Abstract
Sirtuins are a class of enzyme with NAD+-dependent protein lysine deacylase activities. They were initially discovered to regulate transcription and life span via histone deacetylase activities. Later studies expanded their activities to other proteins and acyl lysine modifications. Through deacylating various substrate proteins, they regulate many biological processes, including transcription, DNA repair and genome stability, metabolism, and signal transduction. Here we review recent understandings of the epigenetic functions (broadly defined to include transcriptional, post-transcriptional regulation, and DNA repair) of mammalian sirtuins. Due to the important functions of sirtuins, their own regulation is of great interest and is also discussed.
Introduction
Sirtuins are a class of enzymes that have attracted much interest in the past decades. They were initially discovered as the regulators of aging and epigenetics. The founding member, yeast silencing information regulator 2 (Sir2), was found to be important for gene silencing and calorie restriction-induced life span extension in yeast [1,2]. While studying the gene silencing and longevity roles of Sir2, it was discovered that Sir2 is an NAD+-dependent histone deacetylase [3,4], which stimulated great interest in this class of enzymes. In mammals, there are seven Sir2 homologs or sirtuins (SIRT1–7), found to regulate numerous substrate proteins and biological pathways. Interestingly, several sirtuins later were found to preferentially hydrolyze other acyl lysine modifications, such as succinyl and long-chain fatty acyl groups [5–9]. Although mammalian sirtuins have many other regulatory roles, such as those in metabolism and cell signaling, their function in epigenetics remains of great interest and is highlighted in many excellent review articles [10–13]. Here we focus on the developments in the epigenetic roles of mammalian sirtuins in the last two years.
The epigenetic roles of SIRT1
SIRT1 is the closest mammalian ortholog of the yeast Sir2 and plays a role in many physiological processes. Most are thought to be regulated through its lysine deacetylase activity, yet it was recently found that SIRT1 can efficiently remove fatty acyl chains from lysine residues in vitro [14,15]. It is the largest of the seven mammalian sirtuins with a conserved catalytic domain flanked by the extended N-and C-termini. While SIRT1 shuttles between the cytosol and the nucleus, most activities are done in the nucleus through deacetylation of histones and other transcriptional regulators [13]. In the past several years, more is discovered regarding its epigenetic role via deacetylation of histones and transcriptional regulators. More excitingly, new findings on SIRT1’s role in DNA repair and RNA function and the regulatory mechanisms of SIRT1 activity provide novel and important insights and potential therapeutic strategies.
SIRT1 is tightly regulated to respond to cellular needs.
Understanding how SIRT1 activity is fine-tuned to respond to specific stimuli is of great interest and importance. Recent studies demonstrate that this is through regulatory domains within SIRT1, post-translational modifications (PTMs), transcriptional control, posttranscriptional silencing, and protein-protein interactions.
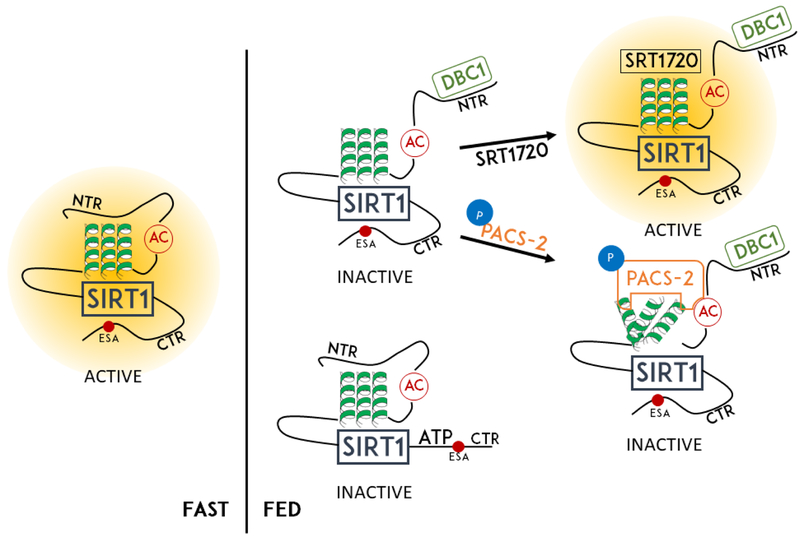
SIRT1 deacetylase activity is regulated by the insulin sensor hidden in the disordered N-terminal region. In the presence of insulin, the sensor engages DBC1 and PACS-2 to inactivate SIRT1 [16,17]. An acidic cluster and a 3-helix bundle within the region are sequestered by a shield that is removed by DBC1, which in turn allows phosphorylated PACS-2 to bind the acidic cluster and one of the helices. This destabilizes the 3-helix bundle and inhibits SIRT1 catalysis and the subsequent PGC-1α and PPARα target gene transcription to promote diet-induced obesity (Figure 1). This work uncovered the mechanism of SIRT1 inhibition by DBC1 and PACS-2 and its role in insulin response [**18].
Figure 1.
Regulation of SIRT1 via its N-terminal and C-terminal extensions by binding to small molecules or proteins. NTR = N-terminal Region, CTR= C-terminal Region, AC = Acidic Cluster, ESA = Essential for SIRT1 Activity region.
The C-terminal domain regulates SIRT1 via ATP. A 25-amino acid peptide in this domain can bind to ATP, causing compaction of the structure of SIRT1, decreasing its affinity for substrates and inhibiting the deacetylase function. Abolishing ATP-binding promotes SIRT1-mediated stress resistance in mice and inhibits adipogenesis in cultured MEFs. Interestingly, transferring this domain to other proteins also puts them under ATP regulation (Figure 1) [**19].
PTMs provide additional regulatory mechanisms of SIRT1 function. SIRT7 and SIRT1 cooperate in regulating adipogenesis through reversible SIRT1 acetylation and the consequent PPARγ transcription in mice. Cell culture, biochemical and mouse studies provided unexpected evidences that SIRT7 antagonizes SIRT1 by preventing its autodeacetylation at K230 through a direct interaction to repress PPARγ [20].
SIRT1 activity is regulated by phosphorylation. During IL-6 stimulation, SIRT1 is phosphorylated by JAK1 at Tyr280 and Tyr301, which is required for SIRT1 interaction with its known deacetylation substrate STAT3 [21,22]. In diet-induced obese but not lean mice, SIRT1 is phosphorylated by CK2 at Ser164, which inhibits its nucleus localization and deacetylase activity. Strongly elevated levels of CK2 and SIRT1 phosphorylation at Ser164 were also observed in liver samples from patients with nonalcoholic fatty liver disease [23]. Furthermore, under oxidative stress SIRT1 is activated by dephosphorylation at Ser47 mediated by the GADD34/PP1α complex [24].
O-linked N-acetyl-β-D-glucosamine (O-GlcNAc) is another PTM recently shown to regulate SIRT1 activity. O-GlcNAc transferase modifies SIRT1 on Ser549, increasing its deacetylase activity. Under genotoxic stress O-GlcNAc-modified SIRT1 but not the S549A mutant robustly deacetylates p53 to promote cell survival [25]. Activation of SIRT1 by sulfhydration enhances its zinc-binding and stability to alleviate atherosclerotic plaque [26].
SIRT1 function in epigenetics is also affected by miRNA and lncRNA targeting, transcriptional regulation by HIF1α, SUV39H and interaction with BRG1 (Table 1) [27–41], [42], [43], [44]). These reports portray the intricate regulation network of SIRT1 enzymatic function and its importance for normal physiology and diseases.
Table 1:
miRNA and lncRNA regulating SIRT1 levels
| Name | Effect on SIRT1 Levels | Functions | References |
|---|---|---|---|
| miR-199a-5p | Decrease | Regulates the pathogenesis of intrauterine growth restriction | [27] |
| miR-361–5p | Decrease | Promotes hepatic triglyceride accumulation and insulin sensitivity during hepatosteatosis | [28] |
| miR-34a | Decrease | 1) Suppresses proliferation and apoptosis of gastric cancer 2) Regulates pro-apoptotic caspase-3/7 activity and p53 levels in cardiac progenitor cells 3) Regulates plasma and hepatic Fgf21 during obesity and insulin resistance |
[29], [32], [35] |
| miR-29b | Decrease | Reverses oxaliplatin-resistance in colorectal cancer by enhancing ROS and JNK phosphorylation to induce apoptosis | [30] |
| miR-30a | Decrease | Suppresses lung cancer progression | [31] |
| miR-221 | Decrease | Promotes white adipose tissue inflammation and insulin-resistance | [33] |
| miR-204 | Decrease | In prostate cancer indices mitochondrial apoptosis through upregulation of Noxa and Puma via acetylated p53 | [34] |
| miR-181a | Decrease | Induces gastric cancer apoptosis by increasing FoxO1 acetylation during oxidative stress | [36] |
| IncRNA NEAT1 |
Increase | Promotes cell proliferation and metastasis in colorectal cancer by competing with miR-34a leading to upregulation of Wnt/βcatenin signaling | [37] |
| IncRNA MALAT1 |
Increase | In high glucose represses SIRT1 transcription by interacting with FoxO1 to induce HK-2 cell injury | [38] |
| IncRNA-PRLB | Increase | Promotes breast cancer by targeting miR-4766–5p to increase SIRT1 levels | [39] |
| LncRNA HNF1A-AS1 |
Increase | Promotes colon cancer metastasis by increasing SIRT1 levels through targeting miR-34a to induce noncanonical Wnt signaling | [40] |
| LncRNA HULC |
Increase | Inhibits SIRT1 degradation to promote protective autophagy in hepatocellular carcinoma | [41] |
SIRT1 controls transcription through epigenetic regulators.
SIRT1 is known to regulate transcription by deacetylating histones and other epigenetic regulators. In the past two years, new transcription factors have been identified as deacetylation substrates of SIRT1 and novel roles have been found for the regulation of TFs and histone marks that are established SIRT1 substrates. Although the regulation of TFs and histone marks is one of the most prominent epigenetic roles of SIRT1, since it has been extensively reviewed, we provide only a brief additional update in Table 2 without further discussion. However, it worth mentioning that SIRT1-catalyzed histone deacetylation often cross-talks with other epigenetic modifications. For example, in MLL-AF9 driven leukemia cells with DOT1L inactivation, SIRT1 deacetylates H3K9 of MLL target genes allowing subsequent H3K9 dimethylation by SUV39H1 to increase chromatin compaction and gene silencing. The combination of SIRT1 activator SRT1720 and DOT1L inhibitor EP24777 was able to suppress MLL-AF9 driven tumor formation in mice, suggesting a potential therapeutic approach against hard-to-treat MLL-rearranged leukemia [45]. SIRT1 has also been reported to associate with KDM2B, an eraser of H3K79 methylation, at gene promoters. The subsequent H4K16 deacetylation by SIRT1 promotes transcriptional repression [46].
Table 2:
Recently reported deacetylation substrates of Sirtuins
| SIRT1 | |||||
|---|---|---|---|---|---|
| Type | Substrate | Full name | Modification Sites | Functions | Reference |
| Transcription Factors | PRRX1 | Paired related homeobox 1 | K160 | Upregulates KLF4 to promote cell sternness and migration through LDH1 upregulation | [122] |
| KLF4 | Kruppel-like factor 4 | Unknown | Regulates the ovarian cancer cell invasion by inducing the expression of Claudin-5 | [123] | |
| NFATcl | Nuclear factor of activated T-cells, cytoplasmic 1 | Unknown | Polarization and recruitment of adipose tissue macrophages | [124] | |
| C/EBPα | CCAAT/enhancer-binding protein alpha | K159, K298 | Increases mitochondrial respiration by upregulating mitochondrial genes | [125] | |
| Others | TET2 | Tet methylcytosine dioxygenase 2 | K1468, K1472, K1473, K1478 | Activates TET2 to ameliorate myelodysplastic syndrome | [126] |
| MDM2 | E3 ubiquitin ligase murine double minute 2 | K182, K185 | Promotes self-ubiquitination and degradation of MDM2, which stabilizes p53 to promote apoptosis in osteosarcoma | [127] | |
| DNMT3I | DNA (cytosine-5)-methyltransferase 3-like | Unknown | Destabilizes DNMT3I protein by direct deacetylation to promote ESCs differentiation | [128] | |
| RPA1 | Replication Protein Al | K163 | Decreases interaction with XPA during DNA repair | [52] | |
| PABP1 | Poly(A)-binding protein 1 | K95 | Retains of PABP1 and mRNA in the nucleus inhibiting protein translation | [57] | |
| 9G8 | SR protein 9G8 | K24 | Promotes the inclusion of tau exon 10. | [58] | |
| SIRT2 | |||||
| Histone | H3 | Histone H3 | K18 | Translocates SIRT2 into nucleus to deacetylate H3K18 to reprogram transcription, when Listeria monocytogenes infected | [75] |
| Transcription Factors | NFAT | Nuclear factor of activated T-cells | unknown | Destabilizes NFATc2 and suppresses its nuclear localization, resulting in decreased transcription activity to cardiac homeostasis | [129] |
| NRF2 | Nuclear factor erythroid-derived 2-related factor 2 | K506, K508 | Reduces NRF2 levels, to reduce ferroportin 1 expression and decrease iron export | [130] | |
| slug | Slug | K116 | Stabilizes slug | [73] | |
| JNK | c-Jun NH2-terminal kinases | K153 | Enhances ATP binding and enzymatic activity of JNK towards c-Jun; favors the phosphorylation of JNK by MKK4 | [131] | |
| p73 | Tumor suppressor p73 | K620, K623, K627 | Suppresses its transcriptional activity, critical for tumorigenicity of glioblastoma cells | [74] | |
| Metabolism Enzyme | ALDA | Aldolase | K322 | Suppresses glycolytic enzyme activities, and regulates metabolic reprogramming during induced pluripotency | [59] |
| ENO1 | Enolase | unknown | Suppresses glycolytic enzyme activities, and regulates metabolic reprogramming during induced pluripotency | [59] | |
| PGK1 | Phosphoglycerate Kinase 1 | unknown | Suppresses glycolytic enzyme activities, and regulates metabolic reprogramming during induced pluripotency | [59] | |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase | unknown | Suppresses glycolytic enzyme activities, and regulates metabolic reprogramming during induced pluripotency | [59] | |
| PKM2 | M2 isoform of pyruvate kinase | K305 | Induces PKM2 activity by promoting tetramerization to the active enzymatic form | [132] | |
| Others | ANKLE2 | Ankyrin and LEM domain-containing protein 2 | K302 | Promotes ANKLE2 phosphorylation and nuclear envelope reassembly | [133] |
| ATRIP | ATR-interacting protein | K32 | Drives ATRIP phopshorylation and accumulation to DNA damage sites in response to replication stress | [76] | |
| GSK3 | Glycogen synthase kinase 3 | K246/K183 | Enhances its binding to ATP | [134] | |
| SIRT6 | |||||
| Histone | H3 | Histone H3 | K9 | Silences Notch1/4, suppresses Notch signaling in podocytes | [86] |
| H3 | Histone H3 | K9 | Allows ADP-ribosylation in response to DNA damage | [98] | |
| H3 | Histone H3 | K56 | Silences IGFBP2, activates AKT signaling in melanoma cell | [87] | |
| H3 | Histone H3 | K56 | Promotes ATF4 destabilization from target genes | [92] | |
| H3 | Histone H3 | K9, K56 | Silences LIN28b, suppresses let-7 target genes in pancreatic cancer | [88] | |
| H3 | Histone H3 | K9, K56 | Transcriptionally suppresses long non-coding RNAH19 | [89] | |
| H3 | Histone H3 | K9, K56 | Silences pluripotency genes in embryonic stem cells | [90] | |
| Transcription Factors | p53 | Tumor suppressor 53 | K381 | Destabilizes p53 | [94] |
| PKM2 | Pyruvate kinase M2 (nuclear) | K433 | Leads to nuclear export of nuclear PKM2, suppressing its transcription activity in hepatocellular carcinoma | [95] | |
| Others | TRF2 | Telomere repeat binding factor 2 | K176, K179, K190 | Destabilizes TRF2 in response to DNA damage | [97] |
| SIRT7 | |||||
| Transcription Factors | SP7/OSX | Osterix | K368 | Promotes its transactivation activity in bone formation | [135] |
| Others | DDX21 | DEAD-box RNA helicase | K18, K137, K600 | Augments helicase activity and overcomes R-loop-mediated stalling of RNA polymerases, safeguards genome stability | [117] |
SIRT1 can promote or inhibit DNA repair.
DNA lesions caused by UV radiation or genotoxins can block transcription leading to apoptosis. SIRT1 is known to regulate the homologous recombination (HR) repair machinery proteins NBS1 and Rad51 by deacetylation and recruitment to DNA damage sites [47,48]. Inactivation of SIRT1 impairs HR repair and enhances lung cancer apoptosis during inhibition of WEE1, a kinase regulator of G2/M checkpoint [49]. In response to the UV-induced DNA damage, SIRT1 deacetylates XPA allowing its phosphorylation by ATR to alleviate the damage via nucleotide excision repair (NER) [50,51]. In addition, the regulator of DNA metabolism RPA1 is acetylated on K163 by PCAF acetyltransferase upon UV-induced DNA damage, which enhances the RPA1 interaction with XPA to promote repair. SIRT1 and HDAC6 erase the acetylation to release XPA from the original DNA damage site at the end of the repair process [52]. SIRT1 also deacetylates BRCA1 to inhibit the intra-S check point and promote DNA replication and cell growth [53,54]. Furthermore, a nucleosome assembly protein TSPY-Like 2 inhibits SIRT1 during DNA damage to induce apoptosis through hyperacetylated p53 [13,55]. These studies underscore the different roles of SIRT1 in DNA repair pathways and stages.
SIRT1 regulates RNA metabolism.
SIRT1 has recently emerged as an RNA regulator. In HPV-infected cervical cancer cells, SIRT1 suppressed the levels of AIM2 inflammasome gene, allowing the cells to escape the immune response. SIRT1 knockdown caused pyroptosis and promoted RelB-dependent AIM2 transcription. Unexpectedly, SIRT1 depletion increased RelB mRNA, but not pre-mRNA, and elevated AIM2 inflammasome-containing exosomes that propagated pyroptosis to neighboring naïve cells. These findings hint to an existing role of SIRT1 in mRNA stability and exosome biogenesis that require further mechanistic delineation [*56].
SIRT1 also protects mRNA transport. During energy starvation, SIRT1 is phosphorylated on T530, allowing SIRT1 to bind and deacetylate K95 of PABP1, a poly(A)-binding protein. This leads to the retention of PABP1 and mRNA in the nucleus, inhibiting protein translation and cell proliferation to conserve energy consumption [*57].
Another RNA regulatory role of SIRT1 is in the alternative splicing of tau. SIRT1 deacetylates K24 of the splicing factor 9G8 to promote the inclusion of tau exon 10. SIRT1 or 9G8 K24R overexpression in HEK293T cells or resveratrol treatment of Htau mice with high levels of tau promote exon 10 inclusion. Of interest is to determine how 9G8 is affected by acetylation to regulate pre-mRNA splicing [58]. These reports open new exciting avenues for understanding the SRT1 function in RNA regulation.
The epigenetic roles of SIRT2
SIRT2 resides mainly in cytoplasm with highly dynamic nucleo-cytoplasmic shuttling and is connected to a wide range of physiological processes, like cell cycle, genome stability, metabolism, and aging. While half of the functions involves deacetylation of cytosolic proteins, including metabolic enzymes [59], SIRT2 regulates many epigenetics processes, including deacetylation of histone marks (H4K16ac [60] and H3K56ac [61]), transcription factors (FoxO1 [62], FoxO3 [63], HIF1α [64]), and chromatin modifying enzymes (p300 [65]).
Recent progress in SIRT2 study highlights the diverse enzymatic activities of SIRT2 and the potential impacts in epigenetics, through its accommodation of other acyl lysine modifications. SIRT2 removes lysine fatty acylation on K-Ras4a, its first identified de-fatty acylation substrate [66,67]. The deacylation leads to subcellular redistribution of K-Ras4a and promotes cellular transformation. SIRT2 also removes lysine γ-oxo-nonanoylation [68] [69], a histone modification derived from lipid peroxidation products, and lysine benzoylation [70].
SIRT2’s epigenetic role has been connected to cancer. Early reports showed that SIRT2 regulates cell cycle checkpoint and acts as a weak tumor suppressor [60,71]. Recently, several new transcription factors were reported to be regulated by SIRT2 and inhibiting SIRT2 confers anticancer activity. Selective inhibition of SIRT2 using a thiomyristoyl-lysine small-molecule, TM, promotes c-Myc degradation and exerts a broad anticancer effect in various human cancer cells and mouse models of breast cancer [72]. This is achieved via an epigenetic mechanism as the mRNA level of several E3 ubiquitin ligases of c-Myc is upregulated by SIRT2 inhibition. SIRT2 deacetylates and stabilizes Slug [73], a transcription factor, important for epithelial to mesenchymal transition (EMT). More recently, in an RNA interference screen in a glioblastoma model, SIRT2 was found to suppress glioblastoma cells through deacetylating and inhibiting of tumor suppressor p73 [74].
Recent reports highlight the function of SIRT2 in stress response, which also involves its epigenetic function. SIRT2 is translocated to the nucleus under Listeria monocytogenes infection and targeted to chromatin to deacetylate H3K18 and reprogram gene expression [75]. Interestingly, SIRT2 is detrimental for controlling the infection. SIRT2 deacetylates K32 of ATR interacting protein (ATRIP) in response to hydroxyurea induced replication stress, which promotes ATR activation and accumulation to DNA damage sites and binds to replication protein A-coated single-stranded DNA [76]. SIRT2 also deacetylates and activates JNK, promoting oxidative stress-induced cell death [77].
SIRT6 in epigenetic regulation
SIRT6 is important in DNA damage response, metabolism, aging, and cancer. SIRT6, as a tumor suppressor, represses HIF1α [78] and c-Myc [79]. SIRT6 knockout mice have severe metabolic defects, genome instability, and premature aging, while Sirt6-transgenic mice have a longer lifespan [80]. This advocates that SIRT6 has a clear longevity role. Those findings draw people’s attention and efforts to develop SIRT6 activator to study its cellular functions and explore the therapeutic potential.
SIRT6 and histone marks.
SIRT6 deacetylates H3K9 [81] and H3K56 [82] to suppress transcription. As a histone deacetylase, SIRT6 represses transcription mediated by many transcription factors, including HIF1α [78], cMyc [79], NF-kB [83], c-Jun [84], and FoxO3 [85].
In the past two years, more physiological functions of SIRT6 as a histone deacetylase are explored. Sirt6 has pleiotropic protective actions in podocytes, through deacetylating H3K9 at the promoter region of Notch1 and Notch4 and suppressing Notch signaling [86]. SIRT6 haploinsufficiency confers melanoma cell resistance to MAPK inhibitors, through increased H3K56 acetylation at IGFBP2 locus, which transcriptionally activates IGF-1 receptor and downstream AKT signaling [87].
SIRT6 also regulates the expression of non-coding RNAs. Loss of SIRT6 increases the protein Lin28b through promoter histone hyperacetylation, leading to induction of downstream let-7 target genes [88]. This epigenetic program defines a distinct subset (30%−40%) of human pancreatic ductal adenocarcinoma with a poor prognosis. SIRT6-null cynomolgus monkeys died hours after birth and exhibited severe prenatal developmental retardation [89]. Mechanistically, SIRT6 deficiency results in imprinting control region histone hyperacetylation and therefore activating long non-coding RNA H19, a developmental repressor.
SIRT6 itself is also regulated by long non-coding RNA. LncPRESS1, a p53-regulated LncRNA, can sequester SIRT6 from chromatin association, leading to high levels of histone acetylation at promoters of pluripotency genes, which maintains human embryonic stem cells pluripotency [90].
SIRT6 and transcription factors.
SIRT6-regulated transcription factors can silence or activate transcription. SIRT6 in adipose tissue is cold-inducible and recruits phospho-ATF2 to activate PGC-1α gene expression and promote thermogenesis [91]. SIRT6 is a co-repressor of ATF4, as deacetylation of H3K56 promotes ATF4 destabilization from target genes. In autophagy deficient cells, glutamine depletion induces SIRT6 dissociation from ATF4 and induces amino acid transporter expression [92]. Palmitate treatment increases the levels and interaction of SIRT6 and p53, activating p53 targeted genes involved in cardiolipin synthesis [93]. As such, SIRT6 serves as a co-activator of p53. Interestingly, a different SIRT6-p53 relationship has also been reported. Haploinsufficiency of p53 in Sirt6-deficient mice rescues several age-related phenotypes, as SIRT6 deacetylates and destabilizes p53 [94]. This could partially explain the extended lifespan of SIRT6 transgenic mice.
SIRT6 deacetylates nuclear PKM2, which leads to nuclear export of PKM2, suppressing its nonmetabolic oncogenic functions in transcriptional regulation. This nuclear export contributes to SIRT6 tumor-suppressor functions in vivo. [95].
SIRT6 and DNA damage repair.
SIRT6 facilitates Ku80/DNA-PKcs interaction and promotes DNAPKcs phosphorylation, leading to efficient non-homologous end joining, which is important in aging and pluripotency [96]. Under DNA damage condition, SIRT6 deacetylates and destabilizes telomere repeat binding factor 2, which is involved in telomere maintenance and DNA damage response [97].
It was recently shown that DNA-damage-induced H3S10 ADP-ribosylation is blocked by H3K9 acetylation [98]. This interesting crosstalk explains the previous finding that SIRT6 often migrates to the DNA damage sites and activates PARP1, a known DNA repair regulator. This work explains how a cell responds to DNA damage in the pre-existing and elaborate chromatin landscape.
The contributions of different activities of SIRT6 in epigenetics.
While the deacetylase activity of SIRT6 is clearly important for epigenetic regulation, SIRT6 can also efficiently remove long-chain fatty acyl groups from TNFα and R-Ras2 [6,99]. Whether the defatty-acylase activity also has epigenetic roles is thus of great interest. Using a SIRT6 G60A mutant that abolishes the deacetylase activity but retains demyristoylase function, it was demonstrated that most of the transcriptional regulation by SIRT6 may come from its deacetylase activity [100].
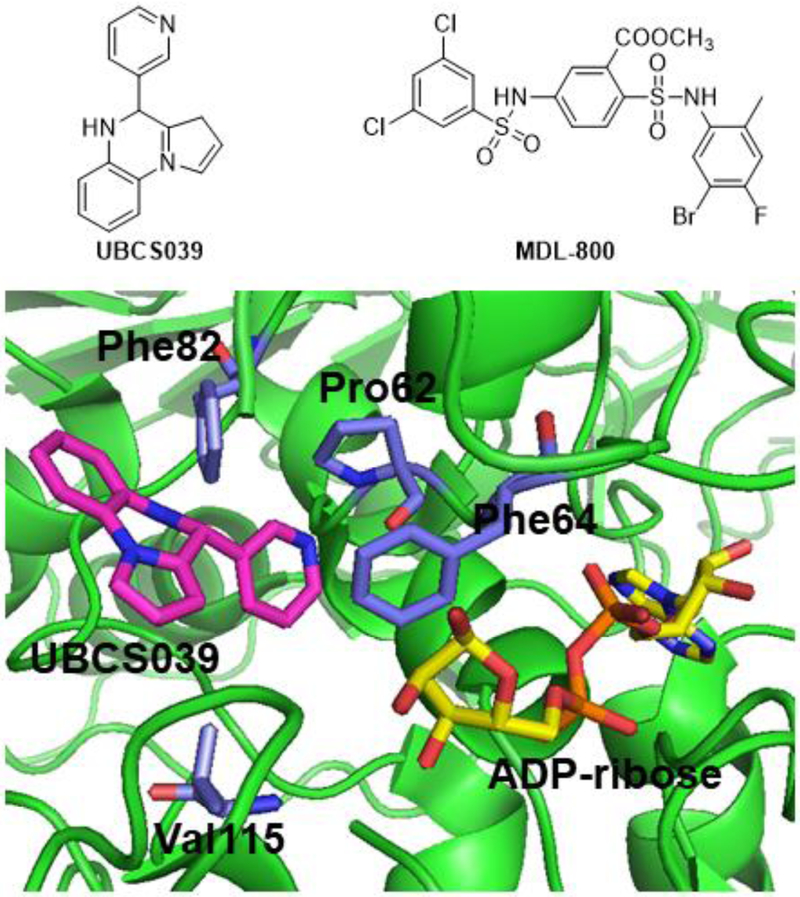
The multiple activities also enable the development of small molecule modulators of SIRT6 that only affect one of the activities. Free fatty acids were found to activate SIRT6’s deacetylase activity but not demyristoylase activity [14]. A pyrrolo[1,2-a]quinoxaline-based compound, UBCS039, which binds to the fatty acyl pocket of SIRT6 (Figure 2), was reported to promote the deacetylation activity of SIRT6 without affecting the demyristoylation activity [101]. UBCS039-treated H1299 cells showed lower acetylation level of H3K9ac and H3K56ac and autophagy-induced cell death [102]. In 2018, MDL-800 was found to activate SIRT6’s deacetylase activity selectively. MDL-800 decreased acetyl H3K9 and H3K56 in cells, two major SIRT6 substrates. In a mouse tumor xenograft model, MDL-800 decreased tumor volumes [103]. These SIRT6 activators, given that they only activate the deacetylase activity of SIRT6, may prove to be useful probes for studying the role of SIRT6’s deacetylation activity in epigenetics in combination with the G60A mutant of SIRT6.
Figure 2.
The structures of two synthetic small molecule activators of SIRT6’s deacetylase activities. One of them, UBCS039 is shown to bind the fatty acyl pocket of SIRT6.
SIRT7 in epigenetic regulation
SIRT7 resides in the nucleus but concentrates in nucleoli and is known to deacetylate H3K18 [104]. It was also reported to desuccinylate histones [105], but the in vitro biochemical evidence is not very strong. The in vitro activity of SIRT7 is weak but can be enhanced dramatically by double stranded DNA and different RNA species [106,107].
SIRT7 is a unique sirtuin as it regulates transcription mediated by all three nuclear RNA polymerases. Proteomics and enzymology studies revealed that SIRT7 regulates Pol I activity and ribosome biogenesis [108–110], Pol II mRNA transcription and metabolic stress response [111–114], and potential regulation of Pol III transcription [115].
The role of SIRT7 in genome stability was recently discovered. SIRT7 maintains rDNA stability and guards against cellular senescence though regulating SNF2H, a component of the nucleolar heterochromatin-silencing complex [116]. SIRT7 deficiency leads to defective rDNA-heterochromatin silencing, causing rDNA instability and cell senescence. SIRT7 also safeguards genome stability through deacetylating and augmenting DDX21, an RNA helicase that unwinds aberrant R loop and maintains genome stability [117].
Concluding remarks
While many studies continue to unveil the epigenetic roles of sirtuins as deacetylases for histones and transcriptional regulators, the studies described above raise new questions and open new research opportunities. The emerging RNA regulatory role of SIRT1 requires more mechanistic delineation. Whether sirtuin’s lysine de-fatty acylase activity has epigenetic roles remains to be determined. Identification of protein substrates of this activity and its downstream effects can shed light on this largely ignored area of study.
It is amazing that recent studies revealed many additional substrates for the seven mammalian sirtuins. This highlights the important biological functions of sirtuins, yet calls for a deeper understanding to simplify the complexity. In this regard, it is becoming increasingly important to understand how the sirtuins themselves are regulated, which will provide key insights to sort out the complexity associated with so many different substrates and functions. SIRT1, the most well studied mammalian sirtuins, is again at the leading edge. As discussed above, SIRT1 is the target of regulation by cellular stimuli necessary to address specific demands. Understanding how other sirtuins are regulated will similarly provide important insights.
At present, the mitochondrial sirtuins, SIRT3–5, seem to have limited direct effects in epigenetics due to their cellular localization. Nevertheless, their nuclear localization should not be completely disregarded. In fact, SIRT3 was reported to localize in the nucleus and regulate histone acetylation [118] and crotonylation [119], but the nuclear localization of SIRT3 was controversial [120]. Histones were known to be succinylated and the first histone H3 succinyltransferase was recently discovered [121]. SIRT5, the only well accepted lysine desuccinylase, could possibly regulate such histone modification and thus have direct epigenetic roles. Future studies should further clarify the epigenetic functions of SIRT3–5.
Sirtuins are increasingly recognized as deacylases instead of deacetylases. However, the epigenetic functions reported have largely attributed to the deacetylase activity. Whether other activities regulate epigenetics is therefore of great interest. It is possible that other acyl lysine modifications are also epigenetic marks. Alternatively, other deacylation function of sirtuins are mainly confined to locations outside of the nucleus.
Acknowledgement:
This work is supported by HHMI, Cornell University, and a grant from NIH/NIDDK R01DK107868.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
*: of special interest
**: of outstanding interest
- 1.Ivy JM, Klar AJ, Hicks JB: Cloning and characterization of four SIR genes of Saccharomyces cerevisiae. Mol. Cell. Biol 1986, 6:688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin S-J, Defossez P-A, Guarente L: Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 2000, 289:2126–2128. [DOI] [PubMed] [Google Scholar]
- 3.Imai S-i, Armstrong CM, Kaeberlein M, Guarente L: Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000, 403:795–800. [DOI] [PubMed] [Google Scholar]
- 4.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R: The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA 2000, 97:5807–5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Du J, Zhou Y, Su X, Yu J, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, et al. : Sirt5 is an NADdependent protein lysine demalonylase and desuccinylase. Science 2011, 334:806–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, et al. : SIRT6 regulates TNF-α secretion through hydrolysis of long-chain fatty acyl lysine. Nature 2013, 496:110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mathias Rommel A, Greco Todd M, Oberstein A, Budayeva Hanna G, Chakrabarti R, Rowland Elizabeth A, Kang Y, Shenk T, Cristea Ileana M: Sirtuin 4 Is a Lipoamidase Regulating Pyruvate Dehydrogenase Complex Activity. Cell 2014, 159:1615–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anderson KA, Huynh FK, Fisher-Wellman K, Stuart JD, Peterson BS, Douros JD, Wagner GR, Thompson JW, Madsen AS, Green MF, et al. : SIRT4 Is a Lysine Deacylase that Controls Leucine Metabolism and Insulin Secretion. Cell Metab. 2017, 25:838–855.e815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bheda P, Jing H, Wolberger C, Lin H: The Substrate Specificity of Sirtuins. Annu Rev Biochem 2016, 85:405–429. [DOI] [PubMed] [Google Scholar]
- 10.Haigis MC, Sinclair DA: Mammalian Sirtuins: Biological Insights and Disease Relevance. Annu. Rev. Pathol 2010, 5:253–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imai S-i, Guarente L: Ten years of NAD-dependent SIR2 family deacetylases: implications for metabolic diseases. Trends Pharmacol. Sci 2010, 31:212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Houtkooper RH, Pirinen E, Auwerx J: Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. Cell Biol 2012, 13:225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jing H, Lin H: Sirtuins in epigenetic regulation. Chem Rev 2015, 115:2350–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feldman JL, Baeza J, Denu JM: Activation of the protein deacetylase SIRT6 by long-chain fatty acids and widespread deacylation by mammalian sirtuins. J Biol Chem 2013, 288:31350–31356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He B, Hu J, Zhang X, Lin H: Thiomyristoyl peptides as cell-permeable Sirt6 inhibitors. Org. Biomol. Chem 2014, 12:7498–7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JE, Chen J, Lou Z: DBC1 is a negative regulator of SIRT1. Nature 2008, 451:583–586. [DOI] [PubMed] [Google Scholar]
- 17.Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W: Negative regulation of the deacetylase SIRT1 by DBC1. Nature 2008, 451:587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krzysiak TC, Thomas L, Choi YJ, Auclair S, Qian Y, Luan S, Krasnow SM, Thomas LL, Koharudin LMI, Benos PV, et al. : An Insulin-Responsive Sensor in the SIRT1 Disordered Region Binds DBC1 and PACS-2 to Control Enzyme Activity. Mol Cell 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]; **A detailed mechanism of SIRT1 inhibition by DBC1 and PACS-2 that promotes diet induced obesity and therefore can be of therapeutic benefit for this disease.
- 19.Kang H, Oka S, Lee DY, Park J, Aponte AM, Jung YS, Bitterman J, Zhai P, He Y, Kooshapur H, et al. : Sirt1 carboxyl-domain is an ATP-repressible domain that is transferrable to other proteins. Nat Commun 2017, 8:15560. [DOI] [PMC free article] [PubMed] [Google Scholar]; **An unexpected regulation of SIRT1 deacetylase activity by ATP.
- 20.Fang J, Ianni A, Smolka C, Vakhrusheva O, Nolte H, Kruger M, Wietelmann A, Simonet NG, Adrian-Segarra JM, Vaquero A, et al. : Sirt7 promotes adipogenesis in the mouse by inhibiting autocatalytic activation of Sirt1. Proc Natl Acad Sci USA 2017, 114:E8352–E8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Li F, Xu Y, Wei J, Zhang Y, Yang H, Gao B, Yu G, Fang D: JAK1-mediated Sirt1 phosphorylation functions as a negative feedback of the JAK1-STAT3 pathway. J Biol Chem 2018, 293:11067–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nie Y, Erion DM, Yuan Z, Dietrich M, Shulman GI, Horvath TL, Gao Q: STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol 2009, 11:492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi SE, Kwon S, Seok S, Xiao Z, Lee KW, Kang Y, Li X, Shinoda K, Kajimura S, Kemper B, et al. : Obesity-Linked Phosphorylation of SIRT1 by Casein Kinase 2 Inhibits Its Nuclear Localization and Promotes Fatty Liver. Mol Cell Biol 2017, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee IC, Ho XY, George SE, Goh CW, Sundaram JR, Pang KKL, Luo W, Yusoff P, Sze NSK, Shenolikar S: Oxidative stress promotes SIRT1 recruitment to the GADD34/PP1alpha complex to activate its deacetylase function. Cell Death Differ 2018, 25:255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han C, Gu Y, Shan H, Mi W, Sun J, Shi M, Zhang X, Lu X, Han F, Gong Q, et al. : O-GlcNAcylation of SIRT1 enhances its deacetylase activity and promotes cytoprotection under stress. Nat Commun 2017, 8:1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du C, Lin X, Xu W, Zheng F, Cai J, Yang J, Cui Q, Tang C, Xu G, Geng B: Sulfhydrated Sirtuin-1 Increasing Its Deacetylation Activity Is an Essential Epigenetics Mechanism of Anti-Atherogenesis by Hydrogen Sulfide. Antioxid Redox Signal 2018. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Gong X, Huang L, Chen P, Wang T, Zhou W, Luo K, Wang J: MiR-199a-5p regulates sirtuin1 and PI3K in the rat hippocampus with intrauterine growth restriction. Sci Rep 2018, 8:13813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Liu X, Xu H, Feng X, Lin Y, Huang Y, Peng Y, Gu M: Obesity-induced upregulation of miR-361–5p promotes hepatosteatosis through targeting Sirt1. Metabolism 2018, 88:31–39. [DOI] [PubMed] [Google Scholar]
- 29.Deng X, Zheng H, Li D, Xue Y, Wang Q, Yan S, Zhu Y, Deng M: MicroRNA-34a regulates proliferation and apoptosis of gastric cancer cells by targeting silent information regulator 1. Exp Ther Med 2018, 15:3705–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Cheng XH: MiR-29b reverses oxaliplatin-resistance in colorectal cancer by targeting SIRT1. Oncotarget 2018, 9:12304–12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guan Y, Rao Z, Chen C: miR-30a suppresses lung cancer progression by targeting SIRT1. Oncotarget 2018, 9:4924–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fomison-Nurse I, Saw EEL, Gandhi S, Munasinghe PE, Van Hout I, Williams MJA, Galvin I, Bunton R, Davis P, Cameron V, et al. : Diabetes induces the activation of pro-ageing miR-34a in the heart, but has differential effects on cardiomyocytes and cardiac progenitor cells. Cell Death Differ 2018, 25:1336–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng J, Zhou Y, Deng Z, Zhang H, Wu Y, Song T, Yang Y, Wei H: miR-221 negatively regulates inflammation and insulin sensitivity in white adipose tissue by repression of sirtuin-1 (SIRT1). J Cell Biochem 2018, 119:6418–6428. [DOI] [PubMed] [Google Scholar]
- 34.Shu Y, Ren L, Xie B, Liang Z, Chen J: MiR-204 enhances mitochondrial apoptosis in doxorubicintreated prostate cancer cells by targeting SIRT1/p53 pathway. Oncotarget 2017, 8:97313–97322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han HS, Choi BH, Kim JS, Kang G, Koo SH: Hepatic Crtc2 controls whole body energy metabolism via a miR-34a-Fgf21 axis. Nat Commun 2017, 8:1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang M, Zhang Q, Hu Y, Xu L, Jiang Y, Zhang C, Ding L, Jiang R, Sun J, Sun H, et al. : miR-181a increases FoxO1 acetylation and promotes granulosa cell apoptosis via SIRT1 downregulation. Cell Death Dis 2017, 8:e3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo Y, Chen JJ, Lv Q, Qin J, Huang YZ, Yu MH, Zhong M: Long non-coding RNA NEAT1 promotes colorectal cancer progression by competitively binding miR-34a with SIRT1 and enhancing the Wnt/beta-catenin signaling pathway. Cancer Lett 2019, 440–441:11–22. [DOI] [PubMed] [Google Scholar]
- 38.Zhou L, Xu DY, Sha WG, Shen L, Lu GY: Long non-coding RNA MALAT1 interacts with transcription factor Foxo1 to regulate SIRT1 transcription in high glucose-induced HK-2cells injury. Biochem Biophys Res Commun 2018, 503:849–855. [DOI] [PubMed] [Google Scholar]
- 39.Liang Y, Song X, Li Y, Sang Y, Zhang N, Zhang H, Liu Y, Duan Y, Chen B, Guo R, et al. : A novel long noncoding RNA-PRLB acts as a tumor promoter through regulating miR-4766–5p/SIRT1 axis in breast cancer. Cell Death Dis 2018, 9:563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang C, Qiu S, Sun F, Li W, Wang Z, Yue B, Wu X, Yan D: Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer Lett 2017, 410:50–62. [DOI] [PubMed] [Google Scholar]
- 41.Xiong H, Ni Z, He J, Jiang S, Li X, Gong W, Zheng L, Chen S, Li B, Zhang N, et al. : LncRNA HULC triggers autophagy via stabilizing Sirt1 and attenuates the chemosensitivity of HCC cells. Oncogene 2017, 36:3528–3540. [DOI] [PubMed] [Google Scholar]
- 42.Qin J, Liu Y, Lu Y, Liu M, Li M, Li J, Wu L: Hypoxia-inducible factor 1 alpha promotes cancer stem cells-like properties in human ovarian cancer cells by upregulating SIRT1 expression. Sci Rep 2017, 7:10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang G, Weng X, Zhao Y, Zhang X, Hu Y, Dai X, Liang P, Wang P, Ma L, Sun X, et al. : The histone H3K9 methyltransferase SUV39H links SIRT1 repression to myocardial infarction. Nat Commun 2017, 8:14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G, Fu Y, Hu F, Lan J, Xu F, Yang X, Luo X, Wang J, Hu J: Loss of BRG1 induces CRC cell senescence by regulating p53/p21 pathway. Cell Death Dis 2017, 8:e2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen CW, Koche RP, Sinha AU, Deshpande AJ, Zhu N, Eng R, Doench JG, Xu H, Chu SH, Qi J, et al. : DOT1L inhibits SIRT1-mediated epigenetic silencing to maintain leukemic gene expression in MLL-rearranged leukemia. Nat Med 2015, 21:335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kang JY, Kim JY, Kim KB, Park JW, Cho H, Hahm JY, Chae YC, Kim D, Kook H, Rhee S, et al. : KDM2B is a histone H3K79 demethylase and induces transcriptional repression via sirtuin-1-mediated chromatin silencing. FASEB J 2018, 32:5737–5750. [DOI] [PubMed] [Google Scholar]
- 47.Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E: SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell 2007, 27:149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oberdoerffer P, Michan S, McVay M, Mostoslavsky R, Vann J, Park SK, Hartlerode A, Stegmuller J, Hafner A, Loerch P, et al. : SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 2008, 135:907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen G, Zhang B, Xu H, Sun Y, Shi Y, Luo Y, Jia H, Wang F: Suppression of Sirt1 sensitizes lung cancer cells to WEE1 inhibitor MK-1775-induced DNA damage and apoptosis. Oncogene 2017, 36:6863–6872. [DOI] [PubMed] [Google Scholar]
- 50.Jarrett SG, Carter KM, Bautista RM, He D, Wang C, D’Orazio JA: Sirtuin 1-mediated deacetylation of XPA DNA repair protein enhances its interaction with ATR protein and promotes cAMPinduced DNA repair of UV damage. J Biol Chem 2018. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 51.Fan W, Luo J: SIRT1 regulates UV-induced DNA repair through deacetylating XPA. Mol Cell 2010, 39:247–258. [DOI] [PubMed] [Google Scholar]
- 52.Zhao M, Geng R, Guo X, Yuan R, Zhou X, Zhong Y, Huo Y, Zhou M, Shen Q, Li Y, et al. : PCAF/GCN5-Mediated Acetylation of RPA1 Promotes Nucleotide Excision Repair. Cell Rep 2017, 20:1997–2009. [DOI] [PubMed] [Google Scholar]
- 53.Jang J, Huh YJ, Cho HJ, Lee B, Park J, Hwang DY, Kim DW: SIRT1 Enhances the Survival of Human Embryonic Stem Cells by Promoting DNA Repair. Stem Cell Reports 2017, 9:629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lahusen TJ, Kim SJ, Miao K, Huang Z, Xu X, Deng CX: BRCA1 function in the intra-S checkpoint is activated by acetylation via a pCAF/SIRT1 axis. Oncogene 2018, 37:2343–2350. [DOI] [PubMed] [Google Scholar]
- 55.Magni M, Buscemi G, Maita L, Peng L, Chan SY, Montecucco A, Delia D, Zannini L: TSPYL2 is a novel regulator of SIRT1 and p300 activity in response to DNA damage. Cell Death Differ 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.So D, Shin HW, Kim J, Lee M, Myeong J, Chun YS, Park JW: Cervical cancer is addicted to SIRT1 disarming the AIM2 antiviral defense. Oncogene 2018, 37:5191–5204. [DOI] [PubMed] [Google Scholar]; *The first implication of SIRT1 in regulation of RNA stability and exosome biogenesis that could be important for immune response.
- 57.Shan P, Fan G, Sun L, Liu J, Wang W, Hu C, Zhang X, Zhai Q, Song X, Cao L, et al. : SIRT1 Functions as a Negative Regulator of Eukaryotic Poly(A)RNA Transport. Curr Biol 2017, 27:2271–2284 e2275. [DOI] [PubMed] [Google Scholar]; *A novel function of SIRT1 as a regulator of mRNA transport to control cellular energy consumption.
- 58.Qian S, Gu J, Dai W, Jin N, Chu D, Huang Q, Liu F, Qian W: Sirt1 enhances tau exon 10 inclusion and improves spatial memory of Htau mice. Aging (Albany NY) 2018, 10:2498–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cha Y, Han MJ, Cha HJ, Zoldan J, Burkart A, Jung JH, Jang Y, Kim CH, Jeong HC, Kim BG, et al. : Metabolic control of primed human pluripotent stem cell fate and function by the miR-200c-SIRT2 axis. Nat Cell Biol 2017, 19:445–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Serrano L, Martinez-Redondo P, Marazuela-Duque A, Vazquez BN, Dooley SJ, Voigt P, Beck DB, Kane-Goldsmith N, Tong Q, Rabanal RM, et al. : The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev 2013, 27:639–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Das C, Lucia MS, Hansen KC, Tyler JK: CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 2009, 459:113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jing E, Gesta S, Kahn CR: SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab 2007, 6:105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang F, Nguyen M, Qin FX, Tong Q: SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell 2007, 6:505–514. [DOI] [PubMed] [Google Scholar]
- 64.Seo KS, Park JH, Heo JY, Jing K, Han J, Min KN, Kim C, Koh GY, Lim K, Kang GY, et al. : SIRT2 regulates tumour hypoxia response by promoting HIF-1alpha hydroxylation. Oncogene 2015, 34:1354–1362. [DOI] [PubMed] [Google Scholar]
- 65.Black JC, Mosley A, Kitada T, Washburn M, Carey M: The SIRT2 deacetylase regulates autoacetylation of p300. Mol Cell 2008, 32:449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jiang H, Khan S, Wang Y, Charron G, He B, Sebastian C, Du J, Kim R, Ge E, Mostoslavsky R, et al. : SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature 2013, 496:110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jing H, Zhang X, Wisner SA, Chen X, Spiegelman NA, Linder ME, Lin H: SIRT2 and lysine fatty acylation regulate the transforming activity of K-Ras4a. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jin J, He B, Zhang X, Lin H, Wang Y: SIRT2 Reverses 4-Oxononanoyl Lysine Modification on Histones. J Am Chem Soc 2016, 138:12304–12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cui Y, Li X, Lin J, Hao Q, Li XD: Histone Ketoamide Adduction by 4-Oxo-2-nonenal Is a Reversible Posttranslational Modification Regulated by Sirt2. ACS Chem Biol 2017, 12:47–51. [DOI] [PubMed] [Google Scholar]
- 70.Huang H, Zhang D, Wang Y, Perez-Neut M, Han Z, Zheng YG, Hao Q, Zhao Y: Lysine benzoylation is a histone mark regulated by SIRT2. Nat Commun 2018, 9:3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim HS, Vassilopoulos A, Wang RH, Lahusen T, Xiao Z, Xu X, Li C, Veenstra TD, Li B, Yu H, et al. : SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell 2011, 20:487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jing H, Hu J, He B, Negron Abril YL, Stupinski J, Weiser K, Carbonaro M, Chiang YL, Southard T, Giannakakou P, et al. : A SIRT2-Selective Inhibitor Promotes c-Myc Oncoprotein Degradation and Exhibits Broad Anticancer Activity. Cancer Cell 2016, 29:767–768. [DOI] [PubMed] [Google Scholar]; **Using thiomyristoyl lysine compound to selectively inhibit SIRT2, the authors demonstrate that SIRT2 inhibition exerts broad anti-cancer effect. This study establishes SIRT2 as a promising anticancer target, especially for certain c-Myc-driven cancers.
- 73.Zhou W, Ni TK, Wronski A, Glass B, Skibinski A, Beck A, Kuperwasser C: The SIRT2 Deacetylase Stabilizes Slug to Control Malignancy of Basal-like Breast Cancer. Cell Rep 2016, 17:1302–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Funato K, Hayashi T, Echizen K, Negishi L, Shimizu N, Koyama-Nasu R, Nasu-Nishimura Y, Morishita Y, Tabar V, Todo T, et al. : SIRT2-mediated inactivation of p73 is required for glioblastoma tumorigenicity. EMBO Rep 2018, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pereira JM, Chevalier C, Chaze T, Gianetto Q, Impens F, Matondo M, Cossart P, Hamon MA: Infection Reveals a Modification of SIRT2 Critical for Chromatin Association. Cell Rep 2018, 23:1124–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang H, Head PE, Daddacha W, Park SH, Li X, Pan Y, Madden MZ, Duong DM, Xie M, Yu B, et al. : ATRIP Deacetylation by SIRT2 Drives ATR Checkpoint Activation by Promoting Binding to RPAssDNA. Cell Rep 2016, 14:1435–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li Y, Zhang M, Dorfman RG, Pan Y, Tang D, Xu L, Zhao Z, Zhou Q, Zhou L, Wang Y, et al. : SIRT2 Promotes the Migration and Invasion of Gastric Cancer through RAS/ERK/JNK/MMP-9 Pathway by Increasing PEPCK1-Related Metabolism. Neoplasia 2018, 20:745–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. : The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell 2010, 140:280–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, et al. : The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 2012, 151:1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY: The sirtuin SIRT6 regulates lifespan in male mice. Nature 2012, 483:218–221. [DOI] [PubMed] [Google Scholar]
- 81.Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, et al. : SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 2008, 452:492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF: Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle 2009, 8:2664–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kawahara TL, Michishita E, Adler AS, Damian M, Berber E, Lin M, McCord RA, Ongaigui KC, Boxer LD, Chang HY, et al. : SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell 2009, 136:62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sundaresan NR, Vasudevan P, Zhong L, Kim G, Samant S, Parekh V, Pillai VB, Ravindra PV, Gupta M, Jeevanandam V, et al. : The sirtuin SIRT6 blocks IGF-Akt signaling and development of cardiac hypertrophy by targeting c-Jun. Nat Med 2012, 18:1643–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tao R, Xiong X, DePinho RA, Deng CX, Dong XC: FoxO3 transcription factor and Sirt6 deacetylase regulate low density lipoprotein (LDL)-cholesterol homeostasis via control of the proprotein convertase subtilisin/kexin type 9 (Pcsk9) gene expression. J Biol Chem 2013, 288:29252–29259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liu M, Liang K, Zhen J, Zhou M, Wang X, Wang Z, Wei X, Zhang Y, Sun Y, Zhou Z, et al. : Sirt6 deficiency exacerbates podocyte injury and proteinuria through targeting Notch signaling. Nat Commun 2017, 8:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Strub T, Ghiraldini FG, Carcamo S, Li M, Wroblewska A, Singh R, Goldberg MS, Hasson D, Wang Z, Gallagher SJ, et al. : SIRT6 haploinsufficiency induces BRAF(V600E) melanoma cell resistance to MAPK inhibitors via IGF signalling. Nat Commun 2018, 9:3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kugel S, Sebastian C, Fitamant J, Ross KN, Saha SK, Jain E, Gladden A, Arora KS, Kato Y, Rivera MN, et al. : SIRT6 Suppresses Pancreatic Cancer through Control of Lin28b. Cell 2016, 165:1401–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This paper demonstrates that SIRT6 can indirectly control non-coding RNA let-7 though histone deacetylation and identifies SIRT6 as an important PDAC tumor suppressor in a molecularly defined pancreas cancer subset.
- 89.Zhang W, Wan H, Feng G, Qu J, Wang J, Jing Y, Ren R, Liu Z, Zhang L, Chen Z, et al. : SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature 2018, 560:661–665. [DOI] [PubMed] [Google Scholar]
- 90.Jain AK, Xi Y, McCarthy R, Allton K, Akdemir KC, Patel LR, Aronow B, Lin C, Li W, Yang L, et al. : LncPRESS1 Is a p53-Regulated LncRNA that Safeguards Pluripotency by Disrupting SIRT6-Mediated De-acetylation of Histone H3K56. Mol Cell 2016, 64:967–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yao L, Cui X, Chen Q, Yang X, Fang F, Zhang J, Liu G, Jin W, Chang Y: Cold-Inducible SIRT6 Regulates Thermogenesis of Brown and Beige Fat. Cell Rep 2017, 20:641–654. [DOI] [PubMed] [Google Scholar]
- 92.Zhang N, Yang X, Yuan F, Zhang L, Wang Y, Wang L, Mao Z, Luo J, Zhang H, Zhu WG, et al. : Increased Amino Acid Uptake Supports Autophagy-Deficient Cell Survival upon Glutamine Deprivation. Cell Rep 2018, 23:3006–3020. [DOI] [PubMed] [Google Scholar]
- 93.Li M, Hou T, Gao T, Lu X, Yang Q, Zhu Q, Li Z, Liu C, Mu G, Liu G, et al. : p53 cooperates with SIRT6 to regulate cardiolipin de novo biosynthesis. Cell Death Dis 2018, 9:941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ghosh S, Wong SK, Jiang Z, Liu B, Wang Y, Hao Q, Gorbunova V, Liu X, Zhou Z: Haploinsufficiency of Trp53 dramatically extends the lifespan of Sirt6-deficient mice. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]; *In SIRT6 deficient mouse, the authors showed partial depletion of p53 could rescue the longevity defects. This work highlights the importance of SIRT6-p53 axis in the regulation of aging.
- 95.Bhardwaj A, Das S: SIRT6 deacetylates PKM2 to suppress its nuclear localization and oncogenic functions. Proc Natl Acad Sci USA 2016, 113:E538–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen W, Liu N, Zhang H, Zhang H, Qiao J, Jia W, Zhu S, Mao Z, Kang J: Sirt6 Promotes DNA End Joining in iPSCs Derived from Old Mice. Cell Rep 2017, 18:2880–2892. [DOI] [PubMed] [Google Scholar]
- 97.Rizzo A, Iachettini S, Salvati E, Zizza P, Maresca C, D’Angelo C, Benarroch-Popivker D, Capolupo A, Del Gaudio F, Cosconati S, et al. : SIRT6 interacts with TRF2 and promotes its degradation in response to DNA damage. Nucleic Acids Res 2017, 45:1820–1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liszczak G, Diehl KL, Dann GP, Muir TW: Acetylation blocks DNA damage-induced chromatin ADPribosylation. Nat Chem Biol 2018, 14:837–840. [DOI] [PMC free article] [PubMed] [Google Scholar]; **The authors identify histone H3 serine 10 as the primary acceptor residue for DNA damage induced chromatin ADP-ribosylation, as well as adjust histone acetylation blocks this modification. This work provides nice mechanistic insights for DNA damage repair.
- 99.Zhang X, Spiegelman NA, Nelson OD, Jing H, Lin H: SIRT6 regulates Ras-related protein R-Ras2 by lysine defatty-acylation. Elife 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang X, Khan S, Jiang H, Antonyak MA, Chen X, Spiegelman NA, Shrimp JH, Cerione RA, Lin H: Identifying the functional contribution of the defatty-acylase activity of SIRT6. Nat Chem Biol 2016, 12:614–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.You W, Rotili D, Li TM, Kambach C, Meleshin M, Schutkowski M, Chua KF, Mai A, Steegborn C: Structural Basis of Sirtuin 6 Activation by Synthetic Small Molecules. Angew Chem Int Ed Engl 2017, 56:1007–1011. [DOI] [PubMed] [Google Scholar]
- 102.Iachettini S, Trisciuoglio D, Rotili D, Lucidi A, Salvati E, Zizza P, Di Leo L, Del Bufalo D, Ciriolo MR, Leonetti C, et al. : Pharmacological activation of SIRT6 triggers lethal autophagy in human cancer cells. Cell Death Dis 2018, 9:996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huang Z, Zhao J, Deng W, Chen Y, Shang J, Song K, Zhang L, Wang C, Lu S, Yang X, et al. : Identification of a cellularly active SIRT6 allosteric activator. Nat Chem Biol 2018, 14:1118–1126. [DOI] [PubMed] [Google Scholar]
- 104.Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K, et al. : SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 2012, 487:114–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li L, Shi L, Yang S, Yan R, Zhang D, Yang J, He L, Li W, Yi X, Sun L, et al. : SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun 2016, 7:12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Tong Z, Wang M, Wang Y, Kim DD, Grenier JK, Cao J, Sadhukhan S, Hao Q, Lin H: SIRT7 Is an RNA-Activated Protein Lysine Deacylase. ACS Chem Biol 2017, 12:300–310. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Interesting in the inconsistence between weak enzymatic activity in vitro and fair histone eraser activity in cells, the authors show that different RNA species could dramatically enhance SIRT7’s catalytic efficiency in vitro. This work provides a novel regulation mechanism, of which future enzymological study on SIRT7 should consider
- 107.Tong Z, Wang Y, Zhang X, Kim DD, Sadhukhan S, Hao Q, Lin H: SIRT7 Is Activated by DNA and Deacetylates Histone H3 in the Chromatin Context. ACS Chem Biol 2016, 11:742–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Tsai YC, Greco TM, Boonmee A, Miteva Y, Cristea IM: Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription. Mol Cell Proteomics 2012, 11:M111 015156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L: Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev 2006, 20:1075–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen S, Blank MF, Iyer A, Huang B, Wang L, Grummt I, Voit R: SIRT7-dependent deacetylation of the U3–55k protein controls pre-rRNA processing. Nat Commun 2016, 7:10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ryu D, Jo YS, Lo Sasso G, Stein S, Zhang H, Perino A, Lee JU, Zeviani M, Romand R, Hottiger MO, et al. : A SIRT7-dependent acetylation switch of GABPbeta1 controls mitochondrial function. Cell Metab 2014, 20:856–869. [DOI] [PubMed] [Google Scholar]
- 112.Shin J, He M, Liu Y, Paredes S, Villanova L, Brown K, Qiu X, Nabavi N, Mohrin M, Wojnoonski K, et al. : SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep 2013, 5:654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yoshizawa T, Karim MF, Sato Y, Senokuchi T, Miyata K, Fukuda T, Go C, Tasaki M, Uchimura K, Kadomatsu T, et al. : SIRT7 controls hepatic lipid metabolism by regulating the ubiquitinproteasome pathway. Cell Metab 2014, 19:712–721. [DOI] [PubMed] [Google Scholar]
- 114.Blank MF, Chen S, Poetz F, Schnolzer M, Voit R, Grummt I: SIRT7-dependent deacetylation of CDK9 activates RNA polymerase II transcription. Nucleic Acids Res 2017, 45:2675–2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tsai YC, Greco TM, Cristea IM: Sirtuin 7 plays a role in ribosome biogenesis and protein synthesis. Mol Cell Proteomics 2014, 13:73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Paredes S, Angulo-Ibanez M, Tasselli L, Carlson SM, Zheng W, Li TM, Chua KF: The epigenetic regulator SIRT7 guards against mammalian cellular senescence induced by ribosomal DNA instability. J Biol Chem 2018, 293:11242–11250. [DOI] [PMC free article] [PubMed] [Google Scholar]; **The authors find SIRT7 protects against senescence though maintaining the adequate silence of rDNA. This work identifies rDNA instability as a driver of mammalian cellular senescence and implicates SIRT7-dependent heterochromatin silencing in protecting against this process.
- 117.Song C, Hotz-Wagenblatt A, Voit R, Grummt I: SIRT7 and the DEAD-box helicase DDX21 cooperate to resolve genomic R loops and safeguard genome stability. Genes Dev 2017, 31:1370–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Scher MB, Vaquero A, Reinberg D: SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev 2007, 21:920–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bao X, Wang Y, Li X, Li XM, Liu Z, Yang T, Wong CF, Zhang J, Hao Q, Li XD: Identification of ‘erasers’ for lysine crotonylated histone marks using a chemical proteomics approach. Elife 2014, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cooper HM, Spelbrink JN: The human SIRT3 protein deacetylase is exclusively mitochondrial. Biochem J 2008, 411:279–285. [DOI] [PubMed] [Google Scholar]
- 121.Wang Y, Guo YR, Liu K, Yin Z, Liu R, Xia Y, Tan L, Yang P, Lee JH, Li XJ, et al. : KAT2A coupled with the alpha-KGDH complex acts as a histone H3 succinyltransferase. Nature 2017, 552:273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shi L, Tang X, Qian M, Liu Z, Meng F, Fu L, Wang Z, Zhu WG, Huang JD, Zhou Z, et al. : A SIRT1-centered circuitry regulates breast cancer stemness and metastasis. Oncogene 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhang X, Chen J, Sun L, Xu Y: SIRT1 deacetylates KLF4 to activate Claudin-5 transcription in ovarian cancer cells. J Cell Biochem 2018, 119:2418–2426. [DOI] [PubMed] [Google Scholar]
- 124.Hui X, Zhang M, Gu P, Li K, Gao Y, Wu D, Wang Y, Xu A: Adipocyte SIRT1 controls systemic insulin sensitivity by modulating macrophages in adipose tissue. EMBO Rep 2017, 18:645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zaini MA, Muller C, de Jong TV, Ackermann T, Hartleben G, Kortman G, Guhrs KH, Fusetti F, Kramer OH, Guryev V, et al. : A p300 and SIRT1 Regulated Acetylation Switch of C/EBPalpha Controls Mitochondrial Function. Cell Rep 2018, 22:497–511. [DOI] [PubMed] [Google Scholar]
- 126.Sun J, He X, Zhu Y, Ding Z, Dong H, Feng Y, Du J, Wang H, Wu X, Zhang L, et al. : SIRT1 Activation Disrupts Maintenance of Myelodysplastic Syndrome Stem and Progenitor Cells by Restoring TET2 Function. Cell Stem Cell 2018, 23:355–369 e359. [DOI] [PMC free article] [PubMed] [Google Scholar]; *TET2, the initiator of DNA demethylation is a novel substrate of SIRT1 that can be activated by SIRT1 to potentially treat MDS.
- 127.Nihira NT, Ogura K, Shimizu K, North BJ, Zhang J, Gao D, Inuzuka H, Wei W: Acetylation-dependent regulation of MDM2 E3 ligase activity dictates its oncogenic function. Sci Signal 2017, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Heo J, Lim J, Lee S, Jeong J, Kang H, Kim Y, Kang JW, Yu HY, Jeong EM, Kim K, et al. : Sirt1 Regulates DNA Methylation and Differentiation Potential of Embryonic Stem Cells by Antagonizing Dnmt3l. Cell Rep 2017, 18:1930–1945. [DOI] [PubMed] [Google Scholar]
- 129.Sarikhani M, Maity S, Mishra S, Jain A, Tamta AK, Ravi V, Kondapalli MS, Desingu PA, Khan D, Kumar S, et al. : SIRT2 deacetylase represses NFAT transcription factor to maintain cardiac homeostasis. J Biol Chem 2018, 293:5281–5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang X, Park SH, Chang HC, Shapiro JS, Vassilopoulos A, Sawicki KT, Chen C, Shang M, Burridge PW, Epting CL, et al. : Sirtuin 2 regulates cellular iron homeostasis via deacetylation of transcription factor NRF2. J Clin Invest 2017, 127:1505–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sarikhani M, Mishra S, Desingu PA, Kotyada C, Wolfgeher D, Gupta MP, Singh M, Sundaresan NR: SIRT2 regulates oxidative stress-induced cell death through deacetylation of c-Jun NH2-terminal kinase. Cell Death Differ 2018, 25:1638–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Park SH, Ozden O, Liu G, Song HY, Zhu Y, Yan Y, Zou X, Kang HJ, Jiang H, Principe DR, et al. : SIRT2-Mediated Deacetylation and Tetramerization of Pyruvate Kinase Directs Glycolysis and Tumor Growth. Cancer Res 2016, 76:3802–3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kaufmann T, Kukolj E, Brachner A, Beltzung E, Bruno M, Kostrhon S, Opravil S, Hudecz O, Mechtler K, Warren G, et al. : SIRT2 regulates nuclear envelope reassembly through ANKLE2 deacetylation. J Cell Sci 2016, 129:4607–4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sarikhani M, Mishra S, Maity S, Kotyada C, Wolfgeher D, Gupta MP, Singh M, Sundaresan NR: SIRT2 deacetylase regulates the activity of GSK3 isoforms independent of inhibitory phosphorylation. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fukuda M, Yoshizawa T, Karim MF, Sobuz SU, Korogi W, Kobayasi D, Okanishi H, Tasaki M, Ono K, Sawa T, et al. : SIRT7 has a critical role in bone formation by regulating lysine acylation of SP7/Osterix. Nat Commun 2018, 9:2833. [DOI] [PMC free article] [PubMed] [Google Scholar]