Abstract
In small RNA (smRNA) sequencing studies, highly abundant molecules such as adapter dimer products and tissue-specific microRNAs (miRNAs) inhibit accurate quantification of lowly expressed species. We previously developed a method to selectively deplete highly abundant miRNAs. However, this method does not deplete adapter dimer ligation products that, unless removed by gel-separation, comprise most of the library. Here, we have adapted and modified recently described methods for CRISPR/Cas9–based Depletion of Abundant Species by Hybridization (‘DASH’) to smRNA-seq, which we have termed miRNA and Adapter Dimer—DASH (MAD-DASH). In MAD-DASH, Cas9 is complexed with single guide RNAs (sgRNAs) targeting adapter dimer ligation products, alongside highly expressed tissue-specific smRNAs, for cleavage in vitro. This process dramatically reduces adapter dimer and targeted smRNA sequences, can be multiplexed, shows minimal off-target effects, improves the quantification of lowly expressed miRNAs from human plasma and tissue derived RNA, and obviates the need for gel-separation, greatly increasing sample throughput. Additionally, the method is fully customizable to other smRNA-seq preparation methods. Like depletion of ribosomal RNA for mRNA-seq and mitochondrial DNA for ATAC-seq, our method allows for greater proportional read-depth of non-targeted sequences.
INTRODUCTION
Small RNAs (smRNAs) are a diverse class of RNA molecules, including microRNAs (miRNAs), transfer RNAs (tRNAs), small nucleolar RNA (snoRNA), Y-RNA and many others, that have diverse roles in biological processes (1,2). miRNAs are particularly well-studied due to their role as post-transcriptional regulators of gene expression in many biological processes (3–5), and altered expression of smRNAs has been implicated in many disease pathologies (6–11). Consequently, there is a need for methods that can precisely and accurately measure smRNAs.
Although smRNAs are measurable with techniques such as quantitative polymerase chain reaction (qPCR) and hybridization based methods, sequencing of smRNA libraries has distinct advantages due to its relative high-throughput and sensitive detection of numerous smRNA species (12–14). Additionally, because it allows for agnostic detection of unknown species, novel smRNAs and sequence variation of known smRNAs (such as miRNA isoMirs) can be assessed. Nevertheless, technical challenges in library preparation limit throughput and can lower library quality. In many protocols, documented ligation biases resulting from the use of adenosine triphosphate (ATP)-turnover deficient truncated T4 RNA Ligase 2 and specific adapter sequences combined with preferential ligation of overabundant sequences can limit accuracy and make the detection of non-favored or lowly expressed smRNAs difficult (15–19). However, it is important to note that because these ligation biases are largely sequence specific, in principle they do not affect inter-sample fold changes with consistent adapter use (16,20,21). Thus, differential expression, and not absolute quantification, is typically the outcome of interest in smRNA sequencing studies. In addition to issues of ligation bias, the formation of large quantities of unwanted adapter dimer ligation product necessitates time- and labor-intensive removal steps via denaturing gel-electrophoresis, as such dimers are only ∼20–30 bp smaller than many desired sequences such as miRNAs.
Targeted reduction of specific sequences from sequencing libraries is frequently employed to enrich for sequences of interest, such as with rRNA- or globin-RNA reduction from mRNA-sequencing libraries. Recently, we and others have demonstrated techniques to deplete-specific smRNA sequences from sequencing libraries by using blocking oligonucleotides that prevent 5′ adapter ligation, effectively preventing further incorporation of these targets in downstream library construction (22,23). While very effective, both strategies have limitations, such as off-targets due to sequence similarity, particularly in the seed region of non-targeted miRNAs with hairpin-oligo blocking. Additionally, neither of these methods address excess adapter dimer ligation products, and thus still require the use of denaturing gel separation of library products before sequencing. Strategies using locked nucleic acid oligos have been employed to prevent adapter dimer formation with some limited success (24). Currently, the most effective means of adapter dimer prevention or removal during smRNA sequencing has been demonstrated with ligation free template-switching protocols or chemically modified adapters that sterically inhibit ligation of the 3′ and 5′ adapters to each other and inhibit adapter dimer reverse transcription (25,26). Because adapter dimer formation is limited, these strategies allow for the use of SPRI-bead based size selection in place of gel separation, which greatly increases library preparation throughput. While very effective at a range of RNA input concentrations, the chemical modifications’ efficacy appears to have some adapter-sequence specificity, which, combined with reported necessity of custom reaction conditions, may limit their use in other custom or commercial smRNA protocols (26). In addition, these methods also do not allow for the targeted removal of endogenous smRNAs, requiring additional experimental methods or greater sequencing depth to allow measurement of lowly abundant smRNAs in the background of a few highly abundant species. Considering the benefits and limitations of these approaches, a single method that can efficiently remove both unwanted smRNAs and adapter dimer from smRNA libraries in a customizable manner would be tremendously useful.
New genome- and epigenome-editing tools based on repurposing the CRISPR/Cas9 (clustered regularly interspersed short palindromic repeats/Cas9) bacterial immune system have many potential uses (27,28). The Cas9 nuclease, when complexed with a short RNA oligonucleotide known as a single guide RNA, or sgRNA, can induce double-stranded breaks (DSBs) at specific sgRNA complementary locations. The low-cost and easily programmable nature of the CRISPR/Cas9 system has led to its use in a variety of applications, such as generation of transgenic animals and cell lines and pooled genome-wide screening (29–31)
Recently, CRISPR/Cas9 has been repurposed as a programmable restriction enzyme to direct cleavage in a more precise and customized manner than conventional restriction enzymes, allowing for innovations in cloning and sequencing of complex repeat regions (32–34). Methods using CRISPR/Cas9 as a restriction enzyme have been used to selectively deplete overabundant sequences in a process termed Depletion of Abundant Sequences by Hybridization (DASH) using CRISPR/Cas9 (35). DASH was used to remove targets such as ribosomal RNA (rRNA) from mRNA-seq and wild-type KRAS background sequence from cancer samples by directing their targeted cleavage and preventing their further amplification and sequencing. Similarly, other groups have applied this process to other assays such as the reduction of mitochondrial DNA from ATAC-seq libraries via CARM (CRISPR-Assisted Removal of Mitochondrial DNA) (36,37). Here, we have adapted the DASH method to deplete adapter dimer and highly abundant miRNAs from smRNA-seq libraries in a process we have termed miRNA and Adapter Dimer - Depletion of Abundant Sequences by Hybridization (MAD-DASH). MAD-DASH effectively removes these sequences either alone or in combination, increasing proportional read depth of non-targeted species and dramatically improving library construction throughput by enabling SPRI bead size-selection instead of denaturing gels with read depth equivalence achieved when using as low as 50 ng RNA input. We identify improvements in the rational design of adapter sequences governing MAD-DASH efficacy and demonstrate the utility of this method for blood based, low-input smRNA-seq biomarker studies with the removal of adapter dimer and a known highly abundant erythrocyte contaminant miRNA from human plasma.
MATERIALS AND METHODS
Total RNA isolation from plasma
Peripheral blood sample collections for the isolation of RNA from human plasma were performed as previously described (22) in accordance with the Institutional Review Board at the University of Alabama at Birmingham. Briefly, each collection 5 ml of blood was drawn, centrifuged at 2200 g for 10 min to isolate plasma and then stored at −80°C until further use. Total RNA was isolated from 1 ml thawed plasma using the Plasma/Serum Circulating and Exosomal RNA Purification Kit (Slurry Format) (Norgen Biotek) and concentrated to 20 μl using the RNA Clean-Up and Concentration Kit (Norgen Biotek). To limit sample variation between MAD-DASH replicate groups, plasma RNA from multiple donors was combined prior to library construction.
Cas9 and MAD-DASH sgRNA preparation
Streptococcus pyogenes Cas9 (5 μg/μl, 30 μM) was purchased from PNABio (CPO2). sgRNAs were designed as described in the main text with full sequences listed in Supplementary Table S1. sgRNAs were constructed following previously described methods (38). Briefly, T7 promoter containing single stranded oligos (Integrated DNA Technologies) corresponding to each sgRNA were annealed to a consistent tracrRNA sequence to generate a double-stranded sgRNA DNA template of the form [TAATACGACTCACTATAGG-N20-GTTTTAGAGCTAGAAATAGCAAGTT-AAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCTTTT] wherein the N20 is the sgRNA sequence. Except where indicated, sgRNAs had three 5′ G nucleotides appended to their sequence for efficient transcription when used as a template for in vitro transcription (IVT) with the MEGAshortscript T7 Transcription Kit (Invitrogen), per manufacturer's instructions. IVT was carried out with 1 μg template at 37°C for 16 h, treated with 1 μl TURBO DNASE (Invitrogen) for 15 min, heat inactivated at 95°C for 10 min and the transcribed RNA was cleaned up using the MEGAclear Transcription Clean-up Kit (Invitrogen). sgRNAs were quantified using Broad Range RNA Qubit (Invitrogen), normalized to 1.5 μg/μl (46.5 μM) and stored in single-use aliquots at −80oC. Multiplex 5′ adapter dimer targeting sgRNAs were pooled evenly before freezing.
MAD-DASH small RNA sequencing
Small RNA sequencing was performed as described previously (22), with modifications to incorporate the MAD-DASH protocol. A full, detailed MAD-DASH smRNA-seq protocol including sgRNA preparation is provided in the Supplemental Methods. Oligos for MAD-DASH smRNA-seq were from Integrated DNA Technologies and are listed in Supplementary Table S1. Briefly, replicate libraries for each treatment condition were generated from 4 μl isolated total RNA from either human plasma (n = 3 or 4 each condition) or 50 ng purchased Human Brain Total RNA (n = 2 each condition) (Invitrogen), with no initial size selection. Except where indicated, no cleanup or dilution steps were performed and in general products from one step in the protocol were used as direct input into the next step. RNA was used as input in a ligation reaction (25°C for 1 h) with 10 μM pre-adenylated 3′ adapter ligation reaction and 1 μl T4 RNA Ligase2, truncated (NEB). This ligation product was then annealed with the 1 μl 10 μM RT primer at 25°C for 1 h. A 5′ adapter ligation was performed at 25°C for 1 h using 1 μl T4 RNA Ligase 1 (NEB) and 1 μl 20 μM multiplex 5′ adapter pool or the modified 5′ adapter (pre-denatured at 70°C for 2 min). The 5′ adapter ligated products were split 1:2 before reverse transcription. Reverse transcription of ligation products (11 μl) was performed using Superscript II Reverse Transcriptase (Invitrogen) before performing four cycles of PCR (PCR1) using Phusion 2× High-Fidelity PCR Master Mix (NEB) with cycling conditions of 94°C for 30 s, four cycles of 94°C for 10 s and 72°C for 45 s and a final extension at 65°C for 5 min. PCR products (50 μl total) were cleaned and concentrated to 10 μl using 1.8× (90 μl) Agencourt AMPure XP SPRI Beads (Beckman-Coulter).
To perform the adapter dimer or hsa-miR-16–5p MAD-DASH, we followed the instructions from PNABio with modification for incorporation into our smRNA-seq workflow. For a MAD-DASH reaction volume of 20 μl, 1 μl 30 μM Cas9 was pre-incubated with 5 μl 46.5 μM of sgRNA, 2 μl 10× NEBuffer3.1 (NEB) and 2 μl 10× bovine serum albumin (NEB) at 37°C for 15 min before combining with the 10 μl sample DNA and incubating at 37°C for 2 h. After 2 h, 1 μl 4 μg/μl RNAseA (NEB) was added to remove sgRNA (37°C for 15 min) followed by 1 μl 800 U/ml (20 μg/μl) Proteinase K (NEB) (37°C for 15 min, 95°C for 15 min) to rapidly inactivate Cas9. We found this incubation with Proteinase K to be critical for library yield, as Cas9 not only has extremely high DNA-binding when not complexed to an sgRNA, but also as described in the main text there was significant non-target competitive-binding when using adapter dimer targeting sgRNAs. Other methods of Cas9 inactivation including heat (65°C or 95°C) or STOP Solution (30% glycerol, 1% sodium dodecyl sulphate SDS), 250 mM ethylenediaminetetraacetic acid (EDTA), pH 8.0) were less effective or resulted in significant reductions in final library yield (Supplementary Figure S1). Proteinase K inactivation at 95°C was critical, as the post-Proteinase K-treated samples were immediately used to perform a second round of PCR amplification (PCR2) for 11 cycles using identical cycling conditions. Proteinase K treatment and inactivation rendered post-MAD-DASH sample cleanup prior to PCR2 unnecessary.
Following PCR2, samples were once again cleaned up with AMPure XP beads. Gel extraction samples were cleaned up with 1.8× (90 μl) beads to concentrate them to facilitate loading into gel electrophoresis wells. Denaturing gel electrophoresis with 10% TBE-Urea Mini-PROTEAN gels (Bio-Rad) and extraction were performed as described previously for miRNA 145 bp band samples. For gel extracted smRNA region samples, the only difference was we extracted the ∼75 bp region between the ∼125 bp adapter dimer band and 200 bp band (rRNA) as opposed to just the 145 bp miRNA-library band. To account for this larger gel slice, we doubled our gel soaking solution (2 ml 5M Ammonium Acetate, 2 ml 1% SDS solution, 4 μl 0.5M EDTA, 16 ml RNAse/DNAse free dH20) step (2 h at 70°C) and subsequent addition of 100% isopropanol volumes to 600 μl to precipitate the DNA overnight. DNA was washed with ice-cold 80% ethanol before resuspending in 10 μl dH20. Samples prepared without gel extraction were cleaned up with 0.9× (45 μl) and 1.8× (90 μl) AMPure bead volumes serially to size select region roughly equivalent to smRNA region gel extraction, i.e. below 200 bp. These samples were eluted in eluting in 20 μl dH20 and we refer to them in text simply as ‘bead cleanup samples’.
Libraries were quantified with the KAPA Library Quantification Kit for Illumina (KAPA Biosystems) and normalized to 5 nM concentration. MAD-DASH smRNA-seq libraries were combined (2 μl for adapter dimer MAD-DASH samples and 1 μl of samples without adapter dimer depletion to prevent adding proportionally too much adapter dimer to the flow cell) to yield a final 5 nM total pool. MAD-DASH smRNA-seq library pools from (i) AMPure bead cleaned-up brain RNA samples (ii) smRNA region gel-extracted brain RNA samples or (iii) smRNA region gel-extracted plasma samples were sequenced on two lanes each on an Illumina HiSeq2500 with single-end 50 bp reads according to standard Illumina protocols. MAD-DASH smRNA-seq library pool consisting of AMPure bead cleaned-up plasma samples and miRNA band gel-extracted plasma samples were sequenced together on an Illumina NextSeq with single-end 75 bp reads according to standard Illumina protocols.
Data analysis
Data analysis was performed as previously described (22) with minor modifications to incorporate MAD-DASH-specific details. FASTQs were demultiplexed using custom index sequences and adapter sequences were trimmed using Cutadapt (39). Sequences <15 bp after adapter trimming were separated into a ‘Short Fail’ FASTQ, from which adapter dimer reads (corresponding to a blank sequence line, i.e. trimmed read length of 0 bp) and non-adapter dimer short fail reads were collected. Trimmed reads were aligned to pre-miRNA sequences (miRBase v19 (40)) using Bowtie2 (41) with only two mismatches allowed and keeping only unique best alignments. Mature miRNA counts were determined by counting the aligned pre-miRNA reads overlapping mature miRNA boundaries using BEDTools (42). Trimmed reads that did not align to miRNAs contained other smRNA sequences and were designated as ‘non-miRNA usable reads’ for purposes of read fraction calculations. Statistical analysis was performed using the R statistical software package (version 3.4.0). For plotting comparisons between groups, replicate libraries were downsampled to equivalent counts and summed before downsampling again with the compared group to yield an equivalent number of aligned reads between groups and then transforming to counts-per-million. Read fraction calculations were composed of all miRNA aligned reads, non-miRNA usable reads, short fail reads and adapter dimer reads and performed using the above process to generate CPM. Average read fractions were used for plotting purposes, while fold changes and Benjamini–Hochberg adjusted P-values (FDR) were determined with DESeq2 (43) with local dispersion estimates and likelihood ratio tests. DESeq2 (with identical parameters) was also used to determine differential miRNA and adapter dimer reads between samples using only miRNA reads and adapter dimer reads with the above downsampling process without summing group replicates prior to between group downsampling. Significant differential read abundance was defined as a BH adjusted P-value (FDR) < 0.05. Pairwise fold-changes and separate treatment group dispersion estimates were also calculated with DESeq2 as above, with dispersions plotted using the smooth.spline() function in R after removing NA values.
Adapter dimer quantitative PCR in human plasma MAD-DASH libraries
qPCR primers were designed to specifically amplify adapter dimer sequences from either multiplex 5′ adapter 1 or the modified 5′ adapter and so they would be incapable of amplifying cleaved adapter dimer sequences (Supplementary Figure S2). Primer sequences are listed in Supplementary Table S1. qPCR was performed using the QuantStudio 6 Flex System (Applied Biosystems) with Power SYBR Green Master Mix (Invitrogen) in 10 μl reactions using post-PCR2 samples diluted 1:10 000 in KAPA diluent (10 mM Tris–HCl, pH 8.0 + 0.05% Tween 20). Absolute adapter dimer concentration in nM was calculated using a standard curve generated from a synthetic adapter dimer library (Supplementary Table S1). Multiplex adapter dimer amounts were multiplied by four to account for the other three adapter dimer sequences resulting from the equimolar multiplex 5′ adapters. Data analysis was performed using R and comparisons between treated and untreated replicate adapter dimer amounts were performed using an unpaired two-sided Wilcox test with significance set as P < 0.05.
RESULTS
Overview of MAD-DASH
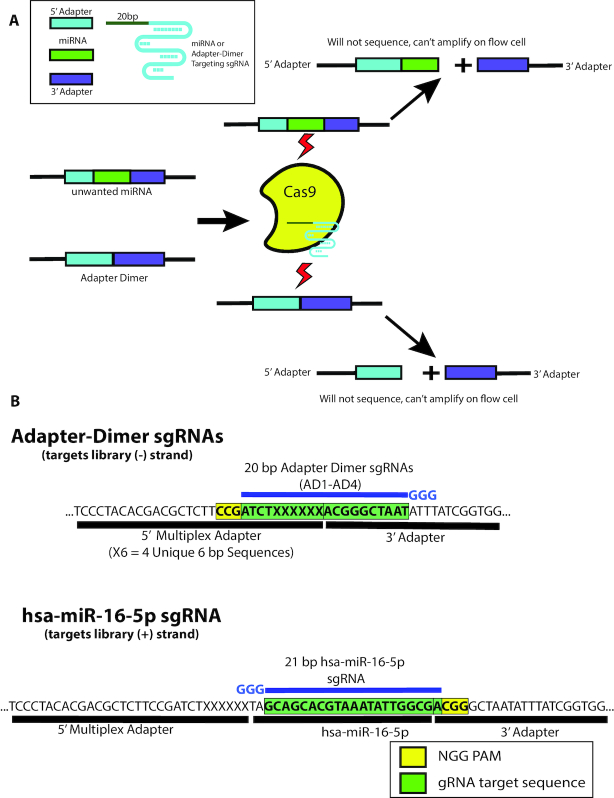
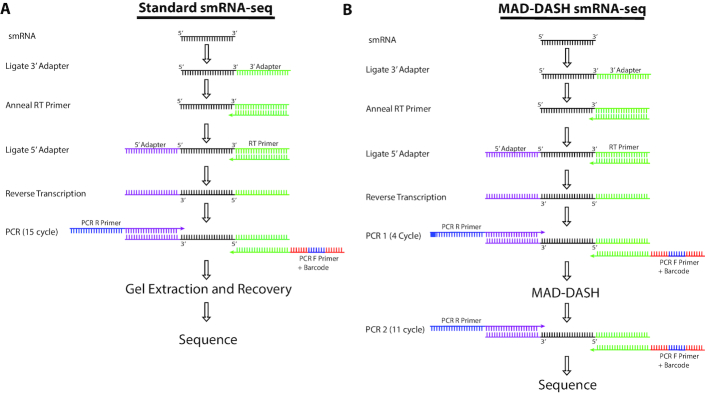
To overcome the experimental limitations and low-throughput of our standard smRNA-seq workflow (22) caused by excessive adapter dimer and overabundant miRNAs, we sought to adapt the DASH procedure to smRNA-seq (Figure 1A). However, technical differences in smRNA-seq library preparation compared to that of RNA-seq used in DASH required several important modifications to the DASH procedure. First, because the Cas9 enzyme almost exclusively depends on double-stranded DNA (dsDNA) for efficient nuclease activity (44), DASH performs CRISPR/Cas9 in vitro digestion after mRNA cDNA second-strand synthesis before library amplification. However, in most smRNA-seq workflows, smRNA cDNA generated in a first-strand synthesis is immediately amplified in PCR to generate dsDNA libraries. Because Cas9 is a single-turnover enzyme (45), it is critical that in vitro digestion occurs before full amplification to ensure sufficient reduction of target sequences. Therefore, to generate dsDNA libraries amenable to CRISPR/Cas9 targeting, we performed a first PCR with a limited number of cycles (four) before performing MAD-DASH and then further amplifying with a second round of PCR (eleven cycles). Given the single-turnover nature of Cas9 enzymatic activity, this approach ensures the presence of dsDNA required for Cas9 in vitro digestion while also maintaining a low DNA-to-Cas9 ratio necessary for efficient targeted sequence removal.
Figure 1.
Depletion of adapter dimer and overabundant miRNAs with MAD-DASH. (A) MAD-DASH employs Cas9 and adapter dimer- or miRNA-specific sgRNAs to selectively deplete these sequences from final libraries. Cleaved sequences will not amplify during the second round of PCR amplification, and are also no longer suitable substrates for bridge amplification during Illumina sequencing (though they will still be able to bind to the flow-cell using the P5/P7 sequences remaining on each cleaved library). (B) Design of adapter dimer and hsa-miR-16–5p sgRNAs using available pre-existing PAM sites on our smRNA-seq 5′- and 3′-adapters. Adapter dimer targeting sgRNAs use a minus strand ‘NGG’ and have 10 bp of homology to both the 5′- and 3′ adapter, ensuring target cleavage specificity for adapter dimer sequences. hsa-miR-16–5p uses a 3′ PAM site one base away from the 3′—adapter ligation junction, which can be generalized to other smRNAs and provides highly specific targeting while minimizing off-targets. Shown are only the plus strand of non-PCR tailed sequences. Green boxes indicate the plus strand sequence corresponding to sgRNA location in the dsDNA library, while the yellow box indicates the plus strand location of the ‘NGG’ PAM in the dsDNA library. sgRNA location is depicted in blue along with the three 5′ ‘G’ nucleotides necessary for high levels of T7 in vitro transcription.
Another issue we considered in developing MAD-DASH is the considerably restricted targetable sequence space. Unlike targeting ribosomal RNA derived cDNA or mitochondrial DNA with hundreds of possible PAM sites, smRNA-seq adapters are usually ∼25–30 bp in length and the location of PAM sites and possible sgRNAs is limited. Critically, successful targeting of adapter dimer sequences must incorporate sufficient base pair sequence from both the 5′ adapter and the 3′ adapter to prevent off-target cleavage of the rest of the library, which has both a 5′ and 3′ adapter ligated to it. In our smRNA-seq workflow, we use an equimolar pool of four 5′ adapters with a consistent region and a unique 3′ 6 bp base-diverse region to improve smRNA ligation efficiency (predicated on improved adapter–RNA hybridization and resulting favorable ligation cofold structure (16)). To deplete adapter dimer sequences, we designed four unique sgRNAs targeting each possible adapter dimer using a ‘CGG’ PAM site found in the consistent region of the 5′ adapters’ minus strand (Figure 1B). We reasoned that these sgRNAs have a balanced 10 bp of sequence complementarity to both their respective 5′ and 3′ adapter sequences and should thus limit off-target cleavage to smRNAs with significant 5′ sequence similarity to the 5′ end of the 3′ adapter. To confirm our sgRNA designs selectively and effectively deplete adapter dimer sequences, we performed Cas9 in vitro digestion of synthetic adapter dimer sequences prior to implementation in the full MAD-DASH protocol (Supplementary Figure S3).
A significant benefit of MAD-DASH relative to other adapter dimer depletion methods is the ability to also deplete specific smRNAs from the library. Similar to depletion of adapter dimer, removal of highly abundant miRNAs such as hsa-miR-16–5p (46) dramatically improves detection of more lowly expressed smRNA species. Utilizing the ‘CGG’ PAM site at the 5′ end of our 3′ adapter allows for highly specific targeting of smRNAs using their 3′-most ∼20 bp sequence (Figure 1B) with minimal dependence on adapter nucleotides. We also devised a modified miRNA MAD-DASH sgRNA targeting strategy that shows similar efficacy to the 3′ adapter PAM sgRNAs on synthetic miRNA libraries using truncated sgRNAs (47) and amenable miRNA internal PAM sites (present in ∼26% of human mature miRNAs annotated in miRBase v21) (Supplementary Figure S4).
Finally, we considered the dynamics of the inverse relationship between RNA input amount and adapter dimer formation and its effect on MAD-DASH design. In general, low RNA inputs lead to greater adapter dimer formation, which can be especially pronounced in the ∼1–10 ng/μl range of RNA from many common clinical biofluids (often comprising >90% of the amplified library in our experience). Frequently, the dilution of adapters prior to ligation reactions is employed as a means of reducing adapter dimer formation, even when using chemically modified adapters. However, we find that high molar excess of both 3′ and 5′ adapters (10 and 20 μM, respectively) is necessary to drive ligation reaction efficiency and more accurate quantification (Supplementary Figure S5). Conversely, there is a direct relationship to RNA input and targeted miRNA species. Therefore, both high- and low-RNA inputs will face specific challenges regarding sufficient removal of abundant miRNAs or adapter dimer, respectively. To demonstrate the effectiveness of adapter dimer- and miRNA-targeting MAD-DASH in the low RNA-input range used in many clinical sequencing projects, we have constructed smRNA-seq libraries from total brain RNA (50 ng) and human plasma RNA (∼1–10 ng), which we term ‘low input’ and ‘very low input’ due to being significantly less than the 1 μg recommended RNA input in many commercial kits. This yielded ∼2 and ∼0.2 nM, respectively, of adapter ligated product prior to MAD-DASH in vitro digestion. For purposes of determining amounts of excess Cas9 and sgRNA relative to target DNA when targeting adapter dimer, we treated the library as if it contained 100% adapter dimer. While this is certainly an overestimation of true adapter dimer concentration, we reasoned that because Cas9/sgRNA DNA-binding activity is more permissive to sequence variation and distinct from its nuclease activity (45,48,49), the presence of significant sgRNA sequence similarity on all adapter-ligated sequences would yield considerable competitive off-target Cas9/sgRNA binding (but not cleavage). We thus used a high molar excess of Cas9 input (5 μg, 30 μM) and adapter dimer targeting sgRNA input (7.5 μg, 232.5 μM), which yielded final molar excess relative to target of ∼1500×/∼6000× and ∼15 000×/∼60 000× in brain and plasma RNA, respectively. Identical Cas9/sgRNA input amounts were used when targeting hsa-miR-16–5p (likely considerably more in excess to target), while multiplexing of adapter dimer and hsa-miR-16–5p targeting sgRNA used an identical Cas9 amount and equal amounts (20%) of each sgRNA in the pool. These ratios represent a roughly 10-fold increase of those used in DASH and other sequencing library CRISPR/Cas9 in vitro digestion methods. However, we found this increase to render the MAD-DASH protocol robust when depleting large amounts of adapter dimer and miRNAs and simplify Cas9/sgRNA-to-target ratios. We expect that varying RNA input during smRNA-seq library construction may require more or potentially less Cas9/sgRNA in the in vitro digestion.
Successful depletion of adapter dimer from smRNA-seq libraries obviates the need to perform low-throughput denaturing gel separation and cleanup, and consequently our MAD-DASH smRNA-seq protocol employs a double-sided SPRI bead size selection for library sequences less than ∼200 bp, corresponding to insert range up to approximately 75 bp. Because all library preparation steps including MAD-DASH and final library cleanup can be done in a 96-well plate using SPRI beads, throughput is dramatically increased and amenable to automation. We estimate that average library construction time for a full plate of 96 samples (requiring six denaturing gels followed by extraction and precipitation in our standard protocol) could be completed in one day for MAD-DASH smRNA-seq compared to 3–4 days with our standard protocol.
Evaluating the quantitative performance of MAD-DASH in brain total RNA
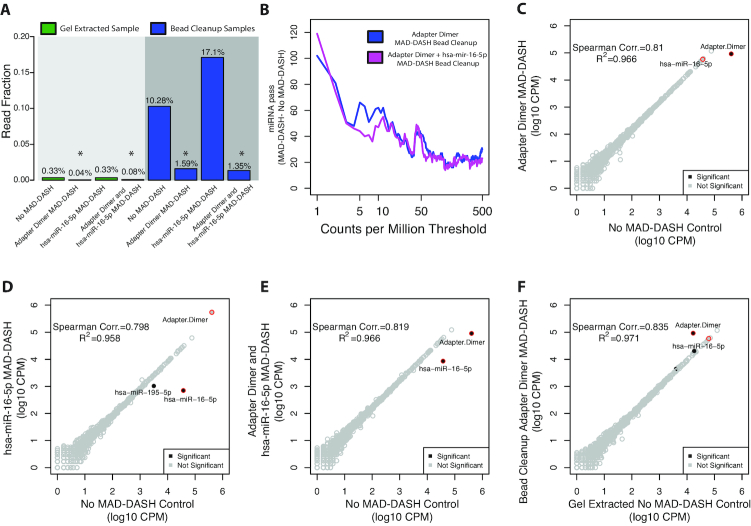
To evaluate the effect of reducing adapter dimer and a representative highly abundant miRNA (hsa-miR-16–5p) in low input total brain RNA (50 ng), both alone and in combination, we generated replicate MAD-DASH no-Cas9/sgRNA control, adapter dimer MAD-DASH, hsa-miR-16–5p MAD-DASH and combined adapter dimer/hsa-miR-16–5p MAD-DASH smRNA-seq libraries and analyzed the normalized abundances of adapter dimer in both gel extracted and bead cleaned up samples (Figure 2A). With smRNA region gel extraction, adapter dimer represents an average 0.33% read fraction, which is reduced further by 8.9× (FDR = 3.74 × 10−10) with adapter dimer MAD-DASH. In bead cleanup samples, MAD-DASH significantly depleted adapter dimer from an average 10.3% to 1.59% read fraction (6.9×, FDR = 6.28 × 10−4) and hsa-mir-16–5p to less than 0.015% read fraction (56.2×, FDR = 2.34 × 10−233). Combination of adapter dimer and hsa mir-16–5p MAD-DASH demonstrated comparable adapter dimer reduction and less (though still significant) hsa mir-16–5p reduction (7.7×, FDR = 4.16 × 10−3 and 7.1×, FDR = 1.38 × 10−33, respectively). MAD-DASH increased non-hsa-miR-16–5p miRNA and non-miRNA usable reads as much as 23% (Supplementary Figure S6). Adapter dimer MAD-DASH in bead cleanup samples was dramatically reduced to within 5.9× of gel extracted samples, which prompted us to further explore the degree to which overall library quality was affected and whether this level of reduction was sufficient to generate high quality libraries. We limited further in-depth differential expression analysis to changes in miRNAs and adapter dimer sequences and not other read fractions primarily because alignment of non-miRNA smRNAs to the full reference assembly can be complicated (50) and was not expected to demonstrate significantly different results.
Figure 2.
MAD-DASH smRNA-seq reduces adapter dimer and hsa-miR-16–5p in human brain total RNA samples. (A) Averaged read fraction of downsampled and CPM normalized adapter dimer sequences in gel extracted and bead cleanup samples. MAD-DASH in bead cleanup samples significantly depletes adapter dimer individually and in combination with depletion of hsa-miR-16–5p simultaneously. Asterisks indicate that a given read fraction was determined to be significantly altered compare to the respective cleanup's no MAD-DASH control using DESeq2 (FDR < 0.05). (B) Plot depicting the difference in the number of mature miRNA species reaching a specified count per million threshold between bead cleanup adapter dimer MAD-DASH smRNA-seq replicates and MAD-DASH control replicates. Downsampling and CPM normalization was performed as described in ‘Materials and Methods’ section. (C–F) Read counts from normalized replicate groups for bead cleanup treated versus control MAD-DASH samples targeting (C) adapter dimer (D) hsa-miR-16–5p and (E) adapter dimer and hsa-miR-16–5p (F) adapter dimer compared to gel extracted no-Cas9/sgRNA control. Significantly different sequences are filled black circles, with those having a log2-fold-change > 1 being labeled with text. Non-significantly different miRNAs are depicted as open gray circles. Adapter dimer and hsa-miR-16–5p are depicted as red circles with the same gray or black fill to denote significance. Significance was determined with DESeq2 and set as a Benjamini–Hochberg corrected P-value < 0.05.
Like the effect observed when using our 5′ hairpin blocking method (22), depletion of highly abundant adapter dimer and hsa-miR-16–5p results in increased sensitivity for lowly abundant species. We detected 116 more miRNAs compared to bead cleanup control replicates, with as many as 62 more at the commonly used threshold of 10 counts per million (CPM) (Figure 2B and Supplementary Figure S7). Comparison of significant differential expression between MAD-DASH treated and control replicates demonstrated highly specific depletion of targeted sequences (Figure 2C–E). No significant off-targets were observed when using adapter dimer sgRNAs while hsa-miR-16–5p sgRNA use resulted in only one significant off target, hsa-mir-195–5p. hsa-mir-195–5p is a member of the hsa-mir-16–5p seed region family and shares nearly identical sequence to hsa-mir-16–5p (Supplementary Table S2). Interestingly, when hybridized to a library sequence containing hsa-mir-195–5p the hsa-mir-16–5p sgRNA contains a 1 bp insertion ‘bulge’ at the PAM -2 site, which appears to be well tolerated and is consistent with prior observations regarding the effect of sgRNA/DNA insertion/deletions on Cas9 nuclease activity (51).
To demonstrate that the MAD-DASH smRNA-seq workflow's implementation of a SPRI bead cleanup did not negatively affect library reproducibility compared to our standard gel extraction method, we next compared bead cleanup adapter dimer MAD-DASH replicates to smRNA region gel extracted MAD-DASH control (no-Cas9/sgRNA) replicates (Figure 2F). Only two miRNAs (hsa-miR-22–3p and hsa-miR-9–3p) were significantly altered but had minimal log2 fold-changes (∼0.2). As discussed above, while the adapter dimer fraction in the MAD-DASH bead cleanup samples was reduced 6.5-fold to <1.6% of the library, it was still ∼5.9× more abundant than adapter dimer in the gel extracted control samples. As correlation may not necessarily be the best metric of reproducibility in smRNA-seq, we employed DESeq2 to estimate the dispersion as a function of read depth in control and MAD-DASH treated replicate groups for both gel extracted and bead cleaned up samples (Supplementary Figure S8). As expected, dispersion was generally higher at lower counts and for both cleanup methods MAD-DASH treatment did not result in greater dispersion amongst replicates and was in fact less dispersed at higher counts.
Taken as a whole, we find that while this iteration of MAD-DASH is very effective at removing adapter dimer and not affecting non-targeted sequences, this iteration was not equivalent to gel extraction at 50 ng RNA input. Nevertheless, this level of reduction is likely to be more than sufficient to take advantage of the throughput improvements of the MAD-DASH protocol with minimal increase in cost and sequencing depth.
Rational design of a modified 5′ adapter improves MAD-DASH adapter dimer depletion
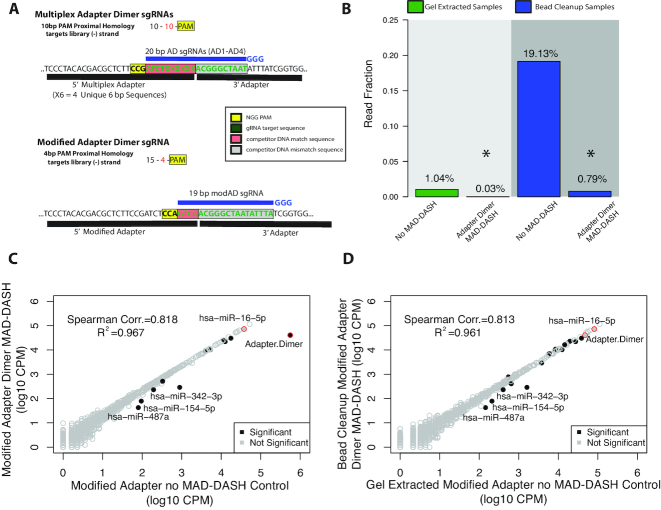
Although our first iteration of the MAD-DASH smRNA-seq method successfully depletes adapter dimer, we sought to optimize the reduction further through rationally modifying the design of our 5′ adapter PAM location. As mentioned, our multiplex adapter dimer sgRNA contains ten base pairs of sequence similarity to both the 3′ adapter and its respective 5′ adapter. Prior work has demonstrated that not only does non-sgRNA bound apo-Cas9 possess considerable nonspecific DNA binding ability, but the Cas9/sgRNA complex is capable of binding competing target–DNA with 12 matching PAM-proximal bases as strongly as perfect target sequence (∼1000× longer than complete mismatch) (45). Our multiplex adapter dimer sgRNAs, which have a 10 bp competitor mismatch—10 bp PAM proximal match—PAM design, are thus expected to have considerable, lasting off-target binding to competitor DNA (every non-target molecule in the library) while not having sufficient sequence similarity to engage the Cas9 HNH/RuvC nuclease domains (48).
With this dynamic in mind, we designed a single 5′ adapter identical to one of our four multiplex 5′ adapters, save for a single extra C at the start of the base diverse region that generated a ‘CCA’ plus strand/minus strand ‘NGG’ PAM site 4 bp from the 5′ adapter/3′ adapter junction (Figure 3A). Our 19 bp modified 5′ adapter sgRNA has a 15 bp competitor mismatch—4 bp PAM proximal match—PAM design, and is thus expected to exhibit considerably greater adapter dimer depletion by limiting competitive binding to all other adapter ligated sequences. We generated replicate modified adapter MAD-DASH control (no-Cas9/sgRNA) and modified adapter dimer MAD-DASH libraries that as predicted demonstrated significantly greater adapter dimer depletion compared to our multiplex 5′ adapter samples. Although our single modified 5′ adapter generated considerably more adapter dimer (likely due to less efficient capture of miRNAs caused by only one sequence dictating hybridization and adapter/RNA cofold structure), in MAD-DASH bead cleanup samples we were able to deplete adapter dimer an average 30.7× (FDR = 2.36 × 10−23) to <0.8% average read fraction and achieve near equivalent read depth of miRNAs (Figure 3B and Supplementary Figure S9). This yields slightly less adapter dimer average read fraction compared with gel extracted samples, but they are statistically equivalent (FDR = 0.998). This represents a substantially greater reduction than in bead cleaned up multiplex adapter dimer MAD-DASH samples. This improvement leads to detection of 27 more miRNAs than in gel extracted control samples, and as many as ∼256 additional miRNAs (29 at 10 CPM) if compared to modified adapter bead cleanup control samples (Supplementary Figure S10).
Figure 3.
Rational design of a modified 5′ adapter with alternate PAM site enhances depletion of adapter dimer with MAD-DASH. (A) Design of the modified 5′ adapter compared to the multiplex 5′ adapter. Compared to the 10 bp competitor mismatch—10 bp competitor match—PAM design used in our first MAD-DASH iteration, the modified adapter uses 15 bp competitor mismatch—4 bp competitor match—PAM design and is predicted to have as much as 100-fold less binding to other non-target sequences in the library. Shown are the plus strands of non-PCR tailed sequences. Red boxes indicate the plus strand sequence corresponding to competitor match sequence in the dsDNA library, while the gray box indicates the sequence corresponding to sequence that drives adapter dimer target specificity, i.e. competitor mismatch sequence. The yellow box indicates the plus strand location of the ‘NGG’ PAM in the dsDNA library. sgRNA location is depicted in blue along with the three 5′ ‘G’ nucleotides necessary for high levels of T7 in vitro transcription. (B) MAD-DASH smRNA-seq using the modified 5′ adapter demonstrates substantially greater depletion of adapter dimer in bead cleanup samples than when using the multiplex adapter, and achieves a lower average read fraction than in modified adapter MAD-DASH control gel extraction samples. Normalized read fraction of downsampled and CPM normalized adapter dimer sequences for both adapter strategies are depicted. Reductions of adapter dimer in both gel extracted and bead cleanup samples were significant (DESeq2 adjusted P-value < 0.05). (C and D) Read counts from normalized replicate treated vs control groups prepared using the modified 5′ adapter. Bead cleanup MAD-DASH samples depleted of (C) modified adapter dimer and (D) bead cleanup modified adapter dimer compared to gel extracted no-Cas9/sgRNA modified adapter control are shown. Significantly different sequences are filled black circles, with those having a log2-fold-change > 1 being labeled with text. Non-significantly different miRNAs are depicted as open gray circles. Adapter dimer and hsa-miR-16–5p are depicted as red circles with the same grey or black fill to denote significance. Significance was determined with DESeq2 and set as a Benjamini–Hochberg corrected P-value < 0.05
Again, comparison of significant differential expression between modified adapter dimer MAD-DASH treated and control replicates demonstrated highly specific depletion, albeit with more off-targets than our multiplex adapter MAD-DASH samples (Figure 3C). This is to be expected due to the reduced number of PAM-proximal bases corresponding to the modified adapter's minus strand ‘CAGC’ needed for off target mismatch (6 versus 12 bp, i.e. NGG + 4 PAM proximal bases or 10 PAM proximal bases). Significantly altered sequences with log2 fold changes >1 (hsa-miR-342–3p, hsa-miR-154–5p and hsa-miR-487–3p) all contained ‘CCN’ corresponding to a target strand ‘NGG’ PAM site in the last 7 bp of their sequence (Supplementary Table S2). Interestingly, hsa-miR-487–3p and hsa-miR-342–3p demonstrate a similar insertion/deletion sgRNA/DNA bulge phenomenon to that seen with hsa-miR-195–5p and our hsa-miR-16–5p targeting sgRNA. As with multiplex adapter dimer MAD-DASH, modified adapter dimer MAD-DASH yielded equivalent or slightly less dispersion among replicates compared to controls (Supplementary Figure S11). Importantly, at 50 ng RNA input the improved MAD-DASH efficiency seen using the modified adapter yields an equivalent reduction in adapter dimer amounts compared to gel extracted control samples with minimal effect on library representation, achieving the desired technical correspondence between our standard smRNA-seq protocol and MAD-DASH smRNA-seq (Figure 3D) while dramatically increasing library throughput.
Evaluating MAD-DASH quantitative performance in human plasma RNA
We next sought to evaluate the utility of MAD-DASH in clinically relevant samples. The detection of circulating smRNAs in human plasma for discovery of diagnostic and prognostic biomarkers is an emerging field. However, as mentioned previously the amount of RNA isolated from patient plasma samples is minimal, often on the order of a few ng RNA/ml plasma, leading to low RNA inputs in smRNA-seq and high levels of adapter dimer formation. Additionally, certain blood cell-specific miRNAs such as hsa-miR-16–5p and hsa-miR-150 are highly abundant contaminants that limit sensitivity (22,46). We therefore first applied MAD-DASH smRNA-seq using both our multiplex 5′ adapter and our modified 5′ adapter strategy to replicate libraries generated from human plasma samples to demonstrate the successful removal of adapter dimer and hsa-mir16–5p and increase detection of lowly abundant species.
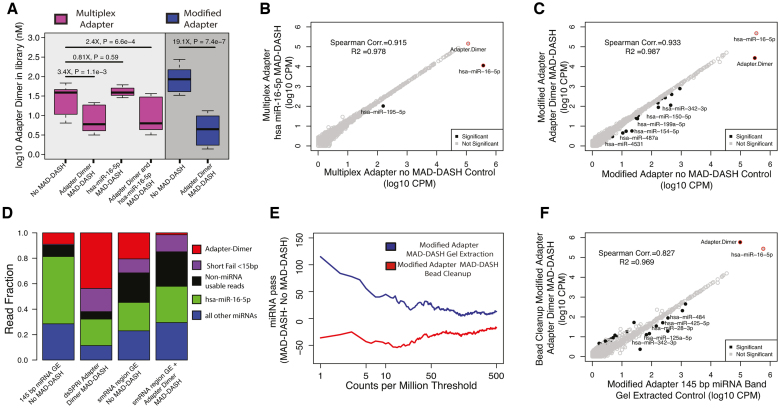
Because no-Cas9/sgRNA MAD-DASH control plasma samples yield >95% adapter dimer, dominating the sequencing reads making interpretation impossible, we instead designed a qPCR assay (Supplementary Figure S3) to quantify the reduction in adapter dimer between MAD-DASH control and treated samples (Figure 4A) before sequencing. We observed significant levels of proportional reduction when using either the multiplex 5′ adapter (3.4×, Wilcox P-value < 1.1 × 10−3) or modified 5′ adapter strategy (20.1×, Wilcox P-value < 7.4 × 10−7), with the modified 5′ adapter MAD-DASH again exhibiting comparatively greater reduction. Similar to our method employed with 50 ng total brain RNA libraries, we confirmed specific depletion of highly abundant hsa-miR-16–5p by sequencing gel extracted plasma control and MAD-DASH samples (54.5×, FDR = 8.86 × 10−225). This demonstrated similar specificity to that of brain RNA samples, with only hsa-miR-195–5p as an off-target (Figure 4B). In the case of the modified adapter MAD-DASH samples, there was still a significant reduction in residual adapter dimer remaining after gel extraction (Figure 4C).
Figure 4.
MAD-DASH smRNA-seq reduces adapter dimer and hsa-miR-16–5p in human plasma samples. (A) Adapter dimer concentrations in control or treated MAD-DASH smRNA-seq replicates (n = 3 in each group) made using the multiplex 5′ adapter (magenta) or the modified 5′ adapter (blue) were determined using qPCR and a synthetic adapter dimer library standard. Multiplex adapter sample concentration were adjusted 4× to account for the three other equimolar multiplex adapters. (B andC) Sequencing read counts from gel extracted, normalized replicate groups are plotted for treated vs control MAD-DASH samples using either (B) multiplex adapters and targeting hsa-miR-16–5p, or using (C) modified adapter and targeting adapter dimer. hsa-miR-16–5p depletion would not expected to vary between multiplex versus modified adapter use due to an identical sgRNA/PAM location. Adapter dimer depletion depicted in (C) represents a depletion of residual adapter dimer remaining after gel extraction, indicating that modified adapter dimer MAD-DASH can yield significant library improvements even when using traditional library cleanup methods. As in previous figures, significantly different sequences are filled black circles, with those having a log2-fold-change > 1 being labeled with text. Non-significantly different miRNAs are depicted as open gray circles. Adapter dimer and hsa-miR-16–5p are depicted as red circles with the same grey or black fill to denote significance. Significance was determined with DESeq2 and set as a Benjamini–Hochberg corrected P-value < 0.05. (D) Averaged read fraction of downsampled and CPM normalized adapter dimer sequences, hsa-miR-16–5p, non-hsa-miR-16–5p miRNAs and non-miRNA usable reads (see ‘Materials and Methods’ section) in modified adapter control miRNA band gel extracted (abbreviated as GE) samples, bead cleanup adapter dimer MAD-DASH samples and control and adapter dimer MAD-DASH smRNA region GE samples (E) Plot depicting the difference in the number of mature miRNA species reaching a specified count per million threshold between gel extracted or bead cleanup MAD-DASH plasma samples targeting adapter dimer. Downsampling and CPM normalization was performed as described in ‘Materials and Methods’ section. (F) Sequencing read counts from normalized replicate groups are plotted for bead cleanup adapter dimer MAD-DASH versus miRNA band gel control extracted samples. Significantly different sequences are filled black circles, with those having a log2-fold-change > 1 and a base mean of 100 being labeled with text to prevent overlapping labels of more lowly abundant significant sequences. Non-significantly different miRNAs are depicted as open gray circles. Adapter dimer and hsa-miR-16–5p are depicted as red circles with the same gray or black fill to denote significance. Significance was determined with DESeq2 and set as a Benjamini–Hochberg corrected P-value < 0.05.
Although these experiments demonstrate the efficacy and specificity of MAD-DASH in combination with gel extraction for very low input samples, we sought to evaluate whether MAD-DASH alone could provide equivalent results to gel extraction of the smRNA region, as demonstrated with 50 ng total brain RNA. We generated an additional four modified adapter dimer MAD-DASH smRNA-seq samples from independent human plasma samples and only performed bead cleanup prior to sequencing. Although adapter dimer was significantly reduced to 43.6% average read fraction, there remained 5.6× (FDR = 5.31 × 10−4) greater adapter dimer when compared to smRNA region gel extracted control samples at 20.3 % average read fraction (Figure 4D). Moreover, there was a greater number of short fail reads and fewer non-miRNA usable reads. Similar results (11.4×, FDR = 1.42 × 10−4) were obtained when comparing average read fractions in libraries prepared from the same plasma samples and subjected to our standard miRNA-seq protocol that gel excises 145 bp miRNA containing libraries, currently the most widely used method for adapter removal and achieving high miRNA read fraction. While the combined MAD-DASH and gel extraction samples yielded a dramatic increase in detection of possibly informative, lowly expressed miRNAs, adapter dimer MAD-DASH alone compared to miRNA band gel extracted samples yielded a small but consistent reduction in the number of detected miRNAs at a given count threshold (Figure 4E). However, bead cleanup with MAD-DASH still generated usable quantity of miRNA reads at an average read fraction of 11.3% versus 22.9% and 32.2% for smRNA region or miRNA band gel extracted samples (Figure 4D), with a high degree of correlation (Spearman rho = 0.827, R2 = 0.987) to miRNA band gel extracted control samples (Figure 4F).
To further assess library quality and reproducibility in these human plasma samples, we again estimated dispersion amongst bead cleanup replicates relative to miRNA-band gel extracted controls which showed overall equivalence, with MAD-DASH bead cleanup samples having slightly less dispersion at higher counts (Supplementary Figure S12A). To more completely ensure that MAD-DASH alone did not significantly affect non-targeted sequences and calculation of differential expression, we calculated fold-changes between all possible sample pairs within MAD-DASH samples and the miRNA-band gel extracted controls (Supplementary Figure S12B). Fold changes were highly concordant among MAD-DASH treated and control matched replicate pairs, with an average pairwise Spearman coefficient of correlation of 0.80, consistent with that seen in our previous work using blocking hairpin oligonucleotides (22). Relative to the smRNA-region extracted MAD-DASH and control plasma samples, bead cleanup MAD-DASH versus miRNA band gel extracted control plasma samples had a slightly greater number of significantly differentially expressed miRNAs with FDR < 0.05 (n = 15 and n = 23, respectively) (Supplementary Table S3), with some of the discrepancy accounted for in miRNAs with lower base mean CPM (<10). Previously identified miRNAs with 3′ proximal ‘NGG’ PAM sites such as hsa-miR-342–3p and hsa-miR-150–5p were amongst the consistently differentially expressed sequences, and as expected differentially expressed miRNAs with base means greater than 10 CPM tended to be consistently altered amongst replicates (Supplementary Figure S13). These analyses indicate that while library cleanup and extraction method impacts library quality, modified adapter dimer MAD-DASH has limited effects on reproducibility and differential expression with generally consistent and predictable off-targets.
Thus, unlike with low input (∼50 ng), with very low input (single ng levels) samples employing our current MAD-DASH protocol alone does not yield technical correspondence to gel separation methods. Nevertheless, it does enable sufficient library quality in cases where the increased throughput afforded is desirable, especially in circumstances using high-sequencing depth. In combination with gel extraction, however, adapter dimer MAD-DASH again yields significant reduction in adapter dimer compared to gel extraction alone. Importantly, we also demonstrate the ability of MAD-DASH to selectively deplete highly abundant hsa-miR-16–5p from plasma samples with the expected increase in non-hsa-miR-16–5p read depth. In principle, extension of MAD-DASH to smRNA-seq samples generated from other biofluids such as cerebrospinal fluid should necessitate only the initial detection of non-informative highly abundant sequences to be depleted along with adapter dimer for greatest effect.
DISCUSSION
In this study, we have adapted CRISPR/Cas9 in vitro digestion for the removal of adapter dimer and highly abundant smRNA species during smRNA-seq library preparation. These sequences limit the ability to detect lowly expressed smRNAs and in the case of adapter dimer necessitate arduous, low-throughput denaturing gel size-selection steps. MAD-DASH is a single, simple workflow that renders the need for gel extraction unnecessary in favor of a SPRI bead based size selection (Figure 5). We estimate that the increased library construction cost when incorporating MAD-DASH is <$10 per sample using commercially purchased Cas9 and in vitro transcription kits. These costs are likely to be easily covered by the dramatic increase in throughput and ensuing time savings in large scale sequencing projects. This may be particularly true if automation is employed, which MAD-DASH makes possible by allowing for all steps to be performed ‘on-plate’. We have designed and optimized the MAD-DASH protocol specifically for smRNA-seq, though like the original DASH CRISPR/Cas9 in vitro digestion methodology it is inherently generalizable to other library preparation methods with significant adapter dimer amounts. Frequently, however, adapter dimers are much smaller and more easily separable from insert-containing libraries in other applications, underscoring the special utility of MAD-DASH in smRNA-seq.
Figure 5.
Illustration of MAD-DASH and standard smRNA-seq workflows. While MAD-DASH involves an extra PCR amplification in addition to the MAD-DASH CRISPR/Cas9 in vitro digestion, the extra ∼3 h to accomplish this relative to finishing the PCR in our standard protocol are more than offset by no longer needing to perform gel extraction, DNA recovery and library concentration. For nearly a hundred samples, we estimate that researcher time for smRNA-seq library construction can be reduced to only 1 day using MAD-DASH smRNA-seq and that this throughput can be further improved with greater protocol familiarity or automation.
We demonstrate that our MAD-DASH smRNA-seq library protocol can deplete adapter dimer and targeted miRNAs with very high specificity that is consistent with known sequence-dependent CRISPR/Cas9 on-/off- target discrimination. We show ability to deplete adapter dimer up to an average 30.7× to <1% of library fraction and reduce abundant hsa-miR-16–5p up to 54.5× in human plasma RNA to below <1%. Additionally, we have demonstrated a rational basis for modifying PAM site and resulting adapter dimer targeting sgRNA location to address the uniquely problematic competitive-binding dynamics present in MAD-DASH smRNA-seq that result from every sequence in the library having significant sequence similarity. While our modified 5′ adapter MAD-DASH exhibits predictably more off-targets, these effects are expected and consistent. While outside the scope of this study, it is expected that further optimizing adapter design with both the dynamics of smRNA ligation efficiency and MAD-DASH depletion efficiency in mind will lead to further improvements in MAD-DASH implementation. Substitution of S. pyogenes Cas9 with other Cas9 variants or Cas9 with engineered alternative PAM specificities (52,53) can expand the limited targeting space in MAD-DASH, allowing for more customizable workflows. Although RNA-targeting CRISPR enzymes have recently been described (54,55) and could provide a substitute for Cas9 in an RNA-targeting MAD-DASH protocol, these enzymes have been shown to have considerable non-specific RNA cleavage when used in vitro which would almost assuredly reduce MAD-DASH specificity and reproducibility. Nevertheless, the most common commercial smRNA-seq library generation kits use ‘NGG’ PAM-site containing adapters that are completely consistent with implementation of MAD-DASH with few changes to their published protocols (Supplementary Table S4). Importantly, because MAD-DASH can not only remove adapter dimer but also other abundant smRNAs, it can also yield improvements in smRNA-seq methods already designed to limit adapter dimer formation by using chemically modified adapters or employing template-switching instead of adapter ligation.
Despite its utility, certain limitations in the use of MAD-DASH exist, primarily driven by the aforementioned dynamics of adapter dimer formation at varying RNA inputs. Although we have demonstrated relative equivalence of MAD-DASH at low RNA input (50 ng, or 20-times less than the 1 μg recommended by many kits, such as Illumina's TruSeq small RNA library kit), sufficient removal of adapter dimer to generate similar equivalence at very-low single ng levels remains challenging. At this time, further optimization of MAD-DASH reaction conditions or sgRNA targeting strategy is necessary to provide a perfect replacement of gel separation at these input levels. Other strategies, such as increasing input (admittedly difficult with RNA isolated from clinical biofluids), improvement in adapter design limiting pre-MAD-DASH adapter dimer formation, or advances in sequencing technology generating more reads per dollar (such as with Illumina's NovaSeq) may help to overcome this issue. Nevertheless, even at very low inputs our MAD-DASH smRNA-seq protocol still yields an average 12% miRNA read fraction (roughly 30–50% that seen in equivalent samples with various ranges of gel extraction), enabling sufficient use-case in many high-volume clinical studies where throughput and not sequencing cost may be the primary concern.
The use of dual randomer sequences in adapters to improve ligation efficiency and thus accurate miRNA representation (16) is likely to hinder the use of MAD-DASH for adapter dimer depletion unless care is taken to design a consistent 17–20 bp sgRNA targetable region. Additionally, the impact of the dynamics of competitive-binding on MAD-DASH when using other adapters remains to be explored. Finally, as described for other CRISPR/Cas9 based depletion methods (35,37), cleaved sequences still contain intact P5 and P7 flow-cell binding sequences, which although not able to be amplified during bridge-amplification can still bind to the flow cell during initial hybridization. While our cleaved adapter sequences are small enough to be partially removed by the lower limit of SPRI bead binding, MAD-DASH samples may require sequencing a greater library concentration relative to conventional methods. The extent to which these possible technical issues impact library quality and their resolution is the subject of future work.
In conclusion, our MAD-DASH smRNA-seq protocol provides a robust, high-throughput method to deplete adapter dimer and unwanted highly abundant smRNAs in a manner analogous to rRNA and globin reduction from mRNA-seq libraries and thus overcomes a significant obstacle in the use of smRNA-seq in large scale sequencing projects.
DATA AVAILABILITY
Sequence data from MAD-DASH smRNA-seq libraries generated in this study have been submitted to GEO and are available under accession number: GSE116029.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the patients who have graciously provided plasma samples for this study and members of the Clinical Research Support Program and the Center for Clinical and Translational Science Laboratory at the University of Alabama at Birmingham for their help with sample collection and processing, in particular C. Mel Wilcox, Meredith B. Fitz-Gerald and Robert P. Kimberly. We thank the Genomic Services Laboratory, led by Dr Shawn Levy, at the HudsonAlpha Institute for Biotechnology for provided sequencing services and data processing. We thank Gregory Cooper and Sara Cooper for critical reading of the manuscript and also thank Kenneth Day for technical advice with qPCR experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Center for Clinical and Translational Science at the University of Alabama in Birmingham [NCATS UL1 TR0001417]; State of Alabama to HudsonAlpha; HudsonAlpha Institute; Anonymous Private Donor; UAB Medical Scientist Training Program [NIGMS 5T32GM008361 to A.A.H., R.C.R.]. Funding for open access charge: HudsonAlpha Institute for Biotechnology; Anonymous Private Donor.
Conflict of interest statement. None declared.
REFERENCES
- 1. Liu Q., Paroo Z.. Biochemical principles of small RNA pathways. Annu. Rev. Biochem. 2010; 79:295–319. [DOI] [PubMed] [Google Scholar]
- 2. Malone C.D., Hannon G.J.. Small RNAs as guardians of the genome. Cell. 2009; 136:656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhao Y., Ransom J.F., Li A., Vedantham V., von Drehle M., Muth A.N., Tsuchihashi T., McManus M.T., Schwartz R.J., Srivastava D.. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007; 129:303–317. [DOI] [PubMed] [Google Scholar]
- 4. Gao M., Wei W., Li M.M., Wu Y.S., Ba Z., Jin K.X., Li M.M., Liao Y.Q., Adhikari S., Chong Z. et al.. Ago2 facilitates Rad51 recruitment and DNA double-strand break repair by homologous recombination. Cell Res. 2014; 24:532–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Michalik K.M., Böttcher R., Förstemann K.. A small RNA response at DNA ends in Drosophila. Nucleic Acids Res. 2012; 40:9596–9603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Di Leva G., Garofalo M., Croce C.M.. MicroRNAs in cancer. Annu. Rev. Pathol. 2014; 9:287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. O’Connell R.M., Rao D.S., Baltimore D.. microRNA regulation of inflammatory responses. Annu. Rev. Immunol. 2012; 30:295–312. [DOI] [PubMed] [Google Scholar]
- 8. Suzuki R., Honda S., Kirino Y.. PIWI expression and function in cancer. Front. Genet. 2012; 3:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Calin G., Croce C.M.. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006; 6:857–866. [DOI] [PubMed] [Google Scholar]
- 10. Bang C., Batkai S., Dangwal S., Gupta S.K., Foinquinos A., Holzmann A., Just A., Remke J., Zimmer K., Zeug A. et al.. Cardiac fibroblast-derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Invest. 2014; 124:2136–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rajasethupathy P., Antonov I., Sheridan R., Frey S., Sander C., Tuschl T., Kandel E.R.. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012; 149:693–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vigneault F., Ter-Ovanesyan D., Alon S., Eminaga S., Chirstodoulou D., Seidman J.G., Eisenberg E., Church G.M.. High-throughput multiplex sequencing of miRNA. Curr. Protoc. Hum. Genet. 2012; doi:10.1002/0471142905.hg1112s73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Alon S., Vigneault F., Eminaga S., Christodoulou D.C., Seidman J.G., Church G.M., Eisenberg E.. Barcoding bias in high-throughput multiplex sequencing of miRNA. Genome Res. 2011; 21:1506–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vigneault F., Sismour A.M., Church G.M.. Efficient microRNA capture and bar-coding via enzymatic oligonucleotide adenylation. Nat. Methods. 2008; 5:777–779. [DOI] [PubMed] [Google Scholar]
- 15. Hafner M., Renwick N., Brown M., Mihailović A., Holoch D., Lin C., Pena J.T.G., Nusbaum J.D., Morozov P., Ludwig J. et al.. RNA-ligase-dependent biases in miRNA representation in deep-sequenced small RNA cDNA libraries. RNA. 2011; 17:1697–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fuchs R.T., Sun Z., Zhuang F., Robb G.B.. Bias in ligation-based small RNA sequencing library construction is determined by adaptor and RNA structure. PLoS One. 2015; 10:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu P., Billmeier M., Mohorianu I., Green D., Fraser W.D., Dalmay T.. An improved protocol for small RNA library construction using High Definition adapters. Methods Next Gen. Seq. 2015; 2:1–10. [Google Scholar]
- 18. Zhuang F., Fuchs R.T., Sun Z., Zheng Y., Robb G.B.. Structural bias in T4 RNA ligase-mediated 3′-adapter ligation. Nucleic Acids Res. 2012; 40:e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jayaprakash A.D., Jabado O., Brown B.D., Sachidanandam R.. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 2011; 39:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sorefan K., Pais H., Hall A.E., Kozomara A., Griffiths-Jones S., Moulton V., Dalmay T.. Reducing ligation bias of small RNAs in libraries for next generation sequencing. Silence. 2012; 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Giraldez M.D., Spengler R.M., Etheridge A., Godoy P.M., Barczak A.J., Srinivasan S., De Hoff P.L., Tanriverdi K., Courtright A., Lu S. et al.. Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling. Nat. Biotechnol. 2018; 36:746–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roberts B.S., Hardigan A.A., Kirby M.K., Fitz-Gerald M.B., Wilcox C.M., Kimberly R.P., Myers R.M.. Blocking of targeted microRNAs from next-generation sequencing libraries. Nucleic Acids Res. 2015; 43:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wickersheim M.L., Blumenstiel J.P.. Terminator oligo blocking efficiently eliminates rRNA from Drosophila small RNA sequencing libraries. Biotechniques. 2013; 55:269–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kawano M., Kawazu C., Lizio M., Kawaji H., Carninci P., Suzuki H., Hayashizaki Y.. Reduction of non-insert sequence reads by dimer eliminator LNA oligonucleotide for small RNA deep sequencing. Biotechniques. 2010; 49:751–754. [DOI] [PubMed] [Google Scholar]
- 25. Dard-Dascot C., Naquin D., d’Aubenton-Carafa Y., Alix K., Thermes C., van Dijk E.. Systematic comparison of small RNA library preparation protocols for next-generation sequencing. BMC Genomics. 2018; 19:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shore S., Henderson J.M., Lebedev A., Salcedo M.P., Zon G., McCaffrey A.P., Paul N., Hogrefe R.I.. Small RNA library preparation method for next-generation sequencing using chemical modifications to prevent adapter dimer formation. PLoS One. 2016; 11:e0167009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al.. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Platt R.J., Chen S., Zhou Y., Yim M.J., Swiech L., Kempton H.R., Dahlman J.E., Parnas O., Eisenhaure T.M., Jovanovic M. et al.. CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014; 159:440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shalem O., Sanjana N.E., Hartenian E., Shi X., Scott D.A., Mikkelsen T.S., Heckl D., Ebert B.L., Root D.E., Doench J.G. et al.. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2013; 343:84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Konermann S., Brigham M.D., Trevino A.E., Joung J., Abudayyeh O.O., Barcena C., Hsu P.D., Habib N., Gootenberg J.S., Nishimasu H. et al.. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2014; 517:583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bennett-Baker P.E., Mueller J.L.. CRISPR-mediated isolation of specific megabase segments of genomic DNA. Nucleic Acids Res. 2017; 45:e165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee N.C.O., Larionov V., Kouprina N.. Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast. Nucleic Acids Res. 2015; 43:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shin G., Grimes S.M., Lee H., Lau B.T., Xia L.C., Ji H.P.. CRISPR-Cas9-targeted fragmentation and selective sequencing enable massively parallel microsatellite analysis. Nat. Commun. 2017; 8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gu W., Crawford E.D., O’Donovan B.D., Wilson M.R., Chow E.D., Retallack H., DeRisi J.L.. Depletion of Abundant Sequences by Hybridization (DASH): Using Cas9 to remove unwanted high-abundance species in sequencing libraries and molecular counting applications. Genome Biol. 2016; 17:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wu J., Huang B., Chen H., Yin Q., Liu Y., Xiang Y., Zhang B., Liu B., Wang Q., Xia W. et al.. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016; 534:652–657. [DOI] [PubMed] [Google Scholar]
- 37. Montefiori L., Hernandez L., Zhang Z., Gilad Y., Ober C., Crawford G., Nobrega M., Sakabe N.J.. Reducing mitochondrial reads in ATAC-seq using CRISPR/Cas9. Sci. Rep. 2017; 7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gagnon J.A., Valen E., Thyme S.B., Huang P., Ahkmetova L., Pauli A., Montague T.G., Zimmerman S., Richter C., Schier A.F.. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014; 9:5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 2011; 17:10–12. [Google Scholar]
- 40. Kozomara A., Griffiths-Jones S.. MiRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014; 42:68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quinlan A.R., Hall I.M.. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ma E., Harrington L.B., O’Connell M.R., Zhou K., Doudna J.A.. Single-stranded DNA cleavage by divergent CRISPR-Cas9 enzymes. Mol. Cell. 2015; 60:398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A.. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014; 507:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pritchard C.C., Kroh E., Wood B., Arroyo J.D., Dougherty K.J., Miyaji M.M., Tait J.F., Tewari M.. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev. Res. (Phila). 2012; 5:492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K.. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014; 32:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sternberg S.H., LaFrance B., Kaplan M., Doudna J.A.. Conformational control of DNA target cleavage by CRISPR-Cas9. Nature. 2015; 527:110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boyle E.A., Andreasson J.O.L., Chircus L.M., Sternberg S.H., Wu M.J., Guegler C.K., Doudna J.A., Greenleaf W.J.. High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:5461–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Roberts B.S., Hardigan A.A., Moore D.E., Ramaker R.C., Jones A.L., Fitz-Gerald M.B., Cooper G.M., Wilcox C.M., Kimberly R.P., Myers R.M.. Discovery and validation of circulating biomarkers of colorectal adenoma by high-depth small RNA sequencing. Clin. Cancer Res. 2018; 24:2092–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lin Y., Cradick T.J., Brown M.T., Deshmukh H., Ranjan P., Sarode N., Wile B.M., Vertino P.M., Stewart F.J., Bao G.. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014; 42:7473–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. et al.. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015; 520:186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kleinstiver B.P., Prew M.S., Tsai S.Q., Topkar V.V., Nguyen N.T., Zheng Z., Gonzales A.P.W., Li Z., Peterson R.T., Yeh J.-R.J. et al.. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015; 523:481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F.. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018; 360:439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A.. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018; 6245:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data from MAD-DASH smRNA-seq libraries generated in this study have been submitted to GEO and are available under accession number: GSE116029.