Abstract
POU3F3, also referred to as Brain-1, is a well-known transcription factor involved in the development of the central nervous system, but it has not previously been associated with a neurodevelopmental disorder. Here, we report the identification of 19 individuals with heterozygous POU3F3 disruptions, most of which are de novo variants. All individuals had developmental delays and/or intellectual disability and impairments in speech and language skills. Thirteen individuals had characteristic low-set, prominent, and/or cupped ears. Brain abnormalities were observed in seven of eleven MRI reports. POU3F3 is an intronless gene, insensitive to nonsense-mediated decay, and 13 individuals carried protein-truncating variants. All truncating variants that we tested in cellular models led to aberrant subcellular localization of the encoded protein. Luciferase assays demonstrated negative effects of these alleles on transcriptional activation of a reporter with a FOXP2-derived binding motif. In addition to the loss-of-function variants, five individuals had missense variants that clustered at specific positions within the functional domains, and one small in-frame deletion was identified. Two missense variants showed reduced transactivation capacity in our assays, whereas one variant displayed gain-of-function effects, suggesting a distinct pathophysiological mechanism. In bioluminescence resonance energy transfer (BRET) interaction assays, all the truncated POU3F3 versions that we tested had significantly impaired dimerization capacities, whereas all missense variants showed unaffected dimerization with wild-type POU3F3. Taken together, our identification and functional cell-based analyses of pathogenic variants in POU3F3, coupled with a clinical characterization, implicate disruptions of this gene in a characteristic neurodevelopmental disorder.
Keywords: POU3F3, Brain-1, de novo variants, intellectual disability, speech/language disorder, POU3F2, FOXP2, luciferase reporter, BRET assay
Main Text
POU3F3 (MIM: 602480) encodes a member of class III of the POU family of transcription factors. These proteins all carry a characteristic POU domain that binds with high affinity to a specific octamer (5′-ATGCAAAT-3′) or closely related DNA sequences in the enhancers and promoters of various different target genes.1 The importance of POU3F3 for the developing brain is reflected in its original name Brain-1 (Brn1).2 Best known as a marker of upper-layer projection neurons in the cortex,3 POU3F3 is implicated in the regulation of many key processes in the development of the central nervous system; these processes include cortical neuronal migration,4 upper-layer specification and production, and neurogenesis.5, 6, 7 However, the phenotypic consequences of pathogenic germline variants in human POU3F3 are currently unknown.
We identified a de novo missense variant disrupting POU3F3 in a female with a severe developmental speech and language disorder, autism spectrum disorder, and mild intellectual disability. This variant was absent in control databases and affected a highly conserved amino acid, and in silico analyses consistently predicted deleterious effects on the function of the encoded protein. In addition, we noted a case report describing a de novo chromosome 2q12.1 deletion in a male with intellectual disability; in the report, POU3F3 haploinsufficiency was discussed as a possible pathogenic mechanism.8 Chromosome 6q16.1 deletions that span the closely related, co-expressed ortholog, POU3F2 (also known as Brn2; MIM: 600494), cause a neurodevelopmental disorder with obesity.9 Moreover, it has been shown that FOXP2 (MIM: 605317), disruptions of which cause a developmental speech disorder (speech language disorder-1; MIM: 602081), contains an intronic binding site for POU3F2.10, 11
We used matchmaking initiatives such as GeneMatcher12 and the Decipher Database13 to identify additional individuals with rare germline variants in POU3F3. Here, we delineate the characteristic phenotypic features and mutational spectrum of a cohort of 19 individuals with pathogenic variants in POU3F3. Nearly all (17 out of 19) individuals had a de novo variant in the gene, and all variants were identified via exome sequencing with a trio approach. One person (individual 18) had inherited the variant from an affected mother (individual 19); in this family, exome sequencing had been performed in the proband and the mother only (duo approach).
The phenotypes of all 19 individuals with POU3F3 variants were systematically assessed and analyzed. A summary of the most common phenotypic characteristics is shown in Table 1, and more details per individual can be found in Table S1. All individuals with a POU3F3 variant in our cohort (19/19; 100%) had developmental delays (DD) and/or an intellectual disability (ID). The level of functioning was broad, ranging from severe ID in two individuals to borderline intellectual functioning (WPPSI-IQ 77) in one individual. For individuals in which the severity of ID and/or DD was known, the majority (8/10; 80%) had a borderline to moderate level of ID and/or DD.
Table 1.
Summary of Phenotypes in Individuals with POU3F3 Variants
| Non-Truncating Variants | Truncating Variants | All Variants Combined | ||
|---|---|---|---|---|
| Feature | Amount | Amount | Amount | Percentage |
| Development | ||||
| Developmental delay (DD) and/or intellectual disability (ID) | 6/6 | 13/13 | 19/19 | 100% |
| Borderline or mild ID | 2/6 | 3/13 | 5/19 | 26% |
| Moderate ID | 1/6 | 2/13 | 3/19 | 16% |
| Severe ID | 2/6 | 0/13 | 2/19 | 11% |
| DD or ID, severity unknown | 1/6 | 8/13 | 9/19 | 47% |
| Speech delay or disorder | 6/6 | 13/13 | 19/19 | 100% |
| Autism Spectrum Disorder (ASD) diagnosis | 1/6 | 6/13 | 7/19 | 37% |
| Neurology | ||||
| Abnormalities reported on brain MRI | 3/4 | 4/7 | 7/11a | 64% |
| Hypotonia | 3/5 | 7/13 | 10/18a | 56% |
| Epilepsy | 2/6 | 0/13 | 2/19 | 11% |
| Drooling | 2/5 | 7/9 | 9/14a | 64% |
| Other features | ||||
| Cupped and/or low-set ears | 3/6 | 13/13 | 16/19 | 84% |
| Vision problems | 2/6 | 8/13 | 10/19 | 53% |
| Sleeping problems (often waking up at night) | 1/5 | 4/9 | 5/14a | 36% |
| Cryptorchidism | 0/1 | 3/10 | 3/11b | 27% of males |
A more detailed overview of phenotypic features per individual can be found in Table S1.
Feature not assessed or not known for all 19 individuals in the cohort.
Feature only applicable to the 11 males in the cohort.
Given that the first proband in our study (individual 3 in Table S1) had a severe developmental speech and language disorder, we paid special attention to the speech and language capacities across the entire cohort. All individuals with POU3F3 variants had delayed speech and language development, often with a delayed onset of producing first words. In two children who spoke their first words at an appropriate age, a halt in development or regression of speech in the first years of life was reported. Although both receptive and expressive language problems were reported, in many children expressive speech capacities were more impaired than language comprehension. Almost all individuals received or had received speech therapy, and commonly reported speech-related problems were oral motor problems, word finding problems, and social communication issues. In addition to this, drooling was reported in 9/14 individuals (64%), and open mouth behavior was seen in four individuals.
Many individuals had autism-like features, and a formal diagnosis of autism spectrum disorder (ASD) was made in 7/19 individuals (37%). Although 3/19 (16%) individuals reached their motor milestones in time, most had delays in both fine and gross motor development. Hypotonia was reported in 10/18 individuals (56%). The two individuals with severe ID each had a form of epilepsy: Lennox-Gastaut syndrome with tonic-clonic seizures in one individual and a severe seizure disorder with myoclonic seizures, drop attacks, absences, and tonic-clonic seizures in the other. In both individuals, capillary hemangiomas were reported; this feature is not present in the rest of the cohort. Magnetic resonance imaging (MRI) revealed cerebral atrophy in both individuals; additionally, white matter cysts were present in one of them. In total, brain anomalies were reported in 7 of the 11 individuals (64%) in which a brain MRI was performed. Anomalies observed in at least two individuals were delayed myelination, cerebral atrophy, and corpus callosum abnormalities.
No significant additional congenital abnormalities were noted in our collected cohort of 19 individuals. Vision problems, which mainly included (mild) refraction errors and strabismus, were reported in 10/19 individuals (53%). Hearing loss was present in one individual, and narrow auditory canals were reported in two other individuals. Although growth parameters were generally normal, small hands with short and broad digits, especially broad thumbs, as well as flat feet and high-arched feet were reported in some of the individuals. Five children (5/14; 36%) had sleeping problems at a young age, waking up several times per night. Three of the eleven males (27%) had cryptorchidism.
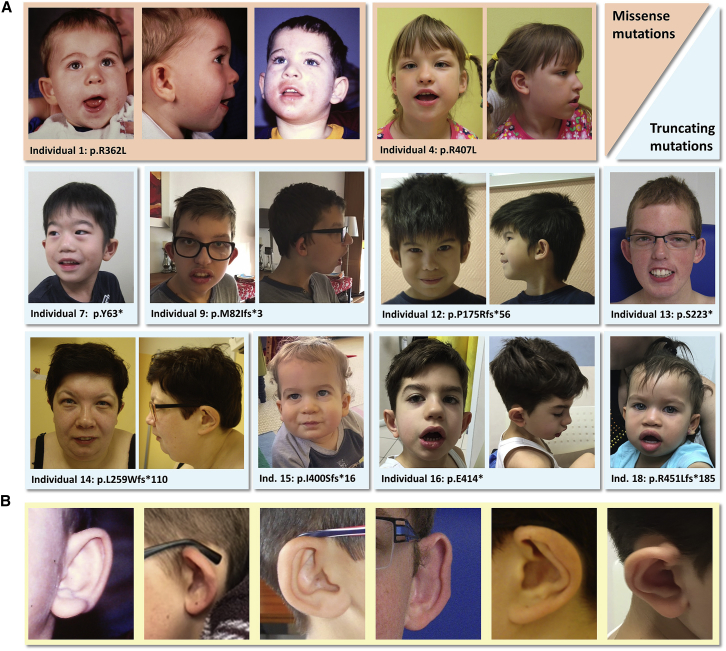
A comparison of facial features revealed a striking overlap in dysmorphisms in individuals with POU3F3 variants. The cupped, prominent, and often low-set ears, present in 16 of the 19 individuals (84%), were most remarkable. Other common facial features included full lips, an open-mouth appearance, a broad and bulbous nasal tip, hypertelorism, epicanthal folds, and peri-orbital fullness (Figure 1).
Figure 1.
Facial Features of Ten Individuals with a Pathogenic POU3F3 Variant
(A) All individuals in this picture have cupped and/or prominent and often low-set ears, except for individual 4. Other overlapping features are full lips, an open-mouth appearance, thick ear helices, a broad and bulbous nasal tip, hypertelorism, epicanthal folds, and peri-orbital fullness.
(B) Magnification of the ear abnormalities in individuals 1, 9, 12, 13, 14, and 16, respectively.
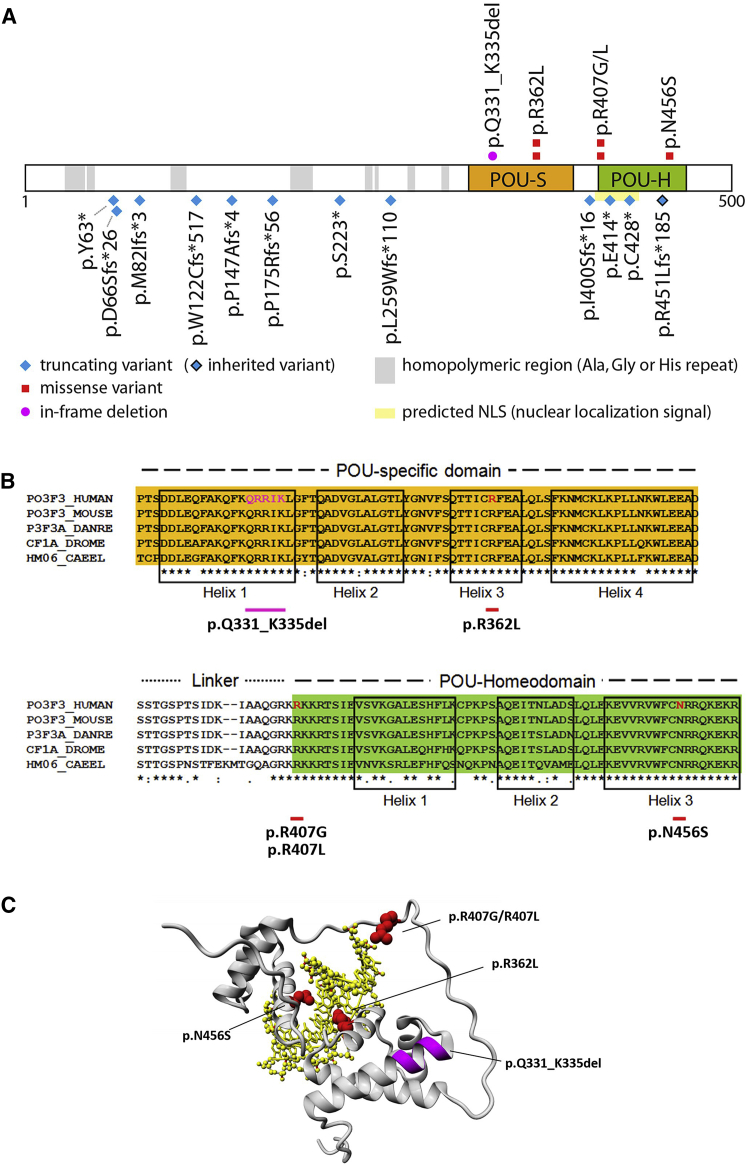
The types of POU3F3 variants in our cohort were diverse and included nonsense variants, frameshift variants, missense variants, and an in-frame deletion of five amino acids. All variants in this study are annotated with respect to the GenBank: NM_006236.2 transcript. None of the identified variants were present in the gnomAD database. The pLI-score of POU3F3 in gnomAD is 0.89, and no high-confidence truncating variants in this gene are present in this dataset, indicating that POU3F3 is especially intolerant for loss-of-function variance. In 13 of the 19 individuals, we found 12 different nonsense or frameshift variants, predicted to truncate POU3F3. For most genes, when such variants arise, the post-transcriptional surveillance mechanism of nonsense-mediated mRNA decay (NMD) helps to prevent translation of aberrant truncated versions of proteins.14 In mammalian cells this surveillance mechanism is tightly linked to pre-mRNA splicing.15 Because POU3F3 is an intronless gene, aberrant transcripts with truncating variants are insensitive to NMD and so will still be expressed. These truncating variants were distributed widely across POU3F3 (Figure 2A) and are thus predicted to yield truncated proteins of a range of different sizes.
Figure 2.
Characterization of POU3F3 Variants
(A) Linear representation of POU3F3 (UniProt: P20264) showing the location of variants from unrelated families in this cohort. There are twelve truncating variants (blue), five missense variants (red), and one in-frame deletion (magenta). POU-S (orange) is the POU-specific domain, and POU-H (green) is the POU-homeodomain. The shown NLS (nuclear localization signal) prediction is derived from cNLS Mapper.27 An overview with mutation details per subject is provided in Table S1.
(B) Alignment of part of the POU3F3 amino acid sequence (using ClustalW) with orthologous sequences from the following species: Mus musculus, Danio rerio, Drosophila melanogaster, and Caenorhabditis elegans. Helix boundaries are defined as previously described.16
(C) Three-dimensional modeling of the functional domains of POU3F3 binding to a target DNA sequence (yellow). Amino acids that are affected by the missense variants are shown in red (wild-type side chains are depicted), and the location of the in-frame deletion is shown in magenta. A more detailed picture for two missense variants, p.Arg362Leu and p.Asn456Ser, can be found in Figure S2.
Five individuals in our cohort had a missense variant. All were located in one of the two known functional domains of POU3F3: the POU-specific (POU-S) domain and the POU-homeobox (POU-H) domain (Figure 2A). Even with this relatively small number of missense variants, a clear clustering was seen: two unrelated individuals had an identical de novo missense variant (c.1085G>T, [p.Arg362Leu]), and another two individuals had missense variants that affected the same amino acid (c.1219C>G, [p.Arg407Gly] and c.1220G>T, [p.Arg407Leu]). One individual had a c.1367A>G, (p.Asn456Ser) substitution. In addition to the missense variants, one individual had an in-frame deletion (c.992_1006del, [p.Gln331_Lys335del]), which removes five amino acids from within the POU-specific domain of the encoded protein. For all the missense variants and the in-frame deletion, highly conserved residues are affected (Figure 2B). All the missense variants of our cohort are predicted to be pathogenic by both PolyPhen-2 and SIFT and have high CADD scores (range 26.9–32.0; Table S1). We also visualized the non-truncating variants in a tolerance landscape of POU3F3 by using the MetaDome web server. The tolerance landscape was computed as a missense over synonymous ratio, on the basis of single nucleotide variants in gnomAD in the protein-coding part of POU3F3. All the non-truncating variants were located in regions of the protein that are extremely intolerant to missense variation (Figure S1).
The two known functional domains of POU3F3, connected via a flexible linker, are both required for site-specific DNA binding with high affinity.16 The POU-S domain forms four alpha-helices; several direct and sequence-specific hydrogen bonds are made between residues in the third helix and the DNA.17 In the POU-H domain, the third helix is also responsible for sequence-specific DNA-binding. Two of the missense variants, c.1085G>T, (p.Arg362Leu) and c.1367A>G, (p.Asn456Ser), affect residues that are located in the third helix of the POU-S domain and the POU-H domain, respectively (Figure 2B).
We used three-dimensional protein modeling to further investigate the potential functional impact of the identified non-truncating variants. The PDB file PDB: 2XSD18 (POU-domain of POU3F1 bound to DNA) provided a template for modeling the DNA-binding region of POU3F3, which spans amino acids 316–466 (Figure 2C). This model revealed that two of the amino acids (Arg362 and Asn456) that are substituted in our cohort are directly involved in binding the major groove of target DNA, consistent with the prior literature on other POU proteins.16, 17 In our model, Arg362 forms hydrogen bonds with a guanine base in the DNA of the transcription-factor binding site (Figure S2). Substitution of a leucine residue at this point of the protein is predicted to abolish DNA-binding. Similarly, Asn456 is directly involved in DNA-binding by forming hydrogen bonds with adenine, an interaction that will be disrupted by a substitution of serine at this position (Figure S2).
The remaining two missense variants that we identified are located at position 407 on the edge of the homeodomain and at the flexible linker in our linear representation of POU3F3 (Figure 2B). In the three-dimensional model, Arg407 lies in the flexible linker between both POU domains, but it is unclear what impact a substitution at this point would have on protein function (Figure 2C). Lastly, the in-frame deletion in our cohort is located in the POU-S domain. Although the amino acids that are deleted do not directly bind to DNA themselves, it is likely that their loss will alter domain structure and indirectly disturb DNA-binding capacities.
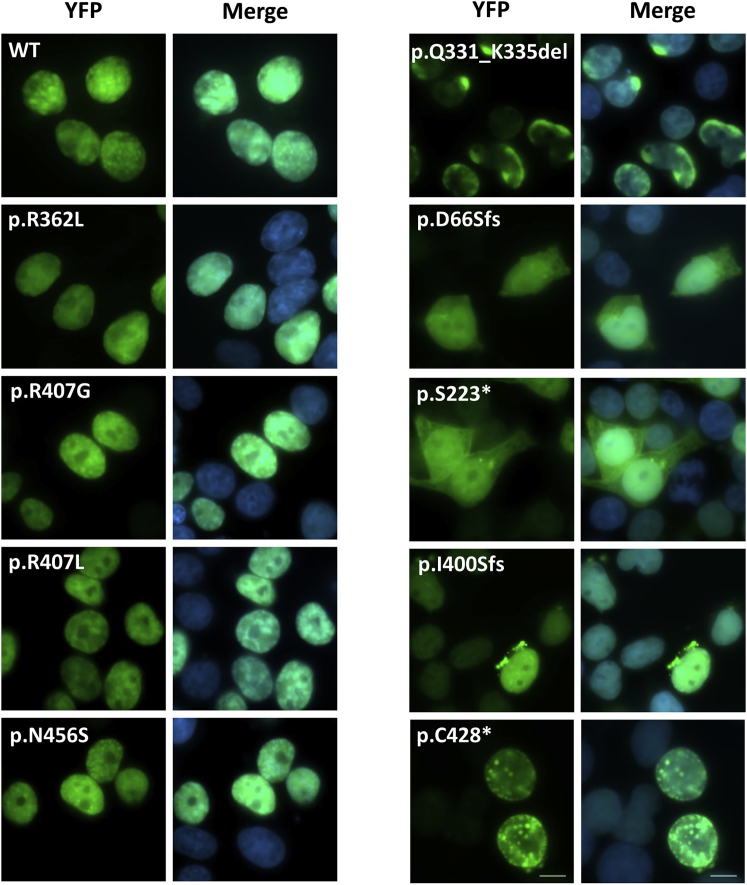
To assess the potential functional effects of the POU3F3 variants, we performed a variety of complementary cell-based assays. We expressed a representative set of nine POU3F3 variants, as well as wild-type (WT) POU3F3, as fusions to YFP tags in HEK293 cells. The set of POU3F3 variants included all four missense variants, the in-frame deletion, and four of the truncating variants. Immunoblot analysis showed that all the expressed YFP-fusion proteins had the expected molecular weights (Figure S3).
We assessed the subcellular localization of the mutant proteins by using direct fluorescence imaging (Figure 3). Although two missense variants (p.Arg407Leu and p.Arg407Gly) map within a computationally predicted nuclear localization signal (NLS) motif (Figure 2A), none of the missense variants disturbed subcellular localization in this assay. All four tested proteins with a missense variant were located in the nucleus in a similar manner to WT. However, all the other tested constructs showed abnormalities in subcellular localization patterns compared to WT. For three truncating constructs (c.196_197delinsT, [p.Asp66Serfs∗26], c.668C>A, [p.Ser223∗], and c.1197delG, [p.Ile400Serfs∗16]), aberrant cytoplasmic expression was noted, in addition to the normal nuclear expression of the protein. For two of these constructs (p.Ser223∗ and p.Ile400Serfs∗16) we observed protein aggregates just around the nuclear membrane in a subset of cells, possibly indicating degradation of mutant protein (Figures 3 and S4). Aberrant localization patterns within the nucleus were observed for the c.1284C>A, (p.Cys428∗) and c.992_1006del, (p.Gln331_Lys335del) proteins, and the former showed small nuclear aggregates in a minority of cells (Figures 3 and S4).
Figure 3.
Subcellular Localization
Direct fluorescence imaging of HEK293 cells expressing YFP-POU3F3 fusion proteins carrying different variants found in our cohort (green). The nuclei are stained with DAPI (blue). The scale bar = 10μm. Pictures showing the aberrant subcellular localization patterns in a larger amount of cells for the variants p.Gln331_Lys335del, p.S223∗, p.Ile400Serfs∗16, and p.Cys428∗ can be found in Figure S4.
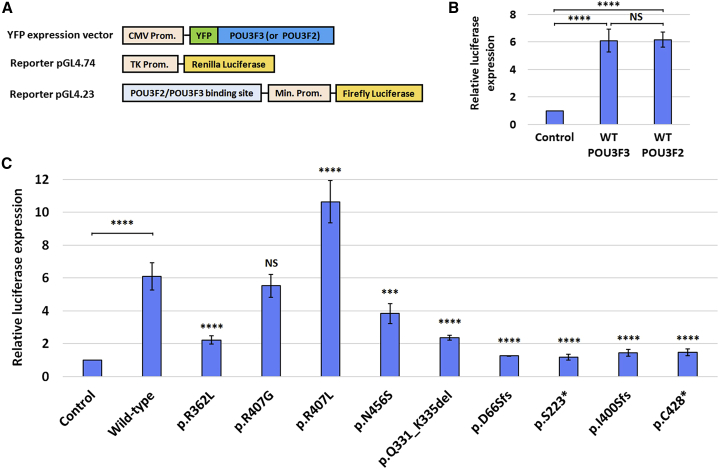
We next investigated whether the variants affect the transcription factor activity of the encoded protein. POU3F3 belongs to the POU family of transcription factors and is known to share important roles in neurodevelopment with its close paralog POU3F2.5, 6 In vitro experiments suggest that POU3F2 is able to activate an intronic binding site in FOXP2,10, 11 a gene that has been implicated in a rare neurodevelopmental disorder mainly characterized by severe speech problems (MIM: 602081).19 We hypothesized that POU3F3 might also be able to activate transcription via this binding site within FOXP2. To test this hypothesis, we performed luciferase assays in which a YFP-fusion protein with POU3F3 or POU3F2 was expressed together with a Firefly luciferase construct containing the conserved FOXP2 binding site (Figure S5), as well as a Renilla luciferase construct, providing a normalization control (Figure 4A). POU3F3 was able to increase luciferase expression as strongly as POU3F2; there was a six-fold increase in expression compared to the negative control (Figure 4B). This finding indicates that the known intronic binding site for POU3F2 can also serve as a functional binding site for POU3F3.
Figure 4.
Luciferase Assays
(A) Expression constructs used in the luciferase assays: a YFP-fused POU3F3 or POU3F2 construct with a CMV promoter; a Firefly luciferase reporter construct with a minimal promoter and a preceding intronic FOXP2-derived binding site; and a control construct with Renilla luciferase under control of a TK promoter.
(B) Results of luciferase assays with WT POU3F3 and WT POU3F2 and the reporter construct with the FOXP2-derived binding site. Values are expressed relative to the control and represent the mean ± SD of three independent experiments, each performed in triplicate (∗∗∗∗ = p < 0.0001 and NS = not significant, using one-way ANOVA and a post-hoc Tukey’s test).
(C) Results of luciferase assay with WT POU3F3 and nine constructs with POU3F3 variants. Values are expressed relative to the control and represent the mean ± SD of three independent experiments, each performed in triplicate (∗∗∗ = p < 0.001; ∗∗∗∗ = p < 0.0001; and NS = not significant when compared to WT POU3F3 using one-way ANOVA and a post-hoc Dunnett’s test).
We used the same luciferase assay to compare our POU3F3 variant constructs with the POU3F3 WT construct. All four POU3F3 constructs with truncating variants showed a severe impairment in transcriptional activation function (Figure 4C). The relative luciferase expression for these variants was similar to that for the negative control (a YFP-expression vector without POU3F3), consistent with the complete or partial loss of the DNA-binding POU domains of POU3F3. Three of the non-truncating variants (p.Arg362Leu, p.Asn456Ser, and p.Gln331_Lys335del) showed partial transactivation capacity that was significantly lower than that of the WT construct. The p.Arg407Leu variant led to a significant increase in relative luciferase expression compared to the WT construct. No significant difference compared to WT was seen for the other missense variant at this position (p.Arg407Gly). In summary, all variants except for the p.Arg407Gly substitution led to significantly disturbed transactivation capabilities in our assays.
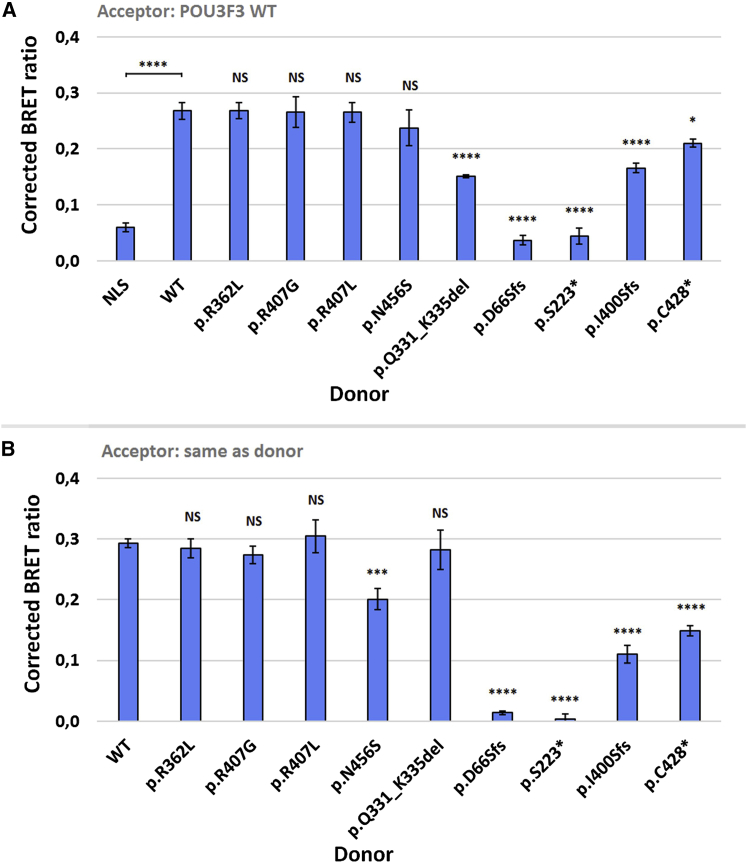
POU proteins are well known to have highly conserved dimerization properties.10 They bind to target genes as monomers or dimers and can form either homo-dimers or hetero-dimers involving other family members.20 To investigate whether the variants in our cohort affected the dimerization capacities of POU3F3, we used bioluminescence resonance energy transfer (BRET), a sensitive live-cell assay, to test putative protein-protein interactions.21 In our assays, the bioluminescent donor construct encodes a Renilla luciferase (RLuc) fusion protein, and the fluorescent acceptor construct encodes a protein fused to YFP. If the proteins of interest are in close proximity, energy transfer can take place from donor to acceptor. We tested the ability of each mutant protein to form dimers with WT POU3F3 (Figure 5A) and with itself (Figure 5B). In these experiments the missense variants showed generally intact dimerization capacity, although the interactions for the p.Asn456Ser variant were slightly decreased. Two variants that are predicted to cause an early truncation of POU3F3 (p.Asp66Serfs∗26 and p.Ser223∗) showed a complete loss of dimerization capacity in both conditions. The two other truncating constructs (p.Ile400Serfs∗16 and p.Cys428∗) showed a less severe decrease, although the dimerization capacity was still significantly different from that of the WT construct. The p.Gln331_Lys335 protein showed impaired dimerization with WT POU3F3 but normal capacities for forming homo-dimers.
Figure 5.
Bioluminescence Resonance Energy Transfer Assays
(A) Bioluminescence resonance energy transfer (BRET) assays to measure interactions between WT POU3F3 and mutant POU3F3 constructs. Bars represent the corrected mean BRET ratio ± SD of three independent experiments performed in triplicate (∗∗∗∗ = p < 0.0001; ∗ = p < 0.05; and NS = not significant when compared to WT using one-way ANOVA and a post-hoc Tukey’s test). The NLS-donor construct is a Renilla luciferase construct with a nuclear localization signal.
(B) BRET assays to measure homodimerization capacity of each mutant POU3F3 construct. Bars represent the corrected mean BRET ratio ± SD of three independent experiments performed in triplicate (∗∗∗∗ = p < 0.0001; ∗∗∗ = p < 0.001; and NS = not significant when compared to WT using one-way ANOVA and a post-hoc Tukey’s test)
All in all, the results of our clinical and molecular characterization show that diverse variants at different locations within POU3F3 lead to a neurodevelopmental disorder with overlapping symptoms. When comparing genotypes and phenotypes within the cohort, several findings are of interest. First, two individuals (individuals 1 and 2) have a distinct and more severe phenotype compared to the rest of the cohort; this phenotype includes severe ID, epilepsy, and capillary hemangiomas. These individuals are unrelated but have an identical p.Arg362Leu variant. In luciferase assays, this variant showed impaired transcription-activation capacity, but it was not lower than that observed for other mutant constructs. Although the reason for the more severe and distinct phenotype associated with the p.Arg362Leu variant remains unclear, a dominant-negative effect is one possibility, given that the mutant protein showed normal subcellular localization and dimerization capacities in our assays.
Second, several pathogenic POU3F3 variants appear to be associated with characteristic facial features, especially the prominent, often cupped, and low-set ears. These ear abnormalities were reported independently in all individuals with a truncating variant and in the individuals with the p.Arg362Leu and p.Gln331_Lys335del variant (Figure 1 and Table S1). Prominent ears were also reported in the previously published individual with a microdeletion that included POU3F3.8 The cupped or prominent ears are not present in the three individuals with the missense variants p.Arg407Leu, p.Arg407Gly, and p.Asn456Ser. The two missense variants affecting amino acid Arg407 did not show loss-of-function effects in our luciferase assay; in fact, p.Arg407Leu showed evidence of a possible gain-of-function. The p.Asn456Ser variant had a mild loss-of-function effect on transcriptional activity. These results suggest that both loss-of-function and gain-of-function mechanisms of different severity might lead to neurodevelopmental disorders with differences on a phenotypic level.
Although the missense variants at positions Arg362 and Asn456 mediate DNA-binding in the major groove, this is not the case for Arg407. This residue is located in the flexible linker between the POU-domains. POU3F2 and POU3F3 are known to be flexible in terms of spacing preference,16 meaning that they can bind to short binding motifs that are separated by 0, 2, or 3 bp, in contrast to other POU proteins that have more fixed preferences. Findings from a study of POU3F2 suggest that the highly conserved arginine residue at a position analogous to POU3F3 residue 407 is one of the residues that form a critical region in regulating the spacing preference of the protein.16 Binding activity experiments showed that mutation of this critical region on the edge of the flexible linker and the homeodomain leads to less flexibility in spacing preference for the POU protein.16 The missense variants p.Arg407Gly and p.Arg407Leu show different effects in our luciferase assay: although p.Arg407Gly did not show any difference compared with the WT POU3F3 construct, the p.Arg407Leu variant showed a gain-of-function effect. It is unclear why these two different variants affecting the same Arg407 residue show different effects on transactivation capacity and how this relates to pathogenic mechanisms. Possibly, the missense variants at Arg407 alter the spacing properties of the encoded POU3F3 protein, and the alteration might affect transcriptional activation depending on the characteristics of the binding site. The architecture of regulatory DNA sites has been shown to influence the structure and organization of POU dimerization, interaction with other proteins, and DNA-binding properties and can therefore be critical in determining the functionality of a transcription factor.18, 22
POU3F3 is highly similar to POU3F2; it has nearly identical (98.7%) amino acid sequences for the POU domains and the flexible linker. The main differences are found within the N-terminal region, which contains homopolymeric repeats that can function as transcriptional activation domains.1, 23 POU3F3 and POU3F2 share some roles and have partially redundant functions in cortical development.4, 5 Nonetheless, our study and the previously published study on POU3F2 haploinsufficiency9 underscore the fact that two functional copies of both POU3F3 and POU3F2 are required for normal neurodevelopment. Microdeletions that span POU3F2 have been shown previously to cause a neurodevelopmental disorder with obesity.9 In contrast to this POU3F2-related disorder, pathogenic variants in POU3F3 do not seem to be associated with obesity, because this feature is only reported in one of the 19 individuals in our cohort. In addition to the microdeletions encompassing POU3F2, a single de novo missense variant in POU3F2 has recently been reported, but the specific location of this variant does not correspond to any variant reported here for POU3F3.24
Our functional data indicate that a known POU3F2 regulatory site mapping within the FOXP2 locus11 can also be bound by POU3F3. By using this binding site, we could develop luciferase-based assays to index the transactivation capacities of POU3F3 proteins carrying different etiological variants. Nevertheless, it remains undetermined whether pathogenic POU3F2 and/or POU3F3 variants actually have a significant impact on FOXP2 expression in the proper genomic context in vivo. Future studies (for example by directly testing for FOXP2 misregulation in individuals with pathogenic POU3F3 variants) might shed light on whether putative functional links between the different genes have physiological relevance for the speech and language impairments observed in the associated neurodevelopmental disorders.25
We emphasize that exome sequencing coverage is variable for POU3F3; the 5′ half of the gene has poor coverage and the 3′ part has good coverage.26 So if the characteristic facial phenotype as shown in Figure 1 is recognized in an individual with an overlapping neurodevelopmental phenotype, it might be prudent to re-assess any existing next-generation-sequencing data and/or perform targeted sequencing of POU3F3. The specific variants identified in this study were all covered by a sufficient number of allele counts in gnomAD, and none of these alleles were found in this large dataset.26
In conclusion, we have shown that pathogenic POU3F3 variants cause a neurodevelopmental disorder with a broad phenotypic spectrum that includes ID and/or DD, speech and language problems, hypotonia, and autism spectrum disorder. Most individuals have mild to moderate delays in neurodevelopment, but a distinct phenotype of severe ID and epilepsy is also reported in two individuals with an identical missense variant. Although most variants result in loss-of-function effects on the transactivation capacities of POU3F3, other possible pathogenic mechanisms cannot be excluded. By showing the effects of POU3F3 dysfunction in humans, our data highlight the essential functions of POU3F3 for normal brain development.
Declaration of Interests
Z.P., S.T., and D.N.S. are full-time employees of Ambry Genetics. Exome sequencing is one of Ambry’s commercially available tests. The authors declare no other competing interests.
Acknowledgments
We thank all individuals and families for their contribution.
This work was supported by the Netherlands Organization for Scientific Research (NWO) Gravitation Grant 24.001.006 to the Language in Interaction Consortium (L.S.B., S.E.F., and H.G.B.), the Netherlands Organization for Health Research and Development (ZonMw grant 91718310 to T.K.), the Max Planck Society (P.D. and S.E.F.), Fondazione Bambino Gesù (Vite Coraggiose) (M.T.), the Italian Ministry of Health (Ricerca Corrente 2019) (M.L.D.), the Japan Agency for Medical Research and Development (AMED) under grant numbers JP18ek0109280, JP18dm0107090, JP18ek0109301, JP18ek0109348, and JP18kk0205001 (N.Ma.), Japan Society for the Promotion of Science Grants-in-Aid for Scientific Research (JSPS KAKENHI) under grant numbers JP17H01539 (N.Ma.) and JP16H05357 (N.Mi.), grants by the Ministry of Health, Labour, and Welfare in Japan (N.Ma.), grants by the Takeda Science Foundation (N.Ma. and N.Mi.), and an Estonian Research Council grant PRG471 (K.Õ.). M.H.W. is supported by grant T32GM007748 (National Institutes of Health).
The Broad Center for Mendelian Genomics (UM1 HG008900) is funded by the National Human Genome Research Institute with supplemental funding provided by the National Heart, Lung, and Blood Institute under the Trans-Omics for Precision Medicine (TOPMed) program and the National Eye Institute.
Individuals 1 and 2 were part of the DDD study cohort. Acknowledgments of the DDD Study are included in the Supplemental Data.
Published: July 11, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.06.007.
Contributor Information
Lot Snijders Blok, Email: lot.snijdersblok@radboudumc.nl.
Simon E. Fisher, Email: simon.fisher@mpi.nl.
Web Resources
ClustalW, https://www.genome.jp/tools-bin/clustalw
Decipher, https://decipher.sanger.ac.uk
MetaDome, https://stuart.radboudumc.nl/metadome
cNLS Mapper, http://nls-mapper.iab.keio.ac.jp
OMIM, https://www.omim.org
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2
UniProt, https://www.uniprot.org
Supplemental Data
References
- 1.Sumiyama K., Washio-Watanabe K., Saitou N., Hayakawa T., Ueda S. Class III POU genes: Generation of homopolymeric amino acid repeats under GC pressure in mammals. J. Mol. Evol. 1996;43:170–178. doi: 10.1007/BF02338824. [DOI] [PubMed] [Google Scholar]
- 2.He X., Treacy M.N., Simmons D.M., Ingraham H.A., Swanson L.W., Rosenfeld M.G. Expression of a large family of POU-domain regulatory genes in mammalian brain development. Nature. 1989;340:35–41. doi: 10.1038/340035a0. [DOI] [PubMed] [Google Scholar]
- 3.Hagino-Yamagishi K., Saijoh Y., Ikeda M., Ichikawa M., Minamikawa-Tachino R., Hamada H. Predominant expression of Brn-2 in the postmitotic neurons of the developing mouse neocortex. Brain Res. 1997;752:261–268. doi: 10.1016/s0006-8993(96)01472-2. [DOI] [PubMed] [Google Scholar]
- 4.McEvilly R.J., de Diaz M.O., Schonemann M.D., Hooshmand F., Rosenfeld M.G. Transcriptional regulation of cortical neuron migration by POU domain factors. Science. 2002;295:1528–1532. doi: 10.1126/science.1067132. [DOI] [PubMed] [Google Scholar]
- 5.Sugitani Y., Nakai S., Minowa O., Nishi M., Jishage K., Kawano H., Mori K., Ogawa M., Noda T. Brn-1 and Brn-2 share crucial roles in the production and positioning of mouse neocortical neurons. Genes Dev. 2002;16:1760–1765. doi: 10.1101/gad.978002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominguez M.H., Ayoub A.E., Rakic P. POU-III transcription factors (Brn1, Brn2, and Oct6) influence neurogenesis, molecular identity, and migratory destination of upper-layer cells of the cerebral cortex. Cereb. Cortex. 2013;23:2632–2643. doi: 10.1093/cercor/bhs252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castro D.S., Skowronska-Krawczyk D., Armant O., Donaldson I.J., Parras C., Hunt C., Critchley J.A., Nguyen L., Gossler A., Göttgens B. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev. Cell. 2006;11:831–844. doi: 10.1016/j.devcel.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Dheedene A., Maes M., Vergult S., Menten B. A de novo POU3F3 deletion in a boy with intellectual disability and dysmorphic features. Mol. Syndromol. 2014;5:32–35. doi: 10.1159/000356060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kasher P.R., Schertz K.E., Thomas M., Jackson A., Annunziata S., Ballesta-Martinez M.J., Campeau P.M., Clayton P.E., Eaton J.L., Granata T. Small 6q16.1 deletions encompassing POU3F2 cause susceptibility to obesity and variable developmental delay with intellectual disability. Am. J. Hum. Genet. 2016;98:363–372. doi: 10.1016/j.ajhg.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhee J.M., Gruber C.A., Brodie T.B., Trieu M., Turner E.E. Highly cooperative homodimerization is a conserved property of neural POU proteins. J. Biol. Chem. 1998;273:34196–34205. doi: 10.1074/jbc.273.51.34196. [DOI] [PubMed] [Google Scholar]
- 11.Maricic T., Günther V., Georgiev O., Gehre S., Curlin M., Schreiweis C., Naumann R., Burbano H.A., Meyer M., Lalueza-Fox C. A recent evolutionary change affects a regulatory element in the human FOXP2 gene. Mol. Biol. Evol. 2013;30:844–852. doi: 10.1093/molbev/mss271. [DOI] [PubMed] [Google Scholar]
- 12.Sobreira N., Schiettecatte F., Valle D., Hamosh A. GeneMatcher: A matching tool for connecting investigators with an interest in the same gene. Hum. Mutat. 2015;36:928–930. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Firth H.V., Richards S.M., Bevan A.P., Clayton S., Corpas M., Rajan D., Van Vooren S., Moreau Y., Pettett R.M., Carter N.P. DECIPHER: Database of chromosomal imbalance and phenotype in humans using Ensembl resources. Am. J. Hum. Genet. 2009;84:524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holbrook J.A., Neu-Yilik G., Hentze M.W., Kulozik A.E. Nonsense-mediated decay approaches the clinic. Nat. Genet. 2004;36:801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 15.Chang Y.F., Imam J.S., Wilkinson M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 16.Li P., He X., Gerrero M.R., Mok M., Aggarwal A., Rosenfeld M.G. Spacing and orientation of bipartite DNA-binding motifs as potential functional determinants for POU domain factors. Genes Dev. 1993;7(12B):2483–2496. doi: 10.1101/gad.7.12b.2483. [DOI] [PubMed] [Google Scholar]
- 17.Phillips K., Luisi B. The virtuoso of versatility: POU proteins that flex to fit. J. Mol. Biol. 2000;302:1023–1039. doi: 10.1006/jmbi.2000.4107. [DOI] [PubMed] [Google Scholar]
- 18.Jauch R., Choo S.H., Ng C.K., Kolatkar P.R. Crystal structure of the dimeric Oct6 (POU3f1) POU domain bound to palindromic MORE DNA. Proteins. 2011;79:674–677. doi: 10.1002/prot.22916. [DOI] [PubMed] [Google Scholar]
- 19.Lai C.S., Fisher S.E., Hurst J.A., Vargha-Khadem F., Monaco A.P. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 20.Smit D.J., Smith A.G., Parsons P.G., Muscat G.E., Sturm R.A. Domains of Brn-2 that mediate homodimerization and interaction with general and melanocytic transcription factors. Eur. J. Biochem. 2000;267:6413–6422. doi: 10.1046/j.1432-1327.2000.01737.x. [DOI] [PubMed] [Google Scholar]
- 21.Deriziotis P., Graham S.A., Estruch S.B., Fisher S.E. Investigating protein-protein interactions in live cells using bioluminescence resonance energy transfer. J. Vis. Exp. 2014;87:e51438. doi: 10.3791/51438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reményi A., Tomilin A., Schöler H.R., Wilmanns M. Differential activity by DNA-induced quarternary structures of POU transcription factors. Biochem. Pharmacol. 2002;64:979–984. doi: 10.1016/s0006-2952(02)01164-4. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell P.J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 24.Westphal D.S., Riedhammer K.M., Kovacs-Nagy R., Meitinger T., Hoefele J., Wagner M. A de novo missense variant in POU3F2 identified in a child with global developmental delay. Neuropediatrics. 2018;49:401–404. doi: 10.1055/s-0038-1669926. [DOI] [PubMed] [Google Scholar]
- 25.Deriziotis P., Fisher S.E. Speech and language: Translating the genome. Trends Genet. 2017;33:642–656. doi: 10.1016/j.tig.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kosugi S., Hasebe M., Tomita M., Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.