Abstract
During autophagy, double-membrane vesicles called autophagosomes capture and degrade the intracellular cargo. The de novo formation of autophagosomes requires several vesicle transport and membrane fusion events which are not completely understood. We studied the involvement of exocyst, an octameric tethering complex, which has a primary function in tethering post-Golgi secretory vesicles to plasma membrane, in autophagy. Our findings indicate that not all subunits of exocyst are involved in selective and general autophagy. We show that in the absence of autophagy specific subunits, autophagy arrest is accompanied by accumulation of incomplete autophagosome-like structures. In these mutants, impaired Atg9 trafficking leads to decreased delivery of membrane to the site of autophagosome biogenesis thereby impeding the elongation and completion of the autophagosomes. The subunits of exocyst, which are dispensable for autophagic function, do not associate with the autophagy specific subcomplex of exocyst.
Keywords: Exocyst, Autophagosome biogenesis, Atg9 trafficking, Autophagy, Tethers
Introduction
Autophagy is an intracellular catabolic process involving capture of cytosolic cargo by double-membrane vesicular structures called autophagosomes. These autophagosomes then fuse with lysosomes (vacuoles in yeast) leading to degradation of the cargo [1]. Studies by various groups have led to the identification of more than 40 ATG proteins (Autophagy Related Genes) and several accessory components. The molecular mechanisms of how these proteins function in the process of autophagy have been deciphered to a substantial extent [2]. Autophagosome biogenesis is a complex process that begins with the assembly of autophagy initiation complex (Atg1 complex) at PAS (Pre-autophagosomal structure, a perivacuolar autophagosome biogenesis site in yeast) followed by activation of VPS34 complex at this site to produce PI3P locally, thereby leading to the recruitment of other core autophagy proteins and nucleation of precursor autophagosome membrane. This nascent structure known as the phagophore further expands into double-membrane vesicle by addition of membrane derived from various sources mediated by Atg9 vesicles [3,4].
Atg9 is an integral membrane protein that appears as multiple puncta in cytoplasm [5]. These puncta represent the peripheral pool of Atg9 containing vesicles, which deliver membrane to PAS allowing for autophagosome expansion. These Atg9 vesicles are known to be derived from various membrane sources including Golgi associated secretory pathway [6–11].
Various secretory pathway proteins were shown to be important for the process of autophagosome biogenesis. The early secretory pathway components consisting of multisubunit protein complexes such as COPII machinery, COG, GARP, TRAPPIII, TRAPP IV, late secretory components such as Sec2, Sec4 and various SNARE proteins such as Sso1, Sso2, Sec9, Tlg2, Ufe1, and others, were shown to be important for autophagosome biogenesis [12–16]. Among these, several tethering complexes like COG, TRAPPIII and exocyst have been associated with autophagy. While TRAPPIII function has been elucidated to an extent, the involvement of other two complexes is not clear [17–21]. Exocyst is a conserved multisubunit protein complex that functions during exocytosis [22,23]. This octameric complex consists of Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo84 and Exo70 subunits [24–26]. It helps in tethering post-Golgi secretory vesicles to the plasma membrane [27]. In yeast, the exocyst subunit Sec3 serves as a landmark for the assembly of exocyst complex at the plasma membrane [28]. This assembly is also regulated by Sec4, a Rab protein present on the secretory vesicle [29,30]. In mammalian cells, Moskalenko and colleagues [31] suggested that the exocyst complex exists as two subcomplexes, one forms a targeting patch at the plasma membrane and the other helps in directing the secretory vesicles to the location marked by targeting patch. This coalition of both the subcomplexes is brought about by Ral-GTPases. This phenomenon of Ral-mediated activation of exocyst complex was also recounted where the activation of Sec5 subunit led to autophagy inhibition while activation of Exo84 subunit led to induction of autophagy by assembly of initiation complex [32]. In yeast, Rho GTPases are known to mediate the assembly of the exocyst complex [33]. Some exocyst subunits are required for autophagy which underscores the overlap between the secretory pathway components and autophagy [16]. It is suggested that the determinants of the membrane flow in the secretory pathway also contribute to autophagosome biogenesis [12]. As exocyst is important for secretory function, it may play a similar role in autophagy where vesicular-based membrane flow is critical. A study suggested that tethering function of exocyst is responsible for SNARE pairing in the secretory pathway [34], and this idea was extended to exocyst function in autophagy [16]. However, the exact role of exocyst in yeast during autophagy is not well understood. Here, we report that several but not all subunits of exocyst are essential for autophagy. Some of the participating subunits affect Atg9 trafficking suggesting a role for this subcomplex in autophagosome biogenesis by contributing to the membrane flow. Our biochemical investigations further provide insights into the function of a distinct subcomplex of exocyst as during autophagosome biogenesis.
Results
Exocyst complex is involved in selective and general autophagy
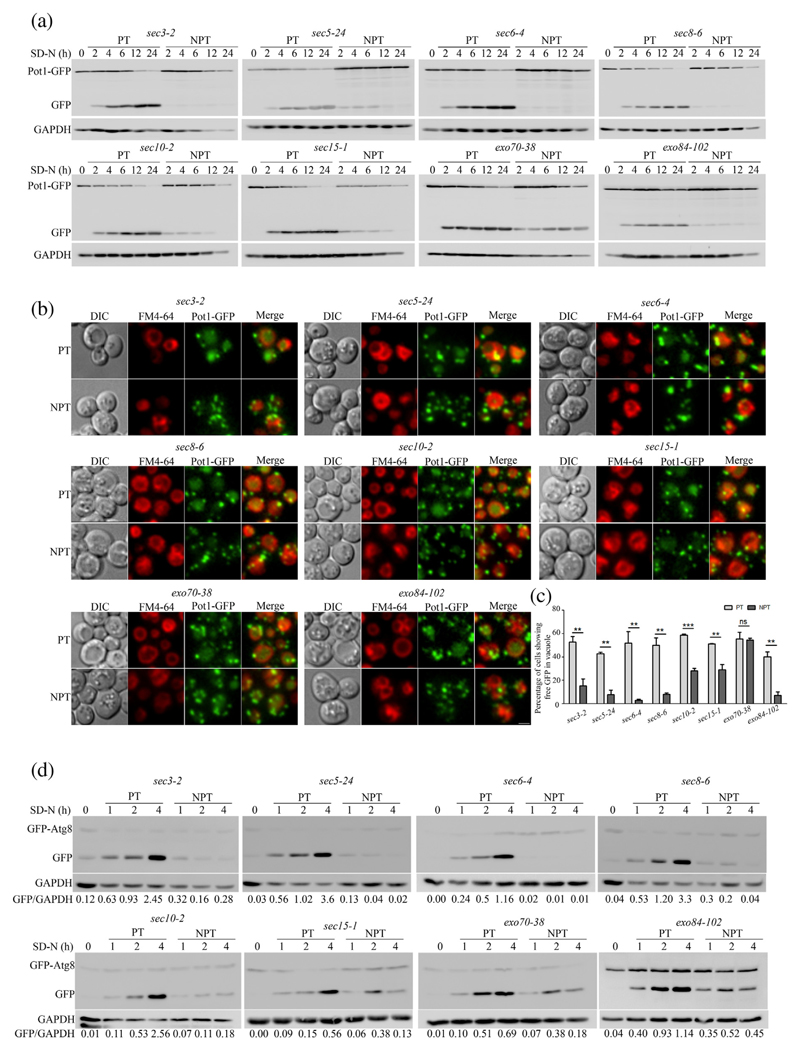
We screened a subset of temperature-sensitive (Ts) Saccharomyces cerevisiae mutants defective in vesicular trafficking for their ability to perform selective autophagy of peroxisomes—pexophagy. We measured pexophagy in these Ts mutants by employing a previously established immunoblotting assay in which accumulation of free GFP with concomitant decrease of fused protein Pot1-GFP is indicative of pexophagy [35]. The cells expressing Pot1-GFP were grown in rich medium (YPD), and actively growing cells were then incubated overnight in fatty acid rich medium (Oleate) to allow build-up of peroxisomes. Pexophagy was induced by moving cells into starvation medium (SD-N). Among the mutants that showed a block in pexophagy, there were several mutants of the exocyst complex. We observed that the Ts mutants of exocyst complex sec3–2, sec5–24, sec6–4, sec8–6, sec8–9, sec10–2 and exo84–102 showed accumulation of free GFP at permissive temperature (PT; 25 °C) but failed to do so at non-permissive temperature (NPT; 37 °C) indicating a block in pexophagy (Figs. 1a, S1a). However, we also observed that not all mutants showed this block. At NPT, while sec15–1 showed reduced levels of pexophagy, exo70–38 was not affected (Fig. 1a).
Fig. 1. Subset of exocyst complex mutants are defective in selective and general autophagy.
(a) Ts exocyst mutants expressing Pot1-GFPwere grown in oleate medium to induce peroxisomes. They were subsequently transferred to nitrogen starvation medium (SD-N) to induce pexophagy under PT or NPT. Samples were collected at indicated time points, processed and subjected to immunoblotting analysis. (b) Cells were treated as in panel A and were imaged 4 h in starvation medium using fluorescence microscopy. Peroxisomes appear green due to the presence of Pot1-GFP. Vacuoles were labeled with FM4-64 dye. Images were deconvolved by nearest neighbor algorithm using softWoRx software (GE Healthcare), and maximum intensity projection images are shown. The scale bar represents 2 μm. (c) Quantitation of pexophagy from images obtained in panel B. About 150–200 cells for each experiment were counted for the presence of GFP in the vacuole and represented as percentage of total cells scored. The bar diagram shows mean of three independent experiments with standard error. Statistical significance was analyzed by Student's unpaired t-test. ns, nonsignificant; **P < 0.01, ***P < 0.001. (d) GFP-Atg8 processing assay for general autophagy. Cells expressing GFP-Atg8 were starved in SD-N medium at PT or NPT. Samples were collected at indicated time points, processed and analyzed by immunoblotting. Numbers indicate ratio of intensity of free GFP/GAPDH.
To further validate these results, we performed fluorescence microscopy-based pexophagy assay with these mutants expressing Pot1-GFP. When grown in oleate medium, peroxisomes appeared as green punctate structures in the cells. These cultures were then resuspended in starvation medium and were incubated at permissive and NPTs. After 4 h of starvation at PTs, all mutants showed accumulation of free GFP inside the vacuoles labeled in red with FM4–64 (Fig. 1b). In agreement with our Western blot data, we observed that there was no free GFP in the vacuole of all the mutants, which showed a pexophagy block at NPT (Figs. 1b, c, S1b and c).
Next, we wanted to check whether this defect is specific to a selective autophagy process like pexophagy or extended to general autophagy as well. Cells expressing GFP-Atg8, an autophagy marker, were grown in selection medium (SD-Ura), transferred to starvation medium and incubated at permissive and non-permissive conditions. Immunoblotting studies revealed accumulation of free GFP over the course of starvation at PTs, while no such accumulation was observed at NPTs in most of the mutants (Figs. 1d and S1d). Wild-type (WT) cells did not show any defect in GFP processing at NPT (Fig. S1d, WT). Similar to the pexophagy results, we observed no defect in general autophagy in sec15–1 and exo70–38 mutants. Surprisingly, we noticed that general autophagy was not blocked in mutant exo84–102. Whether Exo84 is important for only pexophagy or even for general autophagy still needs to be addressed.
Mutants of exocyst complex accumulate immature autophagosomes
To investigate the stage of autophagy in which the mutants were blocked, we used fluorescence microscopy to observe localization of GFP-Atg8 puncta. A single perivacuolar punctum of Atg8 marks the site of autophagosome biogenesis (PAS). During starvation conditions where autophagy is prevalent, autophagosomes formed at PAS are expected to immediately fuse with the vacuole. Atg8, which is also a marker for autophagosome membrane, gets degraded along with the autophagosomal cargo in the vacuole.
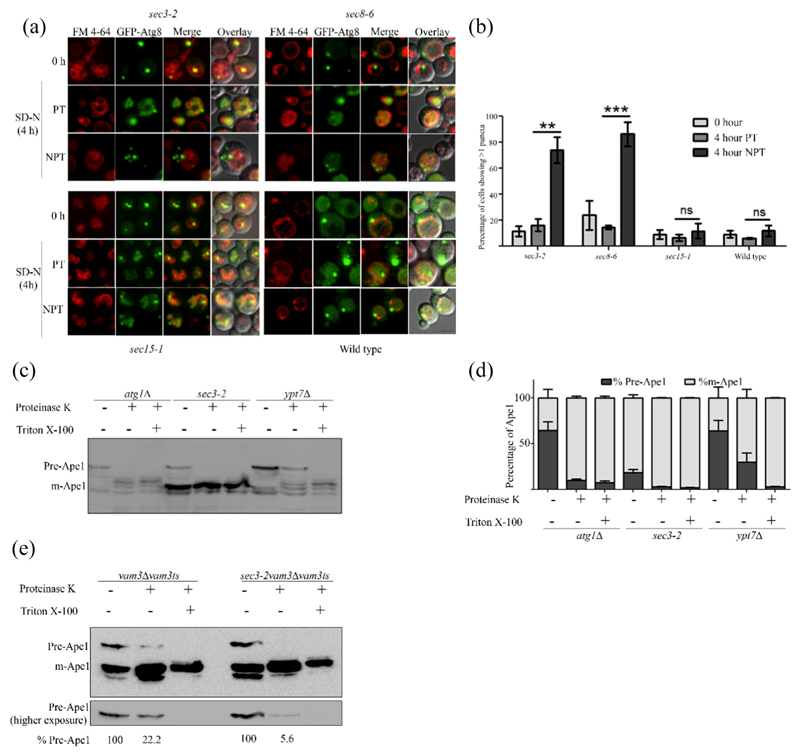
Along with WT cells, we studied two mutants that at NPT showed complete autophagy defect (sec3–2, sec8–6) and one with partial defect (sec15–1). Interestingly, sec3–2 and sec8–6 both showed an increased number of Atg8 puncta in the cells but no diffused GFP in vacuoles, indicating that possibly autophagosomal structures were getting accumulated. In addition, these cells did not display diffused GFP in the vacuole (Fig. 2a). When starved in the presence of PMSF, sec3–2 cells showed accumulation of GFP-Atg8 puncta inside the vacuole at PT, but were present outside the vacuole at NPT, suggesting an autophagy block prior to fusion with vacuole (Fig. S1e). In accordance with our GFP-Atg8 processing assay, sec15–1 and exo70–38 did not show any significant accumulation of multiple Atg8 puncta (Figs. 2a, b, S1f and g).
Fig. 2. Mutants of exocyst complex accumulate incomplete autophagosomes.
(a) Cells expressing GFP-Atg8 were cultured in SD-Ura with FM4–64 and moved to starvation at PT and NPT. Fluorescence microscopy images were acquired at 0 and 4 h in SD-N. Maximum intensity projected images are shown. The scale bar represents 2 μm. (b) Cells showing more than one puncta in panel A were scored, and means of three independent experiments are represented in the bar graph. A minimum of 100 cells were counted per experiment. Error bars represent standard error. Statistical significance was analyzed by Student's unpaired t-test. ns, nonsignificant; **P b 0.01, ***P b 0.001. (c) atg1Δ, ypt7Δ and sec3–2cells were starved at NPT for 4 h. Cells were harvested, and spheroplasts were made and lysed. The clarified lysates were treated with either proteinase K or proteinase K with Triton X-100 and analyzed by immunoblotting using anti-Ape1 antibody. (d) Intensities of precursor (Pr-Ape1) and mature Ape1 (m-Ape1) bands in each lane of c were measured using ImageJ (NIH). The percentages of Pr-Ape1 and m-Ape1 were determined and mean of three independent experiments plotted as bar graphs. Error bars represent S.E.M. (e) vam3Δ Vam3ts and sec3–2 vam3ΔVam3ts strains were starved for 4 h at NPT and treated as in panel c. Precursor Ape1 in proteinase K-treated lane relative to precursor form in the control lane is represented as a percentage below the respective lanes of the blot.
We wanted to test if the GFP-Atg8 puncta represent complete or incompletely formed autophagosomes. For this, we employed the protease protection assay using Ape1 maturation as a readout. Ape1 gets delivered as precursor form to the vacuole by Cvt (cytoplasm to vacuole targeting) vesicles in growth conditions and by autophagosomes during starvation, where it gets cleaved and converted into mature Ape1. If the precursor form of Ape1 is entrapped in vesicles as in the case of ypt7Δ cells, it is resistant to the action of externally added protease such as proteinase K. Addition of detergent Triton X-100 dissolves the membrane of vesicles and exposes the precursor Ape1 to proteinase K. However, if the autophagosomes are not formed as in atg1Δ cells or partially formed, proteinase K can access and cleave precursor Ape1. We asked whether the autophagosomal structures (multiple GFP-Atg8 puncta) observed at NPT in sec3–2 cells were completely formed.
Similar to atg1Δ, sec3–2 showed the presence of pre-Ape1 in untreated fraction but not in the fraction treated with either proteinase K alone or proteinase K with Triton X-100 (Fig. 2c and d). To further investigate this observation, we employed a Vam3ts allele in sec3–2 and vam3Δ strains. vam3Δ was used as a control. Absence of a functional Vam3 at NPT causes accumulation of autophagosomes. In vam3Δvam3ts, out of 100% precursor Ape1 in untreated lane, about 22% of Ape1 was retained in precursor form upon proteinase K treatment, while in sec3–2 only, 5.6% of initial precursor form was observed (Fig. 2e). These data confirm that at NPT, the multiple autophagosomal structures in sec3–2 were incompletely formed and therefore are not able to fuse with the vacuole.
We then investigated if the exocyst complex associates with autophagy components. Live cell microscopy revealed a dynamic interaction of Sec3-GFP and 2XmCherry-Atg8 (Fig. S2a, Video 1). We studied the colocalization of components not affecting autophagy, that is, Exo70 and Sec15 with Atg8. We also performed the colocalization experiments in ypt7Δ expressing Sec3-GFP, Exo70-GFP or Sec15-GFP. We suspected that if exocyst complex is associated with autophagosomes, then accumulation of autophagosomes would increase in colocalization events in these strains. While we noticed an increased colocalization of Sec3 and Atg8 as compared to Exo70 or Sec15, the percentage, however, is very low to be assertive about the interaction of exocyst with Atg8 (Fig. S2b). We also performed colocalization experiments of exocyst subunits Sec3, Exo70 and Sec15 with Atg9 in WT and atg1Δ cells and did not observe any significant change in the colocalization (Fig. 2c and d).
Atg9 vesicle trafficking is perturbed in exocyst Ts mutants
As we observed that the elongation or completion of autophagosomes was impaired in sec3–2 mutant of exocyst, we investigated if dysfunctional Atg9 trafficking attributed to these defects in autophagosome formation. Atg9 is involved in contributing membrane to the growing phagophore aiding in autophagosome elongation [36]. Trafficking of Atg9 vesicles takes place from peripheral membrane sources such as mitochondria, ER and Golgi to the PAS, delivering membrane for autophagosome formation. This shuttling of Atg9 is known to be critical for autophagy progression [11,37].
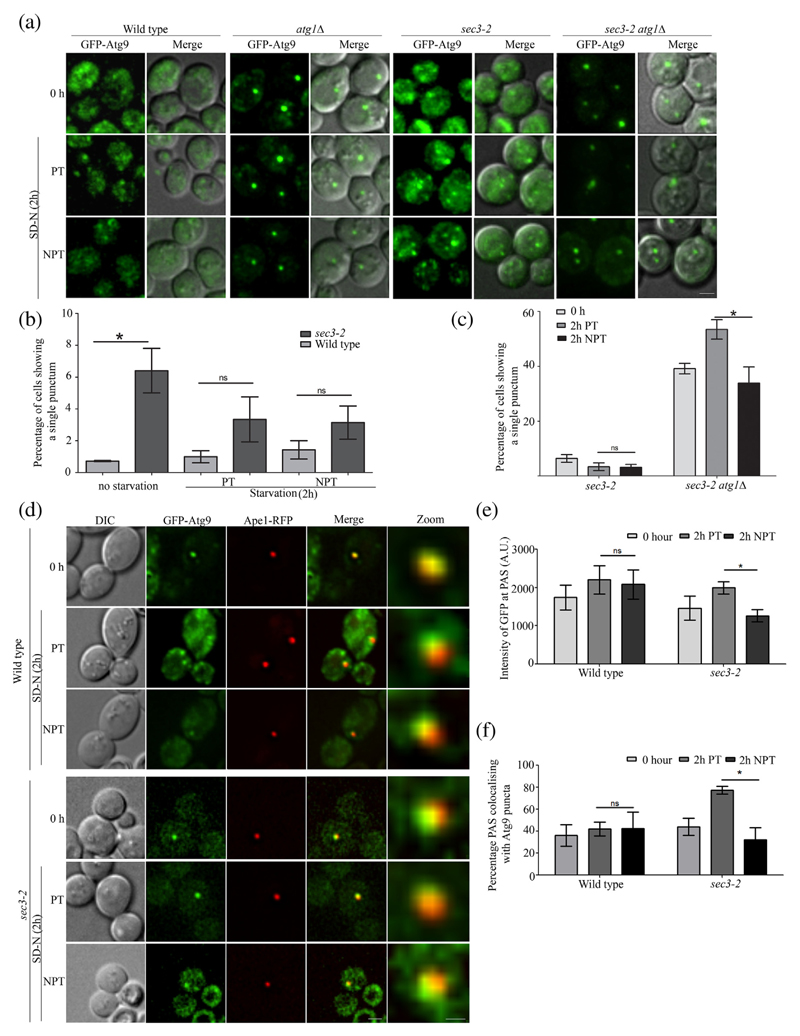
WT, atg1Δ, sec3–2 and exo70–38 cells expressing Atg9-GFP were imaged in growth medium and imaged again 2 h post-starvation. WT cells show a distribution of Atg9 puncta at peripheral membrane sources in the cytoplasm and at the PAS [5,38]. In agreement with these reports, we observed a uniform distribution of Atg9 puncta in WT cells in nutrient-rich medium, under basal autophagy conditions, suggesting dynamic Atg9 trafficking (0 h, Fig. 3a). Interestingly, a small but significant number of sec3–2 cells showed single punctum of Atg9, indicating that the Atg9 trafficking dynamics may be perturbed (Fig. 3b). However, the Atg9 trafficking was completely restored when these mutant cells were transferred to starvation conditions where autophagy is induced (Fig. 3b). It is known that Atg9 vesicles show an increased mobility during starvation conditions [11], and as a result, the mild effect seen during basal autophagy may have been overridden during such autophagy inducing conditions in both permissive and non-permissive conditions. There was no significant change in number of cells showing single puncta in exo70–38 at NPT (Fig. S3b). In addition to showing a prominent Atg9 punctum, sec3–2 cells also display several puncta at NPTs. In order to do a more precise analysis, we resorted to studying Atg9 trafficking using the TAKA (transport of Atg9 in knockout of Atg1) assay. The retrograde trafficking of Atg9 is dependent on the Atg1 complex, and thus in atg1Δ cells, Atg9 vesicles reach but get arrested at PAS (WT versus atg1Δ, Figs. 3a, S3b and c). We also used atg1Δ atg27Δ strain in which anterograde trafficking of Atg9 vesicles to PAS is blocked as a control (Fig. S3a and b). We observed Atg9 trafficking in sec3–2 and exo70–38 cells where ATG1 has been deleted by shifting these cells to PT and NPT conditions in starvation medium. We noticed a significant reduction in number of sec3–2atg1Δ cells showing single puncta at NPT as compared to the PT implying defects in anterograde trafficking (Fig. 3c). No such reduction was observed in exo70–38atg1Δ cells (Fig. S3a and c). In addition, we also studied Atg9 trafficking in exocyst mutants expressing a Ts allele of ATG1 at PT and NPT. We observed a reduction in percentage of cells showing single Atg9 punctum in sec3–2atg1Δatg1ts strain at NPT. Percentage of cells with one punctum of Atg9 in exo70–38atg1-Δatg1ts was comparable to that of atg1Δatg1ts (Fig. S3d and e). Taken together, these results suggest that Atg9 vesicles are being driven to PAS albeit at a reduced rate.
Fig. 3. Anterograde trafficking of Atg9 vesicles is affected in an exocyst mutant.
(a) WT, atg1Δ, sec3–2 and sec3–2atg1Δ cells expressing GFP-Atg9 were grown in SD-Ura and then starved at PT or NPT. Fluorescence imaging was carried out at 0 and 2 h. Deconvolved and maximum intensity projection images are shown. The scale bar represents 2 μm. (b) WT and sec3–2 cells as in panel a were scored for presence of single bright punctum of GFP-Atg9 at 0 and 2 h in starvation at PT and NPT. (c) Comparison of sec3–2 and sec3–2atg1Δ cells showing single punctum of GFP-Atg9 as in panel a. For panels b and c, more than 100 cells per three independent experiments were manually scored and mean values plotted with standard error. (d) WT and sec3–2cells expressing GFP-Atg9 and Ape1-RFP were starved at PT or NPT. Fluorescence images were captured at 0 and 2 h in SD-N. The scale bar represents 2 μm (merge) and 0.5 μm (zoom). Intensity of GFP-Atg9 at PAS (marked by Ape1-RFP) was measured, and average intensity of GFP-Atg9 is plotted in panel e. (f) The percentage of PAS that colocalizes with bright Atg9 puncta was determined from panel d, and mean values were plotted. Error bars represent S.E.M. Statistical significance was analyzed by Student's unpaired t-test. ns, non-significant; *P < 0.05.
To further confirm these observations, we measured the amount of Atg9 at PAS as a read-out of Atg9 trafficking flux. We reasoned that as the retrograde trafficking was unaffected in the exocyst mutant, any changes in Atg9 levels at PAS would reflect a change in anterograde Atg9 dynamics. We co-expressed GFP-Atg9 and Ape1-RFP, which served as a marker for PAS. We imaged cells at 2 h post-starvation in permissive and non-permissive conditions (Fig. 3d). The images with colocalized GFP-Atg9 and Ape1-RFP puncta were then used for intensity measurements. In agreement with our earlier results, we found that the average intensity of Atg9 at PAS was indeed reduced in sec3–2 but not in WT (NPT versus PT, Fig. 3e) and exo70–38 cells (Fig. S3f and g). Furthermore, this mutant showed decreased occurrence of Atg9 at the PAS (PT versus NPT, Fig. 3f). exo70–38 cells did not show this defect (Fig. S3h). These results indicate that in addition to an ineffective anterograde transport of Atg9 vesicles from peripheral sources to PAS, a dysfunctional exocyst component also led to a decreased number of Atg9 vesicles being tethered at PAS.
The autophagy specific exocyst complex is distinct from its secretory pathway counterpart
As described previously, we noticed that mutants of some of the exocyst subunits did not show any effect on autophagy (Fig. 1d). This observation led us to consider the possibility of a disparity in the autophagy function associated exocyst complex than what is known for the secretory associated exocyst complex. To test this hypothesis, we resorted to size exclusion chromatography to analyze the exocyst complex size under starvation conditions.
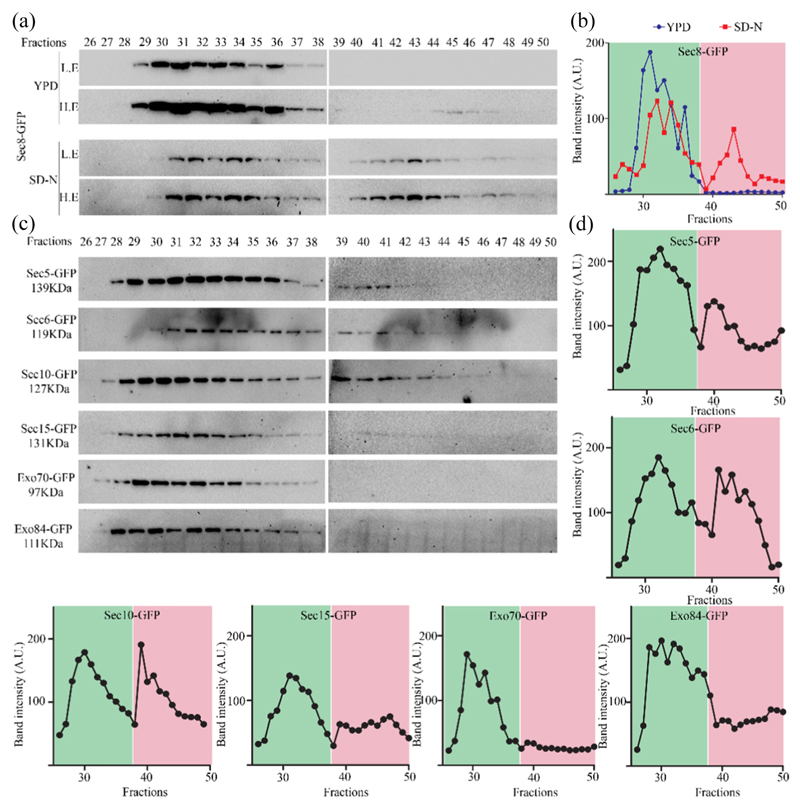
For our initial analysis, we subjected Sec8-GFP cell lysates collected from cells grown in nutrient rich or starvation medium to size exclusion chromatography followed by Western blotting with anti-GFP antibody. Quantification of intensity on Western blot yielded a single peak from cells cultured in rich medium, while those from starvation medium showed two distinct peaks (Fig. 4a and b). To verify these results, we repeated the experiment with other subunits of the exocyst complex in starvation conditions. Interestingly, we find that the lysates of Sec5-GFP, Sec6-GFP and Sec10-GFP showed the presence of two peaks as seen for Sec8-GFP (Fig. 4b, starvation). However, Sec15-GFP, Exo70-GFP and Exo84-GFP showed only the first peak as seen for Sec8-GFP lysates from growth conditions (Fig. 4c and d). These results suggest that the first peak corresponds to the peak seen in growth medium, while the second one suggested the presence of a smaller complex that was unique to autophagy inducing conditions. Importantly, only the exocyst subunits that were affecting autophagy showed the second peak.
Fig. 4. Autophagy prevalent conditions reveal presence of a subcomplex of exocyst comprising of subunits that are required for autophagy.
(a) Sec8-GFP cells were grown in rich medium (YPD) and starved for 4 h. Clarified supernatant (cytosol) from YPD or SD-N grown cells were prepared and subjected to size exclusion chromatography using Superose 6 high load 10/300GL column. Fractions were collected and analyzed by Western blotting using anti-GFP antibody. L.E., lower exposures; H.E., higher exposures. (b) Intensities of bands from panel A were quantitated and plotted against fractions. Peak in the green area represents higher-molecular-weight exocyst complex associated with secretory function, while peak in the pink area represents starvation-specific exocyst subcomplex. (c) Strains expressing exocyst subunits tagged with GFP were starved for 4 h and processed as in panel a. Fractions were analyzed by Western blotting. Intensity of bands from these Western blots is plotted in panel d.
Discussion
Autophagy is a complex catabolic process involving formation of double-membrane vesicles-autophagosomes. The biogenesis of autophagosomes involves several membrane-related transport events in which tethering complexes play an important role. We identified one such tethering complex called the exocyst to be involved in autophagy. The multisubunit exocyst complex has been characterized for its role in the secretory pathway. The secretory exocyst is an octameric complex and has a function of tethering post-Golgi secretory vesicles to the target membrane [39]. Our study reveals the role of a subset of the exocyst complex proteins in the formation of autophagosomes. We systematically address the role of the exocyst subunit proteins for their subunit-specific involvement and their potential role in autophagy.
In our study, we show that not all the subunits of exocyst complex are indispensable for its function in autophagy. Our experiments with Ts strains of S. cerevisiae revealed that the mutants of several subunits of exocyst complex (Sec3, Sec5, Sec6, Sec8 and Sec10), but not others (Sec15, Exo70 and Exo84), showed defect in selective and general autophagy. Further investigation by fluorescence microscopy of these mutants showed presence of more than one GFP-Atg8 puncta in cells at NPT, indicating that the autophagosomes were possibly formed. These puncta accumulated outside the vacuole and therefore confirmed our observation that the autophagy block in these mutants was in a step prior to fusion with the vacuole. Interestingly, these Atg8 puncta represented incompletely formed autophagosomes as revealed by the protease protection assay suggesting a potential role for exocyst in autophagosome expansion and completion as well.
One of the key requirements for autophagosome expansion is delivery of membrane by Atg9 vesicles to the site of autophagosome biogenesis (PAS) [40]. Our investigation in this direction revealed several interesting defects in the trafficking of Atg9 from the peripheral membrane sources to the PAS in the exocyst mutant defective in autophagy. First, we observed less colocalization events between Atg9 and PAS. Second, among these colocalized events, there was decreased occupancy of Atg9 vesicles at PAS. Third, although retrograde trafficking appeared unperturbed, anterograde movement of Atg9 vesicles to PAS from peripheral sources appeared to be compromised, explaining the reason for decreased presence of Atg9 at the PAS. Put together, these observations point at impaired Atg9 trafficking from peripheral membrane sources to the PAS. Thus, this decreased rate of delivery of Atg9 vesicles suggests limited membrane supply. Restricted Atg9 trafficking has been reported to clamp down on autophagosome biogenesis [11]. We propose that diminished supply of Atg9 membrane sources may result in abortive autophagosome biogenesis in these mutants. This limited supply may allow initiation of autophagosome biogenesis but may not be sufficient to fuel expansion and completion of autophagosomes. This may explain the presence of multiple incompletely formed autophagosomes in these mutants.
The next question we addressed was why only certain members of this complex affected autophagy. A previous report suggested that in mammalian cells two subcomplexes of exocyst, a Sec5 containing complex that suppresses autophagy, and an autophagy activating complex consisting of Exo84 exist [32]. RalB—a Ral GTPase—promotes autophagy induction by triggering a switch in the association of autophagy initiation complex (ULK1/Beclin1/VPS34 complex) from the Sec5 subcomplex of exocyst to Exo84 subcomplex. The presence of such distinct subcomplexes has also been reported in yeast, wherein during secretion, exocyst exists as two subcomplexes—one for targeting and other for bridging secretory vesicles with targeting complex [31]. The assembly of complete complex is regulated by the Sec4 and Rho1 GTPases [27,29,41]. Our biochemical investigations using size exclusion chromatography also indicates the possibility of an autophagy specific exocyst complex that differs in composition from that of the conventional secretory exocyst complex components. These results are in agreement with our genetic mutant-based analysis, which suggest a disparity in requirement of the exocyst complex subunits for autophagy function. Absence of functional Sec15, Exo70 and Exo84 did not affect autophagy flux progression. Interestingly, the same three subunits were found to be absent from the autophagy specific exocyst complex. Combined together, these results show that the constituents of exocyst complex functioning in autophagy are different from the one present in secretory pathway and it harbors lesser components (5 out of 8). It is possible that the autophagy specific complex may have additional components.
In conclusion, our study highlights the moonlighting functions of some members of the exocyst machinery that form an autophagy specific subcomplex that catalyzes membrane supply for autophagosome biogenesis.
Materials and Methods
Yeast strains and media
The detailed list of strains used in this study is provided in Table 1.
Table 1. List of strains and plasmids used in this study.
| Sr. No. | Strain/plasmid name | Genotype | Source/Reference |
|---|---|---|---|
| 1 | sSUN28 | sec3–2::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; POT1::POT1-GFP-HIS | This study |
| 2 | sSUN29 | sec5–24::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; POT1::POT1-GFP-HIS | This study |
| 3 | sSUN30 | sec6–4::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; POT1::POT1-GFP-HIS | This study |
| 4 | sSUN31 | sec8–6::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; POT1::POT1-GFP-HIS | This study |
| 5 | sSUN32 | sec8–9::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; POT1::POT1-GFP-HIS | This study |
| 6 | sSUN33 | sec10–2::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; POT1::POT1-GFP-HIS | This study |
| 7 | sSUN34 | sec15–1::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; POT1::POT1-GFP-HIS | This study |
| 8 | sSUN35 | exo70–38::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; POT1::POT1-GFP-HIS | This study |
| 9 | sSUN37 | Exo84–2::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; POT1::POT1-GFP-HIS | This study |
| 10 | sSUN53 | sec3–2::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; GFP-Atg8:: URA 3 [pGFP-Atg8 in pRS316] | This study |
| 11 | sSUN54 | sec5–24::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; GFP-Atg8:: URA 3 [pGFP-Atg8 in pRS316] | This study |
| 12 | sSUN55 | sec6–4::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; GFP-Atg8:: URA 3 [pGFP-Atg8 in pRS316] | This study |
| 13 | sSUN56 | sec8–6::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; GFP-Atg8:: URA 3 [pGFP-Atg8 in pRS316] | This study |
| 14 | sSUN57 | sec8–9::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; GFP-Atg8:: URA 3 [pGFP-Atg8 in pRS316] | This study |
| 15 | sSUN58 | sec10–2::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; GFP-Atg8:: URA 3 [pGFP-Atg8 in pRS316] | This study |
| 16 | sSUN59 | sec15–1::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; GFP-Atg8:: URA 3 [pGFP-Atg8 in pRS316] | This study |
| 17 | sSUN60 | exo70–38::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; GFP-Atg8:: URA 3 [pGFP-Atg8 in pRS316] | This study |
| 18 | sSUN62 | exo84–2::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; GFP-Atg8:: URA 3 [pGFP-Atg8 in pRS316] | This study |
| 19 | sSUN99 | BY4741; Mat a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; ATG1::KanMX4 | Euroscarf |
| 20 | sSUN100 | BY4741; Mat a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; YPT7::KanMX4 | Euroscarf |
| 21 | sSUN105 | BY4741; Mat a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; VAM3::Hph; VAM3ts (p415) | This study |
| 22 | sSUN1 | sec3–2::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; | [43] |
| 23 | sSUN107 | sec3–2::KanR; Mat a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; VAM3::Hph; VAM3ts (p415) | This study |
| 24 | sSUN108 | BY4741; Mat a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; GFP-Atg8:: URA 3 [pGFP-Atg8 in pRS316] | This study |
| 25 | sGRB20 | BY4741; Mat a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; pGFP/N-Aut9 | [44] |
| 26 | sGRB21 | BY4741; Mat a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; ATG1::KanMX4; pGFP/N-Aut9 | [44] |
| 27 | sSUN63 | sec3–2::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; pGFP/N-Aut9 | This study |
| 28 | sSUN101 | sec3–2::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; ATG1::Hph; pGFP/N-Aut9 | This study |
| 29 | sGRB35 | BY4741; Mat a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; pGFP/N-Aut9; pJH1M | [44] |
| 30 | sSUN102 | sec3–2::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; pGFP/N-Aut9; pJH1 | This study |
| 31 | sSUN109 | exo70–38::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; pGFP/N-Aut9 | This study |
| 32 | sSUN110 | exo70–38::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; ATG1::Hph; pGFP/N-Aut9 | This study |
| 33 | sSUN111 | exo70–38::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; pGFP/N-Aut9; pJH1 | This study |
| 34 | sSUN113 | BY4741; Mat a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; ATG1::Hph; ATG27::KanMX; pGFP/N-Aut9 | This study |
| 35 | sSUN73 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; SEC3::SEC3-GFP-HIS | [45] |
| 36 | sSUN74 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; SEC5::SEC5-GFP-HIS | [45] |
| 37 | sSUN75 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; SEC6::SEC6-GFP-HIS | [45] |
| 38 | sSUN76 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; SEC8::SEC8-GFP-HIS | [45] |
| 39 | sSUN77 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; SEC10::SEC10-GFP-HIS | [45] |
| 40 | sSUN78 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; SEC15::SEC15-GFP-HIS | [45] |
| 41 | sSUN79 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; EXO70::EXO70-GFP-HIS | [45] |
| 42 | sSUN80 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; EXO84::EXO84-GFP-HIS | [45] |
| 43 | sSUN103 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; SEC3::SEC3-GFP-HIS; pSUN5 | This study |
| 44 | sSUN104 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; SEC3::SEC3-GFP-HIS; ATG1::Hph; pSUN5 | This study |
| 45 | sSUN114 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; EXO70::EXO70-GFP-HIS; pSUN5 | This study |
| 46 | sSUN115 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; EXO70::EXO70-GFP-HIS; ATG1::Hph; pSUN5 | This study |
| 47 | sSUN116 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; SEC15::SEC15-GFP-HIS; pSUN5 | This study |
| 48 | sSUN117 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; SEC15::SEC15-GFP-HIS; ATG1::Hph; pSUN5 | This study |
| 49 | sSUN81 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; SEC3::SEC3-GFP-HIS; 2XmCherry-Atg8::URA3 [2XmCherry-Atg8 in pRS316] | This study |
| 50 | sSUN118 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; SEC3::SEC3-GFP-HIS; YPT7::Hph; 2XmCherry-Atg8::URA3 [2XmCherry-Atg8 in pRS316] | This study |
| 51 | sSUN121 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; SEC15::SEC15-GFP-HIS; 2XmCherry-Atg8::URA3 [2XmCherry-Atg8 in pRS316] | This study |
| 52 | sSUN122 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; SEC15::SEC15-GFP-HIS; YPT7::Hph; 2XmCherry-Atg8::URA3 [2XmCherry-Atg8 in pRS316] | This study |
| 53 | sSUN119 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; EXO70::EXO70-GFP-HIS; 2XmCherry-Atg8::URA3 [2XmCherry-Atg8 in pRS316] | This study |
| 54 | sSUN120 | MAT a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; EXO70::EXO70-GFP-HIS; YPT7::Hph; 2XmCherry-Atg8::URA3 [2XmCherry-Atg8 in pRS316] | This study |
| 55 | sSUN123 | BY4741; Mat a; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; ATG1::KanMX4; pGFP/N-Aut9; pSUN8 | This study |
| 56 | sSUN124 | sec3–2::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; ATG1::Hph; pGFP/N-Aut9;pSUN8 | This study |
| 57 | sSUN125 | exo70–38::KanR; his3Δ1 leu2Δ0 ura3Δ0 met15Δ0; ATG1::Hph; pGFP/N-Aut9;pSUN8 | This study |
| 58 | GFP-Atg8 in pRS316 | Prof. Yoshinori Ohsumi | |
| 59 | 2XmCherry-Atg8 in pRS316 | Prof. Yoshinori Ohsumi | |
| 60 | pSUN5 | [44] | |
| 61 | pJH1 | Prof. Michael Thumm | |
| 62 | pGFP-N/AUT9 | Prof. Michael Thumm | |
| 63 | Vam3ts in pRS316 | Prof. Rosine Haguenauer-Tsapis [46] | |
| 64 | pSUN8 | Atg1ts allele PCR amplified from pATG1ts 415 (4) and subcloned into pRS315 | This study |
Yeast cells were grown in YPD media (1% yeast extract, 2% peptone and 2% dextrose) or selection media SD-X (0.17% yeast nitrogenous base, 0.5% ammonium sulfate, 2% dextrose, 0.002% uracil, 0.02% histidine, 0.02% methionine, 0.015% lysine and 0.01% leucine), X being the amino acid to be exempted. Cells were starved by incubation in starvation media (SD-N) (0.17% yeast nitrogenous base and 2% dextrose). Oleate medium [2.64 mM K2HPO4, 17.36 mM KH2PO4 (pH 6.0), 0.1% oleic acid with 0.5% Tween-40, 0.25% yeast extract and 0.5% peptone] was used to induce peroxisomes. WT and knockout strains were transformed using lithium acetate method [42]. For Ts strains, the cells were incubated at 25 °C overnight in transformation mix instead of being subjected to a heat shock at 40 °C.
All WT and knockout strains were grown at 30 °C, while Ts strains were grown at 25 °C. Assays were performed at PT (25 °C) or NPT (37 °C).
Pexophagy assay
WT yeast cells expressing Pot1-GFP were a kind gift from Prof. Rachubinski, University of Alberta, Canada. Ts strains were transformed with Pot1-GFP-HIS cassette, which was PCR amplified from the genomic DNA of WT POT1-GFP cells. Actively growing cells expressing Pot1-GFP were transferred to oleate medium (A600 = 1) to induce peroxisome biogenesis [for microscopy, FM4–64 (1 μg/ml) was added to label the vacuoles]. These cells were washed twice with water and starved in SD-N medium (A600 = 3). Cells were collected at mentioned time intervals and either processed for Western blotting or fluorescence microscopy.
GFP-Atg8 processing assay
Cells were transformed with the plasmid GFP-Atg8 cloned in pRS316 vector backbone (a kind gift from Prof. Yoshinori Ohsumi). All strains expressing GFP-Atg8 were grown in SD-Ura medium, and actively growing cells were transferred to starvation medium (SD-N). Cells were harvested at indicated time intervals and TCA precipitated. These precipitates were subjected to SDS-PAGE followed by Western blotting.
Fluorescence microscopy
Cells were mounted on 2% agarose pads on slides. Imaging was performed on DeltaVision microscope (GE Healthcare) using 60× or 100× objective and images were captured with the cool-SNAP HQ camera. Deconvolution and intensity projection of images was done using softWoRx software (GE Healthcare). Colocalization analysis was performed manually or by softWoRx software. Brightness contrast adjustments were made for representation purpose in figures. Quantitation of number of cells showing a particular phenotype or intensity measurements were done using Fiji software (NIH).
Protease protection assay
Cells A600 = 60 were transferred to starvation medium and incubated at 37°Cfor 4 h. Cells were then harvested, washed once and resuspended in 30 ml wash buffer with β-ME (0.6 μl/ml) and incubated for 10 min at 37 °C, 180 rpm. Cells were then resuspended in spheroplasting buffer with zymolyase-20 T (20 mg/ml) and incubated at 37 °C for ~1 h at 70 rpm. The resulting spheroplasts were washed once with spheroplasting buffer and lysed by incubating on ice for 5 min. The lysate was centrifuged at 300g for 10 min at 4 °C. The supernatant was divided into three tubes for control, proteinase K (40 μg/ml) and proteinase K with Triton X-100 (0.2%) treatments. The reactions were incubated on ice for 15 min and then stopped by addition of TCA to a final concentration of 10%, and samples were frozen at −80 °C. The TCA precipitates were washed with acetone, air dried and resuspended in 25 μl SDS-PAGE dye. Samples were separated on 8% SDS-PAGE gels followed by Western blotting.
Western blotting
TCA precipitates were washed with 80% acetone, air dried and boiled with SDS-PAGE dye. A600 = 0.6culture equivalents were loaded onto SDS-PAGE and transferred to PVDF membranes at 100 V for 1–1.5 h. Mouse anti-GFP monoclonal antibody at 1:3000 (Roche Applied Sciences), anti-Ape1 1:5000 (a kind gift from Prof. Yoshinori Ohsumi), anti-mouse and anti-rabbit secondary antibodies conjugated to HRP (Bio-Rad) were used. Blots were developed in G-box Chemi XT4 (Syngene), and band intensities were analyzed using Fiji software (NIH).
Gel filtration
Cultures grown in 2 l YPD to A600 = 0.8–1 were harvested or washed and starved in SD-N at A600 = 3 for 4 h before harvesting. Cell pellets were then resuspended in lysis buffer [20 mM HEPES (pH 7.4), 100 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol, 0.5% NP-40, protease inhibitor cocktail]. Lysis was done by vortexing the cell suspension along with 0.5-mm glass beads (Sigma Aldrich) for 1 min followed by 2-min incubation on ice, for 8–10 times. Lysate was then centrifuged at 13,000g at 4 °C for 10 min, and the resultant supernatant was concentrated before loading onto equilibrated [20 mM Hepes (pH 7.4), 100 mM NaCl) Superose 6 high load 10/300 column (GE Health-care). The flow rate was kept constant at 0.2 ml/min. Fractions (250 μl) were collected and TCA precipitated before loading on 8% SDS-PAGE gels. Western blotting was done, and the relative band intensities were calculated using Fiji software and intensity values were plotted using GraphPad Prism software.
Statistical analysis and image preparation
All statistical analyses were performed by using GraphPad Prism (GraphPad Software). Means of three independent experiments were compared using unpaired two-tailed Student's t-test. Images were processed post-acquisition and prepared using softWoRx software (GE Healthcare). Brightness contrast adjustments were done for visualization purpose during image preparation. Images were then collated using Adobe Photoshop CC.
Supplementary Material
Supplementary data to this article can be found online at 10.1016/j.jmb.2019.04.048.
Acknowledgements
We thank Prof. Charlie Boone (University of Toronto), Prof. Yoshinori Ohsumi (Tokyo Institute of Technology), Prof. Michael Thumm (Georg-August-Universität Göttingen), Dr. Kausik Chakraborty (Institute of Genomics and Integrative Biology), Dr. Richard Rachubinski (University of Alberta), Prof. Daniel Klionsky (University of Michigan), Prof. Kuninori Suzuki (University of Tokyo), Prof. Fulvio Reggiori (University Medical Centre, Groningen), Prof. Scott Emr (Cornell University) and Prof. Rosine Haguenauer-Tsapis (Institut Jacques Monod) for sharing yeast strains, plasmids and antibodies. Thanks to Hariharan and Prem Anand Murugan (IIT-Kanpur) for the technical help in gel filtration experiments. We thank Prof. M.R.S Rao, Aparna Hebbar, Vikas Yadav, Piyush Mishra and Gaurav Barve for critical reading of this manuscript and advice. This work was supported by Wellcome Trust/DBT India Alliance Intermediate Fellowship, Funders: India and UK, Grant number: (500159-Z-09-Z) and intramural funds from JNCASR to R.M.
Abbreviations used
- PAS
pre-autophagosomal structure
- NPT
non-permissive temperature
- Ts
temperature-sensitive
- PT
permissive temperature
- WT
wild-type
- Ape1
Aminopeptidase 1
Footnotes
CRediT authorship contribution statement
Sunaina Singh: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Visualization, Writing - original draft, Writing - review & editing. Ruchika Kumari: Data curation, Formal analysis, Methodology, Software, Validation, Visualization. Sarika Chinchwadkar: Methodology, Software, Validation, Visualization. Amol Aher: Conceptualization, Methodology. Saravanan Matheshwaran: Supervision, Investigation, Methodology, Resources, Writing - review & editing. Ravi Manjithaya: Conceptualization, Formal analysis, Funding acquisition, Project administration, Resources, Supervision, Writing - original draft, Writing - review & editing.
Conflicts of Interest Statement: The authors declare no competing financial interests.
References
- [1].Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290(5497):1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Zhi X, Feng W, Rong Y, Liu R. Anatomy of autophagy: from the beginning to the end. Cellular and molecular life sciences : CMLS. 2018;75(5):815–831. doi: 10.1007/s00018-017-2657-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kim J, Huang WP, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem. 2002;277(1):763–773. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20(21):5971–5981. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, et al. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000;148(3):465–480. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141(4):656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190(6):1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell. 2010;21(22):3998–4008. doi: 10.1091/mbc.E10-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ravikumar B, Moreau K, Jahreiss L, Puri C, Rubinsztein DC. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat Cell Biol. 2010;12(8):747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy. 2005;1(2):101–109. doi: 10.4161/auto.1.2.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198(2):219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hamasaki M, Noda T, Ohsumi Y. The early secretory pathway contributes to autophagy in yeast. Cell Struct Funct. 2003;28(1):49–54. doi: 10.1247/csf.28.49. [DOI] [PubMed] [Google Scholar]
- [13].Tan D, Cai Y, Wang J, Zhang J, Menon S, Chou HT, et al. The EM structure of the TRAPPIII complex leads to the identification of a requirement for COPII vesicles on the macroautophagy pathway. Proc Natl Acad Sci U S A. 2013;110(48):19432–19437. doi: 10.1073/pnas.1316356110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yen WL, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, et al. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol. 2010;188(1):101–114. doi: 10.1083/jcb.200904075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lemus L, Ribas JL, Sikorska N, Goder V. An ER-localized SNARE protein is exported in specific COPII vesicles for autophagosome biogenesis. Cell Rep. 2016;14(7):1710–1722. doi: 10.1016/j.celrep.2016.01.047. [DOI] [PubMed] [Google Scholar]
- [16].Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146(2):290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhao S, Li CM, Luo XM, Siu GK, Gan WJ, Zhang L, et al. Mammalian TRAPPIII complex positively modulates the recruitment of Sec13/31 onto COPII vesicles. Sci Rep. 2017;7:43207. doi: 10.1038/srep43207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Brunet S, Shahrzad N, Saint-Dic D, Dutczak H, Sacher M. A trs20 mutation that mimics an SEDT-causing mutation blocks selective and non-selective autophagy: a model for TRAPP III organization. Traffic. 2013;14(10):1091–1104. doi: 10.1111/tra.12095. [DOI] [PubMed] [Google Scholar]
- [19].Lipatova Z, Majumdar U, Segev N. Trs33-containing TRAPP IV: a novel autophagy-specific Ypt1 GEF. Genetics. 2016;204(3):1117–1128. doi: 10.1534/genetics.116.194910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Taussig D, Lipatova Z, Segev N. Trs20 is required for TRAPP III complex assembly at the PAS and its function in autophagy. Traffic. 2014;15(3):327–337. doi: 10.1111/tra.12145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang J, Menon S, Yamasaki A, Chou HT, Walz T, Jiang Y, et al. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc Natl Acad Sci U S A. 2013;110(24):9800–9805. doi: 10.1073/pnas.1302337110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].TerBush DR, Maurice T, Roth D, Novick P. The exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15(23):6483–6494. [PMC free article] [PubMed] [Google Scholar]
- [23].Pfeffer SR. Transport-vesicle targeting: tethers before SNAREs. Nat Cell Biol. 1999;1(1):E17–E22. doi: 10.1038/8967. [DOI] [PubMed] [Google Scholar]
- [24].Mei K, Li Y, Wang S, Shao G, Wang J, Ding Y, et al. Cryo-EM structure of the exocyst complex. Nat Struct Mol Biol. 2018;25(2):139–146. doi: 10.1038/s41594-017-0016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Munson M, Novick P. The exocyst defrocked, a framework of rods revealed. Nat Struct Mol Biol. 2006;13(7):577–581. doi: 10.1038/nsmb1097. [DOI] [PubMed] [Google Scholar]
- [26].Heider MR, Gu M, Duffy CM, Mirza AM, Marcotte LL, Walls AC, et al. Subunit connectivity, assembly determinants and architecture of the yeast exocyst complex. Nat Struct Mol Biol. 2016;23(1):59–66. doi: 10.1038/nsmb.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Guo W, Sacher M, Barrowman J, Ferro-Novick S, Novick P. Protein complexes in transport vesicle targeting. Trends Cell Biol. 2000;10(6):251–255. doi: 10.1016/s0962-8924(00)01754-2. [DOI] [PubMed] [Google Scholar]
- [28].Finger FP, Hughes TE, Novick P. Sec3p is a spatial landmark for polarized secretion in budding yeast. Cell. 1998;92(4):559–571. doi: 10.1016/s0092-8674(00)80948-4. [DOI] [PubMed] [Google Scholar]
- [29].Guo W, Roth D, Walch-Solimena C, Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 1999;18(4):1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Luo G, Zhang J, Guo W. The role of Sec3p in secretory vesicle targeting and exocyst complex assembly. Mol Biol Cell. 2014;25(23):3813–3822. doi: 10.1091/mbc.E14-04-0907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Moskalenko S, Tong C, Rosse C, Mirey G, Formstecher E, Daviet L, et al. Ral GTPases regulate exocyst assembly through dual subunit interactions. J Biol Chem. 2003;278(51):51743–51748. doi: 10.1074/jbc.M308702200. [DOI] [PubMed] [Google Scholar]
- [32].Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell. 2011;144(2):253–267. doi: 10.1016/j.cell.2010.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].He B, Guo W. The exocyst complex in polarized exocytosis. Curr Opin Cell Biol. 2009;21(4):537–542. doi: 10.1016/j.ceb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Grote E, Carr CM, Novick PJ. Ordering the final events in yeast exocytosis. J Cell Biol. 2000;151(2):439–452. doi: 10.1083/jcb.151.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Manjithaya R, Jain S, Farre JC, Subramani S. A yeast MAPK cascade regulates pexophagy but not other autophagy pathways. J Cell Biol. 2010;189(2):303–310. doi: 10.1083/jcb.200909154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci. 2013;126(Pt 11):2534–2544. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- [37].Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6(1):79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- [38].Lang T, Reiche S, Straub M, Bredschneider M, Thumm M. Autophagy and the cvt pathway both depend on AUT9. J Bacteriol. 2000;182(8):2125–2133. doi: 10.1128/jb.182.8.2125-2133.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Heider MR, Munson M. Exorcising the exocyst complex. Traffic. 2012;13(7):898–907. doi: 10.1111/j.1600-0854.2012.01353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gomez-Sanchez R, Rose J, Guimaraes R, Mari M, Papinski D, Rieter E, et al. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J Cell Biol. 2018;217(8):2743–2763. doi: 10.1083/jcb.201710116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Guo W, Grant A, Novick P. Exo84p is an exocyst protein essential for secretion. J Biol Chem. 1999;274(33):23558–23564. doi: 10.1074/jbc.274.33.23558. [DOI] [PubMed] [Google Scholar]
- [42].Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- [43].Li Z, Vizeacoumar FJ, Bahr S, Li J, Warringer J, Vizeacoumar FS, et al. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol. 2011;29(4):361–367. doi: 10.1038/nbt.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Barve G, Sridhar S, Aher A, Sahani MH, Chinchwadkar S, Singh S, et al. Septins are involved at the early stages of macroautophagy in S. cerevisiae. J Cell Sci. 2018;131(4) doi: 10.1242/jcs.209098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- [46].Erpapazoglou Z, Dhaoui M, Pantazopoulou M, Giordano F, Mari M, Leon S, et al. A dual role for K63-linked ubiquitin chains in multivesicular body biogenesis and cargo sorting. Mol Biol Cell. 2012;23(11):2170–2183. doi: 10.1091/mbc.E11-10-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.