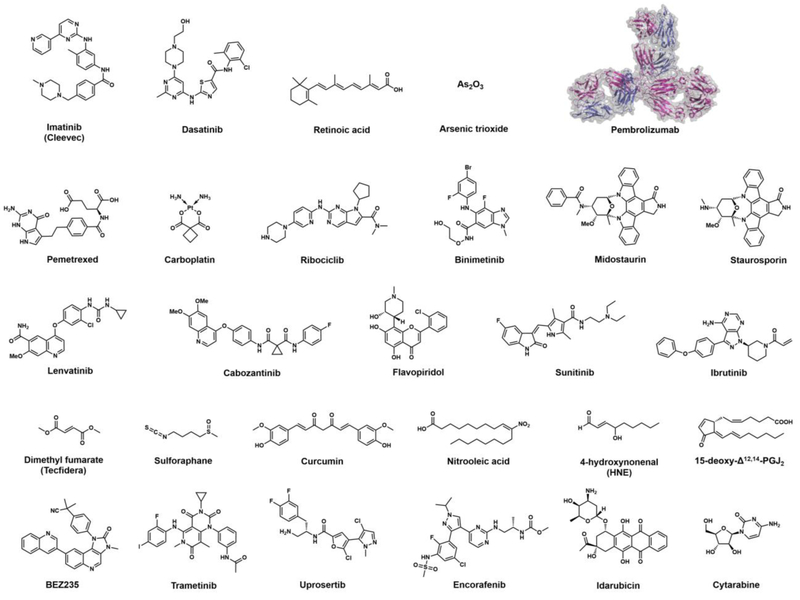
Many drugs have a stereotyped target; Imatinib (Gleevec) (Figure 1) targets the oncogenic kinase Bcr-Abl. Yet, in reality, most drugs actually have multiple targets/interactions[1]. Of course, this promiscuity can cause undesired effects. But, sometimes, secondary targets are beneficial. The second-generation Bcr-Abl-kinase inhibitor, dasatinib, was developed as a Src-kinase inhibitor and also inhibits DDR2-kinase[2]. These additional targets partially explain why dasatinib fares better than imatinib[3], a “monogamous” Bcr-Abl inhibitor[4,5]. Unsurprisingly, many monogamous drugs are too constrained to effect prolonged relapse/cure of cancer. Several concepts have been proposed to combat the issues with monogamous drugs, including combination therapy (CT) and polypharmacology (drugs targeting numerous proteins). CT has been employed against cancer since the 1960’s[6] and a good deal of theory for development and experimental evidence supporting CT’s benefits is available[7,8]. Polypharmacology is arguably not as developed, but recent data are promising[9]. Here we propose a regimen to develop covalent polypharmocological drugs from existing electrophiles.
Figure 1. Small-molecule drugs and antibody discussed in this perspective.
The protein structure of pembrolizumab is displayed in ribbon and surface representation (PDB: 5DK3). The heavy and light chains are shown in magenta and blue, respectively. (The order of the small-molecule drugs and antibody in the figure is based on their first appearance in the corresponding text followed by their appearance in subsequent figures).
Combination therapy
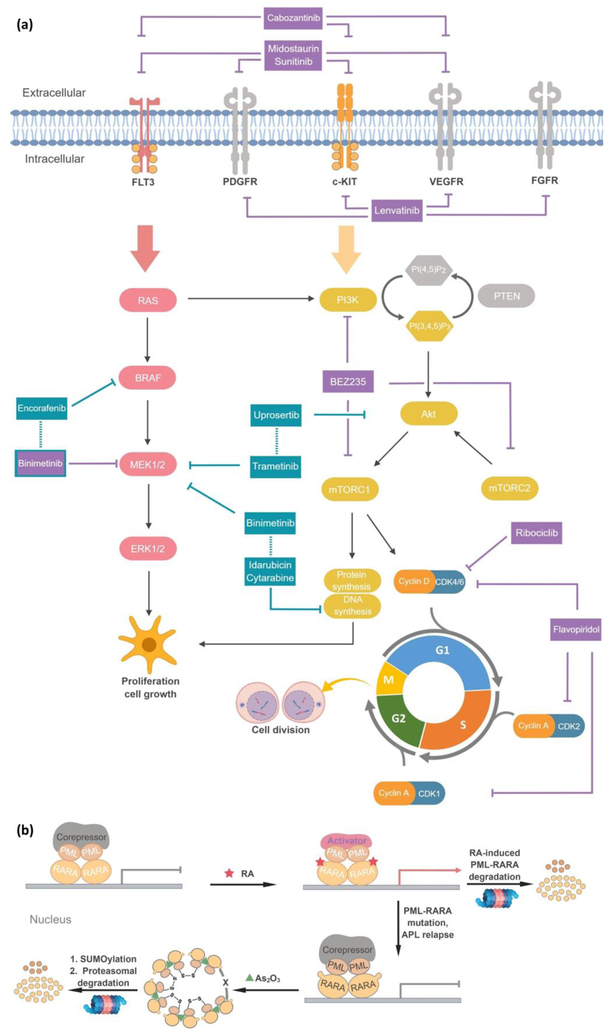
There are many successful small-molecule CT regimens (Figure 2a)[10], including retinoic acid/arsenic trioxide that treats acute promyelocytic leukemia (Figure 2b)[11]. Small-molecule-and-antibody-based CTs are also approved: e.g., lung cancer patients now benefit from a pembrolizumab and pemetrexed/carboplatin combination, twice as effective as chemotherapy alone[12]. However, development of CT is not trivial. It is difficult to identify drugs functioning synergistically[13], and animal models are generally poor predictors of CT efficacy[14]. Combining drugs from different companies can be logistically difficult to coordinate[15]. CT can also change pharmacokinetics of individual drugs and there are multiple pathways to resistance[16]. Mutation is less common for CT than evolution of multiple drug-resistance, although several drug-resistant mutations in the oncogenic fusion-protein target of retinoic acid/arsenic trioxide, PML-RARA, are known (Figure 2b)[11].
Figure 2. (a) Examples of combination therapies (CT) and polypharmacologic drugs[10].
This illustration focuses on combination therapies and polypharmacologic anti-cancer therapeutics (either approved or in clinical trials) that intercept Ras/RAF/MEK/ERK and PI3K/Akt/mTOR signaling. Drugs in aquamarine boxes with dotted connecting lines indicate CT and those in magenta boxes indicate drugs with more than one bioactive target. In particular, the MEK1/2 inhibitor binimetinib in combination with encorafenib has been approved by FDA to treat patients of BRAF-mutated melanoma, whereas the combination therapy (also shown in figure) of binimetinib, idarubicin and cytarabine has recently been terminated owing to logistical problems. Blunt-end and classical arrows (either originating from the drugs or within the pathways) indicate inhibition and activation, respectively. (b) Molecular actions of retinoic acid (RA) and arsenic trioxide (As2O3) CT in the treatment of acute promyelocytic leukemia (APL)[11]. Promyelocytic leukemia/RA receptor alpha (PML-RARA) together with transcription co-repressor mediate transcriptional silencing which impedes APL differentiation. RA induces cell differentiation by promoting the interaction between PML-RARA and its coactivator, eliciting target-gene expression. PML-RARA degradation can also be triggered upon prolonged exposure to RA in high concentrations. However, mutations in the RARA-domain often resist RA-assisted gene expression. As2O3 is used as a second-line of defense, which induces PML-RARA multimerization (denoted by “X” here) of both wild-type and RA-resistant mutants through interprotein disulfide bond formation, with consequent SUMOylation and proteasomal degradation. This CT-approach affords a high cure rate for APL patients.
Non-covalent polypharmacologic anti-cancer drugs
Several non-covalent anti-cancer drugs approved in 2017–2018 are poly- or oligo-pharmacological. These include ribociclib (CDK4/CDK6), binimetinib (MEK1/2) and midostaurin (a “multikinase” inhibitor)[17,18] (Figure 2a). Of these, only midostaurin is truly polypharmacologic; ribociclib and binimetinib target homologous sites within proteins with similar functions. In 2016, lenvatinib, a multireceptor tyrosine kinase inhibitor, and cabozantinib, a multitarget drug, were approved. Flavopiridol, a naturally-derived inhibitor of numerous cyclin-dependent kinases (orphan drug, 2014)[19] and sunitinib, a multiple receptor tyrosine kinase inhibitor (2006), were approved earlier[20]. Notably, many polypharmacologic drugs are inadvertent: imatinib is now approved to treat non-Bcr-Abl-positive tumors, since it targets the mast/stem cell growth factor receptor. Given the paucity of truly polypharmacologic anti-cancer drugs, and since these are predominantly ATP-competitive kinase inhibitors[9,21], new ways to design polypharmacologic drugs[22]—especially with novel target spectra—are needed. Several regimens have been proposed, but there are few guiding principles[23–25].
Polypharmacology and covalent binding
Until recently, pharma has shied away from covalent drugs because their reactive chemotypes were believed to amplify off-target effects, including drug-induced liver injury. We now appreciate that covalent drugs have numerous advantages over non-covalent binders, especially when targeting long-lived proteins. The go-to method to create covalent drugs is appending a low-reactivity electrophilic motif to a specific ligand, giving a more-or-less monogamous drug[1]; other examples of covalent inhibitors include mechanism-based inhibitors and reversible covalent drugs[26], although these are currently much less common.
However, the covalent Bruton-kinase inhibitor, ibrutinib (approved in 2013), which was developed using this common theme, inhibits numerous other kinases[27]. Furthermore, dimethyl fumarate (DMF) (Tecfidera)[28], a highly reactive molecule, is approved to treat relapsing multiple sclerosis, and has potential use in anticancer therapy. DMF—an analog of the oncometabolite fumarate[29,30]—taps into a mechanism called “electrophile signaling”. A thorough/exhaustive analysis of true in vivo pharmacological activities of DMF (or fumarate) is not technologically accessible currently, but it is likely that the targets of DMF and fumarate overlap. Fumarate targets several proteins including GAPDH[31] and Keap1. Additional targets have been proposed for DMF[28,32,33]. However, none of these fully explain the drug’s bioactivities. Sulforaphane—a polypharmacologic reactive compound present in vegetables[34] that is in Phase-II clinical trials—inhibits several deubiquitinating enzymes, and numerous other targets, although the principal target(s) are still debated[35]. Praeternatural sulforaphane analogs have been patented[36]. Similar arguments apply to other electrophilic pharmaceuticals in clinical trials, e.g., curcumin[37] and nitrooleic acid[38].
Thus, the pharmaceutical programs of reactive electrophilic molecules are an amalgam of multiple interactions, some beneficial, others possibly deleterious. How these behaviors fit together to produce a phenotypic output is complex. Improving our understanding of native electrophile signaling both in terms of defining key targets (those most likely/able to engage with the drug)[39] and linking specific ‘target engagement’ to pathway modulation(s) may promote better design of polypharmacologic drugs. Of course, it is not always easy to identify the key causative target(s) of compounds, especially polypharmacologic covalent binders. Indeed, the notion that key targets may exist for reactive electrophilic species (RES) was for a long time contentious. We and others have provided evidence that electrophiles hit specific sensor proteins that are key for their function. We called these sensor residues “privileged sensors” [26,39].
Mining privileged sensors
Insightful work from the Cravatt laboratory has shown that within ~1000 cysteines, 6 were hyperactive to the native lipid-derived RES, 4-hydroxynonenal (HNE)[40]. Only two of these proteins were kinases. 17 cysteines were reactive to an electrophilic prostaglandin (namely, 15-deoxy-Δ12,14-PGJ2). Thus, only ~1% of the targets screened were reactive to these RES. Furthermore, there was little target overlap between the RES-chemotypes, even though they have similar reactive functional groups (enal/enone). Native RES are therefore discriminatory and likely have a built-in diverse repertoire of protein sensors. Despite the thoughtful developments from several laboratories[40–43], state-of-the-art chemoproteomics profiling strategies continue to face limitations as they only cover a fraction of the human cysteome (which contains ~200,000 unique cysteines in total[39]) and are restrained to target identification following bulk RES-exposure to live cells or isolated organelles. Whole-cell flooding with RES can affect redox homeostasis, label multiple proteins that are not typically modified, and/or upregulate apoptosis. Thus, bulk RES-exposure approaches pose formidable challenges in interlinking individual identified targets/sensor residues to compartmentalized/context-specific electrophile signaling[43–46]. Similar issues undermine efforts to understand promiscuous covalent drugs[39]. To understand electrophile signaling at the individual protein level with high spatiotemporal resolution, we developed a complementary target/ligand-pair-specific approach (T-REX)[47]. T-REX evaluates the RES sensitivity of a specific protein to a reactive enal/enone-based RES of choice in a largely unperturbed cell in live culture, worms, or fish, and for those targets that are RES-responsive, defines the precise RES-modification-dependent function directly in vivo [47–50]. Several privileged HNE-sensors that are known or potential drug targets were identified using T-REX. Relatively few were kinases and not all were enzymes. Interesting examples include: Akt3 (an oncogenic kinase; other isoforms were not as HNE-sensitive)[48]; PTEN (a tumor-suppressor phosphatase)[47]; and Ube2V1/Ube2V2 (allosteric stimulators of the ubiquitin-E2-conjugating enzyme, Ube2N)[50]. Importantly, mutation of the specific sensor cysteines [e.g., Akt3(C119), and Ube2V2(C69)] ablated HNE-sensing and signaling downstream. These privileged sensors showed hyper-reactivity to HNE in vitro[26]. Previously-reported HNE-sensors (e.g., glutathione-S-reductase) were folds less reactive to HNE in vitro[26,43]. These data underscore the complexities in pinpointing true electrophile “sensors” in uncontrolled conditions.
Low-occupancy RES-modifications determine cell response
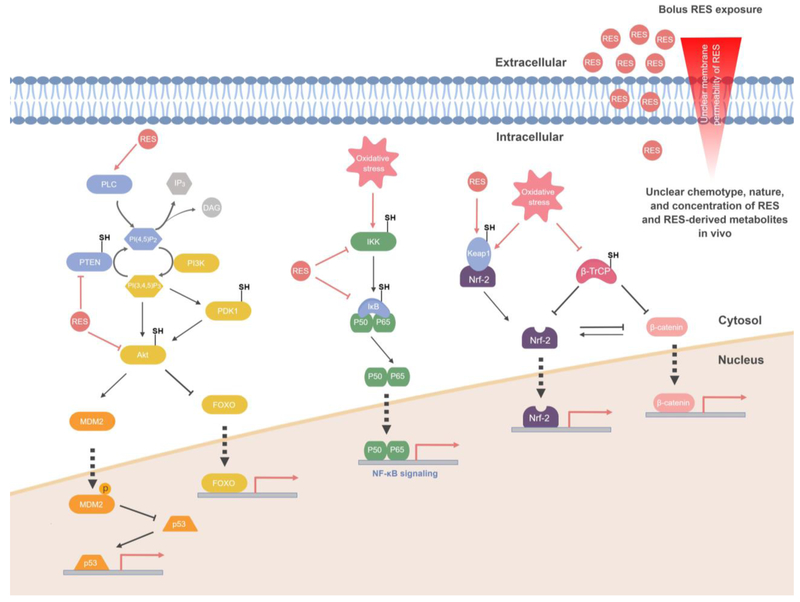
Privileged sensors discovered by targeted RES-delivery in cells or fish altered their canonical activities as a function of RES-modification. By contrast, sensing-defective-but-otherwise-functional mutants did not. ~12% HNEylation of Akt3 elicited ~30% total Akt-inhibition in cells/fish (Akt3 expression was close to endogenous in fish)[48]; similar dominant inhibition occurred with PTEN[47]. Since PTEN and Akt function antagonistically (Figure 3), this result helps explain the difficulty to assign functional roles to HNE (and likely other) RES. The data also emphasize the importance of context to RES-modification-dependent cell responses.
Figure 3. Challenges in deconstructing multicomponent cell responses triggered as a result of uncontrolled RES-exposure.
RES-flooding approaches modify various known and postulated redox-sensitive players (labeled with “–SH”) regulating manifold pathways. Examples shown include PI3K/Akt/mTOR[47,48], NF-κB[44], Keap1/Nrf-2[45] and β-catenin/Wnt signaling[52]. Both cooperative and antagonistic events are elicited: e.g., PTEN and Akt3 are each inhibited by RES adduction, but the effects of each inhibition are antagonistic, suggesting that the change in signaling is dominated by the relative importance of flux through the pathway on PTEN/Akt3 and PTEN/Akt3’s relative RES-sensitivities. RES also modulates PLC activity and accelerates PIP2 hydrolysis[46], further complicating the RES-dependent regulation. Similar issues are encountered in NF-κB stress-responsive pathway. Oxidative stress induced by RES or ROS triggers inflammatory NF-κB signaling through upstream receptors/kinases activation, leading to IKK phosphorylation. The activated IKK phosphorylates IκB to release NF-κB transcription factor (p50/p65-complex), triggering transcription[44]. On the other hand, RES also reportedly covalently modifies (and suppresses the activity of) IKK to impede NF-κB signaling[44]. The key negative regulator of NF-κB signaling IκB also is covalently inactivated by RES upon bulk RES-exposure conditions. These conflicting RES regulatory events in NF-κB signaling occur predominantly at low to modest HNE concentration (1–5 μM). Bolus approaches can also mask RES-regulated pathway intersections, such as the crosstalk between Keap1/Nrf-2 and β-catenin/Wnt pathways[52]. See text and Figure 4 for discussion.
In another example, upregulation of antioxidant response (AR) triggered as a result of Keap1-HNEylation alone under T-REX was as high as that achieved following whole-cell HNE-exposure[47]. Several other α,β-unsaturated electrophiles showed similar behaviors[51]. The parity of AR outputs between T-REX and bolus dosing is surprising, since multiple regulators of AR are RES-sensitive. Subtle differences were observed between T-REX and bolus dosing for HNE, such as signal latency and sub-population-specific response[47]. To us, this suggests that the reasons for this apparent parity are complex and worthy of further study, although by analogy to Akt3 versus PTEN above, antagonistic effects are likely at play. Conversely, Ube2V2-specific HNEylation in cells and fish stimulated activity of its cognate binding-partner, Ube2N, promoting homoallosteric upregulation of the DNA-damage response[50]: sensing-defective-mutant did not show such response. Thus, tractable responses are realizable following low stoichiometry RES-modification of privileged sensors. Such effects are often lost or suppressed in the cacophony of bolus RES-dosing, possibly due to triggering antagonistic pathways.
We thus propose the same muting effect occurs during administration of reactive/bioactive drugs/natural products. Such a dampening of phenotypic response would limit efficacy and render dosing strategies and individual responses difficult to predict, likely decreasing efficacy. Could we therefore “train” native RES to target (a) specific protein(s)? Could these targets be “hand-picked” to give a drug (or drug-like compound) which interacts with target(s) whose modification promotes intended phenotypes/signaling and eschews target(s) triggering phenotypes opposed to the intended outcomes?
Training HNE: monogamy vs. polypharmacology
One strategy is to design hybrid small-molecule modulators housing an attenuated HNE and a high-selectivity ligand. Such a strategy, by analogy to modern covalent-drug design based on “ligandable interactions”, may deliver “monogamous HNEs”. Such a drug should confer benefits of both HNE (covalent binding linked to dominant outputs) and the ligand (specificity). For instance, an Akt3-targeted monogamous HNE would avoid PTEN, and so would not trigger counteracting responses, unlike bolus HNE-exposure. Akt3 selectivity would be instilled by HNE-chemotype-implicit Akt3-isoform-specific reactivity. This proposed pipeline is similar to the development of imatinib, a “Bcr-Abl-monogamous” analog of the promiscuous natural product staurosporin. However, this monogamous-HNE design may not take full advantage of HNE’s intrinsic polypharmocology.
Several semi-selective, non-covalent ligands are known. Some bind similar sites (e.g., ATP-binding site) in a family of enzymes, while others inhibit seemingly-unrelated targets. Such promiscuous ligands could be coupled with an attenuated HNE (or similar covalent handle with tailored target repertoire). The ideal setup would lock in the ligandable interactions to targets, yielding phenotypically-matched outcomes, whereas antagonistic interactions would be transient or not possible. One could consider this pipeline analogous to the polypharmacological staurosporin analog, midostaurin, but because of HNE’s broad binding promiscuity, not necessarily kinase focused. In addition, the inherent mechanism of privileged sensing would elicit phenotypes at modest occupancy on the target and response would be sustained, due to covalent binding. By biasing binding to chosen sensors whose pharmacological outputs are reinforcing/synergistic (critical attributes for polypharmacologic inhibitors)[23,24], and/or by choosing a covalent appendage only capable of interacting with a subset of ligand binders, synergistic polypharmacologic inhibitors could be generated. One element that sets this design apart from traditional therapeutics is that signaling-output is engendered by the electrophilic motif, as well as/or more than the ligand. Thus, chemotype- and function-guided target engagement could be iteratively modeled and an optimized drug motif developed using T-REX.
Interestingly, some cancers are more susceptible to RES-stimulated pathways than normal cells. Furthermore, target-specific responses differ from responses upon bolus RES-dosing[47,48,50,52]. The former can also be masked in studies using bolus approaches[52]. Thus, limiting reactivity to a subset of the targets of the parent compound could engender new and/or potentially more beneficial outputs than have been observed with traditional “untamed” polypharmacologic molecules.
A chemical genetic means to uncover pathway intersections: Achilles heels in cancers
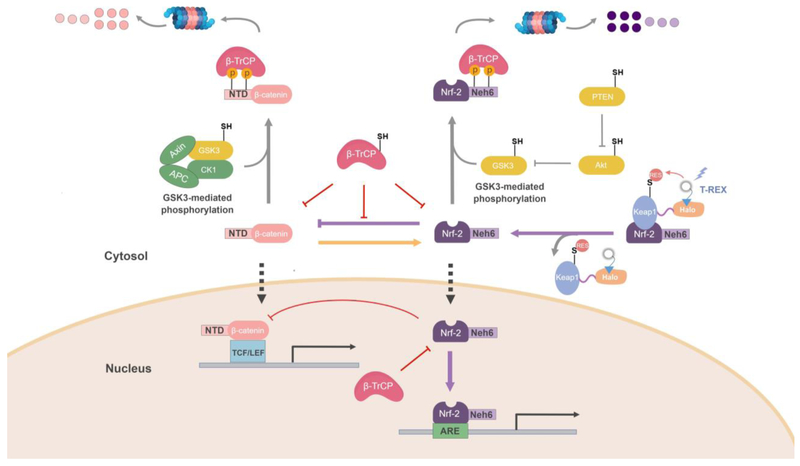
T-REX-specific HNEylation of Keap1 and Akt3 are some of the most selective, effective, and least-invasive ways to modulate canonical signaling pathways. T-REX thus serves as a chemical genetic method to modulate pathway output, or to model target engagement of an oligo-/polypharmacologic RES/electrophilic drug. We illustrate this concept using our newly-uncovered link between AR and Wnt axes, pathways hyperstimulated in cancer. T-REX examined how Keap1-specific AR-signaling was upregulated in lines where proteins that regulate AR orthogonally to Keap1-specific signaling were knocked down. Lines in which β-TrCP1 or GSK3β (proteins in the same pathway) were knocked-down did not mount AR when Keap1 alone was selectively HNEylated. However, these lines upregulated AR upon bolus HNE. We postulated that an HNE-sensitive β-TrCP1/GSK3β-regulated protein may affect AR. One protein that fit this bill was β-catenin, a positive regulator of the Wnt pathway and a driver of cancer.
Wnt upregulation often occurs in cancers through mutation or deletion of the β-catenin N-terminus, preventing GSK3β/β-TrCP1(2) from causing β-catenin degradation (Figure 4). We established that β-catenin/Wnt upregulates AR; however, AR strongly downregulates Wnt. Loss of β-catenin’s N-terminus or knockdown of β-TrCP1 sensitized Wnt signaling to inhibition by upregulated AR. We thus established that prevention of β-TrCP1 binding to β-catenin sensitizes the Wnt-pathway to AR-mediated inhibition. These data uncovered a dichotomy that must be resolved during oncogenesis: Wnt signaling is often upregulated due to loss of β-catenin N-terminus, but AR is also upregulated. We concluded that cancers with mutated β-catenin are likely more susceptible to electrophiles/AR-inducers than cells with wild-type β-catenin. This is because flux through the cancer-promoting Wnt signaling pathway will be more significantly downregulated upon AR in cells in which β-catenin cannot bind β-TrCP1.
Figure 4. Study of AR signaling under Keap1-specific RES-modification reveals an unexpected intersection between β-catenin/Wnt and Keap1/Nrf-2 axes, in which β-TrCP sensitizes Wnt signaling to AR-mediated Wntpathway-inhibition[52].
Nrf-2 strongly inhibits β-catenin/Wnt signaling, whereas β-catenin overexpression upregulates AR. Both Nrf-2 and β-catenin are subject to proteasomal degradation mediated by β-TrCP. β-TrCP’s binding occupancy at the N-terminal domain (NTD) of β-catenin protects β-catenin against Nrf2-mediated inhibition. To prevent β-TrCP-promoted degradation of β-catenin, the β-catenin-NTD is frequently mutated in cancers: these NTD-mutations upregulate Wnt-signaling. However, impeding β-TrCP-binding to β-catenin renders β-catenin signaling more susceptible to Nrf2/AR-mediated inhibition. This novel regulatory mechanism was uncovered only as a result of studying RES-induced AR-upregulation in a Keap1-specific manner. This delicate regulation and crosstalk between β-catenin/Wnt and Keapt1/Nrf-2-signaling pathways are masked during whole-cell RES-stimulation. Known/postulated redox-sensitive players are labeled with “–SH”.
Conclusion
There remains a pressing need for actionable molecular targets and new modes of action. With the growing suite of broad-specificity electrophilic drugs such as DMF, inherently reactive molecules usher a gateway to bona-fide pharmacophores with new and improved properties. The pharmaceutical programs of promiscuous drugs may thus be modified by focusing their specific target spectra. T-REX can parse synergistic vs. antagonistic responses by either executing T-REX simultaneously on two proteins, or measuring outputs that accompany individual protein/electrophile-specific perturbations. By iterative screening, undesired off-target effects may be prevented or winnowed.
Synopsis.
Ability to faithfully report drug—target interactions constitutes a major critical parameter in preclinical/clinical settings. Yet assessment of target engagement remains challenging, particularly for promiscuous and/or polypharmacologic ligands. Drawing from our improved insights into native electrophile signaling and emerging technologies that profile and interrogate these non-enzyme-assisted signaling subsystems, we posit that “trained” polypharmocologic covalent inhibitors can be designed. The accumulating evidence indicates that electrophile-modified states at fractional occupancy can alter cell fate. Thus, by understanding sensing preferences and ligandable regions elected by the seemingly-promiscuous Nature’s electrophilic signals at individual protein–ligand resolution, we can begin to achieve better evaluations of target engagement and function-guided understanding of polypharmacology.
Acknowledgements.
Novartis Foundation for Medical-Biological Research (Switzerland); National Centre of Competence in Research (NCCR): Chemical Biology (Swiss National Science Foundation), NIH innovator award (1DP2GM114850), and Swiss Federal Institute of Technology Lausanne (EPFL) are acknowledged for research support. The authors declare no conflict of interest.
Abbreviations
- Abl
Abelson murine leukemia viral oncogene homolog
- Akt
RAC-alpha serine/threonine-protein kinase
- APC
Adenomatous polyposis coli protein
- ARE
Antioxidant response element
- AXIN1
Axis inhibition protein 1
- Bcr
Breakpoint cluster region protein
- β-TrCP
Beta-transducin repeats-containing proteins
- CDK
Cyclin-dependent kinase
- CK1
Casein kinase 1
- c-KIT
KIT Proto-Oncogene Receptor Tyrosine Kinase
- DAG
Diacylglycerol
- DMF
Dimethyl fumarate
- ERK
Extracellular signal-regulated kinase
- FGFR
Fibroblast growth factor receptor
- FLT3
Fms Related Tyrosine Kinase 3
- FOXO
Forkhead box protein
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GSK3
Glycogen synthase kinase 3
- Halo
Modified bacterial dehalogenase
- HNE
4-Hydroxynonenal
- IKK
IKB kinase
- IP3
Inositol 1,4,5-trisphosphate
- Keap1
Kelch-like ECH-associated protein 1
- MDM2
E3 ubiquitin-protein ligase MDM2
- MEK
Mitogen-activated protein kinase kinase
- mTOR
Mammalian target of rapamycin
- mTORC
Mammalian target of rapamycin complex
- Nrf-2
Nuclear factor (erythroid-derived 2)-like 2
- PDGFR
Platelet-derived growth factor receptor
- PI3K
Phosphatidylinositol-4,5-bisphosphate 3-kinase
- PIP2
Phosphatidylinositol 4,5-bisphosphate
- PIP3
Phosphatidylinositol (3,4,5)-trisphosphate
- PLC
Phospholipase C
- PML-RARA
Promyelocytic leukemia/retinoic acid receptor alpha
- PTEN
Phosphatase and tensin homolog
- RA
Retinoic acid
- RAF
RAF proto-oncogene serine/threonine-protein kinase
- RAS
GTPase RAS protein
- RES
Reactive electrophilic species
- TCF/LEF
Transcription factor TCF/LEF
- T-REX
Targetable reactive electrophiles and oxidants
- Ube2N
Ubiquitin Conjugating Enzyme E2 N
- Ube2V1
Ubiquitin Conjugating Enzyme E2 V1
- Ube2V2
Ubiquitin Conjugating Enzyme E2 V2
- VEGFR
Vascular endothelial growth factor receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading:
Papers of particular interest, published in the past two years have been highlighted as:
* of special interest
** of outstanding interest
- 1.Ferguson FM, Gray NS: Kinase inhibitors: the road ahead. Nature Reviews Drug Discovery 2018, 17:353–377.** This is a comprehensive review that provides insight into the current therapeutic areas covered by kinase targeted therapies. It discusses the opportunities and challenges in these areas and highlights new experimental and computational approaches for rational design of kinase inhibitors.
- 2.Terai H, Tan L, Beauchamp EM, Hatcher JM, Liu Q, Meyerson M, Gray NS, Hammerman PS: Characterization of DDR2 inhibitors for the treatment of DDR2 mutated nonsmall cell lung cancer. ACS Chemical Biology 2015, 10:2687–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciarcia R, Damiano S, Puzio MV, Montagnaro S, Pagnini F, Pacilio C, Caparrotti G, Bellan C, Garofano T, Polito MS, et al. : Comparison of dasatinib, nilotinib, and imatinib in the treatment of chronic myeloid leukemia. Journal of Cellular Physiology 2016, 231:680–687. [DOI] [PubMed] [Google Scholar]
- 4.Cortes JE, Saglio G, Baccarani M, Kantarjian HM, Mayer J, Boqué C, Shah NP, Chuah C, Casanova L, Narayanan G, et al. : Final study results of the phase 3 dasatinib versus imatinib in newly diagnosed chronic myeloid leukemia in chronic phase (CML-CP) Trial (DASISION, CA180–056). Blood 2014, 124:152. [Google Scholar]
- 5.Zhang J, Adrián FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, Sim T, Powers J, Dierks C, Sun F, et al. : Targeting Bcr–Abl by combining allosteric with ATP-binding-site inhibitors. Nature 2010, 463:501–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frei E, Karon M, Levin RH, Freireich EJ, Taylor RJ, Hananian J, Selawry O, Holland JF, Hoogstraten B, Wolman IJ, et al. : The effectiveness of combinations of antileukemic agents in inducing and maintaining remission in children with acute leukemia. Blood 1965, 26:642–656. [PubMed] [Google Scholar]
- 7.Nakagawa F, May M, Phillips A: Life expectancy living with HIV: recent estimates and future implications. Current Opinion in Infectious Diseases 2013, 26:17–25. [DOI] [PubMed] [Google Scholar]
- 8.Bayat Mokhtari R, Homayouni TS, Baluch N, Morgatskaya E, Kumar S, Das B, Yeger H: Combination therapy in combating cancer. Oncotarget 2017, 8:38022–38043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knight ZA, Lin H, Shokat KM: Targeting the cancer kinome through polypharmacology. Nature Reviews Cancer 2010, 10:130–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinicaltrials.gov identifiers: NCT01909453, NCT01935973, NCT01979523, NCT01989598, NCT02049801, NCT00620594 and NCT01343498. https://clinicaltrials.gov/.
- 11.Zhang X, Pan J: Resistance to arsenic trioxide and retinoic acid therapy in acute promyelocytic leukemia. Annals of Hematology 2017, 96:707–708. [DOI] [PubMed] [Google Scholar]
- 12.Basanta D, Gatenby RA, Anderson ARA: Exploiting evolution to treat drug resistance: combination therapy and the double bind. Molecular Pharmaceutics 2012, 9:914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhatia S, Sharma J, Bukkapatnam S, Oweida A, Lennon S, Phan A, Milner D, Uyanga N, Jimeno A, Raben D, et al. : Inhibition of EphB4–Ephrin-B2 signaling enhances response to cetuximab–radiation therapy in head and neck cancers. Clinical Cancer Research 2018, 24:4539–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Day C-P, Merlino G, Van Dyke T: Preclinical mouse cancer models: a maze of opportunities and challenges. Cell 2015, 163:39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humphrey RW, Brockway-Lunardi LM, Bonk DT, Dohoney KM, Doroshow JH, Meech SJ, Ratain MJ, Topalian SL, Pardoll D M: Opportunities and challenges in the development of experimental drug combinations for cancer. JNCI: Journal of the National Cancer Institute 2011, 103:1222–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lopez JS, Banerji U: Combine and conquer: challenges for targeted therapy combinations in early phase trials. Nature Reviews Clinical Oncology 2016, 14:57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peter B, Winter GE, Blatt K, Bennett KL, Stefanzl G, Rix U, Eisenwort G, Hadzijusufovic E, Gridling M, Dutreix C, et al. : Target interaction profiling of midostaurin and its metabolites in neoplastic mast cells predicts distinct effects on activation and growth. Leukemia 2016, 30:464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stone RM, Manley PW, Larson RA, Capdeville R: Midostaurin: its odyssey from discovery to approval for treating acute myeloid leukemia and advanced systemic mastocytosis. Blood Advances 2018, 2:444–453.** This is a multifaceted review delineating the path to clinical approval of midostaurin. This review also reflects the importance of developing a rational design strategy for polypharmacologic drugs.
- 19.Orphan drug status for alvocidib. Oncology Times 2014, 36:91. [Google Scholar]
- 20.Shukla S, Robey RW, Bates SE, Ambudkar SV: Sunitinib (Sutent, SU11248), a small-molecule receptor tyrosine kinase inhibitor, blocks function of the ATP-binding cassette (ABC) transporters P-glycoprotein (ABCB1) and ABCG2. Drug Metabolism and Disposition 2009, 37:359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petrelli A: Polypharmacological kinase inhibitors: new hopes for cancer therapy. Polypharmacology in Drug Discovery 2012. [Google Scholar]
- 22.Apsel B, Blair JA, Gonzalez B, Nazif TM, Feldman ME, Aizenstein B, Hoffman R, Williams RL, Shokat KM, Knight ZA: Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nature Chemical Biology 2008, 4:691–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moya-García A, Adeyelu T, Kruger FA, Dawson NL, Lees JG, Overington JP, Orengo C, Ranea JAG: Structural and functional view of polypharmacology. Scientific Reports 2017, 7:10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lavecchia A, Cerchia C: In silico methods to address polypharmacology: current status, applications and future perspectives. Drug Discovery Today 2016, 21:288–298. [DOI] [PubMed] [Google Scholar]
- 25.Proschak E, Stark H, Merk D: Polypharmacology by design: a medicinal chemist’s perspective on multitargeting compounds. Journal of Medicinal Chemistry 2018.* This is a concise review of the current strategies for rational design of polypharmacologic inhibitors/drugs.
- 26.Parvez S, Long MJC, Poganik JR, Aye Y: Redox signaling by reactive electrophiles and oxidants. Chemical Reviews 2018, 118:8798–8888.** A comprehensive review that covers various aspects of redox signaling events and how these reactive signaling small molecules coordinate cellular activities.
- 27.Lanning BR, Whitby LR, Dix MM, Douhan J, Gilbert AM, Hett EC, Johnson TO, Joslyn C, Kath JC, Niessen S, et al. : A road map to evaluate the proteome-wide selectivity of covalent kinase inhibitors. Nature Chemical Biology 2014, 10:760–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blewett MM, Xie J, Zaro BW, Backus KM, Altman A, Teijaro JR, Cravatt BF: Chemical proteomic map of dimethyl fumarate–sensitive cysteines in primary human T cells. Science Signaling 2016, 9:rs10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wishart DS: Emerging applications of metabolomics in drug discovery and precision medicine. Nature Reviews Drug Discovery 2016, 15:473–484.** This is a comprehensive review of the emerging roles of metabolomics in studies of disease progression and causes, and in the development of precision medicine.
- 30.Kornberg MD, Bhargava P, Kim PM, Putluri V, Snowman AM, Putluri N, Calabresi PA, Snyder SH: Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 2018, 360:449–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blatnik M, Thorpe SR, Baynes JW: Succination of proteins by fumarate. Annals of the New York Academy of Sciences 2008, 1126:272–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersen JL, Gesser B, Funder ED, Nielsen CJF, Gotfred-Rasmussen H, Rasmussen MK, Toth R, Gothelf KV, Arthur JSC, Iversen L, et al. : Dimethyl fumarate is an allosteric covalent inhibitor of the p90 ribosomal S6 kinases. Nature Communications 2018, 9:4344.* This manuscript describes how p90 ribosomal S6 kinase, the effector kinase of Ras/RAF/MEK/ERK signaling pathway, is inhibited through covalent allosteric modulation by dimethyl fumarate. This result provides new insight into the clinical modes of action of dimethyl fumarate.
- 33.Schulze-Topphoff U, Varrin-Doyer M, Pekarek K, Spencer CM, Shetty A, Sagan SA, Cree BAC, Sobel RA, Wipke BT, Steinman L, et al. : Dimethyl fumarate treatment induces adaptive and innate immune modulation independent of Nrf2. Proceedings of the National Academy of Sciences 2016, 113:4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tang L, Paonessa JD, Zhang Y, Ambrosone CB, McCann SE: Total isothiocyanate yield from raw cruciferous vegetables commonly consumed in the United States. Journal of functional foods 2013, 5:1996–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dinkova-Kostova AT, Fahey JW, Kostov RV, Kensler TW: KEAP1 and done? Targeting the NRF2 pathway with sulforaphane. Trends in Food Science & Technology 2017, 69:257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hedstrom LK, Long MJC, Lawson AP: Inhibitors of deubiquitinating proteases. US Patent 2018. [Google Scholar]
- 37.Gupta SC, Patchva S, Aggarwal BB: Therapeutic roles of curcumin: lessons learned from clinical trials. The AAPS Journal 2013, 15:195–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khoo NKH, Li L, Salvatore SR, Schopfer FJ, Freeman BA: Electrophilic fatty acid nitroalkenes regulate Nrf2 and NF-κB signaling: a medicinal chemistry investigation of structure-function relationships. Scientific Reports 2018, 8:2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long MJC, Aye Y: Privileged electrophile sensors: a resource for covalent drug development. Cell chemical biology 2017, 24:787–800.** This perspective depicts how studying redox signaling in precision can provide new insights into rational design of covalent targeted therapy.
- 40.Wang C, Weerapana E, Blewett MM, Cravatt BF: A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nature Methods 2014, 11:79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Qin W, Wang C: Chemoproteomic profiling of protein modifications by lipid-derived electrophiles. Current Opinion in Chemical Biology 2016, 30:37–45. [DOI] [PubMed] [Google Scholar]
- 42.Bak DW, Pizzagalli MD, Weerapana E: Identifying functional cysteine residues in the mitochondria. ACS Chemical Biology 2017, 12:947–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu X, Long MJC, Aye Y: Proteomics and beyond: cell decision-making shaped by reactive electrophiles. Trends in biochemical sciences 2018:article in press.** This review highlights new proteomics profiling and pathway interrogation techniques that enable quantitative understanding of how endogenous redox signals engender specific phenotypic output(s) at the molecular level, and their growing role in pathophysiological research and drug discovery.
- 44.Zhang H, Forman HJ: 4-hydroxynonenal-mediated signaling and aging. Free RadicalBiology and Medicine 2017, 111:219–225.** This review highlights the recent advances of HNE-mediated physiological and pathophysiological signalings and also discusses how bolus RES treatment can lead to misleading phenotypic outputs.
- 45.Hayes JD, Dinkova-Kostova AT: The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends in Biochemical Sciences 2014, 39:199–218. [DOI] [PubMed] [Google Scholar]
- 46.Ayala A, Muñoz MF, Argüelles S: Lipid peroxidation: production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxidative medicine and cellular longevity 2014, 2014:360438–360438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parvez S, Long MJC, Lin H-Y, Zhao Y, Haegele JA, Pham VN, Lee DK, Aye Y: T-REX on-demand redox targeting in live cells. Nature Protocols 2016, 11:2328–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Long MJC, Parvez S, Zhao Y, Surya SL, Wang Y, Zhang S, Aye Y: Akt3 is a privileged first responder in isozyme-specific electrophile response. Nature Chemical Biology 2017, 13:333–338.** This manuscript reports for the first time that HNE engenders isozyme-selective allosteric downregulation of Akt family members in vitro and in vivo. This novel mode of action may be interesting to the kinase drug discovery community.
- 49.Long MJC, Urul DA, Chawla S, Lin H-Y, Zhao Y, Haegele JA, Wang Y, Aye Y: Precision electrophile tagging in caenorhabditis elegans. Biochemistry 2018, 57:216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao Y, Long MJC, Wang Y, Zhang S, Aye Y: Ube2V2 is a rosetta stone bridging redox and ubiquitin codes, coordinating DNA damage responses. ACS Central Science 2018, 4:246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin H-Y, Haegele JA, Disare MT, Lin Q, Aye Y: A Generalizable Platform for Interrogating Target- and Signal-Specific Consequences of Electrophilic Modifications in Redox-Dependent Cell Signaling. Journal of the American Chemical Society 2015, 137:6232–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Long MJC, Lin H-Y, Parvez S, Zhao Y, Poganik JR, Huang P, Aye Y: β-TrCP1 is a vacillatory regulator of Wnt signaling. Cell chemical biology 2017, 24:944–957.e947.** This report shows how Nrf2/AR and β-catenin/Wnt signaling pathways have a “hidden” intersection, wherein Nrf2/AR strongly inhibit β-catenin/Wnt in a β-catenin N-terminus dependent manner. This report also highlights the general barriers surronding the studies of redox-responsive pathways under uncontrolled conditions and how an approach to deliver individual redox signals to individual proteins directly in living systems may resolve some of these issues.