Abstract
Photoreceptor degeneration is a significant cause of visual impairment in the United States and globally. Cell replacement therapy shows great promise in restoring vision by transplanting stem-like cells into the sub-retinal space as substitutes for damaged photoreceptors. However, vision repair via transplantation has been limited, in large part, by low numbers of replacement cells able to migrate into damaged retinal tissue and integrate with native photoreceptors. Projects have used external chemical fields and applied electric fields to induce the chemotaxis and electrotaxis of replacement cells, respectively, with limited success. However, the application of combined electro-chemotactic fields in directing cells within biomaterials and host tissue has been surprisingly understudied. The current work examined the ability of combined electro-chemotactic fields to direct the migration of transplantable retinal progenitor cells (RPCs) in controlled microenvironments. Experiments used our established galvano-microfluidic system (Gal-MµS) to generate tunable chemotactic concentration fields with and without superimposed electric fields. Result illustrate that combination fields increased the distance migrated by RPCs by over three times that seen in either field, individually, and with greater directionality towards increasing gradients. Interestingly, immunofluorescence assays showed no significant differences in the distribution of the total and/or activated cognate receptor of interest, indicating that changes in ligand binding alone were not responsible for the measured increases in migration. Bioinformatics analysis was then performed to identity potential, synergistic mechanistic pathways involved in the electro-chemotaxis measured. Results indicate that increased RPC migration in electro-chemotactic fields may arise from down-regulation of cell adhesion proteins in tandem with up-regulation of cytoskeletal regulation proteins. These comprehensive results point towards a novel migration-targeted treatment that may dramatically improve transplantation outcomes as well as elucidate unreported synergy across biological mechanisms in response to electro-chemotactic fields.
Keywords: Transplantation, chemotaxis, microfluidics, bioinformatics, retinal progenitor cells
1. Introduction
Photoreceptor degeneration is a significant cause of progressive vision loss worldwide (Flaxman, Bourne et al. 2017) from inherited eye diseases, such as retinitis pigmentosa, and age-related diseases like macular degeneration. Several developing therapies to treat vision loss include laser photocoagulation(Luttrull and Dorin 2012), pharmacological inhibition(Martin, Maguire et al. 2012), gene delivery(Dalkara, Byrne et al. 2013), neural implants(da Cruz, Coley et al. 2013) and cell transplantation(Pearson, Hippert et al. 2014). Promising photoreceptor replacement therapies have transplanted a variety of stem-and progenitor-like cells(Lamba, McUsic et al. 2010, Luo, Baranov et al. 2014, Pearson, Hippert et al. 2014) from autologous and heterologous donors(Das, Zhao et al. 2005) into the subretinal space between the retinal pigment epithelium (RPE) and outer segments of native photoreceptors(Luo, Baranov et al. 2014). Transplantation studies have demonstrated that replacement cells can initiate synaptic communication with native photoreceptors via cellular integration, where transplanted cells migrate to position themselves within the retinal outer nuclear layer (ONL) of photoreceptors (Reviewed in Gasparini SJ (Gasparini, Llonch et al. 2018)), or via material transfer, where cells in close proximity share cytoplasmic content with native photoreceptors (Santos-Ferreira 2016). Current and ongoing projects have begun to illustrate that degeneration models favor integration while models of genetic disorders promote material transfer(Waldron, Di Marco et al. 2018).
illustrates an ideal transplantation model of cellular integration where replacement cells: (1) Migrate out of the subretinal space; (2) Navigate directionally into damaged tissue; (3) Position themselves within the retinal outer nuclear layer (ONL); and (4) Begin cellular and synaptic integration with native cells. While the migration of donor cells in this integration model is paramount to successful transplantation outcomes, biological processes of directional migration within adult retinal environments remain incompletely understood.
Previous work from our group has evaluated the migratory behaviors of retinal progenitor cells (RPCs) and photoreceptor precursor cells (PPCs) in response to controlled gradients of chemotactic ligands present in damaged retina(Mishra, Thakur et al. 2015, McCutcheon, Unachukwu et al. 2017, Mishra and Vazquez 2017). We have also demonstrated the influence of electric fields on the extension of RPC processes(Saigal, Cimetta et al. 2013). Several groups have reported an influence of electric fields on neural progenitor process extension and migration(Meng, Li et al. 2012, Chang, Lee et al. 2016, Yao and Li 2016). The current work examined the ability of combined chemotactic and electrical fields to direct the migration of transplantable RPCs, which may help improve the appropriate retinal positioning needed for integration in vivo. This study evaluates the effects of these external fields on the behavior of RPCs derived from the same source as numerous other retinal projects from our group to aid interpretation (Mishra et al. 2015, Unachukwu UJ et al. 2016, McCutcheon S et al. 2017). Experiments utilized a unique microfluidic device, called the Galvano-Macro micro system, or Gal-MµS, to generate tunable chemotactic fields with and without superimposed electric fields. Results illustrate that while chemotactic and electrical fields promoted directed migration of RPCs over long distances, combinatory fields increased the net migration of transplantable cells by three times the amount measured in either field, individually, as well as increased directionality of cell movement.
To decipher the mechanisms responsible for the migratory responses induced by combinatory fields, we performed bioinformatics analyses to identify genes with established roles in cell adhesion and motility (Arikawa, Quellhorst et al. 2010). Results indicated that increases in migration distances are correlated to down-regulation of cell adhesion proteins including β-catenin and cadherin, in conjunction with up-regulation of the cytoskeletal regulator proteins RalGDS and Ral. Building on these data, we generated a list of genes that could be targeted to reduce cell migration for loss-of-function migration analysis. One such target identified was topoisomerase 2B (Top2B), known to affect retinal cell migration through transcriptional regulation and regulation of guidance ques in vivo (Li 2014). Analysis of RPC migration in combinatory electro-chemotactic fields using Top2b inhibition resulted in complete loss of migration. The exciting findings of this work suggest that a strategy to improve the migration of transplantable photoreceptor replacements may be application of controllable electrical and chemical stimuli to synergistically guide migration and retinal integration.
2. Materials and Methods
2.1. System Design and Fabrication
Development of the Gal-MuS device used a two-step optical photolithography process and polydimethylsiloxane (PDMS, Sylgard 184, Dow Corning) micromolding technique previously described by our group (McCutcheon, Unachukwu et al. 2017, Mishra and Vazquez 2017). As shown in Figure 2, the system is comprised of two macro-wells connected by an array of microchannels in a modified H-ladder design(Saadi, Rhee et al. 2007). Controlled volume flow rates on either side of the device establish a stable concentration gradient across the adjoining microfluidic channels. Electrodes imbedded at the center of the device enable superposition of a constant electric field and chemical gradient field across the microarray. Importantly, these superimposed stimuli do not alter the chemical microenvironment of resident cells as previously demonstrated by our group using a variety of neural cells (McCutcheon, Unachukwu et al. 2015, Mishra, Thakur et al. 2015, Rico-Varela, Singh et al. 2015, Unachukwu, Warren et al. 2016).
Figure 2.
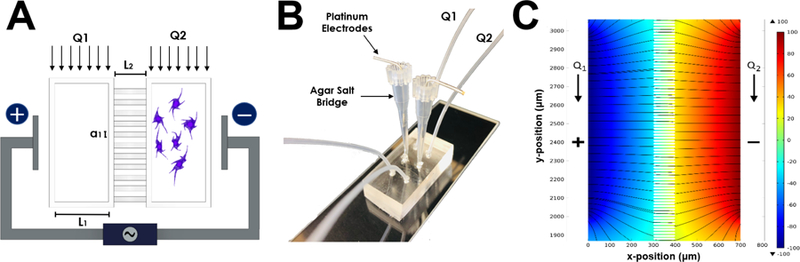
Design and Operation of GalMuS microfluidics device. (A) Schematic illustrating GalMuS operation. Cells are loaded into one side of the device while a desired chemical stimulant is loaded into the other. The chemical gradient generated within the culture chambers is controlled via the volume flow ratio of Q1:Q2. The electrodes placed on either side allow for controlled concurrent electrical field stimulation. (B) Image of fabricated PDMS device demonstrating electrode placement and composition (C) Computer-derived electric field profile within the GalMuS, electric field strength V= 100mV/mm.
2.2. Cell Culture
Multi-passage mouse retinal progenitor cells (RPCs) were cultured as previously described by our group and others (Klassen, Ng et al. 2004, Redenti, Neeley et al. 2009, Unachukwu, Sauane et al. 2013). In brief, RPCs were isolated from eyes of post-natal day 4 (P4) mice cone-rod homeobox (Crx) promoter driven GFP (Crx/GFP) on a C57BL/6J background (Jackson Labs) Crx/GFP+/+ pups maintained at the Lehman College Animal Facility. Culture used polystyrene flasks (T-75, Falcon) in Neurobasal medium (NBM; Invitrogen-Gibco, Rockville, MD) that contained 2 mM L-glutamine, 100mg/ml penicillin–streptomycin, 20 ng/ml epidermal growth factor (EGF; Invitrogen-Gibco) and neural supplement (B27 and N2; Invitrogen-Gibco). Cells were maintained in a biological incubator at 37°C and 5% CO2, and media was refreshed every 3–4 days until cultured cells reached 95% confluency prior to testing.
2.3. Measurement of RPC Migration in Stimulus Fields
RPCs were suspended in media at ~1×105 cells/mL and injected via syringe pump into one chamber of the Gal-Mµ S at a volumetric flow rate of Q1=10 µL/min. Concurrently, an equal volume of cell-less media was injected into the opposite chamber using the same volume flow rate Q1=Q2. This dual injection maintained the pressure balance between the two chambers and enabled even cell seeding into the device. Cells were cultured inside the Gal-Mus for 2–12 hr prior to each stimulus to facilitate cell adhesion. Cells were then subject to stimuli: 1.) Chemotactic gradient field of SDF-1 (100ng/mL), only; 2.) Electric field of 100mV/mm(Mishra and Vazquez 2017), only; and 3.) Superposition of both the chemotactic and electric fields. Each stimulus was performed for 12hrs using a low and constant volume flow rate into both chambers, previously shown to generate minimal shear stress upon adhered cells(Kong, Majeska et al. 2011). This flow was necessary to prevent the accumulation of metabolites and changes in pH due to imposed fields.
Cell trajectories within the microdevice were obtained by imaging the positions of at least n=45 cells every 15 minutes from experiments performed within three independent Gal-MuS devices. The net distances, DN, traveled by cells were calculated using differences between the initial position and final position of the cell center of mass. RPC migration was additionally evaluated using a parameter of cell directedness, DT, previously defined and used by our group and others (Mishra et al, 2017) as the cosine of the angle between a cell’s net migration vector and the external field lines (i.e. electric, chemical, electro-chemical gradients). The average directedness for a sample is then defined as the sum of the directedness of all cells in the sample divided by the total number of cells, as shown in Figure 3. Via this definition, directedness can take values ranging from −1 to 1. Cells moving along field lines in an electrotactic fashion exhibit directedness values between 0.5 and 1, while cells that migrating in non-oriented electrokinetic fashion display directedness values between −0.5 and0.5. Cells undergoing repulsive migration would have a directedness less can −0.5.
Figure 3.
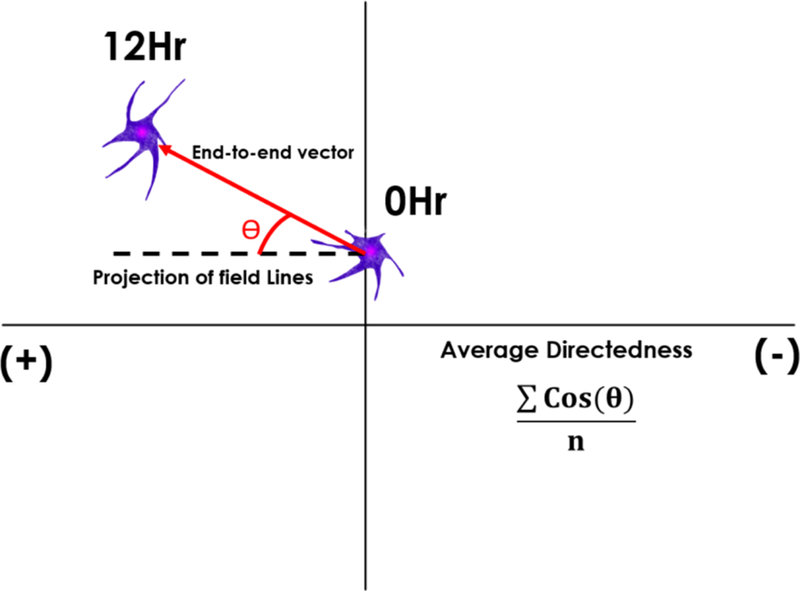
Schematic representation of the directedness parameter, DT. The directedness is defined as the cos(θ), where θ represents the angle between a cell’s end-to-end vector and the projection of electric field lines. The average directedness for a sample is the sum of the directedness of all cells in the sample divided by the total number of cells, n.
2.4. Immunocytochemistry
RPCs were fixed for 10min in ice cold methanol at −20°C directly following experiments. Fixed cells were washed with PBS and a blocking solution containing 1% bovine serum albumin was then applied for 1hr at room temperature (25°C). Cells were subsequently rinsed three times in PBS prior to use with antibodies. Primary antibodies for CXCR4 (Life Technologies, Carlsbad, CA, USA) and phospho-CXCR4 (Life Technologies, Carlsbad, CA, USA) were diluted 1:100 in antibody buffer and applied to cell samples at room temperature for 1hr, followed by 3 rinses with PBS. Note that CXCR4 is the known cognate receptor for SDF-1 (Kucia et al, 2004). The secondary antibody was similarly diluted 1:100 (Life Technologies, Carlsbad, CA, USA), applied at room temperature for 1hr, followed by 3 rinses with PBS. Slides were mounted with ProLong Diamond Antifade Mountant with DAPI (Invitrogen, Carlsbad, CA, USA).
2.5. Gene Expression Analysis by qPCR: CXCR4 and Gene Array
Total cellular RNA was isolated from RPCs directly following exposure to the three experimental stimuli using RNeasy Mini RNA Isolation Kit (Qiagen, 74104 www.qiagen.com), per the manufacturer’s protocol: 1.) Electric Field (EF); 2.) Chemical gradient field (SDF-1); and 3.) Combinatory stimuli of superimposed electro-chemical fields. RNA Quantification was performed via spectrophotometry (Synergy H1, Biotek, Highland Park, VT) using the Taqman RNA-to-Ct One-Step Kit (Life Technologies), per the manufacturer’s protocol. TaqMan Gene expression assay primers for GAPDH and CXCR4 were utilized to generate the PCR product directly from isolated RNA as per Table 1. Negative controls were performed that contained RNA but no primers. Automated PCR was performed in a final volume of 20µL containing 5ng of RNA template, 10µL of TaqMan RT-PCR Mix, 1µL of the Taqman Gene Expression Assay primer and 0.5µL of Taqman RT Enzyme Mix in a StepOne Plus Real-Time PCR system (Applied Biosystems). Reverse transcription was performed at 48°C for 15 min, followed by denaturation of the cDNA and activation of the DNA polymerase at 95°C for 10 min. 40 cycles of 15 sec at 95°C and 1 min at the annealing temperature of 60°C were used to amplify the PCR product. The relative change in expression levels for each product between the unstimulated and experimental conditions was represented as 2−∆∆Ct (Thakur, Mishra et al. 2018).
Table 1.
Primers used for reverse transcription-polymerase chain reaction of retinal progenitor Cells (RPCs).
| GENE | ASSAY ID | UniGene | GenBank | Size (Base pair) |
|---|---|---|---|---|
| GAPDH | Mm99999915 | Mm.304088 | NM_001289726.1 | 107 |
| CXCR4 | Mm01996749 | Mm.1401 | NM_009911.3 | 105 |
Two sets of 96-well array plates (RT2 profiler PCR array gene expression assay, Qiagen) containing SYBER Green primers for 168 different proteins associated with either adherens junctions or cell motility and 5 housekeeping genes, were utilized to generate PCR product. A full listing of the primers used is provided in supplemental Table S1. Automated PCR was performed in a final volume of 25 µL containing 0.5 µg of RNA template and 12.5 µL RT2 SYBR Green Mastermix in a StepOne Plus Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). PCR started with denaturation of the cDNA and activation of the DNA polymerase at 95°C for 10 min. 40 cycles of 15 sec at 95°C and 1 min at the annealing temperature of 60°C were used to amplify the PCR product.
2.6. Differential Expression, Pathway Analysis and Inhibitor Prediction
Differentially expressed genes (DEG) between testing conditions and control (chemokine stimulation) were identified from the qPCR array data utilizing the DESeq2 Bioconductor package(Love, Anders et al. 2014). The DEGs in each group were defined by a fold change >2 and a false discovery rate of 0.1. Fold-change data returned were used in pathway analysis for Parametric Analysis of Gene Set Enrichment (PAGE)(Kim and Volsky 2005). This was performed via the Bioconductor PAGE package using the Kyoto Encyclopedia of Genes and Genomes (KEGG)(Kanehisa and Goto 2000) to measure the predicted perturbations within the overrepresented pathways. The gene sets were then screened against the PAGE-ranked qPCR data to calculate enrichment scores (ES) and p-values for each gene set. The ES represents the degree to which a pathway is overrepresented either by the highest ranked up-regulated and down-regulated genes within the data(Subramanian, Tamayo et al. 2005). Additionally, the DEG were used as input signatures for LINCS L1000CDS2 Characteristic Direction Search Engine (10.1038/npjsba.2016.15) to identify possible small molecular perturbagens for future loss-of-function tests. A flowchart description of these analyses is seen in Figure 4.
Figure 4.

Bioinformatics Pathway. Brief overview of the process flow of the experimental data.
2.7. Inhibitor Loss-of-Function Test
The electro-chemotactic migration of RPC was performed as described and used a syringe pump to flow a constant dosage of 10µM/mL of the small molecule ICRF-193 (Top2B-inhibitor; generously provided by the Cai lab). Cells were stimulated for 12hrs using the same low and constant volume flow rate as before. As described, cell trajectories were imaged to determine net distances traveled, DN, and cell directedness, DT.
2.8. Imaging
Phase contrast images were acquired on a Nikon TE-2000U inverted microscope. The microscope stage was housed in a temperature-controlled, carbon dioxide-controlled and humidity-controlled chamber (Morell Instruments, Melville, NY). The ImageJ plugin, TrackMate, was used to gather cell trajectories by recording positions of cell centroids over time(Tinevez, Perry et al. 2017). High resolution confocal images were obtained with a Leica LCS SP8 STED 3X (Leica, Wetzlar, Germany) using a 63x oil immersion objective and white light laser. Image files were managed using LAS X software (Leica, Wetzlar, Germany).
2.9. Statistical Analysis
Data was evaluated using statistical software (Matlab r2018b). All experiments were performed in triplicate with a sample size of at least n= 3. Analysis of variance (ANOVA) was used to determine statistical significance among the experimental groups. Post-hoc Tukey test revealed where the statistical significance laid on, where p-values <0.05 were denoted by an *, and p<0.01 were marked by **.
3. Results
3.1. Electro-Chemotaxis Enhanced Migration
The migratory behaviors of the RPCs to external stimuli were examined using our established Gal-MuS device. The Gal-MuS unique design enabled experiments to independently stimulate cells with highly-controlled chemical concentration gradients, electric fields, and combinations of both fields. Figure 5 shows that RPCs stimulated with a gradient field of SDF-1 migrated a net distance of DN=38.1 µm ± 3.7 µm toward the cathode, i.e. negative electrode, with a directedness value of DT=0.80 ± 0.04. RPC movement stimulated by external electric fields (EF) resulted in an average distance of DN=48.7 µm ± 5.14 µm and directedness of DT=0.99 ± 0.02. Combinatory stimuli of SDF-1 gradient fields superimposed with electric fields resulted in net RPC migration toward the cathode with an average distance of DN=133.0 µm ± 18.4µm and directedness of DT=.97 ± .03, significantly greater than either condition alone (p<0.01). RPCs demonstrated no measurable motility in any control conditions using media without SDF-1 stimulation or without electric field stimulus. These values are summarized in Table 2.
Figure 5.
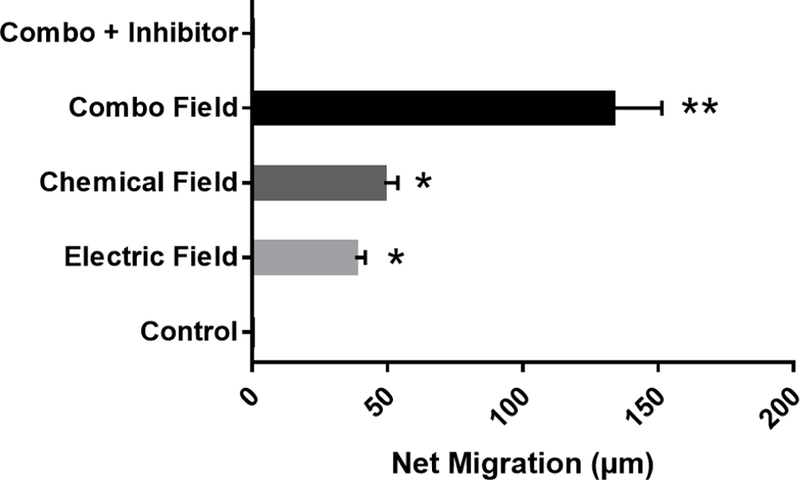
Net migration distance of RPC cells in response to different stimuli: Electric field, Chemotactic field, Combinatory Electro-Chemotactic field and control (no external fields) Electric and chemotactic stimuli are both significantly greater than control, * (p<0.05), but are not significantly different from one another. Combinatory stimulation was larger than either stimulus alone ** (p<0.01) against control. N=3 independent microfluidic devices.
Table 2.
Net Migration and Directionality
| CONDITION | NET MIGRATION DISTANCE – DN (µm) | DIRECTIONALITY – DT |
|---|---|---|
| CONTROL | 0 | N/A |
| CHEMOTACTIC ONLY | 38.1 ± 3.7 | 0.80 ± 0.04 |
| ELECTRIC FIELD ONLY | 48.7 ± 5.14 | 0.99 ± 0.02 |
| COMBO | 133.0 ± 18.4 | 0.97 ± 0.03 |
3.2. CXCR4 Expression and Distribution
The effect of external stimuli on the cognate receptor for SDF-1, known as CXCR4, was measured via RT-PCR. As shown in Figure 6, RPCs treated with exogenous SDF-1 fields produced an approximately 2-fold increase in CXCR4 expression compared to control (no stimulus). By contrast, RPCs treated with EF did not alter CXCR4 expression levels compared to control. Lastly, RPCs exposed to a combinatory field of EF and SDF-1 exhibited CXCR4 expression similar to that recorded in response to SDF-1 stimulus, alone.
Figure 6.
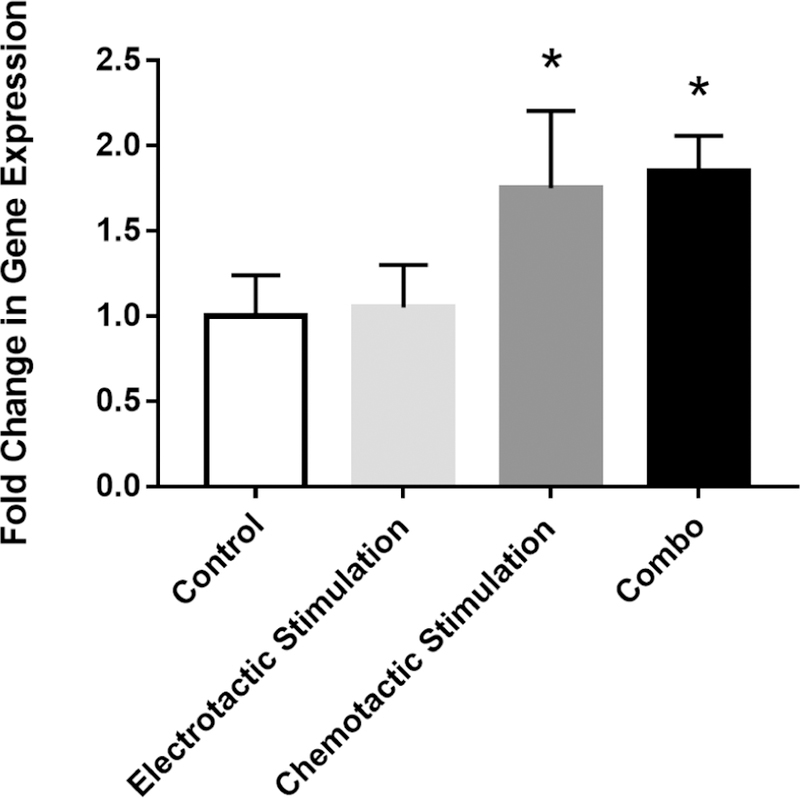
Gene expression of CXCR4 is altered by chemical stimulation but not electrical stimulation. mRNA levels in RPCs were quantified by real time PCR for different stimulation conditions: Electrical, Chemotactic, Combinatory electro-chemotactic and control (no external fields). Chemotactic and combinatory stimulation were significantly greater than control, p<0.05, but electrical stimulation was not.
To investigate whether external stimuli induced redistribution of CXCR4 receptors across RPC membranes, tests immunostained the phosphorylated or activated receptor, p-CXCR4, of cells exposed to each stimulus, independently, in the Gal-MuS. Data in Figure 7 illustrate that external SDF-1 signaling resulted in increased p-CXCR4 expression while external EF did not. Further, both CXCR4 and p-CXCR4 (when present) were found to be evenly distributed across the cytoplasm as measured by fluorescent intensity per average cell surface area in each experimental group.
Figure 7.
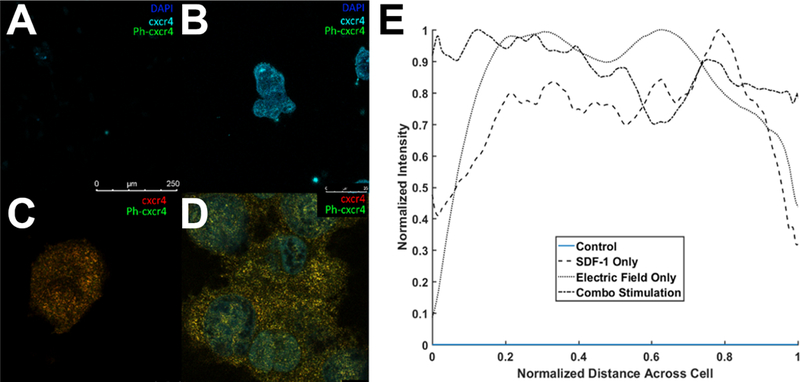
Co-localization of CXCR4 and p-CXCR4 via Stimulated Emission Depletion Confocal Microscopy (STED). Confocal image of (A) Control (B) Electric field stimulation, (C) SDF-1 stimulation. (D) Combined Electric field and SDF-1 stimulation. (E) Average cross-sectional distribution of CXCR4 expression across cell membranes for stimulation conditions.
3.3. Bioinformatics Analysis of Combinatory Stimulation
Experiments next used bioinformatics analyses to identify potential mechanistic pathways downstream of CXCR4 that could regulate the galvano-chemotactic migration observed. Analyses were performed in 3 parts: 1.) Expression levels of the cognate receptor under the stimulus conditions examined were determined using qPCR arrays. 2.) Sets of DEGs were defined as those with a fold change >2 compared to control, as defined and used by multiple projects in the nervous system. and 3.) Parametric Analysis of Gene Set Enrichment was performed to identify the most statistically-significant, overrepresented KEGG pathways in order to target potential underlying pathways.
RT2 Profiler qPCR arrays were first used to identify expression levels of 168 genes of interest. The DESeq2 analysis identified 26 DEGs between RPCs stimulated with electro-chemotactic fields versus stimulation with SDF-1 fields, only. As seen in Table 3, 4 genes were upregulated in the RPCs exposed to electro-chemotactic fields: (1) Talin 2: A protein associated with focal adhesion formation that links integrins to the actin cytoskeleton(Critchley 2000, Debrand, El Jai et al. 2009); (2) lmo7: important in protein-protein interactions involved in ubiquination, shuttling of transcription factors (Holaska et al. (2006)) and retinal development (Semenova et al. (2003))(Ooshio, Irie et al. 2004); (3) JUP: a member of the catenin family that forms distinct complexes with cadherins and desmosomal cadherins linking them to the actin cytoskeleton (Cowin, Kapprell et al. 1986, Lampugnani, Corada et al. 1995); and (4) Svil: a protein tightly-associated with both actin filaments and plasma membranes, suggested for roles as a high-affinity link between the actin cytoskeleton and the membrane (Oh, Pope et al. 2003).
Table 3.
The most differentially expressed genes (DEGs) in chemical stimulation compared to combinatory electro-chemotactic stimulation, as per PAGE (excluding oncogenic pathways), p<0.05
| GENE | FOLD CHANGE | P-VALUE |
|---|---|---|
| TLN2 | −15.88 | 0.000319 |
| LMO7 | −8.1 | 0.000171 |
| SVIL | −4.06 | 0.00091 |
| JUP | −4.02 | 0.004854 |
This list of DEGs was then used for PAGE with the KEGG gene set database to determine enriched or overrepresented metabolic pathways. The most significant pathways are shown in Figure 8. For clarity, the enriched pathways, p-values and associated genes are summarized in Table. PAGE was used to identify the 2 most significantly enriched signaling pathways, the Rap1 signaling pathway and the adherens junction pathway, as shown in Figure 9. Meaningfully, this data identifies the computed total perturbation of each gene within the signaling pathway by considering both the gene’s fold change and the accumulated perturbation propagated from upstream genes. This diagram illustrates significant downregulation of the proteins involved in adherens junctions (cadherins and β-catenin) and focal adhesions (cdc42 and integrins) – highlighted in yellow, and upregulation of genes RalGDS and Ral – highlighted in purple, two proteins that have strong pro-migratory effects through actions on the actin cytoskeleton(Rosse, Hatzoglou et al. 2006).
Figure 8.
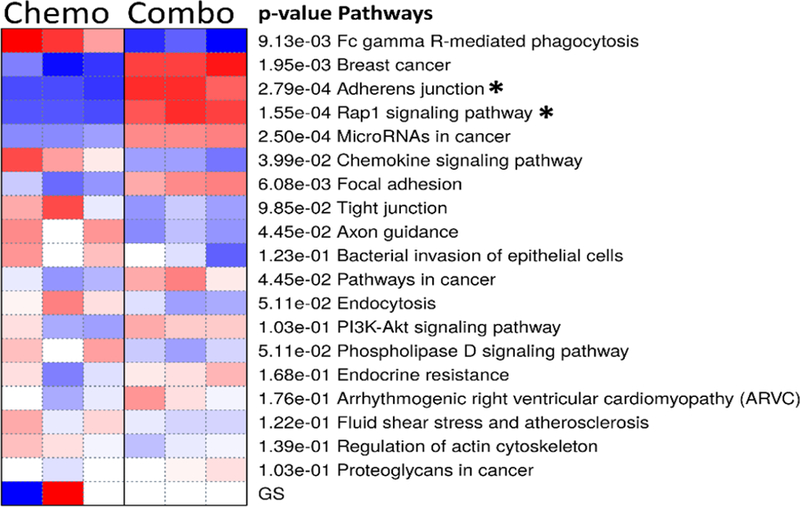
Heat Map of PAGE for the 20 most significantly enriched KEGG pathways between chemotactic (SDF-1 only) and combinatory electro-chemotactic stimulation. Color intensity represents degree by which a pathway is activated (blue) or suppressed (red).
Figure 9.
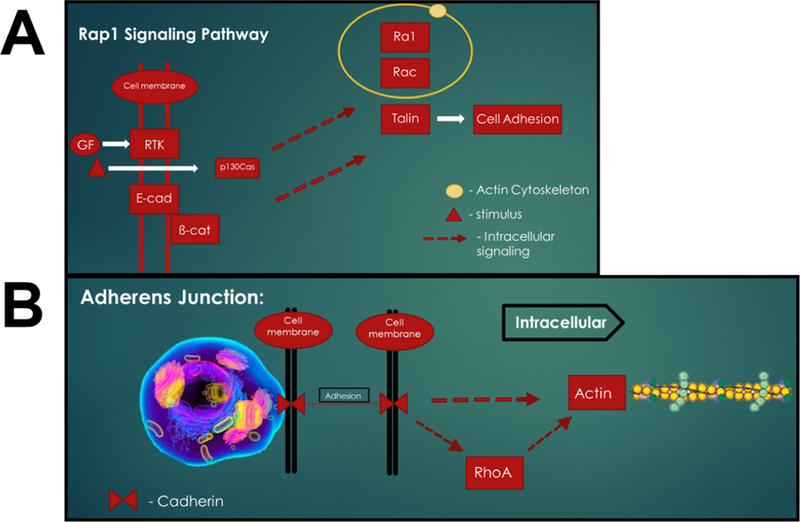
Most significantly-enriched KEG signaling pathways. Pathways have been condensed to most impacted signaling components for clarity. (A) Rap1 signaling pathway and (B) Adherens junction signaling pathway.
Additionally, the DEG were used as input for analysis with the LINC1000CD2 database to identify possible perturbagens that could act as loss-of-function validators of the identified pathways. We identified tens of possible perturbagens and selected the topoisomerase B (Top2B) inhibitor, ICRF-193, as a candidate inhibitor to validate ability of bioinformatics analyses to derive mechanistic targets. A 10µM/mL treatment of RPCs with ICRF-193 during combination stimulation resulted in complete cessation of cell migration.
4. Discussion
Transplantation of retinal neurons shows great promise in treatment of degenerative eye diseases. The migration of transplanted RPC is a critical but incompletely understood component of the transplantation process. This project illustrated that RPC cells can migrate with increased directionality and distance in response to externally imposed electro-chemotactic fields. While our lab and numerous others have previously demonstrated that CNS cells are able to migrate towards signaling from chemoattractants using microfluidic systems(Taylor, Rhee et al. 2006, Mishra, Thakur et al. 2015, McCutcheon, Unachukwu et al. 2017, Mishra and Vazquez 2017, Rothbauer, Zirath et al. 2018), far fewer studies have applied these techniques to retina. Microfluidics enables the precise control of the extracellular environment to, thereby, provide a powerful tool with which to evaluate cell responses to external stimuli, mechanistically (Figure 2). Further, application of the Gal-MuS design provided unique advantages in that the device was able to impose concentration gradients (SDF-1) and electric fields, separately and in tandem, without altering the individual effect of either, but rather inducing synergistic cell migratory responses. Microfluidic data show that the migration of RPCs towards increasing SDF-1 gradient and applied EF is significantly enhanced in the combinatory field compared to either field alone. The net migration during combinatory stimulation remained in the direction of the cathode and with similar levels of directedness as seen during individual stimulus, but with significant increases in distances traveled (Figure 5). Future in vitro studies will incorporate biomimetic materials to act as a physical barrier within the testing system to better mimic retinal tissue. Clinical applications can also use FDA-approved biomaterials and drug-eluding polymers to generate chemotactic gradients in situ during RPC transplantation. Additional studies will also incorporate pathological models of the retinal ex vivo environment (Andjelic, Sofija et al. 2014) to better represent the well-known thickening of the outer membrane in retinal degeneration as well as integrate the intrinsic response of surrounding retinal cells (Cai, Hui et al. 2018, Rettinger, Christina et al. 2018).
To examine biological reasons for such increases, we investigated the expression and distribution of the cognate receptor. Previous work with CNS cells and EF suggested that EF can cause redistribution of cell surface receptors to thereby alter receptor sensitization(Zhao, Dick et al. 1999, McCaig, Rajnicek et al. 2005). In such a scenario, RPCs would exhibit an asymmetric redistribution of CXCR4 activation compared to control or to SDF-1 stimuli. However, immunofluorescence assays revealed no differences in either CXCR4 or p-CXCR4 distribution (Figure 7). Furthermore, data gathered from PCR expression (Figure 6) illustrated that EF stimuli did not cause genetic alterations of the CXCR4 receptor. These findings suggest that a synergistic relationship occurs downstream of the ligand-bound receptors.
We next investigated possible intersections of signaling pathways downstream of CXCR4 via qPCR array to examine the difference between chemotactic signaling and combinatory signaling on the expression of genes associated with cell movement (i.e. cell adhesion and motility). Differentially expressed genes (DEG) that showed enrichment in potential synergistic pathways were first identified and, second, examined via pathway analysis to identify the most significantly perturbed processes downstream of DEG. This bioinformatics analysis (Figure 9) suggest that reduced cell adhesion sites (through the down regulation of their constituent proteins such as cadherin and β-catenin) and cytoskeletal reorganization (through upregulation of RalGDS and Ral1) may represent synergistic points between the electrotactic and chemotactic signaling. The upregulation of proteins like RalGDS and Ral1 indicate that expected increases in cytoskeletal remodeling and organization may result in increased cell migration. This corresponds with findings in the literature, that describe reduced cell adhesion correlated to improved migratory rates (Palecek, Loftus et al. 1997, Friedl and Wolf 2003). Reducing cell adhesion works to reduce the amount of traction forces generated by the cell, and thus, reduce the overall contractile force necessary to pull the cell forward, speeding up cell migration. Focal adhesions act to generate force in opposition to the contractile force that causes the cells to move, by reducing the opposition force, the cells should migrate even more quickly. Further, the upregulation of proteins associated with cytoskeletal organization, including Rac and RalGDS, may indicate increasing lamellipodial projections and extension forces within the RPCs studied(Tapon and Hall 1997, Hall and Nobes 2000). Increasing extensive forces and reducing adhesion forces may, therefore, facilitate enhanced RPC migratory speeds.
We verified our pathway analysis through loss-of-function tests using the Top2B inhibitor, ICRF-193, as predicted from our LINC1000CD2 analysis. Top2B has been shown to play an important role in retinal development and maintenance (Li 2014). Its loss has been linked to reduction in migratory guidance ques within the retina (Cai 2014, Rice and Curran 2001), making Top2B inhibition an effective loss-of-function agent for our study. This study illustrated the potential of activating synergistic chemotactic and electrotactic pathways to dramatically improve the migratory response of retinal progenitor cells. Such innovative strategies can lead to the development of improved migration-targeted cell replacement therapies as well as cell-based reparative treatments more generally.
Supplementary Material
Table S1. The most significantly enriched pathways and associated genes (p <0 .05) per PAGE analysis
Figure 1.
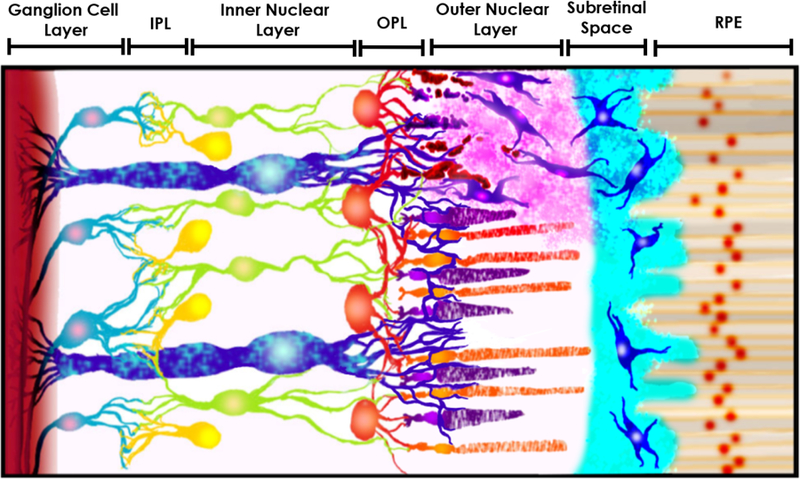
Schematic of transplanted retinal progenitor cells (RPCs) migrating into damaged host retinal tissue. The schematic provides a representative cross-section of retinal tissue from the retinal pigment epithelium (RPE) at the eye posterior to the ganglion cells of the optic nerve (from right to left, not to scale). RPCs (shown in blue) are transplanted in the sub-retinal space between the RPE and outer nuclear layer (ONL), in which the native rod and cone photoreceptors reside. Transplanted RPCs then migrate into retinal tissue to synaptically integrate in the outer plexiform layer (OPL) with native horizontal cells (HCs), bipolar cells (BCs) in the inner nuclear layer (INL) in turn with ganglion cells (GCs) and amacrine cells (ACs) in the inner plexiform layer (IPL) to restore vision. The figure also features the presence of Müller glia cells (MGCs) along all three main retinal layers. Modified after Thakur et al 2018.
While individually-imposed chemotactic and electric fields increased the migration distance of retinal progenitor cells (RPCs), combinatory fields of both chemotactic and electric fields increased RPC migration distance by more than three times that of either field alone.
No changes in the polarization or up-regulation of the cognate receptor were measured during tests of RPC migration.
Bioinformatics analysis implicates Rap1 and Adherins pathways as possible control mechanisms for the enhanced RPC migration
Pathway predictions were validated via loss-of-function assay inhibiting Top2B
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
6. References
- Andjelic S, Lumi X, Veréb Z, Josifovska N, Facskó A, Hawlina M, & Petrovski G (2014). A simple method for establishing adherent ex vivo explant cultures from human eye pathologies for use in subsequent calcium imaging and inflammatory studies. Journal of immunology research, 2014, 232659 435 doi: 10.1155/2014/232659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikawa E, Quellhorst G, Han Y, Pan H and Yang J (2010). “RT2 Profiler PCR Arrays: Pathway-focused gene expression profiling with qRT-PCR” SA Bioscience. [Google Scholar]
- Cai H, Gong J, Del Priore LV, Tezel TH, Fields MA Culturing of Retinal Pigment Epithelial Cells on an Ex Vivo Model of Aged Human Bruch’s Membrane. J. Vis. Exp (134), e57084, 440 doi: 10.3791/57084 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HF, Lee YS, Tang TK and Cheng JY (2016). “Pulsed DC Electric Field-Induced Differentiation of Cortical Neural Precursor Cells.” PLoS One 11(6): e0158133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowin P, Kapprell HP, Franke WW, Tamkun J and Hynes RO (1986). “Plakoglobin: a proteincommon to different kinds of intercellular adhering junctions.” Cell 46(7): 1063–1073. [DOI] [PubMed] [Google Scholar]
- Critchley DR (2000). “Focal adhesions - the cytoskeletal connection.” Curr Opin Cell Biol 12(1): 133–446 139. [DOI] [PubMed] [Google Scholar]
- da Cruz L, Coley BF, Dorn J, Merlini F, Filley E, Christopher P, Chen FK, Wuyyuru V, Sahel J, Stanga P, Humayun M, Greenberg RJ and Dagnelie G (2013). “The Argus II epiretinal prosthesis system allows letter and word reading and long-term function in patients with profound vision loss.” Br J 450 Ophthalmol 97(5): 632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalkara D, Byrne LC, Klimczak RR, Visel M, Yin L, Merigan WH, Flannery JG and Schaffer DV (2013). “In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous.” Sci Transl Med 5(189): 189ra176. [DOI] [PubMed] [Google Scholar]
- Das AM, Zhao X and Ahmad I (2005). “Stem cell therapy for retinal degeneration: retinal neurons from heterologous sources.” Semin Ophthalmol 20(1): 3–10. [DOI] [PubMed] [Google Scholar]
- Debrand E, El Jai Y, Spence L, Bate N, Praekelt U, Pritchard CA, Monkley SJ and Critchley DR (2009). “Talin 2 is a large and complex gene encoding multiple transcripts and protein isoforms.” The 458 FEBS journal 276(6): 1610–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaxman SR, Bourne RRA, Resnikoff S, Ackland P, Braithwaite T, Cicinelli MV, Das A, Jonas JB, Keeffe J, Kempen JH, Leasher J, Limburg H, Naidoo K, Pesudovs K, Silvester A, Stevens GA, Tahhan N, Wong TY and Taylor HR (2017). “Global causes of blindness and distance vision impairment 1990–2020: a systematic review and meta-analysis.” Lancet Glob Health 5(12): e1221–e1234. [DOI] [PubMed] [Google Scholar]
- Friedl P and Wolf K (2003). “Tumour-cell invasion and migration: diversity and escape mechanisms.” Nat Rev Cancer 3(5): 362–374. [DOI] [PubMed] [Google Scholar]
- Gasparini SJ, Llonch S, Borsch O and Ader M (2018). “Transplantation of photoreceptors into the degenerative retina: Current state and future perspectives.” Prog Retin Eye Res [DOI] [PubMed]
- Hall A and Nobes CD (2000). “Rho GTPases: molecular switches that control the organization and dynamics of the actin cytoskeleton.” Philos Trans R Soc Lond B Biol Sci 355(1399): 965–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M and Goto S (2000). “KEGG: kyoto encyclopedia of genes and genomes.” Nucleic acids research 28(1): 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S-Y and Volsky DJ (2005). “PAGE: Parametric Analysis of Gene Set Enrichment.” BMC Bioinformatics 6(1): 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klassen HJ, Ng TF, Kurimoto Y, Kirov I, Shatos M, Coffey P and Young MJ (2004). “Multipotent retinal progenitors express developmental markers, differentiate into retinal neurons, and preserve light-mediated behavior.” Invest Ophthalmol Vis Sci 45(11): 4167–4173. [DOI] [PubMed] [Google Scholar]
- Kong Q, Majeska RJ and Vazquez M (2011). “Migration of connective tissue-derived cells is mediated by ultra-low concentration gradient fields of EGF.” Exp Cell Res 317(11): 1491–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamba DA, McUsic A, Hirata RK, Wang PR, Russell D and Reh TA (2010). “Generation, purification and transplantation of photoreceptors derived from human induced pluripotent stem cells.” 480 PLoS One 5(1): e8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampugnani MG, Corada M, Caveda L, Breviario F, Ayalon O, Geiger B and Dejana E (1995). “The molecular organization of endothelial cell to cell junctions: differential association of plakoglobin, beta-catenin, and alpha-catenin with vascular endothelial cadherin (VE-cadherin).” J Cell Biol 129(1): 484 203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M, Anders S and Huber W (2014). “Differential analysis of count data–the DESeq2 package.” Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Baranov P, Patel S, Ouyang H, Quach J, Wu F, Qiu A, Luo H, Hicks C, Zeng J, Zhu J, Lu J, Sfeir N, Wen C, Zhang M, Reade V, Sinden J, Sun X, Shaw P, Young M and Zhang K (2014). “Human retinal progenitor cell transplantation preserves vision.” J Biol Chem 289(10): 6362–6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttrull JK and Dorin G (2012). “Subthreshold diode micropulse laser photocoagulation (SDM) as invisible retinal phototherapy for diabetic macular edema: a review.” Curr Diabetes Rev 8(4): 274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin DF, Maguire MG, Fine SL, Ying GS, Jaffe GJ, Grunwald JE, Toth C, Redford M and Ferris FL 3rd (2012). “Ranibizumab and bevacizumab for treatment of neovascular age-related macular degeneration: two-year results.” Ophthalmology 119(7): 1388–1398.22555112 [Google Scholar]
- McCaig CD, Rajnicek AM, Song B and Zhao M (2005). “Controlling cell behavior electrically: current views and future potential.” Physiol Rev 85(3): 943–978. [DOI] [PubMed] [Google Scholar]
- McCutcheon S, Unachukwu JU, Redenti S and Vazquez M (2015). “Chemotactic Migration of Clustered Central Nervous System Progenitor Cells”
- McCutcheon S, Unachukwu U, Thakur A, Majeska R, Redenti S and Vazquez M (2017). “In vitro formation of neuroclusters in microfluidic devices and cell migration as a function of stromal-derived growth factor 1 gradients.” Cell Adh Migr 11(1): 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Li W, Young F, Gao R, Chalmers L, Zhao M and Song B (2012). “Electric field-controlled directed migration of neural progenitor cells in 2D and 3D environments.” J Vis Exp(60): 3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S, Thakur A, Redenti S and Vazquez M (2015). “A model microfluidics-based system for the human and mouse retina.” Biomed Microdevices 17(6): 107. [DOI] [PubMed] [Google Scholar]
- Mishra S and Vazquez M (2017). “A Gal-MmicroS Device to Evaluate Cell Migratory Response to Combined Galvano-Chemotactic Fields.” Biosensors (Basel) 7(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Pope RK, Smith KP, Crowley JL, Nebl T, Lawrence JB and Luna EJ (2003). “Archvillin, a muscle-specific isoform of supervillin, is an early expressed component of the costameric membrane skeleton.” J Cell Sci 116(Pt 11): 2261–2275. [DOI] [PubMed] [Google Scholar]
- Ooshio T, Irie K, Morimoto K, Fukuhara A, Imai T and Takai Y (2004). “Involvement of LMO7 in the association of two cell-cell adhesion molecules, nectin and E-cadherin, through afadin and alpha-actinin in epithelial cells.” J Biol Chem 279(30): 31365–31373. [DOI] [PubMed] [Google Scholar]
- Palecek SP, Loftus JC, Ginsberg MH, Lauffenburger DA and Horwitz AF (1997). “Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness.” Nature 516 385(6616): 537–540. [DOI] [PubMed] [Google Scholar]
- Pearson RA, Hippert C, Graca AB and Barber AC (2014). “Photoreceptor replacement therapy: challenges presented by the diseased recipient retinal environment.” Vis Neurosci 31(4–5): 333–344. [DOI] [PubMed] [Google Scholar]
- Redenti S, Neeley WL, Rompani S, Saigal S, Yang J, Klassen H, Langer R and Young MJ (2009). “Engineering retinal progenitor cell and scrollable poly(glycerol-sebacate) composites for expansion and subretinal transplantation.” Biomaterials 30(20): 3405–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettinger CL, & Wang H (2018). Quantitative Assessment of Retina Explant Viability in a Porcine Ex Vivo Neuroretina Model. Journal of ocular pharmacology and therapeutics : the official journal of the Association for Ocular Pharmacology and Therapeutics, 34 7, 521–530. [DOI] [PubMed] [Google Scholar]
- Rico-Varela J, Singh T, McCutcheon S and Vazquez M (2015). “EGF as a New Therapeutic Target for Medulloblastoma Metastasis.” Cell Mol Bioeng 8(4): 553–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosse C, Hatzoglou A, Parrini MC, White MA, Chavrier P and Camonis J (2006). “RalB mobilizes the exocyst to drive cell migration.” Mol Cell Biol 26(2): 727–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer M, Zirath H and Ertl P (2018). “Recent advances in microfluidic technologies for cell-to-cell interaction studies.” Lab Chip 18(2): 249–270. [DOI] [PubMed] [Google Scholar]
- Saadi W, Rhee SW, Lin F, Vahidi B, Chung BG and Jeon NL (2007). “Generation of stable concentration gradients in 2D and 3D environments using a microfluidic ladder chamber.” Biomed 533 Microdevices 9(5): 627–635. [DOI] [PubMed] [Google Scholar]
- Saigal R, Cimetta E, Tandon N, Zhou J, Langer R, Young M, Vunjak-Novakovic G and Redenti S (2013). “Electrical stimulation via a biocompatible conductive polymer directs retinal progenitor cell differentiation.” Conf Proc IEEE Eng Med Biol Soc 2013: 1627–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR and Lander ES (2005). “Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles.” Proceedings of the National Academy of 540 Sciences 102(43): 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapon N and Hall A (1997). “Rho, Rac and Cdc42 GTPases regulate the organization of the actin cytoskeleton.” Curr Opin Cell Biol 9(1): 86–92. [DOI] [PubMed] [Google Scholar]
- Taylor AM, Rhee SW and Jeon NL (2006). “Microfluidic chambers for cell migration and neuroscience research.” Methods Mol Biol 321: 167–177. [DOI] [PubMed] [Google Scholar]
- Thakur A, Mishra S, Pena J, Zhou J, Redenti S, Majeska R and Vazquez M (2018). “Collective adhesion and displacement of retinal progenitor cells upon extracellular matrix substrates of transplantable biomaterials.” J Tissue Eng 9: 2041731417751286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinevez JY, Perry N, Schindelin J, Hoopes GM, Reynolds GD, Laplantine E, Bednarek SY, Shorte SL and Eliceiri KW (2017). “TrackMate: An open and extensible platform for single-particle tracking.” Methods 115: 80–90. [DOI] [PubMed] [Google Scholar]
- Unachukwu UJ, Sauane M, Vazquez M and Redenti S (2013). “Microfluidic generated EGF-gradients induce chemokinesis of transplantable retinal progenitor cells via the JAK/STAT and PI3kinase signaling pathways.” PLoS One 8(12): e83906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unachukwu UJ, Warren A, Li Z, Mishra S, Zhou J, Sauane M, Lim H, Vazquez M and Redenti S (2016). “Predicted molecular signaling guiding photoreceptor cell migration following transplantation into damaged retina.” Sci Rep 6: 22392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron PV, Di Marco F, Kruczek K, Ribeiro J, Graca AB, Hippert C, Aghaizu ND, Kalargyrou AA, Barber AC, Grimaldi G, Duran Y, Blackford SJI, Kloc M, Goh D, Zabala Aldunate E, Sampson RD, Bainbridge JWB, Smith AJ, Gonzalez-Cordero A, Sowden JC, Ali RR and Pearson RA (2018). “Transplanted Donor-or Stem Cell-Derived Cone Photoreceptors Can Both Integrate and Undergo Material Transfer in an Environment-Dependent Manner.” Stem Cell Reports 562 10(2): 406–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao L and Li Y (2016). “The Role of Direct Current Electric Field-Guided Stem Cell Migration in Neural Regeneration.” Stem Cell Rev 12(3): 365–375. [DOI] [PubMed] [Google Scholar]
- Zhao M, Dick A, Forrester JV and McCaig CD (1999). “Electric field-directed cell motility involves up-regulated expression and asymmetric redistribution of the epidermal growth factor receptors and is enhanced by fibronectin and laminin.” Mol Biol Cell 10(4): 1259–1276. 568 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The most significantly enriched pathways and associated genes (p <0 .05) per PAGE analysis


