Abstract
The receptors of the innate immune system detect specific microbial ligands to promote effective inflammatory and adaptive immune responses. Although this idea is well appreciated, studies in recent years have highlighted the complexity of innate immune detection, with multiple host receptors recognizing the same microbial ligand. Understanding the collective actions of diverse receptors that recognize common microbial signatures represents a new frontier in the study of innate immunity, and is the focus of this Review. Here, we discuss examples of individual bacterial cell wall components that are recognized by at least two and as many as four different receptors of the innate immune system. These receptors survey the extracellular or cytosolic spaces for their cognate ligands and operate in a complementary manner to induce distinct cellular responses. We further highlight that, despite this genetic diversity in receptors and pathways, common features exist to explain the operation of these receptors. These common features may help to provide unifying organizing principles associated with host defence.
The detection of microorganisms by the innate immune system is a fundamental aspect of mammalian biology. Microbial detection not only indicates potential threats to the host but also provides training for immune homeostasis in major physiological systems such as the gastrointestinal tract1,2. Bacteria, whether pathogens, commensals or somewhere on the spectrum between the two, produce pathogen-associated molecular patterns (PAMPs) that are recognized by multiple classes of pattern-recognition receptor (PRR)3. These PAMPs are molecules that, when detected by the host, signify the presence of infectious non-self material. PAMP detection leads to inflammatory responses that help to eliminate the invading microorganism.
Some of the most inflammatory PAMPs, such as lipopolysaccharide (LPS), lipoproteins, peptidoglycan and flagellin, are molecules that are derived from the bacterial cell wall. Bacteria can also be detected through their nucleic acids (BOX 1), although this aspect of innate immune detection is not the focus of this Review. Cell-wall-associated PAMPs have long been known to induce inflammation after their detection by Toll-like receptors (TLRs). However, we now know that TLRs are but one subset of a wider family of immune surveillance factors that detect and respond to these ligands4.
Box 1 |. Extracellular and intracellular sensing of microorganisms beyond TLRs and NLRs.
In addition to Toll-like receptors (TLRs) and NOD-like receptors (NLRs), other pattern-recognition receptors (PRRs) sense extracellular and intracellular pathogen-associated molecular patterns (PAMPs)4,14. Complex carbohydrates, which are a major component of bacterial cell walls and exist on fungal and viral surfaces, can be detected by C-type lectin receptors (CLRs), a large family of soluble or membrane-bound receptors134. Many CLRs opsonize microorganisms and promote their phagocytosis14. Several CLRs induce supramolecular organizing centre (SMOC) formation and pro-inflammatory signalling after binding PAMPs. For example, dectin 1, dectin 2 and macrophage-inducible C-type lectin (MINCLE; also known as CLEC4E) use SYK and additional adaptors at the plasma membrane to promote the assembly of multi-protein complexes that induce cytokine production through nuclear factor-κB (NF-κB) activation134–136. Evidence also shows that dectin 1 activation elicits interleukin-1β (IL-1β) in response to Candida albicans and multiple mycobacteria through a non-traditional caspase 8 inflammasome. In this instance, SYK recruits caspase 8 and the adaptor protein ASC to promote inflammasome formation137. Therefore, at least a subset of CLRs use SMOCs to activate an inflammatory response. Intracellular sensing of microorganisms extends beyond cell-wall-associated PAMPs. Microbial nucleic acids are recognized by diverse PRRs, including endosomal TLRs, RIG-I like receptors (RLRs) and cyclic GMP–AMP synthase–stimulator of interferon genes (cGAS–STING). These factors also utilize SMOCs to initiate inflammation. Multiple endosomal TLRs recognize viral or bacterial nucleic acids, including TLR3, TLR7 and TLR8, and TLR9. TLR3 recognizes double-stranded RNA from viral genomes and directly interacts with TIR domain-containing adaptor protein inducing IFNβ (TRIF) to initiate pro-inflammatory cytokine production through NF-κB or interferon (IFN) expression through IFN regulatory factor 3 (IRF3)14,138. TLR7 and TLR8 recognize single-stranded RNA and are likely to use myddosomes to stimulate an NF-κB-mediated cytokine response or an IRF7-driven IFN response14. TLR9 detects non-methylated cytosine-guanine (CpG) motifs in DNA, a hallmark of bacterial and viral genomes139. As for TLR7 and TLR8, TLR9 uses myddosomes to promote inflammation14. TLR3, TLR7 and TLR9 all undergo proteolytic cleavage to initiate inflammation140,141. Murine TLR13 is the only nucleic-acid-binding TLR that recognizes bacterial RNA specifically, as this PRR binds to a specific sequence in bacterial ribosomal RNA142.
RLRs, which sense cytosolic microbial RNAs, use SMOCs after PAMP detection. The RLRs include retinoic acid-inducible gene I protein (RIG-I; also known as DDX58), melanoma differentiation-associated protein 5 (MDA5; also known as IFIH1) and probable ATP-dependent RNA helicase DHX58 (also known as LGP2). On PAMP binding, RIG-I and MDA5 engage the adaptor mitochondrial antiviral-signalling protein (MAVS) — which oligomerizes in a manner analogous to ASC — to recruit ubiquitin ligases and the inhibitor of NF-κB kinase (IKK) family member TANK-binding kinase 1 (TBK1)143,144. These enzymes promote NF-κB, IRF3 and IRF7 activation. Thus, RLR signalling generates NF-κB-dependent cytokines and IFNs and promotes the expression of IFN-stimulated genes138,145. DHX58 lacks signalling domains but promotes IFN responses to viral infection146, perhaps because it potentiates MDA5 interaction with RNA147. Although RLRs are predominantly viral sensors, they also participate in antibacterial responses148.
Nucleic acids are also recognized by cGAS and STING149,150. Unlike many PRRs, cGAS itself is an enzyme. Nevertheless, parallels with canonical PRR signalling exist in the cGAS–STING pathway. On detection of cytosolic DNA (derived from bacterial or viral infection or even self151), cGAS synthesizes cyclic GMP–AMP (cGAMP) from GTP and ATP. cGAMP binding by the adaptor STING results in the formation of a signalling complex that elicits an IFN response via the kinase TBK1 (REF. 151). Thus, cGAS ensures that IFN pathways are initiated on sensing of cytoplasmic DNA and uses an adaptor and a signalling complex to induce this response. Although the role of cGAS–STING in antiviral immunity is well established, it is increasingly clear that cGAS also induces IFN production after detecting bacterial DNA, including from Mycobacterium tuberculosis and other pathogens151.
In this Review, we discuss a collection of structurally diverse PRRs that recognize the same bacterial cell wall component. These PRRs then induce various signalling pathways that depend on structurally diverse signalling components. We discuss how, despite this diversity, themes have emerged that transcend the specific structures of individual regulatory factors. These themes include the ability of diverse PRRs to detect common PAMPs, the ability of diverse proteins to assemble into conceptually similar signalling organelles upon PAMP detection and the ability of diverse enzymes in these organelles to promote inflammation. We therefore propose that, akin to the unifying power of the concepts of PAMPs and PRRs3, common features of the signalling pathways of the innate immune system also exist.
Themes of innate immune signalling
In this section, we describe the best-defined pathways of the innate immune system — those that promote inflammatory gene expression or inflammatory cell death (that is, pyroptosis). Although these two processes (transcription and pyroptosis) are distinct, they are united by common cell biological themes, which result in an inflammatory cell state that is aimed at controlling infection. Here, we focus nearly exclusively on PRRs and receptor-proximal factors that detect extracellular and intracellular bacteria, and introduce the diversity of signalling proteins that detect these microorganisms while highlighting common principles that are associated with their mechanisms of action.
Pathways that promote inflammatory gene expression.
TLRs were first identified as homologues of the Drosophila melanogaster Toll receptor, which regulates development and immunity in the fruitfly5,6, and they are the founding members of a large collection of PRRs that recognize PAMPs (BOX 2). These receptors are transmembrane proteins that localize to the plasma or endosomal membranes. Long leucine-rich repeat (LRR) regions mediate protein–protein or protein–PAMP interactions in the extracellular and lumenal spaces. These distinct subcellular sites correlate with the nature of the PAMPs recognized by individual TLRs. Extracellular TLRs typically detect microbial cell-wall-associated PAMPs, whereas endosomal TLRs usually detect PAMPs that are not displayed on the microbial surface, predominantly nucleic acids (BOX 2).
Box 2 |. Pattern-recognition receptors sense host-derived molecules.
During infection, host cells are damaged, leading to the release of host-specific molecules termed danger-associated molecular patterns (DAMPs), which are also detected by pattern-recognition receptors (PRRs). Classically, Toll-like receptors (TLRs) have been thought to be the predominant sensor of DAMPs, but emerging evidence suggests that DAMPs induce inflammasome assembly as well — both canonical152 and non-canonical153. This observation suggests that multiple signalling pathways evolved to recognize not only signatures of microbial infection but also signatures of dysbiosis: including aberrant host physiology and host tissue disruption that occurs with many microbial infections. The use of the same cellular pathways for PAMPs and DAMPs might be evolutionarily advantageous, as it avoids the cost of developing and maintaining a second system, but it raises many questions about self–non-self discrimination.
Recent evidence suggests that DAMPs can also modulate the activity of a PRR bound to its canonical PAMP. Analysis of caspase 11 binding to PAMPs and DAMPs demonstrated that the recognition of oxidized phospholipids — a type of DAMP that can be derived from the host cell membrane — modulates caspase 11 signalling processes. Analysis of caspase 11 binding to a mixture of oxidized phospholipids (oxidized 1-palmitoyl-2-arachidonyl-sn-glycero-3-phosphorylcholine (oxPAPC)) demonstrated that oxPAPC and lipopolysaccharide (LPS) bind distinct domains153. LPS interacts with the caspase activation and recruitment domain (CARD) of caspase 11, whereas oxPAPC binds its catalytic domain. Some reports suggest that oxidized phospholipids stimulate TLR4-dependent inflammatory signalling153, although oxPAPC was originally defined as an LPS antagonist154. Indeed, oxPAPC does not induce TLR4 endocytosis, pro-inflammatory cytokine production or myddosome formation in macrophages or dendritic cells153. Thus, host-derived oxidized phospholipids may instead modulate LPS-induced inflammatory processes by altering caspase 11 activity.
oxPAPC elicits interleukin-1β (IL-1β) release in primed dendritic cells (but not in macrophages)153, and a recent report demonstrated that a particular oxidized phospholipid, phosphatidylcholine (POVPC), stimulates IL-1β release in macrophages155. IL-1β release after oxPAPC and POVPC treatment requires the NLR family pyrin domain-containing protein 3 (NLRP3) inflammasome153,155. Interestingly, LPS and/or oxPAPC induces the release of IL-1β from living dendritic cells. By contrast, primed cells treated with ATP or LPS readily undergo pyroptosis153. Thus, caspase 11 represents the first example of a PRR that has multiple activation states depending on whether PAMPs or DAMPs are bound, and these have different functional consequences.
Although all cell-surface-localized TLRs detect PAMPs that are present in the bacterial cell wall, the mechanisms by which productive ligand–receptor interactions occur differ. In this discussion, we define productive ligand–receptor interactions as the ability of the TLR to not only bind ligand but also to promote inflammatory gene expression. The simplest mode of interaction seems to be between TLR5 and subunits of flagellin, which may occur directly and without the aid of additional host factors7,8. TLR2 and TLR4, by contrast, require interactions with other host proteins to interact productively with their microbial ligands9–13. TLR2 recognition of di- or triacylated lipoproteins requires the actions of TLR6 or TLR1, respectively12,13. The cell-surface proteins CD14, CD36 and mannose-binding lectin (MBL; also known as MBP) also promote productive TLR2 recognition in a ligand-specific manner14. Unlike TLR2, TLR4 does not require interactions with other TLRs to productively interact with its microbial ligand LPS, but it does depend on LPS-binding protein (LBP), CD14 and MD214. Thus, whereas some TLRs can detect their ligand directly (for example, TLR5), others require the actions of other TLRs (for example, TLR2) or additional PAMP-binding proteins (for example, TLR2 and TLR4). Of these interactions, the interactions between proteins that bind LPS have been best defined. LBP forms a complex with the outer membrane of Gram-negative bacteria (or LPS micelles) and somehow alters the membrane to allow CD14 to extract a monomer of LPS15. CD14 then transfers LPS to a heterodimer of MD2–TLR4, which results in crosslinking of two TLR4 molecules9,16,17. TLR4 crosslinking is the first step in the inflammatory process16,17. This unidirectional flow of LPS from LBP to MD2–TLR4 is explained by the increasing affinity of each LPS receptor for its ligand9. This complex mode of LPS extraction and detection ensures that cells respond robustly to a single bacterium. It is unclear whether similar sensitivity explains why TLR2 recognition and signalling requires additional host factors. Likewise, it is unclear whether any host factors (so far unidentified) facilitate flagellin recognition by TLR5. Answers to these unknowns will allow us to determine whether the apparent heterogeneity of the mechanisms of PAMP detection can be explained by a common fundamental need for signalling sensitivity. Despite this diversity of mechanisms of PAMP detection by TLRs, ligand binding commonly results in receptor crosslinking (or dimerization), which activates the signalling activities of these receptors18.
Although dimerized TLRs display no intrinsic enzymatic activities, they activate enzyme-dependent cellular responses through interactions with downstream effector proteins. The signalling events that TLRs induce lead to the production of various immunoregulatory factors, with the major classes of secreted proteins being pro-inflammatory cytokines and interferons (IFNs)14. All TLRs induce cytokine expression through the activity of nuclear factor-κB (NF-κB) and activator protein 1 (AP1) transcription factors14. By contrast, IFNs are only induced by endosome-localized TLRs through the action of IFN regulatory factor (IRF) family transcription factors14. These cytokines and IFNs then elicit numerous cellular responses, including the recruitment of phagocytes and neutrophils and the activation of dendritic cells, which initiate adaptive immunity14,19.
TLR-induced signals are mediated by the actions of two classes of effector proteins, which are known as signalling adaptors (myeloid differentiation primary response protein 88 (MYD88)20 and TIR domain-containing adaptor protein inducing IFNβ (TRIF)21,22) or sorting adaptors (Toll/interleukin-1 receptor domain-containing adaptor protein (TIRAP)23,24 and TRIF-related adaptor molecule (TRAM)25)14. These factors contain Toll/interleukin-1 (IL-1) receptor (TIR) domains, which allow inter action with TLRs (or each other) to initiate inflammatory responses. Sorting adaptors always act upstream of signalling adaptors, and survey the inner leaflet of the plasma and endosomal membranes for the presence of activated (dimerized) TLRs. This surveillance activity is mediated by the unique ability of sorting adaptors to interact with acidic phosphoinositides or to be myristoylated26. Each of these activities positions sorting adaptors for rapid response to activated TLRs. The most commonly used sorting adaptor is TIRAP23,24, which uses its promiscuous phosphoinositide-binding domain to detect active TLRs at the plasma membrane and endosomes27. On TLR detection, TIRAP promotes the assembly of a large oligomeric complex called the myddosome, which consists of the signalling adaptor MYD88 and several IL-1 receptor-associated kinase (IRAK) family serine/threo-nine kinases27–29. Functionally, the myddosome and the inflammasome (described later in this Review) are supra-molecular organizing centres (SMOCs), which coordinate all inflammatory and immunoregulatory activities that are induced on microbial detection30. The myddosome and inflammasome are only assembled when needed and are not present in resting cells. All TLRs (except TLR3) are expected to assemble a myddosome on microbial detection, although experimental evidence of myddosome assembly has only been reported for TLR4 and TLR9 (REF. 27).
Myddosome-induced inflammatory gene expression proceeds from the plasma membrane and endosomes, whereas signals that lead to IFN production proceed from endosomes exclusively14. IFNs are produced by the activity of both TLR signalling adaptors (MYD88 and TRIF), although TRIF is most commonly associated with this response. The mechanisms of TRIF function are poorly defined, but it is thought that TRIF recruits TNF receptor-associated factor 3 (TRAF3) and downstream kinases that form a SMOC called the triffosome, which consequently activates IRFs26. Because TRIF-dependent IFN expression occurs only from endosomes, it is likely that the putative triffosome is assembled on these organelles. TRIF detects endosomal TLRs either directly (as in the case of TLR3) or indirectly through TRAM (as in the case of TLR4)25,31,32. However, TRIF does not assemble the sole SMOC that initiates IFN signalling. In plasmacytoid dendritic cells, endosomal TLR7 and TLR9 can form distinct endosome-associated myddosomes that signal through TRAF6 and IRF7 to generate IFNs14. Based on this collective knowledge, it seems that common principles associated with TLR signalling include the need for PAMP-induced receptor dimerization to promote downstream cellular responses, and the need to induce the assembly of SMOCs (the myddosome or the putative triffosome) to induce inflammation. As described below, these themes transcend the study of TLRs and also apply to PRRs that survey the cytosol.
Pathways that promote pyroptosis.
As well as being detected by TLRs outside the cell, bacteria are detected by PRRs in the cytosol. Although the regulators and effector responses differ between the systems that survey the extra-cellular and cytosolic environments, both systems detect the same PAMPs and signal through SMOCs to induce their unique cellular responses. A common set of SMOCs that are assembled in response to cytosolic PRRs are collectively known as inflammasomes33 (BOX 3). In contrast to the myddosome, the inflammasome does not induce transcriptional responses. Rather, this SMOC induces inflammation by promoting the release of pre-existing IL-1 family cytokines and other immunoregulatory factors34. An additional difference is based on the fact that most inflammasome-activating PRRs are present in the host cytosol, which is rarely accessed by non-pathogenic microorganisms. Consequently, inflammasomes are most commonly assembled during encounters with pathogens. TLRs at the cell surface, by contrast, cannot determine whether their cognate PAMP originated from pathogenic or non-pathogenic microorganisms. Despite these differences, myddosomes and inflammasomes promote inflammatory responses to control bacterial infection.
Box 3 |. Canonical and non-canonical inflammasomes.
The spectrum of canonical inflammasomes consists of different pattern-recognition receptors (PRRs) that sense diverse pathogen-associated molecular patterns (PAMPs). Despite this diversity, all of these systems use the sensor–adaptor–enzyme chain to effect innate immune responses. The range of sensors allows flexible host responses to various microbial insults, and efficiency is maintained by channelling the activation of these diverse PRRs through the same effector molecules: the adaptor protein ASC and the enzyme caspase 1. Well-studied canonical inflammasomes include the NLR family pyrin domain-containing protein 1 (NLRP1) and NLRP3 inflammasomes, the neuronal apoptosis inhibitory protein 5 (NAIP5)- and NAIP6-dependent NLR family CARD-containing protein 4 (NLRC4) inflammasomes (see main text), and the absent in melanoma 2 (AIM2) and pyrin inflammasomes, which have been expertly reviewed elsewhere34,35. Recent reports suggest that NLRP6, NLRP7 (see main text) and NLRP12 also form canonical inflammasomes34,35. NLRP1 responds to the lethal toxin of Bacillus anthracis, and the NLRP3 inflammasome is the most promiscuous inflammasome described so far, responding to ATP, potassium efflux, crystalline particles and other molecules. The AIM2 inflammasome assembles on recognition of double-stranded DNA from viruses or bacteria156, whereas the pyrin inflammasome indirectly senses microorganisms through bacterial toxin- or effector protein-mediated manipulation of RHO GTPases. The NLRP6 inflammasome has been reported to form in intestinal epithelial cells and may have a role in regulating the microbiota34, and the NLRP12 inflammasome responds to Yersinia species and has a role in bacterial clearance157. These canonical inflammasomes consist of diverse NLR family PRRs, yet all (except the NLRC4 inflammasome in certain instances) use the ASC adaptor to recruit caspase 1 to process interleukin-1 (IL-1) family cytokines and gasdermin D (GSDMD) to effect downstream responses.
The non-canonical inflammasome consists of the PRR caspase 11 in mice (or caspase 4 and caspase 5 in humans), culminating in the activation of GSDMD after lipopolysaccharide binding34. The non-canonical inflammasome also converges on caspase 1 for IL-1β production through the NLRP3 inflammasome (see main text and FIG. 1).
Although the biochemical constituents of myddosomes and inflammasomes differ, the proteins in these complexes have common features. For example, both complexes contain a sensor molecule, an adaptor molecule and an enzyme. As described above, this set of molecules for the myddosome includes a TLR, the sorting adaptor TIRAP, the signalling adaptor MYD88, and IRAK family kinases. For inflammasomes, the sensors are most commonly NLRs (NOD-like receptors), the adaptor protein ASC, and the enzyme caspase 1 (REF. 35). The network of SMOC-associated factors is modular: receptors other than the TLRs can induce myddosome assembly (for example, IL-1 receptor family members), and non-NLRs can induce inflammasome assembly (for example, the PRRs absent in melanoma 2 (AIM2) or pyrin) (BOX 3). In addition, adaptor diversity exists, in that some PRRs do not require the ASC or MYD88 adaptors, but instead use NLR family CARD-containing protein 4 (NLRC4) or TRIF to promote inflammation through caspase 1 or NF-κB14,34. Despite the diversity of proteins that comprise inflammasomes, they all share the feature of utilizing PRRs and adaptors, which converge on inflammatory caspases, such as caspase 1. Activated caspase 1 cleaves the latent cytokines pro-IL-1β and pro-IL-18 and the latent pore-forming protein gasdermin D (GSDMD) into their biologically active forms. Active IL-1β and IL-18 are then released from cells after the pore-forming activity of GSDMD induces pyroptosis34. These IL-1 family cytokines are some of the most inflammatory signals produced by the innate immune system. Thus, signalling by myddosomes and inflammasomes initiates inflammatory programmes through diverse proteins that share common functional characteristics. Interestingly, studies in recent years have revealed additional PRRs that induce cellular responses to bacterial cell wall components36–40. These additional receptors do not induce myddosome or inflammasome assembly, yet they recognize the same microbial ligands as the PRRs that promote assembly of these SMOCs. In the next sections, we describe the specific collections of PRRs that recognize common PAMPs, thereby inducing diverse yet interconnected cellular responses to maximally promote inflammation.
Inflammatory signalling induced by LPS
Four receptors promote inflammatory responses to LPS.
Although TLR4 was long thought to be the only receptor that promotes cellular responses to LPS41, work in recent years has revealed several LPS responses that proceed independently of TLR4. These TLR4-independent responses include LPS-induced endocytosis by CD14 (REF. 10), LPS-induced assembly of inflammasomes by caspase 11 (REFS 42,43) and LPS-induced activation of reactive oxygen species (ROS) synthesis and phagocytosis by brain-specific angiogenesis inhibitor 1 (BAI1)36,37. In this section, we describe the mechanisms underlying these responses to LPS, and highlight how each response may influence the activity of the others.
Detection of LPS in the extracellular space.
In the extracellular space, LPS is detected by the membrane-bound receptors CD14, the TLR4–MD2 heterodimer44 and the G-protein-coupled receptor BAI1 (REFS 36,37) (FIG. 1). Of these receptors, TLR4 has attracted the most attention. TLR4 initiates two signalling cascades that are carried out by different SMOCs, namely the myddosome and the triffosome. At the plasma membrane, TIRAP senses activated TLR4 and assembles the myddosome to induce pro-inflammatory cytokine production27,45. After being internalized in endosomes, TLR4 is detected by the sorting adaptor TRAM, which may seed the formation of the triffosome to induce IFN expression25,31,32. TRAM localizes to the plasma membrane and endosomes25,32, with the former location being the dominant site of TIRAP localization27,45. The overlapping membrane specificities of TIRAP and TRAM could facilitate the progression of TLR4 from TIRAP (and the myddosome) to TRAM (and the triffosome)14. The triffosome would be expected to complement myddosome activities, as TRIF and TRAM are required for prolonged activation of NF-κB and AP1 (REFS 31,46). Thus, the formation of SMOCs at the plasma and endosomal membranes initiates the production of inflammatory cytokines and IFNs downstream of activated TLR4.
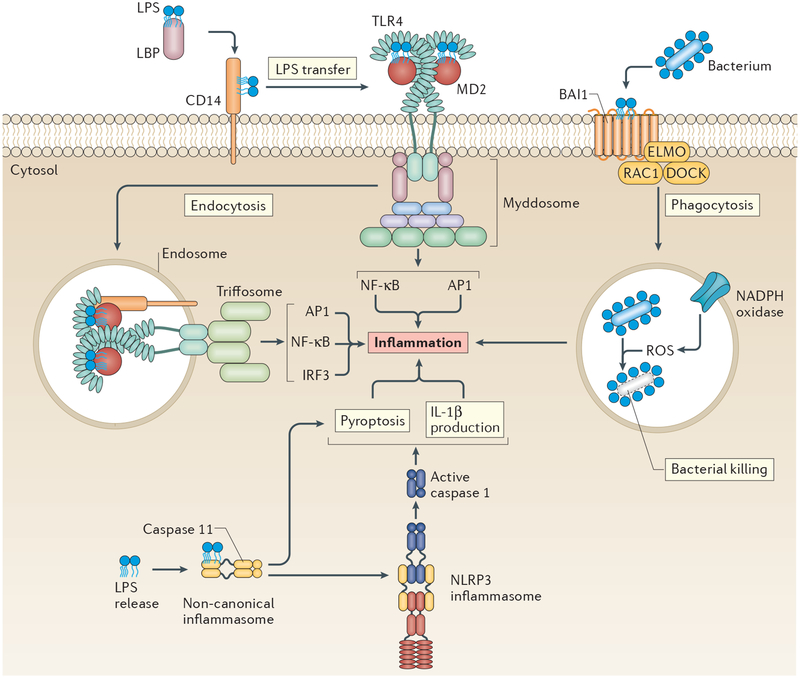
Figure 1 |. Four receptors induce five lipopolysaccharide response pathways to promote inflammation.
Lipopolysaccharide (LPS) is sensed at the plasma membrane initially by LPS-binding protein (LBP), which helps CD14 to extract this pathogen-associated molecular pattern (PAMP) from bacterial cell walls. CD14 then delivers LPS to Toll-like receptor 4 (TLR4)–MD2, prompting the dimerization and activation of TLR4, a process that leads to myddosome assembly. CD14 then delivers dimerized TLR4 to endosomes to promote TIR domain-containing adaptor protein inducing IFNβ (TRIF) signalling through the putative triffosome. Both pathways result in inflammation. Brain-specific angiogenesis inhibitor 1 (BAI1) also detects LPS at the plasma membrane and uses the engulfment and cell motility protein–dedicator of cytokinesis protein–RAC1 (ELMO–DOCK–RAC1) complex to promote phagocytosis, leading to reactive oxygen species (ROS) production and induction of inflammation. LPS that has reached the cytosol is bound by caspase 11, initiating formation of the non-canonical inflammasome and, in turn, pyroptosis. Caspase 11 activity also stimulates the formation of the canonical NLR family pyrin domain-containing protein 3 (NLRP3) inflammasome (solid arrow), which elicits interleukin-1β (IL-1β) processing and release. AP1, activator protein 1; IRF3, interferon regulatory factor 3; NF-κB, nuclear factor-κB.
CD14 is best known for its role in relaying LPS to TLR4, but recent studies have revealed that this PRR induces a cellular response — endocytosis — that is independent of (and upstream of) TLR4–TRIF signalling activity. Studies have shown that CD14 and LPS endocytosis proceeds normally in cells that lack TLR4 (REFS 10,47), whereas cells that lack CD14 cannot induce TLR4 endocytosis in response to LPS10,11. Thus, CD14 is necessary for internalization of the PRR (TLR4) and the PAMP (LPS). Because CD14-dependent endocytosis occurs before TLR4 signalling through the TRIF pathway, one must conclude that CD14 induces an endocytosis pathway upstream of TLR4 signalling, and that the dual actions of independent pathways activated by CD14 and TLR4 promote TRIF signalling. With its dual role in transporting PRRs and PAMPs, CD14 has been classified as a transporter associated with the execution of inflammation (TAXI)11. Multiple factors have been implicated in the regulation of CD14-dependent endocytosis, including the immunoreceptor tyrosine-based activation motif (ITAM)-containing adaptors DNAX-activation protein 12 (DAP12; also known as TYROBP) and FcεRIγ, the tyrosine kinase SYK, phospholipase Cγ2 and inositol-1,4,5-triphosphate receptors10,48. These factors collaboratively regulate CD14 endocytosis, as deletion or inhibition of individual factors diminishes but does not eliminate TRIF signalling, probably owing to delayed kinetics of TLR4 endocytosis10,48. Of the factors that regulate LPS-induced TLR4 endocytosis, MD2 is distinct. Whereas all of the aforementioned factors promote endocytosis of CD14 and TLR4, only MD2 is required for TLR4 endocytosis specifically11. CD14 endocytosis proceeds normally in MD2-deficient cells11. Thus, MD2 is not required for the process of LPS-induced endocytosis, but is the cargo-selection agent for CD14-dependent endocytosis. Its role in TLR4 trafficking caused MD2 to be classed as a member of the TAXI family of proteins alongside CD1411. However, unlike CD14, MD2 has no known ability to signal independently of TLR4. We therefore consider MD2 to be an essential and specific component of the TLR4 pathway. Because the mechanisms of CD14 endocytosis are obscure, it is unclear whether an endocytosis-inducing SMOC exists to promote the movement of LPS and CD14–TLR4.
Although CD14 is the dominant receptor that promotes LPS and TLR4 endocytosis and signalling, the capture of whole bacteria is mediated by a distinct LPS receptor — BAI1 (REFS 36,37). Interestingly, this receptor does not detect the lipid A moiety of LPS, which is detected by all other LPS receptors. Rather, BAI1 recognizes the core oligosaccharide of LPS36,37. BAI1 promotes the phagocytosis of Gram-negative bacteria specifically, and induces ROS production to facilitate bacterial killing. These responses to LPS — namely phagocytosis and ROS production — are not often associated with CD14 or TLR4 signalling, although definitive experimental evidence implicating or discounting their role is lacking36,37. Even though BAI1 detects a different LPS component from that detected by CD14 and TLR4 and elicits distinct responses, the signalling events associated with BAI1 activation are similar to those of other PRRs. Akin to myddosome and inflammasome regulation, BAI1 recruits the engulfment and cell motility protein–dedicator of cytokinesis protein (ELMO–DOCK) adaptor complex and mediates the activation of an enzyme — the GTPase RAC1. RAC1 then promotes phagocytosis37, cytokine production and the assembly and activation of the NADPH oxidase on the phagosomal membrane36,37. The role of BAI1 in Gram-negative bacterial detection is important, as depletion or chemical inhibition of BAI1 reduces phagocytosis, ROS production and bacterial clearance during infection in vivo36,37.
Cytosolic detection of LPS.
LPS can be detected inside the cell as well as in the extracellular space (FIG. 1). The delivery of LPS into the host cytosol is often associated with pathogens that disrupt phagosomal membranes. Key events that underlie how cytosolic LPS is recognized have been identified recently, namely the PRR and the SMOC. Intracellular LPS is detected by caspase 11, a member of the inflammatory caspase family49, and detection stimulates assembly of the non-canonical inflammasome (BOX 3). In a manner similar to LPS recognition by MD2–TLR4, caspase 11 binds the acyl chains of the lipid A moiety of LPS42,43,49. It is currently unclear how similar LPS detection by caspase 11 and MD2–TLR4 is, and the answer to this question will require structural analysis of these interactions. Nevertheless, surface plasmon resonance demonstrates that caspase 11 has sub-nanomolar affinity for lipid A, which it binds through its caspase activation and recruitment domain (CARD)49. Activated caspase 11 then cleaves GSDMD50,51, and the pore-forming activity of the latter promotes pyroptosis and the release of mature IL-1β52–54.
Caspase 11 activation also results in the assembly of a canonical NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3) inflammasome; caspase 1 then cleaves IL-1 family cytokines (and GSDMD) to maximally promote inflammation. Importantly, these caspase 11-dependent events that initiate non-canonical and canonical inflammasomes occur in TLR4-deficient cells42,43. NLRP3 activation downstream of active caspase 11 is dependent on potassium efflux55, probably because of GSDMD-mediated membrane perturbation (a known activator of the NLRP3 inflammasome34). Some evidence also supports a role for caspase 11 in promoting pyroptosis without activating NLRP3 or NLRC4 inflammasomes upon the detection of cytosolic bacteria56. These data suggest that caspase 11-dependent pyroptosis and canonical inflammasome activation can be uncoupled56.
Altogether, we have described four independent signalling pathways that mediate cellular responses to LPS through the PRRs CD14, TLR4–MD2, BAI1 and caspase 11. These diverse PRRs sense LPS and initiate different cellular responses that ultimately converge at a similar end point — an inflammatory state that controls bacterial spread.
Inflammatory responses to flagellin
Three receptors promote responses to flagellin.
LPS is present only on Gram-negative bacteria, but multiple (although not all) classes of bacteria use a flagellum for motility. As such, the detection of flagellar components by PRRs reports on encounters with Gram-positive and Gram-negative bacteria. The flagellin subunit of flagella is recognized by TLR5 in the extracellular space and by the NLRs neuronal apoptosis inhibitory protein 5 (NAIP5; also known as BIRC1E) and NAIP6 (also known as BIRC1F) in the cytosol7,57,58. On flagellin detection, these respective PRRs induce myddosome and inflamma some assembly to promote inflammation. Thus, although flagellin and LPS are distinct entities, common principles govern innate immune detection of these PAMPs.
Detection of flagellin in the extracellular space.
At the cell surface, flagellin is detected by TLR5 (REF. 7) (FIG. 2). TLR5 binds to a conserved site on the flagellin subunit that lies at the interaction interface between two adjacent flagellin subunits that are present in the flagellar apparatus8. No other host factors are known to promote productive interactions between TLR5 and flagellin. Although TLR5 seems to act alone, the fact that the flagellin recognition motif is buried in the flagellar filament8 suggests that some form of flagellin extraction may be necessary for efficient detection.
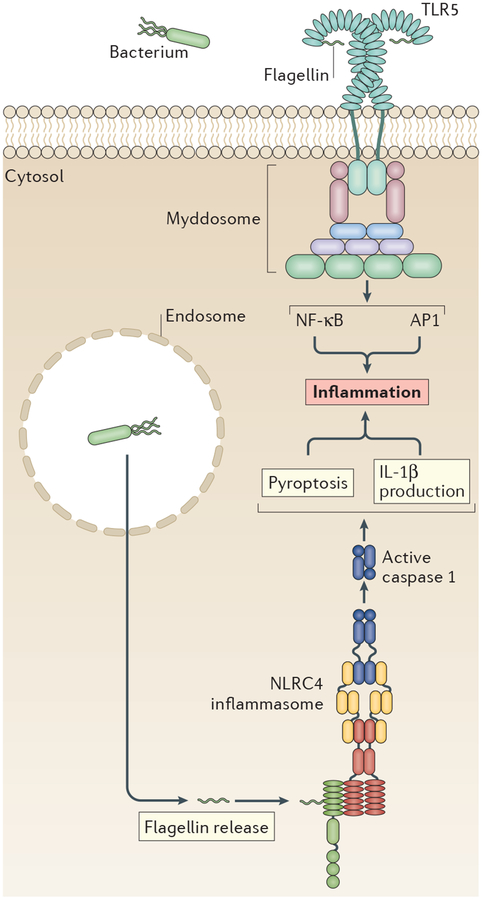
Figure 2 |. Three receptors stimulate two pathways to induce inflammatory responses to flagellin.
Flagellin, a subunit of the bacterial flagellum, is bound by Toll-like receptor 5 (TLR5) at the cell surface. Activated TLR5 is likely to stimulate cytokine production through the formation of a myddosome. TLR5 might be internalized after binding flagellin to elicit interferons (IFNs) through TIR domain-containing adaptor protein inducing IFNβ (TRIF), but further analysis is needed to determine whether this occurs and, if so, whether it is functionally important. Intracellular flagellin is bound by the pattern-recognition receptors (PRRs) neuronal apoptosis inhibitory protein 5 (NAIP5) and NAIP6, which induce assembly of the NLR family CARD-containing protein 4 (NLRC4)-dependent inflammasome. The secondary ASC adaptor is sometimes used by the NLRC4 inflammasome, although it may not always be required. AP1, activator protein 1; IL-1β, interleukin-1β; NF-κB, nuclear factor-κB.
Little is known about TLR5 signalling at the plasma membrane, although some evidence for SMOC formation downstream of PAMP recognition exists in that MYD88 is crucial for TLR5 signalling7, and some reports indicate that TIRAP is important in certain contexts59. For example, TIRAP seems to be required for optimal TLR5 signalling in intestinal epithelial cells59. Similarly, our understanding of the role of TRIF in TLR5 signalling is limited. Evidence suggests that TRIF signalling may occur downstream of TLR5 in intestinal epithelial cells60, which is supported by the observation that, in epithelial cells, flagellin can be internalized in a dynamin-dependent manner61,62. However, further analyses will improve our understanding of flagellin-dependent signalling and may yield unique biology. Indeed, a recent study suggests that TLR5 is transported to the plasma membrane by UNC93 homologue B1 (UNC93B1)63 (which is thought to be required only for endosomal localization of TLR3, TLR7 and TLR8 and TLR9), and this chaperoning is essential for normal TLR5 function64.
Cytosolic detection of flagellin.
In contrast to our limited understanding of flagellin-induced TLR5 signalling, much more is known about the detection of cytosolic flagellin. In mice, cytosolic flagellin stimulates the formation of an inflammasome containing the adaptor NLRC4 (BOX 3; FIG. 2). This inflammasome exhibits the classic features of canonical inflammasomes, including caspase 1 activation and the release of IL-1β65,66. The PRRs that promote NLRC4 inflammasome assembly are NAIP5 and NAIP6, which bind flagellin directly57,58. NLRC4 contains a CARD that can recruit caspase 1 (REF. 57), thereby bypassing the need for ASC. However, there seems to be a role for ASC in NLRC4 activities, as macrophages that are deficient in ASC do not elicit caspase 1 processing downstream of flagellin58,67, although this finding is controversial68.
The similarities between the flagellin-sensory and LPS-sensory systems extend beyond the use of multiple PRRs to induce myddosomes and inflammasomes. For example, as in LPS detection, different receptors detect different regions of flagellin. Detection of flagellin by the NAIPs occurs in a region of the bacterial protein that is distinct from that detected by TLR5 (REFS 69,70). The D0 domain of flagellin is required for assembly of the inflammasome67, and a flagellin mutant lacking D0 does not elicit caspase 1 processing or inflammatory cytokines67. By contrast, loss of the D0 domain does not affect flagellin binding to TLR5. The D1 flagellin domain seems to be the sole domain that directly interacts with TLR5, although the D0 domain is required for efficient signal transduction69. Moreover, flagellin detection occurs in the LRR regions of TLR5, which also exist in NAIPs, but NAIPs recognize flagellin through distinct α-helical domains of the nucleotide-binding domain (NBD) instead71. Binding of flagellin to the NAIP NBD is required for NLRC4 to oligomerize71, which promotes caspase 1 recruitment57,67. Moreover, phosphorylation of NLRC4 advances inflammasome assembly67,72, although this has been disputed73.
In mice and humans, the NLRC4 inflammasome also assembles on detection of the bacterial type three secretion system (T3SS)74; in mice, this occurs through the actions of the PRR NAIP2, which recognizes the T3SS rod component57, and the single NAIP orthologue in humans also recognizes the T3SS rod75. The T3SS and flagella exhibit structural and sequence homology76. By analogy, the rod protein is comparable to the flagellin monomer. Thus, despite being distinct PAMPs, the detection of flagellar and T3SS components by cognate PRRs and the assembly of NLRC4 inflammasomes display molecular symmetry.
Sensing lipoproteins
Three receptors promote responses to lipoproteins.
Lipoprotein PAMPs decorate the surface of Gram-positive and Gram-negative bacteria. Because hexa-acylated LPS is so inflammatory, lipoproteins play a minor part in the host response to Gram-negative bacteria, except when microorganisms alter their LPS to avoid detection (see below). In such cases, the detection of lipoproteins becomes more important77. Lipoproteins have a large role in the detection of Gram-positive bacteria, as they extend out from the surface of the peptidoglycan matrix that surrounds the bacterial cell. Lipoproteins are detected at the plasma membrane by TLR2-containing complexes78,79. Intracellular lipoproteins stimulate the formation of the NLRP7 inflammasome in human cells80, although an analogous mechanism has yet to be identified in murine cells.
Detection of lipoproteins in the extracellular space.
Different bacterial lipoproteins are sensed at the cell surface by distinct TLR2 heterodimers12,13 (FIG. 3). Diacylated lipoproteins are detected by the TLR2–TLR6 heterodimer, whereas triacylated lipoproteins are bound by TLR2–TLR1 (REFS 12,13). In a ligand-specific manner, productive TLR2–PAMP interactions also require the host factors CD14 (REFS 81,82) and CD36 (REFS 14,83). Evidence exists for CD36 affinity for TLR2–TLR6 complexes84. CD14 may signal predominantly with TLR2–TLR1 complexes81, although it can also affect TLR2–TLR6 signalling85. MYD88 and TIRAP are essential for inflammatory cytokine production downstream of lipoprotein stimulation86–88, suggesting that myddosomes form and are responsible for TLR2 signal transduction.
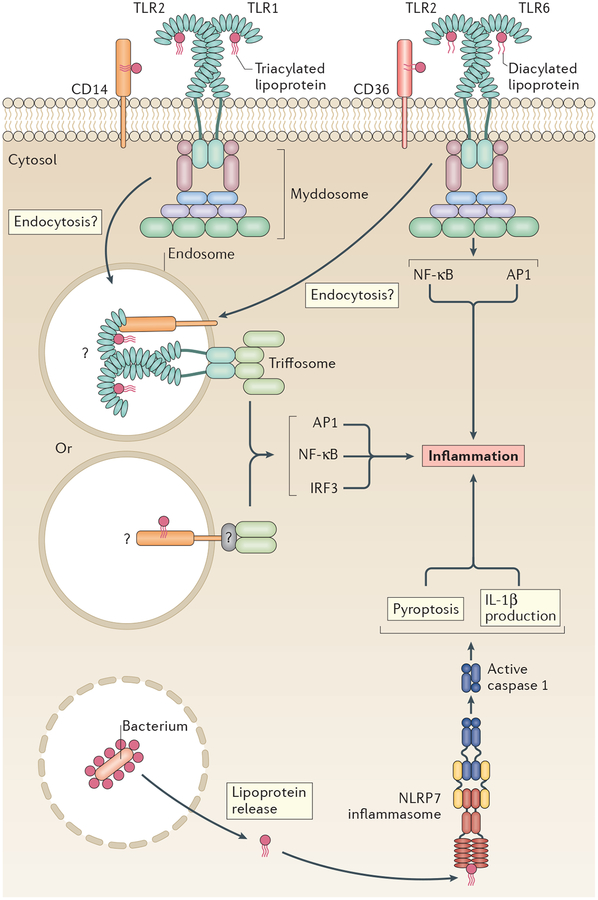
Figure 3 |. Three receptors and pathways induce inflammatory responses to lipoproteins.
Lipoproteins are detected at the plasma membrane by Toll-like receptor 2 (TLR2), TLR1 and TLR6, which form heterodimers. Their recognition of lipoproteins is assisted by the co-receptors CD14 and CD36. TLR2 heterodimers induce pro-inflammatory cytokines through a myddosome. Whether a robust interferon (IFN) response occurs downstream of TLR2 activation remains controversial (indicated by question marks), but it might occur through internalization of ligand bound to the CD14 or CD36 co-receptor, which would require an unidentified adaptor to interact with TIR domain-containing adaptor protein inducing IFNβ (TRIF) (indicated by a question mark). Alternatively, the TLR bound to ligand might be internalized, which could stimulate IFNs through the triffosome. Further experimentation is required to determine whether a robust IFN response occurs downstream of TLR2 (see main text). Intracellular lipoproteins stimulate the formation of an NLR family pyrin domain-containing protein 7 (NLRP7) inflammasome in human, but not murine, cells, leading to interleukin-1β (IL-1β) release and inflammation. AP1, activator protein 1; IRF3, interferon regulatory factor 3; NF-κB, nuclear factor-κB.
Stimulation of IFN by lipoproteins.
Myddosome-dependent cytokine production is the most well defined activity of TLR2, but it is less clear how bacterial lipoproteins stimulate an IFN response (FIG. 3). Early studies showed that TLR2 ligands induce type I IFNs, albeit at a much weaker level than that observed for LPS89,90. Under the conditions examined, these TLR2 and TLR4 ligands induced comparable amounts of mitogen-activated protein kinase (MAPK) activation89. This suggests that TLR2 and TLR4 signal at similar rates and ‘strengths’, but only TLR4 potently induces IFN90. Recent work has re-examined whether TLR2 activation elicits IFN77,91–96 and found that IFNβ or IFN-stimulated genes can be induced downstream of viral91 or bacterial92 infection. Synthetic bacterial lipoproteins can also induce IFN expression93–95, although this finding has been challenged by studies using a comparable TLR2 (REF. 81) synthetic lipopeptide92. Regardless, the IFN response was low in comparison to known stimulators of IFN expression (and could even be due to TLR8 signalling92). Functional assays, such as the inhibition of viral replication, may clarify the physiological relevance of the low-level IFN production induced by TLR2.
If TLR2 does elicit IFN, this would probably occur after receptor endocytosis, as no known receptor can induce IFN production from the cell surface14. Several reports suggest that endocytosis is important for signalling downstream of lipoprotein stimulation83,91,97. Using chemical inhibition of endocytosis, the internalization of fluorescently labelled lipoproteins or lipoteichoic acid (LTA) was blocked83,97, with a concomitant decrease in inflammatory cytokine production97. Chemical inhibition of endocytosis or endosomal maturation eliminated the low-level IFNβ production induced by TLR2 during viral infection91. Although these studies support the PAMP-inducible movement of TLR2 into the endosomal network, it was not until recently that the tools became available to test this hypothesis directly. Monoclonal antibodies raised against endogenous TLR2, TLR1 and TLR6 detected the native proteins in primary macrophages and dendritic cells97. However, the cell-surface population of these receptors was unaltered after lipoprotein stimulation97. The seeming lack of TLR2 movement is in contrast to observations of TLR4, which moves rapidly into the cell within minutes of ligand stimulation10,11. It should be noted, however, that TLR movement was analysed in cells 2 hours post stimulation in REF. 97: thus, TLR2 could have been internalized and then re-populated at the cell surface, as is seen with CD14 (REF. 11). Alternatively, it is possible that a small subpopulation of TLR2 molecules could be endocytosed, which is undetectable by flow cytometry but could be sufficient to initiate low-level IFNβ production.
If it is true that lipoproteins are internalized to elicit low-level IFN but the PRRs remain on the cell surface, how would the ligands be endocytosed? One possibility is that CD14 or CD36 mediates internalization of some TLR2 ligands83 in a process regulated by the SRC and SYK kinases10,98. Another unknown associated with TLR2 signalling from endosomes99,100 is the identity of the corresponding SMOC. The lipoprotein–CD36 (or CD14) co-complex would probably need an adaptor protein to interact with TRIF or MYD88 to elicit IFNs (FIG. 3). Thus, although increasing evidence supports a role for TLR2 in promoting cytokine and low-level IFN expression, the underlying mechanisms and significance of these observations need to be further explored.
Cytosolic detection of lipoproteins.
In a similar way to other cell wall PAMPs, lipopeptides are ligands for multiple receptors, including those that promote inflammasome formation. For example, lipoproteins produced by Mycoplasma species80, Listeria monocytogenes80 and Staphylococcus aureus80,101 stimulate the formation of an NLRP7 inflammasome, which is present in humans but not mice. This process is required for efficient antibacterial defence. NLRP7 may bind lipoproteins directly, as production of NLRP7, ASC, pro-caspase 1 and pro-IL-1β in HEK293 cells was sufficient to induce IL-1β release80. Lipoproteins stimulate ASC oligomerization, caspase 1 processing and ASC-dependent IL-1β release, all of which are hallmarks of inflammasomes, in immortalized and primary human cells80. These studies therefore suggest that NLRP7 recognizes synthetic bacterial lipoproteins and intact bacteria, and elicits an inflammasome-dependent response. Lipoproteins, at least in human cells, are therefore recognized by PRRs at the cell surface and inside the cell, and this recognition stimulates the assembly of diverse SMOCs.
In murine cells, there is some suggestion that lipo-proteins elicit the processing of caspase 1 and release of IL-1β80,102,103. ASC-knockout macrophages secrete a reduced level of IL-1β in response to Pam3CSK4 (REF. 102). Similarly, LTAs, which are also recognized by TLR2, elicit ASC-dependent processing of caspase 1 and IL-1β secretion in murine macrophages104,105, suggesting the formation of an inflammasome. However, a murine intracellular sensor for lipoproteins has not been identified, and so far there is no definitive evidence of lipoprotein-induced inflammasome formation in murine cells. It therefore remains unclear whether lipo-proteins stimulate bona fide inflammasomes in mice. As we discuss below, SMOC formation in response to other Gram-positive PAMPs may bypass the need for lipoproteins to induce inflammasome formation.
Detection of peptidoglycan
Three receptors promote responses to peptidoglycan.
Peptidoglycan is the most abundant component of the Gram-positive bacterial cell wall and, along with its associated lipoproteins, is detected by the innate immune system. Unlike lipoproteins, however, peptidoglycan seems to be detected only in the cytosol. Nevertheless, the PRRs that sense peptidoglycan follow similar principles of PAMP detection and response to other PRRs. Peptidoglycan is composed of long glycan chains of repeating units of N-acetyl muramic acid (NAM) and N-acetyl glucosamine (NAG). These chains are then crosslinked with short pentapeptides that descend from NAM106. Various breakdown products of peptidoglycan stimulate an inflammatory response, including NAG and several dipeptide-containing products. These dipeptides are muramyl-dipeptide (MDP), which is NAM covalently linked to the first two amino acids of the peptide chain (l-Ala-d-Glu), and iE-DAP, which is composed of the d-Glu-mDAP (meso-diaminopimelic acid) residues of the peptide chain. The NLRs nucleotide-binding oligomerization domain-containing protein 1 (NOD1) and NOD2 recognize iE-DAP and MDP107, respectively. Recent work has identified a new factor, the metabolic enzyme hexokinase, which recognizes NAG and stimulates the assembly of the NLRP3 inflammasome108. These peptidoglycan-sensing receptors provide the final example we discuss of complementary sensory systems that operate through SMOCs to promote inflammation in response to a common PAMP.
Cytosolic detection of peptidoglycan.
Cytosolic PRRs detect peptidoglycan breakdown products, including iE-DAP and MDP, that are released or transported out of phagosomes after the degradation of bacteria38 (FIG. 4). MDP can be transported across the endosomal membrane by the peptide transporters solute carrier family 15 member 3 (SLC15A3) and SLC15A4 (REF. 38), but invasive bacteria can also deliver these ligands to the cytosol as part of their infectious cycles107. Cell biological analysis revealed the endolysosomal membrane as the probable site of signal transduction downstream of peptidoglycan detection, as NOD1 or NOD2 colocalize there with their cognate adaptor protein receptor-interacting serine/threonine-protein kinase 2 (RIPK2; also known as RIP2)38,109.
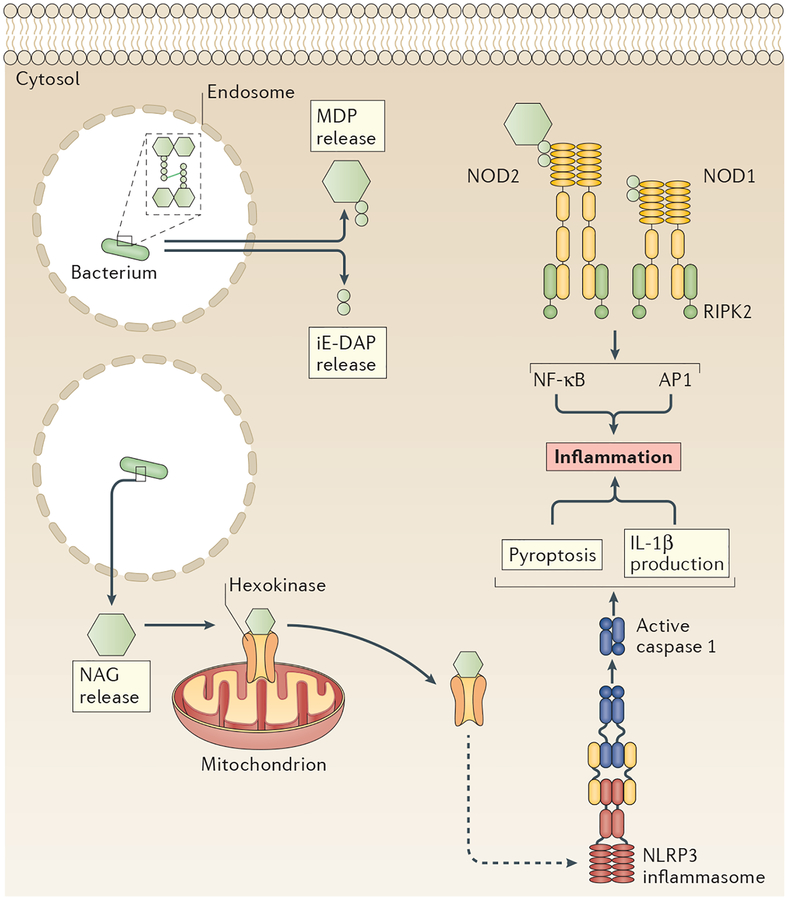
Figure 4 |. Three receptors and pathways induce inflammatory responses to peptidoglycan.
Intracellular peptidoglycan is detected by several pattern-recognition receptors (PRRs), including nucleotide-binding oligomerization domain-containing protein 1 (NOD1) and NOD2, which use a receptor-interacting serine/threonine-protein kinase 2 (RIPK2)-based putative supramolecular organizing centre (SMOC) to promote inflammation. NOD1 detects the iE-DAP dipeptide, whereas NOD2 detects muramyl dipeptide (MDP), both of which are peptidoglycan degradation products. Another peptidoglycan degradation product, N-acetyl glucosamine (NAG), indirectly induces assembly of the NLR family pyrin domain-containing protein 3 (NLRP3) inflammasome. NAG binds to the metabolic enzyme hexokinase and induces its dissociation from the mitochondrial membrane. This dissociation stimulates formation of an NLRP3 inflammasome through an unknown mechanism (dashed arrow). AP1, activator protein 1; IL-1β, interleukin-1β; NF-κB, nuclear factor-κB.
Similarly to TLRs, the respective PAMPs of NOD1 and NOD2 are thought to interact with their LRR domains, and promote their oligomerization110,111. Oligomerized NODs then recruit the kinase RIPK2 through their CARDs. RIPK2 further oligomerizes111 and recruits multiple kinases, assembling a large complex that ultimately activates NF-κB39 or MAPK family kinases (extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinase (JNK) and p38)39,111. Whether this large complex represents a bona fide SMOC is unclear, as there are some conceptual disconnects between NOD-induced signalling complexes and other SMOCs. For example, whereas the adaptors present in other SMOCs are non-enzymes14,30, the NOD1 and NOD2 adaptor, RIPK2, is an enzyme39. Additionally, it remains unclear whether NOD1 and NOD2 bind to PAMPs directly, although recent studies suggest that they do109. On the basis of these disconnects, it is possible that NOD1 and NOD2 simply operate through an unconventional SMOC-dependent signalling pathway. Alternatively, it is possible that a conventional SMOC does exist, and that NOD1 and NOD2 do not serve as PRRs; rather, NOD1 and NOD2 may operate as adaptors that engage the enzyme RIPK2. This latter scenario would require a PRR upstream of an NLR, which is akin to the aforementioned NLRs NAIP5 and NAIP6 acting upstream of NLRC4 (REFS 57,58).
An alternative signalling pathway also exists for NOD1 and NOD2 activity. After sensing a ligand, activated NOD1 or NOD2 can recruit autophagy-related protein 16L (ATG16L) and downstream autophagic machinery to generate autophagosomes40. This alternative NOD signalling pathway enhances bacterial clearance but may also negatively regulate inflammation during infection40. Although many gaps exist in our knowledge of NOD-dependent signalling, the current data suggests that PAMP detection by NOD1 and NOD2 uses a SMOC-dependent pathway to induce inflammation and host defence.
Peptidoglycan and inflammasome assembly.
Cytosolic MDP and NAG stimulate caspase 1 activation and elicit IL-1β production108,112,113, suggesting the formation of an inflammasome (FIG. 4). Indeed, recent work showed that NAG induces NLRP3 inflammasome assembly108. NLRP3 recognition of cytosolic peptidoglycan may compensate for the potential lack of or low-level murine recognition of cytosolic lipopeptides108,112,113. The NLRP3 inflammasome is a broad spectrum SMOC (BOX 3), and peptidoglycan is a recent addition to a long list of ligands that stimulate its formation34. Like most activators of NLRP3, NAG stimulates inflammasome formation indirectly, through a process mediated by the metabolic enzyme hexokinase. NAG binds directly to hexokinase as a competitive inhibitor and induces its dissociation from mitochondria. The disruption of hexokinase–mitochondria interactions leads to NLRP3 inflammasome assembly. Thus, NAG induction of inflammasome assembly requires an additional adaptor protein. It remains to be determined whether NAG induces NLRP3 inflammasome assembly in human cells, although it does elicit IL-1β secretion in human macrophages and dendritic cells108.
The peptidoglycan of Gram-positive bacterial cell walls is substantially thicker than in Gram-negative bacteria106. Thus, peptidoglycan is the most prevalent cell-wall-associated PAMP in Gram-positive bacteria, and innate recognition of peptidoglycan would be a more efficient method of sensing cytosolic Gram-positive bacteria than the recognition of lipoproteins or LTA. However, the level of IL-1β produced downstream of peptidoglycan is much lower than that of other classic activators of the NLRP3 inflammasome108. This finding is probably due to the unusual observation that NAG does not induce pyroptosis downstream of NLRP3 inflammasome assembly108, whereas other PAMPs do35.
Formation of the NLRP3 inflammasome in response to peptidoglycan suggests that, in mice, extracellular versus intracellular Gram-positive bacteria are recognized in distinct ways. Extracellular bacteria are detected by lipoprotein-sensing PRRs, whereas intra-cellular bacteria are detected by peptidoglycan-sensing PRRs. Gram-negative bacteria, by contrast, are recognized extracellularly and intracellularly by a single PAMP, LPS. There may be several reasons why diverse mechanisms evolved to detect Gram-positive bacteria. Lipoproteins extend from the surface of the cell beyond the peptidoglycan matrix, where they can be easily sensed by TLR2, TLR1 and/or TLR6. Peptidoglycan, by contrast, is biophysically excluded from cell-surface PRRs. Inside the cell, however, after phagosomal degradation of bacteria and the release or transport of degradation products into the cytosol, peptidoglycan fragments become accessible and are more ubiquitous than lipoproteins. Thus, it may be a combination of access as well as PAMP prevalence that led to the evolution of distinct PRRs and SMOCs for the detection of Gram-positive bacteria.
Innate detection of commensals and pathogens
PAMPs are generic microbial signatures that are associated with commensal and pathogenic organisms. Consequently, PRRs have evolved to recognize PAMPs without contextual information as to whether the bacteria are virulent or avirulent. Thus, the relationship between microorganism and mammalian host must be regulated to ensure an appropriate immune response. Some have proposed that host sensors detect additional evidence of pathogenicity that helps to distinguish commensals and pathogens114,115. In addition, micro-organisms can modulate their interactions with the host by structurally altering their PAMPs to diminish PRR recognition. This strategy is used by commensals and pathogens alike.
Pathogen evasion of cell surface and cytosolic sensing.
One common PAMP alteration is the modification of LPS. Because LPS is such a potent stimulant, modifications to the structure of LPS are prevalent among bacteria — including extracellular and intracellular pathogens and commensals11,41. Often, these are modifications to the phosphorylation status of the lipid A sugar head group or the number of its acyl chains41; both of these moieties affect LPS interaction with CD14 and, consequently, downstream interactions with TLR4 and MD2 (REFS 11,16,41). Many pathogens, such as Yersinia pestis116 and Shigella flexneri117, alter lipid A acyl chain composition to avoid TLR4 recognition, as these moieties bind inside the MD2-binding pocket41. Decreasing the number of acyl chains destabilizes the lipid A–MD2 interaction as well as the lipid A interaction with the second TLR4 monomer in the TLR4–TLR4 homodimer16,41. Decreasing the number of acyl chains also limits the detection of LPS intra cellularly. Francisella novicida, which dwells in the cytosol, evades caspase 11-mediated detection by enzymatically removing one lipid A acyl chain42. Similarly, caspase 11 recognizes Y. pestis under conditions in which its LPS is hexa-acylated but not when it is tetra-acylated42. Thus, a similar evolutionary strategy of altering LPS structures promotes efficient evasion of cell-surface and cytosolic PRRs.
Pathogens also modify other PAMPs that are recognized by cell surface TLRs70. For example, Helicobacter pylori flagellin is non-stimulatory in comparison with flagellin from other gastrointestinal bacterial pathogens, such as Salmonella enterica subsp. enterica serovar Typhimurium (S. Typhimurium)118. Whereas purified S. Typhimurium flagellin potently induces pro-inflammatory IL-8 production and p38 phosphorylation in gastric epithelial cells, purified H. pylori flagellin elicits neither response118. H. pylori flagella are crucial for bacterial survival in the gastrointestinal tract119, which indicates that flagella are produced during infection but that adaptations to evade detection have evolved. These data suggest that structural changes to flagellin, perhaps locally at the site of TLR5 binding, promote immune avoidance by diminishing PRR detection, and this is another strategy used by pathogens and commensals70 to promote stable mammalian–microbial relationships. Similarly, intracellular detection of flagellin differs between avirulent and virulent bacteria. Flagella derived from S. Typhimurium and non-pathogenic E. coli, although similarly recognized by TLR5, differentially stimulate assembly of the NLRC4 inflammasome. Nonpathogenic E. coli flagellin induces minimal activation of caspase 1, whereas S. Typhimurium flagellin elicits robust caspase 1 processing, resulting in higher levels of IL-1β and cell death120. The specific structural reasons underlying this differential assembly of the NLRC4 inflammasome remain undetermined, but many bacteria have evolved mechanisms to evade intracellular flagellin recognition.
PAMPs have also evolved to evade detection by other PRRs in the cytosol. Bacillus anthracis encodes an N-acetyl glucosamine deacetylase that removes the acetyl group from the NAG constituent (detected by hexokinase) that stimulates assembly of the NLRP3 inflammasome108. NAG inhibits hexokinase, whereas the deacetylated glucosamine does not, and thus does not induce NLRP3 inflammasome formation108. However, in vitro reacetylated NAG elicits NLRP3-dependent IL-1β secretion at a level similar to NAG108. Peptidoglycan deacetylation can also promote PRR recognition. The gene oatA in S. aureus encodes a peptidoglycan acetyl-transferase that acetylates NAM, resulting in a diacetylated muramic acid. This modification mediates resistance to host lysozyme121 and also promotes evasion of NLRP3 detection113. Mutant bacteria that no longer express oatA, and thus generate monoacetylated NAM, elicit substantially higher levels of IL-1β and IL-18 in comparison with wild-type bacteria, and are consequently cleared faster113. Thus, pathogens that avoid PRR detection by changing their peptidoglycan acetylation status limit inflammation and bacterial clearance. Altogether, these data indicate that, for Gram-positive bacteria, structural adaptation of peptidoglycan (their pre dominant PAMP) may be an effective method of avoiding detection. Indeed, for clinically important bacterial pathogens, such as Mycobacterium tuberculosis, modifications to peptidoglycan facilitate bacterial survival during infection106.
Commensal manipulation of PRR signalling.
Similar to pathogens that must evade PRR detection for survival in the host, commensal organisms must manipulate PRR detection to maintain host colonization. Recent work showed that the prominent gut symbiont Bacteroides thetaiotaomicron (B. theta) encodes a phosphatase, LpxF, that removes a phosphate from the lipid A sugar122. Monophosphorylated LPS induces less TLR4 signalling due to decreased electrostatic interactions with CD14 (REFS 41,123). Consequently, TLR4 dimerization, myddosome formation and inflammatory gene expression is inefficient11.
In addition to changing their cell wall structures to avoid PRR detection, bacteria, including commensals, encode various factors that actively interfere with innate immune signal transduction124,125. The best-studied example of commensal perturbation of innate immune signalling is from the gut commensal Bacteroides fragilis. B. fragilis produces a capsule sugar called polysaccharide A (PSA) that is a crucial regulator of the relationship between B. fragilis and the host. PSA induces the proliferation of forkhead box P3 (FOXP3)+ regulatory T cells and the production of the anti-inflammatory cytokine IL-10 (REFS 126,127) to facilitate the residence of B. fragilis in the gastrointestinal tract. To exert these effects, PSA putatively signals through TLR2126,127. There is some evidence that TLR2 is expressed on CD4+ T cells and is functional128,129, although it remains unclear how TLR2 would sense PSA presented on dendritic cells or whether the canonical TLR pathway is functional. Nevertheless, TLR2 seems to be required for dendritic-cell- and self-mediated induction of IL-10 release from T cells127, which is in concordance with the known requirement for TLR signalling in IL-10 production130.
Overall, commensals and pathogens have evolved multiple mechanisms to actively perturb or avoid PRR detection at the cell surface and the cytosol. These adaptations highlight the importance of PRR recognition of PAMPs in mediating appropriate mammalian–microbial relationships — for defence or mutualism.
Limitations and future outlook
In this Review, we discussed several themes that transcend the specifics associated with any given signalling pathway of the innate immune system. These themes include the detection of individual PAMPs by various PRRs, the use of SMOCs to activate unique cellular responses and the ability of these systems to promote inflammation. Although the specific regulators of these responses differ from pathway to pathway, the fundamentals seem to be fixed. The organization of the pathways of the innate immune system may not be constrained by protein structure, but by system organization.
Despite our attempts to provide some unifying features of the immune signalling pathways, our understanding of how well individual PRRs adhere to signalling paradigms is far from complete. Indeed, there are large gaps in our knowledge of most aspects of innate immune signal transduction. Filling these gaps will require tool development and a concerted effort to pair host or bacterial genetics with biochemical and cell biological assays to robustly define molecular steps downstream of PRRs.
One major benefit and limitation of our current approach to studying innate immunity is the use of minimalist experimental systems with a purified PAMP in a single cell type. Although these minimalist systems are crucial for understanding the essential requirements for PAMP sensing and SMOC formation, they do not necessarily recapitulate PAMP detection in physiological contexts. For example, many of the experimentally used TLR2 ligands are chemically synthesized instead of naturally purified forms. Do these synthetic versions stimulate TLR2 signalling at physiological levels? In addition to the potential inconsistencies that arise from using purified or synthetic ligands, some evidence points to discrepancies in signal transduction from PAMPs that are encountered in the context of a whole bacterium as opposed to PAMPs encountered in isolation. For example, why is ASC required for NLRC4 inflammasome assembly upon bacterial infection but is not required for inflammasome assembly in response to purified flagellin67,68,131,132? It could be that encountering flagellin on the surface of a bacterium requires more efficient signal transduction, which ASC may promote in certain contexts. Thus, although synthetic and purified ligands are valuable for determining the molecular mechanisms of signalling, future studies will benefit from physiological experiments that test the minimal signalling requirements defined in simplified systems.
Moreover, PAMP detection by extracellular and intracellular PRRs underlies important processes of mammalian development, homeostasis and protection from infection1,2,14. Although our understanding of these complex molecular pathways continues to expand, there are also gaps in our knowledge of how these processes are regulated spatiotemporally and in complex organismal systems. Mammalian cells encounter a milieu of microbial molecules, particularly in the complex environments of the human gut, lung or skin, where cells naturally encounter simultaneous signals from pathogens and commensals. How do cells integrate these mixed signals? Are the signals even integrated? Our understanding of innate immune signalling is rudimentary in complex microbial eco systems. Single-cell analyses will be useful in deciphering how cells sense and respond in a mixed signal environment. This extends not only to how host cells detect different types of bacteria but also to the metabolites produced by bacteria — many gut commensal metabolites are immunomodulatory133. Discovering how these metabolites regulate innate immune cell function in the presence of commensal or pathogenic bacteria would improve our understanding of cellular responses in a more physiological context.
Acknowledgements
The authors thank members of the Kagan laboratory for helpful discussions and the anonymous referees for insightful critiques. The US National Institutes of Health (grants AI116550, AI093589 and P30 DK34854), the Burroughs Wellcome Fund (Investigator of the Pathogenesis of Infectious Disease Award) and an unrestricted gift from Mead Johnson & Company support research in the laboratory of J.C.K. K.J.K. is supported by a Life Sciences Research Foundation postdoctoral fellowship (Good Ventures Fellow).
Glossary
- Pyroptosis
A form of cell death that is inflammatory and is executed by the inflammatory caspase 1 and caspase 11, as opposed to the apoptotic caspases (for example, caspase 8). During pyroptosis, the plasma membrane blebs, the cell lyses and pro-inflammatory cytokines are released.
- Myddosome
A supramolecular organizing centre that forms to initiate Toll-like receptor (TLR)-dependent inflammatory responses. It is comprised of Toll/interleukin-1 receptor domain-containing adaptor protein (TIRAP) bound to the TIR domain of the TLR, to which myeloid differentiation primary response protein 88 (MYD88) binds. The interleukin-1 receptor-associated kinase (IRAK) kinases IRAK4 and IRAK2 or IRAK1 then associate to initiate a pro-inflammatory signalling cascade. The myddosome can form at the plasma or endosomal membrane.
- Inflammasome
A supramolecular organizing centre that forms to initiate inflammatory responses to cytosolic bacterial products. It is formed by a pattern-recognition receptor, the adaptor protein ASC and caspase 1. It results in the production of interleukin-1β and cleavage of gasdermin D.
- Supramolecular organizing centres
(SMOCs). Multi-protein complexes that are assembled on detection of microorganisms, either at the plasma membrane or on intracellular organelles. These large complexes activate enzymes that initiate inflammatory responses to promote host defence.
- Triffosome
A putative supramolecular organizing centre that assembles on the endosomal membrane in response to activated Toll-like receptor 4 (TLR4) and TLR3. Its main constituent is TIR domain-containing adaptor protein inducing IFNβ (TRIF), which through TNF receptor-associated factor 3 (TRAF3) or TRAF6 can activate interferon regulatory factor 3 (IRF3) or IRF7, respectively, for interferon production. Apart from TRIF-related adaptor molecule (TRAM), which bridges TRIF to TLR4, the adaptor molecules used to assemble the triffosome remain unknown.
- Inflammatory caspases
This group of caspases is composed of caspase 1, caspase 11 and caspase 12. These proteases are involved in the inflammasome pathway and promote an inflammatory form of cell death (pyroptosis). Caspase 1 cleaves immature pro-interleukin-1β (pro-IL-1β) and pro-IL-18 to their mature forms. Caspase 1 and caspase 11 cleave gasdermin D, which is required for pyroptosis. Caspase 4 and caspase 5 are the human homologues of caspase 11. Inflammatory caspases are distinct from apoptotic caspases, which promote the programmed cell death pathway called apoptosis and include caspase 2, caspase 3 and caspase 6 to caspase 10.
- Flagellum
A helical filament that extends from the bacterial cell surface. It promotes bacterial movement and is often essential for flagellated pathogens to be able to colonize their respective hosts.
- Lipoteichoic acid
(LTA). A polymer found in the cell wall of Gram-positive bacteria that is embedded in the bacterial plasma membrane by a glycerol head group. Attached to the head group are long chains of either ribitol or glycerol phosphate. It is a Toll-like receptor 2 ligand.
- Pam3CSK4
A synthetic lipoprotein containing three palmitoyl (Pam) chains that are glycerol-linked to the short peptide CSKKKK (CSK4). It is a classic ligand for Toll-like receptor 2 (TLR2)–TLR1 activity. Pam2CSK4 stimulates TLR2–TLR6.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S & Medzhitov R Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Slack E et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science 325, 617–620 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janeway CA Pillars article: approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol 54, 1–13 (1989). [DOI] [PubMed] [Google Scholar]
- 4.Medzhitov R Approaching the asymptote: 20 years later. Immunity 30, 766–775 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Lemaitre B, Nicolas E, Michaut L, Reichhart JM & Hoffmann JA The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 86, 973–983 (1996). [DOI] [PubMed] [Google Scholar]
- 6.Medzhitov R, Preston-Hurlburt P & Janeway CA A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388, 394–397 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Hayashi F et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410, 1099–1103 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Smith KD et al. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol 4, 1247–1253 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Gioannini TL et al. Isolation of an endotoxin-MD-2 complex that produces Toll-like receptor 4-dependent cell activation at picomolar concentrations. Proc. Natl Acad. Sci. USA 101, 4186–4191 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zanoni I et al. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell 147, 868–880 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work was the first to demonstrate a broadly utilized TLR4-independent response to LPS, mediated by CD14.
- 11.Tan Y, Zanoni I, Cullen TW, Goodman AL & Kagan JC Mechanisms of Toll-like receptor 4 endocytosis reveal a common immune-evasion strategy used by pathogenic and commensal bacteria. Immunity 43, 909–922 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeuchi O et al. Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J. Immunol 169, 10–14 (2002). [DOI] [PubMed] [Google Scholar]
- 13.Takeuchi O et al. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int. Immunol 13, 933–940 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Brubaker SW, Bonham KS, Zanoni I & Kagan JC Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol 33, 257–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright SD, Ramos RA, Tobias PS, Ulevitch RJ & Mathison JC CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science 249, 1431–1433 (1990). [DOI] [PubMed] [Google Scholar]
- 16.Park BS et al. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458, 1191–1195 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Miyake K Innate recognition of lipopolysaccharide by CD14 and toll-like receptor 4-MD-2: unique roles for MD-2. Int. Immunopharmacol 3, 119–128 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Song DH & Lee J-O Sensing of microbial molecular patterns by Toll-like receptors. Immunol. Rev 250, 216–229 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Mellman I Dendritic cells: master regulators of the immune response. Cancer Immunol. Res 1, 145–149 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Medzhitov R et al. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol. Cell 2, 253–258 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto M et al. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-promoter in the Toll-like receptor signaling. J. Immunol 169, 6668–6672 (2002). [DOI] [PubMed] [Google Scholar]
- 22.Oshiumi H, Matsumoto M, Funami K, Akazawa T & Seya T TICAM-1, an adaptor molecule that participates in Toll-like receptor 3–mediated interferon-β induction. Nat. Immunol 4, 161–167 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Horng T, Barton GM & Medzhitov R TIRAP: an adapter molecule in the Toll signaling pathway. Nat. Immunol 2, 835–841 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Fitzgerald KA, Palsson-McDermott EM & Bowie AG Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature 413, 78–83 (2001). [DOI] [PubMed] [Google Scholar]; References 23 and 24 report the discovery of TIRAP, a major factor that regulates TLR signalling. These papers demonstrated that additional factors regulate TLR signalling, leading to successive discoveries of other TLR adaptor proteins.
- 25.Fitzgerald KA et al. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J. Exp. Med 198, 1043–1055 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gay NJ, Symmons MF, Gangloff M & Bryant CE Assembly and localization of Toll-like receptor signalling complexes. Nat. Rev. Immunol 14, 546–558 (2014). [DOI] [PubMed] [Google Scholar]
- 27.Bonham KS et al. A promiscuous lipid-binding protein diversifies the subcellular sites of Toll-like receptor signal transduction. Cell 156, 705–716 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motshwene PG et al. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J. Biol. Chem 284, 25404–25411 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin S-C, Lo Y-C & Wu H Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature 465, 885–890 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kagan JC, Magupalli VG & Wu H SMOCs: supramolecular organizing centres that control innate immunity. Nat. Rev. Immunol 14, 821–826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamoto M et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 301, 640–643 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Kagan JC et al. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-β. Nat. Immunol 9, 361–368 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinon F, Burns K & Tschopp J The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10, 417–426 (2002). [DOI] [PubMed] [Google Scholar]; This report identified the first inflammasome complex and initiated the field of inflammasome biology.
- 34.Broz P & Dixit VM Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol 16, 407–420 (2016). [DOI] [PubMed] [Google Scholar]
- 35.von Moltke J, Ayres JS, Kofoed EM, Chavarría-Smith J & Vance RE Recognition of bacteria by inflammasomes. Annu. Rev. Immunol 31, 73–106 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Das S et al. Brain angiogenesis inhibitor 1 (BAI1) is a pattern recognition receptor that mediates macrophage binding and engulfment of Gram-negative bacteria. Proc. Natl Acad. Sci. USA 108, 2136–2141 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Billings EA et al. The adhesion GPCR BAI1 mediates macrophage ROS production and microbicidal activity against Gram-negative bacteria. Sci. Signal 9, ra14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura N et al. Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature 509, 240–244 (2014). [DOI] [PubMed] [Google Scholar]
- 39.Park JH et al. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J. Immunol 178, 2380–2386 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Sorbara MT et al. The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity 39, 858–873 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Tan Y & Kagan JCA Cross-disciplinary perspective on the innate immune responses to bacterial lipopolysaccharide. Mol. Cell 54, 212–223 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hagar JA, Powell DA, Aachoui Y, Ernst RK & Miao EA Cytoplasmic LPS activates caspase-11: implications in TLR4-independent endotoxic shock. Science 341, 1250–1253 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kayagaki N et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science 341, 1246–1249 (2013). [DOI] [PubMed] [Google Scholar]
- 44.Poltorak A et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282, 2085–2088 (1998). [DOI] [PubMed] [Google Scholar]; References 6 and 44 identified TLR4 as the first human TLR and defined TLR4 as the receptor for LPS, respectively, laying the groundwork for the hypothesis that dedicated receptors for PAMP recognition exist.
- 45.Kagan JC & Medzhitov R Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell 125, 943–955 (2006). [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto M et al. TRAM is specifically involved in the Toll-like receptor 4–mediated MyD88-independent signaling pathway. Nat. Immunol 4, 1144–1150 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Dunzendorfer S, Lee H-K, Soldau K & Tobias PS TLR4 is the signaling but not the lipopolysaccharide uptake receptor. J. Immunol 173, 1166–1170 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Chiang CY, Veckman V, Limmer K & David M Phospholipase C −2 and intracellular calcium are required for lipopolysaccharide-induced Toll-like receptor 4 (TLR4) endocytosis and interferon regulatory factor 3 (IRF3) activation. J. Biol. Chem 287, 3704–3709 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shi J et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature 514, 187–192 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Kayagaki N et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 526, 666–671 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Shi J et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 526, 660–665 (2015). [DOI] [PubMed] [Google Scholar]; References 50 and 51 identified GSDMD as the effector of pyroptosis, one of the main pathways of cell death.
- 52.Aglietti RA et al. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc. Natl Acad. Sci. USA 113, 7858–7863 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu X et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 535, 153–158 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding J et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 535, 111–116 (2016). [DOI] [PubMed] [Google Scholar]
- 55.Rühl S & Broz P Caspase-11 activates a canonical NLRP3 inflammasome by promoting K +efflux. Eur. J. Immunol 45, 2927–2936 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Aachoui Y et al. Caspase-11 protects against bacteria that escape the vacuole. Science 339, 975–978 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kofoed EM & Vance RE Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477, 592–595 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao Y et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477, 596–600 (2011). [DOI] [PubMed] [Google Scholar]
- 59.Choi YJ, Jung J, Chung HK, Im E & Rhee SH PTEN regulates TLR5-induced intestinal inflammation by controlling Mal/TIRAP recruitment. FASEB J 27, 243–254 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi YJ, Im E, Chung HK, Pothoulakis C & Rhee SH TRIF mediates Toll-like receptor 5-induced signaling in intestinal epithelial cells. J. Biol. Chem 285, 37570–37578 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parker D & Prince A Epithelial uptake of flagella initiates proinflammatory signaling. PLoS ONE 8, e59932 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eaves-Pyles T et al. Luminal-applied flagellin is internalized by polarized intestinal epithelial cells and elicits immune responses via the TLR5 dependent mechanism. PLoS ONE 6, e24869 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim Y-M, Brinkmann MM, Paquet M-E & Ploegh HL UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature 452, 234–238 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Huh JW et al. UNC93B1 is essential for the plasma membrane localization and signaling of Toll-like receptor 5. Proc. Natl Acad. Sci. USA 111, 7072–7077 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Franchi L et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in salmonella-infected macrophages. Nat. Immunol 7, 576–582 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Miao EA et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat. Immunol 7, 569–575 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Matusiak M et al. Flagellin-induced NLRC4 phosphorylation primes the inflammasome for activation by NAIP5. Proc. Natl Acad. Sci. USA 112, 1541–1546 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Case CL, Shin S & Roy CR Asc and Ipaf inflammasomes direct distinct pathways for caspase-1 activation in response to Legionella pneumophila. Infect. Immun 77, 1981–1991 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yoon S-I et al. Structural basis of TLR5-flagellin recognition and signaling. Science 335, 859–864 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andersen-Nissen E et al. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc. Natl Acad. Sci. USA 102, 9247–9252 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tenthorey JL, Kofoed EM, Daugherty MD, Malik HS & Vance RE Molecular basis for specific recognition of bacterial ligands by NAIP/NLRC4 inflammasomes. Mol. Cell 54, 17–29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qu Y et al. Phosphorylation of NLRC4 is critical for inflammasome activation. Nature 490, 539–542 (2012). [DOI] [PubMed] [Google Scholar]
- 73.Suzuki S et al. Shigella type III secretion protein MxiI is recognized by Naip2 to induce Nlrc4 inflammasome activation independently of Pkcδ. PLoS Pathog 10, e1003926 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miao EA et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl Acad. Sci. USA 107, 3076–3080 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang J, Zhao Y, Shi J & Shao F Human NAIP and mouse NAIP1 recognize bacterial type III secretion needle protein for inflammasome activation. Proc. Natl Acad. Sci. USA 110, 14408–14403 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Blocker A, Komoriya K & Aizawa S-I Type III secretion systems and bacterial flagella: insights into their function from structural similarities. Proc. Natl Acad. Sci. USA 100, 3027–3030 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cole LE et al. Toll-like receptor 2-mediated signaling requirements for francisella tularensis live vaccine strain infection of murine macrophages. Infect. Immun 75, 4127–4137 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brightbill HD et al. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science 285, 732–736 (1999). [DOI] [PubMed] [Google Scholar]
- 79.Aliprantis AO et al. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science 285, 736–739 (1999). [DOI] [PubMed] [Google Scholar]
- 80.Khare S et al. An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity 36, 464–476 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakata T et al. CD14 directly binds to triacylated lipopeptides and facilitates recognition of the lipopeptides by the receptor complex of Toll-like receptors 2 and 1 without binding to the complex. Cell. Microbiol 8, 1899–1909 (2006). [DOI] [PubMed] [Google Scholar]
- 82.Raby A-C et al. Targeting the TLR co-receptor CD14 with TLR2-derived peptides modulates immune responses to pathogens. Sci. Transl Med 5, 185ra64 (2013). [DOI] [PubMed] [Google Scholar]
- 83.Stuart LM et al. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J. Cell Biol 170, 477–485 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hoebe K et al. CD36 is a sensor of diacylglycerides. Nature 433, 523–527 (2005). [DOI] [PubMed] [Google Scholar]
- 85.Jiang Z et al. CD14 is required for MyD88-independent LPS signaling. Nat. Immunol 6, 565–570 (2005). [DOI] [PubMed] [Google Scholar]
- 86.Takeuchi O et al. Cutting edge: preferentially the R-stereoisomer of the mycoplasmal lipopeptide macrophage-activating lipopeptide-2 activates immune cells through a toll-like receptor 2- and MyD88-dependent signaling pathway. J. Immunol 164, 554–557 (2000). [DOI] [PubMed] [Google Scholar]
- 87.Horng T, Barton GM, Flavell RA & Medzhitov R The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature 420, 329–333 (2002). [DOI] [PubMed] [Google Scholar]
- 88.Yamamoto M et al. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature 420, 324–329 (2002). [DOI] [PubMed] [Google Scholar]
- 89.Hirschfeld M et al. Signaling by toll-like receptor 2 and 4 agonists results in differential gene expression in murine macrophages. Infect. Immun 69, 1477–1482 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Toshchakov V et al. TLR4, but not TLR2, mediates IFN-β–induced STAT1α/β-dependent gene expression in macrophages. Nat. Immunol 3, 392–398 (2002). [DOI] [PubMed] [Google Scholar]
- 91.Barbalat R, Lau L, Locksley RM & Barton GM Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat. Immunol 10, 1200–1207 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cervantes JL & Dunham-Ems SM Phagosomal signaling by Borrelia burgdorferi in human monocytes involves Toll-like receptor (TLR) 2 and TLR8 cooperativity and TLR8-mediated induction of IFN-β. Proc. Natl Acad. Sci. USA 108, 3683–3688 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dietrich N, Lienenklaus S, Weiss S & Gekara NO Murine Toll-like receptor 2 activation induces type I interferon responses from endolysosomal compartments. PLoS ONE 5, e10250 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nilsen NJ et al. A role for the adaptor proteins TRAM and TRIF in toll-like receptor 2 signaling. J. Biol. Chem 290, 3209–3222 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stack J et al. TRAM is required for TLR2 endosomal signaling to type I IFN induction. J. Immunol 193, 6090–6102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Triantafilou M et al. Lipoteichoic acid and Toll-like receptor 2 internalization and targeting to the Golgi are lipid raft-dependent. J. Biol. Chem 279, 40882–40889 (2004). [DOI] [PubMed] [Google Scholar]
- 97.Motoi Y et al. Lipopeptides are signaled by Toll-like receptor 1, 2 and 6 in endolysosomes. Int. Immunol 26, 563–573 (2014). [DOI] [PubMed] [Google Scholar]
- 98.Heit B et al. Multimolecular signaling complexes enable Syk-mediated signaling of CD36 internalization. Dev. Cell 24, 372–383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Underhill DM et al. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature 401, 811–815 (1999). [DOI] [PubMed] [Google Scholar]
- 100.O’Connell CM, Ionova IA, Quayle AJ, Visintin A & Ingalls RR Localization of TLR2 and MyD88 to Chlamydia trachomatis inclusions. Evidence for signaling by intracellular TLR2 during infection with an obligate intracellular pathogen. J. Biol. Chem 281, 1652–1659 (2006). [DOI] [PubMed] [Google Scholar]
- 101.Radian AD, Khare S, Chu LH, Dorfleutner A & Stehlik C ATP binding by NLRP7 is required for inflammasome activation in response to bacterial lipopeptides. Mol. Immunol 67, 294–302 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ozören N et al. Distinct roles of TLR2 and the adaptor ASC in IL-1beta/IL-18 secretion in response to Listeria monocytogenes. J. Immunol 176, 4337–4342 (2006). [DOI] [PubMed] [Google Scholar]
- 103.Zhou Y et al. Virulent Mycobacterium bovis Beijing strain activates the NLRP7 inflammasome in THP-1 macrophages. PLoS ONE 11, e0152853 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mariathasan S et al. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430, 213–218 (2004). [DOI] [PubMed] [Google Scholar]
- 105.Wang L et al. Enterococcus faecalis lipoteichoic acid-induced NLRP3 inflammasome via the activation of the nuclear factor kappa B pathway. J. Endod 42, 1093–1100 (2016). [DOI] [PubMed] [Google Scholar]
- 106.Kieser KJ & Rubin EJ How sisters grow apart: mycobacterial growth and division. Nat. Rev. Microbiol 12, 550–562 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen G, Shaw MH, Kim Y-G & Núñez G NOD-like receptors: role in innate immunity and inflammatory disease. Annu. Rev. Pathol 4, 365–398 (2009). [DOI] [PubMed] [Google Scholar]
- 108.Wolf AJ et al. Hexokinase is an innate immune receptor for the detection of bacterial peptidoglycan. Cell 166, 624–636 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Irving AT et al. The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe 15, 623–635 (2014). [DOI] [PubMed] [Google Scholar]
- 110.Tanabe T et al. Regulatory regions and critical residues of NOD2 involved in muramyl dipeptide recognition. EMBO J 23, 1587–1597 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Philpott DJ, Sorbara MT, Robertson SJ, Croitoru K & Girardin SE NOD proteins: regulators of inflammation in health and disease. Nat. Rev. Immunol 14, 9–23 (2013). [DOI] [PubMed] [Google Scholar]
- 112.Martinon F, Agostini L, Meylan E & Tschopp J Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr. Biol 14, 1929–1934 (2004). [DOI] [PubMed] [Google Scholar]
- 113.Shimada T et al. Staphylococcus aureus evades lysozyme-based peptidoglycan digestion that links phagocytosis, inflammasome activation, and IL-1beta secretion. Cell Host Microbe 7, 38–49 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Vance RE, Isberg RR & Portnoy DA Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe 6, 10–21 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Blander JM & Sander LE Beyond pattern recognition: five immune checkpoints for scaling the microbial threat. Nat. Rev. Immunol 12, 215–225 (2012). [DOI] [PubMed] [Google Scholar]
- 116.Kawahara K, Tsukano H, Watanabe H, Lindner B & Matsuura M Modification of the structure and activity of lipid A in Yersinia pestis lipopolysaccharide by growth temperature. Infect. Immun 70, 4092–4098 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Paciello I et al. Intracellular Shigella remodels its LPS to dampen the innate immune recognition and evade inflammasome activation. Proc. Natl Acad. Sci. USA 110, E4345–E4354 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gewirtz AT et al. Helicobacter pylori flagellin evades toll-like receptor 5-mediated innate immunity. J. Infect. Dis 189, 1914–1920 (2004). [DOI] [PubMed] [Google Scholar]
- 119.Kao C-Y, Sheu B-S & Wu J-J Helicobacter pylori infection: an overview of bacterial virulence factors and pathogenesis. Biomed. J 39, 14–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yang J et al. Flagellins of Salmonella Typhi and nonpathogenic Escherichia coli are differentially recognized through the NLRC4 pathway in macrophages. J. Innate Immun 6, 47–57 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bera A, Biswas R, Herbert S & Götz F The presence of peptidoglycan O-acetyltransferase in various staphylococcal species correlates with lysozyme resistance and pathogenicity. Infect. Immun 74, 4598–4604 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cullen TW et al. Gut microbiota. Antimicrobial peptide resistance mediates resilience of prominent gut commensals during inflammation. Science 347, 170–175 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Maeshima N & Fernandez RC Recognition of lipid A variants by the TLR4-MD-2 receptor complex. Front. Cell. Infect. Microbiol 3, 3 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Rosadini CV & Kagan JC Microbial strategies for antagonizing Toll-like-receptor signal transduction. Curr. Opin. Immunol 32, 61–70 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Baxt LA, Garza-Mayers AC & Goldberg MB Bacterial subversion of host innate immune pathways. Science 340, 697–701 (2013). [DOI] [PubMed] [Google Scholar]
- 126.Mazmanian SK, Liu CH, Tzianabos AO & Kasper DL An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118 (2005). [DOI] [PubMed] [Google Scholar]
- 127.Round JL et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332, 974–977 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Komai-Koma M, Jones L, Ogg GS, Xu D & Liew FY TLR2 is expressed on activated T cells as a costimulatory receptor. Proc. Natl Acad. Sci. USA 101, 3029–3034 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Reynolds JM et al. Toll-like receptor 2 signaling in CD4+ T lymphocytes promotes T helper 17 responses and regulates the pathogenesis of autoimmune disease. Immunity 32, 692–702 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Saraiva M & O’Garra A The regulation of IL-10 production by immune cells. Nat. Rev. Immunol 10, 170–181 (2010). [DOI] [PubMed] [Google Scholar]
- 131.Lage SL, Buzzo CL & Amaral EP Cytosolic flagellin-induced lysosomal pathway regulates inflammasome-dependent and-independent macrophage responses. Proc. Natl Acad. Sci. USA 110, E3321–E3330 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Case CL & Roy CR Asc modulates the function of NLRC4 in response to infection of macrophages by Legionella pneumophila. mBio 2, e00117–11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Belkaid Y & Hand TW Role of the microbiota in immunity and inflammation. Cell 157, 121–141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Geijtenbeek TBH & Gringhuis SI Signalling through C-type lectin receptors: shaping immune responses. Nat. Rev. Immunol 9, 465–479 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rogers NC et al. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity 22, 507–517 (2005). [DOI] [PubMed] [Google Scholar]
- 136.Zhu L-L et al. C-Type lectin receptors Dectin-3 and Dectin-2 form a heterodimeric pattern-recognition receptor for host defense against fungal infection. Immunity 39, 324–334 (2013). [DOI] [PubMed] [Google Scholar]
- 137.Gringhuis SI et al. Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1β via a noncanonical caspase-8 inflammasome. Nat. Immunol 13, 246–254 (2012). [DOI] [PubMed] [Google Scholar]
- 138.Barbalat R, Ewald SE, Mouchess ML & Barton GM Nucleic acid recognition by the innate immune system. Annu. Rev. Immunol 29, 185–214 (2011). [DOI] [PubMed] [Google Scholar]
- 139.Lund J, Sato A, Akira S, Medzhitov R & Iwasaki A Toll-like receptor 9–mediated recognition of herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med 198, 513–520 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ewald SE et al. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature 456, 658–662 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ewald SE et al. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J. Exp. Med 208, 643–651 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Oldenburg M et al. TLR13 recognizes bacterial 23S rRNA devoid of erythromycin resistance-forming modification. Science 337, 1111–1115 (2012). [DOI] [PubMed] [Google Scholar]
- 143.Liu S et al. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. eLife 2, e00785 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu S et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015). [DOI] [PubMed] [Google Scholar]
- 145.Yoneyama M et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol 5, 730–737 (2004). [DOI] [PubMed] [Google Scholar]
- 146.Satoh T et al. LGP2 is a positive regulator of RIG-I− and MDA5-mediated antiviral responses. Proc. Natl Acad. Sci. USA 107, 1512–1517 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bruns AM, Leser GP, Lamb RA & Horvath CM The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5- RNA interaction and filament assembly. Mol. Cell 55, 771–781 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chiu Y-H, MacMillan JB & Chen ZJ RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell 138, 576–591 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Burdette DL et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature 478, 515–518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sun L, Wu J, Du F, Chen X & Chen ZJ Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen Q, Sun L & Chen ZJ Regulation and function of the cGAS–STING pathway of cytosolic DNA sensing. Nat. Immunol 17, 1142–1149 (2016). [DOI] [PubMed] [Google Scholar]
- 152.Schaefer L Complexity of danger: the diverse nature of damage-associated molecular patterns. J. Biol. Chem 289, 35237–35245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zanoni I et al. An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells. Science 352, 1232–1236 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bochkov VN et al. Protective role of phospholipid oxidation products in endotoxin-induced tissue damage. Nature 419, 77–81 (2002). [DOI] [PubMed] [Google Scholar]
- 155.Yeon SH, Yang G, Lee HE & Lee JY Oxidized phosphatidylcholine induces the activation of NLRP3 inflammasome in macrophages. J. Leukoc. Biol 101, 205–215 (2017). [DOI] [PubMed] [Google Scholar]
- 156.Rathinam VAK et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol 11, 395–402 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Vladimer GI et al. The NLRP12 inflammasome recognizes Yersinia pestis. Immunity 37, 96–107 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]