Abstract
The triple-negative breast cancer (TNBC) that comprises approximately 10%–20% of breast cancers is an aggressive subtype lacking effective therapeutics. Among various signaling pathways, mTORC1 and purinergic signals have emerged as potentially fruitful targets for clinical therapy of TNBC. Unfortunately, drugs targeting these signaling pathways do not successfully inhibit the progression of TNBC, partially due to the fact that these signaling pathways are essential for the function of all types of cells. In this study, we report that TRPML1 is specifically upregulated in TNBCs and that its genetic downregulation and pharmacological inhibition suppress the growth of TNBC. Mechanistically, we demonstrate that TRPML1 regulates TNBC development, at least partially, through controlling mTORC1 activity and the release of lysosomal ATP. Because TRPML1 is specifically activated by cellular stresses found in tumor microenvironments, antagonists of TRPML1 could represent anticancer drugs with enhanced specificity and potency. Our findings are expected to have a major impact on drug targeting of TNBCs.
Keywords: Lysosome, TRPML1, mTORC1, Triple negative breast cancer
1. Introduction
Breast cancer is a very common and fatal cancer in women. It is a heterogeneous disease classified into three major subtypes. Two of these are defined by the expression of steroid hormone receptors (estrogen receptor [ER] and progesterone receptor [PR]) and overexpression and/or amplification of the receptor tyrosine kinase HER2 (also known as ERBB2). Agents targeting these proteins have led to a significant increase in patient survival [1–5]. In contrast, the most aggressive and metastatic form of breast cancer is the triple-negative breast cancer (TNBC) subtype, which is defined by the lack of expression of ER, PR and HER2 and its resistance to common therapies targeting these molecules [1–5]. TNBC represents approximately 10%–20% of breast cancers [6,7]. The major cause of TNBC mortality is the propensity of TNBC cells to metastasize to nearby normal tissues and organs and even spread to distant parts of the body through the blood or lymph system. Therefore, understanding the basic cellular and molecular mechanisms of TNBC development is crucial for devising new therapies for this disease.
The mammalian target of rapamycin complex 1 (mTORC1) is crucial for autophagy induction in response to nutrient shortage and cellular stress. Continuous loss of mTORC1 activity during nutrient and energy deprivation leads to cell death [8,9] because mTORC1 is also required for protein synthesis, metabolism, cell growth and proliferation and cellular homeostasis [10–12]. Mounting evidence suggests that mTORC1 represents a major signaling pathway responsible for cancer cell growth and survival [13,14]. Hyperactivation of mTORC1 pathway is common in human cancer including breast cancer [12,14,15]. In particular, activated mTORC1 is more frequently observed in TNBC (36%) than in other subtypes of breast cancer, rendering mTORC1 a potential target for TNBC therapy [16–18]. As such, several molecules that inhibit mTORC1 are currently in clinical trials for the treatment of multiple cancer types, including breast cancer [17,19–23].
Adenosine triphosphate (ATP) is actively released to the extracellular environment in response to tissue damage and cellular stress [24]. Increased levels of extracellular ATP at tumor sites have been suggested to play a key role in host-tumor interaction [24–26], likely acting on their specific cell surface receptors to increase motility, invasion and metastasis of many cancers [27–30], including breast cancer [31,32]. Interestingly, the lysosome plays an important role in mTORC1 signaling and is a major source of ATP. Specifically, mTORC1 is recruited to the surface of the lysosome to be activated by nutrients within the lysosome [33]. Lysosomal ATP (~1 mM) [34,35] can be released into extracellular space via lysosomal exocytosis [35–37], thereby regulating cancer migration and invasion [36,38]. Thus, lysosomes may regulate cancer development by controlling both mTORC1 and ATP signaling pathways.
Recently, we [39], and others [40,41], demonstrated that Transient Receptor Potential Mucolipin 1 (TRPML1 or ML1), the lysosomal Ca2+ release channel, is required for sustained activity of mTORC1 under nutrient stress but not under normal conditions. This regulation of mTORC1 by ML1 is essential for cells to survive extreme conditions [39]. Mounting evidence also suggests that ML1 is a key regulator of lysosomal exocytosis [42–44]. Here we show that ML1 supports tumor development by promoting both mTORC1 activity and lysosomal ATP release. We show that ML1 is markedly upregulated in highly metastatic TNBC when compared with non-metastatic ER+/PR+ MCF-7 breast cancer cell, as well as the non-tumorigenic MCF-10 A epithelial cell line. The mRNA of ML1 from TNBC patients is also significantly higher than that from ER+/PR+ breast cancer patients. Molecular abrogation or pharmacological inhibition of ML1 suppresses TNBC cell proliferation and invasion in vitro, and tumor growth and progression in vivo. In addition, we demonstrate that ML1 regulates TNBC growth by promoting mTORC1 activity and facilitating TNBC invasion through increasing lysosomal ATP release. Our work suggests that inhibiting ML1 could be a potential therapeutic strategy for TNBC.
2. Materials and methods
2.1. Cell culture
MCF10 A breast normal cells were grown in DMEM/F12 (1:1). Human breast cancer cell lines MDA-MB-231, MCF7, and Hs 578 T were grown in DMEM, while SUM159PT were cultured using F12 medium (Life Technologies, Carlsbad, CA) supplemented with 10% FBS (Life Technologies). All cultures were maintained at 37 °C in a 5% CO2 incubator.
2.2. Antibodies, plasmids and reagents
The following primary antibodies were used for western blotting and immunofluorescence staining: anti-p-p70S6K (T389) (9206, Cell Signaling Technology), anti-p70S6K (Cell Signaling Technology, 9202), and anti-GAPDH (H-12) (sc-166574, Santa Cruz Biotechnology), goat-anti-mouse IgG-HRP (Santa Cruz Biotechnology, sc-2005); goat-anti-rabbit IgG-HRP (Thermo Fisher Scientific, PI31460), Ki67 (Santa Cruz, sc-23900), Alexa Fluor®488 goat anti-mouse IgG (Life Technologies, A11001). The chemicals were purchased from Sigma except for ML-SI1 (Enzo Life Sciences Inc) and TO-PRO-3 iodide (T3605, Life Technologies). The human wild-type mTOR, mTOR-S2035 T (active), mTOR-S2035 T/D2357E (kinase dead), RagBGTP (Q99 L) and RagBGDP (T54 L) clones were from Addgene (plasmid # 26603, 26604, 26605, 19315, 19314, respectively) [45–47].
2.3. Western blot
Proteins were separated on SDS–polyacrylamide gel electrophoresis and transferred to a PVDF membrane (Bio-Rad, Hercules, CA, USA). The membrane was blocked with 5% non-fat milk and incubated with specific primary antibody solution at 4 °C overnight with gentle constant shaking. After three thorough TBS-S washes, the membranes were incubated with corresponding HRP-conjugated secondary antibody at room temperature for 1 h. Immunoreactive bands were visualized using Clear ECL (Bio-Rad) and autoradiography.
2.4. Lentivirus production and cell transduction
For knockdown studies, we constructed pLKO shRNA vectors (addgene, 8453) encoding shRNAs that target Syt-VII and ML1. Briefly, the plasmid was linearized using AgeI and EcroR1 to facilitate directional cloning. The sequences for human ML1-shRNA were as follows: #1, 5′-AAAGTATTTGAGACGACGGCG-3′; #2, 5′-GCCCACATCCAGGAGTG TAA-3′ [48]. The sequences for human Syt7 shRNA were as follows: #1, 5′-GAACGAGACCTTCCTCTTTGC-3′; #2, 5′-CAATGACGTCATCGGCAA GAC-3′ [49]. Lentivirus was made using a three-plasmid packaging system. Briefly, shRNAs in the pLKO vector were co-transfected into HEK293 T cells along with psPAX2 and pMD2.G. Lentivirus was harvested 48 h after transfection, and 8 μg/ml polybrene was added. Subconfluent MDA-MB-231 cells were infected with harvested lentivirus, and then were selected in 1 μg/ml puromycin for 3 days.
2.5. Real-time qPCR
Total RNA was extracted using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA). After reverse transcription using iScript™ Advanced cDNA Synthesis Kit (BIO-RAD), real-time qPCR was proformed using GoTaq® qPCR Master Mix (Promega). Gene-specific primers of ML1: Fwd (5′-CGGATGACACCTTCGCAGCCTAC-3′), Rev ( 5′-ACGCATACCGGCCCAGTGACAC -3′). Syt7: Fwd (5′-ACTCCATCA TCG TGAACATCATC −3′), Rev (5′- TCGAAGGCGAAGGACTCATTG-3′). The expression of target gene copies was normalized to GAPDH mRNA expression.
2.6. Cell proliferation
The proliferation of the cells was determined by monitoring their growth over 72 h time periods. For this, the cells were seeded at 5× 104 cells/well in 24-well plates. Cells were incubated overnight at 37 °C/5% CO2 and the three wells of each group were trypsinised and counted. This time point was corresponded to 0 h. The remaining wells were incubated for additional 72 h at 37 °C/5% CO2, and then trypsinised and counted. The cells number at 72 h were calculated relative to the cells number of each group at 0 h.
2.7. Immunostaining
Cells were grown on Poly-d-Lysine (Sigma-Aldrich) coated coverslips and washed with PBS twice before fixation with pre-chilled 4% paraformaldehyde at room temperature for 10 min. Fixed cells were then washed with PBS. Fixed cells were permeabilized with 0.3% Triton X-100 for 7 min and then blocked with 2% BSA in PBS buffer for 30 min at room temperature. After 3 times wash with PBS, cells were incubated with Ki-67 antibody (1: 50, Santa Cruz, sc-23900) at 4 °C overnight. After three more washes with PBS, cells were incubated with Alexa Fluor®488 goat anti-mouse IgG (1: 500, Life technology, A11001) for 60 min at room temperature in the dark. Nuclear stain TO-PRO-3 iodide (1: 500, Life technologies) was added 15 min before imaging. Images were acquired using a confocal microscope (LSM510, Zeiss) with a 40 X plain objective lens and captured using ZEN2009 software (Zeiss).
2.8. ATP measurement
Extracellular ATP was quantified by using the Luminiscent ATP detection assay kit (Abcam, ab113849) that quantifies the amount of light emitted by luciferin when oxidized by luciferase in the presence of ATP and oxygen. Cells were plated into a 12-well plate and grown to 90% confluence in complete medium, washed twice with PBS and then incubated in FBS free medium containing 100 μM ARL67156 for 4 h. Medium was collected and centrifuged at 1000 rpm for 5 min in order to pellet floating cells and used immediately for the ATP assay according to manufacturer protocol. ATP standards were loaded on the same plate as references. Luminiscence was determined by SpectraMax M-3 Multi-Mode Microplate Readers (Molecular Devices). ATP levels were normalized to cell number and compared to corresponding controls.
2.9. Cell viability assays
Cells were stained with propidium iodide (PI, Sigma, P-4170) (10 μg/ml) and incubated at room temperature in PBS for 15 min. Cells were then examined immediately by fluorescence microscopy. For quantitative analysis, adherent cells were collected by trypsinization, and PI+ cells were counted using a hemocytometer under an upright fluorescent microscope.
2.10. Cell invasion assays
5× 104 cells in 500 μl serum-free DMEM were placed into the upper chamber of a 24-well Transwell insert (8 μm pore size; Corning) coated with 25 μg of Matrigel (Corning), and then 750 μl complete medium was added to the lower chamber. After 24 h of incubation, the remaining cells on the upper membrane were removed with cotton wool, whereas the cells that had migrated or invaded through the membrane were stained with the 1% methylene blue. Cells were counted from the digitized images using Image J. All assays were performed in triplicate.
2.11. Cell migration assays
Cell migration was assessed using the wounding-healing assay. Briefly, cells were grown up to 95% confluence. The growth medium was replaced with medium containing 0.5% FBS and the monolayer of cells was scratched using a 200 μl pipette tip. Wound width was monitored over time by microscopy. Percentage wound recovery was expressed as follows: [1-(Width of the wound at a given time/width of the wound at t = 0)] ×100%. For live imaging, cells were grown and wounded as mentioned above. Cells were then incubated in medium containing 0.5% FBS (37 °C, 5% CO2), and cell movement was tracked over 17 h with 3 min intervals under AxioObserver.Z1 wide-field microscope (Zeiss) equipped with a 10 × objective and an Axiocam MRm camera (Zeiss). The total path length for each track was analyzed using Manual Tracking in ImageJ.
2.12. In vivo experimental manipulation
Six-week-old female NOD/SCID mice were purchased from Charles River and separated into two groups: Scramble control (n = 7) and TRPML1 KD (n = 12). MDA-MB-231 cells (2 × 106) expressing ML1 shRNA1 or control shRNA were subcutaneously injected into the mammary fat pads (fat pad number 4) of 8-week-old NOD/SCID mice. Palpable tumors (~ 3 weeks post-injection) were measured every 2 days for a period of 4 weeks using a standard caliper. Mice were euthanized at the selected time point and tumors were extracted. Tumor volume was calculated using the ½ x length x width2 formula. Lymph node metastases was physically assessed at the time of tumor extraction. Data acquired from this experiment was plotted using the GraphPad Prism software. This procedure was performed under the approval of the Dalhousie Animal Ethics Committee, Halifax, NS, Canada.
2.13. Data analysis
Data are presented as mean ± SEM. Statistical comparisons were made using analysis of variance (ANOVA) and Student’s t test. Results presented are mean ± SEM. P values of < 0.05 were considered statistically significant. *: P < 0.05, **: P < 0.01, ***: P < 0.005.
3. Results
3.1. ML1 is highly expressed in metastatic breast cancer cells and breast tumors
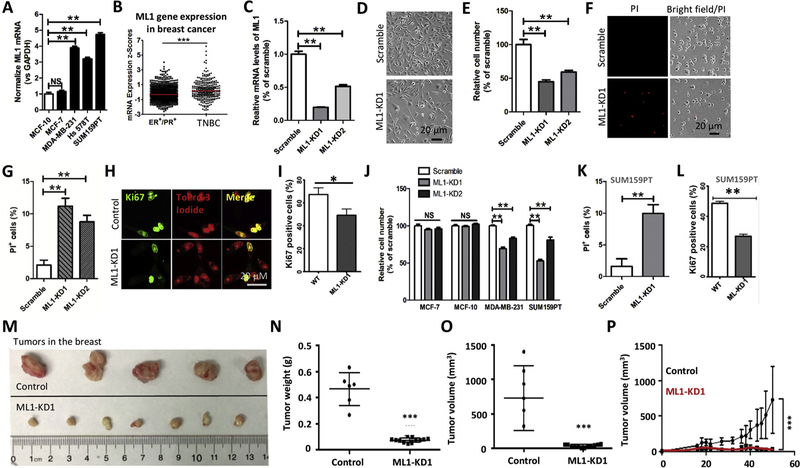
We first investigated the expression profile of ML1 in non-tumorigenic MCF-10 A cells and several breast cancer cell lines, including the non-metastatic ER+/PR+ breast cancer cell line MCF-7 and three metastatic TNBC cell lines MDA-MB-231, Hs 578 T and SUM159PT [50,51]. ML1 expression levels were gauged by quantitative PCR with reverse transcription (RT-PCR). ML1 mRNA was found to be specifically upregulated in aggressive, metastatic TNBC cell lines including MDA-MB-231, Hs 578 T and SUM159PT relative to MCF-7 and MCF10 A (Fig. 1A), suggesting a specific role of ML1 in TNBC development. Analysis of breast cancer dataset (METABRIC) showed higher mRNA expression of ML1 in TNBC compared to ER+/PR+ cancer patients (Fig. 1B). The enhanced expression of ML1 in TNBC cell lines and primary tumors suggested that ML1 might be implicated in TNBC progression.
Fig. 1. ML1 ablation inhibits TNBC development.
(A) ML1 mRNA levels in different breast cancer cell lines were determined using qPCR. ML1 mRNA was highly increased in metastatic breast cancer cell lines relative to MCF10 A and MCF7. (B) Higher ML1 mRNA levels in TNBC patients compared with ER+/PR+ breast cancer patients (METABRIC). (C) ML1 knockdown in MDA-MB-231 cells that were transduced with lentivirus expressing shRNA against ML1. The efficacy for the two shRNAs in reducing ML1 gene expression was measured using qPCR. ML1 mRNA expression were normalized against GAPDH. (D, E) ML1-KD reduced the number of MDA-MB-231 cells. Cell number was measured by direct count of cells at 72 h. Assays were performed in triplicates for each condition. (F, G) ML1 knockdown dramatically increased MDA-MB-231 cell death that was revealed by Propidium Iodide (PI) staining. Cells were cultured in DMEM containing 0.5% FBS for 48 h. (H, I) ML1-KD inhibited MDA-MB-231 cell proliferation as determined by Ki67 immunostaining. (J) ML1-KD reduced the number of TNBC cells but not MCF-7 and MCF-10 cells. Cell number was examined by direct count at 24 h. (K, L) ML1-KD increased cell death while decreasing cell proliferation in SUM159PT cells. (M–P) in vivo characterization of ML1-KD on tumor growth. ML1-KD leads to a smaller tumor and slower growth rate as compared to control tumors. Results presented are mean ± SEM. *: P < 0.05, **: P < 0.01, ***: P < 0.005.
3.2. ML1 knockdown reduces TNBC cell proliferation, cell viability and tumor growth
We generated stable MDA-MB-231 cells with knockdown of ML1 (ML1-KD) [48] (Fig. 1C). Knockdown of ML1 caused a decrease in the overall cell number of MDA-MB-231 cells, as evaluated by direct cell counting (Fig. 1D, 1E). A reduction in cell number in ML1-KD cells could be due to either a decreased cell proliferation or an increased cell death. To differentiate these possibilities, we monitored cell death using Propidium Iodide (PI) staining and cell proliferation using Ki67 staining. We found that knockdown of ML1 caused an increase in cell death (Fig. 1F, 1 G) and a decrease in cell proliferation (Fig. 1H, 1I). The effect of ML1-KD on cell viability and proliferation were recapitulated in another TNBC cell line, SUM159PT cells (Fig. 1J–1 L). However, ML1 deletion in MCF10 A and MCF7 did not affect cell proliferation and/or cell viability (Fig. 1J). Altogether, these data suggest that ML1-KD specifically suppresses cell proliferation and viability in metastatic TNBC cell lines.
To assess whether ML1 regulates TNBC growth in vivo [1,3,52–54], we performed an orthotopic xenograft model of breast cancer cells in SCID/NOD mice [55–57]. MDA-MB-231 cells (2 × 106) expressing ML1 shRNA1 or control shRNA were subcutaneously injected into the mammary fat pads (fat pad number 4) of 8-week-old NOD/SCID mice (51–53). Three weeks later tumor growth was monitored over a period of 4 weeks. ML1-KD reduced tumor volume and tumor weight when compared to control cells expressing scrambled shRNA (Fig. 1M–1 P). These in vivo data are consistent with our in vitro observations, and strongly indicate that ML1 is essential for TNBC tumor growth.
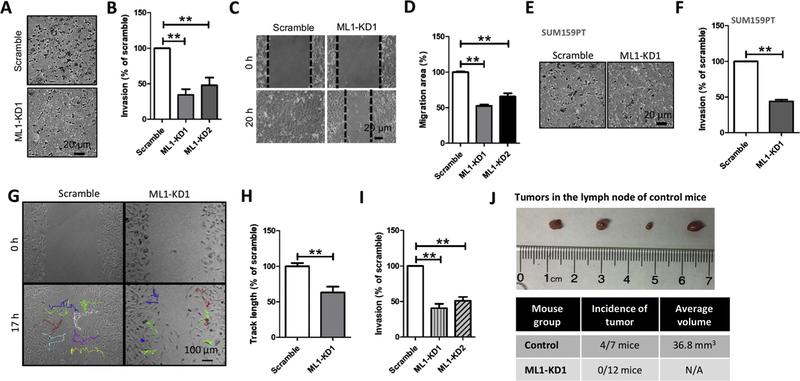
3.3. ML1 knockdown reduces TNBC cell invasion
Despite recent advances in breast cancer therapy, cancer metastasis, which is contributed to enhanced cell invasion, remains the leading cause of breast cancer death [58–61]. We determined the impact of ML1-KD on MDA-MB-231 cell invasion using in vitro Matrigel invasion assays. ML1 deletion significantly suppressed MDA-MB-231 cell invasion (Fig. 2A, 2B). Furthermore, ML1 deletion also inhibited MDA-MB- 231 cell migration examined by wound-healing assays (Fig. 2C, 2D). The reduction in cell invasion by ML1 knockdown was also detected in SUM159PT cells expressing ML1-shRNA1 (Fig. 2E, 2 F). Because knockdown of ML1 inhibited cell proliferation and viability, the effects of ML1-KD on cell migration and invasion could be secondary to its effects on proliferation and survival. To exclude this possibility, we monitored the migration of individual cells using time-lapse cell imaging [62] and found that ML1 knockdown inhibited motility of individual TNBC cells (Fig. 2G, 2 H). Furthermore, despite increasing cell numbers of the ML1-KD group by 15% over the control group (Fig. 1G), significantly less ML1-KD cells passed through Matrigel (Fig. 2I), suggesting that the effect of ML1-KD on cell migration and invasion is not the result of reduced proliferation or survival. Importantly, the xenograft model showed no visible tumors in lymph nodes of the ML1-KD MDA-MB-231-injected group of mice compared to mice injected with control scrambled shRNA-expressing MDA-MB-231 cells (0/12 for ML1-KD vs 4/7 for Scrambled shRNA) (Fig. 2J) [2,5,63,64], suggesting that knockdown of ML1 in MDA-MB-231 cells inhibits cancer cell invasion.
Fig. 2. ML1 knockdown inhibits MDA-MB-231 cells migration and invasion.
(A, B) MDA-MB-231 cells with ML1-KD showed significantly reduced invasion relative to control cells. Cancer cell invasion was examined using Matrigel invasion assays. (C, D) ML1-KD in MDA-MB-231 cells significantly reduced cell migration that was examined using wound healing assay. (E, F) ML1-KD in SUM159PT cells significantly reduced cell invasion. (G, H) time-lapse cell imaging indicated that MDA-MB-231 cell motility was inhibited by ML1-KD. (I) Compared to control, invasion of ML1-KD MDA-MB-231 cells was significantly reduced. To compensate for reduced cell viability of the ML1-KD group, 15% more ML1-KD cells were added relative to the scrambled shRNA control group at time 0. (J) MDA-MB-231 cells with ML1-KD failed to show presence of tumors in Lymph Nodes. Tumors in lymph Node were detected in 4 out of 7 control mice (36.8 mm3, n = 4) but not in mice injected with ML1-KD MDA-MB-231 cells. All graphs represent mean ± SEM with at least three biological replicates. **: P < 0.01.
3.4. ML1 controls cancer development through regulating mTORC1
mTORC1 pathway plays an important role in the regulation of cell proliferation, metabolism and survival. Hyperactivation of mTORC1 is frequently observed in breast cancer and has been linked to cancer growth [13,14,16–19,21,65,66]. Targeted therapy against mTORC1 has been shown to decrease tumor growth in vivo model systems [67,68], including TNBC [5,18] and several agents that inhibit mTORC1 are currently in clinical trials for the treatment of multiple cancer types, including breast cancer [69,70].
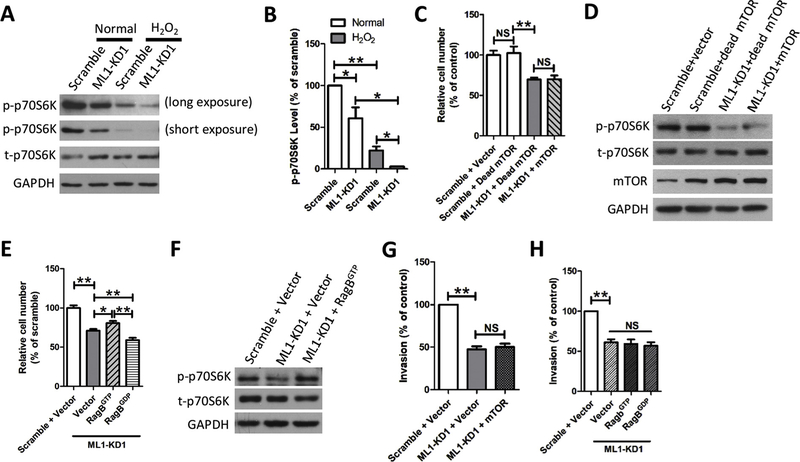
Our previous study demonstrated that ML1 promotes mTORC1 activity under stress but not under normal conditions [39]. This regulation of mTORC1 is essential for cells to survive stressful conditions such as nutrient deprivation known to cause autophagic cell death [8,39,71–73]. Given that tumor growth requires abundance of energy and nutrients, cancer cells constantly face stressful conditions, such as nutrient starvation and enhanced oxidative stress. We tested the idea that ML1 may help cancer cell survival by promoting mTORC1 activity. Indeed, deletion of ML1 in MDA-MB-231 cells reduced mTORC1 activity and increased oxidative stress through H2O2 treatment further reduced mTORC1 activity (Fig. 3A, 3B). Recently, we demonstrated that ML1 is required for mTOR recruitment onto lysosomes [39]. In agreement with this, decreased cell proliferation (Fig. 3C) observed in MDA-MB-231 cells with ML1-KD was not rescued by mTORC1 overexpression (Fig. 3C, 3D). In contrast, RagBGTP (Fig. 3E–3 G), a GTP-bound Rag mutant that retains mTORC1 on the lysosomal membrane [46,74], caused higher cell proliferation in ML1-KD MDA-MB-231 cells compared to control vector and RagBGDP, a GDP-bound Rag mutant that prevents mTORC1 from being recruited to the lysosomal surfaces [46,74,75].
Fig. 3. ML1 controls TNBC cell proliferation through regulating mTORC1.
(A, B) ML1-KD reduced mTORC1 activity in MDA-MB-231 cells. mTORC1 activity was examined by measuring phosphorylation of p70-S6K on Thr-389 (p-p70-S6K) using Western blotting. GAPDH was used as a loading control. ML1 deletion inhibited mTORC1 activity, and this was further suppressed by H2O2 (1 mM, 1 h). (C, D) The reduction in MDA-MB-231 cell number induced by ML1-KD was not rescued by overexpression of either active mTOR or dead mTOR. MDA-MB-231 cells were cultured in DMEM containing 10% FBS for 24 h. (E, F) The reduction in MDA-MB-231 cell number induced by ML1-KD was partially rescued by overexpression of RagBGTP, but not by overexpression of RagBGDP. (G) mTOR overexpression did not rescue the decrease in MDA-MB-231 cell invasion induced by ML1-KD. (H) Comparable ML1-KD MDA-MB-231 cell invasion between RagBGTP and RagBGDP. All graphs represent mean ± SEM. of at least three biological replicates. NS: no significance, *: P < 0.05, **: P < 0.01.
In addition to its role in cancer cell proliferation and growth, suppression of mTORC1 signaling pathway has been suggested to inhibit cancer cell invasion and metastasis [13,14]. However, neither mTORC1 overexpression (Fig. 3H) nor RagBGTP (Fig. 3I) rescued the decrease in the invasion of ML1 deficient MDA-MB-231 cells, suggesting that ML1-dependent mTORC1 activation promotes TNBC cell proliferation and growth but not invasion.
3.5. ML1 controls TNBC invasion by facilitating lysosomal ATP exocytosis
In light of our findings thus far, we reasoned that the mTORC1-independent role of ML1 in invasiveness of TNBCs could be the result of ML1-dependent control of extracellular ATP levels through lysosomal exocytosis. This is based on the following: first, increased levels of extracellular ATP [24–32] and adenosine [76,77] at tumor sites increase migration, invasion and metastasis of some cancer cells; second, lysosomal ATP (~ 1 mM) [34,35,78] and adenosine (~1 mM) [79] can be released into the extracellular space via lysosomal exocytosis to elevate extracellular ATP levels [35–37]; third, lysosomal exocytosis has been implicated in tumor cell migration, invasion and metastasis [36,38]; fourth, ML1 is essential for lysosomal exocytosis [44]; fifth, highly metastatic MDA-MB-231 breast cancer cells release higher levels of ATP compared to MCF-7 breast cancer cells or normal epithelial breast MCF10 A cells [32].
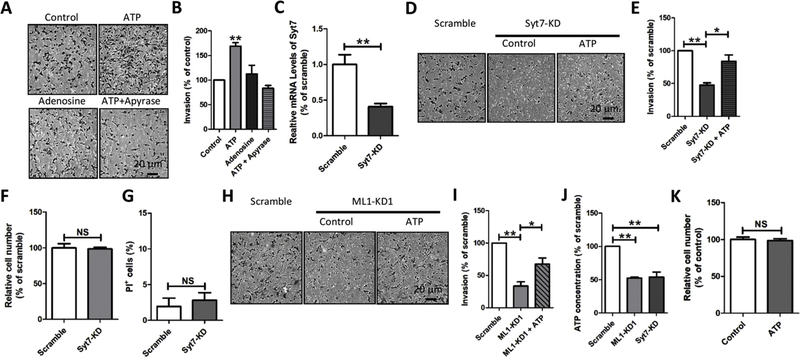
Addition of ATP (200 μM), but not adenosine (200 μM), promoted MDA-MB-231 cell invasion (Fig. 4A, 4B). The effect of ATP on MDA-MB-231 cell invasion was eliminated by promoting ATP degradation using apyrase (Fig. 4A, 4B), suggesting that extracellular ATP, but not adenosine, facilitates MDA-MB-231 cell invasion.
Fig. 4. ML1 controls invasion through regulating lysosomal exocytosis.
(A, B) External ATP (200 μM), but not adenosine (200 μM), dramatically increased the invasion of MDA-MB-231 cells. The effect of ATP was eliminated by apyrase (5 U/ml). (C–E) Synaptotagmin 7 (Syt7) knockdown (Syt7-KD) inhibited MDA-MB-231 cell invasion, and this was partially rescued by ATP (200 μM). (F) Syt7-KD did not alter the number of MDA-MB-231. (G) Syt7-KD had no effect on MDA-MB-231 cell viability. (H, I) ATP (200 μM) partially rescued the reduced invasion of MDA-MB-231 cells caused by ML1-KD. (J) ML1-KD reduced ATP levels in culture medium, suggesting that ML1-KD inhibits lysosomal ATP release. (K) ATP had no effect on number of MDA-MB-231 cells. All graphes show mean ± SEM from triple biological replicates. *: P < 0.05, **: P < 0.01.
Elevated ATP in tumor microenvironments could be attributed to either ATP transport pathways on the plasma membrane (PM) or vesicular exocytosis [80–83]. To test whether ML1 promotes MDA-MB-231 cancer development through facilitating lysosomal ATP release, lysosomal exocytosis was inhibited by deleting synaptotagmin 7 (Syt7) [42,49]. As expected, Syt7 knockdown (Syt7-KD, Fig. 4C) suppressed MDA-MB-231 cell invasion (Fig. 4D, 4E). Interestingly, decreased invasion of MDA-MB-231 cells with Syt7-KD was partially rescued by addition of external ATP (Fig. 4D–4E). Further, the reduced invasion induced by Syt7-KD was not attributed to a defect in either cell proliferation (Fig. 4F) or viability (Fig. 4G). Inhibition of MDA-MB-231 cell invasion by ML1-KD was partially rescued by addition of external ATP (Fig. 4H, 4I) and ML1-KD reduced ATP levels in the culture medium (Fig. 4J), strongly arguing that elevated ML1 in MDA-MB-231 cell promotes cell invasion through enhanced lysosomal ATP release via lysosomal exocytosis.
Because ATP is known to control cell proliferation [84], we considered the possibility that, in addition to enhancement of mTORC1 activity (e.g. Fig. 3), ML1 also promotes MDA-MB-231 cell proliferation through lysosomal ATP release. However, direct application of ATP did not increase MDA-MB-231 cell proliferation (Fig. 4K) in agreement with previous reports [85,86], suggesting that elevated ML1 levels promote TNBC growth by increasing mTORC1 activity while facilitating TNBC invasion through enhanced lysosomal ATP release.
3.6. An ML1 inhibitor impairs TNBC cell proliferation and invasion
To assess the therapeutic relevance of our findings, we investigated whether the proliferation of MDA-MB-231 cells was sensitive to ML1 inhibitors. As shown in Fig. 5A and 5B, the ML1 inhibitor ML-SI1 (20 μM) significantly reduced MDA-MB-231 cell number (Fig. 5C, 5D, 5 G) and increased cell death (Fig. 5E, 5 F, 5 G). The effects of ML-SI1 (20 μM) on proliferation were recapitulated in another TNBC cell line, SUM159TP cells (Fig. 5G). However, ML-SI1 (20 μM) only marginally affected MCF7 cell proliferation (Fig. 5G). Further, ML1 inhibition significantly suppressed TNBC cell invasion (Fig. 5H, 5I) and migration in vitro (Fig. 5J, 5 M). Altogether, our data strongly implicate ML1 as a potential drug target for therapy of TNBCs.
Fig. 5. The ML1 inhibitor ML-SI1 suppresses TNBC cell growth and invasion.
(A, B) ML1 inhibitor ML-SI1 suppressed the number of MDA-MB-231 cells. MDA-MB-231 cells were cultured in DMEM containing 10% FBS and ML-SI1 (20 μM) for 24 h. Assays were performed in duplicates using five independent cultures. (C, D) ML-SI1 (20 μM) significantly inhibited MDA-MB-231 cell proliferation. (E, F) ML-SI1 (20 μM) significantly increased the percentage of MDA-MB-231 cell death. MDA-MB-231 cells were cultured in DMEM containing 0.5% FBS for 48 h. Cell death was examined using PI staining. (G) ML-SI1 (20 μM) reduced the cell number of TNBC cells but not MCF-7 cells. (H, I) ML-SI1 (20 μM) treatment significantly inhibited TNBC cell invasion. (J–M) ML-SI1 (20 μM) significantly decreased migration of MDA-MB-231 cells as determined using wound healing assay. All graphs represent mean ± SEM. of at least three biological replicates. *: P < 0.05, **: P < 0.01.
4. Discussion
Although TNBC is the most aggresive malignant cancercausing high mortality rates among women, therapeutic options for this disease remain limited. This is partially due to the lack of our understanding of key pathways associated with TNBCs. In this study, we report that ML1 is specifically elevated in TNBC cell lines and patients and is associated with increased mTORC1 activity and lysosomal ATP release into the extracellular space to promote TNBC growth and invasion.
In agreement with previous studies [24–32], we also show that increased levels of extracellular ATP promotes cancer cell invasion. While we cannot exclude the involvement of ATP transport pathways across the plasma membrane [80–83], our data shows for the first time that the lysosome is one of the sources of ATP that benefits cancer cell invasion. Intriguingly, some studies suggested that ATP may act by increasing extracellular levels of adenosine [76], the ATP degradation product [87,88]. However, our studies showed that direct application of adenosine or co-application of ATP and apyrase has no effect on TNBC cell invasion, suggesting that adenosine does not play a major role in TNBC cell invasion. We showed that a reduction in MDA-MB-231 cell invasion by Syt7-KD and ML1-KD is bypassed by direct application of ATP. Our data strongly implicate ML1 in promoting MDA-MB-231 cell invasion by facilitating lysosomal ATP release [34,35,78] via lysosomal exocytosis [35–38].
Drugs targeting mTORC1 have attracted great attention recently as anti-cancer therapies. However, mTORC1 inhibitors failed to provide substantial benefits in cancer patients. This is due, at least partially, to the fact that mTORC1 is essential for the function of all cell types. Interestingly, ML1 is normally silent when nutrients are abundant due to its phosphorylation by mTORC1 [39,89]. Cellular stresses such as energy and nutrient starvation [39] and oxidative stress [90] activate ML1, likely through de-phosphorylation [39,89]. Because rapidly growing tumor cells are tightly packed, they quickly exhaust their available supplies of oxygen and nutrients, which leads to increased nutrient depravation and oxidative stress. These may activate ML1 to support cells to survive extreme conditions through increase of mTORC1 activity and enhanced extracellular ATP levels [32,91]. Therefore, ML1 may function as a sensor of the surrounding tumor microenvironment, which initiates adaptive signaling necessary for surviving hostile microenvironments. Because ML1 is specifically activated by cellular stresses that are characteristic of tumor milieu, antagonists of ML1 could represent anticancer drugs with more specificity and potency. In this regard, our research could have a major impact on cancer drug development, particularly for TNBCs. Our studies may also have a broader implication in improving our understanding of the pathogenesis and treatment of other cancers.
Acknowledgements
This work was supported by DMRF Equipment Grant, CIHR grant (PJT-156102), and CFI Leaders Opportunity Fund-Funding for research infrastructure (29291). We thank Mitsunori Fukuda for the Syt-VII-DN plasmid and Haoxing Xu for his constant support. We appreciate the encouragement and helpful comments from other members of the Dong laboratory and El Hiani laboratories.
Footnotes
Conflicts of interest statement
The authors declare that there is no conflict of interest.
References
- [1].Ma L, Teruya-Feldstein J, Weinberg RA, Tumour invasion and metastasis initiated by microRNA-10b in breast cancer, Nature 449 (7163) (2007) 682–688. [DOI] [PubMed] [Google Scholar]
- [2].Lin L, Chen YS, Yao YD, Chen JQ, Chen JN, Huang SY, Zeng YJ, Yao HR, Zeng SH, Fu YS, et al. , CCL18 from tumor-associated macrophages promotes angiogenesis in breast cancer, Oncotarget 6 (33) (2015) 34758–34773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wang Y, Zhang T, Kwiatkowski N, Abraham BJ, Lee TI, Xie S, Yuzugullu H, Von T, Li H, Lin Z, et al. , CDK7-dependent transcriptional addiction in triple-negative breast cancer, Cell 163 (1) (2015) 174–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ooms LM, Binge LC, Davies EM, Rahman P, Conway JR, Gurung R, Ferguson DT, Papa A, Fedele CG, Vieusseux JL, et al. , The inositol polyphosphate 5-phosphatase PIPP regulates AKT1-dependent breast cancer growth and metastasis, Cancer Cell 28 (2) (2015) 155–169. [DOI] [PubMed] [Google Scholar]
- [5].Britschgi A, Andraos R, Brinkhaus H, Klebba I, Romanet V, Muller U, Murakami M, Radimerski T, Bentires-Alj M, JAK2/STAT5 inhibition circumvents resistance to PI3K/mTOR blockade: a rationale for cotargeting these pathways in metastatic breast cancer, Cancer Cell 22 (6) (2012) 796–811. [DOI] [PubMed] [Google Scholar]
- [6].Yam C, Mani SA, Moulder SL, Targeting the molecular subtypes of triple negative breast cancer: understanding the diversity to progress the field, Oncologist 22 (9) (2017) 1086–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, Ollila DW, Sartor CI, Graham ML, Perou CM, The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes, Clin. Cancer Res 13 (8) (2007) 2329–2334. [DOI] [PubMed] [Google Scholar]
- [8].Maiuri MC, Zalckvar E, Kimchi A, Kroemer G, Self-eating and self-killing: crosstalk between autophagy and apoptosis, Nat. Rev. Mol. Cell Biol 8 (9) (2007) 741–752. [DOI] [PubMed] [Google Scholar]
- [9].Liu Y, Levine B, Autosis and autophagic cell death: the dark side of autophagy, Cell Death Differ 22 (3) (2015) 367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Saxton RA, Sabatini DM, mTOR signaling in growth, metabolism, and disease, Cell 169 (2) (2017) 361–371. [DOI] [PubMed] [Google Scholar]
- [11].Bar-Peled L, Sabatini DM, Regulation of mTORC1 by amino acids, Trends Cell Biol 24 (7) (2014) 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Laplante M, Sabatini DM, mTOR signaling in growth control and disease, Cell 149 (2) (2012) 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Hsieh AC, Liu Y, Edlind MP, Ingolia NT, Janes MR, Sher A, Shi EY, Stumpf CR, Christensen C, Bonham MJ, et al. , The translational landscape of mTOR signalling steers cancer initiation and metastasis, Nature 485 (7396) (2012) 55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wander SA, Zhao D, Besser AH, Hong F, Wei J, Ince TA, Milikowski C, Bishopric NH, Minn AJ, Creighton CJ, et al. , PI3K/mTOR inhibition can impair tumor invasion and metastasis in vivo despite a lack of antiproliferative action in vitro: implications for targeted therapy, Breast Cancer Res. Treat 138 (2) (2013) 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cargnello M, Tcherkezian J, Roux PP, The expanding role of mTOR in cancer cell growth and proliferation, Mutagenesis 30 (2) (2015) 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ueng SH, Chen SC, Chang YS, Hsueh S, Lin YC, Chien HP, Lo YF, Shen SC, Hsueh C, Phosphorylated mTOR expression correlates with poor outcome in early-stage triple negative breast carcinomas, Int. J. Clin. Exp. Pathol 5 (8) (2012) 806–813. [PMC free article] [PubMed] [Google Scholar]
- [17].Walsh S, Flanagan L, Quinn C, Evoy D, McDermott EW, Pierce A, Duffy MJ, mTOR in breast cancer: differential expression in triple-negative and non-triple-negative tumors, Breast 21 (2) (2012) 178–182. [DOI] [PubMed] [Google Scholar]
- [18].Chen Y, Wei H, Liu F, Guan JL, Hyperactivation of mammalian target of rapamycin complex 1 (mTORC1) promotes breast cancer progression through enhancing glucose starvation-induced autophagy and Akt signaling, J. Biol. Chem 289 (2) (2014) 1164–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hatem R, El Botty R, Chateau-Joubert S, Servely JL, Labiod D, de Plater L, Assayag F, Coussy F, Callens C, Vacher S, et al. , Targeting mTOR pathway inhibits tumor growth in different molecular subtypes of triple-negative breast cancers, Oncotarget (2016). [DOI] [PMC free article] [PubMed]
- [20].Paplomata E, O’Regan R, The PI3K/AKT/mTOR pathway in breast cancer: targets, trials and biomarkers, Ther. Adv. Med. Oncol 6 (4) (2014) 154–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wander SA, Hennessy BT, Slingerland JM, Next-generation mTOR inhibitors in clinical oncology: how pathway complexity informs therapeutic strategy, J. Clin. Invest 121 (4) (2011) 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Marty B, Maire V, Gravier E, Rigaill G, Vincent-Salomon A, Kappler M, Lebigot I, Djelti F, Tourdes A, Gestraud P, et al. , Frequent PTEN genomic alterations and activated phosphatidylinositol 3-kinase pathway in basal-like breast cancer cells, Breast Cancer Res 10 (6) (2008) R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Shaw RJ, Cantley LC, Ras PI (3)K and mTOR signalling controls tumour cell growth, Nature 441 (7092) (2006) 424–430. [DOI] [PubMed] [Google Scholar]
- [24].Di Virgilio F, Adinolfi E, Extracellular purines, purinergic receptors and tumor growth, Oncogene (2016). [DOI] [PMC free article] [PubMed]
- [25].Di Virgilio F, Purines, purinergic receptors, and cancer, Cancer Res 72 (21) (2012) 5441–5447. [DOI] [PubMed] [Google Scholar]
- [26].Pellegatti P, Raffaghello L, Bianchi G, Piccardi F, Pistoia V, Di Virgilio F, Increased level of extracellular ATP at tumor sites: in vivo imaging with plasma membrane luciferase, PLoS One 3 (7) (2008) e2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang Y, Gong LH, Zhang HQ, Du Q, You JF, Tian XX, Fang WG, Extracellular ATP enhances in vitro invasion of prostate cancer cells by activating Rho GTPase and upregulating MMPs expression, Cancer Lett 293 (2) (2010) 189–197. [DOI] [PubMed] [Google Scholar]
- [28].Shi K, Queiroz KC, Stap J, Richel DJ, Spek CA, Protease-activated receptor-2 induces migration of pancreatic cancer cells in an extracellular ATP-dependent manner, J. Thromb. Haemost 11 (10) (2013) 1892–1902. [DOI] [PubMed] [Google Scholar]
- [29].Li WH, Qiu Y, Zhang HQ, Liu Y, You JF, Tian XX, Fang WG, P2Y2 receptor promotes cell invasion and metastasis in prostate cancer cells, Br. J. Cancer 109 (6) (2013) 1666–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Schumacher D, Strilic B, Sivaraj KK, Wettschureck N, Offermanns S, Platelet-derived nucleotides promote tumor-cell transendothelial migration and metastasis via P2Y2 receptor, Cancer Cell 24 (1) (2013) 130–137. [DOI] [PubMed] [Google Scholar]
- [31].Furlow PW, Zhang S, Soong TD, Halberg N, Goodarzi H, Mangrum C, Wu YG, Elemento O, Tavazoie SF, Mechanosensitive pannexin-1 channels mediate microvascular metastatic cell survival, Nat. Cell Biol 17 (7) (2015) 943–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Jin H, Eun SY, Lee JS, Park SW, Lee JH, Chang KC, Kim HJ, P2Y2 receptor activation by nucleotides released from highly metastatic breast cancer cells increases tumor growth and invasion via crosstalk with endothelial cells, Breast Cancer Res 16 (5) (2014) R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM, mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase, Science 334 (6056) (2011) 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Cao Q, Zhao K, Zhong XZ, Zou Y, Yu H, Huang P, Xu TL, Dong XP: SLC17A9 protein functions as a lysosomal ATP transporter and regulates cell viability, J. Biol. Chem 289 (33) (2014) 23189–23199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang Z, Chen G, Zhou W, Song A, Xu T, Luo Q, Wang W, Gu XS, Duan S: regulated ATP release from astrocytes through lysosome exocytosis, Nat. Cell Biol 9 (8) (2007) 945–953. [DOI] [PubMed] [Google Scholar]
- [36].Liu Y, Zhou Y, Zhu K, Inhibition of glioma cell lysosome exocytosis inhibits glioma invasion, PLoS One 7 (9) (2012) e45910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Takai E, Tsukimoto M, Harada H, Sawada K, Moriyama Y, Kojima S, Autocrine regulation of TGF-beta1-induced cell migration by exocytosis of ATP and activation of P2 receptors in human lung cancer cells, J. Cell. Sci 125 (Pt 21) (2012) 5051–5060. [DOI] [PubMed] [Google Scholar]
- [38].Machado E, White-Gilbertson S, van de Vlekkert D, Janke L, Moshiach S, Campos Y, Finkelstein D, Gomero E, Mosca R, Qiu X, et al. , Regulated lysosomal exocytosis mediates cancer progression, Sci. Adv 1 (11) (2015) e1500603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sun X, Yang Y, Zhong XZ, Cao Q, Zhu XH, Zhu X, Dong XP, A negative feedback regulation of MTORC1 activity by the lysosomal Ca(2+) channel MCOLN1 (mucolipin 1) using a CALM (calmodulin)-dependent mechanism, Autophagy (2018) 1–15. [DOI] [PMC free article] [PubMed]
- [40].Li RJ, Xu J, Fu C, Zhang J, Zheng YG, Jia H, Liu JO, Regulation of mTORC1 by lysosomal calcium and calmodulin, eLife (2016) 5. [DOI] [PMC free article] [PubMed]
- [41].Wong CO, Li R, Montell C, Venkatachalam K, Drosophila TRPML is required for TORC1 activation, Curr. Biol 22 (17) (2012) 1616–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Cao Q, Zhong XZ, Zou Y, Zhang Z, Toro L, Dong XP, BK channels alleviate lysosomal storage diseases by providing positive feedback regulation of lysosomal Ca2+ release, Dev. Cell 33 (4) (2015) 427–441. [DOI] [PubMed] [Google Scholar]
- [43].Samie M, Wang X, Zhang X, Goschka A, Li X, Cheng X, Gregg E, Azar M, Zhuo Y, Garrity AG, et al. , A TRP channel in the lysosome regulates large particle phagocytosis via focal exocytosis, Dev. Cell 26 (5) (2013) 511–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Samie MA, Xu H, Lysosomal exocytosis and lipid storage disorders, J. Lipid Res 55 (6) (2014) 995–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM, Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids, Cell 141 (2) (2010) 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM, The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1, Science 320 (5882) (2008) 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Vilella-Bach M, Nuzzi P, Fang Y, Chen J, The FKBP12-rapamycin-binding domain is required for FKBP12-rapamycin-associated protein kinase activity and G1 progression, J. Biol. Chem 274 (7) (1999) 4266–4272. [DOI] [PubMed] [Google Scholar]
- [48].Miao Y, Li G, Zhang X, Xu H, Abraham SN, A TRP channel senses lysosome neutralization by pathogens to trigger their expulsion, Cell 161 (6) (2015) 1306–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hoshino D, Kirkbride KC, Costello K, Clark ES, Sinha S, Grega-Larson N, Tyska MJ, Weaver AM, Exosome secretion is enhanced by invadopodia and drives invasive behavior, Cell Rep 5 (5) (2013) 1159–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S, Gibb WJ, Wang NJ, Ziyad S, Tong F, et al. , Subtype and pathway specific responses to anticancer compounds in breast cancer, Proc. Natl. Acad. Sci. U. S. A 109 (8) (2012) 2724–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA, Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies, J. Clin. Invest 121 (7) (2011) 2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Liu J, Liao S, Diop-Frimpong B, Chen W, Goel S, Naxerova K, Ancukiewicz M, Boucher Y, Jain RK, Xu L, TGF-beta blockade improves the distribution and efficacy of therapeutics in breast carcinoma by normalizing the tumor stroma, Proc. Natl. Acad. Sci. U. S. A 109 (41) (2012) 16618–16623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Jessani N, Humphrey M, McDonald WH, Niessen S, Masuda K, Gangadharan B, Yates JR 3rd, Mueller BM, Cravatt BF, Carcinoma and stromal enzyme activity profiles associated with breast tumor growth in vivo, Proc. Natl. Acad. Sci. U. S. A 101 (38) (2004) 13756–13761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Marcato P, Dean CA, Liu RZ, Coyle KM, Bydoun M, Wallace M, Clements D, Turner C, Mathenge EG, Gujar SA, et al. , Aldehyde dehydrogenase 1A3 influences breast cancer progression via differential retinoic acid signaling, Mol. Oncol 9 (1) (2015) 17–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sharif T, Dai C, Martell E, Ghassemi-Rad MS, Hanes MR, Murphy PJ, Kennedy BE, Venugopal C, Subapanditha M, Giacomantonio CA, et al. , TAp73 modifies metabolism and positively regulates growth of Cancer Stem-Like cells in a redox-sensitive manner, Clin. Cancer Res (2018). [DOI] [PubMed]
- [56].Sultan M, Vidovic D, Paine AS, Huynh TT, Coyle KM, Thomas ML, Cruickshank BM, Dean CA, Clements DR, Kim Y, et al. , Epigenetic silencing of TAP1 in Aldefluor(+) breast Cancer stem cells contributes to their enhanced immune evasion, Stem Cells 36 (5) (2018) 641–654. [DOI] [PubMed] [Google Scholar]
- [57].Muscella A, Vetrugno C, Migoni D, Biagioni F, Fanizzi FP, Fornai F, DePascali SA, Marsigliante S, Antitumor activity of [Pt(O,O’-acac)(gamma-acac) (DMS)] in mouse xenograft model of breast cancer, Cell Death Dis 5 (2014) e1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Huang X, Jan LY, Targeting potassium channels in cancer, J. Cell Biol 206 (2) (2014) 151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chaffer CL, Weinberg RA, A perspective on cancer cell metastasis, Science 331 (6024) (2011) 1559–1564. [DOI] [PubMed] [Google Scholar]
- [60].Gupta GP, Massague J, Cancer metastasis: building a framework, Cell 127 (4) (2006) 679–695. [DOI] [PubMed] [Google Scholar]
- [61].Fidler IJ, The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited, Nat. Rev. Cancer 3 (6) (2003) 453–458. [DOI] [PubMed] [Google Scholar]
- [62].Cao Q, Zhong XZ, Zou Y, Murrell-Lagnado R, Zhu MX, Dong XP, Calcium release through P2X4 activates calmodulin to promote endolysosomal membrane fusion, J. Cell Biol 209 (6) (2015) 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Sharifi MN, Mowers EE, Drake LE, Collier C, Chen H, Zamora M, Mui S, Macleod KF, Autophagy promotes focal adhesion disassembly and cell motility of metastatic tumor cells through the direct interaction of Paxillin with LC3, Cell Rep 15 (8) (2016) 1660–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Maes H, Kuchnio A, Peric A, Moens S, Nys K, De Bock K, Quaegebeur A, Schoors S, Georgiadou M, Wouters J, et al. , Tumor vessel normalization by chloroquine independent of autophagy, Cancer Cell 26 (2) (2014) 190–206. [DOI] [PubMed] [Google Scholar]
- [65].Yao D, Wang P, Zhang J, Fu L, Ouyang L, Wang J, Deconvoluting the relationships between autophagy and metastasis for potential cancer therapy, Apoptosis 21 (6) (2016) 683–698. [DOI] [PubMed] [Google Scholar]
- [66].Murugan AK, Alzahrani A, Xing M, Mutations in critical domains confer the human mTOR gene strong tumorigenicity, J. Biol. Chem 288 (9) (2013) 6511–6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Chresta CM, Davies BR, Hickson I, Harding T, Cosulich S, Critchlow SE, Vincent JP, Ellston R, Jones D, Sini P, et al. , AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity, Cancer Res 70 (1) (2010) 288–298. [DOI] [PubMed] [Google Scholar]
- [68].Lu CH, Wyszomierski SL, Tseng LM, Sun MH, Lan KH, Neal CL, Mills GB, Hortobagyi GN, Esteva FJ, Yu D, Preclinical testing of clinically applicable strategies for overcoming trastuzumab resistance caused by PTEN deficiency, Clin. Cancer Res 13 (19) (2007) 5883–5888. [DOI] [PubMed] [Google Scholar]
- [69].Courtney KD, Corcoran RB, Engelman JA, The PI3K pathway as drug target in human cancer, J. Clin. Oncol 28 (6) (2010) 1075–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Alvarez RH, Valero V, Hortobagyi GN, Emerging targeted therapies for breast cancer, J. Clin. Oncol 28 (20) (2010) 3366–3379. [DOI] [PubMed] [Google Scholar]
- [71].Fulda S, Kogel D, Cell death by autophagy: emerging molecular mechanisms and implications for cancer therapy, Oncogene (2015). [DOI] [PubMed]
- [72].Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. , Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition), Autophagy 12 (1) (2016) 1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Levine B, Kroemer G, Autophagy in the pathogenesis of disease, Cell 132 (1) (2008) 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Cang C, Zhou Y, Navarro B, Seo YJ, Aranda K, Shi L, Battaglia-Hsu S, Nissim I, Clapham DE, Ren D: mTOR regulates lysosomal ATP-sensitive two-pore Na(+) channels to adapt to metabolic state, Cell 152 (4) (2013) 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Xu H, Ren D, Lysosomal physiology, Annu. Rev. Physiol 77 (2015) 57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Zhou JZ, Riquelme MA, Gao X, Ellies LG, Sun LZ, Jiang JX, Differential impact of adenosine nucleotides released by osteocytes on breast cancer growth and bone metastasis, Oncogene 34 (14) (2015) 1831–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Fernandez-Gallardo M, Gonzalez-Ramirez R, Sandoval A, Felix R, Monjaraz E, Adenosine stimulate proliferation and migration in triple negative breast Cancer cells, PLoS One 11 (12) (2016) e0167445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Huang P, Zou Y, Zhong XZ, Cao Q, Zhao K, Zhu MX, Murell-Lagnado R, Dong XP, P2X4 forms functional ATP-activated cation channels on lysosomal membranes regulated by luminal pH, J. Biol. Chem (2014). [DOI] [PMC free article] [PubMed]
- [79].Zhong XZ, Zou Y, Sun X, Dong G, Sr Q Cao A. Pandey, Rainey JK, Zhu X, Dong XP, Inhibition of TRPML1 by lysosomal adenosine involved in severe combined immunodeficiency diseases, J. Biol. Chem (2017). [DOI] [PMC free article] [PubMed]
- [80].Corriden R, Insel PA, Basal release of ATP: an autocrine-paracrine mechanism for cell regulation, Sci. Signal 3 (104) (2010) re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Haanes KA, Novak I, ATP storage and uptake by isolated pancreatic zymogen granules, Biochem. J 429 (2) (2010) 303–311. [DOI] [PubMed] [Google Scholar]
- [82].Sawada K, Echigo N, Juge N, Miyaji T, Otsuka M, Omote H, Yamamoto A, Moriyama Y, Identification of a vesicular nucleotide transporter, Proc. Natl. Acad. Sci. U. S. A 105 (15) (2008) 5683–5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Suadicani SO, Brosnan CF, Scemes E, P2X7 receptors mediate ATP release and amplification of astrocytic intercellular Ca2+ signaling, J. Neurosci 26 (5) (2006) 1378–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Ledur PF, Villodre ES, Paulus R, Cruz LA, Flores DG, Lenz G, Extracellular ATP reduces tumor sphere growth and cancer stem cell population in glioblastoma cells, Purinergic Signal 8 (1) (2012) 39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Zhang JL, Liu Y, Yang H, Zhang HQ, Tian XX, Fang WG, ATP-P2Y2-beta-catenin axis promotes cell invasion in breast cancer cells, Cancer Sci 108 (7) (2017) 1318–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Zhang Y, Chin-Quee K, Riddle RC, Li Z, Zhou Z, Donahue HJ, BRMS1 sensitizes breast Cancer cells to ATP-Induced growth suppression, Biores. Open Access 2 (2) (2013) 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Arsenis C, Gordon JS, Touster O, Degradation of nucleic acids by lysosomal extracts of rat liver and Ehrlich ascites tumor cells, J. Biol. Chem 245 (1) (1970) 205–211. [PubMed] [Google Scholar]
- [88].Zimmermann H, 5’-Nucleotidase: molecular structure and functional aspects, Biochem. J 285 (Pt 2) (1992) 345–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Onyenwoke RU, Sexton JZ, Yan F, Diaz MC, Forsberg LJ, Major MB, Brenman JE, The mucolipidosis IV Ca2+ channel TRPML1 (MCOLN1) is regulated by the TOR kinase, Biochem. J 470 (3) (2015) 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zhang X, Cheng X, Yu L, Yang J, Calvo R, Patnaik S, Hu X, Gao Q, Yang M, Lawas M, et al. , MCOLN1 is a ROS sensor in lysosomes that regulates autophagy, Nat. Commun 7 (2016) 12109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Joo YN, Jin H, Eun SY, Park SW, Chang KC, Kim HJ, P2Y2R activation by nucleotides released from the highly metastatic breast cancer cell MDA-MB-231 contributes to pre-metastatic niche formation by mediating lysyl oxidase secretion, collagen crosslinking, and monocyte recruitment, Oncotarget 5 (19) (2014) 9322–9334.l [DOI] [PMC free article] [PubMed] [Google Scholar]