Abstract
Sulfated peptides are plant hormones that are active at nanomolar concentrations. The sulfation at one or more tyrosine residues is catalysed by tyrosylprotein sulfotransferase (TPST), which is encoded by a single-copy gene. The sulfate group is provided by the co-substrate 3´-phosphoadenosine 5´-phosphosulfate (PAPS), which links synthesis of sulfated signaling peptides to sulfur metabolism. The precursor proteins share a conserved DY-motif that is implicated in specifying tyrosine sulfation. Several sulfated peptides undergo additional modification such as hydroxylation of proline and glycosylation of hydroxyproline. The modifications render the secreted signaling molecules active and stable. Several sulfated signaling peptides have been shown to be perceived by leucine-rich repeat receptor-like kinases (LRR-RLKs) but have signaling pathways that, for the most part, are yet to be elucidated. Sulfated peptide hormones regulate growth and a wide variety of developmental processes, and intricately modulate immunity to pathogens. While basic research on sulfated peptides has made steady progress, their potential in agricultural and pharmaceutical applications has yet to be explored.
Keywords: Casparian strip integrity factor, LRR-RLK, phytosulfokine, plant peptide containing sulfated tyrosine, root meristem growth factor, sulfated peptide hormone, tyrosine sulfation, tyrosylprotein sulfotransferase
Signaling peptides are ubiquitous in higher plants and regulate growth, development, and environmental interactions. Sulfation provides a specific modification that is catalysed by an enzyme unique to these secreted signals.
Introduction
The identification of systemin, a peptide induced by wounding in Solanaceae (Pearce et al., 1991) led to the discovery of a whole new class of phytohormones, the peptide hormones. Unlike the previously known classical hormones that are small metabolite molecules, peptide hormones have an amino acid backbone that is derived from a functional or non-functional precursor. The peptide hormones have been classified into 43 peptide types so far, but the actual number is expected to be higher due to their low native concentrations and technical difficulties regarding identification of peptides. Post-translationally modified peptides, which possess post-translational modifications (PTMs) such as proline hydroxylation or glycosylation, are classified in eight peptide hormone groups (Tavormina et al., 2015). The isolation of the disulfated pentapeptide phytosulfokine (PSK) from cultured Asparagus officinalis cells (Matsubayashi and Sakagami, 1996) was the first evidence of an activating PTM of a plant hormone, tyrosine sulfation, that had previously only been reported for animal hormones (Moore, 2009). Unlike other PTMs, tyrosine sulfation is not present in metabolite hormones and it leads to activation of peptide hormones. By contrast, glycosylation, hydroxylation, or conjugation to amino acids of metabolite hormones such as indole-3-acetic acid, abscisic acid, and others by a specific set of enzymes leads to reversible or irreversible inactivation (Nambara and Marion-Poll, 2005). Tyrosine sulfation links peptide hormone synthesis to sulfate metabolism via the common intermediate 3´-phosphoadenosine 5´-phosphosulfate (PAPS). The sulfate moiety provides stability of the peptide hormone that is secreted to the apoplast and specificity with regard to receptor recognition (Wang et al., 2015; Song et al., 2016). To achieve tyrosine sulfation, a unique sulfotransferase has evolved in plants, namely tyrosylprotein sulfotransferase. In this review, we summarize our current knowledge on the known groups of sulfated peptide hormones, their modifications, and their processing with a focus on sulfation, known peptide receptors, and physiological activities of peptide hormones. Long-distance transport of sulfated peptides has not been reported to date, but is easily conceivable. For example, the C-TERMINALLY ENCODED PEPTIDE involved in the regulation of nitrogen acquisition (Ohkubo et al., 2017) and CLAVATA3 (CLV3)/ENDOSPERM SURROUNDING REGION-related (CLE) 12 from Medicago trunculata and MtCLE13 that regulates root nodule formation have been shown to be transported from root to shoot (Okamoto et al., 2013; Kassaw et al., 2017). The recent discovery of sulfated peptides in xylem sap fuels the idea of functions in long-distance organ-to-organ communication (Okamoto et al., 2015; Patel et al., 2018).
Tyrosine sulfation requires the co-substrate PAPS
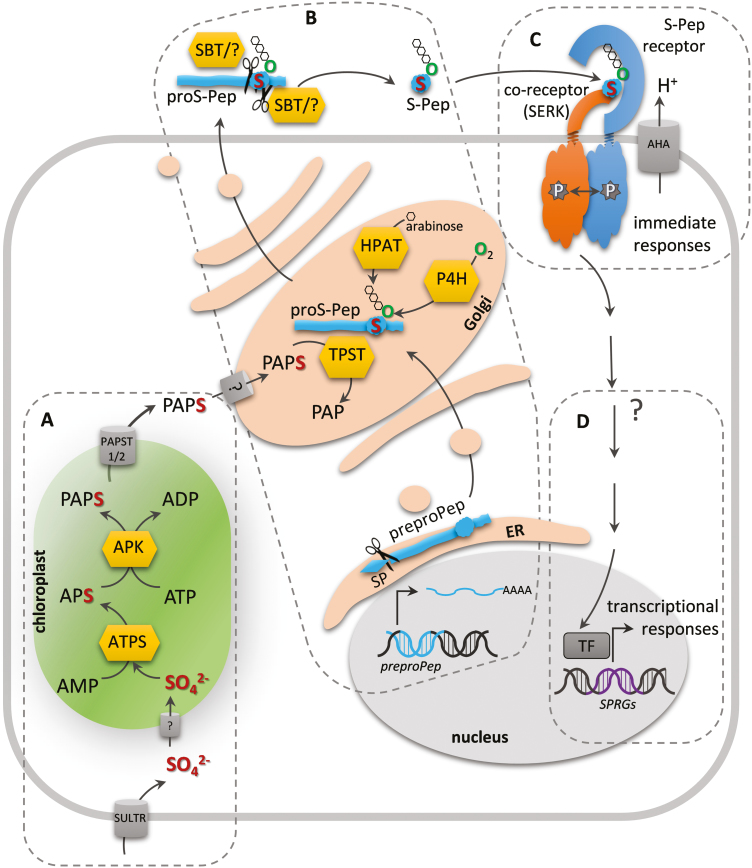
Sulfur that is taken up by the plant as inorganic sulfate is activated by ATP sulfurylase (ATPS, encoded by AtATPS1-4 in Arabidopsis, to adenosine 5´-phosphosulfate (APS) (Fig. 1A). APS kinases, located in plastids (AtAPK1/2/4) or in the cytosol (APK3), phosphorylate APS to 3´-phosphoadenosine 5´-phosphosulfate (PAPS). The fact that none of the apk single-mutants displays a phenotype whereas the apk1 apk2 double-mutant has a dwarf phenotype (Mugford et al., 2009) suggests that APK3 and APK4 only contribute to PAPS synthesis to a minor degree. In Arabidopsis, export of PAPS from the chloroplast occurs through PAPS transporter 1 (AtPAPST1) and probably also a second as yet unknown transporter (Gigolashvili et al., 2012). The transporter(s) responsible for the import of PAPS into the Golgi, where peptide-precursor sulfation occurs, have not been identified.
Fig. 1.
Activation of sulfate, maturation of sulfated peptides, and peptide perception. (A) Sulfate uptake occurs via sulfate transporters (SULTR) into the cytoplasm, but the transporter into the chloroplast is unknown. ATP sulfurylase (ATPS) and APS kinase (APK) produce the tyrosylprotein sulfotransferase (TPST) co-substrate 3´-phosphoadenosine 5´-phosphosulfate (PAPS) (Mugford et al., 2009). The PAPS transporter (PAPST) exports it into the cytoplasm, but how PAPS enters the Golgi is not known (Gigolashvili et al., 2012). (B) Maturation of sulfated peptides includes cleavage of the signal peptide (SP) in the ER during preproprotein synthesis, tyrosine sulfation by TPST in the Golgi (Komori et al., 2009), proline hydroxylation by prolyl-4-hydroxylase (P4H) and triarabinosylation by hydroxyproline O-arabinosyltransferase (HPAT) in the case of PSYs in the Golgi (Amano et al., 2007), and cleavage of the N- and C-terminals by subtilases (SBT) and/or other unknown proteases probably in the apoplast to release the mature sulfated peptide (S-Pep) (Srivastava et al., 2008; Ghorbani et al., 2016). (C) PSKR, PSY1R, and RGFRs are perceived at the plasma membrane by a receptor/co-receptor pair that mutually transphosphorylate each other and activate the receptor (Hohmann et al., 2017). The proton-pumping H+-ATPase (AHA) has been identified as a direct target of the S-Pep receptors PSKR1 and PSY1R (Fuglsang et al., 2014; Ladwig et al., 2015) (D) S-Peps are predicted to regulate S-Pep-responsive genes (SPRGs) via unknown signaling intermediates and transcription factors (TF).
A unique tyrosylprotein sulfotransferase catalyses tyrosine sulfation of peptides in the cis-Golgi
The transfer of sulfate to the hydroxyl group of a substrate is catalysed by sulfotransferases (SOTs). In Arabidopsis, the SOT family (EC 2.8.2.-) consists of 22 members, most of which catalyse sulfation of glucosinolates, flavonols, and brassinosteroids (Hirschmann et al., 2014). The identification of PSK was the first evidence for tyrosine sulfation in plants (Matsubayashi and Sakagami, 1996; Moore, 2009). It was reported earlier in animals and is catalysed by tyrosylprotein sulfotransferases (TPSTs; EC 2.8.2.20). The Arabidopsis TPST, also termed ACTIVE QUIESCENT CENTER 1 (AQC1) (Zhou et al., 2010), HYPERSENSITIVE TO Pi STARVATION 7 (Kang et al., 2014), or SCHENGEN2 (SGN2) (Doblas et al., 2017), transfers sulfate from the co-substrate PAPS to the hydroxyl group of one or more defined tyrosine residue(s) of a peptide precursor to produce a tyrosine O4-sulfate ester (Moore, 2003) (Fig. 1B). Plant-specific TPSTs were first identified in rice (Oryza sativa), carrot (Daucus carota), Asparagus officinalis, and subsequently in Arabidopsis (Hanai et al., 2000; Komori et al., 2009). Aside from their shared catalytic activity, plant TPSTs are distinct from other plant SOTs or animal TPSTs in that they have an overall low sequence identity and differ in their domain structure. Human TPSTs, with 370 and 377 residues, are type II membrane proteins with a short N-terminal domain in the cytoplasm and a putative stem region between the transmembrane and catalytic domains, whereas the Arabidopsis TPST has 500 residues and is a type I membrane protein with a short C-terminus in the cytoplasm, an N-terminal signal peptide, and a heparan sulfate 6-O-sulfotransferase 2 homology domain (Moore, 2009). TPST is located in the cis-Golgi where sulfation occurs, as shown by co-localization with the cis-Golgi marker SYP31 (Komori et al., 2009). The gene is ubiquitously expressed, with highest transcript levels in the root apical meristem, lateral root primordia, and the vasculature (Komori et al., 2009). To our current knowledge, TPST is a unique gene in plants and the TPST enzyme is responsible for sulfation. TPST not only sulfates PSK but also other sulfated peptide hormones including PLANT PEPTIDE CONTAINING SULFATED TYROSINE 1 (PSY1) and ROOT GROWTH MERISTEM FACTOR 1 (RGF1), as shown by incubation of TPST-containing microsomal protein fractions with the radiolabeled TPST-substrate [35S]PAPS and affinity purification of TPST with a PSY precursor column (Amano et al., 2007; Komori et al., 2009; Matsuzaki et al., 2010). Knockout of TPST in Arabidopsis tpst-1, aqc1-1, and sgn2-1 causes a pleiotropic phenotype with shorter roots, a reduced root apical meristem size, early senescence, a reduced number of reproductive organs (Komori et al., 2009; Zhou et al., 2010), a defective Casparian strip (Doblas et al., 2017), increased metaxylem cell numbers (Holzwart et al., 2018), and an altered immunity to specific pathogens (Mosher et al., 2013). An altered immune response has also been observed in TPST-silenced Solanum lycopersicum (tomato) plants (Zhang et al., 2018). Some tpst phenotypes can be restored by specific sulfated peptides, for example root growth is partially restored in tpst-1 by exogenous application of PSK and PSY1, which promote cell elongation, and RGF1, which stimulates cell division in the root apical meristem, and is fully restored by applying all three peptides (Matsuzaki et al., 2010). Application of CASPARIAN STRIP INTEGRITY FACTOR (CIF) rescues the defect in the Casparian strip diffusion barrier. The altered immunity responses of tpst-1 (partially) rely on the loss of PSK and PSY signaling (Mosher and Kemmerling, 2013). While a recognition motif for TPST has not been identified so far, it has been shown that adjacent and distant acidic amino acids in the flanking sequence of the proPSK sequence are important for recognition and that an aspartate residue N-terminal of the targeted tyrosine is required (Hanai et al., 2000). Indeed, all sulfated peptides known to date (Fig. 2) contain a DY-motif except GLV9 (Golven), whose tyrosine is preceded by the acidic amino acid glutamate (Fig. 2B). Whether or not GLV9 is actually sulfated has not been determined but its biological activity in meristem maintenance that is required for root elongated growth is massively reduced compared to RGF1-6 (Matsuzaki et al., 2010).
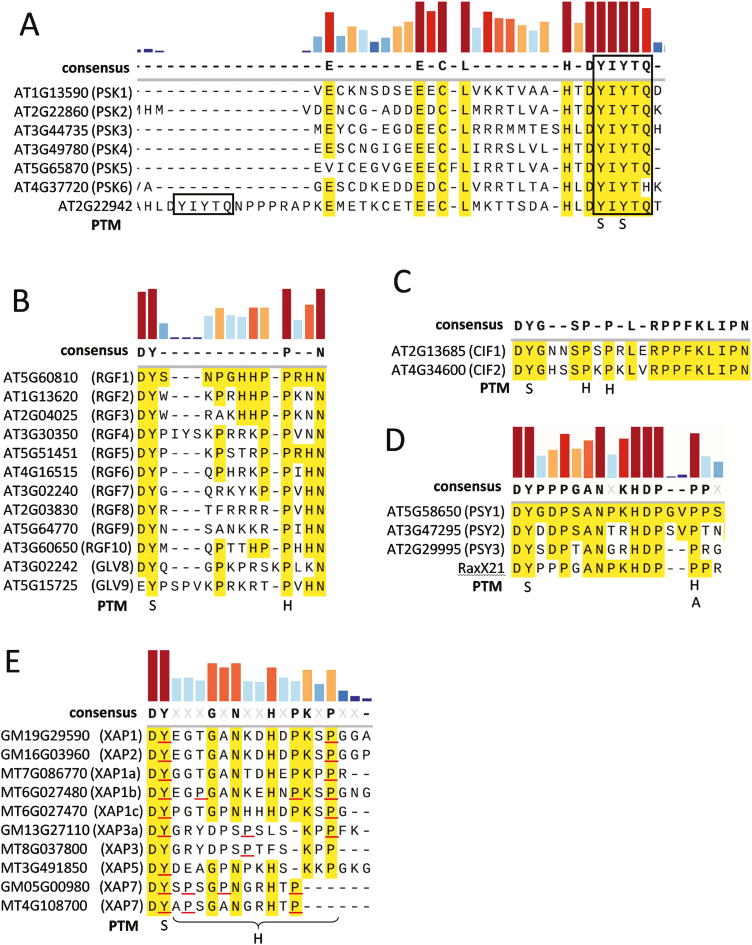
Fig. 2.
Protein sequence alignments (MUSCLE) of sulfated peptide families of the mature peptide motif regions. All sulfated peptides, except GLV9, share a DY-motif. Hydroxylated and glycosylated amino acids are present in the majority of sequences. (A) The PSK sequences are N-terminally extended to align a functionally undescribed ORF with two PSK motifs (black boxes). In (A–E) coloured bars indicate sequence conservation between all sequences with a gradient (in 10 % steps) between dark-blue (low, <10%) and dark-red (high, 90–100%). Yellow highlighting indicates consensus to the identified peptides: (B) RGF1, (C) CIF1, and (D) PSY1. (E) Uncharacterized sulfated peptides (XAPs) were identified from the xylem sap of Glycine max and Medicago trunculata (Okamoto et al., 2015; Patel et al., 2018). Experimentally verified post-translational modifications (PTMs) are indicated at the bottom of each figure (if underlined in red in the case of XAPs): S = tyrosine sulfation; H = hydroxylation of proline residues; A = triple L-arabinosylation of hydroxyproline.
Why tyrosine sulfation?
The transfer of a sulfate group from PAPS to the tyrosine side-chain alters the chemical properties of the peptide by increasing hydrophilic binding. This alters the affinity to proteins that bind sulfated peptides and act as peptide receptors. Binding of PSK to its receptor PSKR1 (PSK RECEPTOR1) occurs mainly by hydrogen-bond formation whereby the sulfate moieties of PSK interact with specific residues in the perception domain of PSKR1 (Wang et al., 2015). It is therefore not surprising that both the amino acid backbone (Bahyrycz et al., 2004, 2005) and sulfation of both tyrosine residues are required for full biological activity of PSK (Matsubayashi et al., 1996; Matsubayashi and Sakagami, 1996; Kobayashi et al., 1999; Kwezi et al., 2011; Stührwohldt et al., 2011; Igarashi et al., 2012; Mosher et al., 2013). Similarly, unsulfated RGF1 has a 185-fold reduced affinity to RGF RECPTOR1 (RGFR1), one of its three receptors (Shinohara et al., 2016). Song et al., (2016) identified an RxGG motif in RGFRs that is responsible for specific recognition of the sulfate group. Some studies have reported activity of the unmodified peptide, which raises the question as to how crucial this PTM actually is (Kutschmar et al., 2009; Meng et al., 2012; Doblas et al., 2017). A requirement of sulfation for bioactivity has controversially been considered for the RGF family member RGF1/CLEL8/GLV1. Meng et al. (2012) showed that CLEL8 (CLE-like) peptide without sulfation and hydroxylation alters root growth direction and lateral root development whilst the sulfated RGF1 peptide but not the unmodified peptide backbone is able to rescue the defective root apical meristem of tpst-1 (Matsuzaki et al., 2010). In some studies it is possible that the activity of unmodified peptide has been due to the high concentration applied (Kutschmar et al., 2009; Meng et al., 2012; Whitford et al., 2012). When investigating modified peptides, a dose–response analysis and titration curves to determine the physiological activity and binding affinity of the non-/modified peptide are advisable, as has been shown for RGF1 (Matsuzaki et al., 2010), PSK (Kutschmar et al., 2009; Stührwohldt et al., 2011), PSY1 (Amano et al., 2007), and CIFs (Doblas et al., 2017). Unsulfated peptides are active in the micromolar range whereas binding affinity assays with the ectodomains of the corresponding peptide receptors and growth assays have shown a KD or half-maximal biological activity of the sulfated peptides in the nanomolar range (Matsubayashi et al., 2002, 2006; Stührwohldt et al., 2011; Whitford et al., 2012; Fernandez et al., 2013; Doblas et al., 2017). Aside from increasing the specificity and affinity of peptides to their receptors, tyrosine sulfation as well as proline hydroxylation are very stable and irreversible modifications (Moore, 2003; Gorres and Raines, 2010). To date, desulfation by enzymes has not been reported for either plant-derived or for plant pathogen-derived sulfatases, demonstrating that sulfated peptides are stable signaling molecules even in a highly biochemically active environment. In mammals, arylsulfatases, present in lysosomes, may be involved in tyrosine-sulfated protein degradation (Parenti et al., 1997). By contrast, phosphorylation and acetylation are reversible PTMs catalysed by phosphatases and deacetylases, which allows for dynamic regulation of signaling pathways. Aside from enzymatic inactivation, tyrosine-sulfated peptides are fairly stable even in non-physiological, harsh acidic conditions of pH 1–3, as shown for the model peptides gastrin-17, caerulein, and drosulfokinin (Balsved et al., 2007), which suggests that sulfated peptides are stable in the acidic apoplast. The same holds true for neutral and alkaline conditions (Huttner, 1984).
Glycosylation and hydroxylation of sulfated peptides
Sulfated peptides can undergo proline hydroxylation and glycosylation (Fig. 1B). Proline hydroxylation is catalysed by prolyl-4-hydroxylases (P4Hs), which are encoded by 13 genes in Arabidopsis. The specific P4Hs involved in maturation of sulfated peptides have not been identified (Myllyharju, 2003; Matsubayashi, 2012). PSY1 is a glycosylated sulfated peptide. L-arabinosylation (L-Ara3) at the hydroxylated proline of PSY1 is catalysed by hydroxyproline O-arabinosyltransferases (HPATs) (Amano et al., 2007; Ogawa-Ohnishi et al., 2013). Triarabinosylation has been shown to be important for the biological activity of AtCLV3 (Ohyama et al., 2009; Shinohara and Matsubayashi, 2013), MtCLE12 and 13 (Kassaw et al., 2017; Imin et al., 2018), as well as CLE ROOT SIGNAL 1 and 2 in Lotus japonicus (Okamoto et al., 2013).
Proteolytic cleavage of sulfated peptide precursors
Sulfated peptides are encoded as non-functional precursors. The modified precursors are proteolytically cleaved by endo- and probably exopeptidasen to release the mature and active peptides, most likely after secretion from the Golgi into the apoplast, as suggested for example for peptide hormones from M. trunculata and AtIDA (Djordjevic et al., 2011; Schardon et al., 2016; Patel et al., 2018) (Fig. 1B). In animals, proteolytic processing has been reported for precursor molecules of peptide hormones and neuropeptides such as insulin and melanocyte-stimulating hormones (Seidah and Prat, 2012). Precursor processing catalysed by prohormone convertases is essential to form the bioactive entities. The enzymes are considered as therapeutic targets, which highlights the importance of this activating step. In plants, less is known about proteolytic enzymes of secreted hormone peptides. Partial cleavage of the N-terminus of Arabidopsis proPSK4 has been demonstrated in vivo by SUBTILASE (SBT) 1.1, a prohormone convertase of the subtilisin-like serine protease family, with the highest specificity observed in vitro for proPSK4 over other PSK precursors (Srivastava et al., 2008). Since the cleavage product still has additional N- and C-terminal residues, further trimming must be predicted. PSK derivatives with additional amino acids attached to the N- or C-terminus of the disulfated YIYTQ-sequence have a 1000-fold lower bioactivity compared to PSK (Matsubayashi et al., 1996). For the RGF/GLV/CLEL peptide family, SBT 6.1 and 6.2 have been shown to cleave the RGF6/CLEL6/GLV1 and RGF9/CLEL9/GLV2 precursors and to be required for peptide activation (Ghorbani et al., 2016), For the majority of the 56 AtSBTs, targeting to the secretory pathway is predicted by bioinformatics tools (Rautengarten et al., 2005) and has been shown experimentally as well for SBT1.1, 4.12, 4.13, and 5.2 (Srivastava et al., 2008; Schardon et al., 2016). Interestingly, the target predictions for S1P/SBT6.1 and SBT6.2 localization are mitochondria and chloroplasts (Rautengarten et al., 2005), but S1P/SBT6.1 co-localizes with the Golgi-marker BODIPY TR ceramide (Liu et al., 2007) and interacts with the protease inhibitor Serpin1 in the apoplast, as shown by BiFC analysis (Boruc et al., 2010; Ghorbani et al., 2016). SBT6.1 targets not only RGF precursors but also the precursor of RAPID ALKALINIZATION FACTOR 23 (Srivastava et al., 2009; Schaller et al., 2018). It is as yet unclear whether the tyrosine sulfation in the cis-Golgi is required for recognition by the protease. For SBTs it has been shown that these proteases are kept inactive by an auto-inhibiting prodomain until they reach the acidic trans-Golgi where the prodomain is released in a pH-dependent manner (Meyer et al., 2016; Schaller et al., 2018), further indicating that proteolytic cleavage occurs in the extracellular space, or at least upstream of the trans-Golgi.
PSK
Phytosulfokine-α (PSK) is a Tyr-disulfated pentapeptide that displays hormone-like activities at nanomolar concentrations (Stührwohldt et al., 2011). PSK is encoded as a 75–123-amino-acid preproprotein by nuclear genes (Lorbiecke and Sauter, 2002). The conserved PSK protein family PF06404 (http://pfam.xfam.org/family/PF06404) consists of 352 homologous sequences found in 53 species from both mono- and dicotyledonous plant species, including Arabidopsis thaliana, Glycine soja, Brassica napus, Zea mays, and Oryza sativa. Sequence conservation and the presence of multiple genes encoding PSK in each species examined to date and an in silico conservation mapping for ligand-binding sites for PSKR1 orthologs suggest a conserved ligand–receptor interaction (Orr and Aalen, 2017). In Arabidopsis, seven loci have been annotated as putative PSK precursor genes. AtPSK1-5 contains the canonical PSK domain YIYTQ (Fig. 2A). Transcriptome analysis by RNA-seq and reporter gene expression analysis has revealed PSK expression at varying levels throughout all tissues and developmental stages, resulting in a ubiquitous supply of the PSK precursor (Stührwohldt et al., 2011; Cheng et al., 2017). The pseudogene AtPSK6 (At4g37720) encodes the PSK-related sequence YIYTH. As the result of a new Arabidopsis genome annotation (Araport 11; Cheng et al., 2017), the locus At2g22942 has been identified as a previously unrecognized putative phytosulfokine precursor gene. Interestingly, the encoded protein sequence contains two canonical PSK sequences (Fig. 2A). For AtPSK6, no expressed sequence tags have been reported and RNA-seq data show very low expression levels for AtPSK6 in pollen and for At2g22942 in the receptacle (Cheng et al., 2017), possibly suggesting minor or highly specific biological functions of these genes that have yet to be described.
As with metabolite plant hormones, PSK has been shown to have multiple physiological functions. It stimulates somatic embryogenesis (Kobayashi et al., 1999; Igasaki et al., 2003) and promotes adventitious root formation in cucumber hypocotyls (Yamakawa et al., 1998). Furthermore, PSK promotes the differentiation of mesophyll cells of Zinnia elegans to tracheary elements in cell cultures (Matsubayashi et al., 1999) by down-regulating stress-response genes at the onset of the trans-differentiation process (Motose et al., 2009). In tpst-1 knockout mutants and in the PSK receptor null mutant the metaxylem cell number is increased, which suggests a role for PSK in the control of xylem cell fate (Holzwart et al., 2018). Interestingly, the modified peptide hormone TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR acts in an opposite manner (Etchells et al., 2016). In plant reproduction, PSK promotes pollen germination (Chen et al., 2000) and pollen-tube elongation (Stührwohldt et al., 2015), and it acts as a short-distance signal that helps to guide the pollen tube from the transmission tract along the funiculus to the embryo sac. Plants that lack PSK receptors produce fewer seeds (Stührwohldt et al., 2015).
With the use of synthetic PSK and genetic knockout of PSK receptors, PSK signaling has been shown to promote hypocotyl elongation (Stührwohldt et al., 2011), leaf (Hartmann et al., 2014) and root growth (Kutschmar et al., 2009), and cotton fiber-cell elongation (Han et al., 2014). The growth-promoting activity is mainly driven by increased cell expansion rather than cell division, as is evident from increased leaf epidermal cell size, longer hypocotyl cells, and longer differentiated root cells.
Finally, PSK signaling differentially affects plant immunity depending on the type of the invading pathogen. The contribution of PSK signaling to plant immunity seems to be dependent on the lifestyle rather than the phylogenetic origin of the pathogen (Rodiuc et al., 2016). Arabidopsis plants deficient in PSK perception are more susceptible to pathogens with a necrotrophic lifestyle such as the fungi Alternaria brassicicola and Sclerotinia sclerotiorum and the bacterium Ralstonia solanacearum, whereas their resistance against hemi-/biotrophs such as the bacterium Pseudomonas syringae and the oomycete Hyaloperonospora arabidopsidis is increased (Loivamäki et al., 2010; Igarashi et al., 2012; Mosher et al., 2013; Rodiuc et al., 2016). PSK signaling reduces the expression of microbe-associated molecular pattern-inducible genes elicited by elf18 or flg22, as determined by quantitative PCR after 8 h (Igarashi et al., 2012). The expression of PSK precursor genes and of PSK receptor 1 (PSKR1) is induced by wounding, the fungal elicitor E-Fol, elf18, and following infection with necrotrophic fungi, whereas infection with the hemibiotrophic bacterium P. syringae does not change the expression (Loivamäki et al., 2010; Igarashi et al., 2012), which is suggestive of a role for PSK signaling in specific host–pathogen interactions. Recently, Zhang et al. (2018) revealed that PSK signaling induces auxin-dependent immune responses in tomato when it is infected with the necrotrophic fungus Botrytis cinerea. Besides growth promotion through cell expansion, PSK contributes to the regulation of quiescent center cell division and the differentiation of distal stem cells (Heyman et al., 2013; Kong et al., 2018).
RGFs/GLVs/CLELs
The sulfated and hydroxylated peptide ROOT GROWTH MERISTEM FACTOR 1 (RGF1) was identified as the first member of the RGF group in a search for sulfated peptides that can rescue the stunted root growth phenotype of the tpst-1 mutant (Matsuzaki et al., 2010). Like all sulfated peptides, the RGF members contain the typical DY-motif, except for GLV9, and a highly conserved hydroxylated proline residue. Unlike the mature PSK peptide that is fully conserved within and across species, RGFs have conserved and non-conserved amino acids (Fig. 2B). PSK and PSY have been shown to partially restore root growth in tpst-1 (Matsuzaki et al., 2010). RGF1 affects root development by increasing the abundance of the transcription factor PLETHORA, which leads to maintenance of the root stem-cell niche. Application of synthetic RGF1 restores meristematic activity in tpst-1. RGF1, PSK, and PSY together fully restore root growth (Matsuzaki et al., 2010). RGF1 is a member of the RGF/GOLVEN(GLV)/CLE-LIKE(CLEL) peptide family (Fig. 2B) (Matsuzaki et al., 2010; Meng et al., 2012) with partially redundant functions. RGF7–9 have reduced or no biological activity in root elongation growth (Matsuzaki et al., 2010). The expression patterns differ but, taking all members together, RGF peptides seem to be present throughout the plant (Fernandez et al., 2013). Aside from their activity in the root apical meristem, RGFs control root gravitropism (Meng et al., 2012; Whitford et al., 2012), lateral root development (Meng et al., 2012; Fernandez et al., 2015), and root hair development (Fernandez et al., 2013).
CIFs
Recently, a new group of tyrosine-sulfated peptides, the CASPARIAN STRIP INTEGRITY FACTORs 1 and 2 (CIF1 and CIF2), have been identified (Doblas et al., 2017; Nakayama et al., 2017). The peptides consist of 21 amino acids with one sulfated tyrosine in the typical DY-motif and two hydroxylated proline residues as post-translational modifications (Nakayama et al., 2017), whilst the often-accompanying glycosylation of hydroxyprolines has not been reported (Fig. 2C). In contrast to other tyrosine-sulfated peptides, CIFs appear to have one highly specific function: CIF peptide signaling is required for the formation of the Casparian strip diffusion barrier of endodermal cells (Doblas et al., 2017; Nakayama et al., 2017). CIFs are predominately expressed in the stele of primary (CIF1 and CIF2) and lateral roots (CIF2) (Nakayama et al., 2017). They act locally, as only tyrosine-sulfation activity in the stele is able to rescue the diffusion-barrier defective phenotype. This conclusion was based on an extensive analysis of tissue-specific TPST-expression mutants and propidium iodide permeability (Doblas et al., 2017). Mutants lacking CIF signaling due to mutations in the genes encoding for the peptide precursors cif1-1 and cif2-1, the tyrosylprotein-sulfotransferase (TPST) sgn2-1, or the CIF receptors gso1/sgn3-3 and gso2-1 display mislocalized CASPARIAN STRIP MEMBRANE DOMAIN PROTEIN 1 and lignin in the endodermis cell layer, resulting in a discontinuous casparian strip (Doblas et al., 2017; Nakayama et al., 2017).
PSYs
PLANT PEPTIDE CONTAINING SULFATED TYROSINE 1 (PSY1) is a sulfated and triarabinosylated 18-amino-acid peptide (Fig. 2D) that promotes cell expansion and cell division. It originates from a 75-amino-acid precursor, encoded in Arabidopsis by three nuclear genes (Amano et al., 2007). PSY1 is expressed in a wide range of tissues but high expression is found in the root elongation zone and the shoot apical meristem. Wounding enhances PSY1 expression, as shown by northern blot analysis (Amano et al., 2007), which points to a role in biotic stress (Matsuzaki et al., 2010). Indeed, PSY signaling promotes susceptibility to the hemi-biotrophic fungus Fusarium oxysporum (Shen and Diener, 2013) and increases resistance to the necrotrophic fungus Alternaria brassicicola (Mosher et al., 2013). PSY1 and PSK are therefore also classified as damage-associated molecular pattern factors (Wang et al., 2014; Tang et al., 2017) that modulate immunity responses.
Perception and downstream signaling of tyrosine-sulfated peptides
A common factor of all sulfated peptides is that they are perceived by plasma membrane-localized leucine-rich repeat receptor-like kinases (LRR-RLKs) from classes X and XI (Diévart and Clark, 2003) (Fig. 1C and Table 1), in most cases by more than one receptor per peptide, for example PSK by PSKR1 and PSKR2 (Matsubayashi et al., 2002, 2006; Amano et al., 2007), RGFs by RGFR1–5 (Ou et al., 2016; Shinohara et al., 2016; Song et al., 2016), and CIFs by GSO1 and GSO2 (Doblas et al., 2017; Nakayama et al., 2017), which leaves PSY1R (PSY RECEPTOR1) for PSYs as the only single-copy receptor known so far (Amano et al., 2007). Several peptide receptors heterodimerize with co-receptors of the SOMATIC EMBRYOGENESIS RECEPTOR KINASE family, as shown for the sulfated peptide receptors PSKR1, PSY1R, and RGFRs (Table 1), resulting in subsequent mutual transphosphorylation of the cytoplasmic kinase domains within the activation loops. Kinase activity of the receptor is essential for peptide signaling (Hartmann et al., 2014, 2015; Ladwig et al., 2015; Wang et al., 2015; Ma et al., 2016; Song et al., 2016, 2017; Hohmann et al., 2017; Kaufmann et al., 2017; Oehlenschlæger et al., 2017). In the case of CIF signaling, it has not yet been reported that SGN3 forms a dimer with a co-receptor; however, based on the extremely high ligand affinity to SGN3 as measured by isothermal titration calorimetry in the absence of a possible co-receptor, Doblas et al. (2017) speculated that the paradigm of peptide receptor–co-receptor formation might not apply for SGN3 activation. Only few downstream signaling components of peptide receptors have been identified (Table 1), most of which are localized to the plasma membrane, such as the cytoplasmic receptor-like kinase SGN1 that acts in CIF signaling (Doblas et al., 2017) and RECEPTOR-LIKE PROTEIN 44 that participates in PSK signaling (Holzwart et al., 2018). RECEPTOR-LIKE PROTEIN 44 controls procambial cell identity and connects the PSK and brassinosteroid signaling pathways through interactions with PSKR1 and the brassinosteroid receptor BRASSINOSTEROID INSENSITIVE 1 (Holzwart et al., 2018). For PSKR1, formation of a nanocluster at the plasma membrane has been shown with the ARABIDOPSIS H+-ATPases 1 and 2 (AHA1 and AHA2) and the CYCLIC NUCLEOTIDE GATED CHANNEL 17 (Ladwig et al., 2015) (Fig. 1C). Aside from being an active kinase, PSKR1 is a calmodulin-binding protein and has been described to possess guanylyl cyclase activity, making the PSK receptor a versatile protein both with respect to regulation of its activity and to downstream signaling through phosphorylation, Ca2+-calmodulin signaling, and cGMP synthesis (Kwezi et al., 2011; Hartmann et al., 2014; Kaufmann et al., 2017). Regulation of root elongation by PSY1 depends on the interaction with PSY1R and subsequent activation of AHA2 through transphosphorylation (Fuglsang et al., 2014). A similar mechanism may apply for PSK, which induces protoplast swelling within minutes (Stührwohldt et al., 2011) (Fig. 1C). Evidence for crosstalk between sulfated peptides and hormone signaling has also been reported. Auxin increases TPST expression, which probably promotes the maturation of sulfated peptides (Zhou et al., 2010). Furthermore, PSK promotes auxin synthesis (Zhang et al., 2018) whereas RGFs change auxin flux by altering the trafficking dynamics of the auxin efflux carrier PIN2 (Whitford et al., 2012) (Table 1). Transcriptional regulation by sulfated peptides for a few S-Pep responsive genes (SPRGs) has been shown (Fig. 1D). PSK signaling inhibits ethylene production by regulation of ACC SYNTHASEs (ACSs) and ACC OXIDASEs (ACOs) (Wu et al., 2015; Zhang et al., 2018), and CIFs regulate CASPARIAN STRIP MEMBRANE DOMAIN PROTEIN LIKE (CASPL) and MYB DOMAIN PROTEIN 15 (MYB15), resulting in lignification (Drapek et al., 2018).
Table 1.
Overview of sulfated peptides, peptide receptors, molecular signaling components, and physiological responses
| Peptide | PTM | Receptor | LRR-RLK class* | Co-receptor | Downstream signaling | Physiological responses |
|---|---|---|---|---|---|---|
| RGFs/GLVs/CLELs (Matsuzaki et al., 2010) | Tyr-sulfation Pro-hydroxylation | RGFR1/RGI1 RGFR2/RGI2 RGFR3/RGI3 RGI4 RGI5 (Ou et al., 2016; Shinohara et al., 2016) | XI | SERKs (Ou et al., 2016; Song et al., 2016) | PIN2 (Whitford et al., 2012) PLT2 (Matsuzaki et al., 2010; Ou et al., 2016; Shinohara et al., 2016) | Root apical meristem homeostasis (Matsuzaki et al., 2010) Root hair formation (Fernandez et al., 2013) Lateral root formation (Fernandez et al., 2013, 2015) Gravitropic responses of root and shoot (Whitford et al., 2012) |
| PSK (Matsubayashi and Sakagami, 1996) | Tyr-sulfation | PSKR1 PSKR2 (Matsubayashi et al., 2002, 2006; Amano et al., 2007) | X | SERKs (Ladwig et al., 2015; Wang et al., 2015) | AHA1/2 and CNGC17 (Ladwig et al., 2015) RLP44 (Holzwart et al., 2018) CaMs (Hartmann et al., 2014; Kaufmann et al., 2017) cGMP (Kwezi et al., 2011) YUCs → auxin ↑ (Zhang et al., 2018) Ca2+ influx (Zhang et al., 2018) ACSs, ACOs → ethylene ↓ (Wu et al., 2015; Zhang et al., 2018) | Cell expansion in leaves, hypocotyls and primary root (Kutschmar et al., 2009; Stührwohldt et al., 2011; Han et al., 2014; Hartmann et al., 2014) Quiescent center cell division (Heyman et al., 2013) Reproduction: pollen germination (Chen et al., 2000), pollen-tube growth (Stührwohldt et al., 2015), funicular pollen tube guidance (Stührwohldt et al., 2015) Immunity response to necrotrophic fungi (Zhang et al., 2018) Xylem formation (Holzwart et al., 2018) Adventitious root formation (Yamakawa et al., 1998) Production of secondary metabolite taxol (Kim et al., 2006) |
| PSYs (Amano et al., 2007) | Tyr-sulfation Pro-hydroxylation Hyp-triarabinosylation | PSY1R (Amano et al., 2007) | XI | SERKs (Oehlenschlæger et al., 2017) | AHA2 (Fuglsang et al., 2014) | Cell expansion and cell division (Amano et al., 2007) Immunity response to necrotrophic fungi (Mosher et al., 2013) |
| CIFs (Doblas et al., 2017; Nakayama et al., 2017) | Tyr-sulfation Pro-hydroxylation | GSO1/SGN3 (Nakayama et al., 2017) | XI | – | SGN1 (Doblas et al., 2017) CASPL, MYB15 → CASP+lignin ↑ (Drapek et al., 2018) | Casparian strip formation (Doblas et al., 2017; Nakayama et al., 2017) |
| XAPs (Okamoto et al., 2015; Patel et al., 2018) | Tyr-sulfation Pro-hydroxylation | – | – | – | – | Lateral root formation (Patel et al., 2018) |
* LRR-RLK, leucine rich repeat receptor-like kinase class according to Diévart and Clark (2003).
PTM, post-translational modification.
Sulfated peptide hormones produced by microbes
Tyrosine sulfation was first reported for plants and animals (Moore, 2009). Interestingly, RaxST, a tyrosylprotein sulfotransferase, was recently identified from the biotrophic plant pathogenic bacterium Xanthomonas oryzae pv. oryzae (Han et al., 2012; Ronald, 2014). This bacterium produces the sulfated peptide RaxX, which mimics PSY1 (Fig. 2D) and triggers PSY1 responses, suggesting that RaxX can hijack the PSY1 signaling pathway. RaxX is perceived by the Xa21 immune receptor and modulates host defense (Pruitt et al., 2017). As mentioned above, tyrosine sulfation is a non-reversible modification that results in the formation of highly stable signaling molecules that are targets in the arms race between plant hosts and pathogens.
Future perspectives
Research on sulfated peptides has so far mainly focused on the model plant Arabidopsis, whereas genes encoding for sulfated peptides and their signaling components are highly conserved throughout the higher plant kingdom (Lorbiecke and Sauter, 2002; Hartmann et al., 2015). Beneficial effects of sulfated peptides on crop plant yields is an emerging topic that has great potential. Examples for agricultural exploitation of sulfated peptides have been described for an RGF in peach fruit ripening (Busatto et al., 2017) and for PSK in the development of longer cotton fibers (Han et al., 2014). Sulfated peptides seem to also affect secondary metabolism, as shown for PSK that synergistically acts with methyljasmonate to promote production of taxol (Kim et al., 2006), a diterpenoid used as a therapeutical drug against cancer, thus revealing a great potential in pharmaceutical applications. Recently, a group of sulfated peptides were discovered from the xylem sap of G. max and M. trunculata, namely the XYLEM ASSOCIATED PEPTIDES (XAPs) (Fig. 2E) (Okamoto et al., 2015; Patel et al., 2018). Their functions are largely unknown and their characterization and elucidation as potential long-distance signals will be an exciting topic in the coming years.
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft through grant SA495/13-2. The authors declare no conflict of interest.
Glossary
Abbreviations:
- AHA
Arabidopsis H+-ATPase
- APK
APS kinase
- APS
adenosine 5´-phosphosulfate
- ATPS
ATP sulfurylase
- CIF
Casparian strip integrity factor
- CLE
CLAVATA3/EMBRYO SURROUNDING REGION-RELATED
- CLEL
CLE-like
- GLV
Golven
- HPAT
hydroxyproline O-arabinosyltransferases
- LRR-RLK
leucine rich repeat receptor-like kinase
- P4H
prolyl-4-hydroxylases
- PAPS
3´-phosphoadenosine 5´-phosphosulfate
- PAPST
PAPS transporter
- PSK
phytosulfokine
- PSKR
PSK receptor
- PSY
peptide containing sulfated tyrosine
- PSY1R
PSY1 receptor
- PTM
post-translational modification
- RGF
root meristem growth factor
- RGFR
RGF receptor
- SBT
subtilase
- TPST
tyrosylprotein sulfotransferase
References
- Amano Y, Tsubouchi H, Shinohara H, Ogawa M, Matsubayashi Y. 2007. Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proceedings of the National Academy of Sciences, USA 104, 18333–18338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahyrycz A, Matsubayashi Y, Ogawa M, Sakagami Y, Konopińska D. 2004. Plant peptide hormone phytosulfokine (PSK-alpha): synthesis of new analogues and their biological evaluation. Journal of Peptide Science 10, 462–469. [DOI] [PubMed] [Google Scholar]
- Bahyrycz A, Matsubayashi Y, Ogawa M, Sakagami Y, Konopińska D. 2005. Further analogues of plant peptide hormone phytosulfokine-alpha (PSK-alpha) and their biological evaluation. Journal of Peptide Science 11, 589–592. [DOI] [PubMed] [Google Scholar]
- Balsved D, Bundgaard JR, Sen JW. 2007. Stability of tyrosine sulfate in acidic solutions. Analytical Biochemistry 363, 70–76. [DOI] [PubMed] [Google Scholar]
- Boruc J, Van den Daele H, Hollunder J, Rombauts S, Mylle E, Hilson P, Inzé D, De Veylder L, Russinova E. 2010. Functional modules in the Arabidopsis core cell cycle binary protein–protein interaction network. The Plant Cell 22, 1264–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busatto N, Salvagnin U, Resentini F, Quaresimin S, Navazio L, Marin O, Pellegrini M, Costa F, Mierke DF, Trainotti L. 2017. The peach RGF/GLV signaling peptide pCTG134 is involved in a regulatory circuit that sustains auxin and ethylene actions. Frontiers in Plant Science 8, 1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YF, Matsubayashi Y, Sakagami Y. 2000. Peptide growth factor phytosulfokine-alpha contributes to the pollen population effect. Planta 211, 752–755. [DOI] [PubMed] [Google Scholar]
- Cheng CY, Krishnakumar V, Chan AP, Thibaud-Nissen F, Schobel S, Town CD. 2017. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. The Plant Journal 89, 789–804. [DOI] [PubMed] [Google Scholar]
- Diévart A, Clark SE. 2003. Using mutant alleles to determine the structure and function of leucine-rich repeat receptor-like kinases. Current Opinion in Plant Biology 6, 507–516. [DOI] [PubMed] [Google Scholar]
- Djordjevic MA, Oakes M, Wong CE, Singh M, Bhalla P, Kusumawati L, Imin N. 2011. Border sequences of Medicago truncatula CLE36 are specifically cleaved by endoproteases common to the extracellular fluids of Medicago and soybean. Journal of Experimental Botany 62, 4649–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doblas VG, Smakowska-Luzan E, Fujita S, Alassimone J, Barberon M, Madalinski M, Belkhadir Y, Geldner N. 2017. Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science 355, 280–284. [DOI] [PubMed] [Google Scholar]
- Drapek C, Sparks EE, Marhavy P, Taylor I, Andersen TG, Hennacy JH, Geldner N, Benfey PN. 2018. Minimum requirements for changing and maintaining endodermis cell identity in the Arabidopsis root. Nature Plants 4, 586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchells JP, Smit ME, Gaudinier A, Williams CJ, Brady SM. 2016. A brief history of the TDIF-PXY signalling module: balancing meristem identity and differentiation during vascular development. New Phytologist 209, 474–484. [DOI] [PubMed] [Google Scholar]
- Fernandez A, Drozdzecki A, Hoogewijs K, Nguyen A, Beeckman T, Madder A, Hilson P. 2013. Transcriptional and functional classification of the GOLVEN/ROOT GROWTH FACTOR/CLE-like signaling peptides reveals their role in lateral root and hair formation. Plant Physiology 161, 954–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A, Drozdzecki A, Hoogewijs K, Vassileva V, Madder A, Beeckman T, Hilson P. 2015. The GLV6/RGF8/CLEL2 peptide regulates early pericycle divisions during lateral root initiation. Journal of Experimental Botany 66, 5245–5256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuglsang AT, Kristensen A, Cuin TA, et al. 2014. Receptor kinase-mediated control of primary active proton pumping at the plasma membrane. The Plant Journal 80, 951–964. [DOI] [PubMed] [Google Scholar]
- Ghorbani S, Hoogewijs K, Pečenková T, et al. 2016. The SBT6.1 subtilase processes the GOLVEN1 peptide controlling cell elongation. Journal of Experimental Botany 67, 4877–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigolashvili T, Geier M, Ashykhmina N, Frerigmann H, Wulfert S, Krueger S, Mugford SG, Kopriva S, Haferkamp I, Flügge UI. 2012. The Arabidopsis thylakoid ADP/ATP carrier TAAC has an additional role in supplying plastidic phosphoadenosine 5´-phosphosulfate to the cytosol. The Plant Cell 24, 4187–4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorres KL, Raines RT. 2010. Prolyl 4-hydroxylase. Critical Reviews in Biochemistry and Molecular Biology 45, 106–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Tan J, Tu L, Zhang X. 2014. A peptide hormone gene, GhPSK promotes fibre elongation and contributes to longer and finer cotton fibre. Plant Biotechnology Journal 12, 861–871. [DOI] [PubMed] [Google Scholar]
- Han SW, Lee SW, Bahar O, Schwessinger B, Robinson MR, Shaw JB, Madsen JA, Brodbelt JS, Ronald PC. 2012. Tyrosine sulfation in a Gram-negative bacterium. Nature Communications 3, 1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanai H, Nakayama D, Yang H, Matsubayashi Y, Hirota Y, Sakagami Y. 2000. Existence of a plant tyrosylprotein sulfotransferase: novel plant enzyme catalyzing tyrosine O-sulfation of preprophytosulfokine variants in vitro. FEBS Letters 470, 97–101. [DOI] [PubMed] [Google Scholar]
- Hartmann J, Fischer C, Dietrich P, Sauter M. 2014. Kinase activity and calmodulin binding are essential for growth signaling by the phytosulfokine receptor PSKR1. The Plant Journal 78, 192–202. [DOI] [PubMed] [Google Scholar]
- Hartmann J, Linke D, Bönniger C, Tholey A, Sauter M. 2015. Conserved phosphorylation sites in the activation loop of the Arabidopsis phytosulfokine receptor PSKR1 differentially affect kinase and receptor activity. The Biochemical Journal 472, 379–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyman J, Cools T, Vandenbussche F, et al. 2013. ERF115 controls root quiescent center cell division and stem cell replenishment. Science 342, 860–863. [DOI] [PubMed] [Google Scholar]
- Hirschmann F, Krause F, Papenbrock J. 2014. The multi-protein family of sulfotransferases in plants: composition, occurrence, substrate specificity, and functions. Frontiers in Plant Science 5, 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann U, Lau K, Hothorn M. 2017. The structural basis of ligand perception and signal activation by receptor kinases. Annual Review of Plant Biology 68, 109–137. [DOI] [PubMed] [Google Scholar]
- Holzwart E, Huerta AI, Glöckner N, Garnelo Gómez B, Wanke F, Augustin S, Askani JC, Schürholz A-K, Harter K, Wolf S. 2018. BRI1 controls vascular cell fate in the Arabidopsis root through RLP44 and phytosulfokine signaling. Proceedings of the National Academy of Sciences, USA 115, 11838–11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttner WB. 1984. Determination and occurrence of tyrosine O-sulfate in proteins. Methods in Enzymology 107, 200–223. [DOI] [PubMed] [Google Scholar]
- Igarashi D, Tsuda K, Katagiri F. 2012. The peptide growth factor, phytosulfokine, attenuates pattern-triggered immunity. The Plant Journal 71, 194–204. [DOI] [PubMed] [Google Scholar]
- Igasaki T, Akashi N, Ujino-Ihara T, Matsubayashi Y, Sakagami Y, Shinohara K. 2003. Phytosulfokine stimulates somatic embryogenesis in Cryptomeria japonica. Plant & Cell Physiology 44, 1412–1416. [DOI] [PubMed] [Google Scholar]
- Imin N, Patel N, Corcilius L, Payne RJ, Djordjevic MA. 2018. CLE peptide tri-arabinosylation and peptide domain sequence composition are essential for SUNN-dependent autoregulation of nodulation in Medicago truncatula. New Phytologist 218, 73–80. [DOI] [PubMed] [Google Scholar]
- Kang J, Yu H, Tian C, Zhou W, Li C, Jiao Y, Liu D. 2014. Suppression of photosynthetic gene expression in roots is required for sustained root growth under phosphate deficiency. Plant Physiology 165, 1156–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassaw T, Nowak S, Schnabel E, Frugoli J. 2017. ROOT DETERMINED NODULATION1 is required for M. truncatula CLE12, but not CLE13, peptide signaling through the SUNN receptor kinase. Plant Physiology 174, 2445–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann C, Motzkus M, Sauter M. 2017. Phosphorylation of the phytosulfokine peptide receptor PSKR1 controls receptor activity. Journal of Experimental Botany 68, 1411–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BJ, Gibson DM, Shuler ML. 2006. Effect of the plant peptide regulator, phytosulfokine-alpha, on the growth and taxol production from Taxus sp. suspension cultures. Biotechnology and Bioengineering 95, 8–14. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Eun C-H, Hanai H, Matsubayashi Y, Sakagami Y, Kamada H. 1999. Phytosulphokine-α, a peptidyl plant growth factor, stimulates somatic embryogenesis in carrot. Journal of Experimental Botany 50, 1123–1128. [Google Scholar]
- Komori R, Amano Y, Ogawa-Ohnishi M, Matsubayashi Y. 2009. Identification of tyrosylprotein sulfotransferase in Arabidopsis. Proceedings of the National Academy of Sciences, USA 106, 15067–15072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong X, Tian H, Yu Q, et al. 2018. PHB3 maintains root stem cell niche identity through ROS-responsive AP2/ERF transcription factors in Arabidopsis. Cell Reports 22, 1350–1363. [DOI] [PubMed] [Google Scholar]
- Kutschmar A, Rzewuski G, Stührwohldt N, Beemster GT, Inzé D, Sauter M. 2009. PSK-α promotes root growth in Arabidopsis. New Phytologist 181, 820–831. [DOI] [PubMed] [Google Scholar]
- Kwezi L, Ruzvidzo O, Wheeler JI, Govender K, Iacuone S, Thompson PE, Gehring C, Irving HR. 2011. The phytosulfokine (PSK) receptor is capable of guanylate cyclase activity and enabling cyclic GMP-dependent signaling in plants. The Journal of Biological Chemistry 286, 22580–22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladwig F, Dahlke RI, Stührwohldt N, Hartmann J, Harter K, Sauter M. 2015. Phytosulfokine regulates growth in Arabidopsis through a response module at the plasma membrane that includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H+-ATPase, and BAK1. The Plant Cell 27, 1718–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH. 2007. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. The Plant Journal 51, 897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loivamäki M, Stührwohldt N, Deeken R, Steffens B, Roitsch T, Hedrich R, Sauter M. 2010. A role for PSK signaling in wounding and microbial interactions in Arabidopsis. Physiologia Plantarum 139, 348–357. [DOI] [PubMed] [Google Scholar]
- Lorbiecke R, Sauter M. 2002. Comparative analysis of PSK peptide growth factor precursor homologs. Plant Science 163, 321–332. [Google Scholar]
- Ma X, Xu G, He P, Shan L. 2016. SERKing coreceptors for receptors. Trends in Plant Science 21, 1017–1033. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y. 2012. Recent progress in research on small post-translationally modified peptide signals in plants. Genes to Cells 17, 1–10. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Hanai H, Hara O, Sakagami Y. 1996. Active fragments and analogs of the plant growth factor, phytosulfokine: structure–activity relationships. Biochemical and Biophysical Research Communications 225, 209–214. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Ogawa M, Kihara H, Niwa M, Sakagami Y. 2006. Disruption and overexpression of Arabidopsis phytosulfokine receptor gene affects cellular longevity and potential for growth. Plant Physiology 142, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y, Ogawa M, Morita A, Sakagami Y. 2002. An LRR receptor kinase involved in perception of a peptide plant hormone, phytosulfokine. Science 296, 1470–1472. [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y, Sakagami Y. 1996. Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proceedings of the National Academy of Sciences, USA 93, 7623–7627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y, Takagi L, Omura N, Morita A, Sakagami Y. 1999. The endogenous sulfated pentapeptide phytosulfokine-alpha stimulates tracheary element differentiation of isolated mesophyll cells of zinnia. Plant Physiology 120, 1043–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki Y, Ogawa-Ohnishi M, Mori A, Matsubayashi Y. 2010. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science 329, 1065–1067. [DOI] [PubMed] [Google Scholar]
- Meng L, Buchanan BB, Feldman LJ, Luan S. 2012. CLE-like (CLEL) peptides control the pattern of root growth and lateral root development in Arabidopsis. Proceedings of the National Academy of Sciences, USA 109, 1760–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Leptihn S, Welz M, Schaller A. 2016. Functional characterization of propeptides in plant subtilases as intramolecular chaperones and inhibitors of the mature protease. The Journal of Biological Chemistry 291, 19449–19461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KL. 2003. The biology and enzymology of protein tyrosine O-sulfation. The Journal of Biological Chemistry 278, 24243–24246. [DOI] [PubMed] [Google Scholar]
- Moore KL. 2009. Protein tyrosine sulfation: a critical posttranslation modification in plants and animals. Proceedings of the National Academy of Sciences, USA 106, 14741–14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher S, Kemmerling B. 2013. PSKR1 and PSY1R-mediated regulation of plant defense response. Plant Signaling & Behavior 8, e24119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher S, Seybold H, Rodriguez P, et al. 2013. The tyrosine-sulfated peptide receptors PSKR1 and PSY1R modify the immunity of Arabidopsis to biotrophic and necrotrophic pathogens in an antagonistic manner. The Plant Journal 73, 469–482. [DOI] [PubMed] [Google Scholar]
- Motose H, Iwamoto K, Endo S, Demura T, Sakagami Y, Matsubayashi Y, Moore KL, Fukuda H. 2009. Involvement of phytosulfokine in the attenuation of stress response during the transdifferentiation of zinnia mesophyll cells into tracheary elements. Plant Physiology 150, 437–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford SG, Yoshimoto N, Reichelt M, et al. 2009. Disruption of adenosine-5´-phosphosulfate kinase in Arabidopsis reduces levels of sulfated secondary metabolites. The Plant Cell 21, 910–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllyharju J. 2003. Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biology 22, 15–24. [DOI] [PubMed] [Google Scholar]
- Nakayama T, Shinohara H, Tanaka M, Baba K, Ogawa-Ohnishi M, Matsubayashi Y. 2017. A peptide hormone required for Casparian strip diffusion barrier formation in Arabidopsis roots. Science 355, 284–286. [DOI] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. 2005. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology 56, 165–185. [DOI] [PubMed] [Google Scholar]
- Oehlenschlæger CB, Gersby LBA, Ahsan N, Pedersen JT, Kristensen A, Solakova TV, Thelen JJ, Fuglsang AT. 2017. Activation of the LRR receptor-like kinase PSY1R requires transphosphorylation of residues in the activation loop. Frontiers in Plant Science 8, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa-Ohnishi M, Matsushita W, Matsubayashi Y. 2013. Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nature Chemical Biology 9, 726–730. [DOI] [PubMed] [Google Scholar]
- Ohkubo Y, Tanaka M, Tabata R, Ogawa-Ohnishi M, Matsubayashi Y. 2017. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nature Plants 3, 17029. [DOI] [PubMed] [Google Scholar]
- Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. 2009. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nature Chemical Biology 5, 578–580. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Shinohara H, Mori T, Matsubayashi Y, Kawaguchi M. 2013. Root-derived CLE glycopeptides control nodulation by direct binding to HAR1 receptor kinase. Nature Communications 4, 2191. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Suzuki T, Kawaguchi M, Higashiyama T, Matsubayashi Y. 2015. A comprehensive strategy for identifying long-distance mobile peptides in xylem sap. The Plant Journal 84, 611–620. [DOI] [PubMed] [Google Scholar]
- Orr RJS, Aalen RB. 2017. In silico prediction of ligand-binding sites of plant receptor kinases using conservation mapping. In: Aalen RB, ed. Plant receptor kinases. New York: Humana Press, 93–105. [DOI] [PubMed] [Google Scholar]
- Ou Y, Lu X, Zi Q, et al. 2016. RGF1 INSENSITIVE 1 to 5, a group of LRR receptor-like kinases, are essential for the perception of root meristem growth factor 1 in Arabidopsis thaliana. Cell Research 26, 686–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenti G, Meroni G, Ballabio A. 1997. The sulfatase gene family. Current Opinion in Genetics & Development 7, 386–391. [DOI] [PubMed] [Google Scholar]
- Patel N, Mohd-Radzman NA, Corcilius L, et al. 2018. Diverse peptide hormones affecting root growth identified in the Medicago truncatula secreted peptidome. Molecular & Cellular Proteomics 17, 160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA. 1991. A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253, 895–897. [DOI] [PubMed] [Google Scholar]
- Pruitt RN, Joe A, Zhang W, Feng W, Stewart V, Schwessinger B, Dinneny JR, Ronald PC. 2017. A microbially derived tyrosine-sulfated peptide mimics a plant peptide hormone. New Phytologist 215, 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautengarten C, Steinhauser D, Büssis D, Stintzi A, Schaller A, Kopka J, Altmann T. 2005. Inferring hypotheses on functional relationships of genes: analysis of the Arabidopsis thaliana subtilase gene family. PLoS Computational Biology 1, e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodiuc N, Barlet X, Hok S, et al. 2016. Evolutionarily distant pathogens require the Arabidopsis phytosulfokine signalling pathway to establish disease. Plant, Cell & Environment 39, 1396–1407. [DOI] [PubMed] [Google Scholar]
- Ronald PC. 2014. The role of RaxST, a prokaryotic sulfotransferase, and RaxABC, a putative type I secretion system, in activation of the rice XA21-mediated immune response. Scientifica 2014, 532816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A, Stintzi A, Rivas S, et al. 2018. From structure to function – a family portrait of plant subtilases. New Phytologist 218, 901–915. [DOI] [PubMed] [Google Scholar]
- Schardon K, Hohl M, Graff L, Pfannstiel J, Schulze W, Stintzi A, Schaller A. 2016. Precursor processing for plant peptide hormone maturation by subtilisin-like serine proteinases. Science 354, 1594–1597. [DOI] [PubMed] [Google Scholar]
- Seidah NG, Prat A. 2012. The biology and therapeutic targeting of the proprotein convertases. Nature Reviews Drug Discovery 11, 367–383. [DOI] [PubMed] [Google Scholar]
- Shen Y, Diener AC. 2013. Arabidopsis thaliana resistance to FUSARIUM OXYSPORUM 2 implicates tyrosine-sulfated peptide signaling in susceptibility and resistance to root infection. PLoS Genetics 9, e1003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Matsubayashi Y. 2013. Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity. Plant & Cell Physiology 54, 369–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H, Mori A, Yasue N, Sumida K, Matsubayashi Y. 2016. Identification of three LRR-RKs involved in perception of root meristem growth factor in Arabidopsis. Proceedings of the National Academy of Sciences, USA 113, 3897–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Han Z, Wang J, Lin G, Chai J. 2017. Structural insights into ligand recognition and activation of plant receptor kinases. Current Opinion in Structural Biology 43, 18–27. [DOI] [PubMed] [Google Scholar]
- Song W, Liu L, Wang J, et al. 2016. Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth. Cell Research 26, 674–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Liu JX, Guo H, Yin Y, Howell SH. 2009. Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. The Plant Journal 59, 930–939. [DOI] [PubMed] [Google Scholar]
- Srivastava R, Liu JX, Howell SH. 2008. Proteolytic processing of a precursor protein for a growth-promoting peptide by a subtilisin serine protease in Arabidopsis. The Plant Journal 56, 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stührwohldt N, Dahlke RI, Kutschmar A, Peng X, Sun MX, Sauter M. 2015. Phytosulfokine peptide signaling controls pollen tube growth and funicular pollen tube guidance in Arabidopsis thaliana. Physiologia Plantarum 153, 643–653. [DOI] [PubMed] [Google Scholar]
- Stührwohldt N, Dahlke RI, Steffens B, Johnson A, Sauter M. 2011. Phytosulfokine-α controls hypocotyl length and cell expansion in Arabidopsis thaliana through phytosulfokine receptor 1. PLoS ONE 6, e21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang G, Zhou JM. 2017. Receptor kinases in plant–pathogen interactions: more than pattern recognition. The Plant Cell 29, 618–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavormina P, De Coninck B, Nikonorova N, De Smet I, Cammue BP. 2015. The plant peptidome: an expanding repertoire of structural features and biological functions. The Plant Cell 27, 2095–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Li H, Han Z, Zhang H, Wang T, Lin G, Chang J, Yang W, Chai J. 2015. Allosteric receptor activation by the plant peptide hormone phytosulfokine. Nature 525, 265–268. [DOI] [PubMed] [Google Scholar]
- Wang X, Jiang N, Liu J, Liu W, Wang GL. 2014. The role of effectors and host immunity in plant–necrotrophic fungal interactions. Virulence 5, 722–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford R, Fernandez A, Tejos R, et al. 2012. GOLVEN secretory peptides regulate auxin carrier turnover during plant gravitropic responses. Developmental Cell 22, 678–685. [DOI] [PubMed] [Google Scholar]
- Wu T, Kamiya T, Yumoto H, Sotta N, Katsushi Y, Shigenobu S, Matsubayashi Y, Fujiwara T. 2015. An Arabidopsis thaliana copper-sensitive mutant suggests a role of phytosulfokine in ethylene production. Journal of Experimental Botany 66, 3657–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa S, Sakuta C, Matsubayashi Y, Sakagami Y, Kamada H, Satoh S. 1998. The promotive effects of a peptidyl plant growth factor, phytosulfokine-α, on the formation of adventitious roots and expression of a gene for a root-specific cystatin in cucumber hypocotyls. Journal of Plant Research 111, 453–458. [Google Scholar]
- Zhang H, Hu Z, Lei C, et al. 2018. A plant phytosulfokine peptide initiates auxin-dependent immunity through cytosolic Ca2+ signaling in tomato. The Plant Cell 30, 652–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Wei L, Xu J, et al. 2010. Arabidopsis tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. The Plant Cell 22, 3692–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]