Abstract
Vision is underpinned by phototransduction, a signaling cascade that converts light energy into an electrical signal. Among insects, phototransduction is best understood in Drosophila melanogaster. Comparison of D. melanogaster against three insect species found several phototransduction gene gains and losses, however, lepidopterans were not examined. Diurnal butterflies and nocturnal moths occupy different light environments and have distinct eye morphologies, which might impact the expression of their phototransduction genes. Here we investigated: 1) how phototransduction genes vary in gene gain or loss between D. melanogaster and Lepidoptera, and 2) variations in phototransduction genes between moths and butterflies. To test our prediction of phototransduction differences due to distinct visual ecologies, we used insect reference genomes, phylogenetics, and moth and butterfly head RNA-Seq and transcriptome data. As expected, most phototransduction genes were conserved between D. melanogaster and Lepidoptera, with some exceptions. Notably, we found two lepidopteran opsins lacking a D. melanogaster ortholog. Using antibodies we found that one of these opsins, a candidate retinochrome, which we refer to as unclassified opsin (UnRh), is expressed in the crystalline cone cells and the pigment cells of the butterfly, Heliconius melpomene. Our results also show that butterflies express similar amounts of trp and trpl channel mRNAs, whereas moths express ∼50× less trp, a potential adaptation to darkness. Our findings suggest that while many single-copy D. melanogaster phototransduction genes are conserved in lepidopterans, phototransduction gene expression differences exist between moths and butterflies that may be linked to their visual light environment.
Keywords: opsin, trp, DAGL, wunen, Calx, Nckx30C
Introduction
Vision has intrigued scientists for many years. One of the earliest steps in vision involves the conversion of light into an electrical signal, a process known as phototransduction (Shichida and Matsuyama 2009). Phototransduction is one of the best-studied signaling pathways. In Drosophila melanogaster, phototransduction genes have been investigated for over 40 years (Hardie 2001; Hardie and Raghu 2001; Katz and Minke 2009; Montell 2012; Hardie and Juusola 2015). However, studies of phototransduction genes in other insects are largely lacking. A comparison of vision-related genes in four insect genomes (mosquito, red flour beetle, honeybee, and fruit fly) found gains and losses across lineages (Bao and Friedrich 2009). Drosophila melanogaster had by far the largest number of gene gains compared with the other insects. This implies that those insects missing D. melanogaster orthologs may differ in the genes underlying phototransduction.
Phototransduction takes place in specialized neurons known as photoreceptor cells whose microvilli incorporate light-sensitive opsin proteins bound to a retinal-derived molecule called a chromophore (Fain et al. 2010). Phototransduction begins when light is absorbed by the chromophore (11-cis-3-hydroxyretinal in D. melanogaster) causing the chromophore to change its conformation from cis- to all-trans (von Lintig et al. 2010). In D. melanogaster, this change in conformation triggers a G-protein-coupled cascade (similar to fig. 1) that activates phospholipase C (PLC) (Bloomquist et al. 1988). PLC hydrolyzes phosphatidylinositol 4,5-bisphosphate (PIP2) to produce inositol 1,4,5-trisphosphate (InsP3) and diacylglycerol (DAG) (Bloomquist et al. 1988; Hardie 2001). Concurrently, by a mechanism that is not well understood, there is an opening of Ca2+-permeable light-sensitive transient receptor potential (TRP) and transient receptor potential-like (TRPL) channels, which causes depolarization of the cell (Montell and Rubin 1989; Hardie and Minke 1992; Niemeyer et al. 1996; Shieh and Zhu 1996; Montell 2005). Finally, phototransduction is terminated when the activated rhodopsin (metarhodopsin) binds arrestin (Dolph et al. 1993; Stavenga and Hardie 2011).
Fig. 1.
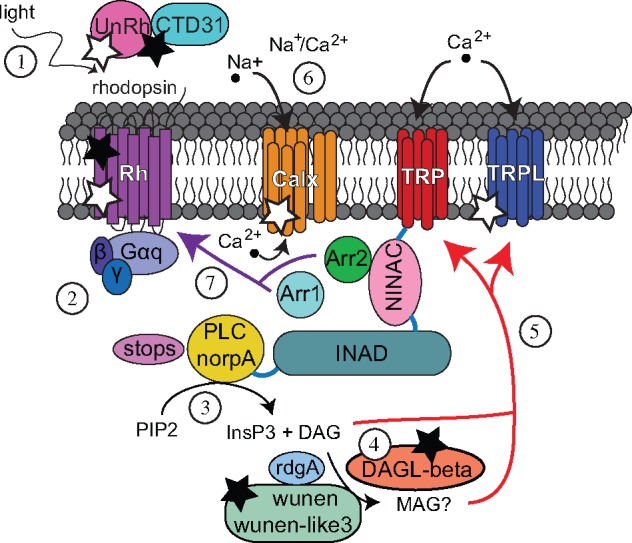
—Speculative model of the lepidopteran phototransduction cascade. Most lepidopteran vision genes are single-copy orthologs of D. melanogaster genes except in chromophore binding, photoisomerization, and diacylglycerol metabolism. 1) Light activates rhodopsin by a conformational change of the chromophore from 11-cis 3-hydroxyretinal to all-trans. The chromophore is transported by Hme CTD31 and photoisomerized from all-trans 3-hydroxyretinal to 11-cis by the unclassified opsin (UnRh), orthologs of which are not found in D. melanogaster. 2) Gαq is released from a G-protein complex of three subunits (α, β, and γ) and activates phospholipase C (PLC). 3) PLC hydrolyzes PIP2 to produce inositol 1,4,5-trisphosphate (InsP3) and diacylglycerol (DAG). 4) Diacylglycerol lipase (DAGLβ) hydrolyzes DAG to produce MAG. In D. melanogaster, DAGLα (inaE) hydrolyzes DAG. DAG levels are also regulated by rdgA and, perhaps, wunen or wunen-like 3. In D. melanogaser, lazaro plays the latter role. 5) DAG and MAG may activate TRP and TRPL by a mechanism that has not been established. A signaling complex that includes TRP and PLC is coordinated by INAD. 6) Na+/Ca2+ exchanger channel (Calx) pumps Ca2+ out of the photoreceptor cell. 7) Arrestin 1 and 2 bind rhodopsin to terminate the cascade with Arrestin 2 as the dominant arrestin in both D. melanogaster and butterflies. INAD and NinaC bind to each other, and both bind calmodulin, to accelerate arrestin binding rhodopsin. STOPS is another protein that terminates phototransduction. Black stars signify differences in phototransduction between D. melanogaster and Lepidoptera due to gene duplication. White stars represent differences in relative expression of UnRh, Rh7, and Calx between moths and butterflies, and in trp and trpl between flies, moths, and butterflies. Further description of these genes can be found in supplementary Table S11, Supplementary Material online.
A plethora of studies have focused on characterizing the opsins including their expression in photoreceptor cells and the arrangement of those photoreceptor cells across the compound eye (Spaethe and Briscoe 2005; Henze et al. 2012; Futahashi et al. 2015; McCulloch et al. 2016; Perry et al. 2016; Giraldo-Calderón et al. 2017; McCulloch et al. 2017). Opsin phylogenies have been used to understand the evolutionary history of light detection (Arendt 2003; Raible et al. 2006; Plachetzki et al. 2007; Suga et al. 2008; Porter et al. 2012; Ramirez et al. 2016; Vöcking et al. 2017). These studies have reconstructed opsins present in the ancestor of bilaterian animals (Ramirez et al. 2016) and have described new opsin types (Vöcking et al. 2017). However, despite the large focus on opsins, changes in the downstream pathway in which opsins function undoubtedly contribute to differences in vision (Plachetzki et al. 2010). Fewer studies have investigated the downstream phototransduction cascade in non-D. melanogaster insects. Studies of phototransduction in other insects have focused on presence, absence, or relative expression of genes in head transcriptomes. In the troglobiont beetle, Ptomaphagus hirtus, for example, 20 genes were identified from adult head mRNA (Friedrich et al. 2011). Exposure of the oriental armyworm, Mythimna separata, to different light environments resulted in differential expression of phototransduction genes in adult heads (Duan et al. 2017). Similarly, phototransduction genes were also differentially expressed (DE) between seasonal forms in heads of the butterfly Bicyclus anynana (Macias-Muñoz et al. 2016). One study quantified opsin and TRP channel gene expression and used RNAi to determine that trpl has the largest effect on phototransduction in the nocturnal cockroach Periplaneta americana (French et al. 2015). Yet, it remains largely unknown how variable the phototransduction cascade is between insect species.
Lepidoptera provides an interesting group in which to investigate the molecular evolution and expression of phototransduction genes in insects adapted to different light environments (Yagi and Koyama 1963; Horridge et al. 1972; Nilsson et al. 1984; Yack et al. 2007; Warrant and Dacke 2016). Unlike D. melanogaster, in which an ommatidium consists of eight photoreceptors with an open rhabdom, the microvillar stacks where light is absorbed by the rhodopsins (Wernet et al. 2015), butterflies have nine photoreceptor cells and a fused rhabdom (Wernet et al. 2015). Interestingly, moths and butterflies also differ from each other in eye morphology related to their light environments. Most butterflies have apposition-type eyes, where light from each lens is processed by one rhabdom and each ommatidium is separated by a sheath of light-absorbing screening pigment which blocks stray light from other ommatidia (Yack et al. 2007; Warrant and Dacke 2016; Conversely, moths have superposition-type eyes where rhabdoms are separated from the crystalline cones by a translucent area allowing light to reach each rhabdom from hundreds of lenses (Yack et al. 2007; Warrant and Dacke 2016).
We predicted that we would find variation in phototransduction gene gains and losses between D. melanogaster and Lepidoptera, and between moths and butterflies due to differences in eye morphology. In fact, phylogenetic analyses have revealed numerous duplications of lepidopteran opsin genes (Spaethe and Briscoe 2004; Sison-Mangus et al. 2008; Briscoe et al. 2010). A survey of 23 vision-related gene families in 19 metazoan genomes revealed that eye development and phototransduction genes have higher rates of retention and duplications in pancrustaceans (Rivera et al. 2010). Because only the nocturnal domesticated silkmoth Bombyx mori was used in the pancrustacean study and only five gene families involved in phototransduction were examined (r-opsin, TRP, PLC, Gq-alpha, and arrestin) (Rivera et al. 2010), it remains to be seen if there are additional differences in phototransduction genes between D. melanogaster and moth and butterfly species. In our present study, we expand the genes surveyed thus far by looking at 76 phototransduction-related genes. Phylogenetic analyses of phototransduction genes in Lepidoptera may reveal: 1) the extent to which D. melanogaster phototransduction genes are duplicated or deleted in Lepidoptera, 2) lepidopteran-specific phototransduction features, and 3) differences between moths and butterflies.
Although gene trees tell the probable evolutionary history of gene families, gene expression data provide a step toward inferring gene function. Genes involved in vision should be highly expressed in photoreceptor cells and upregulated in the eyes relative to other tissue types. Visualizing or quantifying where phototransduction genes are expressed will reveal whether they have a potential role in vision. As an example, the horseshoe crab Limulus polyphemus has 18 opsins, some of which are expressed only in the eyes, in eyes and central nervous system, exclusively in the central nervous system, and some not expressed in either (Battelle et al. 2016). It is possible that the opsins missing from the eyes and central nervous system are expressed in other tissue types and have nonvisual functions (Feuda et al. 2016) or are not expressed at all. Similarly, the reference genome of the butterfly Heliconius melpomene (Davey et al. 2016) has a UVRh duplication but mRNA levels of one copy are downregulated in adult eyes compared with the other copy, and no protein expression of the downregulated copy is detectable in the eye (McCulloch et al. 2017). Studies such as these highlight the importance of quantifying gene expression in candidate tissues before inferring gene function based on sequence alone. Furthermore, it is also possible that a paralog has assumed the predicted visual function. As an example, H. melpomene is missing an ortholog of D. melanogaster chromophore-binding pinta (Smith and Briscoe 2015; Wang and Montell 2005). Instead, a lepidopteran paralog (CTD31) appears to carry out a similar function to that of the missing gene (Macias-Muñoz et al. 2017). Moreover, as observed in the cockroach, whereas genes such as trp and trpl are conserved and expressed, one gene copy (trpl) might have a greater impact on phototransduction than the other (French et al. 2015). Consequently, investigating both gene gain/loss and the expression of phototransduction genes in Lepidoptera might uncover differences in visual processing that helps moths and butterflies function in different light environments.
In this study, we combined transcriptomics and phylogenetics to perform an extensive investigation of candidate phototransduction genes in Lepidoptera. We used RNA-Sequencing data from four tissues of the butterfly H. melpomene to identify genes upregulated in heads. We hypothesized that genes upregulated in heads might have eye and vision-related functions. A functional enrichment analysis suggested that many of the genes upregulated in H. melpomene heads function in phototransduction. To identify gene gain or loss between D. melanogaster and Lepidoptera, and between moths and butterflies, we extracted 76 phototransduction-related gene sequences from reference genomes of eight insect species including the moth, Manduca sexta, and the butterflies, Danaus plexippus and H. melpomene (Zhan et al. 2011; Davey et al. 2016; Kanost et al. 2016). Then we generated 32 phylogenetic trees. In case any genes were missing annotations in the reference assemblies, we searched de novo transcriptome assemblies from M. sexta, H. melpomene, and D. plexippus. We found that most of the phototransduction pathway is conserved between Lepidoptera and D. melanogaster, with some exceptions (see stars in fig. 1). Our methods allowed us to uncover two lepidopteran opsin genes that lack a homolog in D. melanogaster. One of the opsins was highly expressed in butterfly eyes so we used antibodies to locate its expression in pigment cells. In addition, DAG regulation appears to differ between Lepidoptera and D. melanogaster, where a paralogous gene in lepidopterans, DAGβ, may be taking on a role of a lost ortholog of D. melanogaster, DAGα. Although we found a few gene duplication differences between moth and butterfly species, we did not find any consistent differences in gene duplications between the moths and butterflies investigated. Instead, we discovered an intriguing difference between moths and butterflies in their expression of vision-related ion channels, trp, Calx, and Nckx30C.
Materials and Methods
Transcriptome-Wide Differential Expression Analysis
RNA-sequencing data for H. melpomene male and female heads, antennae, legs, and mouth parts were obtained from ArrayExpress projects E-MTAB-1500 and E-MTAB-6249 (supplementary Table S1, Supplementary Material online). A four tissue de novo transcriptome made from one library per tissue type per sex was used as reference (see Macias-Muñoz et al. 2017). Reads from each sample were mapped to the transcriptome using bwa (Li and Durbin 2009) and RSEM (Li and Dewey 2011) was used to quantify mapped raw reads. We used edgeR (Robinson et al. 2010) to perform three pairwise comparisons for differential expression analysis: Heads versus antennae, heads versus legs, and heads versus mouth parts. For each comparison, a generalized linear model was used to include terms for batch, tissue, sex, the interaction of sex, and tissue (∼batch + tissue + sex + sex*tissue). Each analysis also included filtering to remove contigs with low expression (<1 count per million for at least four groups). Samples were normalized using a trimmed mean of the log expression ratios (TMM) (Robinson and Oshlack 2010). After each comparison, P-values were further corrected using a Bonferroni false discovery rate (FDR) correction. Contigs were considered significantly DE when the FDR was <0.05 and the log fold change (logFC) was >1.
Of these DE contigs, we identified those which were upregulated in heads for each comparison. The resulting gene lists were merged to identify contigs commonly upregulated in heads. Patterns of expression for significant contigs and those commonly upregulated in heads were visualized using heatmaps (Ploner 2012). Contigs were annotated with D. melanogaster gene IDs (Marygold et al. 2012) by using command-line BLAST+ to compare H. melpomene transcriptome sequences to D. melanogaster gene sequences (Camacho et al. 2009). We used batch download in Flybase to acquire gene ontology (GO) terms for our DE and head upregulated contigs. DE contigs with unique annotations were enriched for function using a Database for Annotation, Visualization, and Integrated Discovery (DAVID) (Huang et al. 2009). Contigs upregulated in heads were also assigned GO terms and protein classification by NCBI BLAST and InterProScan in BLAST2GO to uncover additional annotations potentially missing from a comparison to D. melanogaster only (Conesa et al. 2005; Conesa and Götz 2008; Götz et al. 2008).
Phototransduction Genes in Insect Genomes
To identify phototransduction genes in Lepidoptera and explore their evolutionary history, we used D. melanogaster sequences to search for homologs in published genomes. We began with a compilation of sequences by Bao and Friedrich (2009) but expanded it to include Lepidoptera species and additional phototransduction genes (supplementary Table S2, Supplementary Material online). We used BLAST to search the genomes of Anopheles gambiae, Apis mellifera, Tribolium castaneum, B. mori, M. sexta, H. melpomene, and D. plexippus. Sequences with identity of more than 20% and an E-value greater than 1E−10 were tested for homology using reciprocal blastp to the NCBI database. The search for D. melanogaster homologs in eight insect genomes resulted in a list of 76 unique genes from phototransduction gene families in insects. In addition to searching lepidopteran reference genomes, we searched de novo transcriptomes to improve annotations and find duplicates that are not found in genomes. We searched a H. melpomene four tissue transcriptome (Macias-Muñoz et al. 2017) and a M. sexta head transcriptome (Smith et al. 2014). We used Trinity to generate a de novo transcriptome using two D. plexippus adult whole heads. The de novo transcriptome was used in addition to the genome to confirm gene duplications (supplementary Tables S3–S5, Supplementary Material online). The nucleotide sequences recovered from de novo transcriptomes were translated using OrfPredictor with the blastx option before testing them by reciprocal blast hits (Min et al. 2005).
Sequence corrections were accomplished by aligning sequences in molecular evolutionary genetics analysis (MEGA) software and manually correcting missing pieces. BLAST was then used to recover the segment from the genome. To obtain the consensus sequences, we inputted corrected sequences to CLC Genomics (CLCBio) and mapped reads against them. With some exceptions, we recovered the entire sequence for all phototransduction genes in H. melpomene (supplementary Table S3, Supplementary Material online), M. sexta (supplementary Table S4, Supplementary Material online), and D. plexipplus (supplementary Table S5, Supplementary Material online). Phototransduction genes for H. melpomene, M. sexta, and D. plexippus were annotated and deposited in GenBank with accession numbers MK983015–MK983088, MK983089–MK983165, and MN037884–MN037955 (supplementary Tables S3–S5, Supplementary Material online). In addition, to examine the evolution of the inaE gene in D. melanogaster and the DAGLβ-like gene in lepidopterans in a wider context, we searched NCBI for insect sequence matches as well as matches to Homo sapiens, Mus musculus, and Hydra vulgaris sequences.
Protein sequences for each gene family were aligned in MEGA 7.0 using the Multiple Sequence Comparison by Log-Expectation (MUSCLE) algorithm (Edgar 2004; Kumar et al. 2016). The alignments were further corrected manually. Before generating maximum likelihood trees, we calculated Bayesian Information Criterion values to assess which substitution model would best fit our data (Schwarz 1978; Kumar et al. 2016). We used the best fit model to generate phylogenies using 100 bootstrap replicates (supplementary Table S6, Supplementary Material online).
Expression of Candidate Genes
To study expression patterns among homologs, we looked at the expression of all genes found in 32 phototransduction gene families in M. sexta heads and in H. melpomene heads, antennae, legs, and mouth parts (i.e., labial palps+proboscis). Rearing conditions for M. sexta are described in Smith et al. (2014) and for H. melpomene in Briscoe et al. (2013) and Macias-Muñoz et al. (2017). We began by adding our corrected H. melpomene and M. sexta sequences (supplementary Tables S3 and S4, Supplementary Material online) to the de novo transcriptome assembly. We uniquely mapped trimmed and parsed reads from four male and four female M. sexta heads (E-MTAB-2066; Smith et al. 2014) to the corrected M. sexta transcriptome using bowtie v. 1.0 (Langmead et al. 2009). We also mapped processed reads from H. melpomene heads, antennae, legs, and mouth parts (E-MTAB-1500, E-MTAB-6249, E-MTAB-6342; Macias-Muñoz et al. 2017) to the corrected H. melpomene transcriptome. RSEM was used to count raw reads mapped (Li and Dewey 2011). We visualized expression levels by graphing Transcripts Per Million (TPM) for each gene of interest using ggplot2 (Wickham 2009). Differential expression between tissue types for H. melpomene was repeated as outlined above in edgeR using uniquely mapped reads to the transcriptome with corrected sequences. However, for this data set to allow for less stringency, we used q-values (Dabney and Storey 2013) to correct P-values rather than Bonferroni.
Immunohistochemistry
An antibody was generated against the peptide N-CKGARTVDEDKKKE-C of the H. melpomene unclassified opsin (UnRh) in guinea pig and was immunoaffinity purified (New England Peptide, Gardner, MA, USA). We also used an antibody against the long-wavelength sensitive opsin (LWRh) of Limenitis astyanax (Frentiu et al. 2007; 2015) which labels LWRh expressing cells in Heliconius (McCulloch et al. 2016). Eyes were fixed, sucrose protected, cryosectioned, and immunolabeled according to methods in McCulloch et al. (2016). Following washes with 1× Phosphate-buffered saline and block (McCulloch et al. 2016; Macias-Muñoz et al. 2017), slides were incubated with 1:15 rabbit anti-LWRh and 1:30 guinea pig anti-UnRh antibodies in blocking solution overnight at 4°C. After washing in 1× Phosphate-buffered saline, slides were incubated with 1:500 goat anti-rabbit Alexafluor 555 and 1:250 goat anti-guinea pig Alexafluor 633 secondary antibodies in blocking solution for 2 h at room temperature in the dark. Slides were washed once more in 1× PBS in the dark and stored for imaging in Aqua Poly/Mount (Polysciences, Inc. Cat. No. 18606). Images were taken at the UC Irvine Optical Biology Core Facility using a Zeiss LSM700 confocal microscope under a 20× objective. Two-channel composites were generated using Fiji and brightness was adjusted for clarity using Adobe Photoshop.
Results and Discussion
Transcriptome-Wide Differential Expression Analysis
To determine the possible functions of genes expressed in butterfly heads, we used H. melpomene RNA-Seq data to identify contigs upregulated in head tissues relative to antennae, legs, and mouth parts. We predicted that head upregulated contigs would be annotated with GO terms associated with vision. A multidimensional scaling plot showed that head RNA-Seq profiles group together and away from other tissue types (supplementary fig. S1A, Supplementary Material online). Differential expression analysis comparing heads versus antennae yielded 1,173 DE contigs (supplementary fig. S2 and Table S7, Supplementary Material online), 561 of these were upregulated in heads (Table 1). Analysis of head versus legs mRNAs gave 1,472 DE contigs (supplementary fig. S2 and Table S8, Supplementary Material online). Of these contigs, 928 were upregulated in heads. Heads versus mouth parts comparison yielded 1,486 DE contigs (supplementary fig. S2 and Table S9, Supplementary Material online); 914 of these were upregulated in heads (Table 1). DE contigs from each of the three pairwise comparisons matched 576, 730, and 685 unique gene FlyBase gene IDs (Table 1).
Table 1.
Summary of Heliconius melpomene Transcriptome-Wide Analysis
| Bonferroni | Upregulated in Heads | Commonly Upregulated in Heads | Unique FlyBase Gene ID | |
|---|---|---|---|---|
| Head versus antennae | 1,173 | 561 | 576 | |
| Head versus legs | 1,472 | 928 | 730 | |
| Head versus mouth | 1,486 | 914 | 685 | |
| Mergeda | 281 | 154 |
aMerged are genes commonly upregulated in heads after merging results of pairwise comparisons.
Most of the genes enriched in the DE analyses between heads and other tissues have vision-associated functions (supplementary Results and Table S10, Supplementary Material online), as has been found in a transcriptomic analysis of M. sexta adult head tissue alone (Smith et al. 2014). This could be because more transcription is actively occurring in the adult butterfly head and the head is mostly composed of the eye and optic lobe (Girardot et al. 2006). Heliconius butterflies have large eyes due to selective pressures that favor development of big eyes relative to body size. The optic lobe accounts for ∼64% of the total brain volume (Seymoure et al. 2015; Montgomery et al. 2016).
Head Upregulated Genes
We merged the lists of contigs upregulated in heads in each pairwise comparison to obtain 281 contigs commonly upregulated in heads across the three comparisons (Table 1). Head upregulated contigs annotated using BLAST2GO level 2 analysis showed that 78 of the annotated genes were involved in cellular processes and 32 were involved in responses to stimulus (supplementary fig. S1C, Supplementary Material online) (Conesa et al. 2005; Conesa and Götz 2008; Götz et al. 2008, 2011). A multilevel analysis of all head upregulated contigs shows that ∼33% are involved in ion transmembrane transport and 23% in G protein coupled receptor signaling pathways (supplementary fig. S1D, Supplementary Material online).
The 281 commonly upregulated and annotated contigs in heads across the three comparisons corresponded to 154 unique D. melanogaster FlyBase gene IDs (Table 1; supplementary Table S11, Supplementary Material online). These 154 contigs were grouped into eleven annotation clusters using the highest stringency in DAVID (supplementary fig. S1E; Huang et al. 2009). The top three annotation clusters were: 1) detection of light stimulus, 2) regulation of rhodopsin-mediated signaling pathway, and 3) detection of light stimulus involved in visual perception (supplementary fig. S1E, Supplementary Material online). The genes grouped within these clusters were annotated with phototransduction functions due to homology with D. melanogaster genes, Rh3, Rh5, Gbeta76, norpA, ninaC, ninaA, INAD, Calx, trpl, Arr1, Arr2, and stops (further discussed below; supplementary fig. S1E, Supplementary Material online). Of the remaining eight annotation clusters, clusters 9 and 10 are also directly associated with vision and are enriched for homeobox and rhabdomere development, respectively. Two genes in common between these two clusters include PvuII-PstI homology 13 (Pph13) and ocelliless (oc) that function in ocellus and compound eye photoreceptor development (Fichelson et al. 2012; Mahato et al. 2014).
Some of the genes enriched in other annotation clusters also have a role in vision. One gene in common between annotation clusters 4, 5, and 6 is ora transientless (ort), a gene that is necessary for vision as it encodes a postsynaptic chlorine channel gated by the photoreceptor neurotransmitter, histamine (Gengs et al. 2002). Annotation clusters 4, 5, and 8 include resistant to dieldrin (Rdl), a gene that has a role in the circuits underlying visual processing, odor coding, learning and memory, sleep, and courtship behavior (Brotz et al. 2001; Liu et al. 2007; Chung et al. 2009; Yuan et al. 2014).
Conservation of Phototransduction Genes in Lepidoptera
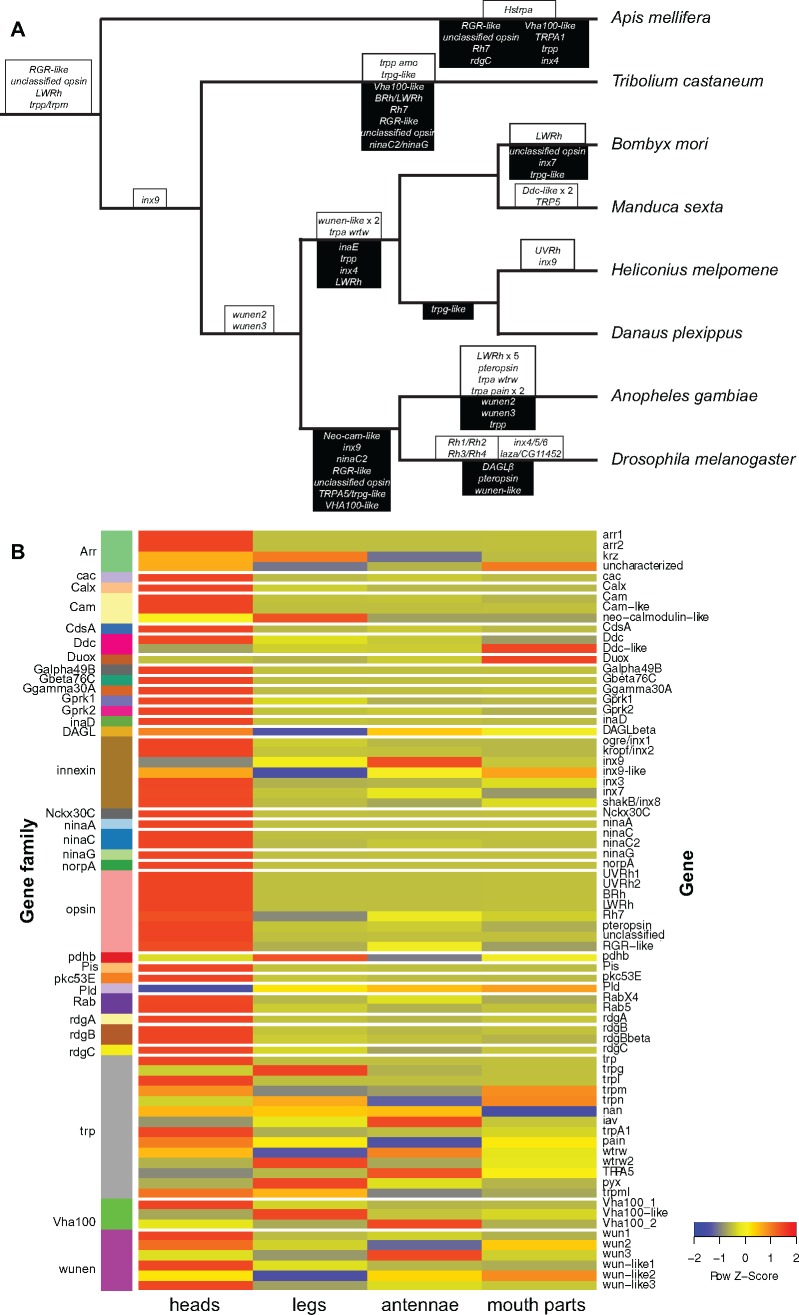
Genes commonly upregulated in H. melpomene heads were annotated with functions relating to vision and phototransduction in Drosophila (supplementary fig. S1, Supplementary Material online). Yet their evolutionary history and potential functional conservation requires further validation. To evaluate whether phototransduction genes were lost or expanded in Lepidoptera relative to D. melanogaster, we generated 32 insect phylogenies for 76 phototransduction-related genes (supplementary Tables S2–S5, Supplementary Material online). For each phylogeny, we searched eight insect genomes including two moth species (M. sexta and B. mori) and two butterfly species (H. melpomene and D. plexippus). Across all eight insect genomes we detected gene gains and losses in gene families such as opsin, trp, innexin, and lazaro/wunen (fig. 2A). Between D. melanogaster and lepidopterans, differences in gene gain and loss occur in the gene families opsin, innexin, lazaro/wunen, and DAGL. We did not detect any conserved differences in gene gain or loss between moths and butterflies (fig. 2A). Yet, an interesting gene family to note is Vha100, which has a Vha100-like gene that is lost in nonlepidopteran insects (supplementary Results and fig. S6G, Supplementary Material online) and also innexin 9, which is duplicated in H. melpomene (supplementary Results and fig. S7, Supplementary Material online).
Fig. 2.
—Phototransduction gene gains, losses, and expression. Most changes across the insect phylogeny occur in the opsin and trp gene families. (A) Insect phylogeny showing gains in white boxes above branches and losses in black boxes below branches. (B) Heatmap of expression of genes orthologous to D. melanogaster phototransduction genes in Heliconius melpomene heads, antennae, legs, and mouth parts. Red signifies high expression while blue signifies low expression. Gene names are listed on the right while gene family names are listed on the left and assigned a different block color per gene family. Most vision-related genes have elevated expression in the butterfly head.
Because many genes seem to be conserved between D. melanogaster and Lepidoptera, we visualized their expression in H. melpomene heads, antennae, legs, and mouth parts. Upregulation of orthologs in H. melpomene heads would suggest a conserved role in vision for genes annotated with phototransduction function. Conversely, upregulation of a paralog suggests that butterflies are using a different member of the gene family to perform a visual function. We found 32 genes upregulated in heads relative to other tissue types (fig. 2B;Table 2; supplementary figs. S3–S7, Supplementary Material online). Most of the main genes involved in D. melanogaster phototransduction were found as single copies in Lepidoptera and were upregulated in H. melpomene heads such as Gqα, β and γ, norpA, inaD, ninaC, Calx, trp, trpl, Arr1, Arr2, and stops (fig. 1; Table 2, for additional orthocluster analysis in butterflies see Catalán et al. 2018). These results suggest that second messengers, ion channels, and termination of phototransduction are conserved between D. melanogaster and Lepidoptera (see below). The main differences in the phototransduction cascade between H. melpomene and D. melanogaster are in the opsins which initiate phototransduction and in DAG regulation (discussed further below; fig. 1). Although there is no consistent difference between moths and butterflies in gene gains and losses, we found large differences in trp gene expression (see below).
Table 2.
Q-Values for Four Tissue Pair-Wise Comparisons in Heliconius melpomene
| Gene Family | Gene Symbol | Head Versus Antennae | Head Versus Legs | Head Versus Mouth |
|---|---|---|---|---|
| Arr | Arr2 | 2.33E−15 | 1.33E−32 | 2.03E−19 |
| Arr | Arr1 | 2.56E−11 | 1.16E−12 | 5.22E−14 |
| Arr | krz | 0.467 | 0.023 | 0.061 |
| Arr | uncharacterized | 0.192 | 1.60E−04 | 0.455 |
| cac | cac | 0.225 | 1.81E−04 | 1.07E−09 |
| Calx | Calx | 6.76E−04 | 8.48E−10 | 5.46E−07 |
| Cam | Cam-like | 1.99E−11 | 1.24E−18 | 1.96E−16 |
| Cam | neo-calmodulin-like | 0.003 | 0.003 | 7.63E−07 |
| Cam | Cam | 0.043 | 1.39E−04 | 0.005 |
| CdsA | CdsA | 0.003 | 7.53E−18 | 5.21E−14 |
| DAGL | DAGLbeta | 2.37E−04 | 0.489 | 0.409 |
| Ddc | Ddc-like | 1.20E−05 | 1.46E−06 | 4.16E−13 |
| Ddc | Ddc | 0.132 | 0.240 | 0.278 |
| Duox | Duox | 0.028 | 0.009 | 9.77E−07 |
| Galpha49B | Galpha49B | 7.24E−25 | 3.51E−34 | 2.87E−22 |
| Gbeta76C | Gbeta76C | 5.3E−20 | 1.69E−20 | 5.55E−20 |
| Ggamma30A | Ggamma30A | 2.83E−05 | 5.89E−10 | 7.20E−07 |
| Gprk1 | Gprk1 | 0.176 | 0.146 | 0.097 |
| Gprk2 | Gprk2 | 0.544 | 0.093 | 0.124 |
| inaD | inaD | 1.40E−07 | 3.40E−07 | 1.68E−07 |
| innexin | shakB/inx8 | 8.05E−10 | 2.69E−10 | 0.003 |
| innexin | ogre/inx1 | 0.064 | 0.061 | 0.021 |
| innexin | kropf/inx2 | 0.070 | 0.061 | 0.037 |
| innexin | inx9-like | N/A | N/A | N/A |
| innexin | inx9 | 0.038 | 0.142 | 0.273 |
| innexin | inx3 | 0.355 | 0.425 | 0.416 |
| innexin | inx7 | N/A | N/A | N/A |
| Nckx30C | Nckx30C | 0.007 | 3.75E−09 | 1.16E−10 |
| ninaA | ninaA | 8.24E−16 | 4.54E−18 | 1.72E−17 |
| ninaC | ninaC | 1.18E−25 | 4.57E−29 | 7.34E−27 |
| ninaC | ninaC2 | 7.7E−09 | 7.71E−16 | 9.16E−16 |
| ninaG | ninaG | 1.22E−12 | 8.15E−20 | 8.72E−14 |
| norpA | norpA | 4.55E−24 | 5.01E−22 | 1.82E−18 |
| opsin | LWRh | 4.1E−21 | 3.54E−27 | 5.63E−26 |
| opsin | BRh | 3.9E−28 | 1.77E−22 | 3.41E−20 |
| opsin | unclassified | 2.13E−17 | 1.86E−18 | 6.64E−12 |
| opsin | UVRh1 | 1.62E−14 | 7.53E−18 | 3.17E−09 |
| opsin | UVRh2 | 1.28E−09 | 1.48E−06 | 1.78E−07 |
| opsin | RGR-like | 0.022 | 0.159 | 0.058 |
| opsin | Rh7 | 0.610 | 0.276 | 0.244 |
| opsin | pteropsin | N/A | N/A | 0.124 |
| Pdhb | Pdhb | 0.374 | 0.007 | 0.439 |
| Pis | Pis | 0.002 | 6.47E−09 | 3.17E−10 |
| pkc53E | pkc53E | 2.19E−07 | 1.40E−07 | 3.17E−09 |
| Pld | Pld | 0.167 | 0.020 | 0.044 |
| Rab | Rax4 | 0.232 | 7.75E−04 | 1.95E−06 |
| Rab | Rab5 | 0.267 | 0.035 | 0.166 |
| rdgA | rdgA | 4.06E−04 | 0.001 | 0.002 |
| rdgB | rdgB | 1.07E−04 | 1.97E−08 | 1.40E−04 |
| rdgB | rdgBbeta | 0.488 | 0.127 | 0.060 |
| rdgC | rdgC | 0.083 | 0.024 | 0.248 |
| trp | trpl | 8.17E−20 | 6.25E−24 | 6.21E−19 |
| trp | TRPA5 | 4.64E−21 | 3.41E−08 | 2.29E−12 |
| trp | trp | 7.05E−05 | 2.96E−05 | 9.93E−06 |
| trp | TrpA1 | 0.200 | 0.002 | 0.002 |
| trp | nan | 0.479 | 0.469 | 0.002 |
| trp | Trpm | 0.350 | 0.384 | 0.047 |
| trp | trpn | 0.514 | 0.039 | 0.166 |
| trp | trpml | 0.190 | 0.370 | 0.204 |
| trp | pain | 0.437 | 0.143 | 0.217 |
| trp | trpg | 0.565 | 0.364 | 0.297 |
| trp | iav | 2.82E−04 | 0.089 | 0.343 |
| trp | wtrw | 0.455 | 0.420 | 0.490 |
| trp | pyx | N/A | 1.98E−04 | N/A |
| trp | wtrw2 | N/A | N/A | N/A |
| Vha100 | Vha100-1 | 0.146 | 0.010 | 0.011 |
| Vha100 | Vha100-like | 0.187 | 2.51E−04 | 0.085 |
| Vha100 | Vha100-2 | 2.45E−05 | 0.045 | 0.119 |
| wunen | wun-like3 | 0.062 | 0.002 | 0.011 |
| wunen | wun1 | 0.365 | 0.104 | 0.013 |
| wunen | wun3 | 0.211 | 0.466 | 0.307 |
| wunen | wun-like1 | 0.299 | 0.288 | 0.310 |
| wunen | wun-like2 | 0.582 | 0.307 | 0.331 |
| wunen | wun2 | 0.001 | 0.159 | 0.473 |
Bold numbers represent significance at a level of P-value <0.05.
Opsins in Lepidoptera
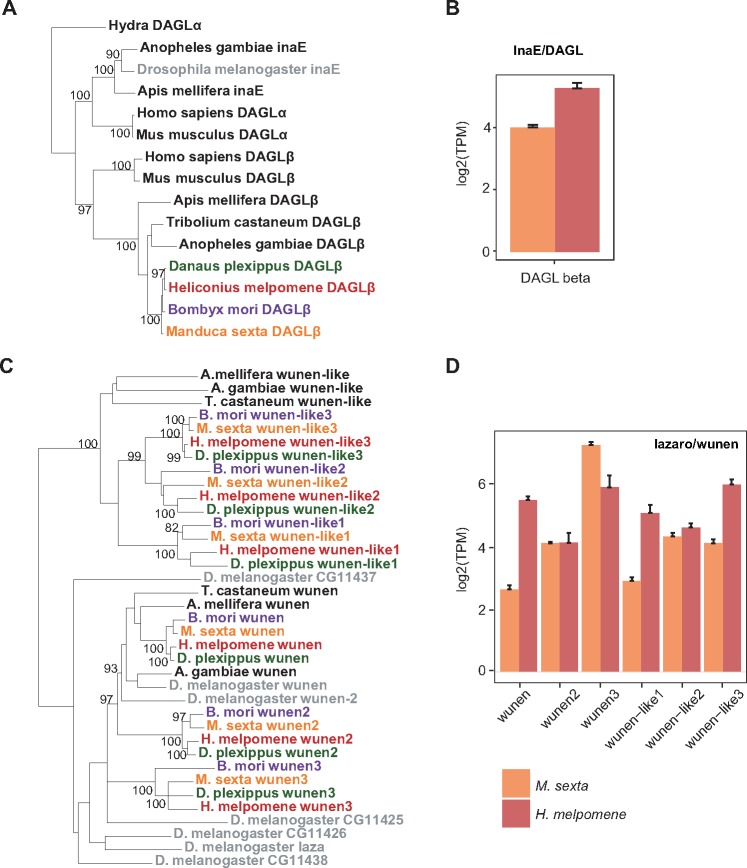
We began our survey of phototransduction genes in Lepidoptera by investigating the molecular evolution and expression of opsin genes typically responsible for initiating the phototransduction cascade (fig. 1). To inspect the phylogenetic history of the opsins, we added H. melpomene sequences from the reference genome and a four-tissue de novo transcriptome (Macias-Muñoz et al. 2017) to a set of sequences used in Kanost et al. (2016). We recovered the previously described Heliconius-specific UVRh duplication and orthologs for all other known opsins (fig. 3A) (Briscoe et al. 2010; Yuan et al. 2010; McCulloch et al. 2017). We also found two opsin genes: An unclassified opsin (UnRh) first described in Kanost et al. (2016) and RGR-like that both lack a D. melanogaster ortholog but are found in our butterfly genomes (fig. 3A).
Fig. 3.
—Insect opsin phylogeny and opsin gene expression in a moth and butterfly. Lepidopterans have two opsin genes related to squid retinochrome, unclassified and RGR-like, not found in D. melanogaster. (A) Opsin phylogenetic tree generated using amino acid sequences from Kanost et al. (2016) and from Heliconius melpomene and Danaus plexippus. D. melanogaster is in gray while lepidopteran species are in different colors, Bombyx mori (purple), Manduca sexta (orange), D. plexippus (green), and H. melpomene (red). Maximum-likelihood tree was generated using opsin amino acid sequences from 17 species with an LG+G + I+F model. Lepidopteran opsin clades are indicated by black labeled arcs. (B) Expression of opsin genes in M. sexta heads (n = 8, orange) and H. melpomene heads (n = 8, red) measured using RNA-Seq. The y axis is in transcripts per million on a log2 scale. Bars indicate standard errors.
To determine a role for all opsin genes we looked at their expression profile in M. sexta and H. melpomene. We expected opsins involved in vision to be highly expressed in heads. In M. sexta, all opsins had expression in head tissue (fig. 3B). In H. melpomene, our functional enrichment showed that homologs of D. melanogaster rhodopsin genes Rhodopsin 3 (Rh3) and Rhodopsin 5 (Rh5), which correspond to UVRh1/Rh2 and BRh, respectively, were upregulated in H. melpomene heads (supplementary fig. S1E and Table S11, Supplementary Material online) (Briscoe et al. 2010; Yuan et al. 2010). LWRh and the unclassified opsin (UnRh) are also upregulated in H. melpomene heads (Table 2; fig. 3B). LWRh was the most highly expressed opsin gene probably due to the amount of LW photoreceptor cells per ommatidium. Heliconius ommatidia have nine photoreceptor cells each where at least six cells express LWRh and two express short wavelength BRh, UVRh1, or UVRh2 (McCulloch et al. 2016, 2017).
Upregulation of UnRh was intriguing because Kanost et al. (2016) noted the unclassified opsin lacks a lysine at the typical location where the chromophore necessary to initiate phototransduction is bound in opsins, yet the gene is highly expressed in H. melpomene eyes and brain suggesting a role in vision (fig. 3; Table 2). A recent study found that alternative amino acid sites may be used in some G-protein coupled receptors for chromophore-binding (Faggionato and Serb 2017). Furthermore, cephalopods have a photosensitive pigment called retinochrome, studied biochemically, that lacks a conserved rhodopsin glutamic acid base (Terakita et al. 1989, 2000). Retinochrome, unlike rhodopsin, binds an all-trans retinal and acts as a photoisomerase converting the chromophore to 11-cis to regenerate the photosensitive rhodopsin (Sperling and Hubbard 1975). By adding a squid retinochrome sequence to our opsin phylogeny we found that the lepidopteran-specific unclassified opsin and RGR-like opsin are more closely related to retinochrome than they are to other opsins with known functions (fig. 3A).
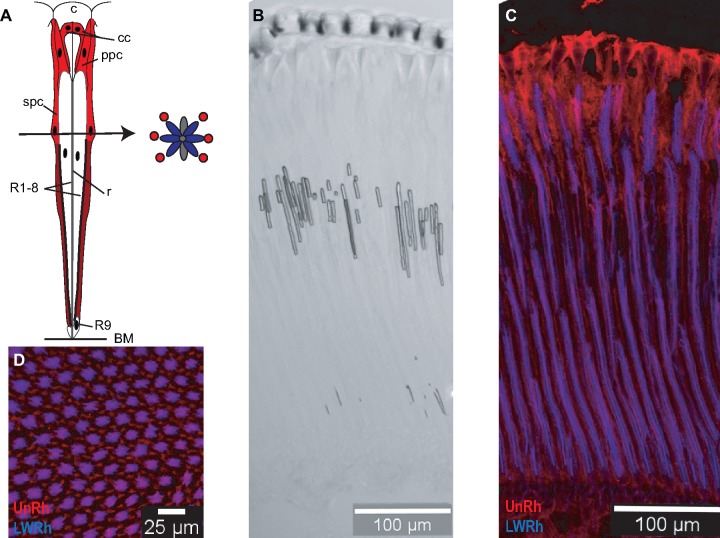
As UnRh has high expression in eyes and is phylogenetically similar to retinochrome, both proteins may have related enzymatic roles in vision if UnRh is expressed near the photoreceptor cells. To localize where in the butterfly eye UnRh is expressed, we made an antibody against one of its unique domains. We visualized UnRh expression alongside that of LWRh in H. melpomene. In Heliconius, LWRh is expressed in photoreceptor cells R3-8 (McCulloch et al. 2016). Intriguingly, we found UnRh abundantly expressed in crystalline cone cells, in primary pigment cells, and in the six secondary pigment cells surrounding the ommatidium (fig. 4). Staining is brighter in the distal part of the retina presumably because the secondary pigment cells decrease in size as they approach the basement membrane (fig. 4A). If UnRh had a function similar to that of the color vision opsins, we would expect it to be expressed in the photoreceptor cells. However, this protein is expressed in other retina cells adjacent to the photoreceptor cells. In squid, retinochrome is expressed in inner segment cells while the rhodopsin that it interchanges chromophore with is in the outer segment, separated by the basement membrane (Kingston et al. 2015; Chung and Marshall 2017). Taken together, these results suggest that UnRh might have a role similar to squid retinochrome in photoisomerization of the butterfly chromophore. This mechanism could be required for fast regeneration of an active rhodopsin necessary to quickly process visual information during flight.
Fig. 4.
—Immunohistochemistry of a butterfly retinochrome, unclassified opsin (UnRh). UnRh is expressed in several kinds of cells found in the distal retina but not in photoreceptor cells. (A) Drawing of a butterfly ommatidium showing the cornea (c), crystalline cone (cc), rhabdom (r), photoreceptor cells (R1-9), primary pigment cells (ppc), secondary pigment cells (spc), and basement membrane (bm) based on Kolb (1985). Red represents areas where UnRh expression is detected, dark red indicates where the cell presumably narrows and staining is not as bright. A drawing of a cross section shows cells R1-8, blue cells represent LWRh staining and red circles represent UnRh staining. (B) Brightfield image of a longitudinal section of a Heliconius melpomene eye showing the anatomy of each ommatidium and an intact cornea. (C) Fluorescent image of the same section stained for opsins using rabbit anti-LWRh (blue) and guinea pig anti-UnRh (red) antibodies. (D) Transverse section stained for opsins LWRh (blue) and UnRh (red).
In flies, the presence of 11-cis 3-hydroxyretinal is necessary for the synthesis of rhodopsin, suggesting a mechanism needs to be in place to rapidly convert the all-trans form into a reactive molecule. In Drosophila, all-trans 3-hydroxyretinal is transported to the pigment cells where a photoisomerase converts it back into the 11-cis configuration by blue light (Stavenga et al. 2017). Light intensity and wavelength affect the rate of 11-cis 3-hydroxyretinal synthesis in blowflies meaning that photoregeneration maintains levels of rhodopsin (Schwemer 1984). Interestingly, Lepidoptera are thought to rely more on enzymatic regeneration of 11-cis 3-hydroxyretinal than is the case in Diptera (Bernard 1983a, 1983b; Stavenga and Hardie 2011). Furthermore, a retinal-binding protein (RBP) was found in honeybees that binds the all-trans retinal that is isomerized in light (Pepe and Cugnoli 1980). Studies of honey bee RBP-A and RBP-B found that RBP-B binds all-trans retinal and also catalyzes the photoisomerization into the 11-cis conformation (Schwemer et al. 1984). Honey bee RBP-B function is similar to squid retinochrome but is unlikely to be a member of the same gene family due to its size (Pepe and Cugnoli 1980). Like these proteins, UnRh in butterflies may also have the ability to photoisomerize the chromophore molecule.
Regulation of DAG
After phototransduction is triggered by photon absorption, Gαq is released from a G-protein complex of three subunits (α, β, and γ) and activates PLC (is encoded by norpA) to produces DAG (Bloomquist et al. 1988; Lee et al. 1994). DAG has been implicated in the activation of TRP and TRPL channels (Chyb et al. 1999; Leung et al. 2008). DAG is hydrolyzed by the actions of DAG lipase (DAGL) encoded by the gene inaE (Leung et al. 2008). InaE mutants in D. melanogaster have defective responses to light, demonstrating that DAGL activity is required for photoreceptor responses (Leung et al. 2008). Although this gene is crucial for D. melanogaster phototransduction, an ortholog of inaE is missing in Lepidoptera (fig. 2A;supplementary Table S2, Supplementary Material online). We found that Lepidoptera retains DAGLβ, D. melanogaster retains DAGLα (inaE), and A. mellifera, A. gambiae, T. castaneum, and mammals retain both (fig. 5A). Both DAGLα and DAGLβ encode an Sn-1 DAGL that generates a monoacylglycerol (MAG) product. Note that for T. castaneum, DAGLα is not included in the phylogeny because the sequence was too short to generate a correct alignment. We predict that DAGLβ carries out the phototransduction function of hydrolyzing DAG in moth and butterfly vision because Lepidoptera have lost an ortholog of D. melanogaster inaE and have retained DAGLβ. DALGβ was expressed in M. sexta heads and in H. melpomene heads (fig. 5B). Although we confirm expression in heads, DAGLβ is not upregulated in heads relative to other tissue types. DAGLβ may have a role in vision in Lepidoptera, but it might also be used in other tissues for other functions. The role of DAGLβ in other insects is not clear. However, in humans and mice DAGLα and DAGLβ are necessary for axonal growth and synaptic signaling and inhibiting these proteins results in changes in brain signaling (Bisogno et al. 2003; Ogasawara et al. 2016). Interestingly, although both DAGLα and DAGLβ are expressed in axonal tracts and the developing spinal cord, only DAGLβ is expressed in the retinal ganglion layer and the optic lobe (Bisogno et al. 2003).
Fig. 5.
—Molecular evolution and expression of DAGL and lazaro/wunen. In lepidopterans, orthologs of Drosophila melanogaster DAGLα and lazaro are missing. Other gene family members may be playing a similar role in lepidopterans. (A) DAGL phylogenetic tree generated using amino acid sequences from eight insect genomes and Homo sapiens and M. musculus. Phylogenetic label colors follow those of figure 3. (B) Expression of DAGL genes in Manduca sexta heads (n = 8) and Heliconius melpomene heads (n = 8). (C) Wunen phylogenetic tree generated using amino acid sequences from eight insect genomes. (D) Expression of wunen genes in M. sexta heads (n = 8, orange) and H. melpomene heads (n = 8, red) measured using RNA-Seq. The y axis is in transcripts per million on a log2 scale. Bars indicate standard errors.
DAG level is also regulated by degeneration A (RDGA) (conserved in moths and butterflies; supplementary fig. S6, Supplementary Material online) and Lazaro (LAZA) (Garcia-Murillas et al. 2006; Bao and Friedrich 2009). Lazaro is a lipid phosphate phosphatase and is found in D. melanogaster photoreceptors (Garcia-Murillas et al. 2006). Lazaro is a member of the wunen subfamily (fig. 5C). Wunen helps regulate the level of bioactive phospholipids, has a role in germ line migration and is necessary for tracheal development (Zhang et al. 1997; Ile et al. 2012). We found seven sequences belonging to the wunen gene family in D. melanogaster; Lazaro is a D. melanogaster-specific duplication, as previously noted (Bao and Friedrich 2009). Although other non-D. melanogaster insects have one copy of wunen, lepidopterans have three copies (fig. 5C). In addition, although other insects have one copy of wunen-like, Lepidoptera have three copies of wunen-like that arose after lepidopteran divergence from other insects (fig. 5C). All copies of wunen and wunen-like are expressed in M. sexta and H. melpomene heads (fig. 5D). Wunen and wunen-like3 are the two copies most highly expressed in H. melpomene heads. Taken together, the above results suggest a difference in the gene family members involved in DAG regulation between D. melanogaster and lepidopterans.
TRP Channels
TRP and TRPL channels are essential in D. melanogaster phototransduction. They allow the influx of Ca2+ and cause cell depolarization (Montell and Rubin 1989). Trp is the dominant light-sensitive channel in Drosophila rhabdomeres (∼10× more abundant than trpl), and flies with mutated trp behave as though they are blind (Montell and Rubin 1989). The TRP superfamily contains more than 20 cation channels (Montell et al. 2002). Although trp and trpl function in D. melanogaster vision, other trp genes sense pain, vanilloid compounds, and heat, among other stimuli (Montell et al. 2002; Montell 2005). In our examination of the TRP gene family, we found 14 members in H. melpomene and 17 in M. sexta.
Differences between D. melanogaster and Lepidoptera include a duplication of trpa wtrw and a loss of trpp in moths and butterflies. The function of trpa wtrw (encoding TRP channel water witch) has not been characterized in any insect species but trpa genes, related gene family members, have been shown to function in temperature sensitivity, fructose aversion, and sexual receptivity in D. melanogaster (Xu et al. 2008; Sakai et al. 2009; Peng et al. 2016). Trpa wtrw is expressed in M. sexta heads and in H. melpomene heads whereas trpa wtrw2 has very low expression. In the trp family, M. sexta retains a trpg-like gene that is lost in D. melanogaster and butterflies H. melpomene and D. plexippus (figs. 2A and 6). Trpg encodes a protein that is found in D. melanogaster photoreceptors and has been speculated to form a heteromultimeric channel with TRPL (Montell 2005). The role of trpg in Drosophila vision is uncertain. It is expressed in H. melpomene heads, but trpg and trpg-like have low expression in M. sexta heads (fig. 6B). Furthermore, M. sexta also has three TRPA5 genes. Other lepidopterans have one copy. D. melanogaster and A. gambiae do not have any copies (fig. 6A). All three TRPA5 genes are expressed in M. sexta heads as is TRPA5 in H. melpomene heads (fig. 6B).
Fig. 6.
—Phylogeny and expression of the transient receptor potential (trp) cation channel gene family. Lepidopterans have a duplication of the wtrw trpa gene and Manduca sexta has a duplication of TRPA5. Expression of trp is ∼50× less than trpl in M. sexta, a lepidopteran which is active under dim light conditions. (A) Trp phylogenetic tree generated using amino acid sequences inferred from eight insect genomes using a WAG+G + F model. Lepidopteran trp clades are indicated by black labeled arcs. Phylogenetic label colors follow those of figure 3. (B) Expression of trp, Calx, and Nckx30C genes in M. sexta heads (n = 8) and Heliconius melpomene heads (n = 8) measured using RNA-Seq. The y axis is in transcripts per million on a log2 scale. Bars indicate standard errors. Trp and trpl shown in bold are used in high light and low light conditions, respectively, indicated by arrows.
Ion Channels Used in Diurnal and Nocturnal Insects
A transcriptome study in cockroaches found that trpl was ∼10 times more abundant than trp (French et al. 2015). RNAi of trpl reduced electroretinogram responses much more than RNAi of trp after 21 days suggesting that, as opposed to D. melanogaster, cockroach TRPL rather than TRP has a larger contribution to phototransduction (French et al. 2015). The authors suggested that differences in visual ecology are responsible for differential functions of the ion channels: Daylight-active D. melanogaster rely on fast responsive TRP and dark- or dim-light active cockroaches rely on TRPL (French et al. 2015). We found that trp and trpl are both highly expressed in H. melpomene heads which is different from either D. melanogaster or cockroach. Like cockroaches, we found that trp and trpl both have expression in M. sexta heads, but trp is expressed at a much lower level compared with trpl (fig. 6B). Our results suggest that the TRPL ion channel is also used by Lepidoptera in low light conditions.
TRP and TRPL channels allow Ca2+ and Na+ into the photoreceptor cell and are co-localized with a Na+/Ca2+ exchanger encoded by Calx which allows Ca2+ out of the cell (Montell 2005). Mutations of Calx result in a transient light response and a decrease in signal amplification implying a role for this gene in Ca2+ maintenance for proper TRP signaling (Wang et al. 2005). Overexpression of Calx can suppress retinal degeneration due to TRP constitutive activation (Montell 2005; Wang et al. 2005). Calx is upregulated in H. melpomene heads and is found as a single copy in all insect genomes (fig. 6B;Table 2; supplementary fig. S3C, Supplementary Material online). We detected a lower expression of Calx in M. sexta heads compared with the expression in H. melpomene heads potentially correlated with the lower expression of trp compared with trpl in M. sexta (supplementary fig. S3C, Supplementary Material online). A similar pattern of expression was also observed for another Na+/Ca2+ exchanger encoded by Nckx30C. Nckx30C was upregulated in H. melpomene heads yet expression of this ion channel was lower in M. sexta heads compared with H. melpomene heads (Table 2; fig. 6B;supplementary fig. S5B, Supplementary Material online). Nckx30C has a similar role to Calx in moving Ca2+ out of the cell (Haug-Collet et al. 1999). Both Nckx30C and Calx are expressed in the embryonic nervous system of D. melanogaster and in the adult eye and brain (Haug-Collet et al. 1999). Our results suggest that decreased expression of trp in nocturnal moths is accompanied by a decrease in Calx and Nckx30C expression. We conclude that one difference between moth and butterfly phototransduction is in the expression of ion channels used for calcium exchange.
Proposed Phototransduction Cascade in Lepidoptera
Based on phylogenetic relationships and gene expression analyses we propose a model of phototransduction in Lepidoptera (fig. 1). Phototransduction initiation requires an opsin to be bound to a chromophore to initiate the cascade. We propose that in Lepidoptera, the chromophore is transported by CTD31 rather than the ortholog of D. melanogaster PINTA, which has been lost in lepidopterans (Macias-Muñoz et al. 2017). Similar to D. melanogaster, visual opsins (BRh, LWRh, and UVRh) initiate the phototransduction cascade by a change in conformation when the chromophore molecule absorbs light energy (von Lintig et al. 2010). We note that lepidopterans vary in opsin number (Frentiu et al. 2007; Briscoe 2008; Pirih et al. 2010; Xu et al. 2013). Photoisomerized 11-cis-3-hydroxyretinal is supplied to light-activated rhodopsin by retinochrome (UnRh) proteins found in pigment cells. Change in opsin conformation due to light absorption triggers the G-protein signaling cascade. Gαq, β, and γ are present as single copies and highly expressed in heads suggesting a conserved function in PLC activation, encoded by norpA, when Gαq is released (supplementary figs. S1E, S4D–S4F, and S5E, Supplementary Material online) (Bloomquist et al. 1988; Lee et al. 1994).
PLC produces InsP3 and DAG (Bloomquist et al. 1988; Hardie 2001). However, the regulation of DAG levels appears to differ between lepidopterans and D. melanogaster due to the absence of laza and the loss of inaE. We propose that in Lepidoptera the actions of inaE are undertaken by a lepidopteran paralog DAGLβ and those of laza by other members of the gene family, wunen or wunen-like3. LAZA acts in opposition to DAG kinase encoded by rdgA (Garcia-Murillas et al. 2006). In D. melanogaster, DAG is converted into PIP2 by the phosphoinositide pathway which gives photoreceptor cells sensitivity and fast response (Hardie 2001; Garcia-Murillas et al. 2006). The actions of this pathway seem conserved in Lepidoptera because rdgA, cdsA, and rdgB are upregulated in H. melpomene heads. Although phosphatidic acid (PA) is likely converted into DAG by a laza paralog (wunen or wunen-like3), kinase rgdA maintains a role in converting DAG into PA. CDP-diacylglycerol synthase encodes a protein that converts PA into cytidine diphosphate DAG (CDP-DAG). Phosphatidyl inositol (PI) synthase then changes CDP-DAG into PI which is transported by phosphatidylinositol transfer protein encoded by rdgB. Phosphorylation converts PI into PIP2. The actions by which DAG functions in phototransduction are not well understood. DAGL produces the metabolite polyunsaturated MAG (Montell 2012). DAG might activate TRP and TRPL channels, although its role in phototransduction is debated (Chyb et al. 1999; Leung et al. 2008).
TRP and TRPL allow Ca2+ and Na+ into the cell that causes the photoreceptor cell to depolarize (Montell and Rubin 1989). We propose that the phototransduction cascade varies between moths and butterflies in the deployment of TRP and Na+/Ca+ channels. According to our expression data, butterflies use TRP and TRPL in similar amounts, whereas moths downregulate their TRP channel mRNAs. Because moths presumably have fewer TRP channels allowing in Ca2+, they also downregulate Na+/Ca2+ channels encoded by Calx and Nckx30C.
Phototransduction requires protein complexes to transduce and terminate the signal. One such complex is a target of Gαq and is formed by INAD, TRP, PLC, and protein kinase C (Shieh et al. 1989; Chevesich et al. 1997; Tsunoda et al. 1997; Bähner et al. 2000; Montell 2005). InaD is required to localize and coordinate proteins in the phototransduction cascade to the microvillar membrane (Bähner et al. 2000). INAD and ninaC bind to each other, and individually bind calmodulin, which accelerates arrestin binding to rhodopsin to terminate phototransduction (Liu et al. 2008; Venkatachalam et al. 2010). Arrestin 1 and Arrestin 2 bind light-activated rhodopsin and discontinue cascade signaling in D. melanogaster (Dolph et al. 1993; Stavenga and Hardie 2011). Our data suggest that Arrestin 2 might be the major arrestin in butterfly phototransduction; it is more highly expressed than Arrestin 1 in moths as well (supplementary fig. S3A, Supplementary Material online). Phototransduction is also terminated by a protein with a suppressor of cytokine signaling box encoded by stops. The stops phenotype is associated with slow termination of phototransduction due to a decrease in norpA (PLC) (Wang et al. 2008). We find these genes to be upregulated in butterfly heads (Table 2), suggesting the actions of these complexes remain conserved. Lastly, Lepidoptera have a ninaC2 gene, missing in D. melanogaster, which is upregulated in H. melpomene heads (supplementary fig. S5G, Supplementary Material online).
Conclusions
Most studies of phototransduction in insects extrapolate from what is known in D. melanogaster to assign potential functions to genes based on sequence similarity. In our study, we used transcriptomics and phylogenetics to explore the conservation of phototransduction genes between D. melanogaster and Lepidoptera. We found that many orthologs of key D. melanogaster phototransduction genes were upregulated in H. melpomene heads relative to legs, antennae, and mouth parts. Our results suggest that many features of the D. melanogaster phototransduction cascade are conserved in lepidopteran vision. However, we found instances where lepidopteran paralogs are implicated in carrying out a visual role when an ortholog is lost. Differences in phototransduction between D. melanogaster and Lepidoptera occur in chromophore transport, chromophore regeneration, opsins, and DAG regulation. Although we found no conserved differences between moths and butterflies in gene gains and losses, quantifying gene expression in M. sexta and H. melpomene allowed us to detect differences in phototransduction between moths and butterflies. Notably, we found evidence that butterflies use both TRP and TRPL channels for phototransduction while moths downregulate trp, which is used for high light conditions (French et al. 2015). Along with decreased expression of trp, Na+/Ca2+ exchange channel mRNAs show decreased expression in nocturnal moths. We have thus completed the most extensive investigation of the evolution of the phototransduction cascade in Lepidoptera and have found that differences between Lepidoptera and D. melanogaster are due to gene gains and losses while differences between moths and butterflies are due to gene expression changes.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Ali Mortazavi, Kevin Thornton, Jorge Llorente-Bousquets and Pablo Vinuesa for advice on data analysis. We thank Zachary Johnston, JP Lawrence, and Andrew Dang for comments on the manuscript, and Roger Hardie and Simon Laughlin for helpful discussions. This work was supported in part by National Science Foundation grants DEB-1342759 and IOS-1656260 to A.D.B., a Ford Foundation Predoctoral Fellowship to AMM, and a CNBES grant to AGRO.
Data deposition: Phototransduction genes for H. melpomene, M. sexta, and D. plexippus were annotated and deposited in GenBank with accession numbers MK983015–MK983088, MK983089–MK983165, and MN037884–MN037955.
Literature Cited
- Arendt D. 2003. Evolution of eyes and photoreceptor cell types. Int J Dev Biol. 47(7–8):563–571. [PubMed] [Google Scholar]
- Bähner M, Sander P, Paulsen R, Huber A.. 2000. The visual G protein of fly photoreceptors interacts with the PDZ domain assembled INAD signaling complex via direct binding of activated Gα(q) to phospholipase Cβ. J Biol Chem. 275(4):2901–2904. [DOI] [PubMed] [Google Scholar]
- Bao R, Friedrich M.. 2009. Molecular evolution of the Drosophila retinome: exceptional gene gain in the higher Diptera. Mol Biol Evol. 26(6):1273–1287. [DOI] [PubMed] [Google Scholar]
- Battelle BA, et al. 2016. Opsin repertoire and expression patterns in horseshoe crabs: evidence from the genome of Limulus polyphemus (Arthropoda: chelicerata). Genome Biol Evol. 8(5):1571–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard GD. 1983. Dark-processes following photoconversion of butterfly rhodopsins. Biophys Struct Mech. 9(4):277–286. [Google Scholar]
- Bernard GD. 1983. Bleaching of rhabdoms in eyes of intact butterflies. Science 219(4580):69–71. [DOI] [PubMed] [Google Scholar]
- Bisogno T, et al. 2003. Cloning of the first sn1-DAG lipases points to the spatial and temporal regulation of endocannabinoid signaling in the brain. J Cell Biol. 163(3):463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomquist BT, et al. 1988. Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell 54(5):723–733. [DOI] [PubMed] [Google Scholar]
- Briscoe AD. 2008. Reconstructing the ancestral butterfly eye: focus on the opsins. J Exp Biol. 211:1805–1813. [DOI] [PubMed] [Google Scholar]
- Briscoe AD, et al. 2010. Positive selection of a duplicated UV-sensitive visual pigment coincides with wing pigment evolution in Heliconius butterflies. Proc Natl Acad Sci U S A. 107(8):3628–3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe AD et al. 2013. Female behaviour drives expression and evolution of gustatory receptors in butterflies. PLoS Genet. 9:e1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotz TM, Gundelfinger ED, Borst A.. 2001. Cholinergic and GABAergic pathways in fly motion vision. BMC Neurosci. 2(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, et al. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10(1):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalán A, Briscoe AD, Höhna S.. 2018. Drift and directional selection are the evolutionary forces driving gene expression divergence in eye and brain tissue of Heliconius butterflies. biorxiv, doi:10.1101/463174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevesich J, Kreuz AJ, Montell C.. 1997. Requirement for the PDZ domain protein, INAD, for localization of the TRP store-operated channel to a signaling complex. Neuron 18(1):95–105. [DOI] [PubMed] [Google Scholar]
- Chung BY, Kilman VL, Keath JR, Pitman JL, Allada R.. 2009. The GABAA receptor RDL acts in peptidergic PDF neurons to promote sleep in Drosophila. Curr Biol. 19(5):386–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W, Marshall NJ.. 2017. Complex visual adaptations in squid for specific tasks in different environments. Front Physiol. 8: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chyb S, Raghu P, Hardie RC.. 1999. Polyunsaturated fatty acids activate the Drosophila light-sensitive channels TRP and TRPL. Nature 397(6716):255–259. [DOI] [PubMed] [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21(18):3674–3676. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S.. 2008. Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008: 619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabney A, Storey JD.. 2013. qvalue: Q-value estimation for false discovery rate control. R package version 1.36.0.
- Davey JW, et al. 2016. Major improvements to the Heliconius melpomene genome assembly used to confirm 10 chromosome fusion events in 6 million years of butterfly evolution. G3 6(3):695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolph PJ, et al. 1993. Arrestin function in inactivation of G protein-coupled receptor rhodopsin in vivo. Science 260(5116):1910–1916. [DOI] [PubMed] [Google Scholar]
- Duan Y, et al. 2017. Transcriptome analysis of molecular mechanisms responsible for light-stress response in Mythimna separata (Walker). Sci Rep. 7:45188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faggionato D, Serb JM.. 2017. Strategy to identify and test putative light-sensitive non-opsin G-protein-coupled receptors: a case study. Biol Bull. 233(1):70–82. [DOI] [PubMed] [Google Scholar]
- Fain GL, Hardie R, Laughlin SB.. 2010. Phototransduction and the evolution of photoreceptors. Curr Biol. 20(3):R114–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuda R, Marle F, Bentley MA, Holland P.. 2016. Conservation, duplication, and divergence of five opsin genes in insect evolution. Genome Biol Evol. 8(3):579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichelson P, Brigui A, Pichaud F.. 2012. Orthodenticle and Kruppel homolog 1 regulate Drosophila photoreceptor maturation. Proc Natl Acad Sci U S A. 109(20):7893–7898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AS, Meisner S, Liu H, Weckström M, Torkkeli PH.. 2015. Transcriptome analysis and RNA interference of cockroach phototransduction indicate three opsins and suggest a major role for TRPL channels. Front Physiol. 6:00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentiu FD, et al. 2007. Adaptive evolution of color vision as seen through the eyes of butterflies. Proc Natl Acad Sci. 104:8634–8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentiu FD, et al. 2015. Opsin clines in butterflies suggest novel roles for insect photopigments. Mol Biol Evol. 32(2):368–379. [DOI] [PubMed] [Google Scholar]
- Frentiu FD, Bernard GD, Sison-Mangus MP, Van Zandt Brower A, Briscoe AD.. 2007. Gene duplication is an evolutionary mechanism for expanding spectral diversity in the long-wavelength photopigments of butterflies. Mol Biol Evol. 24(9):2016–2028. [DOI] [PubMed] [Google Scholar]
- Friedrich M, et al. 2011. Phototransduction and clock gene expression in the troglobiont beetle Ptomaphagus hirtus of Mammoth cave. J Exp Biol. 214(Pt 21):3532–3541. [DOI] [PubMed] [Google Scholar]
- Futahashi R, et al. 2015. Extraordinary diversity of visual opsin genes in dragonflies. Proc Natl Acad Sci U S A. 112(11):E1247–E1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Murillas I, et al. 2006. Lazaro encodes a lipid phosphate phosphohydrolase that regulates phosphatidylinositol turnover during Drosophila phototransduction. Neuron 49(4):533–546. [DOI] [PubMed] [Google Scholar]
- Gengs C, et al. 2002. The target of Drosophila photoreceptor synaptic transmission is a histamine-gated chloride channel encoded by ort (hclA). J Biol Chem. 277(44):42113–42120. [DOI] [PubMed] [Google Scholar]
- Giraldo-Calderón GI, Zanis MJ, Hill CA.. 2017. Retention of duplicated long-wavelength opsins in mosquito lineages by positive selection and differential expression. BMC Evol Biol. 17(1):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girardot F, Lasbleiz C, Monnier V, Tricoire H.. 2006. Specific age related signatures in Drosophila body parts transcriptome. BMC Genomics 7(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz S, et al. 2011. B2G-FAR, a species-centered GO annotation repository. Bioinformatics 27(7):919–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz S, et al. 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36(10):3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie RC. 2001. Phototransduction in Drosophila melanogaster. J Exp Biol. 204(Pt 20):3403–3409. [DOI] [PubMed] [Google Scholar]
- Hardie RC, Juusola M.. 2015. Phototransduction in Drosophila. Curr Opin Neurobiol. 34:37–45. [DOI] [PubMed] [Google Scholar]
- Hardie RC, Minke B.. 1992. The trp gene is essential for a light-activated Ca2+ channel in Drosophila photoreceptors. Neuron 8(4):643–651. [DOI] [PubMed] [Google Scholar]
- Hardie RC, Raghu P.. 2001. Visual transduction in Drosophila. Nature 413(6852):186–193. [DOI] [PubMed] [Google Scholar]
- Haug-Collet K, et al. 1999. Cloning and characterization of a potassium-dependent sodium/calcium exchanger in Drosophila. J Cell Biol. 147(3):659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henze MJ, Dannenhauer K, Kohler M, Labhart T, Gesemann M.. 2012. Opsin evolution and expression in arthropod compound eyes and ocelli: insights from the cricket Gryllus bimaculatus. BMC Evol Biol. 12: 163.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horridge GA, Giddings C, Stange G.. 1972. The superposition eye of skipper butterflies. Proc R Soc Lond B Biol Sci. 182:457–495. [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4(1):44–57. [DOI] [PubMed] [Google Scholar]
- Ile KE, Tripathy R, Goldfinger V, Renault AD.. 2012. Wunen, a Drosophila lipid phosphate phosphatase, is required for septate junction-mediated barrier function. Development 139(14):2535–2546. [DOI] [PubMed] [Google Scholar]
- Kanost MR, et al. 2016. Multifaceted biological insights from a draft genome sequence of the tobacco hornworm moth, Manduca sexta. Insect Biochem Mol Biol. 76:118–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz B, Minke B.. 2009. Drosophila photoreceptors and signaling mechanisms. Front Cell Neurosci. 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston ACN, Wardill TJ, Hanlon RT, Cronin TW.. 2015. An unexpected diversity of photoreceptor classes in the longfin squid, Doryteuthis pealeii. PLoS One 10(9):e0135381–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K.. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL.. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YJ, et al. 1994. The Drosophila dgq gene encodes a G alpha protein that mediates phototransduction. Neuron 13(5):1143–1157. [DOI] [PubMed] [Google Scholar]
- Leung H, et al. 2008. DAG lipase activity is necessary for TRP channel regulation in Drosophila photoreceptors. Neuron 58(6):884–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12(1):323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lintig J, Kiser PD, Golczak M, Palczewski K.. 2010. The biochemical and structural basis for trans-to-cis isomerization of retinoids in the chemistry of vision. Trends Biochem Sci. 35(7):400–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CH, et al. 2008. Ca2+-dependent metarhodopsin inactivation mediated by calmodulin and NINAC myosin III. Neuron 59(5):778–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Krause WC, Davis RL.. 2007. GABAA receptor RDL inhibits Drosophila olfactory associative learning. Neuron 56(6):1090–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias-Muñoz A, McCulloch KJ, Briscoe AD.. 2017. Copy number variation and expression analysis reveals a non-orthologous pinta gene family member involved in butterfly vision. Genome Biol Evol. 9(12):3398–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias-Muñoz A, Smith G, Monteiro A, Briscoe AD.. 2016. Transcriptome-wide differential gene expression in Bicyclus anynana butterflies: female vision-related genes are more plastic. Mol Biol Evol. 33(1):79–92. [DOI] [PubMed] [Google Scholar]
- Mahato S, et al. 2014. Common transcriptional mechanisms for visual photoreceptor cell differentiation among Pancrustaceans. PLoS Genet. 10(7):e1004484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marygold SJ, et al. 2012. FlyBase: improvements to the bibliography. Nucleic Acids Res. 41(D1):D751–D757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch KJ, et al. 2017. Sexual dimorphism and retinal mosaic diversification following the evolution of a violet receptor in butterflies. Mol Biol Evol. 34(9):2271–2284. [DOI] [PubMed] [Google Scholar]
- McCulloch KJ, Osorio D, Briscoe AD.. 2016. Sexual dimorphism in the compound eye of Heliconius erato: a nymphalid butterfly with at least five spectral classes of photoreceptor. J Exp Biol. 219(15):2377–2387. [DOI] [PubMed] [Google Scholar]
- Min XJ, Butler G, Storms R, Tsang A.. 2005. OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res. 33:677–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C. 2012. Drosophila visual transduction. Trends Neurosci. 35(6):356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C. 2005. TRP channels in Drosophila photoreceptor cells. J Physiol. 567(1):45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montell C, Birnbaumer L, Flockerzi V.. 2002. The TRP channels, a remarkably functional family. Cell 108(5):595–598. [DOI] [PubMed] [Google Scholar]
- Montell C, Rubin GM.. 1989. Molecular characterization of the Drosophila trp locus: a putative integral membrane protein required for phototransduction. Neuron 2(4):1313–1323. [DOI] [PubMed] [Google Scholar]
- Montgomery SH, Merrill RM, Ott SR.. 2016. Brain composition in Heliconius butterflies, posteclosion growth and experience-dependent neuropil plasticity. J Comp Neurol 524(9):1747–1769. [DOI] [PubMed] [Google Scholar]
- Niemeyer BA, Suzuki E, Scott K, Jalink K, Zuker CS.. 1996. The Drosophila light-activated conductance is composed of the two channels TRP and TRPL. Cell 85(5):651–659. [DOI] [PubMed] [Google Scholar]
- Nilsson DE, Land MF, Howard J.. 1984. Afocal apposition optics in butterfly eyes. Nature 312(5994):561–563. [Google Scholar]
- Ogasawara D, et al. 2016. Rapid and profound rewiring of brain lipid signaling networks by acute diacylglycerol lipase inhibition. Proc Natl Acad Sci U S A. 113(1):26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, et al. 2016. TRPA1 channels in Drosophila and honey bee ectoparasitic mites share heat sensitivity and temperature-related physiological functions. Front Physiol. 7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepe IM, Cugnoli C.. 1980. Isolation and characterization of a water-soluble photopigment from honeybee compound eye. Vision Res. 20(2):97–102. [DOI] [PubMed] [Google Scholar]
- Perry M, et al. 2016. Molecular logic behind the three-way stochastic choices that expand butterfly colour vision. Nature 535(7611):280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirih P, Arikawa K, Stavenga DG.. 2010. An expanded set of photoreceptors in the Eastern Pale Clouded Yellow butterfly, Colias erate. J Comp Physiol A 196(7):501–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachetzki DC, Degnan BM, Oakley TH.. 2007. The origins of novel protein interactions during animal opsin evolution. PLoS One 2(10):e1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachetzki DC, Fong CR, Oakley TH.. 2010. The evolution of phototransduction from an ancestral cyclic nucleotide gated pathway. Proc R Soc B 277(1690):1963–1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploner A. 2012. Heatplus: heatmaps with row and/or column covariates and colored clusters. version 2.6.0
- Porter ML, et al. 2012. Shedding new light on opsin evolution. Proc R Soc B Biol Sci. 279(1726):3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raible F, et al. 2006. Opsins and clusters of sensory G-protein-coupled receptors in the sea urchin genome. Dev Biol. 300(1):461–475. [DOI] [PubMed] [Google Scholar]
- Ramirez MD, et al. 2016. The last common ancestor of most bilaterian animals possessed at least nine opsins. Genome Biol Evol. 8(12):3640–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera AS, et al. 2010. Gene duplication and the origins of morphological complexity in pancrustacean eyes, a genomic approach. BMC Evol Biol. 10(1):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M, Oshlack A.. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Mccarthy DJ, Smyth GK.. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Kasuya J, Kitamoto T, Aigaki T.. 2009. The Drosophila TRPA channel, painless, regulates sexual receptivity in virgin females. Genes Brain Behav. 8(5):546–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G. 1978. Estimating the dimension of a model. Ann Statist. 6(2):461–464. [Google Scholar]
- Schwemer J. 1984. Renewal of visual pigment in photoreceptors of the blowfly. J Comp Physiol. 154(4):535–547. [Google Scholar]
- Schwemer J, Pepe I, Paulson R, Cugnoli C.. 1984. Light-induced trans-cis isomerization of retinal by a protein from honeybee retina. J Comp Physiol. 154(4):549–554. [Google Scholar]
- Seymoure BM, Mcmillan WO, Rutowski R.. 2015. Peripheral eye dimensions in Longwing (Heliconius) butterflies vary with body size and sex but not light environment nor mimicry ring. J Res Lepid. 48:83–92. [Google Scholar]
- Shichida Y, Matsuyama T.. 2009. Evolution of opsins and phototransduction. Phil Trans R Soc B 364(1531):2881–2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh BH, Stamnes MA, Seavello S, Harris GL, Zuker CS.. 1989. The ninaA gene required for visual transduction in Drosophila encodes a homologue of cyclosporin A-binding protein. Nature 338(6210):67–70. [DOI] [PubMed] [Google Scholar]
- Shieh BH, Zhu MY.. 1996. Regulation of the TRP Ca2+ channel by INAD in Drosophila photoreceptors. Neuron 16(5):991–998. [DOI] [PubMed] [Google Scholar]
- Sison-Mangus MP, Briscoe AD, Zaccardi G, Knuttel H, Kelber A.. 2008. The lycaenid butterfly Polyommatus icarus uses a duplicated blue opsin to see green. J Exp Biol. 211(3):361–369. [DOI] [PubMed] [Google Scholar]
- Smith G, Briscoe AD. 2015. Molecular evolution and expression of the CRAL_TRIO protein family in insects. Insect Biochem. Mol. Biol. 62:168–173. [DOI] [PubMed] [Google Scholar]
- Smith G, Chen YR, Blissard GW, Briscoe AD.. 2014. Complete dosage compensation and sex-biased gene expression in the moth Manduca sexta. Genome Biol Evol. 6(3):526–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaethe J, Briscoe AD.. 2004. Early duplication and functional diversification of the opsin gene family in insects. Mol Biol Evol. 21(8):1583–1594. [DOI] [PubMed] [Google Scholar]
- Spaethe J, Briscoe AD.. 2005. Molecular characterization and expression of the UV opsin in bumblebees: three ommatidial subtypes in the retina and a new photoreceptor organ in the lamina. J Exp Biol. 208: 2347–2361. [DOI] [PubMed] [Google Scholar]
- Sperling L, Hubbard R.. 1975. Squid retinochrome. J Gen Physiol. 65(2):235–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavenga DG, Hardie RC.. 2011. Metarhodopsin control by arrestin, light-filtering screening pigments, and visual pigment turnover in invertebrate microvillar photoreceptors. J Comp Physiol A 197(3):227–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavenga DG, Wehling MF, Belušič G.. 2017. Functional interplay of visual, sensitizing and screening pigments in the eyes of Drosophila and other red-eyed dipteran flies. J Physiol. 595(16):5481–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga H, Schmid V, Gehring WJ.. 2008. Evolution and functional diversity of jellyfish opsins. Curr Biol. 18(1):51–55. [DOI] [PubMed] [Google Scholar]
- Terakita A, Hara R, Hara T.. 1989. Retinal-binding protein as a shuttle for retinal in the rhodopsin-retinochrome system of the squid visual cells. Vision Res. 29(6):639–652. [DOI] [PubMed] [Google Scholar]
- Terakita A, Yamashita T, Shichida Y.. 2000. Highly conserved glutamic acid in the extracellular IV-V loop in rhodopsins acts as the counterion in retinochrome, a member of the rhodopsin family. Proc Natl Acad Sci U S A. 97(26):14263–14267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunoda S, et al. 1997. A multivalent PDZ-domain protein assembles signalling complexes in a G- protein-coupled cascade. Nature 388(6639):243–249. [DOI] [PubMed] [Google Scholar]
- Venkatachalam K, et al. 2010. Dependence on a retinophilin/myosin complex for stability of PKC and INAD and termination of phototransduction. J Neurosci. 30(34):11337–11345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vöcking O, Kourtesis I, Tumu S, Hausen H.. 2017. Co-expression of xenopsin and rhabdomeric opsin in photoreceptors bearing microvilli and cilia. Elife 6:e23435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, et al. 2005. Light activation, adaptation, and cell survival functions of the Na+/Ca2+ exchanger CalX. Neuron 45(3):367–378. [DOI] [PubMed] [Google Scholar]
- Wang T, Montell C. 2005. Rhodopsin formation in Drosophila is dependent on the PINTA retinoid-binding protein. J. Neurosci. 25:5187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wang X, Xie Q, Montell C.. 2008. The SOCS box protein STOPS is required for phototransduction through its effects on phospholipase C. Neuron 57(1):56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrant E, Dacke M.. 2016. Visual navigation in nocturnal insects. Physiology 31(3):182–192. [DOI] [PubMed] [Google Scholar]
- Wernet MF, Perry MW, Desplan C.. 2015. The evolutionary diversity of insect retinal mosaics: common design principles and emerging molecular logic. Trends Genet. 31(6):316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. 2009. ggplot2: elegant graphics for data analysis. New York: Springer-Verlag. [Google Scholar]
- Xu J, Sornborger AT, Lee JK, Shen P.. 2008. Drosophila TRPA channel modulates sugar-stimulated neural excitation, avoidance and social response. Nat Neurosci. 11(6):676–682. [DOI] [PubMed] [Google Scholar]
- Xu P, et al. 2013. The evolution and expression of the moth visual opsin family. PLoS One 8(10):e78140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yack JE, Johnson SE, Brown SG, Warrant EJ.. 2007. The eyes of Macrosoma sp. (Lepidoptera: hedyloidea): a nocturnal butterfly with superposition optics. Arthropod Struct Dev. 36(1):11–22. [DOI] [PubMed] [Google Scholar]
- Yagi N, Koyama N.. 1963. The compound eye of Lepidoptera: approach from organic evolution. Tokyo, Japan: Shinkyo-Press 318 pp.
- Yuan F, Bernard GD, Le J, Briscoe AD.. 2010. Contrasting modes of evolution of the visual pigments in Heliconius butterflies. Mol Biol Evol. 27(10):2392–2405. [DOI] [PubMed] [Google Scholar]
- Yuan Q, Song Y, Yang C-H, Jan LY, Jan YN.. 2014. Female contact modulates male aggression via a sexually dimorphic GABAergic circuit in Drosophila. Nat Neurosci. 17(1):81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S, Merlin C, Boore JL, Reppert SM.. 2011. The Monarch butterfly genome yields insights into long-distance migration. Cell 147(5):1171–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Zhang J, Purcell KJ, Cheng Y, Howard K.. 1997. The Drosophila protein wunen repels migrating germ cells. Nature 385(6611):64–67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.