(The American Journal of Human Genetics 105, 65–77; July 3, 2019)
In the originally published version of this article, Figure 3 erroneously appeared in place of Figure 4, and Figure 4 erroneously appeared in place of Figure 3. The article has been corrected online, and the authors apologize for this error.
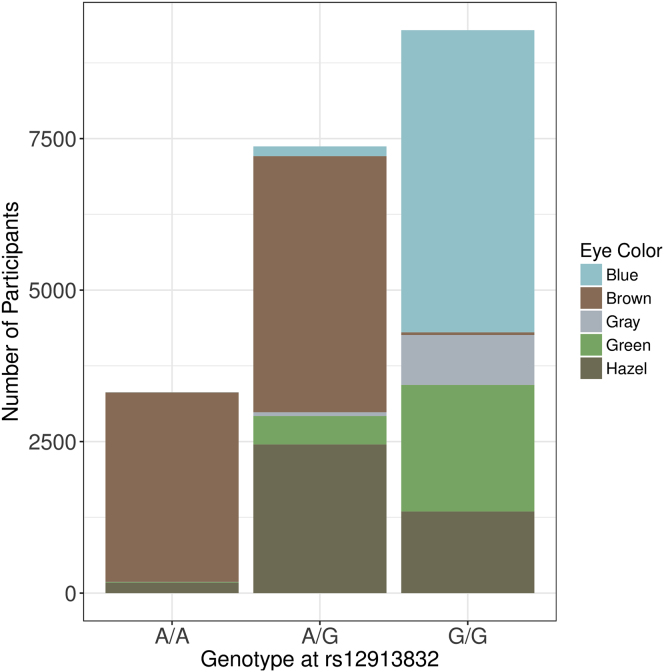
Figure 3.
Eye Color Distribution
Distribution of eye color among participants with different genotypes at rs12913832 (the top signal when performing GWAS using blue eye color in Genes for Good participants), a marker in HERC2 known to play a role in eye color determination.
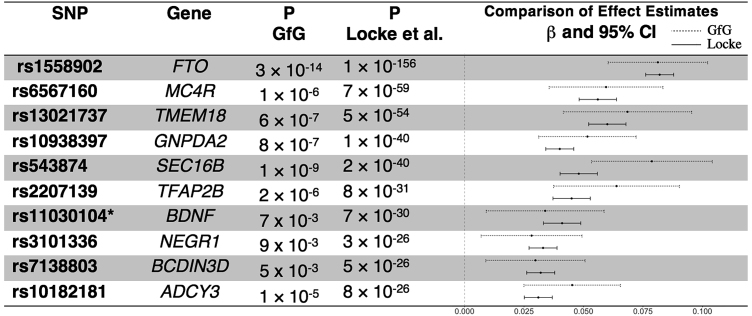
Figure 4.
Effect Size Estimates of a GWAS for BMI in Our Study Sample Compared to Findings from a Meta-analysis
We compare effect estimates from Genes for Good to published findings from the Locke et al. meta-analysis of BMI GWAS.24 Specifically, we looked at the top ten reported signals and were able to replicate all of these effects in direction and nominal significance (p < 0.05). The forest plot on the right compares effect size estimates across studies; the dashed lines represent the confidence intervals around the Genes for Good estimates, while the solid lines represent results from Locke et al. Given the relatively small sample size available in this data freeze, our estimates have fairly wide confidence limits. However, Locke’s estimates are completely contained within our limits for eight of ten SNPs. Asterisk indicates imputed variant.
Contributor Information
Anita Pandit, Email: anitapan@umich.edu.
Gonçalo R. Abecasis, Email: goncalo@umich.edu.