Fischer D, Prochiantz A, Moya KL, Joshi RL. Engrailed signaling in axon guidance and neuron survival. Eur J Neurosci 2012;35:1837–45. [DOI] [PubMed] [Google Scholar]
- 6.Yang J, Brown A, Ellisor D, Paul E, Hagan N, Zervas M. Dynamic temporal requirement of Wnt1 in midbrain dopamine neuron development. Development 2013;140:1342–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, et al. Beta-catenin activation is necessary and sufficient to specify the dorsal dermal fate in the mouse. Dev Biol 2006;296:164–76. [DOI] [PubMed] [Google Scholar]
- 8.Yeung KY, Fraley C, Murua A, Raftery AE, Ruzzo WL. Model-based clustering and data transformations for gene expression data. Bioinformatics 2001;17:977–87. [DOI] [PubMed] [Google Scholar]
- 9.Baudry JP, Raftery AE, Celeux G, Lo K, Gottardo R. Combining Mixture Components for Clustering. J Comput Graph Stat 2010;9:332–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scrucca L, Fop M, Murphy TB, Raftery AE. mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. R J 2016;8:289–317. [PMC free article] [PubMed] [Google Scholar]
- 11.Su Y, Subedee A, Bloushtain-Qimron N, Savova V, Krzystanek M, Li L, et al. Somatic Cell Fusions Reveal Extensive Heterogeneity in Basal-like Breast Cancer. Cell Rep 2015;11:1549–63. [DOI] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ringner M, Fredlund E, Hakkinen J, Borg A, Staaf J. GOBO: gene expression-based outcome for breast cancer online. PLoS One 2011;6:e17911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alves dos Santos MT, Smidt MP. En1 and Wnt signaling in midbrain dopaminergic neuronal development. Neural Dev 2011;6:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alvarez-Fischer D, Fuchs J, Castagner F, Stettler O, Massiani-Beaudoin O, Moya KL, et al. Engrailed protects mouse midbrain dopaminergic neurons against mitochondrial complex I insults. Nat Neurosci 2011;14:1260–6. [DOI] [PubMed] [Google Scholar]
- 17.Ekins S, Nikolsky Y, Bugrim A, Kirillov E, Nikolskaya T. Pathway mapping tools for analysis of high content data. Methods Mol Biol 2007;356:319–50. [DOI] [PubMed] [Google Scholar]
- 18.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell 2013;155:934–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saint-Andre V, Federation AJ, Lin CY, Abraham BJ, Reddy J, Lee TI, et al. Models of human core transcriptional regulatory circuitries. Genome Res 2016;26:385–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snel B, Lehmann G, Bork P, Huynen MA. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res 2000;28:3442–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stemmler MP, Eccles RL, Brabletz S, Brabletz T. Non-redundant functions of EMT transcription factors. Nat Cell Biol 2019;21:102–12. [DOI] [PubMed] [Google Scholar]
- 22.Veenvliet JV, Dos Santos MT, Kouwenhoven WM, von Oerthel L, Lim JL, van der Linden AJ, et al. Specification of dopaminergic subsets involves interplay of En1 and Pitx3. Development 2013;140:3373–84. [DOI] [PubMed] [Google Scholar]
- 23.Bachar-Dahan L, Goltzmann J, Yaniv A, Gazit A. Engrailed-1 negatively regulates beta-catenin transcriptional activity by destabilizing beta-catenin via a glycogen synthase kinase-3beta-independent pathway. Mol Biol Cell 2006;17:2572–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papachristou EK, Kishore K, Holding AN, Harvey K, Roumeliotis TI, Chilamakuri CSR, et al. A quantitative mass spectrometry-based approach to monitor the dynamics of endogenous chromatin-associated protein complexes. Nat Commun 2018;9:2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agarwal M, Kumar P, Mathew SJ. The Groucho/Transducin-like enhancer of split protein family in animal development. IUBMB Life 2015;67:472–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fong KW, Zhao JC, Song B, Zheng B, Yu J. TRIM28 protects TRIM24 from SPOP-mediated degradation and promotes prostate cancer progression. Nat Commun 2018;9:5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herquel B, Ouararhni K, Khetchoumian K, Ignat M, Teletin M, Mark M, et al. Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to form regulatory complexes that suppress murine hepatocellular carcinoma. Proc Natl Acad Sci U S A 2011;108:8212–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rueda OM, Sammut SJ, Seoane JA, Chin SF, Caswell-Jin JL, Callari M, et al. Dynamics of breast-cancer relapse reveal late-recurring ER-positive genomic subgroups. Nature 2019;567:399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venkatesh HS, Johung TB, Caretti V, Noll A, Tang Y, Nagaraja S, et al. Neuronal Activity Promotes Glioma Growth through Neuroligin-3 Secretion. Cell 2015;161:803–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danielian PS, McMahon AP. Engrailed-1 as a target of the Wnt-1 signalling pathway in vertebrate midbrain development. Nature 1996;383:332–4. [DOI] [PubMed] [Google Scholar]
- 31.McGrath SE, Michael A, Morgan R, Pandha H. EN2: a novel prostate cancer biomarker. Biomark Med 2013;7:893–901. [DOI] [PubMed] [Google Scholar]
- 32.Martin TA, Goyal A, Watkins G, Jiang WG. Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann Surg Oncol 2005;12:488–96. [DOI] [PubMed] [Google Scholar]
- 33.Beltran AS, Graves LM, Blancafort P. Novel role of Engrailed 1 as a prosurvival transcription factor in basal-like breast cancer and engineering of interference peptides block its oncogenic function. Oncogene 2014;33:4767–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alberi L, Sgado P, Simon HH. Engrailed genes are cell-autonomously required to prevent apoptosis in mesencephalic dopaminergic neurons. Development 2004;131:3229–36. [DOI] [PubMed] [Google Scholar]
- 35.Rinkevich Y, Walmsley GG, Hu MS, Maan ZN, Newman AM, Drukker M, et al. Skin fibrosis. Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science 2015;348:aaa2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arese M, Serini G, Bussolino F. Nervous vascular parallels: axon guidance and beyond. Int J Dev Biol 2011;55:439–45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.