Abstract
Background:
Current in vitro human lung epithelial cell models derived from adult tissues may not accurately represent all attributes that define homeostatic and disease mechanisms relevant to the pediatric lung.
Methods:
We report methods for growing and differentiating primary Pediatric Human Lung Epithelial (PHLE) cells from organ donor infant lung tissues. We use immunohistochemistry, flow cytometry, quantitative RT-PCR, and single cell RNA sequencing (scRNAseq) analysis to characterize the cellular and transcriptional heterogeneity of PHLE cells.
Results:
PHLE cells can be expanded in culture up to passage 6, with a doubling time of approximately 4 days, and retain attributes of highly enriched epithelial cells. PHLE cells can form resistant monolayers, and undergo differentiation when placed at air-liquid interface. When grown at Air-Liquid Interface (ALI), PHLE cells expressed markers of airway epithelial cell lineages. scRNAseq suggests the cultures contained 4 main sub-phenotypes defined by expression of FOXJ1, KRT5, MUC5B and SFTPB. These cells are available to the research community through the Developing Lung Molecular Atlas Program Human Tissue Core.
Conclusion:
Our data demonstrate that PHLE cells provide a novel in vitro human cell model that represents the pediatric airway epithelium, which can be used to study perinatal developmental and pediatric disease mechanisms.
INTRODUCTION
Primary lung epithelial cell culture can be used as a model to understand cellular responses to challenge, and related homeostatic and disease mechanisms. Adult human airway epithelial cell cultures can be traced back to the 1980s, and have been grown from tissue explants of adult human bronchi (1–3), explants of bronchial brushings (4), and explants of adult nasal polyps (5). These cells can be cultured on a permeable support, differentiated toward an airway phenotype by placing them at the Air-Liquid Interface (ALI), and these differentiated cells provide a powerful tool to investigate human airway epithelial biology (6, 7). Studies using human embryonic and early fetal tissue explants and cells have been completed, but have been fairly limited (8, 9). Pediatric bronchial epithelial cells isolated from proximal airway tissues of cadaveric lungs, and differentiated in vitro as a model to study mechanisms involved in pediatric asthma have also been reported (4, 10). Nasal epithelial cells have been assessed as a benchmark for evaluating the effect of environmental challenge on lung function (11, 12), and nasal epithelial cells from children can be grown and differentiated in vitro (13, 14). Limited pediatric bronchial epithelial cell cultures have been established previously by using bronchial epithelial samples from children who undergo elective surgical procedure (15–17). However, there is no widely available cell model derived from the distal portion of human lung epithelia in the newborn, infant or pediatric age range, limiting research into perinatal mechanisms of human airway cell differentiation, and the response of neonatal and pediatric lung epithelium to environmental challenges.
The Developing Lung Molecular Atlas Program (LungMAP) has obtained 200 human organ donor lungs, primarily ranging in age from 1 day to 10 years of age (with limited collection of older organs), and processed these lungs into dissociated cells (18). Here, we describe the growth and differentiation of primary infant and pediatric lung epithelial (PHLE) cells from these organ donor lung tissues. We demonstrated that PHLE differentiated at ALI express common airway markers such as Forkhead box J1(FOXJ1), Keratin 5 (KRT5), Mucin 5B (MUC5B) and Surfactant protein B (SFTPB ) at the population level. Single cell RNA sequencing (scRNAseq) analysis revealed these cultures contained clusters of cells that could be distinguished by expression of these same markers. We conclude that PHLE cells are an age-appropriate in vitro cell model that represents human infant and pediatric airway epithelium.
MATERIALS AND METHODS
Materials
Collagenase type A (Roche, Basel, Switzerland); Dispase II (Gibco/ThermoFisher, Waltham, MA); Elastase (Worthington-Biochem, Lakewood, NJ); DNAase (Sigma-Aldrich, St. Louis, MO); Absolute RNA Microprep kit (Agilent, #400805, Stratagene, La Jolla, CA); bronchial epithelial basal medium (BEBM, Lonza, Mapleton, IL); small airway epithelial cell growth medium (SAGM; Lonza); Dulbecco’s modified Eagle medium (DMEM; GIBCO, Rockville, MD); PneumaCult-ALI medium (Stemcell Technologies, Vancouver, Canada); iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA); SYBR Green (Applied Biosystems, Foster City, CA. SCGB1A1/CCSP (Secretoglobin family 1A, member 1) antibodies (LifeSpan BioScience, Seattle, WA); FOXJ1/HFH4 (Novus Biologicals, Littleton, CO).
Cell Culture
A detailed cell isolation protocol was published by Bandyopadhyay et al. (18) and a detailed protocol for growing PHLE cells is provided in the supplemental material. Briefly, fresh right upper and right middle lobes were separated into proximal and distal segments. Proximal lung tissue was obtained by first identifying the lobar bronchus, and dissecting out airway (using scissors) up to approximately the fourth branch point. The remaining tissue was considered to be distal. Tissues were digested with a protease cocktail containing collagenase type A (2 mg/ml), dispase II (1 mg/ml), elastase (0.5 mg/ml) and DNAase (2 mg/ml). Single cell suspensions were washed 2 times with Dulbecco’s Phosphate buffered saline (DPBS) supplemented with 1% Penicillin-Streptomycin (Gibco), 50 μg/ml Gentamicin and 0.25 μg/ml amphotericin B, and centrifuged with 800 xg for 10 minutes. As reported elsewhere (18), the viability of freshly dissociated cells is >95% by trypan blue exclusion. Approximately 10 million cells were grown in a 75-cm2 tissue culture flask with Small Airway Growth Medium (SAGM) supplemented with 1% Fetal Bovine Serum (FBS; for distal lung tissues) or Bronchial Epithelial Basal Medium (BEBM; for proximal digests). After 24 hours, non-adherent cells were removed. Fibroblast-like cells were removed from cultures by brief treatment (1–2 minutes) with low concentration trypsin in DPBS (0.0125% with EDTA) at room temperature, as described previously (29, 30).
For establishing Air-Liquid Interface (ALI) cultures, 100,000 cells were seeded on rat tail collagen I (34.5 μg/mL) coated Transwell inserts (12 well PET membrane, 0.4 μm pore size, 12 mm diameter), with mediuam in both the apical and baso-lateral chambers. After 24–48 hours, medium was changed to 1:1 mixture of BEBM and Dulbecco’s Modified Eagle Medium (DMEM). When cells reached confluence (approximately 10–15 days), and established transepithelial resistance (≥ 300 Ohms*cm2), they were transferred to ALI by removing the apical medium. Upon transition to ALI, cells were maintained in PneumaCult-ALI medium containing supplements and hydrocortisone, according to manufacturer’s instructions. ALI cultures were differentiated for 7–12 days at ALI before examination. Normal Human Bronchial Epithelial (NHBE) cell cultures were established and maintained as we previously described (19, 20), were grown and analyzed contemporaneously with the PHLE cells, and were used at passage 2.
Immunocytological analysis
Cytospin slides, for digested lung cells prior to culture and PHLE cells growing in non-ALI conditions, were fixed with 4% paraformaldehyde and washed twice with PBS. ALI cell monolayers were washed in PBS 2 times and directly fixed in 4% paraformaldehyde for 10 minutes at room temperature and washed with PBS, 2 times for 10 minutes. Cells were permeabilized with 0.2% Triton-X-100 (Fisher Scientific) in PBS (PBS-TX) for 10 minutes then blocked for 1 hour with 10% goat serum (Gibco) in PBS-TX. Primary antibodies to pan-cytokeratin (mouse monoclonal anti-PanCytokeratin; C11; Thermo Fisher Scientific), cytokeratin 5 (rabbit monoclonal anti-cytokeratin 5; Abcam, Cambridge, MA ) or acetylated tubulin (mouse monoclonal anti-acetylated tubulin; Sigma-Aldrich), diluted to 1:200 with 1% BSA in PBS-TX were added and incubated overnight at 4 ⁰C. Slides were washed with PBS-TX (5 minutes X 3) and incubated with Alexa Fluor-488-cojugated goat anti-mouse IgG secondary antibody (Thermo Fisher Scientific) for 1 hour. Slides were washed with PBS-TX (5 minutes X 3). Cells were counter-stained with 300 nM DAPI (Invitrogen/ThermoFisher), washed 3 times with PBS-TX, and mounted on slides with ProLong Gold antifade reagent (Invitrogen/ThermoFisher). Slides were dried overnight in dark and imaged with fluorescence microscope (Olympus IX71 and Olympus DP control software).
RNA Isolation, Reverse Transcription and Real-Time Quantitative RT-PCR (qPCR)
DNA free-total RNA was isolated using the Absolutely RNA Microprep Kit. Total RNA was reverse-transcribed using the iScript cDNA Synthesis Kit. qPCR analysis was performed using SYBR Green chemistry. Gene expression levels were calculated relative to PPIA (cyclophilin A) using the ΔΔCT method as we have previously described (19) . Primer sequences were selected from PrimerBank (http://pga.mgh.harvard.edu/primerbank/).
Flow Cytometry
Cells from ALI and submerged culture were stained in 96 well plates. Cells were divided evenly for controls (unstained, heat killed, and Fluorescence Minus One Control (FMO)) and for full stains. After washing with DPBS, BD Fixable Viability Stain 510 (BD Biosciences) was applied at a concentration of 1:1000 for 30 minutes. Live/dead staining was neutralized using 2% BSA with DPBS, the cells were centrifuged (1000xg, 10min, 4 degree), and re-suspended and blocked with appropriate non-specific sera for 15 minutes. Cells were stained for surface staining of Epithelial cell adhesion molecule (EPCAM) (eBioscience) at 4⁰C for 90 minutes. After neutralizing, centrifugation and washing, the cells were fixed and permeabilized (BD fixation and permeabilization kit; BD Biosciences) for 40 minutes. Cells were washed and blocked again with non-specific sera, and stained with SCGB1A1/CCSP antibodies overnight at 4 ⁰C. Cells were washed, re-suspended in 10% neutral buffered formalin and analyzed on a 4-laser, 18 channel BD LSR II (Blue, Red, Violet, and Green). Gating was applied using FlowJo software (version 10).
Single cell RNAseq
PHLE cells grown at ALI were recovered by trypsinization, stained for viability (Calcien AM/EthD-1 LIVE/DEAD assay; Life Technologies) and captured using the Fluidigm C1 platform (Fluidigm Corporation). cDNAs were prepared from wells containing single, live cells using the SMARTer Ultra Low RNA kit for Illumina (Clontech, Mountain View, CA). Libraries were constructed using the NexteraXT DNA Sample Preparation kit (Illumina), and sequenced on an Illumina HiSeq. Sequencing data were cleaned using Trimmomatic (21), and aligned to Genome Reference Consortium Human Build 38 (GRCh38.7) library (Genocde-25) using Spliced Transcript Alignment to a Reference (STAR) algorithm (v2.5.2b)(22). Uniquely aligned reads were quantified (23) and quality-assessed using picardtools (v1.114). Outlier cells were removed based on the number of genes expressed (<2,000 or >10,000), percent mitochondrial (0–0.2) or ribosomal (0–0.1) reads. Normalized expression values were computed, hyper-variable genes were identified, and data were converted into Monocle (v2.2.0) (24, 25) specific objects using the scran/scater (v1.4.5/v1.4.0)(26, 27) framework. For “supervised” analyses, individual cells were assigned a ‘cell type’ based on expression (>=1) of specific hyper-variable “marker” genes: KRT5, FOXJ1, SFTPB, and MUC5B. A cell not expressing any of the 4 marker genes was defined as “Unknown” while any cell expressing multiple genes was designated “Ambiguous”.
Influenza virus infection in PHLE cells
PHLE cells were infected with Pandemic 2009 H1N1 Influenza Venus reporter virus (28), by placing 0.2 ml of virus-containing medium on the apical side of ALI PHLE cultures for 1 hour at 37 °C. After the 1 hour of incubation, virus was removed and cells were washed apically with DPBS. Fluorescence microscopy was used to determine infectivity, by counting fluorescent foci at 24 and 48 hours post infection. RNA was isolated at 48 hours post infection to test for PHLE cell responses and viral gene transcripts, as a measure of productive infection.
Statistical analysis
Statistical assessments for differences in population mean expression by qPCR were performed using the student’s T test, implemented in Excel, with significance defined as unadjusted p<0.05. For tests of changes in expression across donor age, we used a linear regression model in R, with significance defined as unadjusted p<0.05. Donor age was calculated as a whole number, and/or fraction thereof, in months at the time of donation. Sex was not evaluated as a variable in the regression model. For single cell RNAseq analysis, genes were selected for clustering that significantly (q-value < 0.01) co-vary with the “supervised” marker genes (KRT5, FOXJ1, SFTPB, and MUC5B), and filtered to the top 500 most specific for each cell type. Dimensionality reduction was performed using T-distributed Stochastic Neighbor Embedding (tSNE) clustering. Visualizations were generated using the first two components generated by tSNE. Plots were colored based on cell cluster membership and previously defined cell type assignment.
RESULTS
Isolation and growth of PHLE cells.
Pediatric lungs were obtained from donors at 1 day to 10 years of age (Supplemental Table 1) by the developing lung molecular atlas program (LungMAP)-supported Human Tissue Core repository at the University of Rochester. Procurement of lung tissue from the International Institute for the Advancement of Medicine and the National Disease Research Interchange was approved by the University of Rochester Medical Center Research Subjects Review Board. Single cell suspensions were generated from lung tissues by digestion with a protease cocktail as recently described (18).
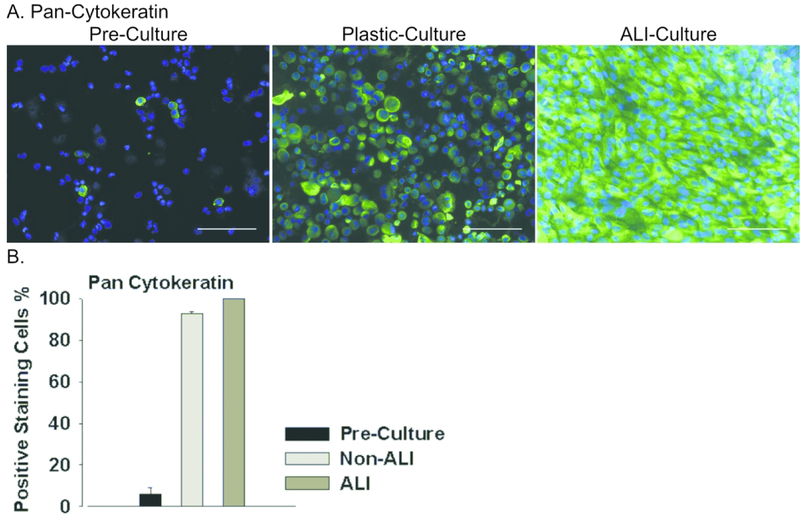
We expanded epithelial cells from these single cell suspensions using epithelial cell-selective growth media (see Methods and Supplemental Methods). Small clusters of epithelioid cells were apparent after 2–3 days in culture, proliferated rapidly and reached near confluence within 10–14 days. Mesenchyme-like cells occurred in these cultures, but were removed by brief treatment with low concentration trypsin (29, 30). While prior to culture (pre-culture), only 5% cells stained positive for the epithelial marker pan cytokeratin, cells expanded under appropriate conditions were nearly 100% (97.12 % ± 1.4) positive (Figure 1). The epithelial cultures could be routinely expanded up to passage 6, were capable of forming confluent, resistant monolayers (≥ 300 Ohms*cm2) when grown on type I collagen atop a permeable membrane (Supplemental Table 1), and could be stably maintained at ALI. We refer to these primary cultures as Pediatric Human Lung Epithelial (PHLE) cells.
Figure 1. Assessing general epithelial lineage marker expression in PHLE cells.
A. Cultures of PHLE cells were assessed for expression of pan-cytokeratin by immunofluorescence staining. Cells were examined while in submerged culture (Non-ALI) or following differentiation at air-liquid interface (ALI), and compared to expression of these markers in freshly prepared single cell suspensions (Pre-Culture). Green indicates antibody specific staining, blue indicates nuclear (DAPI) staining. Scale bar is 100 μm. B. Summary of pan-cytokeratin staining in each condition for PHLE cells from 7 donor lungs.
Characterization of lineage marker expression in PHLE cells.
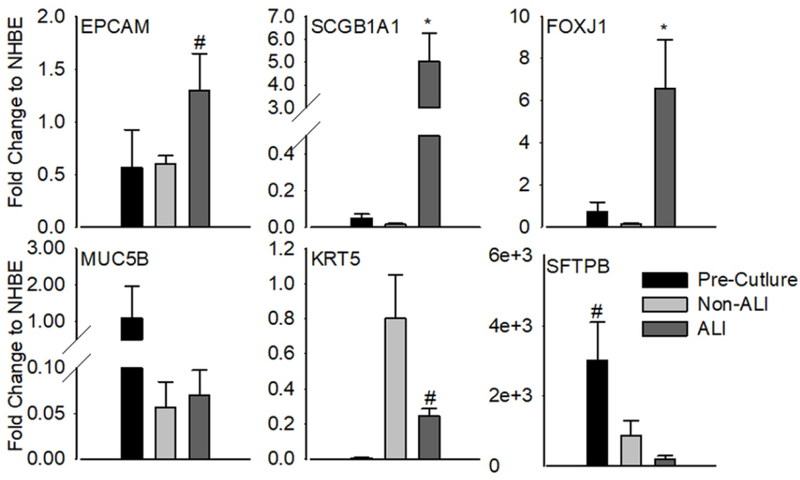
We characterized non-specific epithelial lineage and lung-specific epithelial lineage marker mRNA expression in PHLE cell cultures. We present expression results relative to ALI cultures of adult human airway epithelial cells (NHBE; Figure 2). Expression of the general epithelial lineage marker EPCAM in cultured PHLE cells was comparable to NHBE. High levels of the airway-specific lineage markers including SCGB1A1 and FOXJ1 were noted in PHLE differentiated at ALI. The basal cell marker KRT5 was highly expressed in cells expanding in culture, but reduced at ALI. While MUC5B and SFTPB expressing cells were apparent in single cell suspension, expression of these genes was reduced in culture. Distal lung epithelial markers such as SFTPA and HOPX were highly expressed prior to culture, but reduced dramatically during culture, similar to NHBE (data not shown). Similarly, while there was a detectable level of SFTPC in freshly isolated cells prior to culture (data not show), expression was not detected in cultured PHLE cells. Expanded and ALI-differentiated PHLE cells in culture, therefore, resemble human airway epithelial cells.
Figure 2. Population-based characterization of airway epithelial cell marker gene expression.
Cultures of PHLE cells were assessed for expression of general epithelial cell and lung-specific lineage markers at the RNA level. Cells were examined while in submerged culture (Non-ALI) or following differentiation at air-liquid interface (ALI), and compared to freshly prepared single cell suspensions (Pre-Culture). Data are expressed relative to adult human airway epithelial cells (NHBE) differentiated at ALI. Differentiation of PHLE at ALI is associated with a significant increase in the expression of airway lineage markers SCGB1A1 and FOXJ1. Data are from 10 donor lungs, including multiple passages (n=18). “*” P< 0.05, “#”, 0.05<P<0.10 by paired t-test.
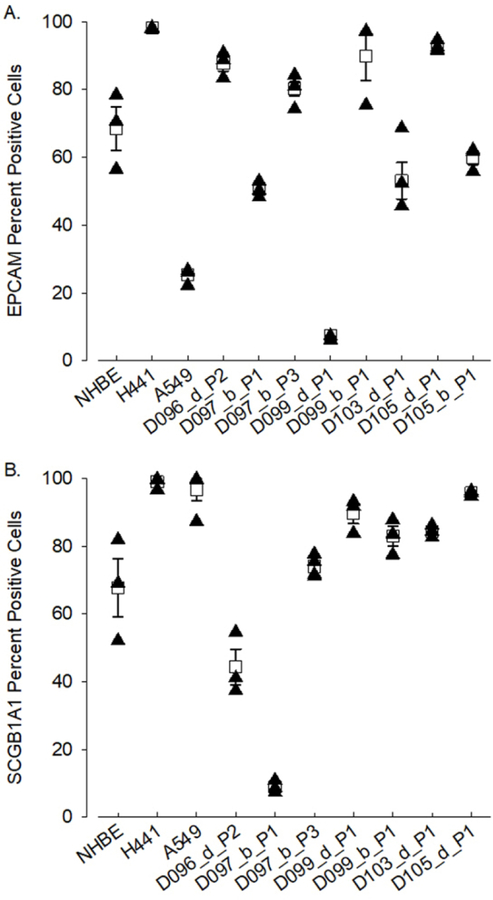
We confirmed expression of epithelial cell markers at the protein level in ALI cultures of PHLE cells (Figure 3). Flow cytometry analysis demonstrated robust expression of EPCAM and SCGB1A1, detected on average in 68.8 ± 10.5% and 66.9 ± 12.4% of PHLE cells, respectively. As can be seen, occasionally cultures expressed very low levels of these proteins.
Figure 3. Characterization of airway epithelial cell marker protein expression by flow cytometry.
Expression of epithelial cell marker proteins EPCAM (A) and SCGB1A1 (B) were assessed in PHLE by flow cytometry. Data from transformed human lung epithelial cell lines (H441 and A549) are included for reference. Shown are mean and standard error of positive cells from replicate cultures. Expression of both EPCAM and SCGB1A1 are variable across cells derived from different donor lungs, but evident in a majority of cells in most cultures. DXX=donor ID, d/b=distal/bronchus, PX=passage number.
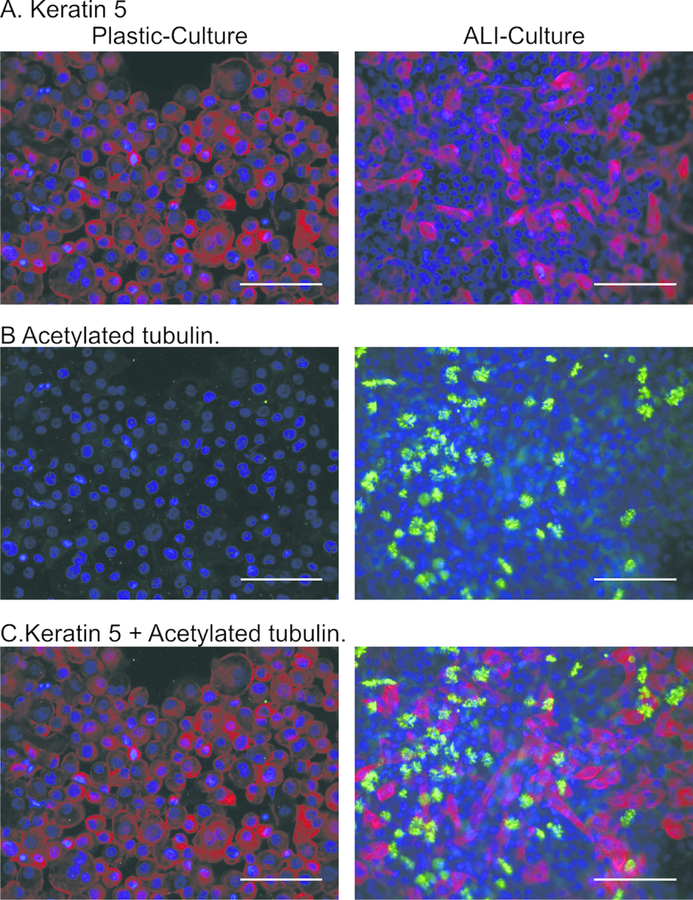
Immuno-cytochemistry for keratin 5 (KRT5; Figure 4A) demonstrated widespread expression (82.7 ± 7.9%) during PHLE cell expansion on plastic, which was decreased upon differentiation at ALI (28.0 ± 12.1 %). Immunocytochemistry for acetylated tubulin (αTub; Figure 4B) showed classic cilia-patterned staining dispersed throughout the differentiated cultures at ALI, with an average of 15.0 ± 2.7% positive cells. Interestingly, cells containing αTub appeared to grow as small colonies, similar to NHBE (data not shown), and did not co-stain for KRT5 (Figure 4C).
Figure 4. Characterization of airway epithelial cell marker protein expression by immunostaining.
Expression of KRT5 (red; basal cells) and acetylated tubulin (green; ciliated cells) were assessed in PHLE by immunocytochemistry. Counter-stain is DAPI (blue). During non-ALI culture, a majority of cells express KRT5 but not acetylated tubulin. Upon differentiation at ALI, the number of KRT5 expressing cells decreases, and acetylated tubulin staining appears in cells not expressing KRT5. Scale bar is 100 μm.
Assessment of expression variability among PHLE cells.
We tested for potential relationships between marker gene expression in ALI cultures with time of cell outgrowth and age of donor (Supplemental Figure S1 and Supplemental Table 2). The expression of most genes was not significantly affected by these variables. However, the expression of both FOXJ1 and (Dynein Axonal heavy chain 5 (DNAH5; 5-fold; p<0.05) declined significantly with the number of times cells were passed in culture. Likewise, SCGB1A1, SFTPB and SFTPA showed modest, but non-significant declines with increasing passage (data not shown). EPCAM (r2=0.447; p=0.001), SCGB1A1 (r2=0.278; p<=0.025), DNAH5 (r2= 0.398; p=0.009) and SFTPB (r2=0.404; P=0.002) expression were significantly, albeit modestly, positively correlated with donor age (Supplemental Table 2).
While a majority of cultures were established from cells recovered by digestion of parenchymal lung tissues, we also studied cultures derived from single cell suspensions of dissected bronchial airways. We tested for differences in expression of lung lineage markers between distal-and bronchial-derived cells in ALI cultures (Supplemental Figure S1). The expression level for most markers was not statistically different. However, we noted a significant increase in MUC5B (4.15 fold, p<0.05) in cultures established from proximal, bronchial cells. There was a highly variable, large magnitude, but non-significant decrease in SFTPB expression (189 fold; p<0.10) for cells derived from bronchus digests. We also noted a trend for increased FOXJ1 (4.45 fold; p<0.10) in these cells.
Single cell molecular profiling of PHLE cells.
Single cell RNA sequencing (scRNASeq) has been used to identify and characterize cellular subpopulations and gene expression heterogeneity in many tissues and organs, including the lung (31, 32). Our data suggested PHLE cell cultures contained heterogeneous cell populations. In order to more comprehensively define cell phenotypes within PHLE cell cultures, we conducted scRNASeq on a total of 168 cells differentiated at ALI, from 4 separate donors ranging in age from 11 to 36 months. Interestingly, all cells were found to express SCGB1A1, albeit at variable levels. EPCAM expression was observed in 139 cells (83%), SFTPB in 116 cells (69%), KRT5 in 109 cells (65%), MUC5B in 89 cells (53%) and FOXJ1 in 43 cells (26%). Taken together, these data suggest that these cultures represent differentiating pulmonary epithelial cells.
We used Monocle to further assess cellular heterogeneity. Twenty cells were removed from this Monocle analysis, due to quality control filtering. We used KRT5 as a marker for “basal” lineage, FOXJ1 for “ciliated” lineage, SFTPB for “secretory” lineage and MUC5B for “goblet” lineage. Cells were assigned lineage based upon unambiguous expression of one of these marker genes, and differentially expressed genes among cell populations were identified. Among the 148 cells tested, we defined 35 cells (24%) as unambiguously basal, 10 (7%) as ciliated, 25 (17%) as secretory and 25 (17%) as goblet. Approximately one-third of cells (55) were of unknown (expressing no markers) or ambiguous (expressing more than 1 marker) phenotype, possibly representing cells not yet differentiated to a clear lineage. When cells are displayed in tSNE space defined by the top 500 most differentially expressed genes for each cell type (Figure 5), basal and ciliated cells appear clearly distinct, while secretory and goblet cells are highly similar to one-another, and group closely with unambiguous/undefined cells. Further, this representation of the data demonstrates considerable selectivity for markers for these cell lineages (FOXJ1, KRT5, MUC5B and SFTPB), as opposed to the widespread expression of SCGB1A1 (Supplemental Figure S2). Importantly, there was no apparent effect of donor or age on cell type clustering (data not shown).
Figure 5. Single cell RNAseq characterization of differentiated PHLE cells.
PHLE cells differentiated at ALI (n=168, from 4 donors) were captured on the C1 platform and subjected to single cell RNA sequencing. Cells passing quality filtering (n=148) were assigned to a lineage based upon expression of single markers (FOXJ1, KRT5, MUC5B and SFTPB) using Monocle. Gene expression diversity among the cells is displayed in 2D tSNE space where each plot element represents a cell. Four separate plots are presented, one for each lineage marker. Plot elements are colored to represent individual cell lineages, with the size of the dot representing the relative enrichment in expression for that lineage marker. Olive dots represent cilated lineage (FOXJ1), green represents basal lineage (KRT5), turquoise represents goblet lineage (MUC5B), blue represents secretory lineage (SFTPB), red dots represent ambiguous cell lineage (expression of multiple markers) and pink represents unknown lineage.
Influenza virus infection of PHLE cells
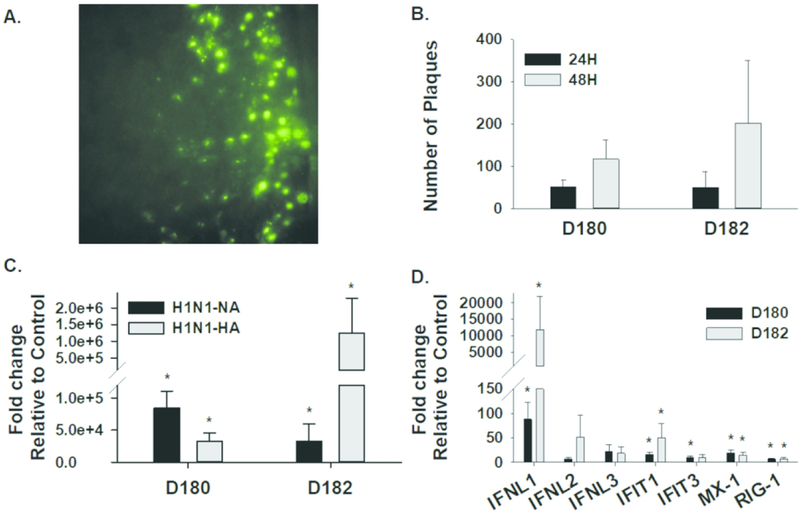
The influenza virus can only produce productive infection in differentiated airway epithelial cells (33, 34). Therefore, we tested the susceptibility of ALI-differentiated PHLE cells to apically-presented influenza virus, and monitored their ability to generate productive infections (Figure 6). The existence of viral infection was evident as plaques after 24 hours, and the size and number of the plaques expanded at 48 hours. qPCR detection of viral transcripts for Neuraminidase (NA) and Hemagglutinin (HA) 48 hours after infection further demonstrated the evidence for productive infection. Furthermore, PHLE cells showed increased expression of interferon ligands (particularly Interferon Lambda 1 (IFNL1)), and genes indicative of interferon signaling (Interferon Induced Protein With Tetratricopeptide Repeats 1(IFIT1), Interferon Induced Protein With Tetratricopeptide Repeats 3 (IFIT3), MX Dynamin Like GTPase 1 (MX1), Retinoic Acid Receptor Responder 3 (RIG1)), at 48 hours after infection.
Figure 6. Influenza virus infection of PHLE cells.
PHLE cells differentiated at ALI were challenged apically with infleunza virus. A. Representative image of fluorescent plaques indicative of viral infection. B. The number of individual plaques were counted at 24 and 48 hours, and show an increase over time after infection. C. Expression of two viral transcripts was assessed at 48 hours after infection by qPCR, and compared to uninfected cells (control). Expression of viral transcripts is indicative of a productive infection. D. Expression of interferon ligands (IFNA, IFNB, IFNL½/3) and interferon response genes (IFIT1, IFIT3, MX1, RIG1) was assessed at 48 hours after infection by qPCR, and compared to uninfected cells (control). Evidence of interferon production and signaling indicate a physiological response to infection. “*” P< 0.05,
DISCUSSION
The primary goal for this study was to develop an in vitro system for the growth and differentiation of primary human lung epithelial cells from pediatric lungs. Our morphological and molecular studies demonstrate that cells isolated from digests of whole lung lobes can be grown in culture, enriched for epithelial cells by growth in selective media, can form resistance monolayers and can be differentiated at ALI culture. These primary cultures of pediatric human lung epithelial cells (PHLE) express general epithelial cell lineage markers and lung-specific epithelial cell lineage markers. PHLE appear to expand as KRT5 expressing basal-like cells, and show increased expression of markers for airway, but not alveolar, epithelial differentiation when placed at ALI.
We purposely chose to emulate the methods and procedures commonly used to generate similar culture models of adult human trachea-bronchial airway epithelial cells (NHBE (19, 20);), and find PHLE cells to share many of the morphological and molecular features of NHBE cells. One limitation of our studies relates to the challenges of determining how much PHLE differ from NHBE cells. Interestingly, we could find limited, modest, but statistically significant associations between organ donor age and the expression of some genes. Conversely, PHLE showed evidence for decreasing ability to undergo differentiation with higher passage, as evidence by reductions in population-level expression of FOXJ1 and DNAH5 (Supplemental Figure S1), similar to NHBE. Although many similarities and some differences exist, we feel PHLE cells are likely to be a superior model for studies focused on responses in infants and children. These cells, and protocols for their growth and differentiation, are available to the research community upon request from LungMAP (www.brindl.urmc.rochester.edu).
We performed studies to examine differences in phenotype of PHLE cells established from whole lung lobe digests, or from proximal/bronchial airway tissue. However, due to limited availability of proximal airway tissues, most cultures were established with distal lung tissue composed mainly of parenchyma. There was no significant differences in the expression of lineage marker genes when comparing distal and bronchus derived cells, with the exception of MUC5B. Other, non-significant differences in gene expression from PHLE cells derived from proximal/bronchus tissues were noted (more FOXJ1 and MUC5B, less SFTPB).
We also tested different culture media compositions in an effort to expand and differentiate PHLE cells, with the intention of generating ALI cultures with distinct proximal versus distal phenotypes. Others have demonstrated growth of ATII-like cells from fetal lungs (9, 35) and adult lungs cells (36). We used three different media: SAGM+1%FBS (36), DMEM with KGF, Dexamethasone, IBMX and cAMP supplement (KDCI) (35) and Waymouth’s media with DCI (9, 37). However, PHLE cells showed poor growth characteristics and failed to maintain transepithelial resistance when grown under conditions promoting distal/alveolar cell differentiation. Additional efforts to grow alveolar-like cells, as well as non-epithelial lineages, are a focus of current investigation.
Another limitation of the current studies is the restricted number of subjects represented in the current data set, due to related time and cost. Small to moderate variation in the phenotype of cells obtained between organs is to be expected. We find evidence for differences, such as modest but significant associations between organ donor age and expression of some genes. However, these differences appear to be limited, as evidenced by the lack of donor-specific clustering in scRNAseq data (data not shown). Additional studies will be required to further characterize cells derived from different organs, and how they may relate to organ donor age or other demographic variables.
Finally, in order to further demonstrate that PHLE cells are a physiologically-relevant airway epithelial cell model, we demonstrated that they can be infected with influenza virus. It has been documented that only airway epithelial cells can produce a productive infection under physiological conditions (28, 33, 34), and our PHLE cultures consistently showed evidence of productive viral infection. Not only did PHLE cells produce viral transcript, they responded to infection by increased expression of interferon-related genes, consistent with physiological responses in airway epithelial cells (38). In summary, PHLE cells provide a unique in vitro airway epithelial cell model that can be used to future study the homeostasis and disease of the pediatric lung epithelium.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to acknowledge the following for assistance; Christopher Slaunwhite, Chin-Yi Chu, Yue Ren, Dr. Siva Kumar Solleti, Claire Wyman, Michelle Zanche, and the UR Genomic Research Center, the UR Flow Cytometry Core facility, the developing lung molecular atlas program and members of University of Rochester Medical Center Human Tissue Core for assistance. We also thank Dr. Luis Martinez and Antony Depiazza for providing pH1N1 virus. We particularly thank our donor families for their generosity.
GRANTS
This work was supported by the NIH HL122700 and DA037447.
Footnotes
DISCLOSURES
None of the authors have any conflicts of interest, financial or otherwise, to declare.
REFERENCES
- 1.Lechner JF, Haugen A, Autrup H, McClendon IA, Trump BF, Harris CC 1981. Clonal growth of epithelial cells from normal adult human bronchus. Cancer Res 41:2294–2304. [PubMed] [Google Scholar]
- 2.Lechner JF, Haugen A, McClendon IA, Pettis EW 1982. Clonal growth of normal adult human bronchial epithelial cells in a serum-free medium. In Vitro 18:633–642. [DOI] [PubMed] [Google Scholar]
- 3.Lechner JF, McClendon IA, LaVeck MA, Shamsuddin AM, Harris CC 1983. Differential control by platelet factors of squamous differentiation in normal and malignant human bronchial epithelial cells. Cancer Res 43:5915–5921. [PubMed] [Google Scholar]
- 4.Hackett NR, Shaykhiev R, Walters MS, et al. 2011. The human airway epithelial basal cell transcriptome. PLoS One 6:e18378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chevillard M, Hinnrasky J, Zahm JM, Plotkowski MC, Puchelle E 1991. Proliferation, differentiation and ciliary beating of human respiratory ciliated cells in primary culture. Cell Tissue Res 264:49–55. [DOI] [PubMed] [Google Scholar]
- 6.Fulcher ML, Gabriel S, Burns KA, Yankaskas JR, Randell SH 2005. Well-differentiated human airway epithelial cell cultures. Methods Mol Med 107:183–206. [DOI] [PubMed] [Google Scholar]
- 7.Whitcutt MJ, Adler KB, Wu R 1988. A biphasic chamber system for maintaining polarity of differentiation of cultured respiratory tract epithelial cells. In Vitro Cell Dev Biol 24:420–428. [DOI] [PubMed] [Google Scholar]
- 8.Messina E, Muhlhauser J, Giuliano M, Pandolfi A, Morgese G, Procopio A 1996. Surfactant protein A-producing cells in human fetal lung are good targets for recombinant adenovirus-mediated gene transfer. Pediatr Res 40:142–147. [DOI] [PubMed] [Google Scholar]
- 9.Gonzales LW, Angampalli S, Guttentag SH, et al. 2001. Maintenance of differentiated function of the surfactant system in human fetal lung type II epithelial cells cultured on plastic. Pediatr Pathol Mol Med 20:387–412. [DOI] [PubMed] [Google Scholar]
- 10.Devalia JL, Sapsford RJ, Wells CW, Richman P, Davies RJ 1990. Culture and comparison of human bronchial and nasal epithelial cells in vitro. Respir Med 84:303–312. [DOI] [PubMed] [Google Scholar]
- 11.Chu CY, Qiu X, Wang L, et al. 2016. The Healthy Infant Nasal Transcriptome: A Benchmark Study. Sci Rep 6:33994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spann KM, Baturcam E, Schagen J, Jet al. 2014. Viral and host factors determine innate immune responses in airway epithelial cells from children with wheeze and atopy. Thorax 69:918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thavagnanam S, Parker JC, McBrien ME, Skibinski G, Shields MD, Heaney LG 2014. Nasal epithelial cells can act as a physiological surrogate for paediatric asthma studies. PLoS One 9:e85802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller L, Brighton LE, Carson JL, Fischer WA 2nd, Jaspers I 2013. Culturing of human nasal epithelial cells at the air liquid interface. J Vis Exp [DOI] [PMC free article] [PubMed]
- 15.Parker J, Sarlang S, Thavagnanam S, et al. 2010. A 3-D well-differentiated model of pediatric bronchial epithelium demonstrates unstimulated morphological differences between asthmatic and nonasthmatic cells. Pediatr Res 67:17–22. [DOI] [PubMed] [Google Scholar]
- 16.Villenave R, Thavagnanam S, Sarlang S, et al. 2012. In vitro modeling of respiratory syncytial virus infection of pediatric bronchial epithelium, the primary target of infection in vivo. Proc Natl Acad Sci U S A 109:5040–5045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villenave R, Touzelet O, Thavagnanam S, et al. 2010. Cytopathogenesis of Sendai virus in well-differentiated primary pediatric bronchial epithelial cells. J Virol 84:11718–11728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bandyopadhyay G, Huyck HL, Misra RS, et al. 2018. Dissociation, Cellular Isolation and Initial Molecular Characterization of Neonatal and Pediatric Human Lung Tissues. Am J Physiol Lung Cell Mol Physiol [DOI] [PMC free article] [PubMed]
- 19.Solleti SK, Bhattacharya S, Ahmad A, et al. 2017. MicroRNA expression profiling defines the impact of electronic cigarettes on human airway epithelial cells. Sci Rep 7:1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Solleti SK, Simon DM, Srisuma S, et al. 2015. Airway epithelial cell PPARgamma modulates cigarette smoke-induced chemokine expression and emphysema susceptibility in mice. Am J Physiol Lung Cell Mol Physiol 309:L293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bolger AM, Lohse M, Usadel B 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobin A, Davis CA, Schlesinger F, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao Y, Smyth GK, Shi W 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930. [DOI] [PubMed] [Google Scholar]
- 24.Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C 2017. Single-cell mRNA quantification and differential analysis with Census. Nat Methods 14:309–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C, Cacchiarelli D, Grimsby J, et al. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy DJ, Campbell KR, Lun AT, Wills QF 2017. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33:1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lun AT, McCarthy DJ, Marioni JC 2016. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res 5:2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DiPiazza A, Nogales A, Poulton N, Wilson PC, Martinez-Sobrido L, Sant AJ 2017. Pandemic 2009 H1N1 Influenza Venus reporter virus reveals broad diversity of MHC class II-positive antigen-bearing cells following infection in vivo. Sci Rep 7:10857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez O, Montesino R, Toledo JR, et al. 2007. The goat mammary glandular epithelial (GMGE) cell line promotes polyfucosylation and N,N’-diacetyllactosediaminylation of N-glycans linked to recombinant human erythropoietin. Arch Biochem Biophys 464:322–334. [DOI] [PubMed] [Google Scholar]
- 30.Liu X, Ory V, Chapman S, et al. 2012. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol 180:599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Treutlein B, Brownfield DG, Wu AR, et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509:371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu Y, Mizuno T, Sridharan A, et al. 2016. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI Insight 1:e90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ilyushina NA, Ikizler MR, Kawaoka Y, et al. 2012. Comparative study of influenza virus replication in MDCK cells and in primary cells derived from adenoids and airway epithelium. J Virol 86:11725–11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Endo Y, Carroll KN, Ikizler MR, Wright PF 1996. Growth of influenza A virus in primary, differentiated epithelial cells derived from adenoids. J Virol 70:2055–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mariani TJ 2015. Update on Molecular Biology of Lung Development--Transcriptomics. Clin Perinatol 42:685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hersh CP, Silverman EK, Gascon J, et al. 2011. SOX5 is a candidate gene for chronic obstructive pulmonary disease susceptibility and is necessary for lung development. Am J Respir Crit Care Med 183:1482–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daugherty BL, Mateescu M, Patel AS, et al. 2004. Developmental regulation of claudin localization by fetal alveolar epithelial cells. Am J Physiol Lung Cell Mol Physiol 287:L1266–1273. [DOI] [PubMed] [Google Scholar]
- 38.Mindaye ST, Ilyushina NA, Fantoni G, Alterman MA, Donnelly RP, Eichelberger MC 2017. Impact of Influenza A Virus Infection on the Proteomes of Human Bronchoepithelial Cells from Different Donors. J Proteome Res 16:3287–3297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.