Abstract
Genome-wide association studies (GWAS) associate single nucleotide polymorphisms (SNPs) to complex phenotypes. Most human SNPs fall in non-coding regions and are likely regulatory SNPs, but linkage disequilibrium (LD) blocks make it difficult to distinguish functional SNPs. Therefore, putative functional SNPs are usually annotated with molecular markers of gene regulatory regions and prioritized with dedicated prediction tools. We integrated associated SNPs, LD blocks and regulatory features into a supervised model called TAGOOS (TAG SNP bOOSting) and computed scores genome-wide. The TAGOOS scores enriched and prioritized unseen associated SNPs with an odds ratio of 4.3 and 3.5 and an area under the curve (AUC) of 0.65 and 0.6 for intronic and intergenic regions, respectively. The TAGOOS score was correlated with the maximal significance of associated SNPs and expression quantitative trait loci (eQTLs) and with the number of biological samples annotated for key regulatory features. Analysis of loci and regions associated to cleft lip and human adult height phenotypes recovered known functional loci and predicted new functional loci enriched in transcriptions factors related to the phenotypes.
In conclusion, we trained a supervised model based on associated SNPs to prioritize putative functional regions. The TAGOOS scores, annotations and UCSC genome tracks are available here: https://tagoos.readthedocs.io.
INTRODUCTION
Complex human phenotypes arise from a contribution of genetic and environmental factors. Genome-wide association studies (GWAS) is a key technique to link genetic loci to human phenotypes (1). For instance, a single GWAS detected more than 400 loci associated to adult human height (2). Given the large amount and importance of GWAS, several databases have compiled GWAS results (3–5)
The large majority of associated SNPs fall in intronic and intergenic regions (6). This association is likely mediated through gene regulatory regions and enhancers, as for instance, non-coding associated loci are enriched in eQTLs and DNase I hypersensitive sites (DHS) (7,8). Enhancers of gene expression are non-coding regions at any distance of regulated genes with particular chromatin configurations that facilitate transcription factor (TF) binding and gene expression (9). The chromatin of enhancers is DNase I hypersensitive, marked by particular histone modifications such as histone H3 lysine 4 monomethylation (H3K4me1) and H3K4me2, histone H3 lysine 27 acetylation (H3K27ac) marks and produces enhancer RNAs. The three dimensional structure of the chromosome then brings closer enhancers and transcription start sites, for the TFs to recruit RNA polymerase and promote gene transcription (10).
The resolution of associated loci is limited by linkage disequilibrium (LD) blocks of correlated SNPs. In addition, mechanistic understanding of the association requires the analysis of the loci at the molecular level. To prioritize and understand functional non-coding associated loci, the simplest option is to annotate the loci with key regulatory features such as H3K27ac or ChromHMM chromatin states (11). Regarding dedicated computational tools, there are two broad families to prioritize functional loci in LD blocks associated to complex diseases (Supplementary Tables S1 and S2). The first family of computational tools, which we call GWAS loci priorization approaches, link GWAS loci, LD blocks and regulatory annotations. Tools like FunciSNP or HaploReg retrieve SNPs above an LD cutoff and annotate them with gene regulatory annotations (12,13) (Supplementary Table S1). More elaborated methods such as GWAS3D, GREGOR and GenoWAP calculate probabilities for the set of GWAS loci, which take into account the enrichment of the GWAS loci annotated with regulatory features within the LD blocks (Supplementary Table S1) (4,14,15). These methods usually do not provide pre-calculated scores genome-wide and therefore they cannot be easily compared to each other. The second family of computational tools, which we call regulatory variant priorization approaches, use statistical or classification methods to learn models from regulatory variants with known medical impact based on signatures of regulatory annotations. An important difference is that regulatory variant priorization approaches do not take into account the LD structure of reference populations. Regulatory variant priorization approaches can be divided again in supervised and unsupervised approaches. In supervised methods such as CADD, GWAVA, DANN, FATHMM-MKL, DIVAN, REMM and LINSIGHT, a set of known functional variants is used as input to create a model (Supplementary Table S2) (16–22). In unsupervised priorization methods such as FunSeq2, DeepSea, FitCons, GenoCanyon, EIGEN, IW-Scoring and PINES, the score is calculated in the absence of known functional variants (Supplementary Table S2) (15,23–28). Regulatory variant priorization approaches usually provide pre-computed scores genome-wide and have been tested on GWAS loci with moderate to low success (AUC ≤0.6) (27).
Here, we train a supervised model called TAGOOS (TAG SNP bOOsting) based on non-coding associated loci and regulatory features that takes into account the LD structure of the reference population. The TAGOOS score is able to prioritize and enrich unseen non-coding associated loci at higher levels than most existing variant priorization approaches. We use the TAGOOS score to analyze two loci associated to nonsyndromic cleft lip and adult human height. The TAGOOS score recovers known gene regulatory regions and SNPs within the two loci and make new predictions.
METHODS
Supervised classification using XGBOOST
XGBOOST is a software library for supervised classification based on the gradient boosting technique. The gradient boosting technique uses ensembles of regression trees constructed with the boosting meta-algorithm. The XGBOOST software has several advantages over othe gradient boosting libraries. Regularization has been formalized in the objective function to better control over-fitting. The XGBOOST library has been optimized for large data sets. We used the python framework with the binary logistic objective function and parameters colsample_bytree 1, eta 0.3, max_delta_step 1, max_depth 6, min_child_weight 1, num_boost_round 10 and subsample 1 for both intronic and intergenic models.
Training
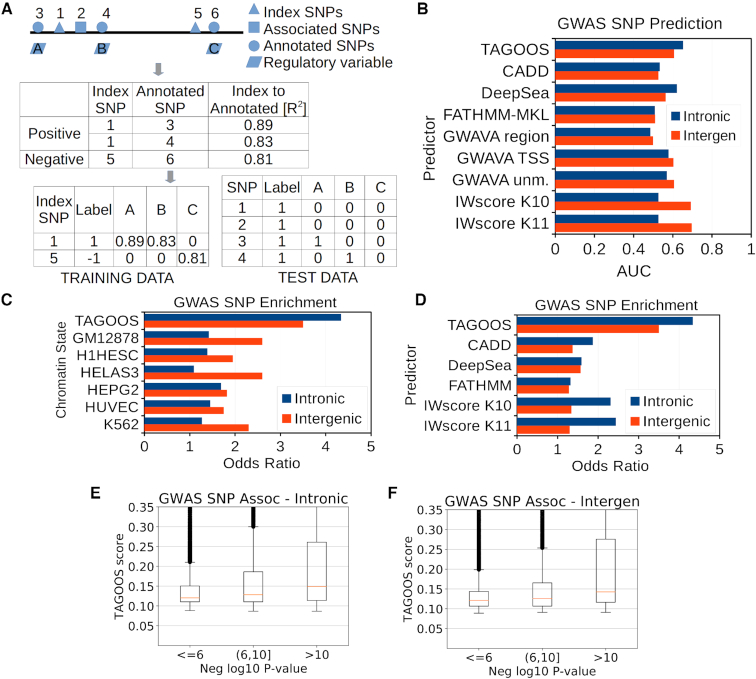
Training SNPs that were common (MAF > 1%), belonged to the European population and to chromosomes 1–22 were selected from the 1000 Genome data set using plink (v1.90b4.4 64-bit) (29). Intronic and intergenic SNPs were defined relative to RefSeq gene annotations. For intergenic SNPs, 1 kb gene upstream regions were excluded to avoid gene promoters during the training. The following steps were carried out to create and train the model. Index SNPs were generated with plink indep option: 5, 1, 100. Linkage disequilibrium (LD) between training SNPs was computed using plink ld-snp-list limited to windows of 1000 kb, 1 000 000 SNPs and r2 of 0.8. SNPs were defined as associated if the SNP showed at least one significant association (P < 5 × 10−8). Expression phenotypes present in the GRASP database were removed before the training step. Index SNPs were labelled as positive if they were in LD with an associated SNP and negative otherwise (Figure 1A). The training matrix contained 2 834 469 index SNPs (2.2% positive) in the intronic regions and 3 454 116 (1.6% positive) in the intergenic regions. Index SNPs labels were annotated with the LD r2 value whenever r2 ≥ 0.8 to annotated SNPs or 0 otherwise (Figure 1A). In Figure 2A, B and Supplementary Tables S5 and S6, the feature gain importance type corresponds to the average accuracy gain brought by a feature to the branches it is on (http://xgboost.readthedocs.io, accessed 14 February 2018). Performance of the training data set was evaluated using a leave-one-chromosome-out cross-validation strategy using each of the 22 somatic chromosomes (Supplementary Figure S2).
Figure 1.
A GWAS SNP prediction model based on gene regulatory annotations. (A) Computation of training and test data sets from LD pruned index SNPs, associated SNPs from the GRASP database and annotated SNPs with regulatory features. (B) AUC values of intronic and intergenic GWAS Catalog SNPs calculated with TAGOOS, CADD, DeepSea, FATHMM-MKL, GWAVA and IW-Scoring scores. (C) Odds ratio of GWAS Catalog SNP enrichment in significant regions (P<0.05) computed with TAGOOS, CADD, DeepSea, FATHMM and IW-Scoring. (D) Odds ratio of GWAS Catalog SNP enrichment in TAGOOS-significant or cell-specific regulatory (promoter flanking region, weak or strong enhancer) chromatin state regions. (E, F) Box plots of TAGOOS scores of GWAS Catalog SNPs split by the negative decimal logarithm of the GWAS P-value in intronic and intergenic regions.
Figure 2.
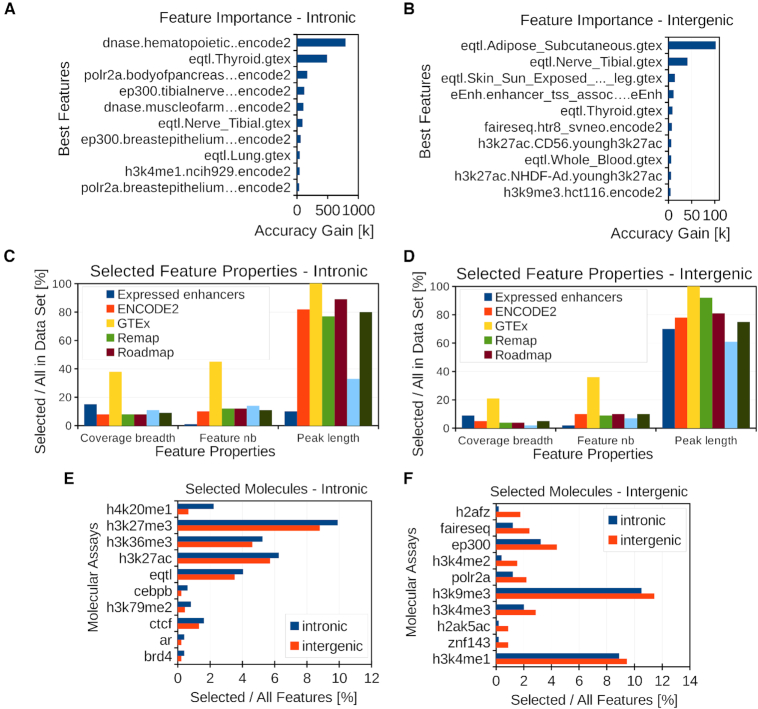
Analysis of selected features. (A, B) Best ten predictor features in the intronic and intergenic models assessed by the XGBOOST gain feature importance type. The gain feature importance represents the average accuracy gain brought by a feature to the branches it is on. (C, D) Percentage of total length, feature number and average peak length between selected and initial features for given data sets in the intronic and intergenic regions. (E, F) Percentage of selected features for given molecular assays with higher percentage in intronic (E) or intergenic (F).
Genome-wide TAGOOS scores and P-values
The selected features of the intronic (495 features) and intergenic (455 features) models were used to annotate intronic and intergenic genomic regions, respectively (Supplementary Tables S5 and S6). The genomics regions were binned based on genomic annotation groups and scored. One million intronic or intergenic positions were sampled to calculate the TAGOOS P-value as one minus the empirical cumulative distribution function of the random scores (Supplementary Figure S3).
Analysis of binding of transcriptional regulators and mouse ontology
SNPs were split according to the TAGOOS significance and annotated using non-redundant TFs from the ReMap database (30,31). For each TFs, the percentage of bound SNPs with either TAGOOS-significant or non-significant scores was calculated and assessed with a paired Wilcoxon test for transcriptional regulators (Figures 4C, F, 5C, D and 6C–F). The TFs were sorted according to the difference of percentage of bound SNPs with significant and non-significant scores and plotted (Figures 4C, F, 5C and 6C, D). In Figures 5D and 6E, F, only the first 10 TFs were plotted. This list of TFs in Figures 4C, F, 5C and 6C, D) sorted by the binding difference between TAGOOS-significant and non-significant SNPs was split in two equal groups and submitted separately or together to the EnrichR web site (32). The EnrichR combined scores of mouse MGI phenotypes related to orofacial or body size phenotypes were plotted (Figures 5E and 6G, H).
Figure 4.
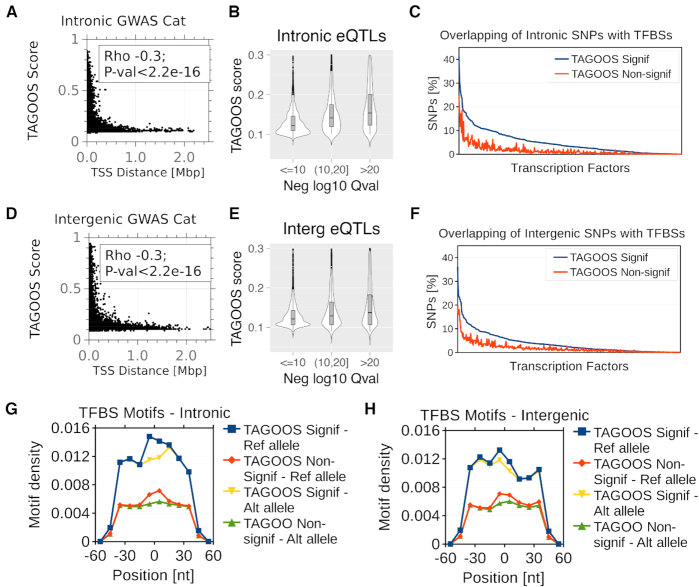
Functional properties of the TAGOOS scores. (A, D) Scatter plots and Spearman correlation coefficient and P-value of the distance to the nearest transcription start sites (TSS) and the TAGOOS scores for unseen GWAS Catalog SNPs in intronic (A) and intergenic (D) regions. (B, E) Violin plots of TAGOOS scores of unseen GTEX eQTLs split by the negative decimal logarithm of the maximal Q-value association in intronic (b) and intergenic (e) regions. (C, F) Percentage of intronic (C) and intergenic (F) unseen GWAS Catalog SNPs with significant and non-significant TAGOOS scores annotated with each transcriptional regulator from the ReMap database ordered by the decreasing difference between the blue (TAGOOS Signif) and red (TAGOOS non-significant) lines (PWilcoxpaired = 2.2 × 10−16). (G, H) Motif density in 10 nt bins in a 100 nt window around intronic (G) and intergenic (H) unseen GWAS Catalog SNPs split according to the TAGOOS significance and the reference or most common alternative allele.
Figure 5.
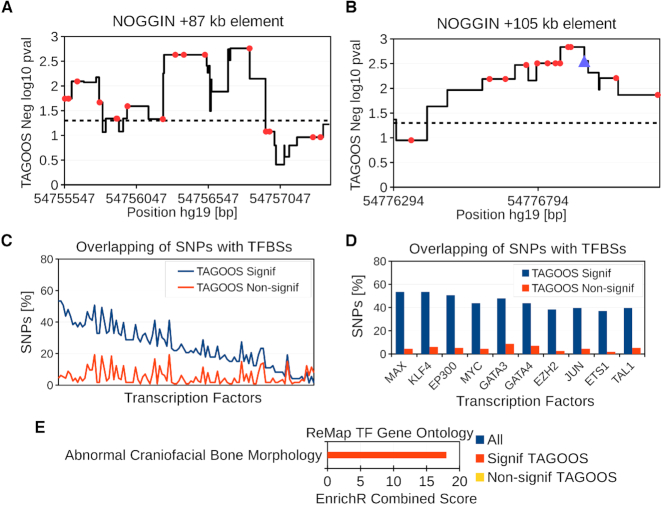
Analysis of the rs227727 linkage disequilibrium (LD) block (hg19, chr17:54,752,926-54,778,620). (A, B) Negative decimal logarithm of the TAGOOS P-value for the +87 kb NOGGIN (chr17:54,754,994-54,757,864) and +105 kb NOGGIN regions (hg19,chr17:54,776,180-54,777,328) with common SNPs (Red points) and the previously validated SNP rs227727 (Blue triangle). The horizontal dashed line stands for the significance threshold. (C) Percentage of SNPs in the rs227727 LD block with significant and non-significant TAGOOS scores annotated with each transcriptional regulator from the ReMap database ordered by the decreasing difference between the blue (TAGOOS Signif) and red (TAGOOS Non-signif) lines (PWilcoxpaired = 2.2 × 10−16). (D) Zoom into the first ten transcription factors of subfigure (C). (E) Mouse craniofacial phenotype annotation enrichment of TFs with highest (Signif TAGOOS) or lowest (Non-signif TAGOOS) binding difference between TAGOOS-significant and non-significant SNPs.
Figure 6.
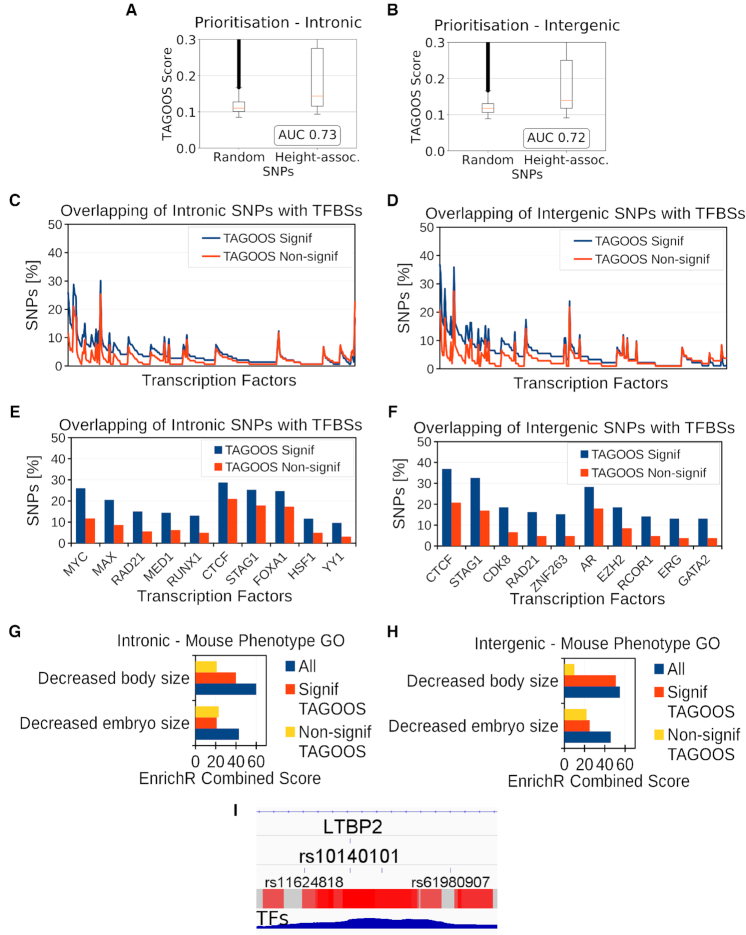
Height GWAS analysis with the TAGOOS scores. (A, B) Box plot and AUC performance of unseen height GWAS SNPs in intronic (A) and intergenic (B) regions. (C, D) Percentage of unseen intronic (C) and intergenic (D) height SNPs with significant and non-significant TAGOOS scores annotated with each transcriptional regulator from the ReMap database ordered by the decreasing difference between the blue (TAGOOS Signif) and red (TAGOOS Non-signif) lines (PWilcoxpaired = 2.2 × 10−16). (E, F) Zoom into the first ten transcription factors of subfigures (C, D). (G, H) Mouse body size phenotype annotation analysis of ReMap TFs with with highest (Signif TAGOOS) or lowest (Non-signif TAGOOS) binding difference between intronic (G) and intergenic (H) TAGOOS-significant and non-significance SNPs. (I) Screenshot of the IGV genome browser showing the TAGOOS score and ReMap TF coverage around the height associated SNP rs10140101. The trackers show the LTBP2 intron, the rs10140101 SNP, surrounding SNPs as reference, and non-redundant ReMap TF coverage.
Motif analysis
Input sequences were built with a 100 bp sequence around each SNP with either the RefSeq allele or the most common alternative allele. Sequences of TAGOOS-significant or non-significant SNPs with the RefSeq allele were used to create the background model with Markov parameter 1. A library of non-redundant motifs based on RSAT matrix-clustering and the HOCOMOCO database was used (33,34). The matrix-scan tool of the RSAT suite was run to detect significant motifs (P < 1 × 10−4) (35). The motif density was plotted in 10 bp bins of the 100 bp sequences (Figure 4G, H).
Data sets, scores and software
SNPs were downloaded from here: GRASP (3), NCBI dbSNP (36), 1000 Genomes databases (37), GWAS Catalog (5), ClinVar (38), a height GWAS (2) and a cleft lip association study (11) (Supplementary Table S3). Annotations were downloaded from here: expressed enhancers (39), eQTLs (40), ReMap (30,31), RoadMap (41) and H3K27ac from the Young laboratory (42) (Supplementary Table S3).
Training and test pipelines were implemented with Snakemake (Supplementary Figure S1) (43). SNP RS identifiers were converted to coordinate bed files using the UCSC mysql server (44). Coordinate bed files were converted to fasta sequences using the UCSC twoBitToFa tool (44). Peak bed files were manipulated using bedtools (v2.26.0) (45). The Integrative Genomics Viewer was used to browse the genome (46). Heatmaps (pheatmap package), correlogram (corrplot package), factor analysis and statistical tests were carried out with the R software.
The TAGOOS scores, P-values, negative decimal logarithms of the P-values, annotations and UCSC tracks are provided here: https://tagoos.readthedocs.io. The developer documentation and scripts to download the data sets, annotate the SNPs, train the model and score the genome can be found here: https://github.com/aitgon/tagoos.
RESULTS
Training data set and model performance
First, we computed a set of pruned index SNPs for chromosomes 1–22 that are in approximate linkage equilibrium with each other (Figure 1A, Supplementary Figure S1). Index SNPs were labelled as positive or negative depending on whether index SNPs were in LD (r2 > 0.8) or not with associated SNPs from the GRASP database (3). Then, we annotated 1000 genome database SNPs with 4684 molecular features from various public databases related to non-coding gene regulatory sequences such as eQTLs, H3K27ac or transcription factors (Figure 1, Table 1, Supplementary Tables S3 and S4). Index SNPs were annotated with the linkage disequilibrium (LD) r2 between the index SNP and the annotated SNP and the annotation (Figure 1A). This so-called training data set was used to train the XGBOOST algorithm (47). Preliminary analysis showed that the most relevant chromatin marks were different in intronic and intergenic regions. Therefore we created two models for intronic and intergenic regions based on the 22 somatic chromosomes.
Table 1.
Size of the annotation data sets
| Peak Number [M] | Total Length [Mb] | Feature Number | Peak Length Average [bp] | |
|---|---|---|---|---|
| Expressed enhancers | 0.2 | 12,407 | 115 | 58,505 |
| ENCODE | 264.3 | 122,143 | 2,864 | 462 |
| GTEx | 16.4 | 16 | 44 | 1 |
| ReMap | 13 | 4,598 | 550 | 354 |
| Roadmap | 160 | 67,906 | 1,025 | 424 |
| Young H3K27ac | 1.1 | 7,454 | 86 | 6,817 |
| TOTAL | 455 | 214,525 | 4,684 | 417 |
To find optimal parameters and calculate the performance, we carried out a leave-one-chromosome-out cross-validation procedure where a model was trained based on a set of chromosomes and the model was tested in a different chromosome. The average area under the curve (AUC) performance for the associated SNPs (P < 5 × 10−8) from the GRASP database versus random SNPs was ∼0.65 for both the intronic and intergenic SNPs (Supplementary Figure S2). This result suggests that the training data set is able to produce models that enrich unseen associated SNPs.
Then we annotated intronic and intergenic regions genome-wide with selected features and computed the TAGOOS scores (Figure 1A). To evaluate the TAGOOS score with unseen GWAS SNPs, we downloaded the GWAS Catalog and removed SNPs present in the GRASP database (5). Prediction of associated SNPs in the GWAS Catalog SNPs versus random DBSNP SNPs reached AUC values of ∼0.65 for the intronic and 0.60 for the intergenic model (Figure 1B, C). We compared the AUC values of TAGOOS with other priorization tools of functional SNPs, namely CADD, DeepSea, FATHMM-MKL, GWAVA and IW-Scoring (16,17,19,24,27). Generally, TAGOOS showed better AUCs than the other priorization tools. A notable exception is IW-Scoring for intergenic regions, which reached AUC 0.7 in intergenic regions, while TAGOOS showed only AUC 0.6 (Figure 1B). In the same region, GWAVA and TAGOOS performances were roughly equivalent with AUC 0.6 (Figure 1B). These AUC values were generally consistent with a recent survey of functional SNP priorization tools for priorization of GWAS SNPs that included IW-Scoring, Eigen/EigenPC, DeepSea, Funseq2, LINSIGHT, FATHMM-NC, GWAVA, CADD, ReMM and FitCons tools. In this survey, none of the tools reached AUC values of 0.6 for priorization of GWAS versus randomly selected noncoding SNPs from the 1000 genomes database (27). The difference between the IW-Scoring AUC performance here (AUC 0.7) (Figure 1B) and the original reported AUC 0.6 (27) might arise, because in the original publication of IW-Scoring, no difference between intergenic and intronic SNPs was made and the negative data set came from the 1000 genome database (27).
Another strategy to enrich functional SNPs within a linkage disequilibrium (LD) block is to annotate the LD blocks with gene regulatory annotations (11). A particularly popular method is to learn so-called chromatin states such as active enhancers based on annotations related to gene regulation using Hidden Markov Models (11,48). To compare TAGOOS with these approaches, we computed TAGOOS P-values across intronic and intergenic regions (See Methods) (Supplementary Figure S3). These P-values allow us to partition the genome in significantly 5% functional versus 95% non-functional regions based on a P < 0.05 threshold. First, we generated 50 000 regions of 1000 bp and split them according to the TAGOOS P-value. Then we computed the probability of finding an unseen associated SNP from the GWAS Catalog in any SNP within the TAGOOS-significant or non-significant regions and calculated an odds ratio. We also carried out the same protocol using as functional regions the union of predicted TSS, promoter flanking regions and weak and strong enhancers chromatin states generated by a combination of the ChromHMM and Segway tools in the GM12878, H1hesc, HelaS3, HEGP2, HUVEC and K562 cell types (48). In the case of intronic regions, we found an odds ratio of 4.3 (PFisher < 2.2 × 10−16) using the TAGOOS P-value compared to odds ratios of around 2 for the chromatin state annotations (Figure 1C). In the case of intergenic regions, we found an odds ratio of ∼3.5 (PFisher < 2.2 × 10−16) with the TAGOOS P-value compared to values of ∼1.5 for chromatin state annotations (Figure 1C).
We also evaluated the enrichment of GWAS signals in functional regions predicted by significant scores for regulatory variant prioritization methods that provide P-values, such as TAGOOS, CADD, DeepSea, FATHMM and IW-Scoring (Figure 1D). We defined functional regions based on scores at a significance threshold P < 0.05. We found that TAGOOS is able to enrich better at selected significance thresholds than the other tools (Figure 1D).
It has been suggested that ENCODE overrepresented tissues such as blood and immune cell types create a prediction bias for related diseases (49). To evaluate this bias, we split associated SNPs by GWAS traits that belonged to five different experimental factor ontology (EFO) categories (Supplementary Figure S4). We computed AUC values of trait-dependent associated SNPs against random SNPs. We found that traits related to immune system diseases and hematological measurements showed higher median AUC values than other diseases such as nervous system diseases (Supplementary Figure S4). This observation agreed with a systematic bias towards overrepresented cell types in ENCODE that merits special attention in particular when using phenotype-dependent priorization methods such as PINES (28).
We next examined whether there exists a correlation between the significance of genetic associations and the TAGOOS scores. We found a positive correlation in both intronic (ρSpearman = 0.19, PSpearman < 2.2 × 10−16) and intergenic regions (ρSpearman = 0.15, PSpearman < 2.2 × 10−16) (Figure 1E, F). We split SNPs according to the negative decimal logarithm of the most significant association P-value and examined the distribution of the TAGOOS scores in different groups of SNPs. We observed that the distribution of TAGOOS scores increases with the negative decimal logarithm of the association P-values (Figure 1E, F). This suggests that the maximal association significance observed genome-wide across phenotypes is a predictable consequence of the molecular context.
ClinVar is a golden standard for benchmarking predictors of highly penetrant SNPs. Priorization of ClinVar SNPs showed low AUC values 0.56 and 0.65 for intronic and intergenic values suggesting that other tools trained on this data set are more appropriate to prioritize these type of SNPs (Supplementary Figure S5).
These results showed that the TAGOOS method achieved higher priorization and enrichment performances than other popular bioinformatics tools for GWAS SNPs.
Analysis of the features of the TAGOOS model
In the TAGOOS model, a feature is composed of the molecular assay (e.g. a DNase-seq, eQTL or H3K27ac), the biological sample (e.g. a tissue or cell type) and the data set name (e.g. GTEx or ENCODE). We used 4684 annotations composed of 718 molecular assays in 1284 biological samples and 6 data sets (Table 1 and Supplementary Tables S3 and S4). The molecular assays comprised eQTLs, transcription factors, histone modifications, DNA accessibility and expressed enhancers (Supplementary Table S4). For the intronic model, the learning algorithm selected 495 features composed of 155 molecular assays in 311 biological samples (Supplementary Table S5). For the intergenic model, the learning algorithm selected 455 features composed of 125 molecular assays in 289 biological samples (Supplementary Table S6). Among the 10 most predictive features, there were eQTLs, open chromatin regions and H3K27ac annotations (Figure 2A, B). These molecular annotations are found in non-coding functional SNPs and gene regulatory regions (7,50,51). To further gain insight into the molecular properties of the model, we examined the percentage of selected feature number, coverage breadth and average peak length in each data set (Figure 2C, D). We found high percentage of selected eQTL features and coverage breadth in both intronic and intergenic regions (Figure 2C, D).
To look for differences between the intronic and intergenic models, we compared the percentage of selected biological samples for each molecular assay relative to the total sample number in each model. We found that the percentage of biological samples with H3K36me3, H3K79me2 and H4K20me1 was higher for the intronic model (Figure 2E). This agrees with their known enrichment in the gene bodies and promoters (52,53). By contrast, the intergenic model had a higher percentage of biological samples related to DNA accessibility (FAIRE-Seq) and transcription (POLR2A) (Figure 2F).
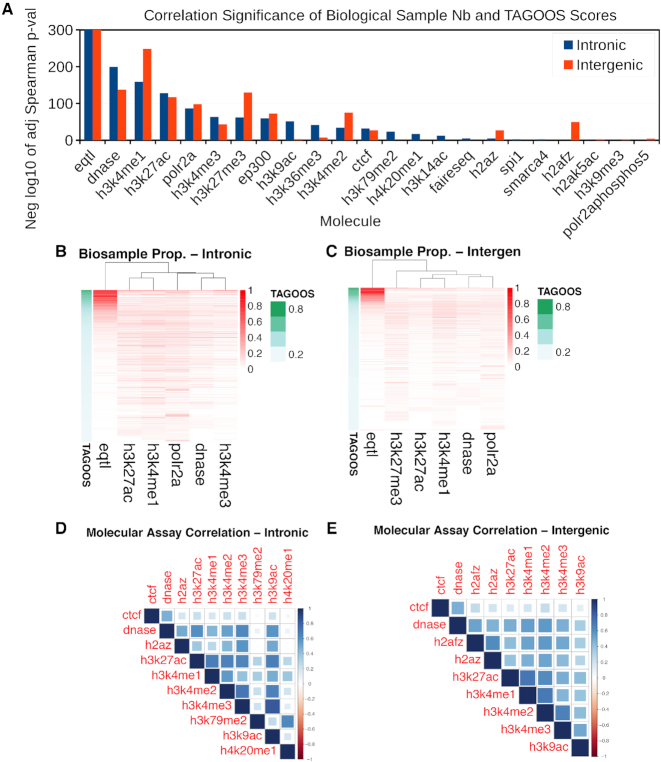
To further gain insights into the contribution of different features to the models, we annotated unseen GWAS catalog SNPs with the TAGOOS scores and selected features. We calculated for each SNP the proportion of biological samples out of the maximal sample number of a given molecular assay. Then we selected and plotted heatmaps of molecular assays with a significant correlation (Bonferroni-corrected PSpearman < 5 × 10−2) between the sample proportion and the TAGOOS score (Figure 3B, C). We found significant correlations between the TAGOOS score and the number of biological samples with typical markers of regulatory regions such as eQTLs, DNAse-seq, H3K4me1 or H3K27ac (Figure 3A). Typical markers of intronic regions such H3K36me3, H3K79me2 and H4K20me1 showed higher correlation in intronic regions (Figure 3A). On the other hand, the correlation of H3K4me1 and H3K27me3 with the TAGOOS score was more significant in intergenic regions (Figure 3A). These molecular markers were consistent with known molecular markers of functional intronic and intergenic regulatory regions. eQTLs were clearly found in many biological samples for SNPs with high TAGOOS scores (Figure 3B, C). Other markers such as DNAse-seq and H3K4me1 were correlated statistically but not visually (Figure 3A–C).
Figure 3.
Correlation of biological sample number for given molecular assays with the TAGOOS score in unseen GWAS Catalog SNPs. We created a matrix with the proportion of biological samples out of the maximal sample number for given molecular assays. (A) Negative decimal logarithm of the adjusted (Bonferroni) PSpearman-value (PSpearman < 0.05) of the Spearman correlation between biological sample proportion and the TAGOOS score ordered by the maximal significance. (B, C) Heatmap of the biological sample proportion matrix with the SNPs in the rows ordered by the TAGOOS score (Green column). In the columns, the six more correlated assays from subfigure (A) were selected for intronic and intergenic regions. (D, E) Pairwise correlation between molecular asays based on the biological sample proportions.
Next we carried out the same analysis using random SNPs from the DBSNP databases. We randomly sampled 105 common SNPs from the dbSNP database and annotated them with the TAGOOS scores and annotations. Then we looked for significant correlations between the biological sample proportion of each molecular assay and the TAGOOS scores. We also found that markers of regulatory regions such as eQTLs, H3K27ac and H3K4me1 were correlated with the TAGOOS score (Supplementary Figure S6). Altogether, these results suggested that the number of biological samples with given annotations such as eQTL in a SNP correlated with the TAGOOS score of that SNP. This also implied that the number of biological samples positive for an annotation such as eQTL at given SNP was also a predictor of the maximal association significance for that SNP.
To look for groups of correlated features, we carried out a factor analysis with four factors. Then we selected and plotted ∼6–10 features with highest contributions to at least one of the factors (Supplementary Figure S7). We also plotted pairwise correlations between the same features (Figure 3D, E). We found the markers of active gene regulatory regions such as H3K27ac and H3K4me1 form strong correlated groups in both regions (Supplementary Figure S7 and Figure 3D, E). CTCF and DNase signals were also strongly correlated in agreement with previous observations (54,55). On the other hand, H3K79me2 and H4K20me1 belonged to a correlated group in the intronic SNPs (Supplementary Figure S7A and Figure 3D). This agrees with the enrichment of H3K79me2 and H4K20me1 in the gene bodies of actively transcribed genes (52).
Functional properties of TAGOOS scores
We expected TAGOOS-significant SNPs to show more functional molecular properties, such as being closer to transcription start sites (TSS). Therefore we hypothesized that the TAGOOS score negatively correlates with the distance to genes and TSSs. To test this hypothesis, we plotted TAGOOS scores as a function of the distance to the TSS for a number of random intronic and intergenic SNPs (Figure 4A, D). We found a negative correlation between the TAGOOS score and the distance to the TSS for intronic (ρSpearman = −0.3, PSpearman < 2.2 × 10−16) and intergenic SNPs (ρSpearman = −0.3, PSpearman < 2.2 × 10−16) (Figure 4A, D).
We found that there is a relationship between the number of biological samples with given eQTLs and the TAGOOS scores of the eQTLs (Figure 3A–C). Priorization of eQTLs was shown to involve markers of active regulatory sequences such as DNase I hypersensitive sites (DHS) and histone marks similarly to the TAGOOS score (56). Therefore, we hypothesized that the TAGOOS score correlates with the maximal eQTL significance. We recall that even though the TAGOOS model used eQTLs as predictive features, the strength of the eQTL associations was not seen during the training. We found a positive correlation of TAGOOS scores and unseen GTEx eQTL significance in both intronic (ρSpearman = 0.52, PSpearman < 2.2 × 10−16) and intergenic regions (ρSpearman = 0.4, PSpearman < 2.2 × 10−16). To visualize this correlation, we split unseen GTEx eQTLs in three groups according to the most significant Q-value and plotted them against the TAGOOS score (Figure 4B, E). We found that more significant eQTLs showed higher TAGOOS score median (Figure 4B, E). Altogether, these results showed that there was a positive correlation between the TAGOOS score and the eQTL significance.
Transcription factors (TFs) are very important components of the gene regulatory machinery in non-coding functional regions (9,57). Therefore, we hypothesized that SNPs with high TAGOOS are more often bound by TFs. To test this hypothesis, we took unseen GWAS Catalog SNPs and split them in two groups according to the TAGOOS score significance. Then we annotated both groups of SNPs with transcription factor binding sites (TFBSs) from the ReMap database (30,31) and plotted the percentage of SNPs annotated for each TF (Figure 4E, F). We found that higher percentage of TAGOOS-significant SNPs are annotated with TFs in intronic and intergenic regions (PWilcoxpaired = 2.2 × 10−16) (Figure 4C, F).
More TFBSs in TAGOOS-significant SNPs could be due to enriched DNA motifs for TFBSs. To test this hypothesis, we took the GWAS Catalog SNPs and a library of 127 non-redundant position frequency matrices based on the Hocomoco TF database (33,34). Then we scanned sequences of 100 bp around the SNPs with either the reference or most frequent alternative alleles and plotted the motif densities in 10 bp bins. We found local higher densities of motifs in the middle of the sequences for the reference and lower density for the alternative alleles (Figure 4G, H). We also found that the TAGOOS-significant SNPs had higher motif density in agreement with our hypothesis (Figure 4G, H). This analysis showed that SNPs with higher TAGOOS scores were richer in DNA motifs of TFBSs.
Altogether these results showed that the TAGOOS-significant SNPs showed functional properties of regulatory regions and SNPs.
Case study 1: the cleft lip locus rs227727
In this case study, we analyzed the LD block around the SNP rs227727 (hg19, chr17:54 752 926–54 778 620), which was shown to be associated to nonsyndromic cleft lip and functional as a regulatory region (11). In an initial study, rs227731 was associated to nonsyndromic cleft lip in the European population (58). Subsequent sequencing of the 17q22 region found highest association significance at rs227727, which was in complete linkage disequilibrium with rs227731 (11). Annotation of this region with ChromHMM and other regulation related annotations pointed at two regions called the NOGGIN +87 kb and +105 kb elements with putative enhancer activity within the rs227727 LD block (11). The NOGGIN +87 kb (hg19, chr17:54 755 547–54 757 398) and +105 kb regulatory elements (hg19, chr17: 54 776 294–54 777 215) were found to show an additive gene regulatory activity, which depended on the rs227727 allele (11).
We annotated the LD block around the rs227727 region with the negative decimal logarithm of the TAGOOS P-value and common SNPs from the DBSNP database (Figure 5A, B; Supplementary Figure S8; Supplementary Table S7). The rs227727 showed one of the most significant TAGOOS values in the region (Figure 5B). Furthermore, the two +87 kb and +105 kb NOGGIN regulatory elements showed uninterrumpted significant TAGOOS scores consistent with their known regulatory activities (Figure 5A, B; Supplementary Figure S8; Table S7). Based on these results, we conclude that the TAGOOS score was able to recover known regulatory regions and functional SNPs.
Then we looked for predicted functional regions and SNPs. A zoom of the TAGOOS score in these two regions showed that the predicted functional region extends beyond the tested constructs (Supplementary Figure S8; Table S7). In addition to the two +87 kb and +105 kb NOGGIN elements, our analysis uncovered two new long regions of 1031 bp and 894 bp (chr17:54 753 743–54 754 774 and chr17:54 758 876–54 759 770 in hg19) with uninterrupted significant TAGOOS scores around the +87 kb element (P < 0.05) (Supplementary Figure S8; Table S7). We also found that seven SNP loci showed more significant TAGOOS scores than rs227727 including four loci in the +87 kb NOGGIN element (rs138753947, rs192133406, rs73992081, rs141875137), two loci in the +105 NOGGIN element (rs116625135, rs538735669) and a seventh SNP between both elements (rs139384573) (Figure 5A, B; Supplementary Table S7).
Then we evaluated whether we could use the TAGOOS score to enrich TF binding events in the rs227727 LD block. We annotated TAGOOS-significant and non-significant SNPs in the rs227727 LD block with the ReMap catalog of transcriptional regulators (30,31). We found a higher percentage of TAGOOS-significant SNPs annotated with TFs (PWilcoxpaired < 2.2 × 10−16) (Figure 5C). Among these TFs, GATA3 mutations are known to correlate with craniofacial defects in human, mouse and zebrafish (59–61) (Figure 5D).
Therefore, we examined whether TFs with a largest binding difference between TAGOOS-significant SNPs and non-signficant SNPs were enriched for craniofacial phenotypes (Methods). We found TFs with largest binding differences between TAGOOS-significant SNPs and non-signficant SNPs to be enriched in mouse craniofacial bone morphology (Figure 5E). By contrast, TFs with lowest binding difference were not enriched in any mouse craniofacial phenotype (Figure 5E).
Alltogether these results demostrated that the TAGOOS score was able to recover known functional SNPs and genomic regions. In addition, the TAGOOS score presents new predictions of common SNPs and gene regulatory regions that could contribute to the phenotype in addition to the known rs227727 SNP.
Case study 2: Human adult height GWAS
In this case study, we analysed 697 loci associated to adult human height with the TAGOOS score (2). We evaluated whether the TAGOOS score can prioritize unseen height SNPs compared to random SNPs. We observed AUC performance values 0.73 and 0.72 for intronic and intergenic SNPs, respectively (Figure 6A, B).
Then we evaluated whether we could use the TAGOOS score to enrich TF binding events in height-associated loci. We found that TFs bind preferentially to TAGOOS-significant SNPs in intronic and intergenic regions (PWilcoxpaired < 2.2 × 10−16) (Figure 6C, D). We sorted TFs according to the difference of percentage of bound SNPs with TAGOOS-significant or non-significant scores. Among TFs bound preferentially to TAGOOS-significant SNPs, MYC, MED1, RUNX1 in intronic and AR and GATA2 in intergenic regions, are annotated as related to body size in the Mouse Genome Informatics (MGI) and EnrichR databases (32,62) (Figure 6E, F). Therefore we hypothesized that we could focus on TFs that bind more often to TAGOOS-significant SNPs to look for relevant functions using the EnrichR server (Methods). The list of sorted TFs in Figure 6C, D were ordered and split in two equal groups and submitted to the EnrichR online tool (Figure 6G, H) (Methods). The EnrichR online tool showed higher enrichment of genes related to the mouse body size phenotypes for TFs with highest difference of binding between TAGOOS-significant and non-significant SNPs (Figure 6G, H).
To further ilustrate the use of TAGOOS, we focused on the LTBP2 locus. We annotated the LTBP2 locus with the negative decimal logarithm of the P-value of the intronic TAGOOS, common SNPs and P-values from the height GWAS associations (Supplementary Table S8). Among the two height-associated SNPs in this locus (rs862034, rs10140101), only rs10140101 showed a significant TAGOOS score (Supplementary Table S8). However, four other common SNPs (rs862037, rs862036, rs862035, rs3784030) at a distance of <400 bp to rs862034 showed significant TAGOOS scores (Supplementary Table S8) and are functional candidate SNPs for the height association of rs862034. The other height-associated SNP rs10140101 belonged to a large region of uninterrupted significant TAGOOS scores with a length of 665 bp (hg19, chr14:75 038 463–75 039 127) that could act as a gene regulatory region for LTBP2 (Supplementary Table S8). In this region chr14:75 038 463–75 039 127, there were 12 other common SNPs that could contribute to the effect of rs10140101 (Supplementary Table S8). In the LTBP2 locus, there were three other regions larger than 400 bp with uninterrupted significant TAGOOS scores, chr14:74 981 188–74 981 684, chr14:74 992 079–74 992 846 and chr14:75 075 601–75 076 114 (hg19) that could contribute to the height association. These three regions contained 17 common SNPs that could also contribute to the height phenotype (Supplementary Table S8).
To further ilustrate the use of TAGOOS, we focused on the rs10140101 SNP located in a LTBP2 intron. This SNP showed a significant TAGOOS P-value of 0.001 and 71 annotations where 62% of them belonged to DNAse, H3K27ac and H3K4me1 molecular assays. Interestingly this location was not annotated with eQTLs or SNPs associated to complex diseases. This means that interesting SNPs could be predicted by the TAGOOS SNP in the absence of known eQTLs or association data. As shown in the IGV browser, the rs10140101 was surrounded by a peak of bound TFs (Figure 6I). More precisely, the SNP was bound to 101 ReMap TFs with 35 of these factors related to decreased mouse body size phenotype. The UCSC browser also showed that the C allele was highly conserved with other mammals (Supplementary Figure S9). In addition, the integrative IW-score predicts significant molecular effects for this SNP (27). The LTBP2 gene (OMIM ID 602091) was involved in the Weill-Marchesani syndrome, glaucoma and microspherophakia diseases (OMIM IDs 614819, 613086 and 251750), which also show abnormal stature phenotypes (63).
Our analysis of the LTBP2 locus predicts four gene regulatory regions with regions larger than 400 bp that contain 31 putation functional common SNPs. The height-associated SNP rs10140101 and the surrounding region were particularly interesting for further analysis.
DISCUSSION
Here we develop a supervised learning approach that predicts functional scores genome-wide based on SNPs associated to complex phenotypes and regulatory annotations that predict functional loci. Compared to previous tools, the TAGOOS method lies between so-called GWAS priorization and regulatory variants priorization tools (Supplementary Tables S1 and S2). The TAGOOS method takes GWAS signals, LD blocks and gene regulatory features and outputs functional scores genome-wide for regulatory variants. Previous prioritization strategies for GWAS signals that take into account LD blocks such as GWAS3D, GREGOR or GenoWAP were not evaluated genome-wide (4,14,15). On the other hand, most predictors of regulatory variants such as GWAVA are trained with highly penetrant SNPs such as those from the ClinVar databases (17,38). Moreover, the performance of these tools to predict SNPs associated to complex phenotypes was moderate (27). The TAGOOS method is different from previous predictors, because the model is trained with common variants associated to complex diseases and with correlated SNPs in linkage disequilibrium in the reference population. The resulting model has been used to generate genome-wide gene regulation scores with better priorization and enrichment performances than most other methods (27). We have shown that the TAGOOS score was able to recover known functional gene regulatory regions (+87 kb and +105 kb NOGGIN elements) and SNPs (rs227727). In addition we have also demonstrated that the TAGOOS score was able to predict new gene regulatory regions and functional SNPs, which could contribute to the cleft lip and height phenotypes.
There are three main sources of annotation data to predict gene regulation potential: (i) open chromatin and chromatin modifications such as DHSs and H3K27ac, (ii) eQTLs and (iii) transcription factor binding and motifs (7,8,30,31,56). Unlike previous approaches, we use all three types of regulatory annotations. Therefore even though eQTLs are strong predictive features of the TAGOOS model (Figure 3A–C), the TAGOOS score can still prioritize SNPs without eQTL annotation such as the SNP rs10140101 based on chromatin and transcription factor annotations. We also did not select particular cell types. The advantage was that the TAGOOS score was able to prioritize associated SNPs from different disease types (Supplementary Figure S4). Nevertheless, the performance seems to be better in diseases and traits with an overrepresentation of tissues in the public databases such as blood and immune cell types (Supplementary Figure S4).
Including DNA methylation in our model could be interesting, because it is an important property of inactive regions (64). Transcription is another important property of enhancers and RNA-seq data is available in the ENCODE project, so that it could be included in our model (39). We do not provide scores for the X and Y chromosomes because some data sets do not provide information for these chromosomes. It is however possible to generate scores despite the missing data by using relative performance and significance measures for these chromosomes.
Two models for intronic and intergenic regions were created, because in principle, the molecular signatures of functional regions in these two regions were only partially overlapping. Our results support this choice, because for instance, gene body typical modifications such as H3K36me3 were more often found among the features in the intronic model.
In the present model, we found that eQTLs, DNA accessibility and H3K27ac are very predictive of GWAS SNPs, which agrees with known properties of gene regulatory regions (7,8,56). We were surprised not to find many TFs as features, because TFs are usually enriched in gene regulatory regions (10). However, subsequent analysis found that TAGOOS-significant SNPs are more frequently bound by TFs (Figure 4C, F, 5C, D and 6C–F). The reason might be that particular TFs are implicated in specific phenotypes and that no TF is generally involved in all phenotypes. By contrast, the number of biological samples annotated with an eQTL or H3K27ac in a given SNP correlated with the TAGOOS score and thus indirectly with the maximal GWAS significance of that SNP (Figures 1E, F and 3A–C). This means that the TAGOOS scores potentially predict functional regions, whereas a more precise phenotype-specific regulation is exerted through phenotype-specific molecules such as transcription factors.
In conclusion, the TAGOOS score is a new score to predict functional intronic and intergenic regions. The TAGOOS score has been trained on associated loci to complex phenotypes and achieves better priorization performances than most other methods.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to J. Castro (NCMM, Oslo, Norway) for technical help and D. Puthier (TAGC, Marseille, France), S. Spicuglia (TAGC, Marseille, France), J. van Helden (TAGC, Marseille, France), L. Perrin (TAGC, Marseille, France) and B. Ballester (TAGC, Marseille, France) for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: INSERM and Aix-Marseille University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Visscher P.M., Wray N.R., Zhang Q., Sklar P., McCarthy M.I., Brown M.A., Yang J.. 10 Years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 2017; 101:5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wood A.R., Esko T., Yang J., Vedantam S., Pers T.H., Gustafsson S., Chu A.Y., Estrada K., Luan J., Kutalik Z. et al.. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 2014; 46:1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eicher J.D., Landowski C., Stackhouse B., Sloan A., Chen W., Jensen N., Lien J.-P., Leslie R., Johnson A.D.. GRASP v2. 0: an update on the Genome-wide repository of associations between SNPs and phenotypes. Nucleic Acids Res. 2014; 43:D799–D804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li M.J., Liu Z., Wang P., Wong M.P., Nelson M.R., Kocher J.-P.A., Yeager M., Sham P.C., Chanock S.J., Xia Z. et al.. GWASdb v2: an update database for human genetic variants identified by genome-wide association studies. Nucleic Acids Res. 2015; 44:D869–D876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., Hastings E., Junkins H., McMahon A., Milano A., Morales J. et al.. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog. Nucleic Acids Res. 2017; 45:D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A.. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U.S.A. 2009; 106:9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nicolae D.L., Gamazon E., Zhang W., Duan S., Dolan M.E., Cox N.J.. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010; 6:e1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J. et al.. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012; 1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Spitz F., Furlong E.E.. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012; 13:613–626. [DOI] [PubMed] [Google Scholar]
- 10. Andrey G., Mundlos S.. The three-dimensional genome: regulating gene expression during pluripotency and development. Development. 2017; 144:3646–3658. [DOI] [PubMed] [Google Scholar]
- 11. Leslie E.J., Taub M.A., Liu H., Steinberg K.M., Koboldt D.C., Zhang Q., Carlson J.C., Hetmanski J.B., Wang H., Larson D.E. et al.. Identification of functional variants for cleft lip with or without cleft palate in or near PAX7, FGFR2, and NOG by targeted sequencing of GWAS loci. Am. J. Hum. Genet. 2015; 96:397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ward L.D., Kellis M.. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2011; 40:D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coetzee S.G., Rhie S.K., Berman B.P., Coetzee G.A., Noushmehr H.. FunciSNP: an R/bioconductor tool integrating functional non-coding data sets with genetic association studies to identify candidate regulatory SNPs. Nucleic Acids Res. 2012; 40:e139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmidt E.M., Zhang J., Zhou W., Chen J., Mohlke K.L., Chen Y.E., Willer C.J.. GREGOR: evaluating global enrichment of trait-associated variants in epigenomic features using a systematic, data-driven approach. Bioinformatics. 2015; 31:2601–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lu Q., Yao X., Hu Y., Zhao H.. GenoWAP: GWAS signal prioritization through integrated analysis of genomic functional annotation. Bioinformatics. 2016; 32:542–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014; 46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ritchie G.R.S., Dunham I., Zeggini E., Flicek P.. Functional annotation of noncoding sequence variants. Nat. Methods. 2014; 11:294–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Quang D., Chen Y., Xie X.. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2014; 31:761–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shihab H.A., Gough J., Mort M., Cooper D.N., Day I.N.M., Gaunt T.R.. Ranking non-synonymous single nucleotide polymorphisms based on disease concepts. Hum. Genomics. 2014; 8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen L., Jin P., Qin Z.S.. DIVAN: accurate identification of non-coding disease-specific risk variants using multi-omics profiles. Genome Biol. 2016; 17:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smedley D., Schubach M., Jacobsen J.O.B., K”ohler S., Zemojtel T., Spielmann M., Jäger M., Hochheiser H., Washington N.L., McMurry J.A. et al.. A Whole-genome analysis framework for effective identification of pathogenic regulatory variants in Mendelian disease. Am. J. Hum. Genet. 2016; 99:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Huang Y.-F., Gulko B., Siepel A.. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat. Genet. 2017; 49:618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fu Y., Liu Z., Lou S., Bedford J., Mu X.J., Yip K.Y., Khurana E., Gerstein M.. FunSeq2: a framework for prioritizing noncoding regulatory variants in cancer. Genome Biol. 2014; 15:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou J., Troyanskaya O.G.. Predicting effects of noncoding variants with deep learning-based sequence model. Nat. Methods. 2015; 12:931–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gulko B., Hubisz M.J., Gronau I., Siepel A.. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat. Genet. 2015; 47:276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ionita-Laza I., McCallum K., Xu B., Buxbaum J.D.. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 2016; 48:214–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang J., Ullah A.Z.D., Chelala C.. IW-Scoring: an Integrative weighted scoring framework for annotating and prioritizing genetic variations in the noncoding genome. Nucleic Acids Res. 2018; 46:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bodea C.A., Mitchell A.A., Bloemendal A., Day-Williams A.G., Runz H., Sunyaev S.R.. PINES: phenotype-informed tissue weighting improves prediction of pathogenic noncoding variants. Genome Biol. 2018; 19:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J.. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015; 4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Griffon A., Barbier Q., Dalino J., Van Helden J., Spicuglia S., Ballester B.. Integrative analysis of public ChIP-seq experiments reveals a complex multi-cell regulatory landscape. Nucleic Acids Res. 2014; 43:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chèneby J., Gheorghe M., Artufel M., Mathelier A., Ballester B.. ReMap 2018: an updated atlas of regulatory regions from an integrative analysis of DNA-binding ChIP-seq experiments. Nucleic Acids Res. 2017; 46:D267–D275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. et al.. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016; 44:W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kulakovskiy I.V., Vorontsov I.E., Yevshin I.S., Sharipov R.N., Fedorova A.D., Rumynskiy E.I., Medvedeva Y.A., Magana-Mora A., Bajic V.B., Papatsenko D.A. et al.. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 2017; 46:D252–D259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Castro-Mondragon J.A., Jaeger S., Thieffry D., Thomas-Chollier M., van Helden J.. RSAT matrix-clustering: dynamic exploration and redundancy reduction of transcription factor binding motif collections. Nucleic Acids Res. 2017; 45:e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Medina-Rivera A., Defrance M., Sand O., Herrmann C., Castro-Mondragon J.A., Delerce J., Jaeger S., Blanchet C., Vincens P., Caron C. et al.. RSAT 2015: regulatory sequence analysis tools. Nucleic Acids Res. 2015; 43:W50–W56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smigielski E.M., Sirotkin K., Ward M., Sherry S.T.. dbSNP: a database of single nucleotide polymorphisms. Nucleic acids Res. 2000; 28:352–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Consortium G.P. et al.. A global reference for human genetic variation. Nature. 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Landrum M.J., Lee J.M., Benson M., Brown G., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Hoover J. et al.. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016; 44:D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. et al.. An atlas of active enhancers across human cell types and tissues. Nature. 2014; 507:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N. et al.. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roadmap E.C., Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J. et al.. Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A.. Super-enhancers in the control of cell identity and disease. Cell. 2013; 155:934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Köster J., Rahmann S.. Snakemake-a scalable bioinformatics workflow engine. Bioinformatics. 2012; 28:2520–2522. [DOI] [PubMed] [Google Scholar]
- 44. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Quinlan A.R. BEDTools: the Swiss-army tool for genome feature analysis. Curr. Protoc. Bioinformatics. 2014; 47:11.12.1–11.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P.. Integrative genomics viewer. Nat. Biotechnol. 2011; 29:24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen T., He T.. Higgs Boson discovery with boosted trees. Proceedings of the 2014 International Conference on High-Energy Physics and Machine Learning. 2014; 42:69–80. [Google Scholar]
- 48. Hoffman M.M., Ernst J., Wilder S.P., Kundaje A., Harris R.S., Libbrecht M., Giardine B., Ellenbogen P.M., Bilmes J.A., Birney E. et al.. Integrative annotation of chromatin elements from ENCODE data. Nucleic Acids Res. 2013; 41:827–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Beer M.A. Predicting enhancer activity and variant impact using gkm-SVM. Hum. Mutat. 2017; 38:1251–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schuster-B”ockler B., Lehner B.. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012; 488:504–507. [DOI] [PubMed] [Google Scholar]
- 51. Heinz S., Romanoski C.E., Benner C., Glass C.K.. The selection and function of cell type-specific enhancers. Nat. Rev. Mol. Cell Biol. 2015; 16:144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Z., Zang C., Rosenfeld J.A., Schones D.E., Barski A., Cuddapah S., Cui K., Roh T.-Y., Peng W., Zhang M.Q. et al.. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008; 40:897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Song Q., Smith A.D.. Identifying dispersed epigenomic domains from ChIP-Seq data. Bioinformatics. 2011; 27:870–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xi H., Shulha H.P., Lin J.M., Vales T.R., Fu Y., Bodine D.M., McKay R.D.G., Chenoweth J.G., Tesar P.J., Furey T.S. et al.. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007; 3:e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gaulton K.J., Nammo T., Pasquali L., Simon J.M., Giresi P.G., Fogarty M.P., Panhuis T.M., Mieczkowski P., Secchi A., Bosco D. et al.. A map of open chromatin in human pancreatic islets. Nat. Genet. 2010; 42:255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zeng H., Edwards M.D., Guo Y., Gifford D.K.. Accurate eQTL prioritization with an ensemble-based framework. Hum. Mutat. 2017; 38:1259–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bass J. I.F., Sahni N., Shrestha S., Garcia-Gonzalez A., Mori A., Bhat N., Yi S., Hill D.E., Vidal M., Walhout A.J.. Human gene-centered transcription factor networks for enhancers and disease variants. Cell. 2015; 161:661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mangold E., Ludwig K.U., Birnbaum S., Baluardo C., Ferrian M., Herms S., Reutter H., de Assis N.A., Al Chawa T., Mattheisen M. et al.. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nat. Genet. 2010; 42:24–26. [DOI] [PubMed] [Google Scholar]
- 59. Lim K.C., Lakshmanan G., Crawford S.E., Gu Y., Grosveld F., Engel J.D.. Gata3 loss leads to embryonic lethality due to noradrenaline deficiency of the sympathetic nervous system. Nat. Genet. 2000; 25:209–212. [DOI] [PubMed] [Google Scholar]
- 60. Bernardini L., Sinibaldi L., Capalbo A., Bottillo I., Mancuso B., Torres B., Novelli A., Digilio M.C., Dallapiccola B.. HDR (Deafness, Renal dysplasia) syndrome associated to GATA3 gene duplication. Clin. Genet. 2009; 76:117–119. [DOI] [PubMed] [Google Scholar]
- 61. Sheehan-Rooney K., Swartz M.E., Zhao F., Liu D., Eberhart J.K.. Ahsa1 and Hsp90 activity confers more severe craniofacial phenotypes in a zebrafish model of hypoparathyroidism, sensorineural deafness and renal dysplasia (HDR. Dis. Models Mech. 2013; 6:1285–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Smith C.L., Blake J.A., Kadin J.A., Richardson J.E., Bult C.J., Group M.G.D.. Mouse Genome Database (MGD)-2018: knowledgebase for the laboratory mouse. Nucleic Acids Res. 2018; 46:D836–D842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hamosh A., Scott A.F., Amberger J.S., Bocchini C.A., McKusick V.A.. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005; 33:D514–D517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Suzuki M.M., Bird A.. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008; 9:465. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.