Abstract
Antibody-based drugs are a leading class of biologics used to treat a variety of diseases, including cancer. However, wide antibody implementation is hindered by manufacturing challenges and high production cost. Use of in-vitro-transcribed mRNA (IVT-mRNA) for endogenous protein expression has the potential to circumvent many of the shortcomings of antibody production and therapeutic application. Here, we describe the development of an IVT-mRNA system for in vivo delivery of a humanized anti-HER2 (also known as ERBB2) antibody, trastuzumab, and demonstrate its anticancer activity. We engineered the IVT-mRNA sequence to maximize expression, then formulated the IVT-mRNA into lipid-based nanoparticles (LNPs) to protect the mRNA from degradation and enable efficient in vivo delivery. Systemic delivery of the optimized IVT-mRNA loaded into LNPs resulted in antibody serum concentrations of 45 ± 8.6 μg/mL for 14 days after LNP injection. Further studies demonstrated an improved pharmacokinetic profile of the produced protein compared to injection of trastuzumab protein. Finally, treatment of tumor-bearing mice with trastuzumab IVT-mRNA LNPs selectively reduced the volume of HER2-positive tumors and improved animal survival. Taken together, the results of our study demonstrate that using IVT-mRNA LNPs to express full-size therapeutic antibodies in the liver can provide an effective strategy for cancer treatment and offers an alternative to protein administration.
Keywords: mRNA therapeutics, mRNA design, mRNA delivery, lipid nanoparticles, breast cancer, antibody-based drugs, Herceptin, trastuzumab
mRNA-induced host protein expression has the potential to overcome challenges associated with the application of antibody therapeutics and to improve treatment efficacy without hampering safety. This study demonstrates that multiple injections of mRNA encoding the anticancer antibody trastuzumab can safely and efficiently inhibit tumor growth, even outperforming trastuzumab protein therapy.
Introduction
Antibody-based drugs are a leading class of biologics for treatment of cancer, chronic inflammation, and autoimmune diseases.1 However, the process of large-scale antibody production has many challenges: purification protocols are complex and often have to be tailored for each individual antibody; high frequency of aberrant post-translational modifications necessitate extensive and costly quality control; mammalian antibody-producing cell lines are prone to mutations and expensive to maintain.2, 3, 4, 5, 6 Furthermore, antibodies themselves are susceptible to rapid clearance and degradation by serum enzymes. In some cases, this rapid elimination requires frequent drug dosing, which additionally increases treatment cost and can lead to unwanted side effects, including immune system activation.7
As an alternative approach, antibodies can be produced in vivo by delivery of in-vitro-transcribed mRNA (IVT-mRNA). IVT-mRNA has demonstrated the potential to generate high levels of circulating protein in vivo as compared to DNA plasmids and viral vectors.8, 9 IVT-mRNA production protocol is uniform for several antibodies and does not require mammalian cell culture. Using IVT-mRNA, antibody production is transferred inside the body, allowing better control over post-translational modifications. Moreover, the process of antibody structure optimization can be simplified by using IVT-mRNA to express protein therapeutics, rather than delivering protein directly.10 In addition, IVT-mRNA delivery allows therapeutic antibody expression within the host cell and can therefore aid in directing therapeutics toward intracellular targets. Although RNA is very unstable, the introduction of base modifications such as 5-methoxyuridine (5 moU) or N1-methyl-pseudouridine (N1Ψ) has recently been reported to improve IVT-mRNA stability and reduce the associated immunogenicity.11, 12, 13, 14 Finally, because there is no need for IVT-mRNA to enter the cell nucleus to be functional, the risk of insertional mutagenesis is minimized, and transfection efficacy is improved in non-dividing cells.
Currently, lipid-based nanoparticles (LNPs) are the most advanced non-viral platform for IVT-mRNA delivery to the liver. LNPs composed of ionizable lipids and lipid-like materials have demonstrated efficacy for delivery of therapeutic mRNA in both rodents and non-human primates.15, 16, 17, 18 By formulating mRNA into LNPs, IVT-mRNA is protected from degradation, and its transfer across cell membranes is facilitated. Besides, LNPs enable selective mRNA to the liver and facilitate the escape of IVT-mRNA from endosomal compartments into the cell cytoplasm.19, 20, 21, 22, 23, 24 Nanoparticle-formulated IVT-mRNA is therefore well suited to overcoming many difficulties associated with the production and administration of antibody-based drugs.
Here, we describe the development of an LNP-based IVT-mRNA system for in vivo delivery of an anti-human epidermal growth factor receptor 2 (HER2, also known as ERBB2) antibody, trastuzumab, and demonstrate functionality of the produced protein. Trastuzumab is an antibody-based cancer drug targeting HER2,25, 26 which is overexpressed in approximately 30% of breast cancer patients.27 The binding of trastuzumab to HER2 prevents both receptor cleavage and dimerization and results in abrogation of downstream proliferative and pro-survival pathways (reviewed by Hudis28). In addition, trastuzumab-induced, antibody-dependent, cell-mediated cytotoxicity has been shown to be very important in the trastuzumab antitumor effect (F. Hotaling et al., 1996, Proc. Am. Assoc. Cancer Res., abstract; S. Pegram et al., 1997, Proc. Am. Assoc. Cancer Res., abstract). Knockout of Fc receptors in mice significantly diminished the ability of trastuzumab to arrest tumor growth in vivo.29
Based on a publicly available trastuzumab amino acid sequence, we designed an IVT-mRNA sequence and codon optimized it for expression in mammalian cells. We used liver-targeting LNPs to achieve IVT-mRNA in vivo delivery and production of trastuzumab. We showed that optimization of signal peptide and antibody heavy-to-light-chain ratio results in an average serum trastuzumab concentration of ∼40 μg/mL 24 h after injection. Furthermore, we performed pharmacodynamic and pharmacokinetic studies and demonstrated the anticancer efficacy of the expressed antibody, using in vitro assays and a mouse xenograft model. We showed that using IVT-mRNA LNPs to express full-size therapeutic antibody trastuzumab in the liver is an effective strategy for cancer treatment. This approach may offer an alternative to the administration of a wide range of antibody therapeutics in cancer treatment.
Results
Design and Optimization of Trastuzumab mRNA for LNP Delivery
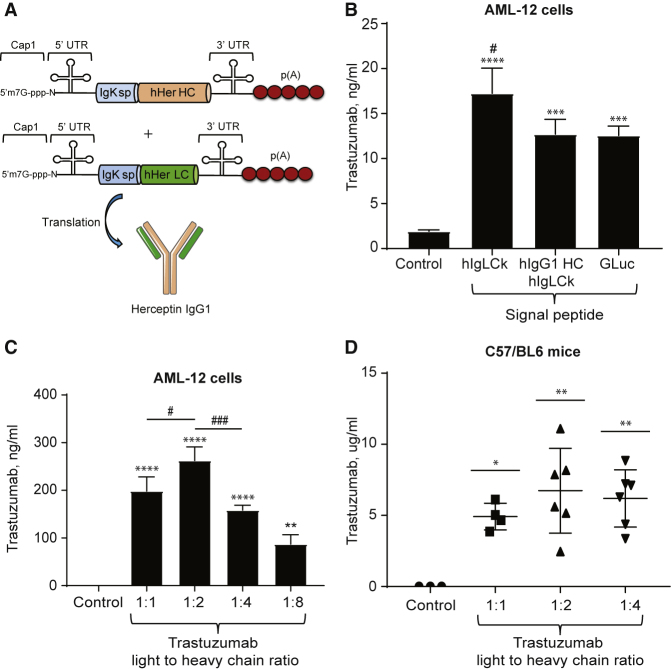
An mRNA molecule consists of a cap, 5′ and 3′ UTRs, a protein-coding sequence, and a polyA tail (Figure 1A). It has been shown that 2′-O-methylation of naturally occurring Cap 0 (Cap1) significantly enhances mRNA translation.30 Cap 1 was therefore used for all IVT-mRNA synthesized in the study. We used a 5′ UTR consisting of a partial sequence of the cytomegalovirus (CMV) immediate early 1 (IE1) gene and 3′ UTR consisting of a partial sequence of the human growth hormone (hGH) gene. These two regions have been shown to induce efficient protein expression in different tissues in vivo.24, 31 A 3′ polyA tail was estimated to be approximately 120 nt long. From this basic sequence we continued RNA construct optimization by varying the signal peptide sequence and the heavy-to-light-chain ratio to improve trastuzumab expression.
Figure 1.
Optimization of the RNA Construct
(A) Schematic of mRNAs encoding the heavy (HC) and light (LC) chains of the trastuzumab. sp, signal peptide; p(A), poly A tail. (B) Trastuzumab levels (mean ± SD) in medium and lysates 24 h after AML-12 cell transfection with trastuzumab. IVT-mRNAs containing signal peptide from human immunoglobulin light chain kappa (hIgLCk), human immunoglobulin heavy chain G1 and light chain kappa (hIgG1 HC/hIgLCk) or Gaussia luciferase (GLuc). ****p ≤ 0.0001 or ***p ≤ 0.001 compared to control, #p ≤ 0.05 compared to hIgG1 HC/hIgLCk SP and GLuc SP, n = 3. (C) Trastuzumab levels (mean ± SD) in AML-12 cell medium 24 h after transfection with IVT-mRNA for trastuzumab light and heavy chain at 1:1, 1:2, 1:4, or 1:8 ratio, respectively. ****p ≤ 0.0001, ***p ≤ 0.001 or **p ≤ 0.01 compared to control; #p ≤ 0.05 and ###p ≤ 0.001, n ≥ 3. (D) Trastuzumab levels (mean ± SD) in C57BL/6 mouse serum 24 h after injection of 0.5 mg/kg LNPs with IVT-mRNA for trastuzumab light and heavy chain in 1:1, 1:2, or 1:4 ratios. *p ≤ 0.05, **p ≤ 0.01, n ≥ 3. (B–D) One-way ANOVA with Tukey’s post hoc test.
The majority of secretory proteins contain a short amino acid sequence (16–30 amino acids long) at the N terminus called the signal peptide (SP) sequence. The SP is important for secretory protein translocation into the endoplasmic reticulum and the subsequent secretion of proteins from the cell. Despite substantial variation in SP sequences between different proteins and species, they are often functionally interchangeable. For example, in human endostatin, the replacement of native SP with Gaussia luciferase (GLuc) SP enhanced protein expression and secretion. Interestingly, in that study, GLuc SP increased protein production more than SP from human albumin or interleukin-2, which are both well-known proteins secreted at high levels in the serum.32 Similarly, replacement of the original SP in cytokine-like 1 (CYTL1) protein with human immunoglobulin light chain kappa (hIgLCk) SP significantly increased the expression of this protein.33 Moreover, using a bioinformatics approach based on analysis and clustering of existing human IgG1 heavy and light chain SP sequences, an H5/L1 SP combination has previously been identified and validated as the most optimal synthetic SP combination for trastuzumab production.34
Protein sequences for the majority of recombinant therapeutic antibodies can be found in public databases; however, these sequences often do not include an SP. To identify the most efficient SP for trastuzumab expression, we compared IVT-mRNA synthesized with GLuc, H5/L1, or hIgLCk SP (SP sequences are listed in Table S1). We used a protein sequence available at DrugBank to synthesize IVT-mRNA for trastuzumab’s (trastuzumab mRNA) heavy and light chains. The protein sequence was converted into an mRNA sequence optimized for expression in mammalian cells. Trastuzumab mRNA with different SPs was then transfected into mouse hepatocytes (AML-12 cells), and trastuzumab concentrations were measured by ELISA in cell lysates and conditioned media. Although intracellular levels of trastuzumab were not affected by the SP sequence, we found that the mRNA containing hIgLCk SP resulted in higher levels of trastuzumab in the cell medium compared to GLuc and H5/L1 SPs (Figure 1B). Trastuzumab mRNA with hIgLCk SP was used in all subsequent studies.
It has been reported that protein translation rates decrease as mRNA length increases.35, 36 Since the heavy chain (HC) of human IgG1 is two times longer than the light chain (LC), we hypothesized that slower expression rates of the HC compared to the LC were limiting IgG1 assembly and secretion. We next investigated whether increasing the HC-to-LC ratio could enhance protein synthesis. We transfected AML-12 cells with trastuzumab’s heavy and LC mRNA in different ratios (wt:wt): 1:1, 1:2, 1:4, and 1:8. We found that transfection using constructs containing 1:2 LC:HC resulted in significantly higher expression of trastuzumab compared to the other ratios (Figure 1C). We also observed that further increasing the LC-to-HC mRNA ratio decreased protein production, possibly because of the stoichiometric limit of LC molecules required to form a complete antibody. Surprisingly, in vivo we did not find a significant difference between the different ratios, although average protein expression was highest in animals injected with the 1:2 LC:HC ratio compared to the 1:1 or 1:4 ratios (Figure 1D). We chose to use an LC:HC ratio of 1:2 in all subsequent studies. The full trastuzumab sequence used in the study is presented in Figure S1.
In Vivo Validation of Trastuzumab Production
Next, we explored delivery of the optimized trastuzumab mRNA in vivo. For this study we used cKK-E12 (also known as MD-1) lipid-like nanoparticles that have demonstrated potent and selective small interfering RNA (siRNA) delivery to parenchymal liver cells in non-human primates37 and high expression of EPO IVT-mRNA in rodents.38
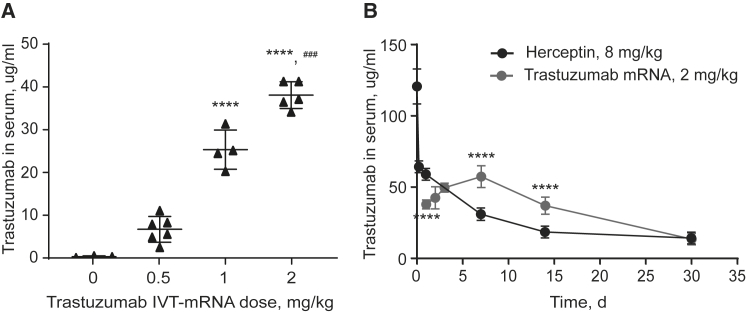
To evaluate whether the optimized trastuzumab mRNA is capable of producing protein in vivo, we measured trastuzumab levels in mouse serum after intravenous administration of a single-dose of trastuzumab mRNA encapsulated in cKK-E12 nanoparticles. C57BL/6 mice were injected with 0.5, 1, or 2 mg/kg (based on encapsulated mRNA) of trastuzumab mRNA. All doses were well tolerated, and no adverse events were observed. As shown in Figure 2A, we found a dose-dependent increase in trastuzumab protein levels in mouse serum 24 h after administration. Upon injection with 2 mg/kg of the mRNA, approximately 40 μg trastuzumab/mL serum was produced (Figure 2A). Further monitoring revealed that trastuzumab serum concentration continued to increase during the following 7 days and reached 57.5 ± 7.6 μg/mL at 7 days after the injection (Figure 2B).
Figure 2.
In Vivo Validation of Trastuzumab mRNA
(A) Trastuzumab concentrations in C57BL/6 mouse serum 24 h after injection of cKK-E12 LNPs with trastuzumab mRNA via the tail vein in different doses. Mean ± SD. ****p ≤ 0.0001 compared with control, ###p ≤ 0.001 compared to 1 mg/kg. (B) Pharmacokinetics of trastuzumab in C57BL/6 mouse serum after a single i.v. dose of 8 mg/kg Herceptin (Genentech) or of 2 mg/kg cKK-E12 LNPs with trastuzumab mRNA. Mean ± SD. Two-way repeated-measures ANOVA with the Sidak multiple-comparisons test was used to compare trastuzumab serum levels on day 1, 7, 14, and 30, after administration of trastuzumab mRNA versus Herceptin. ****p ≤ 0.0001, ns, p ≤ 0.05, n = 5.
To compare the pharmacokinetic profiles of our endogenously expressed trastuzumab with the current standard of treatment (trastuzumab protein [Herceptin]), we administered a single injection of 8 mg/kg Herceptin (Genentech) to C57BL6 mice and monitored trastuzumab serum levels for 1 month. We used 8 mg/kg because it is the highest Herceptin dose used in patients.39, 40 As shown in Figure 2B, 30 min after administration, 120.5 ± 12 μg/mL Herceptin antibody was detected in the serum. These levels dramatically decreased within the next few hours and reached 64 ± 4 μg/mL at 6 h after administration. One day after injection, the trastuzumab serum concentration was measured at 59 ± 4 μg/mL and continued to decrease slowly over the next few weeks. Analysis of these trastuzumab pharmacokinetics (Table 1) revealed a comparable half-life for both mRNA-LNPs and Herceptin treatment. The mRNA-based delivery resulted in a 64% higher antibody serum concentration over the course of 30 days, as reflected by the area under the curve (AUC0–t). The serum clearance (Cl) and steady-state volume of distribution (Vss) were approximately 5-fold higher after injection with Herceptin than after injection of trastuzumab mRNA-LNPs.
Table 1.
Analysis of Trastuzumab Pharmacokinetics in Mouse Serum after a Single i.v. Dose of Herceptin or Trastuzumab mRNA-LNPs
| Parameter | Unit | Trastuzumab mRNA (2 mg/kg) | Herceptin (8 mg/kg) |
|---|---|---|---|
| t1/2 | h | 241 | 251 |
| AUC0–t | μg/mL*h | 28,608 | 17,422.7 |
| Cl_obs | (mg/kg)/(μg/mL)/h | 6.991E-05 | 0.0003549 |
| Vss_obs | (mg/kg)/(μg/mL) | 0.0275 | 0.151 |
Calculations were performed using PKSolver 2.0. Cl, plasma clearance; Vss, steady-state volume of distribution; t1/2, half-life; AUC0–t, the area under the curve; obs, observed.
Overall, a single dose of LNPs with IVT-mRNA encoding trastuzumab at 2 mg/kg showed favorable serum pharmacokinetics of the antibody over the course of 1 month, as compared to a single injection of Herceptin at 8 mg/kg.
Functional In Vitro Validation of the Optimized Trastuzumab RNA Construct
To verify the primary sequence of the protein produced upon delivery of trastuzumab mRNA-loaded LNPs, we performed mass spectrometry analysis of the IgG isolated from the mouse serum. Liquid chromatography-tandem mass spectometry (LC-MS/MS) confirmed that the primary sequence of the produced protein matched trastuzumab’s amino acid sequence (Figure S2; Table S2).
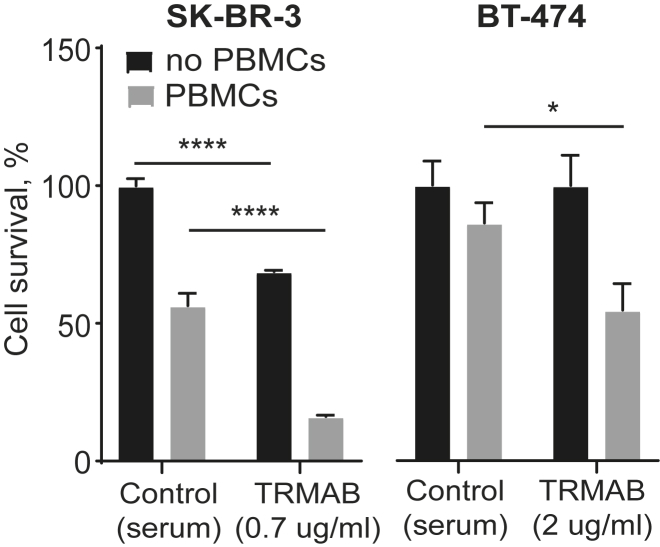
To investigate whether trastuzumab produced using optimized mRNA could retain its anticancer activity, we studied its effects on breast cancer cell survival and antibody-dependent cellular cytotoxicity (ADCC). We used two HER2-positive cell lines: SKBR3 and BT-474.41, 42 Trastuzumab was isolated from the serum of mice injected with trastuzumab mRNA on a Protein A column. To control for mouse IgG binding to the column along with trastuzumab, the process was also performed on serum isolated from control mice that did not receive trastuzumab mRNA. The isolate from control mice was used as a control for the ADCC assay. Cell survival was measured using a Cell Titer Glo assay. Analysis of breast cancer cell survival after treatment with the isolated trastuzumab revealed a decrease in SK-BR-3 cell survival, as compared to the untreated control (Figure 3, black bars). We did not observe a decrease in BT-474 cell survival, which may have been due to the reduced sensitivity of BT-474 cells to the cell growth inhibitory effects of trastuzumab.
Figure 3.
The ADCC Assay
Survival (mean ± SD) of HER2-positive breast cancer cells (BT-474 and SK-BR-3) after incubation with trastuzumab isolated from the serum of mice injected with 2 mg/kg trastuzumab mRNA in combination with human PBMCs. The ratio of breast cancer cells to PBMCs, 1:5. Two-way ANOVA with Tukey’s post hoc test, *p ≤ 0.05, ****p ≤ 0.0001, n = 3.
Trastuzumab-mediated ADCC has been shown to be crucial for trastuzumab’s antitumor effects. ADCC is triggered by the interaction between the antibody constant region (Fc) and the Fc-receptor present on the effector cells or on the components of the complement system. The binding of trastuzumab to HER2 receptors on the surface of target cells (HER2-positive breast cancer cells) and the subsequent recognition of the antibody Fc by effector cells results in target cell lysis. To test if mRNA-produced trastuzumab preserves its potency to induce ADCC, human peripheral blood mononuclear cells (PBMCs) were used as effector cells. We measured BT-474 and SK-BR-3 cell survival after treatment with trastuzumab isolated from mouse serum in combination with PBMCs. As shown in Figure 3 (gray bars), incubation with trastuzumab in the presence of PBMCs significantly decreased breast cancer cell survival compared with cells incubated only with PBMCs. SK-BR-3 cells were more sensitive to the treatment, most likely because of higher levels of HER2 expression.43, 44 Taken together, these data demonstrate the ability of trastuzumab produced using mRNA to induce HER2-positive breast cancer cell death via both interactions with the HER2-receptor and activation of the ADCC pathway in the presence of PBMCs.
Functional In Vivo Validation of the Optimized Trastuzumab mRNA Construct
To study whether trastuzumab produced using the optimized mRNA could retain its anticancer activity in vivo, we assessed its effects on tumor growth and animal survival in a mouse model using HER2-positive and -negative human breast cancer xenografts. The MDA-MB-231 cell line is HER2-negative and therefore is insensitive to trastuzumab. We transduced MDA-MB-231 cells with a human HER2-expressing plasmid to create an MDA-MB-231-HER2 cell line sensitive to trastuzumab. HER2 expression was confirmed by qPCR (Figure S3). The HER2-negative cell line was used to control for non-specific side effects associated with RNA, such as production of inflammatory cytokines (TNFa, IL-12, IL-8, and type I interferons) after RNA recognition by innate immune receptors (TLR3, TLR7, TLR8, RIG-I, MDA5, and others) and triggering of antitumor immune response via mechanisms independent of the RNA sequence.11, 45, 46 In addition, these two cell lines allow us to assess HER2-specific efficacy of trastuzumab mRNA.
We have previously demonstrated that IVT-mRNA from cKK-E12 LNPs is predominantly translated in the liver with minimal translation in the spleen.38 However, tumors often have abnormal blood supply because of high vascular permeability and ineffective lymphatic drainage. This phenomenon can lead to nanoparticle accumulation and retention (reviewed in Nagy et al.47 and Kanasty et al.48). To evaluate whether cKK-E12 LNPs deliver IVT-mRNA to xenografts, we injected xenograft-bearing mice with 2 mg/kg firefly luciferase (FLuc) IVT-mRNA formulated into cKK-E12 LNPs and measured bioluminescence levels, using IVIS (Figure S4). Whole-body IVIS showed that 6 h after the injection, LNPs accumulated mainly in the liver (Figure S4A). Analysis of organs excised from athymic nude mice xenografted with MDA-MB-231-HER2 cells revealed that a bioluminescent signal was also present in tumors (Figure S4B and S4C).
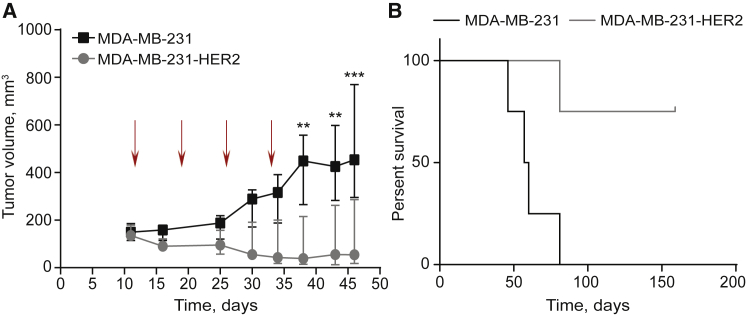
To study the effects of trastuzumab mRNA on tumor growth, mice with established MDA-MB-231 or MDA-MB-231-HER2 xenografts were treated with weekly injections of 2 mg/kg trastuzumab mRNA LNPs for 4 weeks and xenograft growth was monitored (Figure 4A). At the beginning of treatment, tumor volume in both groups was ∼150 mm3 (Figure 4A, day 11). We found a significant delay in HER2-positive tumor growth compared to HER2-negative tumors (Figure 4A). One week after the final injection the average volume of HER2-positive tumors was ∼100 mm3, whereas HER2-negative xenografts were, on average, four times larger (Figure 4A, day 38). Calculation of survival fractions using the Kaplan-Meier method revealed that the treated mice with HER2-positive tumors had significantly longer morbidity-free survival (p = 0.0169, log-rank test) compared to the treated mice with HER2-negative tumors. One mouse in each group gnawed their tumors, and they were removed from the survival study.
Figure 4.
Investigation of the In Vivo Effects of Trastuzumab Therapy
(A) Growth (median ± interquartile range [IQR]) of HER2-negative (MDA-MB-231) and HER2-positive (MDA-MB-231-HER2) tumors in mice treated with the trastuzumab mRNA. Arrows indicate the days of mRNA-LNP injections. Two-way repeated-measures ANOVA with the Sidak multiple-comparisons test, **p ≤ 0.01, ***p ≤ 0.001, compared with the volume of HER2-negative tumors, n = 5. (B) Kaplan-Meier analysis of animal survival after treatment with trastuzumab mRNA. Log-rank test, p = 0.0169, n = 4.
In addition, we studied whether the trastuzumab mRNA injections, per se, can contribute to the tumor growth reduction. Mice with HER2-negative tumors were treated with saline, and the tumor growth was monitored (Figure S6A). When we compared those mice with those treated with the trastuzumab mRNA, we observed no difference in tumor growth between the two groups. Finally, to compare the antitumor effects of the trastuzumab mRNA and Herceptin, mice with HER2-positive tumors received one injection of 8 mg/kg Herceptin or saline per week for 4 weeks (Figure S6B). Although we did not observe a significant difference, the average tumor volume in mice treated with Herceptin was lower than in mice treated with saline. The low efficacy of Herceptin in our study may have been the result of infrequent injections, as we administered Herceptin only once a week instead of the standard two times per week. Importantly, when compared to mice treated with the trastuzumab mRNA, we still observed a significant reduction in tumor growth compared to saline. In addition, the average tumor volume in mice treated with the mRNA was lower than in mice treated with Herceptin.
Toxicity was evaluated 1 week after the final injection by analyzing blood and organs. Blood count revealed an approximately 2-fold decrease in lymphocyte levels in all tumor-bearing mice, which may have been associated with tumor formation (Table S3). We also observed slight variations in several serum chemistry parameters, but the changes were less than 2-fold (Table S4). Importantly, no significant changes in liver enzymes were found after 4 weeks of treatment with the LNPs, as compared to the untreated tumor-bearing mice. Histological analysis did not reveal any morphological changes in the liver and spleen of treated mice (Figure S5). Together, the data suggest that treatment with trastuzumab mRNA LNPs significantly delayed the growth of HER2-positive breast cancer in athymic nude mice, with no apparent toxicity.
Discussion
In recent years, RNA therapeutics have been broadly used in a variety of clinical applications, including vaccines, protein replacement therapies, and gene editing technologies. Therapeutic anticancer vaccines designed to target tumor-associated antigens and mobilize T cell responses are being currently evaluated in the clinic (reviewed in Pastor et al.,49 Sahn and Türeci,50 and Kowalski et al.51). The use of IVT-mRNA for the initiation of endogenous antibody expression can overcome the challenges associated with the manufacture and delivery of highly demanded monoclonal-antibody-based drugs.52, 53 Specifically, the use of LNP-formulated IVT-mRNA, rather than protein, can address the limitations found with bi-specific antibodies, such as short serum half-life, manufacturing challenges, and the requirement for repeated administration.52 In addition, using host translational machinery for continuous expression of high levels of properly folded and modified proteins makes mRNA-based antibody production simple and cost effective compared to protein-based therapeutics.53 Furthermore, the use of IVT-mRNA simplifies the process of antibody structure optimization and enables localized antibody expression that is unavailable for conventional therapeutics.10
Prior efforts to use monoclonal mRNA for endogenous antibody expression have been mainly situated at the level of passive antiviral immunization.10, 53, 54 In our study, we broaden application of mRNA-based delivery for monoclonal antibody production to treatment of solid tumors. We designed and optimized an IVT-mRNA delivery system for potent in vitro and in vivo expression of trastuzumab. Trastuzumab is a widely used and successful anticancer monoclonal antibody, with a well-understood mechanism of action. We developed a strategy for the rational design of an IVT-mRNA structure to maximize antibody production. Further, we demonstrated that this optimization can significantly improve in vivo trastuzumab protein expression, maintain both HER2 specificity and ADCC functionality, and extend survival of animals with HER2-positive breast cancer.
We identified hIgLCk as the most efficient signal peptide and optimized the antibody light-to-heavy-chain ratio to obtain maximal trastuzumab expression upon liver delivery in mice. We also demonstrated improved trastuzumab pharmacokinetics when produced using IVT-mRNA compared to Herceptin and confirmed that trastuzumab isolated from mouse serum maintained both HER2 specificity and ADCC functionality.
Mice injected with 2 mg/kg (40 μg per animal) of trastuzumab mRNA demonstrated serum levels comparable with injection of the highest clinically relevant protein dose (8 mg/kg, 160 μg per animal).39, 40 Notably, in our study, we observed potent protein expression and longer antibody persistence in mouse serum after delivery of trastuzumab mRNA, compared to previously published studies.52, 53, 54 After delivery of 40 μg mRNA-LNPs or 160 μg protein, we demonstrated that the serum concentration of endogenously produced trastuzumab is at 24 h, comparable with the levels of the injected antibody. Moreover, trastuzumab levels in mice that received Herceptin continued to decline, while trastuzumab levels in mice injected with mRNA-LNP remained within 45 ± 8.6 μg/mL for 14 days. In fact, 2 weeks after mRNA injection, trastuzumab serum levels were still above 10 μg/mL, which is considered the minimum therapeutic plasma concentration needed for antiproliferative effects.40 These data also support findings from previous studies,52, 53 where administration of mRNA-based drugs resulted in similar or higher antibody plasma levels compared to the corresponding purified antibody. The differences in peak concentrations may be a result of differences in the antibody sequence and delivery method. In general, plasma levels of the endogenously translated antibodies tend to be sustained longer than levels of the protein. Extended plasma circulation time in our study compared to the previously published studies using the mRNA-based approach52, 53 can be a result of differences in antibody sequence or mechanism of action. The standard administration protocol for trastuzumab in mice requires injections of 1–10 mg/kg of the protein twice a week. We showed that a prolonged presence of the endogenously expressed therapeutic antibody in the plasma has the potential to reduce administration frequency, dosing, and, subsequently, treatment cost.
Finally, our study is the first demonstration of the inhibition of solid tumor growth by a full-size antibody produced in the liver upon delivery of IVT-mRNA formulated into LNPs. We did not observe any toxic effects during our treatment regimen. In vivo validation of anticancer activity showed that weekly intravenous (i.v.) administration of 2 mg/kg of trastuzumab mRNA LNPs for 4 weeks inhibited breast cancer growth in athymic nude mice and improved animal survival, thus outperforming the standard of treatment, Herceptin. Importantly, we demonstrated that the inhibition of tumor growth was specific to HER2 receptor presence in the tumor cells by using MDA-MB-231 and MDA-MB-231-HER2 xenografts. Interestingly, we found that LNPs can deliver IVT-mRNA directly to the tumor. Expression of therapeutic antibodies within the tumor microenvironment or directly in tumor cells can potentially enhance therapeutic efficacy and reduce off-target effects.
In summary, our study demonstrates that using IVT-mRNA loaded into LNPs to express full-size therapeutic antibodies, such as trastuzumab, in the liver can provide an effective strategy for cancer treatment and offers an alternative to protein administration. This study also provides a guideline for the optimization of an IVT-mRNA construct, which can be used for the development of other monoclonal antibody mRNA therapies.
Materials and Methods
Cell Culture
The mouse hepatocytes AML-12 and the human HER-positive breast carcinoma cells BT-474 and MDA-MB-231 were obtained from ATCC. SK-BR-3 cell line was a gift from the Hammond Lab (MIT). MDA-MB-231 cells expressing human HER2 (MDA-MB-231-HER2) were created in our lab. AML-12 cells were grown in a 1:1 mixture of DMEM and Ham’s F12 with Insulin-Transferrin-Selenium (ITS-G) medium supplement, 100 U/mL penicillin-streptomycin (Pen/Strep), and 10% fetal bovine serum (FBS) (Gibco). The BT-474 cells were cultured in ATCC Hybri-Care Medium supplemented with 1.5 g/L sodium bicarbonate, 100 U/mL Pen/Strep, and 10% FBS. MDA-MB-231, MDA-MB-231-HER2, and SK-BR-3 cells were grown in DMEM with 100 U/mL Pen/Strep and 10% FBS. All cell lines were cultured at 37°C in 5% CO2. When the cells reached 70%–80% monolayer, they were detached from the flask using 0.25% Trypsin-EDTA solution and split 1:5.
Mice
All animal studies were approved by the MIT Institutional Animal Care and Use Committee and were consistent with local, state, and federal regulations, as applicable. RNA/LNPs (0.5–2 mg/kg) were administered to female C57BL/6 mice (20–25 g; Charles River Laboratories) or to female athymic nude mice (20–22 g Charles River Laboratories) intravenously via tail vein injection. Mouse blood was collected from the lateral tail vein or the submandibular vein in a sterile 1.5 mL microcentrifuge tube, incubated on ice for 10 min, and centrifuged at 2,000 g for 20 min at 4°C. Supernatants (serum) were collected and used for ELISA or protein isolation.
Mouse Xenograft Models
Athymic nude mice (female, aged 7–8 weeks) received subcutaneous implants in the right flank of 4 × 106 MDA-MB-231 or MDA-MB-231-HER2 cells in Dulbecco’s PBS (DPBS)-Matrigel (v/v, 1:1). Treatment was started 11 days later, when xenografts reached 100–200 mm3. Both groups of mice received one i.v. injection of 2 mg/kg trastuzumab mRNA per week for 4 weeks, formulated into cKK-E12 nanoparticles. Tumors were measured with a digital caliper, and volumes were calculated by the formula tumor volume (mm3) = length (mm) × (width [mm])2/2. Termination criteria were a tumor volume of 1,000 mm3 or weight loss of 20%.
Protein Isolation
Trastuzumab was isolated from mouse blood using the Pierce Protein A IgG Purification Kit according to the manufacturer’s instructions. Briefly, mouse serum was diluted with the Protein A Binding Buffer 1:1 and applied to the equilibrated Protein A column. Then, the column was washed by adding 10–15 mL of Binding Buffer. The bound IgG was eluted with 5 mL of the IgG Elution Buffer. Ultracel-10K centrifugal filters (Millipore) were used to substitute the Binding Buffer with PBS and concentrate the eluent. Trastuzumab concentration in the eluent was measured using ELISA.
In Vitro ADCC Assay
Buffy coats collected from human peripheral blood were purchased from Research Blood Components, LLC (Brighton, MA, USA) and PBMCs were isolated by density gradient centrifugation. Briefly, buffy coats were diluted 1:1 in RPMI-1640 and carefully layered over a sterile, endotoxin tested (<0.12 EU/mL) solution of Ficoll Paque Plus (density 1.007 g/mL; GE Healthcare). Layered solutions were centrifuged at 500 g for 40 m at room temperature to sediment unwanted cells, and the interfacial layer of PBMCs was manually isolated. PMBCs were then washed twice in RPMI-1640 before further use. After they were washed with the RPMI-1640 cell culture medium (Gibco), the cells were immediately subjected to ADCC or frozen, using Gibco Recovery Cell Culture Freezing Medium. BT-474 and SK-BR-3 cells were seeded in a 96-well plate at 15,000 cells per well and incubated overnight. The next day, 50 μL trastuzumab was added to the plates and incubated for 30 min to allow opsonization. After incubation, 50 μL of PBMCs were added to each well in 1:2, 1:5, or 1:10 ratio. The PBMCs were removed from the wells 72 h later by washing with 100 μL DPBS (Gibco). The number of survived breast cancer cells was measured by using the Cell Titer Glo assay.
Statistical Analysis
One-way ANOVA followed by Tukey’s post hoc test was used for multiple-comparisons analysis. Two-way ANOVA followed by the Sidak multiple-comparisons test was used for analysis of repeated measures. The GraphPad Prism 7 package was used.
Author Contributions
Y.R. designed the study, executed the experiments, analyzed the data, and wrote the manuscript; P.S.K. was responsible for the mRNA design, synthesis, and delivery and edited the manuscript; Y.H. and J.T.G. assisted with the experiments; M.W.H. and F.D. contributed to the mRNA design and edited the manuscript; D.D. contributed to the ADCC experimental design and execution and edited the manuscript; and D.G.A. supervised the study and edited the manuscript.
Conflicts of Interest
D.G.A., P.S.K., and Y.R. filed a patent for the design and development of the trastuzumab mRNA delivery system. The remaining authors declare no competing interests.
Acknowledgments
This work was supported by Translate Bio (Lexington, MA, USA), the MIT Skoltech Initiative, Defense Advanced Research Projects Agency (W32P4Q-13-1-001), S. Leslie Misrock Frontier Research Fund for Cancer Nanotechnology, Juvenile Diabetes Research Foundation (JDRF) postdoctoral fellowship grant 3-PDF-2017-383-A-N, and a Koch Institute Support (core) grant P30-CA14051 from the National Cancer Institute. We thank the Koch Institute Swanson Biotechnology Center for technical support, specifically the Animal Imaging and Preclinical Testing Core, the Histology Core, and the Biopolymers and Proteomics Core. We also thank Prof. Oliver Jonas (Brigham and Women’s Hospital) and Dr. Marion Paolini (MIT), who provided insight into the development of a HER2-positive breast cancer model, and Dr. Kevin McHugh (MIT) for comments that improved the manuscript.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.ymthe.2019.05.012.
Supplemental Information
References
- 1.Urquhart L. Market watch: Top drugs and companies by sales in 2017. Nat. Rev. Drug Discov. 2018;17:232. doi: 10.1038/nrd.2018.42. [DOI] [PubMed] [Google Scholar]
- 2.Samaranayake H., Wirth T., Schenkwein D., Räty J.K., Ylä-Herttuala S. Challenges in monoclonal antibody-based therapies. Ann. Med. 2009;41:322–331. doi: 10.1080/07853890802698842. [DOI] [PubMed] [Google Scholar]
- 3.Kunert R., Reinhart D. Advances in recombinant antibody manufacturing. Appl. Microbiol. Biotechnol. 2016;100:3451–3461. doi: 10.1007/s00253-016-7388-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chusainow J., Yang Y.S., Yeo J.H., Toh P.C., Asvadi P., Wong N.S., Yap M.G. A study of monoclonal antibody-producing CHO cell lines: what makes a stable high producer? Biotechnol. Bioeng. 2009;102:1182–1196. doi: 10.1002/bit.22158. [DOI] [PubMed] [Google Scholar]
- 5.Li F., Vijayasankaran N., Shen A.Y., Kiss R., Amanullah A. Cell culture processes for monoclonal antibody production. MAbs. 2010;2:466–479. doi: 10.4161/mabs.2.5.12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Hoecke L., Roose K. How mRNA therapeutics are entering the monoclonal antibody field. J. Transl. Med. 2019;17:54. doi: 10.1186/s12967-019-1804-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Awwad S., Angkawinitwong U. Overview of antibody drug delivery. Pharmaceutics. 2018;10:1–24. doi: 10.3390/pharmaceutics10030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zangi L., Lui K.O., von Gise A., Ma Q., Ebina W., Ptaszek L.M., Später D., Xu H., Tabebordbar M., Gorbatov R. Modified mRNA directs the fate of heart progenitor cells and induces vascular regeneration after myocardial infarction. Nat. Biotechnol. 2013;31:898–907. doi: 10.1038/nbt.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahiny A.J., Dewerth A., Mays L.E., Alkhaled M., Mothes B., Malaeksefat E., Loretz B., Rottenberger J., Brosch D.M., Reautschnig P. In vivo genome editing using nuclease-encoding mRNA corrects SP-B deficiency. Nat. Biotechnol. 2015;33:584–586. doi: 10.1038/nbt.3241. [DOI] [PubMed] [Google Scholar]
- 10.Tiwari P.M., Vanover D., Lindsay K.E., Bawage S.S., Kirschman J.L., Bhosle S., Lifland A.W., Zurla C., Santangelo P.J. Engineered mRNA-expressed antibodies prevent respiratory syncytial virus infection. Nat. Commun. 2018;9:3999. doi: 10.1038/s41467-018-06508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karikó K., Buckstein M., Ni H., Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Holtkamp S., Kreiter S., Selmi A., Simon P., Koslowski M., Huber C., Türeci O., Sahin U. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108:4009–4017. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- 13.Andries O., Mc Cafferty S., De Smedt S.C., Weiss R., Sanders N.N., Kitada T. N(1)-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J. Control. Release. 2015;217:337–344. doi: 10.1016/j.jconrel.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 14.Li B., Luo X., Dong Y. Effects of Chemically Modified Messenger RNA on Protein Expression. Bioconjug. Chem. 2016;27:849–853. doi: 10.1021/acs.bioconjchem.6b00090. [DOI] [PubMed] [Google Scholar]
- 15.Kauffman K.J., Dorkin J.R., Yang J.H., Heartlein M.W., DeRosa F., Mir F.F., Fenton O.S., Anderson D.G. Optimization of Lipid Nanoparticle Formulations for mRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- 16.DeRosa F., Guild B., Karve S., Smith L., Love K., Dorkin J.R., Kauffman K.J., Zhang J., Yahalom B., Anderson D.G., Heartlein M.W. Therapeutic efficacy in a hemophilia B model using a biosynthetic mRNA liver depot system. Gene Ther. 2016;23:699–707. doi: 10.1038/gt.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J. A Novel Amino Lipid Series for mRNA Delivery: Improved Endosomal Escape and Sustained Pharmacology and Safety in Non-human Primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J.B., Karmali P.P., Chivukula P., Verma I.M. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. USA. 2017;114:E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sahin U., Karikó K., Türeci Ö. mRNA-based therapeutics--developing a new class of drugs. Nat. Rev. Drug Discov. 2014;13:759–780. doi: 10.1038/nrd4278. [DOI] [PubMed] [Google Scholar]
- 20.Yin H., Kanasty R.L., Eltoukhy A.A., Vegas A.J., Dorkin J.R., Anderson D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 21.Kauffman K.J., Webber M.J., Anderson D.G. Materials for non-viral intracellular delivery of messenger RNA therapeutics. J. Control. Release. 2016;240:227–234. doi: 10.1016/j.jconrel.2015.12.032. [DOI] [PubMed] [Google Scholar]
- 22.Hajj K.A., Whitehead K.A. Tools for translation: Non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2017;2:17056. [Google Scholar]
- 23.Guan S., Rosenecker J. Nanotechnologies in delivery of mRNA therapeutics using nonviral vector-based delivery systems. Gene Ther. 2017;24:133–143. doi: 10.1038/gt.2017.5. [DOI] [PubMed] [Google Scholar]
- 24.Kowalski P.S., Capasso Palmiero U., Huang Y., Rudra A., Langer R., Anderson D.G. Ionizable Amino-Polyesters Synthesized via Ring Opening Polymerization of Tertiary Amino-Alcohols for Tissue Selective mRNA Delivery. Adv. Mater. 2018;30:e1801151. doi: 10.1002/adma.201801151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hudziak R.M., Lewis G.D., Winget M., Fendly B.M., Shepard H.M., Ullrich A. p185HER2 monoclonal antibody has antiproliferative effects in vitro and sensitizes human breast tumor cells to tumor necrosis factor. Mol. Cell. Biol. 1989;9:1165–1172. doi: 10.1128/mcb.9.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carter P., Presta L., Gorman C.M., Ridgway J.B., Henner D., Wong W.L., Rowland A.M., Kotts C., Carver M.E., Shepard H.M. Humanization of an anti-p185HER2 antibody for human cancer therapy. Proc. Natl. Acad. Sci. USA. 1992;89:4285–4289. doi: 10.1073/pnas.89.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hudis C.A. Trastuzumab: mechanism of action and use in clinical practice. N. Engl. J. Med. 2007;357:39–51. doi: 10.1056/NEJMra043186. [DOI] [PubMed] [Google Scholar]
- 29.Clynes R.A., Towers T.L., Presta L.G., Ravetch J.V. Inhibitory Fc receptors modulate in vivo cytotoxicity against tumor targets. Nat. Med. 2000;6:443–446. doi: 10.1038/74704. [DOI] [PubMed] [Google Scholar]
- 30.Kuge H., Brownlee G.G., Gershon P.D., Richter J.D. Cap ribose methylation of c-mos mRNA stimulates translation and oocyte maturation in Xenopus laevis. Nucleic Acids Res. 1998;26:3208–3214. doi: 10.1093/nar/26.13.3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kauffman K.J., Mir F.F., Jhunjhunwala S., Kaczmarek J.C., Hurtado J.E., Yang J.H., Webber M.J., Kowalski P.S., Heartlein M.W., DeRosa F., Anderson D.G. Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials. 2016;109:78–87. doi: 10.1016/j.biomaterials.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knappskog S., Ravneberg H., Gjerdrum C., Trösse C., Stern B., Pryme I.F. The level of synthesis and secretion of Gaussia princeps luciferase in transfected CHO cells is heavily dependent on the choice of signal peptide. J. Biotechnol. 2007;128:705–715. doi: 10.1016/j.jbiotec.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 33.Wang X., Liu H., Yuan W., Cheng Y., Han W. Efficient production of CYTL1 protein using mouse IgGκ signal peptide in the CHO cell expression system. Acta Biochim. Biophys. Sin. (Shanghai) 2016;48:391–394. doi: 10.1093/abbs/gmw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haryadi R., Ho S., Kok Y.J., Pu H.X., Zheng L., Pereira N.A., Li B., Bi X., Goh L.T., Yang Y., Song Z. Optimization of heavy chain and light chain signal peptides for high level expression of therapeutic antibodies in CHO cells. PLoS ONE. 2015;10:e0116878. doi: 10.1371/journal.pone.0116878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arava Y., Wang Y., Storey J.D., Liu C.L., Brown P.O., Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valleriani A., Zhang G., Nagar A., Ignatova Z., Lipowsky R. Length-dependent translation of messenger RNA by ribosomes. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2011;83:042903. doi: 10.1103/PhysRevE.83.042903. [DOI] [PubMed] [Google Scholar]
- 37.Dong Y., Love K.T., Dorkin J.R., Sirirungruang S., Zhang Y., Chen D., Bogorad R.L., Yin H., Chen Y., Vegas A.J. Lipopeptide nanoparticles for potent and selective siRNA delivery in rodents and nonhuman primates. Proc. Natl. Acad. Sci. USA. 2014;111:3955–3960. doi: 10.1073/pnas.1322937111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fenton O.S., Kauffman K.J., Kaczmarek J.C., McClellan R.L., Jhunjhunwala S., Tibbitt M.W., Zeng M.D., Appel E.A., Dorkin J.R., Mir F.F. Synthesis and Biological Evaluation of Ionizable Lipid Materials for the In Vivo Delivery of Messenger RNA to B Lymphocytes. Adv. Mater. 2017;29:1–7. doi: 10.1002/adma.201606944. [DOI] [PubMed] [Google Scholar]
- 39.Goldenberg M.M. Trastuzumab, a recombinant DNA-derived humanized monoclonal antibody, a novel agent for the treatment of metastatic breast cancer. Clin. Ther. 1999;21:309–318. doi: 10.1016/S0149-2918(00)88288-0. [DOI] [PubMed] [Google Scholar]
- 40.Leyland-Jones B., Arnold A., Gelmon K., Verma S., Ayoub J.P., Seidman A., Dias R., Howell J., Rakhit A. Pharmacologic insights into the future of trastuzumab. Ann. Oncol. 2001;12(Suppl 1):S43–S47. doi: 10.1093/annonc/12.suppl_1.s43. [DOI] [PubMed] [Google Scholar]
- 41.Holliday D.L., Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011:13–215. doi: 10.1186/bcr2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai X., Cheng H., Bai Z., Li J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer. 2017;8:3131–3141. doi: 10.7150/jca.18457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subik K., Lee J.F., Baxter L., Strzepek T., Costello D., Crowley P., Xing L., Hung M.C., Bonfiglio T., Hicks D.G., Tang P. The Expression Patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by Immunohistochemical Analysis in Breast Cancer Cell Lines. Breast Cancer (Auckl.) 2010;4:35–41. [PMC free article] [PubMed] [Google Scholar]
- 44.Pasleau F., Grooteclaes M., Gol-Winkler R. Expression of the c-erbB2 gene in the BT474 human mammary tumor cell line: measurement of c-erbB2 mRNA half-life. Oncogene. 1993;8:849–854. [PubMed] [Google Scholar]
- 45.Karikó K., Ni H., Capodici J., Lamphier M., Weissman D. mRNA is an endogenous ligand for Toll-like receptor 3. J. Biol. Chem. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 46.Lee S., Margolin K. Cytokines in cancer immunotherapy. Cancers (Basel) 2011;3:3856–3893. doi: 10.3390/cancers3043856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagy J.A., Chang S.-H., Dvorak A.M., Dvorak H.F. Why are tumour blood vessels abnormal and why is it important to know? Br. J. Cancer. 2009;100:865–869. doi: 10.1038/sj.bjc.6604929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanasty R., Dorkin J.R., Vegas A., Anderson D. Delivery materials for siRNA therapeutics. Nat. Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- 49.Pastor F., Berraondo P., Etxeberria I., Frederick J., Sahin U., Gilboa E., Melero I. An RNA toolbox for cancer immunotherapy. Nat. Rev. Drug Discov. 2018;17:751–767. doi: 10.1038/nrd.2018.132. [DOI] [PubMed] [Google Scholar]
- 50.Sahin U., Türeci Ö. Personalized Vaccines for Cancer Immunotherapy. Science. 2018;359:1355–1360. doi: 10.1126/science.aar7112. [DOI] [PubMed] [Google Scholar]
- 51.Kowalski P.S., Rudra A., Miao L., Anderson D.G. Delivering the Messenger: Advances in Technologies for Therapeutic mRNA Delivery. Mol. Ther. 2019;27:710–728. doi: 10.1016/j.ymthe.2019.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stadler C.R., Bähr-Mahmud H., Celik L., Hebich B., Roth A.S., Roth R.P., Karikó K., Türeci Ö., Sahin U. Elimination of large tumors in mice by mRNA-encoded bispecific antibodies. Nat. Med. 2017;23:815–817. doi: 10.1038/nm.4356. [DOI] [PubMed] [Google Scholar]
- 53.Pardi N., Secreto A.J., Shan X., Debonera F., Glover J., Yi Y., Muramatsu H., Ni H., Mui B.L., Tam Y.K. Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat. Commun. 2017;8:14630. doi: 10.1038/ncomms14630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Thran M., Mukherjee J., Pönisch M., Fiedler K., Thess A., Mui B.L., Hope M.J., Tam Y.K., Horscroft N., Heidenreich R. mRNA mediates passive vaccination against infectious agents, toxins, and tumors. EMBO Mol. Med. 2017;9:1434–1447. doi: 10.15252/emmm.201707678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.