Abstract
The CRISPR system is widely used in genome editing for biomedical research. Here, using either dual paired Cas9D10A nickases or paired Cas9 nuclease we characterize unintended larger deletions at on-target sites that frequently evade common genotyping practices. We found that unintended larger deletions are prevalent at multiple distinct loci on different chromosomes, in cultured cells and mouse embryos alike. We observed a high frequency of microhomologies at larger deletion breakpoint junctions, suggesting the involvement of microhomology-mediated end joining in their generation. In populations of edited cells, the distribution of larger deletion sizes is dependent on proximity to sgRNAs and cannot be predicted by microhomology sequences alone.
INTRODUCTION
CRISPR/Cas9-based genome engineering approaches are widely used to generate deletions or insertions at genomic regions of interest for biomedical research purposes. Cas9 nuclease (derived from Streptococcus pyogenes) generates double strand DNA breaks (DSBs) when targeted to a locus by a single-guide RNA (sgRNA) that facilitates base pairing with the DNA template, recruiting the nuclease on-target (1). Although targeting is primarily specific, off-target sites, which differ by one or more bases, may also be recognised and cut (1). To reduce off-target editing, modified approaches have been developed including high-fidelity Cas9 (hfCas9) (2), and Cas9 nickase (3). Two different Cas9 nickase enzymes were engineered, Cas9D10A and Cas9H840A, which each harbour inactivating mutations in one of the two Cas9 nuclease domains and generate single stranded DNA (ssDNA) nicks upon recruitment to the DNA (3). To generate a DSB at the locus of interest, Cas9 nickase is targeted using two sgRNA molecules (paired sgRNAs) that yield two ssDNA nicks in close proximity on opposite strands, resulting in staggered DSB formation (3). At off-target sites, the likelihood of two sgRNAs binding is small, thus resulting in ssDNA nicks that are repaired with high efficiency by the mismatch repair pathway (4). Because of its reduced probability of off-target mutations, Cas9 nickase-mediated genome editing has been suggested to be favourable for gene therapy approaches (5).
For the deletion of a specific genomic region using Cas9 nuclease or Cas9 nickase, two (paired) or four (dual paired) sgRNAs are typically used, respectively, to generate DSBs flanking a target region. DSBs can be repaired through several endogenous repair pathways including non-homologous-end-joining (NHEJ), homologous recombination (HR), microhomology-mediated end joining (MMEJ) or single strand annealing (SSA) (1,6,7). In some alleles, DNA repair will result in loss of the intervening DNA between flanking DSBs. Such deletions are commonly detected by short-range (S-R) polymerase chain reaction (PCR) and Sanger sequencing (8,9). In this approach, flanking primers adjacent to the target region (typically ≤200 bp away from sgRNA cut sites) are used to amplify genomic DNA (gDNA) of targeted cells or tissues (10,11). DSB repair can also result in additional insertions or deletions (indels). Indels generated by Cas9 are typically small in size (<50 bp) (12–16), but larger deletions (LDs) have also been reported. For example, larger than expected indels from one sgRNA spanned from ∼300 to 9.5 kb in HEK-293T or mouse embryonic stem cells (mESCs) (12,17). Paired sgRNAs also induced LDs in MEL cells (18). When genome editing in vivo, LDs ranging from hundreds of bp to several kb were seen in up to 45% of mouse or rat embryos edited with single or paired sgRNAs (8,19–27). The majority of LDs previously identified were induced by Cas9 nuclease but have also been observed at a single locus targeted with dual Cas9 nickase complexes in mouse embryos (21). Such unanticipated LDs may be difficult to detect using S-R PCR screening methods, as primer binding sites may be lost, resulting in a failure to amplify these alleles (17,24). It remains unclear how prevalent or widespread LDs are at different loci, or when using different CRISPR/Cas9 genome editing modalities such as Cas9 nickase. Moreover, the DNA repair mechanisms at play for LD generation remain unclear.
Here, we performed genome editing using dual paired Cas9D10A nickase, paired Cas9 nuclease, and single Cas9 nuclease complexes in mESCs and a haematopoietic progenitor cell line in vitro, and in mouse embryos in vivo. Sixteen separate regions ranging from 100 bp to 1.5 kb corresponding to both coding and non-coding regions were targeted at nine different genomic loci on seven chromosomes. Using PCR and droplet digital PCR, we identified LDs of up to 7 kb from paired sgRNA target sites and sequenced the breakpoint junctions. Significant microhomologies consistent with MMEJ were detected at almost all LD breakpoint junctions. Our findings show that LDs are not repaired at the most proximal microhomologies. Instead, using computational approaches we show that the distribution of LDs in a cell population can be modelled based on proximity to sgRNA cut sites.
MATERIALS AND METHODS
Cell culture
The 416B myeloid progenitor cell line (28) was grown in Fischers medium (Gibco) supplemented with 20% horse serum (Gibco), 2 mM l-glutamine. Cells were maintained at 37°C and 5% CO2, at densities of between 2 × 105 and 8 × 105 cells/ml. E14-TG2a (29) and E14-TG2a-RV mESCs (stably transfected with a Venus reporter at the 3′ end of Runx1 and a hsp68-mCherry-Runx1+23 enhancer-reporter transgene in the Col1a1 locus; L Greder, unpublished data) were cultured in GMEM medium (Gibco) supplemented with 10% FCS (Gibco), 2% Leukemia Inhibitory Factor (LIF) conditioned medium, 2 mM l-glutamine (Gibco) and 100 μM β-mercaptoethanol (Sigma). Cells were passaged every 2–3 days.
Genome editing in cultured cells
Dual-Cas9D10A nickase and Cas9 nuclease-based knock-out strategies were designed using the Zhang lab online tool (crispr.mit.edu). Single guide RNAs (Supplementary Table S1) were ordered as oligonucleotides (IDT) and cloned into the BbsI site of pX335-Neo (30) or pX459 (Addgene plasmid #62988, (31)). Gibson assembly was used to create tandem constructs that contained one, two or four sgRNAs in one plasmid. DH10β chemically competent Escherichia coli (Invitrogen) were transformed with plasmid DNA according to the manufacturer's instructions. Correct sgRNA inserts were confirmed by Sanger sequencing. Purified plasmids (Qiagen plus Midi prep kit (Qiagen)) were transfected into mESCs using lipofectamine 2000 (Invitrogen, 5 μg/well of a 6-well plate). 1 × 107 416B cells were electroporated with 10 μg of a modified pX335 plasmid co-expressing eGFP with a Bio-Rad gene pulser (Bio-Rad, 40mm cuvette, 220 mV, 960 μFD). Cells were either FACS sorted based on GFP expression or selected using 1 μg/ml puromycin or 175 μg/ml G418 (Gibco). Colony picking of mESCs was performed as previously described (32). DNA was purified from bulk populations of selected or sorted mESCs or sorted 416B cells using a Qiagen DNeasy blood and tissue kit (Qiagen).
Mice
All animals were housed and maintained in the Mary Lyon Centre, MRC Harwell Institute under specific-pathogen-free (SPF) conditions, in individually ventilated cages adhering to environmental conditions as outlined in the Home Office Code of Practice. Mice were euthanized by Home Office Schedule 1 methods.
Reagents for microinjection, delivery to embryos and germline transmission
Guide sequence selection was carried out using two online tools: CRISPOR (33) and Wellcome Trust Sanger Institute (WTSI) Genome Editing (WGE) (34). sgRNA sequences were selected with as few predicted off-target events as possible, particularly on the same chromosome as the intended modification (Sequences shown in Supplementary Table S1). Two sgRNAs for each side of the critical regions to be deleted were synthesized and co-injected with Cas9 mRNA as previously described (27). Injected embryos were re-implanted in CD-1 pseudopregnant females. Host females were allowed to litter and rear G0s. G0 animals where the presence of a desired allele was detected by PCR were mated to wild-type animals to obtain G1 animals for germline transmission of the allele of interest and definitive validation of its integrity.
PCR analysis of cultured cells
PCR products were amplified from gDNA isolated from clones or pools of cells as indicated. PCR was performed using a HotStar Taq master mix kit (Qiagen), with 100 ng gDNA and primers at 200 nM (Primers listed in Supplementary Table S2). PCR products were analysed by agarose gel electrophoresis and a 1 kb plus ladder (Thermo Fischer). PCR products were gel extracted using a Zymoclean gel extraction kit (Zymo Research). TA cloning was performed according to the manufacturer's instructions (Invitrogen). Plasmids were isolated using a Qiagen Spin mini-prep kit (Qiagen) followed by Sanger sequencing (Source Bioscience, Oxford, UK).
PCR analysis of mice
Genomic DNA was extracted from ear clip biopsies using the DNA Extract All Reagents Kit (Applied Biosystems) according to the manufacturer's instructions. Genotyping primers for SR-PCR assays were chosen to be at least 200 bp away from the sequences targeted by sgRNA, depending on available sequences for design. PCR assays were optimised and performed as previously described (27). The PCR products were purified employing a QIAquick Gel Extraction Kit (Qiagen) and sent for Sanger sequencing.
Alignment of larger deletion alleles
All deletions were aligned to the mm9 reference genome using UCSC BLAT and visualised using the UCSC genome browser (35,36). Fine mapping was performed by subsequent local alignment using MUSCLE (37). Repetitive elements were mapped using UCSC RepeatMasker (38).
Droplet digital PCR
Droplet Digital PCR was used to determine copy number variation in genome-edited mice and to quantify deletions in edited mESCs. Experiments were performed as duplex reactions, where the sequence employed as a donor was amplified using a fluorescein amidite (FAM)-labelled assay selected from a Universal Probe Library (UPL) set (Human, sourced from Roche, Basel, SZ). Suitable probes and primers were identified using the ProbeFinder software at the Roche assay design centre (accessible from www.universalprobelibrary.com, Supplementary Table S2). In cases where a UPL set was not available, custom assays were ordered from LGC Biosearch Technologies (Novato, USA). UPL or custom assays were used in parallel with a VIC-labelled reference gene assay (Dot1l, sourced from ThermoFisher) set at two copies (CNV2) on the Bio-Rad QX200 ddPCR System (Bio-Rad) as per Codner et al. (27). Reaction mixes (22 μl) contained 2 μl crude DNA lysate or 50 ng of phenol/chloroform purified genomic DNA, 1× ddPCR Supermix for probes (Bio-Rad), 225 nM of each primer (two primers per assay) and 50 nM of each probe (one VIC-labelled probe for the reference gene assay and one FAM-labelled for the target genomic region assays). For deletion quantification in pools of cells, 100 ng purified DNA was used. These reaction mixes were loaded either into DG8 cartridges together with 70 μl droplet oil per sample and the droplets generated using the QX100 Droplet Generator or loaded in plate format into the Bio-Rad QX200 AutoDG and the droplets generated as per the manufacturer's instructions. Post droplet generation, the oil/reagent emulsion was transferred to a 96-well semi-skirted plate (Eppendorf), and the samples were amplified on a Bio-Rad C1000 Touch thermocycler (95°C for 10 min, followed by 40 cycles of 94°C for 30 s and 58°C for 60 s, with a final elongation step of 98°C for 10 min, where all temperature ramping was set to 3°C/s). The plate containing the droplet amplicons was subsequently loaded into the QX200 Droplet Reader (Bio-Rad). Standard reagents and consumables supplied by Bio-Rad were used, including cartridges and gaskets, droplet generation oil and droplet reader oil. Copy numbers were assessed using the QuantaSoft Analysis Pro™ software using at least 10 000 accepted droplets per sample. The copy numbers (mice) or ratio compared to internal control (mESCs) were calculated by applying Poisson statistics to the fraction of end-point positive reactions, and the 95% confidence interval of this measurement is shown. When visualizing ddPCR quantification in pools of cells the mean and 95% confidence intervals are shown.
Linear regression modelling of ddPCR data
Multiple linear regression was performed in R using the ‘lm’ and ‘predict’ functions. The model was fit using the formula ‘y ∼ log(x) + a’, where ‘y’ (frequency of deletion) was the response variable, ‘x’ (proximity to sgRNAs) was explanatory variable one and ‘a’ (sgRNA cutting efficiency) explanatory variable two. Data used to fit the model were empirically determined by ddPCR in eight different targeted cell populations and at two different loci. Frequency of deletion at a particular region was calculated by dividing the relative concentration determined by ddPCR in the transfected sample by the corresponding non-targeting control. Proximity to sgRNA binding sites was determined from the midpoints of the ddPCR amplicon and sgRNA. The sgRNA cutting efficiency in each sample was the highest frequency of deletion directly at a sgRNA target site as determined by ddPCR. For model estimates of deletion frequency, cutting efficiency was either set to the average cutting efficiency for all samples (when visualising general estimates) or a known cutting efficiency when estimating values in a subset of the experiments. Goodness of fit testing was performed by plotting a histogram of residuals (where residual = observed value - predicted value), and a Q–Q plot (quantile–quantile plot) to check that residuals are normally distributed (which is an assumption of the linear regression model). The distribution of estimated relative allele frequencies (1 – deletion frequency) was plotted across a 3 kb window up and downstream of a simulated sgRNA cut site.
Microhomology scoring and quantification
Deletions were considered LDs if they spanned >200 bp from sgRNA target sites and ablated at least one S-R primer binding site. All of the larger deletions that could be resolved by PCR and Sanger sequencing were quantified (Supplementary Table S5). Deletions were considered to be of expected size if indels reached ≤25 bp beyond the protospacer adjacent motif (PAM) site of the nearest sgRNA binding site. LDs previously described in the literature (8,19–24) were considered if they spanned >80 bp away from predicted sgRNA cut sites. Microhomology scoring was performed using a custom R script and the package Biostrings. 10 bp in the 5′ and 3′ direction at up and downstream breakpoints was used to search for microhomologies. Identical bases were given a score of 1 and mismatches were given a score of 0. The highest scoring homology at each breakpoint site (either upstream or downstream) was considered. Bases were considered homologous if they were identical and directly abutted the deletion, as previously described (39). To simulate the background distribution of microhomologies in the genome, random genomic locations were selected using a custom R script and the package BSgenome. The length of the simulated LDs was set to the average length of LDs identified. Chance expectation of homology at any two locations for a k-mer of a given length was calculated as previously described (40) using the equation P(x) = (x + 1)(1/4)x(3/4)2. Alternative microhomologies were counted within deleted sequences (excluding sequences between paired sgRNAs) using R and Biostrings. Statistical analysis was performed in graph pad prism and R using χ2 test, Kruskal–Wallis with Dunn's multiple comparison test, or two-tailed Mann–Whitney test as indicated. All custom scripts are freely available on GitHub (https://github.com/d0minicO/mhscanR).
Deep sequencing of short deletions in CRISPR/Cas9 edited cells
Locus specific next-generation sequencing primers (NGS, Supplementary table S2) were designed to amplify a 225 bp amplicon centred on each sgRNA cut site. Primers were modified to contain Illumina Truseq adapter sequences at the 5′ end. PCR was performed (25 cycles, Herculase II PCR kit (Agilent)) on a pool of genomic DNA harvested from cells 3–7 days post transfection with one sgRNA. Truseq indices (NEB E7335) were added to the PCR amplified fragments by using a further 6 cycles of PCR with Herculase II PCR kit. The size distribution of libraries was analysed using a D1000 Tapestation (Agilent) and library concentration was quantified using KAPA library quantification kit (Roche) both according to the manufacturer's instructions. Libraries were normalised to 4nM and pooled before sequencing using Illumina MiSeq v2 300 cycle paired end kit. Raw fastq files have been deposited in NCBI’s Gene Expression Omnibus (41) and are accessible through GEO Series accession number GSE130621 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE130621). Fastq files were trimmed using trim_galore. For visualization, trimmed fastq files were flashed (42) and mapped using bwa mem v 0.7.12. Sam files were converted to bam using samtools (43), converted to bigwig using deeptools bamCoverage (option -bs 1) (44), and visualised using UCSC genome browser. For analysis of individual alleles trimmed fastq files were analysed using CRISPResso v1.0.8 (45). A custom R script was used to quantify microhomologies in simple deletions (spanning a contiguous region and free of insertions or mutations within 10 bp of the deletion) using the same microhomology scoring criteria as at LDs. Reads containing insertions were quantified against the background of all modified reads. Graphing and χ2 tests were performed in R and graph pad prism.
Analysis of GC content at microhomologies
GC content of microhomologies was analysed as previously described (46). Briefly, in the absence of GC bias, the GC content of a microhomology is assumed equal to the background GC content over the region containing the deletions. The background GC content in deep sequencing data was calculated over the region containing 93–95% of the reads. The observed GC base pair content of microhomologies was then compared to the expected probability using the χ2 test (Graph Pad Prism, or R).
RESULTS
Genome editing with Cas9D10A nickase in mESCs causes larger than expected on-target deletions
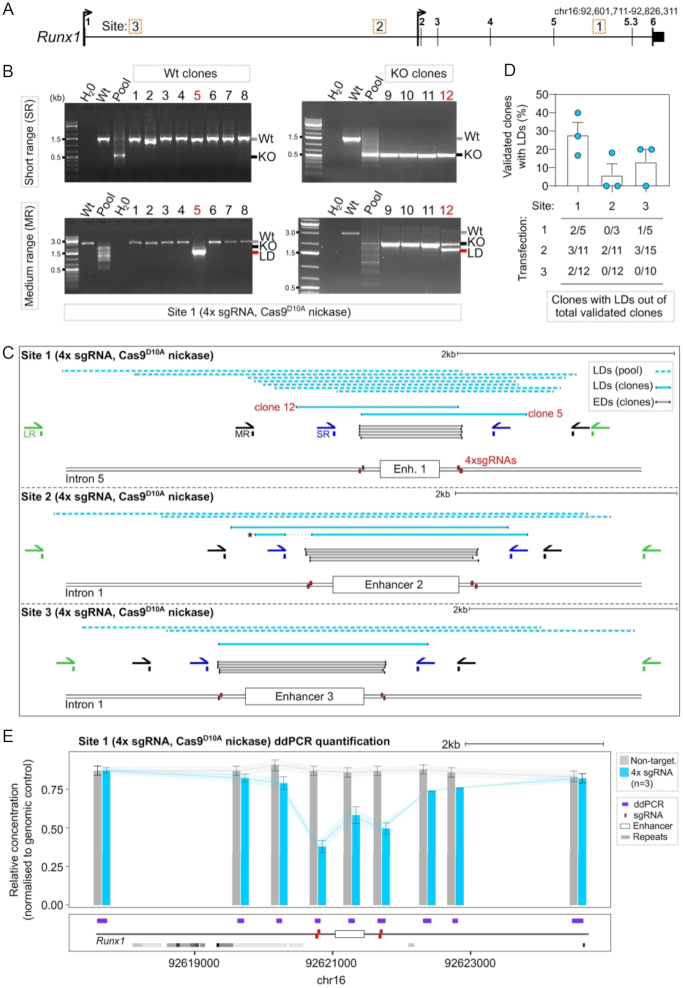
We used a dual paired sgRNA (4x sgRNA) CRISPR/Cas9D10A nickase strategy in mESCs that had been stably transfected with fluorescent reporters (E14-TG2a-RV) to delete evolutionarily conserved Runx1 cis-regulatory elements (47,48) (Figure 1A). Intended deletions ranged from 1 to 1.5 kb. Individual clones were analysed for the desired genotype using S-R PCR (Figure 1B, C). Out of 445 clones analysed, an average of 35% and 20% of the total isolated clones for each of the three targeted sites appeared to be homozygous knock-out or wild type, respectively (Supplementary Figure S1A–D). Several clones with unique alleles harbouring deletions of expected size (EDs, spanning <25 bp from expected sgRNA cut sites) were mapped using Sanger sequencing of PCR products (Figure 1B, C, grey lines). Sanger sequencing often generated a single sequencing trace, indicative either of an iso-allelic HR event (both alleles carrying the same deletion) or loss of a primer binding site, leading to failure to amplify one of the alleles (allelic drop-outs) (Supplementary Figure S1E).
Figure 1.
Characterisation of larger deletions at three sites targeted by CRISPR/Cas9D10A nickase. (A) Locus map of the Runx1 gene showing the positions of evolutionarily conserved cis-regulatory elements (Site 1–3, corresponding to Enhancer 1–3) that were targeted in E14-TG2a-RV mESCs using CRISPR/Cas9D10A nickase. (B) Example gel images from one experiment targeting Site 1. Gel images show PCR amplification from gDNA of isolated wild type (wt) clones (left hand gels) and knock-out (KO) clones (right hand gels) with SR primers (top gels) and MR primers (bottom gels). Wt next to the gel image indicates the size of the wild type allele, KO indicates the size of alleles harbouring the expected deletion, and LD indicates the size of alleles in clones identified as harbouring LDs. (C) Schematic showing the positions of S-R PCR primers (SR, blue), M-R PCR primers (MR, black), L-R PCR primers (LR, green), sgRNAs (red boxes), and LDs isolated from clones (dark blue lines) and pools of cells (light blue dashed lines) at each of the three sites. The allele marked with a star contained a secondary deletion at Site 2 distal to the primary cut site that destroyed a primer binding site. (D) Quantification of clone frequencies with homozygous wild type or knock-out genotypes by S-R PCR (validated clones) that were later found to contain a LD on another allele by M-R PCR. Quantification of clone numbers for each transfection that were homozygous knock-out or wild type and contained a LD on the other allele (n = 3 independent transfections per site, each dot is one independent experiment). (E) ddPCR quantification of deletions across a 7 kb window centred on Enhancer 1. Each bar represents the mean relative concentration of the target DNA sequence (±95% confidence interval). mESCs were targeted with 4× sgRNA (blue bars, n = 3) and a non-targeting control (grey bar).
To investigate the genotypes of clones with possible allelic drop-outs we performed PCR screening using medium-range (M-R) PCR, with primers located >600 bp away from the sgRNA cut sites (Figure 1B, C). We found several clones that harboured a LD that was not detected using S-R primers (Figure 1B, C). Indeed, multiple deleted alleles not detected using S-R PCR were observed in a pool of targeted and selected mESCs (Figure 1B). Out of a total of 84 clones that were assigned a homozygous knock-out or wild type genotype based on S-R PCR, 13 (15%) harboured a LD on one allele only detected by M-R PCR (Figure 1B, D). Five clones were further analysed using Sanger sequencing which confirmed bona fide LDs on one allele (Figure 1C). The deletions spanned 300–600 bp from either of the sgRNA cut sites at each of the three distinct genomic sites. Interestingly, one clone contained a secondary deletion upstream from the original cut site that removed one of the S-R PCR primer binding sites (Figure 1C, *, mid panel). Longer-range (L-R) PCR amplifying 5.5 kb fragments revealed even larger LDs spanning up to 2.7 kb away from sgRNA cut sites in pools of selected cells (Figure 1C, light blue dashed lines).
Importantly, PCR and sequencing based approaches may be impacted by biases including over amplification of shorter alleles containing LDs. To accurately quantify LDs in a Cas9D10A nickase-targeted cell population without amplification bias, we utilized droplet digital PCR (ddPCR). Using ddPCR amplicons spaced at 500 bp, 1 kb and 3 kb intervals away from 4× sgRNA cut sites, we found that relative target DNA concentration was significantly reduced in pools of selected cells compared to non-targeting controls (Figure 1E). These findings reveal that LDs are readily detectable in pools of targeted cells irrespective of biases due to PCR primer position or design.
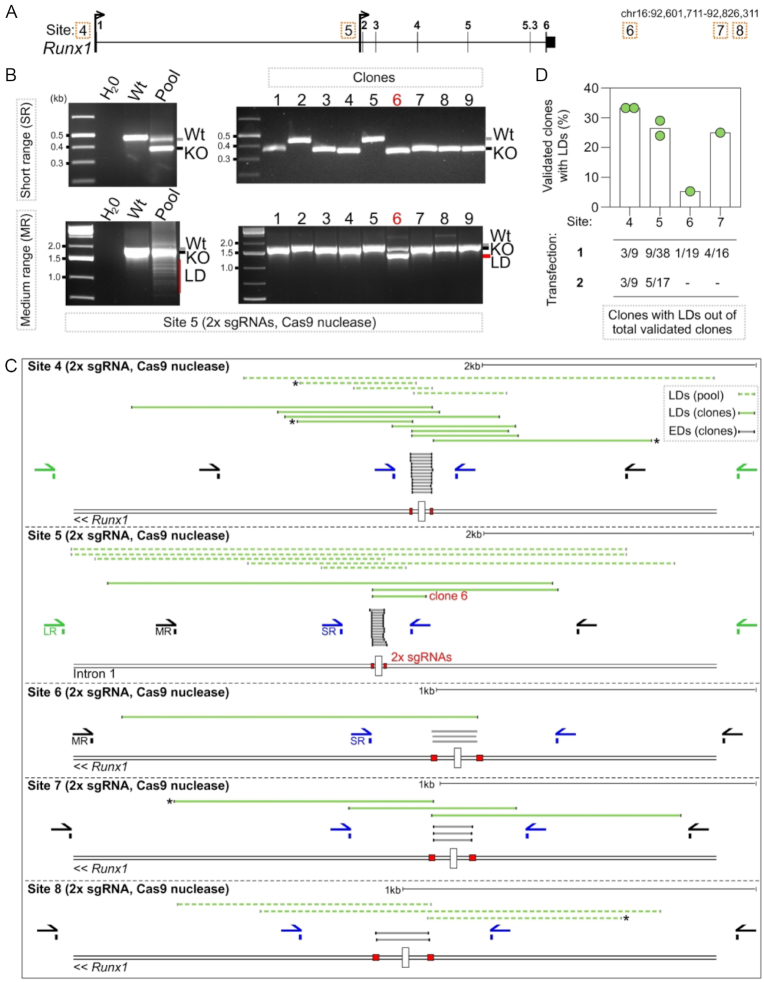
Cas9D10A nickase generates larger deletions at a similar frequency to Cas9 nuclease
To compare the occurrence of LDs between Cas9D10A nickase and Cas9 nuclease, we targeted five further sites (Sites 4–8) at the Runx1 locus using two sgRNAs (2x sgRNAs) to delete 100–150 bp per site (Figure 2A). Out of a total of 506 clones analysed by S-R PCR, 13% and 46% of individual clones targeted using Cas9 nuclease were assigned homozygous wild type or knock-out genotypes, respectively (Figure 2B, C). However, M-R and L-R PCR analysis of several apparent homozygous knock-out or wild type clones revealed that on average 23% (25/108) of these contained a LD on one allele (Compare Figure 2B, D with Figure 1C, D). Sanger sequencing of PCR products amplified from fifteen of these clones confirmed that they harboured alleles carrying LDs that abolished primer binding sites, spanning up to 2 kb away from sgRNA target sites (Figure 2C). M-R and L-R PCR analysis of pools of selected cells targeted with Cas9 nuclease revealed similar LDs to those seen at Sites 1–3 targeted with Cas9D10A nickase (Figure 2C).
Figure 2.
Frequency of larger deletions when genome editing with Cas9 nuclease. (A) Locus maps of CRISPR/Cas9 nuclease strategies to delete Sites 4–8, corresponding to transcription factor binding sites at the Runx1 locus. (B) Gel images showing PCR amplification of gDNA isolated from a pool of selected cells (left-hand gels) or isolated clones (right-hand gels) targeted with Cas9 nuclease at Site 5. PCR screening was performed with SR primers (top gels) and MR primers (bottom gels). Wt next to the gel image indicates the size of the wild type allele, KO indicates the size of alleles harbouring the expected deletion, and LD indicates the size of alleles identified harbouring larger deletions. (C) Schematic showing the positions of S-R PCR primers (SR blue), M-R PCR primers (MR, black), L-R PCR primers (LR, green), sgRNAs (red boxes), and LDs isolated from clones (dark green lines) and pools of cells (light green dashed lines). (D) Quantification of clone frequencies with homozygous wild type or knock-out genotypes by S-R PCR (validated clones) that contained a LD on one allele only detected by medium-range or longer-range PCR (n = 1–2 independent transfections per site, each dot is one independent experiment).
Larger deletions are generated in a variety of genome editing contexts
It has been suggested that previous genetic modification employing the CRISPR/Cas9 system may select for cells with defective p53-mediated DNA repair responses (49,50). We next investigated whether previous exposure to CRISPR/Cas9 might explain the LDs that were induced by Cas9D10A nickase cleavage, as the E14-TG2a-RV mESC line used had previously been targeted with Cas9. Using the same strategy as above we found that targeting Runx1 Site 1 with Cas9D10A nickase in wild type parental E14-TG2a mESCs (that had not previously undergone genome editing) generated alleles with similar LDs to those seen in cells that were previously exposed to Cas9 (Compare Supplementary Figure S2A, with Figure 1C).
To assess whether mESCs are especially susceptible to Cas9 nickase-induced LDs or whether this occurs also in other cell types, we deleted Runx1 Site 1 in the 416B haematopoietic progenitor cell line (28) using a 4× sgRNA CRISPR/Cas9D10A nickase approach (Supplementary Figure S2A). M-R PCR on gDNA isolated from a pool of electroporated (GFP-positive) 416B cells revealed shorter than expected amplicons indicative of LDs that were not detected with S-R PCR (Supplementary Figure S2A). Sanger sequencing of isolated PCR products again confirmed that the shorter PCR amplicons corresponded to LDs up to 980 bp that destroyed one of the S-R primer binding sites (Supplementary Figure S2A).
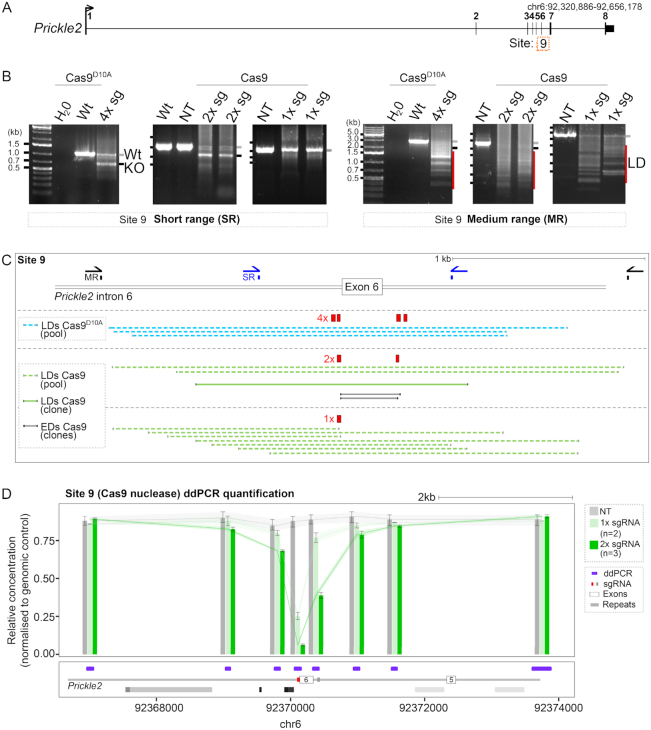
As all targeted sites were located in Runx1, we investigated whether LDs induced by Cas9 could reflect a locus-specific feature of this gene that might not be applicable to other loci. RUNX1 is frequently translocated in acute-myeloid leukaemia (51) and has previously been shown to be exquisitely susceptible to DSBs (52). To explore this possibility, we targeted a gene on another chromosome that has not been associated with DSBs (52) (Prickle2 on chromosome 6). We designed a 4× sgRNA CRISPR/Cas9D10A nickase and 2x sgRNA Cas9 nuclease strategy to delete 300 bp corresponding to exon 6 (Site 9) (53) (Figure 3A). PCR screening of gDNA from pools of transfected and selected E14-TG2a mESCs revealed smaller than expected PCR products that were only detected using M-R primers and were indicative of LDs when using either Cas9 nuclease or Cas9D10A nickase (Figure 3B). M-R PCR screening of isolated mESC clones targeted using 2x sgRNA and Cas9 nuclease revealed a similar frequency of clones (6/17) harbouring LDs as clones targeted at Runx1. Sanger sequencing of M-R PCR products from isolated clones and pools of cells revealed deletions spanning up to 1.2 kb in Prickle 2 beyond expected cut sites that abolished S-R primer binding sites (Figure 3C).
Figure 3.
Larger deletions are generated in a variety of genome-editing contexts. (A) Locus schematic showing Site 9 (Prickle2 exon 6) on mouse chromosome 6. (B) Gel images showing PCR amplification products from gDNA harvested from a pool of transfected cells targeted using the CRISPR/Cas9 strategies indicated. Left gel images correspond to SR primers and right gel images correspond to MR primers. Wt and a grey line next to the gel image indicates the size of the wild type allele, KO and a black line indicates the size of alleles harbouring the expected deletion (based on the location of 2× or 4× sgRNAs), and LD and a red line indicates the size of alleles identified harbouring LDs. (C) Schematic showing the 4× sgRNA CRISPR/Cas9D10A nickase, 1× and 2× sgRNA Cas9 nuclease strategies targeting Prickle2 exon 6. Sequenced PCR products amplified from pools of cells (light blue and light green dashed lines) and one isolated clone (dark green line). Mapped deletions of expected size (EDs) based on the location of the 2× sgRNA cut sites are shown (grey lines). (D) ddPCR quantification of deletions targeting exon 6 with Cas9 nuclease and 1x sgRNA (red box) or 2× sgRNAs (red and grey boxes). Each bar represents the mean ±95% confidence interval. mESCs were targeted with 1× sgRNA (light green bars, n = 2), 2× sgRNA (dark green bars, n = 3) and non-targeting control (grey bar).
It has previously been suggested that end resection may be favoured when two DSBs are located in close proximity to each other (54), which might increase the likelihood of LD formation. However, when generating two proximal DSBs in mESCs, five alleles containing LDs spanned from only one of the two sgRNA target sites (Figure 2C, starred alleles) suggesting that either one DSB was repaired to outside of the LD in these instances, or that a single DSB is sufficient for LD formation. To examine the ability for a single DSB to generate LDs, Site 9 at Prickle2 and Site 7 at Runx1 were targeted with just one sgRNA each. M-R PCR and sequencing identified LDs at both sites (Figure 3B, C, Supplementary Figure S2B), implying that LD formation is independent of two adjacent DSBs. Quantitatively, ddPCR in pools of selected cells showed a slight increase in LDs when cells were targeted using 2× instead of 1× sgRNA and Cas9 nuclease (Figure 3D). Collectively, the above results show that LDs are formed in several different Cas9-mediated genome editing scenarios in cultured cells.
Cas9-induced larger deletions also occur when genome editing in vivo in mouse embryos
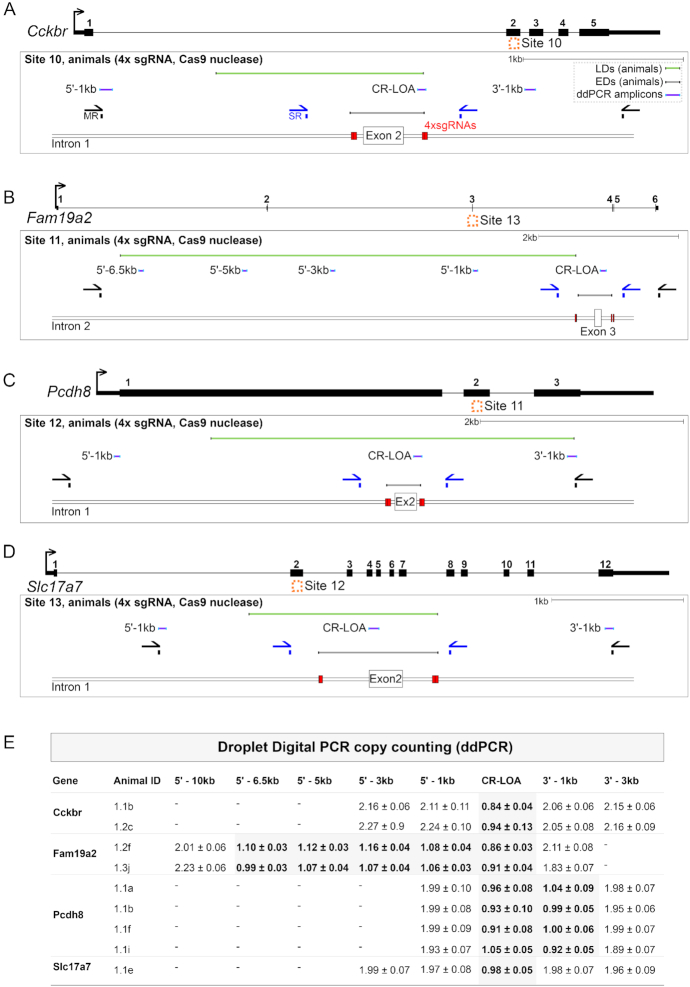
We next extended the study to the generation of Cas9-induced deletions with paired sgRNAs in vivo (International Mouse Phenotyping Consortium, (8)). We surveyed 32 projects aiming to create null alleles where the transmission of a deletion of an expected size had been detected by S-R PCR in the G1 generation, demonstrating sgRNA efficiency. We further validated the positive G1 animals using copy-counting of the deleted fragment by ddPCR. We ran in parallel a small number of control littermates where S-R PCR had not identified any deletion (Supplementary Table S3, Figure 4). We found control animals (without deletion detected by the S-R PCR assay) that nevertheless showed the loss of one copy of the targeted locus by ddPCR in seven out of the 32 surveyed projects. We mapped the extent of the deletion in these animals by running copy counting assays at regular intervals upstream and downstream of the intended deletion interval. Thus, we narrowed down the deletion span in these animals to a 1 kb window to each side of the paired sgRNA target sites (Figure 4). We amplified by PCR and sequenced seven of these genomic loci (Sites 10–16, targeting Cckbr, Fam19a2, Pcdh8, Slc17a7, Elavl4, Scn11a and Trpm2 respectively), and found LDs that destroyed at least one S-R PCR primer binding site and extended up to an additional 7 kb from the intended sgRNA cut sites (Figure 4, Supplementary Figure S3).
Figure 4.
Larger deletions when genome editing in mouse embryos. (A–D) Locus maps of CRISPR/Cas9 strategies to delete Sites 10–13, corresponding to the genes Cckbr, Fam19a2, Pcdh8 and Slc17a7 respectively. Schematics show the positions of S-R PCR primers (SR, blue), M-R PCR primers (MR, black), sgRNAs (red boxes), ddPCR amplicons (purple lines) and LDs (green lines). (E) Copy counting results from ddPCR experiments. Assays against the wild type genomic sequence were designed in the critical region (CR-LOA) and at 1–3 kb intervals in the 5′ or 3′ direction distal to sgRNA cut sites (e.g. a 5′-1 kb amplicon is located 1 kb in the 5′ direction of the sgRNA cut sites). Each row corresponds to an animal where no deletion was detected by S-R PCR.
Microhomologies consistent with MMEJ are prevalent at larger deletions
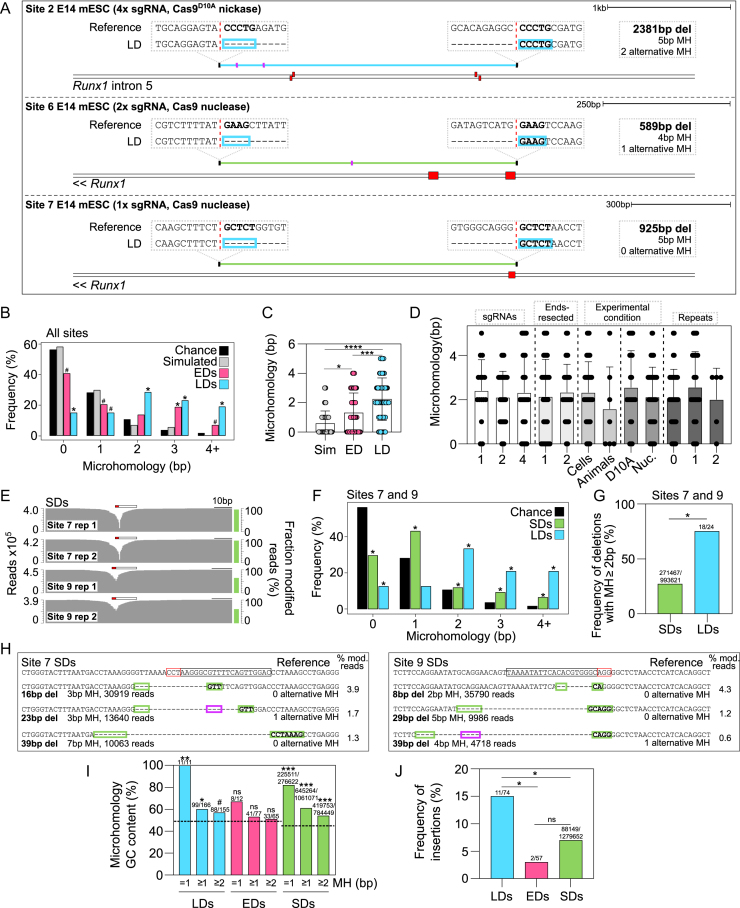
We next explored whether the new DNA sequences that were created after DNA breaks could inform on potential DNA repair mechanisms associated with LDs. Microhomologies of 2–5 bp in length were found at 52 out of 74 (70%) of the LD breakpoint junctions we identified (Figure 5A; Supplementary Tables S4 and S5). Homologous base pair scoring identified significantly more microhomology at LDs compared to simulated LDs, microhomology expected by chance for a k-mer of a given length, and microhomology found at EDs (Figure 5B, C). LDs contained microhomologies irrespective of whether LDs were generated with single or multiple sgRNAs, exhibited DNA end-resection at one or two adjacent DSBs, were generated in vitro or in vivo, by Cas9D10A nickase or Cas9 nuclease (Figure 5D). There was no difference between the length or frequency of microhomologies found at LD breakpoints associated with zero, one or two annotated repeats (Figure 5D), nor were annotated repeats enriched at LD breakpoint junctions (Supplementary Figure S4). In addition to our own data, we analysed 69 Cas9-induced LDs from the literature that were previously generated by single or pairs of sgRNAs (8,19–24). These 69 distinct LDs also contained a significant over-representation of microhomologies compared to the chance expectation or simulated deletions (Supplementary Figure S5; Supplementary Table S5). Altogether, these data suggest that MMEJ is active during the repair of LD alleles, as MMEJ depends on short (<20 bp) microhomologies that are shared between both breakpoints, with some tolerance for mismatches (7).
Figure 5.
Microhomologies consistent with MMEJ are prevalent at Cas9-induced larger deletions. (A) Examples of LDs (blue and green lines) with microhomologies and corresponding reference sequences shown (mm9). Sequences outlined with blue boxes represent microhomologies. Red dashed vertical lines represent the exact breakpoint junctions in the repaired alleles and sgRNAs are shown (red boxes). Total deletion size, microhomology amount, and number of alternative (more proximal) microhomologies are shown (pink lines in deleted sequence). (B) Frequency distribution histogram of microhomologies at 74 LD breakpoint junctions (LDs) across 16 sites, 59 EDs across 16 sites (EDs), 74 simulated deletions (Simulated), and the chance expectation of finding at two locations a k-mer of a given length (Chance) (χ2 test, *, P< 6−7, #, P< 0.02). (C) Microhomology at 74 LDs compared to 59 EDs, and 74 simulated deletions (Sim) (two-tailed Kruskal–Wallis test, ****, P<0.0001, ***, P = 0.0007, *, P = 0.0105). (D) Comparison of microhomology at LDs generated with one, two or four sgRNAs, with ends resected in one or two directions, generated under different experimental conditions, or intersecting with 0, 1 or 2 repeat elements (two-tailed Kruskal–Wallis test, P> 0.9999). (E) Short-amplicon sequencing from pools of mESCs targeted using one sgRNA. Fraction of modified reads and read counts are shown. Protospacer (black outlined bar) and PAM (red outlined bar) are indicated. (F) Microhomology quantification in 24 LDs and all SDs mapped at Sites 7 and 9 compared to the chance expectation of finding a k-mer of a given length (χ2 test, *, P< 0.0003). (G) Quantification of deletions containing microhomology ≥2 bp in all SDs and LDs generated at Sites 7 and 9 using one sgRNA (χ2 test, *, P = 5.4−7). (H) Reference sequence and Cas9-induced deletion alleles containing significant microhomologies at their breakpoints. The total number of reads and the percentage of modified reads is shown. Protospacer (black outlined bar) and PAM (red outlined bar) are indicated. Short microhomologies that abut the deletion (green boxes) and alternative microhomologies located within the deleted region (pink boxes) are shown. (I) Quantification of microhomology GC base pair content in microhomologies of different lengths at all LDs, EDs, and SDs at Site 7 and 9. The expected background GC base pair content is shown as a black dashed line. (χ2 test, ns, P> 0.2, #, P = 0.059, *, P< 0.01, **, P< 0.001, ***, P< 10−10). (J) The number of total LDs, total EDs and SDs at Sites 7 and 9 containing a short insertion (χ2 test, *, P< 0.003, ns, P = 0.2395).
MMEJ was previously implicated in the repair of Cas9-induced DSBs at shorter deletion alleles (SDs) of <60 bp (15,39,55–61). To directly compare the prevalence of microhomologies at Cas9-induced SDs with LDs, we performed short-amplicon deep sequencing after targeting two different chromosomes with 1x sgRNA each (Figure 5E). LDs at Sites 7 and 9 (characterised in Figure 2 and Figure 3) were significantly enriched for microhomologies compared to SDs quantified at the same sites (Figure 5F, G). Still, microhomologies were significantly over-represented at SDs compared to the chance expectation of two sequences containing a k-mer of a given length (Figure 5F, H). MMEJ has previously been shown to favour thermostable microhomologies with elevated GC content (46,55). Microhomologies at all Cas9-induced LDs and SDs at Sites 7 and 9 were both significantly enriched for GC base pairing compared to background (Figure 5I, Supplementary Figure S6), while microhomologies across all EDs were observably but not significantly enriched (Figure 5I). Interestingly, GC bases were always the most enriched in microhomologies of 1 bp, compared to longer microhomologies (Figure 5I). MMEJ is also known to frequently generate small non-templated insertions (62,63). In line with this, Cas9-induced LDs were enriched for small insertions compared to EDs and SDs (Figure 5J, (Supplementary Figure S7). Collectively these data show that the majority of larger Cas9-induced deletions contain microhomologies consistent with MMEJ at their breakpoints.
Larger deletion distribution is dependent on proximity to sgRNAs and cannot be predicted by microhomology sequences alone
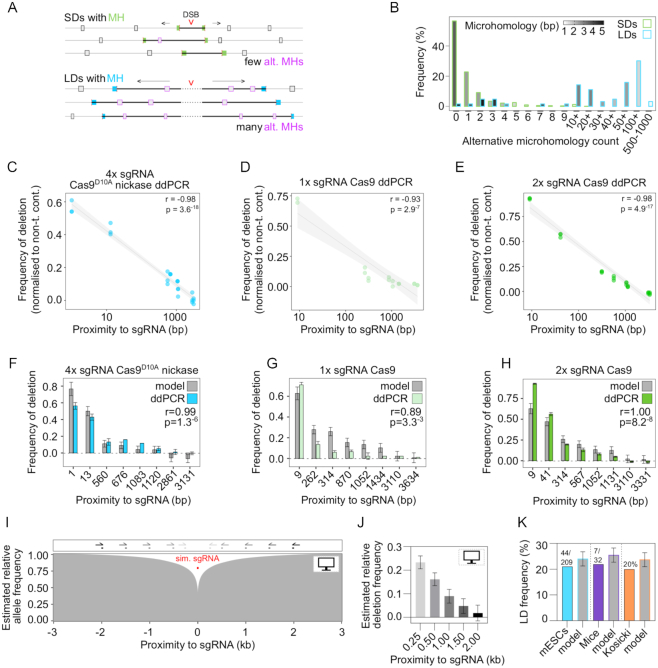
Recent work has suggested that DNA repair outcomes are predictable at Cas9-induced DSBs based on the presence of microhomologies in cut site-proximal DNA sequences (39,55–58). We asked whether the distribution of LDs was similarly dependent on the proximity of microhomologies to cut sites. At LDs, deletion size was independent of microhomology length, unlike at SDs (Supplementary Figure S8A). For all but one LD, the intervening sequence between deletion ends and sgRNA cut sites contained several alternative (more proximal) microhomologies that were bypassed during repair (median = 49, Figure 6A, B). In contrast, microhomologies used for repair at SDs were predominantly (but not exclusively) the most proximal to the cut site (Figure 6A, B, Figure 5H). The number of alternative microhomologies present in the deleted sequence was dependent on deletion length and microhomology length (Supplementary Figure S8B, C, D), reflecting the random distribution of microhomology sequences throughout the genome. Together this indicates that in contrast to SDs, LDs are not repaired to the closest microhomology.
Figure 6.
Larger deletion breakpoints do not occur at proximal microhomology sequences but are dependent on proximity to sgRNAs. (A) Schematic representation of LDs and SDs undergoing end-resection and bypassing alternative more proximal microhomologies during DNA repair. SD microhomologies are shown in green, LD microhomologies are shown in blue, and alternative microhomologies are indicated by pink boxes. The sequence included in the deletion is shown as a bold black line. (B) Quantification of alternative microhomologies that were found in the deleted sequences at Sites 7 and 9 SDs and across all LDs with microhomologies at their breakpoints. The colour gradient represents the mean microhomology score of all deletions within each bin. (C–E) Correlation between frequency of deletion determined by ddPCR and sgRNA proximity. Pearson correlation r and P values are indicated and a linear regression with 95% confidence interval is shown. (F–H) Deletion frequencies of real ddPCR data and model estimates with Pearson correlation r and P values indicated. (I) Model estimate of deletion frequency over a 6 kb window around a simulated sgRNA cut site with simulated PCR primers indicated as grey to black half arrows above the plot. (J) Relative predicted deletion frequencies at each of the simulated primer sites with 95% confidence intervals indicated. (K) Comparison between estimated and empirically determined deletion frequencies in two of our own independent data sets and one recent experiment reported in the literature (17).
Given the fact that LD sizes are independent of cut site proximal microhomology sequences, we examined what other factors might influence LD formation. At the population level, the distribution of deletion sizes as inferred from ddPCR was negatively correlated with proximity to sgRNA target sites (Figure 6C–E). We modelled this relationship using multiple linear regression and found that over 80% of the variance in the distribution of deletion sizes depended on proximity to sgRNAs and sgRNA cutting efficiency determined by ddPCR (Figure 6F–H, Supplementary Figure S9, adjusted R2 = 0.8275, P < 2−16). Interestingly, the model based on empirical ddPCR measurements estimated that in general 22 ± 3% of alleles were deleted 250 bp from sgRNA target sites (Figure 6I, J). This agrees with the 21% (44 out of 209) of our isolated mESC clones that harboured LDs abolishing S-R PCR primers (Figure 1D, Figure 2D, Figure 6K, mean sgRNA proximity = 243 bp). Furthermore, S-R primer binding sites with a mean sgRNA proximity of 211 bp were abolished in 22% (7 out of 32) mouse projects (Figure 4, Supplementary Figure S3, Figure 6K). A recent study also found that LDs >250 bp occurred in up to 20% of alleles (17,64) (Figure 6K).
DISCUSSION
Unintended larger Cas9-induced deletions are common with both Cas9 nuclease and nickases
We have characterised unintended larger than expected on-target deletions induced by CRISPR/Cas9 genome editing. These deletions were found at sixteen sites across nine gene loci on seven different chromosomes. They were not linked to previous exposure to Cas9 nor restricted to one particular cell type. The frequency of larger than expected deletions in response to Cas9-induced DSBs was in keeping with earlier reports (8,12,17–21,24–27). Notably, LDs occurred at a comparable frequency with Cas9 nuclease and Cas9D10A nickases and were found in vitro and in vivo. Thus, although Cas9D10A nickase is favourable over Cas9 nuclease for use in gene therapy based on its reduced off-target effects (5), it is equally prone to generating LDs. The largest unintended on-target deletion we identified spanned an additional 7 kb away from the target sgRNA sites. Often LDs coincided with the location of the PCR primer used in a particular screen, making it likely that even larger deletions are generated. In one previous study, an edited allele was shown to harbour a deletion spanning 42 kb, the largest deletion reported so far (21). We showed that LDs cannot be predicted by proximal regions of microhomology. Instead the distribution of LDs in a cell population could be modelled using computational approaches based on proximity to sgRNA cut sites and sgRNA cutting efficiency. This provides important information for the future design of genome editing experiments. Moreover, it lays the foundation to build more robust models that may be able to accurately predict LDs.
Considerations for the use of genome editing
Failure to detect LDs could lead to the misidentification of a heterozygous deletion as a wild type or homozygous knock-out genotype and misinterpretation of experimental results. Critically, such oversight would also lead to failure to detect potential oncogenic mutations when editing for gene therapy (17,65). Of note, LDs may also be generated at off-target sites. Given the scale and frequency of potentially deleterious LDs, robust screening methods need to be employed to ensure their timely detection. To limit allelic drop-outs when using PCR-based methods, primers should be designed as far away as possible from sgRNA binding sites. Complementary screening methods can be used to increase confidence in genome editing outcomes. Compound heterozygote clones or animals (with different deletions on each allele) can be identified by a convoluted Sanger sequencing read beginning at the sgRNA cut site. Because two copies of the target region are detected, they should be less likely to contain a LD or allelic drop-out (27). Southern blotting has also been used to investigate targeted deletions and insertions by genome editing (66). However, LDs extending beyond a hybridization probe would still be undetectable and copy number analysis would need to be used to alleviate this. Whole-genome sequencing (WGS) is a robust method to detect genome editing outcomes (67) but may be prohibitively expensive for most applications of CRISPR/Cas9 as specialized mate-pair or paired-end sequencing approaches and high coverage must be used to reliably detect chromosomal rearrangements (68,69). However, DNA target capture has been used to reduce sequencing costs (59). Alternatively, fluorescence in situ hybridisation (FISH) (70), and chromosome conformation capture (3C)-based methods have been used to detect structural variants (71,72). In contrast to all these methods, copy number counting by ddPCR is much simpler to implement and does not require prior knowledge about the deletions being screened. It also allows quantification of the frequency of events in populations of cells with a mix of complex genotypes and distinguishes between an iso-allelic deletion (two copies of an identical allele that were repaired by HR), and the presence of LDs (this study and ref. (8)), which is not possible by Sanger sequencing or targeted next-generation sequencing (73).
As well as considering different methods of detection, it may be beneficial to develop methods to reduce the occurrence of LDs. It has been suggested that long-term exposure to sgRNA either through plasmid or lentivirus delivery may increase the frequency of larger indels (>6 bp) (74). Some of our experiments used plasmid delivery leading to relatively prolonged sgRNA and Cas9 expression, which might contribute to the observed high frequency of LDs. Both RNP-delivery (75) and conditional Cas9 approaches (76) limit the time that Cas9 is active within cells and, therefore, might reduce off-target effects and be favourable for gene therapy. However, we also detected LDs when using Cas9 mRNA to perform genome editing, demonstrating that plasmid exposure is not a prerequisite for larger Cas9 deletions, in line with recent results (17,24).
Possible mechanisms for larger deletion generation
Microhomologies were significantly overrepresented at LDs irrespective of site, Cas9 used, whether targeting cultured cells or embryos, or whether DNA end-resection occurred in one or both directions. This may hint at a common molecular mechanism for LD generation downstream of DSB formation. Since we found no association between annotated repeat elements and LDs, SSA is an unlikely mechanism for LD generation. The size of LDs was also outside the normal range for NHEJ (12–16), making this an unlikely mechanism. Since we found microhomologies were significantly overrepresented at LD breakpoint junctions, this suggests that microhomologies are preferentially used as part of the repair mechanism that generates LDs, possibly through MMEJ. MMEJ has previously been implicated in targeted insertions (77–80) as well as deletions, but in previous studies was associated with smaller deletions than the LDs we observed (7,15,39,46,55–61). MMEJ has also been implicated in chromosomal translocations (7,81,82), which by their nature take place over large genomic sequence scales. A recent study found that MMEJ was most active with 5 bp microhomologies (81), which is in line with the overrepresented short microhomologies we observed at Cas9-induced LDs. Moreover, MMEJ repair is thought to favour GC base paring (46), which we also observed at Cas9-induced LDs. Together, these findings are consistent with a possible role for MMEJ in LD formation.
In the context of genome editing, what could cause cells to undergo extensive end resection at Cas9-induced DSBs? One possibility is that during the repair of a DSB, the HR pathway is compromised by both alleles being targeted by Cas9. In preparation for HR, extensive 5′ to 3′ resection occurs at DSB DNA ends, inhibiting NHEJ (83,84). However, because both alleles are likely to be targeted and cut by Cas9 concomitantly, productive HR may fail. It is possible that after extensive end resection and abortive HR, MMEJ then repairs the allele, generating LDs. Indeed, it has previously been suggested that if NHEJ fails, alternative end joining pathways such as MMEJ or SSA are favoured (61,84–86). Whether MMEJ is a back-up survival or primary DNA repair mechanism also remains uncertain (7). Future genetic studies will be needed to determine a critical dependency of Cas9-induced LDs on MMEJ.
Despite finding significant microhomologies at Cas9-induced LDs, we cannot rule out other DSB repair mechanisms also playing a role. Another possible mechanism for LD generation could involve interference with the normal functions of the NHEJ pathway by Cas9 residing on the DNA template. Cas9 has a residence time on DNA of >3 h (87), and might interfere with the normal function of the Ku70/80 heterodimer, which binds to DNA ends and is required for NHEJ (6). In support of this, it has been shown previously that inactivating the Ku80 protein favours larger deletions (7,88,89). In order to test this, a weaker binding genome-editing modality (such as hfCas9 (2)) that might interfere less with Ku binding or function could be tested for its ability to generate LDs. Alternatively, small molecule inhibition of NHEJ (90) could elucidate whether LDs are NHEJ-dependent. Gaining a full molecular understanding of LD generation may be challenging, in part because multiple repair pathways may be active at the same locus (61).
In summary, our findings emphasize the fact that larger than expected deletions are generated at a high frequency when genome editing. LD breakpoint junctions occur at regions with significant microhomology, implicating MMEJ as a possible DSB repair pathway in their formation. In contrast to SDs, the LDs cannot be predicted by proximal microhomology sequences. Instead, the distribution of LD sizes can be modelled in cell populations based on proximity to sgRNA cut sites.
DATA AVAILABILITY
Raw fastq files have been deposited in NCBI’s Gene Expression Omnibus (41) and are accessible through GEO Series accession number GSE130621. All custom scripts are freely available on GitHub (https://github.com/d0minicO/mhscanR).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yavor Bozhilov and Damien Downes for technical advice and critical reading of the manuscript. We thank Mohsin Badat, Joost Van Haasteren, and Ron Schwessinger for technical advice. We thank Jim Hughes, Emanuele Azzoni, Christina Rode, Ruth Williams and Tatjana Sauka-Spengler for helpful discussions and advice.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
MRC Molecular Hematology Unit Core award [MC_UU_12009/2 to MdB]; MRC Unit studentship as part of the MRC Molecular Hematology Unit Core award [MC_UU_12009/2 to D.O.]; Medical Research Council IMPC Strategic Award [53658 to L.T.]; National Institute for Health [U42OD011174 to L.T.]; WIMM Flow Cytometry facility is supported by the MRC HIU, MRC MHU [MC_UU_12009], NIHR Oxford BRC and John Fell Fund [131/030 and 101/517], EPA fund [CF182 and CF170], WIMM Strategic Alliance awards [G0902418 and MC_UU_12025]. Funding for open access charge: MRC [Oxford's RCUK Open Access Block Grant].
Conflict of interest statement. None declared.
REFERENCES
- 1. Sander J.D., Joung J.K.. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014; 32:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F.. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016; 351:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y. et al.. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013; 154:1380–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caldecott K.W. DNA single-strand break repair. Exp. Cell Res. 2014; 329:2–8. [DOI] [PubMed] [Google Scholar]
- 5. Dai W.J., Zhu L.Y., Yan Z.Y., Xu Y., Wang Q.L., Lu X.J.. CRISPR-Cas9 for in vivo gene therapy: promise and hurdles. Mol. Ther. Nucleic Acids. 2016; 5:e349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Symington L.S., Gautier J.. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011; 45:247–271. [DOI] [PubMed] [Google Scholar]
- 7. McVey M., Lee S.E.. MMEJ repair of double-strand breaks (director's cut): deleted sequences and alternative endings. Trends Genet.: TIG. 2008; 24:529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mianne J., Codner G.F., Caulder A., Fell R., Hutchison M., King R., Stewart M.E., Wells S., Teboul L.. Analysing the outcome of CRISPR-aided genome editing in embryos: Screening, genotyping and quality control. Methods. 2017; 121–122:68–76. [DOI] [PubMed] [Google Scholar]
- 9. Hendel A., Fine E.J., Bao G., Porteus M.H.. Quantifying on- and off-target genome editing. Trends Biotechnol. 2015; 33:132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hendel A., Kildebeck E.J., Fine E.J., Clark J., Punjya N., Sebastiano V., Bao G., Porteus M.H.. Quantifying genome-editing outcomes at endogenous loci with SMRT sequencing. Cell Rep. 2014; 7:293–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Komor A.C., Kim Y.B., Packer M.S., Zuris J.A., Liu D.R.. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016; 533:420–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cradick T.J., Fine E.J., Antico C.J., Bao G.. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013; 41:9584–9592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010; 79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koike-Yusa H., Li Y., Tan E.P., Velasco-Herrera Mdel C., Yusa K.. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat. Biotechnol. 2014; 32:267–273. [DOI] [PubMed] [Google Scholar]
- 15. van Overbeek M., Capurso D., Carter M.M., Thompson M.S., Frias E., Russ C., Reece-Hoyes J.S., Nye C., Gradia S., Vidal B. et al.. DNA repair profiling reveals nonrandom outcomes at Cas9-Mediated breaks. Mol. Cell. 2016; 63:633–646. [DOI] [PubMed] [Google Scholar]
- 16. Tan E.P., Li Y., Velasco-Herrera Mdel C., Yusa K., Bradley A.. Off-target assessment of CRISPR-Cas9 guiding RNAs in human iPS and mouse ES cells. Genesis. 2015; 53:225–236. [DOI] [PubMed] [Google Scholar]
- 17. Kosicki M., Tomberg K., Bradley A.. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018; 36:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Canver M.C., Bauer D.E., Dass A., Yien Y.Y., Chung J., Masuda T., Maeda T., Paw B.H., Orkin S.H.. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J. Biol. Chem. 2014; 289:21312–21324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma Y., Shen B., Zhang X., Lu Y., Chen W., Ma J., Huang X., Zhang L.. Heritable multiplex genetic engineering in rats using CRISPR/Cas9. PLoS One. 2014; 9:e89413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhou J., Shen B., Zhang W., Wang J., Yang J., Chen L., Zhang N., Zhu K., Xu J., Hu B. et al.. One-step generation of different immunodeficient mice with multiple gene modifications by CRISPR/Cas9 mediated genome engineering. Int. J. Biochem. Cell Biol. 2014; 46:49–55. [DOI] [PubMed] [Google Scholar]
- 21. Parikh B.A., Beckman D.L., Patel S.J., White J.M., Yokoyama W.M.. Detailed phenotypic and molecular analyses of genetically modified mice generated by CRISPR-Cas9-mediated editing. PLoS One. 2015; 10:e0116484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang H., Yang H., Shivalila C.S., Dawlaty M.M., Cheng A.W., Zhang F., Jaenisch R.. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013; 153:910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang L., Jia R., Palange N.J., Satheka A.C., Togo J., An Y., Humphrey M., Ban L., Ji Y., Jin H. et al.. Large genomic fragment deletions and insertions in mouse using CRISPR/Cas9. PLoS One. 2015; 10:e0120396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adikusuma F., Piltz S., Corbett M.A., Turvey M., McColl S.R., Helbig K.J., Beard M.R., Hughes J., Pomerantz R.T., Thomas P.Q.. Large deletions induced by Cas9 cleavage. Nature. 2018; 560:E8–E9. [DOI] [PubMed] [Google Scholar]
- 25. Shin H.Y., Wang C., Lee H.K., Yoo K.H., Zeng X., Kuhns T., Yang C.M., Mohr T., Liu C., Hennighausen L.. CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat. Commun. 2017; 8:15464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Birling M.C., Schaeffer L., Andre P., Lindner L., Marechal D., Ayadi A., Sorg T., Pavlovic G., Herault Y.. Efficient and rapid generation of large genomic variants in rats and mice using CRISMERE. Scientific Rep. 2017; 7:43331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Codner G.F., Mianne J., Caulder A., Loeffler J., Fell R., King R., Allan A.J., Mackenzie M., Pike F.J., McCabe C.V. et al.. Application of long single-stranded DNA donors in genome editing: generation and validation of mouse mutants. BMC Biol. 2018; 16:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dexter T.M., Allen T.D., Scott D., Teich N.M.. Isolation and characterisation of a bipotential haematopoietic cell line. Nature. 1979; 277:471–474. [DOI] [PubMed] [Google Scholar]
- 29. Handyside A.H., O’Neill G.T., Jones M., Hooper M.L.. Use of BRL-conditioned medium in combination with feeder layers to isolate a diploid embryonal stem cell line. Roux's Arch. Dev. Biol. 1989; 198:48–56. [DOI] [PubMed] [Google Scholar]
- 30. Logan C.V., Cossins J., Rodriguez Cruz P.M., Parry D.A., Maxwell S., Martinez-Martinez P., Riepsaame J., Abdelhamed Z.A., Lake A.V., Moran M. et al.. Congenital myasthenic syndrome type 19 Is caused by mutations in COL13A1, encoding the atypical non-fibrillar collagen type XIII alpha1 chain. Am. J. Hum. Genet. 2015; 97:878–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Anastassiadis K., Schnutgen F., von Melchner H., Stewart A.F.. Gene targeting and site-specific recombination in mouse ES cells. Methods Enzymol. 2013; 533:133–155. [DOI] [PubMed] [Google Scholar]
- 33. Haeussler M., Schonig K., Eckert H., Eschstruth A., Mianne J., Renaud J.B., Schneider-Maunoury S., Shkumatava A., Teboul L., Kent J. et al.. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016; 17:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hodgkins A., Farne A., Perera S., Grego T., Parry-Smith D.J., Skarnes W.C., Iyer V.. WGE: a CRISPR database for genome engineering. Bioinformatics. 2015; 31:3078–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kent W.J. BLAT-the BLAST-like alignment tool. Genome Res. 2002; 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D.. The human genome browser at UCSC. Genome Res. 2002; 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004; 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr. Protoc. Bioinformatics. 2004; Chapter 4:Unit 4 10. [DOI] [PubMed] [Google Scholar]
- 39. Bae S., Kweon J., Kim H.S., Kim J.S.. Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods. 2014; 11:705–706. [DOI] [PubMed] [Google Scholar]
- 40. Simsek D., Jasin M.. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation. Nat. Struct. Mol. Biol. 2010; 17:410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Edgar R., Domrachev M., Lash A.E.. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Magoc T., Salzberg S.L.. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011; 27:2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. 1000 Genome Project Data Processing Subgroup . The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ramirez F., Dundar F., Diehl S., Gruning B.A., Manke T.. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014; 42:W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pinello L., Canver M.C., Hoban M.D., Orkin S.H., Kohn D.B., Bauer D.E., Yuan G.C.. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016; 34:695–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Glover L., Jun J., Horn D.. Microhomology-mediated deletion and gene conversion in African trypanosomes. Nucleic Acids Res. 2011; 39:1372–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nottingham W.T., Jarratt A., Burgess M., Speck C.L., Cheng J.F., Prabhakar S., Rubin E.M., Li P.S., Sloane-Stanley J., Kong A.S.J. et al.. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood. 2007; 110:4188–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schutte J., Wang H., Antoniou S., Jarratt A., Wilson N.K., Riepsaame J., Calero-Nieto F.J., Moignard V., Basilico S., Kinston S.J. et al.. An experimentally validated network of nine haematopoietic transcription factors reveals mechanisms of cell state stability. eLife. 2016; 5:e11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Haapaniemi E., Botla S., Persson J., Schmierer B., Taipale J.. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018; 24:927–930. [DOI] [PubMed] [Google Scholar]
- 50. Ihry R.J., Worringer K.A., Salick M.R., Frias E., Ho D., Theriault K., Kommineni S., Chen J., Sondey M., Ye C. et al.. p53 inhibits CRISPR-Cas9 engineering in human pluripotent stem cells. Nat. Med. 2018; 24:939–946. [DOI] [PubMed] [Google Scholar]
- 51. de Bruijn M., Dzierzak E.. Runx transcription factors in the development and function of the definitive hematopoietic system. Blood. 2017; 129:2061–2069. [DOI] [PubMed] [Google Scholar]
- 52. Crosetto N., Mitra A., Silva M.J., Bienko M., Dojer N., Wang Q., Karaca E., Chiarle R., Skrzypczak M., Ginalski K. et al.. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods. 2013; 10:361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tao H., Manak J.R., Sowers L., Mei X., Kiyonari H., Abe T., Dahdaleh N.S., Yang T., Wu S., Chen S. et al.. Mutations in prickle orthologs cause seizures in flies, mice, and humans. Am. J. Hum. Genet. 2011; 88:138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guirouilh-Barbat J., Gelot C., Xie A., Dardillac E., Scully R., Lopez B.S.. 53BP1 protects against CtIP-Dependent capture of ectopic chromosomal sequences at the junction of distant double-strand breaks. PLoS Genet. 2016; 12:e1006230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shen M.W., Arbab M., Hsu J.Y., Worstell D., Culbertson S.J., Krabbe O., Cassa C.A., Liu D.R., Gifford D.K., Sherwood R.I.. Predictable and precise template-free CRISPR editing of pathogenic variants. Nature. 2018; 563:646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ata H., Ekstrom T.L., Martinez-Galvez G., Mann C.M., Dvornikov A.V., Schaefbauer K.J., Ma A.C., Dobbs D., Clark K.J., Ekker S.C.. Robust activation of microhomology-mediated end joining for precision gene editing applications. PLoS Genet. 2018; 14:e1007652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Allen F., Crepaldi L., Alsinet C., Strong A.J., Kleshchevnikov V., De Angeli P., Palenikova P., Khodak A., Kiselev V., Kosicki M. et al.. Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol. 2018; 37:64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Taheri-Ghahfarokhi A., Taylor B.J.M., Nitsch R., Lundin A., Cavallo A.L., Madeyski-Bengtson K., Karlsson F., Clausen M., Hicks R., Mayr L.M. et al.. Decoding non-random mutational signatures at Cas9 targeted sites. Nucleic Acids Res. 2018; 46:8417–8434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chakrabarti A.M., Henser-Brownhill T., Monserrat J., Poetsch A.R., Luscombe N.M., Scaffidi P.. Target-Specific precision of CRISPR-Mediated genome editing. Mol. Cell. 2018; 73:699–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kim S.I., Matsumoto T., Kagawa H., Nakamura M., Hirohata R., Ueno A., Ohishi M., Sakuma T., Soga T., Yamamoto T. et al.. Microhomology-assisted scarless genome editing in human iPSCs. Nat. Commun. 2018; 9:939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brinkman E.K., Chen T., de Haas M., Holland H.A., Akhtar W., van Steensel B.. Kinetics and fidelity of the repair of Cas9-induced double-strand DNA breaks. Molecular cell. 2018; 70:801–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yousefzadeh M.J., Wyatt D.W., Takata K., Mu Y., Hensley S.C., Tomida J., Bylund G.O., Doublie S., Johansson E., Ramsden D.A. et al.. Mechanism of suppression of chromosomal instability by DNA polymerase POLQ. PLoS Genet. 2014; 10:e1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sfeir A., Symington L.S.. Microhomology-Mediated end Joining: A Back-up survival mechanism or dedicated pathway. Trends Biochem. Sci. 2015; 40:701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Thomas M., Burgio G., Adams D.J., Iyer V.. Collateral damage and CRISPR genome editing. PLoS Genet. 2019; 15:e1007994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Teboul L., Greenfield A.. CRISPR-Cas9-mediated mutagenesis: mind the gap. CRISPR J. 2018; 1:263–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Glenn G., Andreou L.V.. Analysis of DNA by Southern blotting. Methods Enzymol. 2013; 529:47–63. [DOI] [PubMed] [Google Scholar]
- 67. Smith C., Gore A., Yan W., Abalde-Atristain L., Li Z., He C., Wang Y., Brodsky R.A., Zhang K., Cheng L. et al.. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell. 2014; 15:12–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Korbel J.O., Urban A.E., Affourtit J.P., Godwin B., Grubert F., Simons J.F., Kim P.M., Palejev D., Carriero N.J., Du L. et al.. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007; 318:420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Johnson S.H., Smadbeck J.B., Smoley S.A., Gaitatzes A., Murphy S.J., Harris F.R., Drucker T.M., Zenka R.M., Pitel B.A., Rowsey R.A. et al.. SVAtools for junction detection of genome-wide chromosomal rearrangements by mate-pair sequencing (MPseq). Cancer Genet. 2018; 221:1–18. [DOI] [PubMed] [Google Scholar]
- 70. Boroviak K., Fu B., Yang F., Doe B., Bradley A.. Revealing hidden complexities of genomic rearrangements generated with Cas9. Scientific Rep. 2017; 7:12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Simonis M., Klous P., Homminga I., Galjaard R.J., Rijkers E.J., Grosveld F., Meijerink J.P., de Laat W.. High-resolution identification of balanced and complex chromosomal rearrangements by 4C technology. Nat. Methods. 2009; 6:837–842. [DOI] [PubMed] [Google Scholar]
- 72. Harewood L., Kishore K., Eldridge M.D., Wingett S., Pearson D., Schoenfelder S., Collins V.P., Fraser P.. Hi-C as a tool for precise detection and characterisation of chromosomal rearrangements and copy number variation in human tumours. Genome Biol. 2017; 18:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bell C.C., Magor G.W., Gillinder K.R., Perkins A.C.. A high-throughput screening strategy for detecting CRISPR-Cas9 induced mutations using next-generation sequencing. BMC Genomics. 2014; 15:1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kosicki M., Rajan S.S., Lorenzetti F.C., Wandall H.H., Narimatsu Y., Metzakopian E., Bennett E.P.. Dynamics of indel profiles induced by various CRISPR/Cas9 Delivery Methods. Prog. Mol. Biol. Transl. Sci. 2017; 152:49–67. [DOI] [PubMed] [Google Scholar]
- 75. Kim S., Kim D., Cho S.W., Kim J., Kim J.S.. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014; 24:1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhou W., Deiters A.. Conditional control of CRISPR/Cas9 function. Angew. Chem. 2016; 55:5394–5399. [DOI] [PubMed] [Google Scholar]
- 77. Yao X., Wang X., Hu X., Liu Z., Liu J., Zhou H., Shen X., Wei Y., Huang Z., Ying W. et al.. Homology-mediated end joining-based targeted integration using CRISPR/Cas9. Cell Res. 2017; 27:801–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yao X., Wang X., Liu J., Shi L., Huang P., Yang H.. CRISPR/Cas9-mediated targeted integration in vivo using a Homology-mediated End Joining-based strategy. J. Visual. Exp.: JoVE. 2018; 12:doi:10.3791/56844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Suzuki K.T., Sakane Y., Suzuki M., Yamamoto T.. A simple Knock-In system for xenopus via microhomology mediated end joining repair. Methods Mol. Biol. 2018; 1865:91–103. [DOI] [PubMed] [Google Scholar]
- 80. Nakade S., Tsubota T., Sakane Y., Kume S., Sakamoto N., Obara M., Daimon T., Sezutsu H., Yamamoto T., Sakuma T. et al.. Microhomology-mediated end-joining-dependent integration of donor DNA in cells and animals using TALENs and CRISPR/Cas9. Nat. Commun. 2014; 5:5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yanik M., Ponnam S.P.G., Wimmer T., Trimborn L., Muller C., Gambert I., Ginsberg J., Janise A., Domicke J., Wende W. et al.. Development of a reporter system to explore MMEJ in the context of replacing large genomic fragments. Mol. Ther. Nucleic Acids. 2018; 11:407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Simsek D., Brunet E., Wong S.Y., Katyal S., Gao Y., McKinnon P.J., Lou J., Zhang L., Li J., Rebar E.J. et al.. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011; 7:e1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. San Filippo J., Sung P., Klein H.. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008; 77:229–257. [DOI] [PubMed] [Google Scholar]
- 84. Wang H., Perrault A.R., Takeda Y., Qin W., Wang H., Iliakis G.. Biochemical evidence for Ku-independent backup pathways of NHEJ. Nucleic Acids Res. 2003; 31:5377–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. DiBiase S.J., Zeng Z.C., Chen R., Hyslop T., Curran W.J. Jr, Iliakis G.. DNA-dependent protein kinase stimulates an independently active, nonhomologous, end-joining apparatus. Cancer Res. 2000; 60:1245–1253. [PubMed] [Google Scholar]
- 86. Guirouilh-Barbat J., Rass E., Plo I., Bertrand P., Lopez B.S.. Defects in XRCC4 and KU80 differentially affect the joining of distal nonhomologous ends. Proc. Natl. Acad. Sci. U.S.A. 2007; 104:20902–20907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Ma H., Tu L.C., Naseri A., Huisman M., Zhang S., Grunwald D., Pederson T.. CRISPR-Cas9 nuclear dynamics and target recognition in living cells. J. Cell Biol. 2016; 214:529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Boulton S.J., Jackson S.P.. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996; 24:4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liang F., Romanienko P.J., Weaver D.T., Jeggo P.A., Jasin M.. Chromosomal double-strand break repair in Ku80-deficient cells. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:8929–8933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Perrault R., Wang H., Wang M., Rosidi B., Iliakis G.. Backup pathways of NHEJ are suppressed by DNA-PK. J. Cell. Biochem. 2004; 92:781–794. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw fastq files have been deposited in NCBI’s Gene Expression Omnibus (41) and are accessible through GEO Series accession number GSE130621. All custom scripts are freely available on GitHub (https://github.com/d0minicO/mhscanR).