Abstract
Epstein-Barr virus (EBV) is a ubiquitous human γ-herpesvirus that infects over 90% of the global population. EBV is considered a contributory factor in a variety of malignancies including nasopharyngeal carcinoma, gastric carcinoma, Burkitt lymphoma, and Hodgkin’s lymphoma. Notably, EBV was the first virus found to encode microRNAs (miRNAs). Increasing evidence indicates that EBV-encoded miRNAs contribute to the carcinogenesis and development of EBV-associated malignancies. EBV miRNAs have been shown to inhibit the expression of genes involved in cell proliferation, apoptosis, invasion, and immune signaling pathways. Therefore, EBV miRNAs perform a significant function in the complex host-virus interaction and EBV-driven carcinogenesis. However, the integrated mechanisms underlying the roles of EBV miRNAs in carcinogenesis remain to be further explored. In this review, we describe recent advances regarding the involvement of EBV miRNAs in the pathogenesis of EBV-associated malignancies and discuss their potential utility as cancer biomarkers. An in-depth investigation into the pro-carcinogenic role of EBV miRNAs will expand our knowledge of the biological processes associated with virus-driven tumors and contribute to the development of novel therapeutic strategies for the treatment of EBV-associated malignancies.
Keywords: EBV, microRNA, carcinogenesis, cancer pathogenesis, EBV-associated malignancies, cancer biomarker
Main Text
Epstein-Barr virus (EBV), also called human herpesvirus 4 (HHV-4), is a ubiquitous member of the human herpesvirus family and infects over 90% of the global population.1 EBV was the first human tumor virus, and its infection is an etiologic factor in various epithelial and lymphoid malignancies, such as nasopharyngeal carcinoma (NPC), gastric carcinoma (GC), Burkitt lymphoma (BL), and Hodgkin’s lymphoma (HL).2, 3, 4 Despite its tight association with a variety of malignancies, primary EBV infection commonly causes no or slight symptoms in most lifelong carriers.5, 6 In infected cells, EBV can establish two alternative modes of infections known as latent or lytic.7 Upon primary infection, EBV transiently undergoes a short lytic program in the epithelial cells of the oropharynx and salivary glands, and subsequently establishes lifelong latent infection in memory B cells.8, 9 During latency, EBV expresses a limited number of viral latent genes, which is advantageous for its long-term persistence in infected cells.10 Infected B cells would switch from latency into the lytic stage upon stimulation.11 During the lytic stage, a repertoire of over 80 viral genes is expressed, accompanied by viral DNA replication, resulting in the production of progeny viruses.12 Both EBV latent and lytic infections have been shown to contribute to the carcinogenesis and development of EBV-associated tumors.13, 14, 15, 16, 17
MicroRNAs (miRNAs) are endogenous small non-coding RNAs (ncRNAs) that exert a powerful effect on gene regulation through inducing mRNA degradation or translational inhibition.18, 19 Accumulating evidence demonstrates that miRNAs are engaged in diverse physiological and pathological processes, including development, metabolism, and cancer.20, 21, 22 More importantly, miRNAs function as crucial players in the onset and progression of cancer.23, 24, 25 Intriguingly, EBV was the first virus in which viral miRNAs were detected.26 EBV can encode 44 mature miRNAs from 25 miRNA precursors.27 A growing body of evidence underlines the regulatory roles of EBV-encoded miRNAs in the carcinogenesis and progression of EBV-associated malignancies.28, 29 EBV miRNAs are capable of interfering with a wide range of biological processes, such as cell proliferation, apoptosis, transformation, and invasion. Moreover, EBV miRNAs can directly target immune-related genes and thus enable infected cells to evade immune destruction. In this review, we summarize the current understanding of the functional roles of EBV-encoded miRNAs in the pathogenesis of human malignancies. Increasing understanding of the involvement of EBV miRNAs in cancer progression may accelerate our efforts to comprehensively reveal the molecular mechanisms underlying EBV-induced carcinogenesis and propel the development of novel therapeutic measures for EBV-associated malignancies.
The Life Cycle of EBV
In infected cells, EBV adopts a biphasic life cycle consisting of latent and lytic phases.30 During the lytic cycle, the full repertoire of viral genes is expressed, and progeny viruses are produced.31 The lytic cycle allows the dissemination of virions among host cells. During the course of latency, EBV expresses a limited quantity of viral genes essential for the maintenance of its genome.32 Because only a few viral proteins that can be targeted by the host immune system are produced, the latent infection of EBV facilitates its survive and pathogenesis.33 According to the expression of latent genes, EBV latency can be categorized into four different types.5 Latency 0 is found only in healthy carriers, in which no viral proteins are generated or low levels of EBV nuclear antigen 1 (EBNA1) and latent membrane protein 2A (LMP2A) can be detected in B lymphocytes.34 BL generally presents a latency I type, in which only EBNA1 is expressed.35 In latency II, three LMPs (LMP1, LMP2A, and LMP2B) are expressed in addition to EBNA1.36 Moreover, latency II is tightly associated with NPC, HL, and natural killer (NK)/T cell lymphoma. Latency III is characterized by expression of all nuclear proteins (EBNA1, 2, 3A, 3B, 3C, and -LP), the three LMPs, and viral ncRNAs.37 Latency III is the least restrictive latency type and is commonly found in immunodeficiency-associated lymphoma, such as post-transplant lymphoproliferative disorder (PTLD) and AIDS-related lymphoma.
EBV-Encoded miRNAs
miRNAs are a highly conserved group of small ncRNAs, which are 21–25 nt in length.38 miRNAs can be derived from exons of non-coding genes, introns of coding and non-coding genes, and the intragenic regions.39 The canonical miRNA biogenesis pathway involves two processing steps that have been extensively defined in previous studies.40, 41, 42 In brief, the miRNA genes are transcribed to generate long primary miRNA (pri-miRNA) molecules by RNA polymerase II (Pol II) or Pol III.43, 44 The nascent pri-miRNAs are first processed into ∼70-nt precursor miRNAs (pre-miRNAs) by a microprocessor complex consisting of Drosha and DiGeorge syndrome critical region 8 (DGCR8) in the nucleus. Pre-miRNAs, characterized by hairpin structures, are then exported to the cytoplasm by exportin-5, where they are further processed by Dicer to yield mature miRNA duplexes (21–25 nt).45 One strand of the duplex is preferentially incorporated into the RNA-induced silencing complex (RISC) containing Argonaute (Ago), and is guided to target mRNAs.46 miRNAs mainly recognize complementary sequences in the 3′ UTRs of their target mRNAs.47 The interaction between miRNAs and mRNAs causes mRNA degradation or translation suppression, contributing to altered gene expression. miRNAs play a significant role in various biological processes including proliferation, apoptosis, and differentiation.48 They are also implicated in the occurrence and progression of cancer.49, 50 So far, the function and mechanisms of miRNAs in cancer pathogenesis have been disclosed. Based on their abundance and target genes, miRNAs are mainly divided into two categories: oncogenic and tumor-suppressive miRNAs.51
EBV is the first virus in which miRNAs are discovered.26 EBV encodes 44 miRNAs that are transcribed from two regions: BamHI-A region rightward transcript (BART) and BamHI fragment H rightward open reading frame 1 (BHRF1) clusters.52 BART transcript encodes 22 miRNA precursors (miR-BART1-22) with 40 mature miRNAs, whereas BHRF1 transcript expresses three miRNA precursors (miR-BHRF1-1, -2, and -3) producing four mature miRNAs.27 Intriguingly, EBV miRNAs are differentially expressed in an infection stage-dependent manner. The expression of BART miRNAs takes place in all types of latency and is more restricted to the latency types I and II.53 In contrast, BHRF1 miRNAs are abundantly expressed in type III latency and lytically infected cells, such as B lymphoma cells, but are nearly undetectable in cells under the latency types I and II.54, 55, 56 The expression difference between BART and BHRF1 miRNAs is attributed to distinct promoters responsible for their transcription.
Likewise, EBV miRNAs are differentially expressed in different types of cancer. Several studies were previously conducted to profile the expression of EBV-encoded miRNAs in cancer. miR-BART7, miR-BART9, miR-BART14*, miR-BART17-3p, and miR-BART22 were found to be the most abundant viral miRNAs in NPC samples.57 Zhu et al.58 revealed that miR-BART1, miR-BART4, miR-BART6, miR-BART7, and miR-BART11 were abundantly expressed in NPC. High levels of miR-BART3-3p, miR-BART4-5p, miR-BART5-5p, miR-BART7-3p, and miR-BART9-3p were detected in NPC samples.59 Microarray-based viral miRNA profile in NPC demonstrated that miR-BART1-3p, miR-BART3, miR-BART5, miR-BART7, and miR-BART8* showed high abundance.60 Moreover, EBV miRNAs accounted for approximately 19% of total miRNAs in one NPC sample. Another study also reported the expression profile of EBV miRNAs in NPC.61 A total of nine BART miRNAs (miR-BART1-5p, 3, 4, 5, 6-3p, 7, 8, 10, and 18-3p) were identified to be upregulated. Notably, all of these studies verified a significant upregulation of miR-BART7 in EBV-positive NPC, suggesting that miR-BART7 might play an important role in the carcinogenesis and progression of NPC. EBV miRNAs were demonstrated to account for about 15% of total miRNAs in EBV-infected AGS GC cell lines.62 The most abundant viral miRNAs in AGS cells were miR-BART6-3p, miR-BART7-5p, miR-BART8-5p, miR-BART10-3p, and miR-BART22. Expression of EBV miRNAs in EBV-associated GC (EBVaGC) samples and cell lines was further profiled by deep sequencing.63 miR-BART1-3p, miR-BART5-5p, miR-BART7-3p, miR-BART9-3p, and miR-BART10-3p had high abundance. Tsai et al.64 found that miR-BART2-5p, miR-BART4-5p, miR-BART6-3p, miR-BART9-3p, and miR-BART1s1-3p were the most abundant in EBVaGC samples and cell lines. Collectively, BART miRNAs account for a large proportion of total viral miRNAs in EBV-positive epithelial tumors, suggesting that BART miRNAs may be associated with the development of epithelial malignancies.
In EBV-positive diffuse large B cell lymphoma (DLBCL) samples, all BART miRNAs were detected with the exception of miR-BART15 and miR-BART22.65 Remarkably, miR-BART7, miR-BART10, miR-BART11-5p, miR-BART16, and miR-BART22 were the most abundantly expressed miRNAs. The most highly expressed viral miRNAs in NK/T cell lymphoma included miR-BART1-5p, miR-BART5, miR-BART7, miR-BART11-5p, and miR-BART19-3p.66 In endemic BL, miR-BART6-3p, miR-BART7, miR-BART10, miR-BART11-3p, and miR-BART17-5p were the most abundant EBV miRNAs.67 It seems that miR-BART7 and miR-BART11 act as vital participants in the pathogenesis of EBV-associated lymphoma. However, BHRF1 miRNAs were not found in all these types of lymphoma but could be detected in PTLD and AIDS-related DLBCL.68, 69 The deregulation of EBV miRNAs might contribute to viral infection and pathogenesis. It has been reported that the strain and type of EBV, and the background of infected cells may affect the expression level of viral miRNAs.70, 71 The transcription factors in infected cells can also modulate EBV miRNA expression. For instance, the nuclear factor-κB (NF-κB) signaling was able to upregulate the expression of BART miRNAs in NPC.72 Nevertheless, further studies are required to disclose the regulatory mechanisms of EBV miRNA expression. Furthermore, the association between aberrantly expressed viral miRNAs and EBV-associated malignancies remains to be systemically explored.
The Role of EBV miRNAs in Cancer Pathogenesis
EBV is a well-recognized carcinogen that is directly implicated in the etiology of human epithelial and lymphoid malignancies.73, 74 It is believed that EBV miRNAs perform a crucial role in the pathobiology of EBV-associated malignancies.75 In terms of mechanism, EBV miRNAs are able to modulate various cellular pathways such as cell proliferation, apoptosis, invasion, metastasis, and immune signaling by targeting cellular or viral mRNAs.
EBV miRNAs Control Cancer Cell Growth
EBV miRNAs are able to regulate cell proliferation or apoptosis, contributing to the carcinogenesis of EBV-associated malignancies. The previous report indicated that BART miRNAs could potentiate the seeding and growth of EBVaGC in vivo.76 Deregulated BART miRNAs were implicated in NPC tumorigenesis by interfering with signaling pathways associated with proliferation, apoptosis, invasion, and metastasis.77 Upregulated BART miRNAs (e.g., miR-BART3 and miR-BART5) were predicted to target genes involved in the p53, transforming growth factor-β (TGF-β), and Wnt signaling pathways, thus modulating NPC apoptosis and transformation.61 miR-BART7 could enhance the proliferation of NPC cells by affecting oncogenic pathways including the TGF signaling pathway.78 Collectively, these studies underlined the vital role of BART miRNAs in the tumorigenesis of EBV-associated epithelial malignancies. The contributions of EBV miRNAs to lymphomagenesis have also been investigated. Aberrantly expressed BART miRNAs in infected B cells could regulate genes associated with cell-cycle control, apoptosis, and the Wnt signaling.79 Specifically, most of the viral miRNAs co-targeted mRNAs with host miRNAs, such as oncogenic miRNAs (the miR-17∼92 cluster) and immunoregulatory miRNAs (miR-142-3p and miR-155). The viral miRNA targetome was identified in EBV-infected lymphoblastoid cell lines (LCLs).80 The results demonstrated that EBV miRNAs participated in various cellular processes that were directly associated with innate immunity, cell proliferation, and survival. The comprehensive survey of viral miRNA targetome would provide novel opportunities to discover effective therapeutic targets for EBV-associated malignancies.
One of the mechanisms underlying the oncogenic capability of EBV miRNAs involves the regulation of tumor suppressors. For instance, miR-BART10 could inhibit the expression of β-transducin repeat-containing E3 ubiquitin protein ligase (β-TrCP).81 The downregulation of β-TrCP by miR-BART10 might lead to reduced degradation of its substrates, including Snail and β-catenin, thus contributing to NPC development.82 miR-BART7-3p, a highly expressed miRNA in NPC, could promote tumor growth.83 In terms of mechanism, miR-BART7-3p stimulated the phosphatase and tensin homolog (PTEN)/phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt) pathway and induced the expression of oncogenic transcription factors c-Myc and c-Jun. miR-BART3* promoted the growth and transformation of NPC by downregulating the tumor suppressor deleted in cancer 1 (DICE1).84 miR-BART19-3p was shown to reduce the expression of Wnt inhibitory factor 1 (WIF1) and two tumor suppressors, Nemo-like kinase (NLK) and adenomatous polyposis coli (APC).85 Resultantly, miR-BART19-3p promoted GC cell proliferation. Both miR-BART6-3p and miR-142 decreased the expression of interleukin-6 receptor (IL-6R) and PTEN, thus promoting the proliferation of EBV-positive BL cells.86 Another study also verified that miR-BART6-3p could target IL-6R and PTEN in BL cells, thereby affecting their respective downstream pathways such as NF-κB and PI3K/Akt signalings.87 Thus, miR-BART-6-3p might serve as a novel therapeutic target for the treatment of EBV-positive BL. PR domain zinc-finger protein 1 (PRDM1/Blimp1), a master regulator of B cell terminal differentiation, worked as a tumor suppressor.88, 89 miR-BHRF1-2 promoted cell-cycle progression and inhibited cell apoptosis in LCL cells by downregulating PRDM1.90
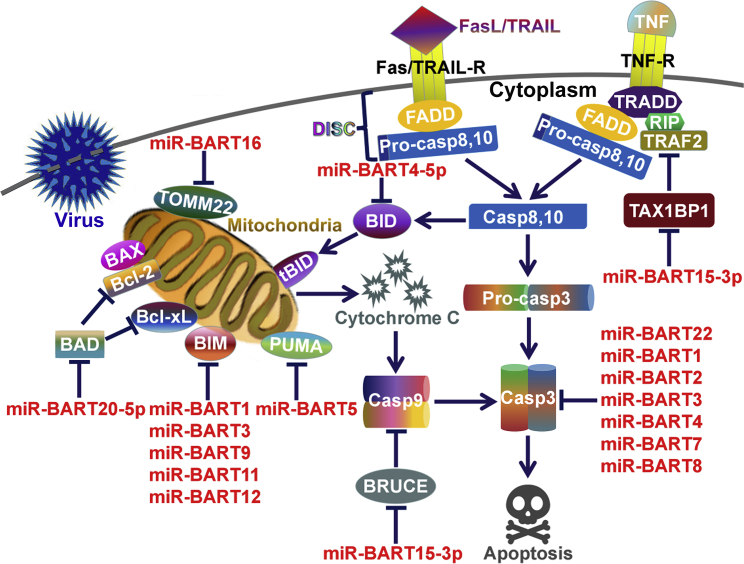
Alternatively, EBV BART miRNAs are able to target a variety of pro-apoptotic genes to promote tumor cell survival (Figure 1). miR-BART4-5p reduced GC cell apoptosis by inhibiting the expression of the pro-apoptotic protein BH3-interacting domain death agonist (BID).63 miR-BART16 interacted with the translocase of outer mitochondrial membrane 22 (TOMM22), leading to dissociation of the B cell lymphoma 2 (Bcl-2)-associated X protein (BAX) from mitochondria.91, 92 As a result, miR-BART16 functioned to inhibit BAX-induced apoptosis. miR-BART20-5p inhibited GC cell apoptosis by directly targeting Bcl-2-associated death promoter (BAD).93 Another study also uncovered the anti-apoptotic function of miR-BART20-5p in EBVaGC.94 Multiple BART miRNAs, such as miR-BART1, miR-BART3, miR-BART9, miR-BART11, and miR-BART12, could downregulate the Bcl-2-interacting mediator of cell death (BIM) in GC cells.95 EBV takes advantage of various mechanisms to downregulate BIM. It is proposed that inhibition of BIM-induced apoptosis is pivotal for viral persistence in cancer cells. miR-BART5 was upregulated in NPC and EBVaGC cells, and suppressed cancer cell apoptosis by negatively regulating the expression of p53 upregulated modulator of apoptosis (PUMA).96 miR-BART1, miR-BART2, miR-BART3, miR-BART4, miR-BART7, miR-BART8, and miR-BART22 were found to significantly repress the expression of the central executioner of apoptosis, caspase-3.97 Altogether, EBV miRNAs widely interfere with the apoptotic pathway in cancer cells.
Figure 1.
EBV miRNAs Control Cell Apoptosis
Death receptors Fas, tumor necrosis factor-related apoptosis-inducing ligand receptors (TRAIL-R), and tumor necrosis factor receptors (TNF-R) form death-inducing signaling complex (DISC) upon interacting with their ligands, consequently inducing the caspase cascade. Caspase-8 activation causes the cleavage of BID to tBID, which initiates the mitochondria-mediated intrinsic apoptosis pathway. EBV miRNAs regulate intrinsic apoptosis mainly through their gene-regulatory function. EBV miRNAs can repress the expression of pro-apoptotic proteins (BAD, BIM, TOMM22, BID, PUMA, and CASP3). One viral miRNA, miR-BART15-3p, is found to inhibit the expression of anti-apoptotic proteins (BRUCE and TAX1BP1). BAD, Bcl-2-associated death promoter; BAX, Bcl-2-associated X protein; Bcl-2, B cell lymphoma 2; Bcl-xL, B cell lymphoma-extra large; BID, the BH3-interacting domain death agonist; BIM, Bcl-2-interacting mediator of cell death; BRUCE, BIR repeat-containing ubiquitin-conjugating enzyme; FADD, Fas-associated protein with death domain; PUMA, p53 upregulated modulator of apoptosis; RIP, receptor-interacting protein; TAX1BP1, Tax1-binding protein 1; tBID, truncated BID; TOMM22, translocase of outer mitochondrial membrane 22; TRADD, TNFR1-associated death domain protein; TRAF2, TNF-R-associated factor 2.
However, EBV miRNAs can inhibit tumor cell growth by controlling anti-apoptotic proteins. For instance, miR-BART15-3p repressed GC cell proliferation and promoted apoptosis by descending the protein level of the BIR repeat-containing ubiquitin-conjugating enzyme (BRUCE).98 miR-BART15-3p also downregulated anti-apoptotic Tax1-binding protein 1 (TAX1BP1) in GC cells, leading to increased cell apoptosis.99 Because caspase-mediated cleavage of viral proteins favors viral replication and spreading,100 miR-BART15-3p might promote EBV lytic replication or release of progeny viruses by inducing cell apoptosis. EBV miRNAs may possess multifaceted functions in cell apoptosis. The functional roles of EBV miRNAs in cell apoptosis need more systematic survey. Moreover, the effects of EBV miRNA-regulated cell apoptosis on viral infection and carcinogenesis deserve deep investigation.
In addition, EBV miRNAs alter the expression of their own genes to control cancer cell proliferation. For instance, miR-BART9 promoted the proliferation of nasal NK/T cell lymphoma (NNKTL) cells by upregulating the oncogenic LMP1.101 EBV-encoded BHRF1 was a Bcl-2-related protein and prevented cell apoptosis partially by interacting with BIM.102 miR-BART10-3p could directly target BHRF1.79 Paradoxically, miR-BART10-3p suppressed BL cell apoptosis, which might be attributed to its inhibitory effect on apoptosis-associated proteins. Recently, EBV miRNAs have been found to be involved in the establishment of a favorable microenvironment for cancer growth. EBV infection and chronic inflammation were tightly linked with the progression of NPC and GC.103 miR-BART11 could directly target forkhead box P1 (FOXP1) to promote monocyte differentiation into macrophage.104 Accordingly, miR-BART11 enhanced the inflammatory response in NPC and GC, leading to accelerated proliferation of tumor cells. This study revealed a new EBV-dependent mechanism underlying the carcinogenesis of NPC and GC.
Collectively, EBV miRNAs are capable of modulating tumor cell proliferation through diverse mechanisms, such as silencing tumor suppressor genes, regulating cell apoptosis, controlling the function of viral oncogenic proteins, and establishing a tumor-promoting milieu. Based on the above studies, it can be concluded that EBV miRNAs can target multiple genes that are involved in a wide range of cellular pathways. Therefore, EBV miRNAs and their target genes constitute complicated regulatory networks during cancer progression. The profound impact of EBV miRNA-target gene-regulatory networks on the growth of EBV-driven malignancies is worthy of further investigation. An in-depth understanding of the effects of EBV miRNAs on cancer cell proliferation may provide insights into novel therapeutic strategies for EBV-associated malignancies.
EBV miRNAs Are Involved in Cell Transformation and Malignancy
Malignant transformation is a complex process by which normal cells convert into cancerous cells.105 Aberrant cell growth and immortalization are pivotal landmarks of malignant transformation and common characteristics of cancer cells. EBV infects human primary B cells and efficiently transforms them into actively proliferating, immortal LCLs.106 EBV BHRF1 miRNAs promoted cell-cycle progression and prevented apoptosis in B cells during the early infection phase.107 Feederle et al.108 found that B cells exposed to EBV mutant that lacked the BHRF1 miRNA cluster displayed reduced transforming capacity. The three BHRF1 miRNAs (miR-BHRF1-1, -2, and -3) were synergistically involved in EBV-induced B cell transformation.109 Mechanistically, BHRF1 miRNAs promoted B cell expansion by lowering the expression of latent genes, hence facilitating viral persistence in infected cells.108 Bernhardt et al.110 confirmed that EBV BHRF1 miRNA cluster promoted B cell transformation. They found that B cells infected with the virus lacking the BHRF1 miRNAs underwent more serious apoptosis and grew more slowly than cells infected with an intact virus. Moreover, the effect of BHRF1 miRNA cluster on cell proliferation was partly mediated by the viral protein BHRF1. At the early phase of EBV infection, BHRF1 miRNAs promoted cell division by stimulating BHRF1 transcription and lowering PTEN expression. During the late infection stage, these miRNAs induced the cleavage of primary BHRF1 transcripts to reduce BHRF1 expression and indirectly elevated p27 expression. This regulation resulted in a decreased propensity to neoplastic cell transformation. Reportedly, EBNA leader protein (EBNA-LP) was essential for B cell transformation.111 It was assumed that BHRF1 miRNAs could modulate EBNA-LP expression to a level optimum for B cell transformation.112 Surprisingly, a recent study indicated that the inhibition of BHRF1 miRNAs enhanced the adhesion and growth of EBV-infected B cells.113 EBV BHRF1 miRNAs may play dual roles in expansion and transformation of infected B cells. EBV has evolved its own miRNAs to modulate its well-known transforming capacity.
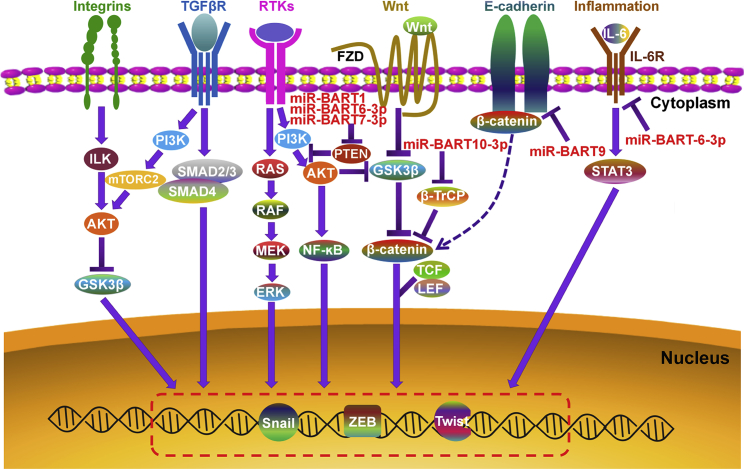
During transformation, epithelial cancer cells can acquire the mesenchymal phenotype via a process defined as epithelial-mesenchymal transition (EMT).114 EMT is associated with tumor invasion, dissemination, and metastasis.115, 116 EMT acts as a key driver of tumor malignancy. Intriguingly, EBV miRNAs are able to regulate the EMT process in cancer cells (Figure 2). miR-BART9 could suppress the expression of E-cadherin, thus promoting the EMT process in EBVaGC cells.64 Moreover, miR-BART9 negatively regulated the expression of the EMT-related miRNA, miR-200a.117 As expected, miR-BART9 played a crucial role in NPC invasion and migration by targeting E-cadherin.118 Therefore, miR-BART9 serves as a pro-metastatic miRNA contributing to the aggressiveness of EBV-associated epithelial tumors. miR-BART10-3p elevated the expression of the EMT markers β-catenin and Snail by targeting BTRC gene that encoded β-TrCP.82 miR-BART1 was able to drive the EMT program in NPC cells by activating PTEN-dependent pathways.119 The BART miRNA cluster 2 was responsible for the downregulation of N-myc downstream regulated gene 1 (NDRG1), an epithelial differentiation marker and inhibitor of tumor metastasis.120 miR-BART20-5p enhanced the invasion of invasive NNKTL cells by inhibiting T-bet translation and repressing p53 expression.121 On the contrary, some EBV miRNAs negatively modulate the EMT program in cancer cells. For instance, miR-BART6-3p suppressed the migration and invasion of NPC and GC cells by reversing the EMT process.122 The long ncRNA (lncRNA), LOC553103, might mediate the anti-migratory effects of miR-BART6-3p on cancer cells.
Figure 2.
EBV miRNAs Can Modulate the Cellular EMT Program
Multiple signaling pathways play a pivotal role in inducing EMT: (1) the integrin signaling can inhibit GSK3β via the ILK/AKT signaling. (2) TGF-βR is capable of activating SMAD2 and SMAD3. The activated SMAD2/3 interacts with SMAD4 to form a complex. The trimeric SMAD complex translocates to the nucleus and induces the transcription of EMT transcription factors (EMT-TFs). (3) RTKs can activate the RAS/RAF/MEK/ERK signaling cascade. Activated ERK can facilitate EMT by enhancing the expression of EMT-TFs. (4) The Wnt signaling promotes EMT by suppressing GSK3β to stabilize β-catenin. In the absence of Wnt, GSK3β constitutively induces the phosphorylation of cytoplasmic β-catenin, resulting in its degradation by the proteasome. Activation of the Wnt signaling causes the inhibition of GSK3β activity. Consequently, the stabilized β-catenin accumulates in the cytoplasm and enters the nucleus, where it forms a transcriptional-activating complex with TCF/LEF to initiate EMT transcriptional programs. (5) The cytoplasmic tails of E-cadherin are connected to β-catenin. Inhibition of E-cadherin causes a redistribution of β-catenin from the membrane to the nucleus, resulting in the activation of pro-EMT genes. (6) During inflammation, IL-6 is able to promote EMT by activating STAT3. EBV miRNAs interfere with the PI3K/Akt and IL-6 signaling to regulate the expression of pro-EMT genes. EBV miRNAs also activate β-catenin to promote EMT by inhibiting β-TrCP and E-cadherin. Akt, protein kinase B; ERK, extracellular signal-regulated kinase; FZD, frizzled receptor; GSK3β, glycogen synthase kinase-3β; IL-6, interleukin-6; ILK, integrin-linked kinase; LEF, lymphoid enhancer-binding factor; MEK, mitogen-activated protein kinase kinase; mTORC2, mammalian target of rapamycin (TOR) complex 2; NF-κB, nuclear factor-κB; PI3K, phosphoinositide 3-kinase; PTEN, phosphatase and tensin homolog; RTK, receptor tyrosine kinase; SMAD, small mothers against decapentaplegic homolog; STAT3, signal transducer and activator of transcription 3; TCF, T cell factor; TGF-βR, transforming growth factor-β family receptor; β-TrCP, β-transducin repeat-containing E3 ubiquitin protein ligase; ZEB, zinc-finger E-box-binding homeobox.
In summary, EBV miRNAs are widely involved in cell transformation, EMT, invasiveness, and migration of EBV-associated malignancies. These studies shed light on novel mechanisms adopted by the virus to facilitate cell malignant transformation. EBV miRNAs may act synergistically or antagonistically to influence the progression of EBV-associated malignances. However, it is unclear how the virus controls the dynamic balance between the promotive and inhibitory effects of its miRNAs on cancer cell invasion. Therefore, the function of EBV miRNAs in cell transformation and malignancy needs to be investigated in larger detail.
EBV miRNAs Contribute to Immune Escape
In addition to affecting the behaviors of cancer cells, EBV miRNAs can manipulate the host immune system. TGF-β is a potent immunosuppressive cytokine that modulates host immune responses to EBV infection by inhibiting the proliferation of activated B lymphocytes and preventing viral latency.123, 124 Multiple components of the TGF-β signaling pathway, including SMAD3, CREB-binding protein (CREBBP), FOS, and JUN, were confirmed to be inhibited by EBV BHRF1 miRNAs.125 A specific cluster of BART miRNAs (miR-BART2, miR-BART4, miR-BART5, miR-BART18, and miR-BART22) might exert their destructive activity via induction of cytokines that blocked the immune responses during EBV-induced carcinogenesis.126 miR-BART16 blocked IFN-α-mediated anti-proliferative function in BL cells by lowering the expression of CREBBP.127 The IL-1 signaling performs a significant role in inflammation and early activation of innate immune responses against viral infection.128, 129 miR-BHRF1-2-5p directly targeted IL-1 receptor 1 (IL-1R1), which disrupted cellular responsiveness to IL-1 stimulation and blocked cytokine expression within infected cells.130 miR-BHRF1-2-5p might attenuate excessive inflammation that could be detrimental to virus-infected cells, and thus facilitated cell survival for long-term viral persistence. miR-BART6-3p directly targeted IL-6R and dampened the host immune response, thus favoring virus-induced lymphomagenesis.87, 131 miR-BART3 lowered the expression of IL-6 by targeting importin 7 (IPO7).91 miR-BHRF1-3 enabled AIDS-related DLBCL cells to evade host immune surveillance by targeting C-X-C motif chemokine ligand 11 (CXCL-11).68 These findings suggest that EBV miRNAs have a broad impact on host cytokine signaling.
EBV miRNAs can govern the innate immune responses. Retinoic acid-inducible gene-I (RIG-I) functions as a pattern recognition receptor (PRR) to sense viral nucleic acids, thus playing an important role in initiating host innate immune response against viral infection.132 miR-BART6-3p inhibited EBV infection-triggered RIG-I-like receptor signaling and the type I interferon (IFN) response, favoring EBV infection and pathogenesis.133 NK cells can produce the prominent cytotoxic cytokine IFN-γ to inhibit viral proliferation and tumor growth.134, 135 NK cell-secreted IFN-γ activates the IFN-γ receptor to induce the phosphorylation of signal transducer and activator of transcription 1 (STAT1).136 miR-BART20-5p directly targeted IFN-γ, and miR-BART8 downregulated STAT1 in nasal NK cell lymphoma (NNL).137 BART miRNA-mediated inhibition of the IFN-γ/STAT1 pathway led to secondary suppression of TP53. In summary, miR-BART20-5p and miR-BART8 promoted NNL progression by targeting the IFN-γ/STAT1 pathway. Major histocompatibility complex (MHC) class I chain-related molecule B (MICB) is a stress-induced NK cell ligand and would be upregulated at the cell surface in the case of viral infection or tumor transformation.138 EBV miR-BART2-5p reduced the protein level of MICB, thus permitting infected cells to escape from immune recognition and NK cell-mediated killing.139 NF-κB could enhance the expression of BART miRNAs in NPC cells by inducing the BART promoters.72 The viral LMP1 could upregulate BART miRNAs by activating the NF-κB signaling. miR-BART5-5p in turn decreased the expression of LMP1. It was likely that deregulated NF-κB signaling and aberrant expression of BART miRNAs constituted an auto-modulatory loop for the maintenance of EBV latency and NPC progression. miR-BART15 was reported to limit inflammation by directly targeting NLR family pyrin domain-containing 3 (NLRP3).140
EBV miRNAs inhibit T cell responses to impair the host immune surveillance. EBV miRNAs can block CD4+ T cell response in infected B cells.141 miR-BART1, miR-BART2, and miR-BHRF1-2 reduced the release of IL-12 from infected cells and prevented the differentiation of T helper type 1 (Th1) cells. These miRNAs also attenuated the expression of IFN-γ-inducible protein 30 (IFI30), legumain (LGMN), and cathepsin B (CTSB), all of which are lysosomal enzymes implicated in MHC class II peptide processing.142 miR-BART3 and miR-BART16 were identified as viral miRNA inhibitors of LMP1 to suppress surface expression of adhesion molecules and immune co-receptors. Collectively, EBV miRNAs could inhibit MHC class II-mediated antigen processing and presentation, thus repressing the activation of CD4+ T cell response. On the other hand, EBV miRNAs abrogated CD8+ T cell response. For instance, miR-BART17 and miR-BHRF1-3 markedly inhibited the expression of transporter associated with antigen processing 2 (TAP2), which was involved in loading antigenic peptides onto MHC class I molecules.143, 144 In addition, EBV miRNAs could inhibit the expression of TAP1, the major MHC class I molecule, and EBNA1, a target of EBV-specific CD8+ T cells. LMP2A is a potent immunogenic antigen that can be recognized by cytotoxic T cells.145 miR-BART22 allowed infected cells to evade host immune surveillance and facilitated NPC tumorigenesis by targeting LMP2A.146 EBV miRNAs inhibit T cell-mediated immunity to promote immune evasion.
Undoubtedly, EBV miRNAs are critical regulators in the virus-host interaction. EBV miRNAs play a vital role in immune evasion by promoting the survival of infected cells and suppressing host immune responses. Nevertheless, identifying the targets of EBV miRNAs is only an initial step in understanding their function. The genuine impact of EBV miRNAs on the host immune system remains to be further validated. More studies are needed to comprehensively elucidate how EBV miRNAs function in immune evasion.
Extracellular Vesicle-Shuttled EBV miRNAs Play an Important Role in Cancer Pathogenesis
Extracellular vesicles (EVs) including exosomes can transfer nucleic acids and proteins among cells to govern multiple cellular processes.147 EBV infection can alter the nucleic acid cargo of EVs released from infected cells. The virus manages to load its own sets of miRNAs into EVs.148 EBV BART miRNAs could be secreted via EVs by LCLs and NPC cells.148, 149 Of note, BART1-5p, BART5, BART7-3p, BART12, and BART13 could be found in the circulating EVs from NPC patients.150 In recent years, the contributions of EV-carried EBV miRNAs to cancer pathogenesis have been studied. EBV miRNAs in EVs favor viral immune escape and provide a favorable microenvironment for the progression of EBV-associated malignancies by being transmitted to nearby immune cells. For example, EBV BHRF1 miRNAs could be transferred from infected B cells to T cells via EVs, thus leading to the suppression of target gene expression in recipient cells.151 EBV miR-BART15-3p was secreted by infected GC cells via exosomes and induced the apoptosis of recipient immune cells by targeting BRUCE.98 It will be intriguing to investigate how EBV miRNAs are incorporated into EVs. The mechanism behind the uptake of EVs by host cells remains unclear. Reportedly, exosomes derived from EBV-infected cells were internalized by recipient cells via caveola-dependent endocytosis.152 It is likely that receptor-mediated pathways may participate in the internalization of EBV-associated EVs. Additional studies are necessary to elucidate the EV internalization pathway. EBV miRNAs can regulate a wide variety of cellular targets and signaling pathways that are associated with carcinogenesis and cancer progression. Thus, EBV miRNAs delivered by EVs may control similar targets or signaling cascades in recipient cells. The EV-mediated intercellular cross-talk between EBV-positive tumor cells and immune cells merits further investigation.
EBV miRNAs could be transported from tumor cells to the bloodstream.150 EV-encapsulated EBV miRNAs possess various requisite characteristics of ideal biomarkers. Because of the wide existence of EVs in body fluids, these miRNAs are easily accessible. EBV miRNAs incorporated into EVs can be protected from ribonuclease degradation and stay stable in body fluids. Circulating EBV miRNAs could offer a variety of information about tumor progression and stage.153 Despite the great potential of circulating EBV miRNAs as cancer biomarkers, this field is still in its early stages. The abundance and clinical value of circulating EBV miRNAs in cancer patients deserve profound exploration. The age and physiological state of cancer patients, as well as medical intervention, may affect the expression profile of circulating EBV miRNAs. Thus, the kinetics of circulating viral miRNAs should be comprehensively monitored to correct for these alterations in future studies. Collectively, well-designed clinical studies with larger cohorts are urgently needed to support the clinical utility of circulating EBV miRNAs. It is expected that further studies on EV-shuttled EBV miRNAs will contribute to the development of better diagnostic or prognostic biomarkers for EBV-associated malignancies.
EBV miRNAs Serve as Potential Biomarkers in EBV-Associated Malignancies
EBV-related antibodies have been widely utilized as diagnostic markers for EBV-associated tumors.154 Nevertheless, it is still urgently needed to develop novel biomarkers for the early detection of EBV-associated tumors owing to the low positive predictive rate of EBV-related antibodies. Because BART miRNAs display high abundance in tumor cells, they hold great promise as novel biomarkers for early diagnosis of EBV-associated malignancies. A total of 12 BART miRNAs (miR-BART1-3p, 2-5p, 5, 6-3p, 6-5p, 7, 8, 9, 14, 17-5p, 18-5p, and 19-3p) were validated to be upregulated in NPC tissues versus their non-tumorous biopsies.155 Moreover, the abundance of these miRNAs in the sera of NPC patients was positively associated with their copy numbers in tumor cells. More detailed studies are warranted to elaborately assess the diagnostic value of EBV BART miRNAs in NPC. Another study showed that miR-BART3, miR-BART7, and miR-BART13 were highly expressed in EBV-positive NPC.156 miR-BART7 and miR-BART13 could distinguish NPC patients from healthy controls with high specificity and accuracy. These two miRNAs might represent effective serological biomarkers for NPC diagnosis and prognosis. Zheng et al.157 found that miR-BART1-5p displayed remarkably higher expression in NPC tissues than in controls. Specifically, the expression level of miR-BART1-5p was reflective of NPC progression. Detection of miR-BART1-5p was effective for diagnosing NPC at an early stage, even in cases that were misdiagnosed as negative based on conventional detection methods. miR-BART2-5p was found to be able to discriminate NPC patients from healthy controls.158 In addition, the high level of circulating miR-BART2-5p was correlated with tumor progression and poor prognosis in NNKTL patients.153 Therefore, miR-BART2-5p may be a promising biomarker for different types of EBV-associated tumor. Additional investigations are required to verify the clinical value of miR-BART2-5p in EBV-associated malignancies.
The expression levels of miR-BART1-5p, miR-BART4-5p, and miR-BART20-5p were remarkably higher in EBVaGC tumor tissues than those in paired normal tissues.159 Moreover, miR-BART20-5p upregulation was correlated with worse recurrence-free survival of EBVaGC patients. Therefore, miR-BART20-5p had the potential to be used as a biomarker for predicting recurrence-free survival in EBVaGC patients. Although a large number of studies have demonstrated the diagnostic value of EBV miRNAs in several cancers, EBV miRNAs have yet to be utilized as biomarkers for the clinical detection of EBV-associated malignancies. The major challenges in the clinical translation of EBV miRNAs are identifying specific or common viral miRNAs that could be utilized as authentic biomarkers for different types of cancer. Therefore, the clinical significance of EBV miRNAs in cancer awaits further verification. Large-scale clinical studies are demanded to impel their translational application. Moreover, it is necessary to develop standardized methods for the isolation and quantification of EBV miRNAs in cancer patients. In addition, combined detection of circulating EBV miRNAs and conventional biomarkers may be a promising approach for the diagnosis and prognosis of EBV-associated malignancies.
Strategies for EBV miRNA-Based Therapeutics in EBV-Associated Malignancies
EBV BART miRNAs are abundantly expressed in EBV-associated epithelial tumors and have been found to target multiple genes of both viral and cellular origins.96, 146, 159 Thus, BART miRNAs are tightly associated with the carcinogenesis and progression of EBV-associated epithelial tumors. Blocking the activity of BART miRNAs may specifically suppress the development of EBV-associated epithelial tumors. Currently, miRNA-targeting therapeutics is being developed. For example, the miRNA sponges, using EBV promoters such as EBER2 promoter, were reported to effectively silence specific genes in EBV-infected cells and might be useful in the targeting of EBV-positive NPC cells.160 The anti-miRNA oligonucleotides can be used to target EBV miRNAs. EBV miR-BART7-3p was highly expressed in NPC and promoted tumor growth in animal models.83 A therapeutic experiment by using gold nanoparticles carrying anti-EBV-miR-BART7-3p was conducted to evaluate the therapeutic efficacy of anti-EBV-miR-BART7-3p in vivo. The result showed that depletion of miR-BART7-3p inhibited in vivo growth of EBV-positive NPC cells, demonstrating the feasibility of utilizing nanoparticles to deliver therapeutic anti-miRNAs and silence endogenous viral miRNAs. EBV miR-BART7-3p might present a novel target for miRNA-based therapies. Thus, the miRNA-sponge or anti-miRNA oligonucleotide therapeutics may be suitable for targeting individual viral miRNA whose expression greatly contributes to the carcinogenesis of EBV-associated tumors. Although miRNA-based therapeutics is still in its infancy, these studies are encouraging and provide a direction for developing new therapeutic strategies for EBV-associated malignancies.
Concluding Remarks and Future Perspectives
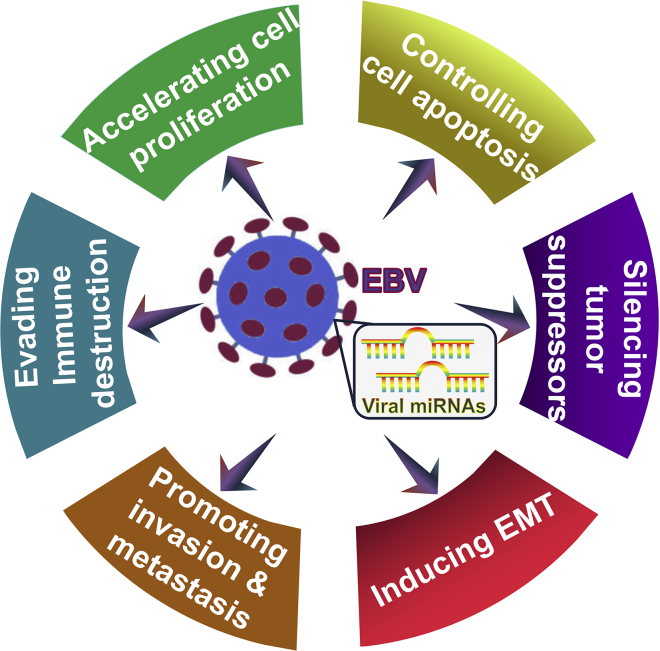
EBV-encoded miRNAs play an important role in the onset and progression of virus-associated malignancies by manipulating target gene expression. Accumulating evidence shows that EBV miRNAs exert a significant influence on cell proliferation and apoptosis, EMT, tumor invasion and metastasis, and immune escape (Figure 3). Although the targets and functional roles of EBV miRNAs have been disclosed, the detailed mechanisms of EBV miRNAs in carcinogenesis remain unclear. A single EBV miRNA can synchronously regulate diverse target genes, suggesting that complicated EBV miRNA-target gene-regulatory networks may exist during viral infection and pathogenesis. Comprehensive computational prediction together with experimental verification is essential to characterize the full targetome of EBV miRNAs, which will help to uncover the entire regulatory network underlying EBV-driven carcinogenesis. The oncogenes and tumor suppressor genes may be targeted by an identical viral miRNA. Therefore, the complex causative relationship between EBV miRNAs and carcinogenesis needs to be further studied. Collectively, increasing knowledge of the pivotal role of EBV miRNAs in cancer pathogenesis will provide a more detailed view of their contributions in EBV-driven malignancies and facilitate the identification of novel therapeutic targets for the clinical intervention of EBV-associated malignancies.
Figure 3.
Graphical Representation of Possible Mechanisms by which EBV miRNAs Contribute to the Progression of EBV-Associated Malignancies
EBV miRNAs can govern multiple cellular and signaling machineries. EBV miRNAs function in regulation of cell proliferation and apoptosis, silencing of tumor suppressors, promotion of the EMT program, invasion and metastasis, as well as subversion of host immune surveillance. Therefore, EBV miRNAs play an important role in the etiology of EBV-associated malignancies.
Author Contributions
M.W. and K.W. conceived and designed the study. M.W. wrote the manuscript. B.G. and X.C. prepared the figures. Y.W., P.L., and K.W. edited the manuscript. All authors have read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81701991) and Applied Basic Research Programs of Qingdao, China (17-1-1-59-jch).
Contributor Information
Man Wang, Email: wangman@qdu.edu.cn.
Kun Wang, Email: wangk696@163.com.
References
- 1.Cohen J.I., Fauci A.S., Varmus H., Nabel G.J. Epstein-Barr virus: an important vaccine target for cancer prevention. Sci. Transl. Med. 2011;3:107fs7. doi: 10.1126/scitranslmed.3002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jha H.C., Pei Y., Robertson E.S. Epstein-Barr Virus: Diseases Linked to Infection and Transformation. Front. Microbiol. 2016;7:1602. doi: 10.3389/fmicb.2016.01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Javier R.T., Butel J.S. The history of tumor virology. Cancer Res. 2008;68:7693–7706. doi: 10.1158/0008-5472.CAN-08-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ko Y.H. EBV and human cancer. Exp. Mol. Med. 2015;47:e130. doi: 10.1038/emm.2014.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young L.S., Rickinson A.B. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 6.Vockerodt M., Yap L.F., Shannon-Lowe C., Curley H., Wei W., Vrzalikova K., Murray P.G. The Epstein-Barr virus and the pathogenesis of lymphoma. J. Pathol. 2015;235:312–322. doi: 10.1002/path.4459. [DOI] [PubMed] [Google Scholar]
- 7.Li H., Hu J., Luo X., Bode A.M., Dong Z., Cao Y. Therapies based on targeting Epstein-Barr virus lytic replication for EBV-associated malignancies. Cancer Sci. 2018;109:2101–2108. doi: 10.1111/cas.13634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murata T., Tsurumi T. Switching of EBV cycles between latent and lytic states. Rev. Med. Virol. 2014;24:142–153. doi: 10.1002/rmv.1780. [DOI] [PubMed] [Google Scholar]
- 9.Cohen J.I. Epstein-Barr virus infection. N. Engl. J. Med. 2000;343:481–492. doi: 10.1056/NEJM200008173430707. [DOI] [PubMed] [Google Scholar]
- 10.Kenney S.C., Mertz J.E. Regulation of the latent-lytic switch in Epstein-Barr virus. Semin. Cancer Biol. 2014;26:60–68. doi: 10.1016/j.semcancer.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H., Liu S., Hu J., Luo X., Li N., M Bode A., Cao Y. Epstein-Barr virus lytic reactivation regulation and its pathogenic role in carcinogenesis. Int. J. Biol. Sci. 2016;12:1309–1318. doi: 10.7150/ijbs.16564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ressing M.E., van Gent M., Gram A.M., Hooykaas M.J., Piersma S.J., Wiertz E.J. Immune Evasion by Epstein-Barr Virus. Curr. Top. Microbiol. Immunol. 2015;391:355–381. doi: 10.1007/978-3-319-22834-1_12. [DOI] [PubMed] [Google Scholar]
- 13.Elgui de Oliveira D., Müller-Coan B.G., Pagano J.S. Viral Carcinogenesis Beyond Malignant Transformation: EBV in the Progression of Human Cancers. Trends Microbiol. 2016;24:649–664. doi: 10.1016/j.tim.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah K.M., Young L.S. Epstein-Barr virus and carcinogenesis: beyond Burkitt’s lymphoma. Clin. Microbiol. Infect. 2009;15:982–988. doi: 10.1111/j.1469-0691.2009.03033.x. [DOI] [PubMed] [Google Scholar]
- 15.Hong G.K., Gulley M.L., Feng W.H., Delecluse H.J., Holley-Guthrie E., Kenney S.C. Epstein-Barr virus lytic infection contributes to lymphoproliferative disease in a SCID mouse model. J. Virol. 2005;79:13993–14003. doi: 10.1128/JVI.79.22.13993-14003.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiorini S., Ooka T. Secretion of Epstein-Barr virus-encoded BARF1 oncoprotein from latently infected B cells. Virol. J. 2008;5:70. doi: 10.1186/1743-422X-5-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang C.Y., Lee C.H., Wu C.C., Chang Y.T., Yu S.L., Chou S.P., Huang P.T., Chen C.L., Hou J.W., Chang Y. Recurrent chemical reactivations of EBV promotes genome instability and enhances tumor progression of nasopharyngeal carcinoma cells. Int. J. Cancer. 2009;124:2016–2025. doi: 10.1002/ijc.24179. [DOI] [PubMed] [Google Scholar]
- 18.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 19.Grundhoff A., Sullivan C.S. Virus-encoded microRNAs. Virology. 2011;411:325–343. doi: 10.1016/j.virol.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wahid F., Shehzad A., Khan T., Kim Y.Y. MicroRNAs: synthesis, mechanism, function, and recent clinical trials. Biochim. Biophys. Acta. 2010;1803:1231–1243. doi: 10.1016/j.bbamcr.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 21.Wei F., Yang S., Wang S. MicroRNAs: a critical regulator under mechanical force. Histol. Histopathol. 2018;33:335–342. doi: 10.14670/HH-11-924. [DOI] [PubMed] [Google Scholar]
- 22.Jansson M.D., Lund A.H. MicroRNA and cancer. Mol. Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balatti V., Pekarky Y., Croce C.M. Role of microRNA in chronic lymphocytic leukemia onset and progression. J. Hematol. Oncol. 2015;8:12. doi: 10.1186/s13045-015-0112-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Bryan S., Dong S., Mathis J.M., Alahari S.K. The roles of oncogenic miRNAs and their therapeutic importance in breast cancer. Eur. J. Cancer. 2017;72:1–11. doi: 10.1016/j.ejca.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 25.Chen E., Xu X., Liu R., Liu T. Small but Heavy Role: MicroRNAs in Hepatocellular Carcinoma Progression. BioMed Res. Int. 2018;2018:6784607. doi: 10.1155/2018/6784607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfeffer S., Zavolan M., Grässer F.A., Chien M., Russo J.J., Ju J., John B., Enright A.J., Marks D., Sander C., Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 27.Barth S., Meister G., Grässer F.A. EBV-encoded miRNAs. Biochim. Biophys. Acta. 2011;1809:631–640. doi: 10.1016/j.bbagrm.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 28.Navari M., Etebari M., Ibrahimi M., Leoncini L., Piccaluga P.P. Pathobiologic Roles of Epstein-Barr Virus-Encoded MicroRNAs in Human Lymphomas. Int. J. Mol. Sci. 2018;19:1168. doi: 10.3390/ijms19041168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vojtechova Z., Tachezy R. The Role of miRNAs in Virus-Mediated Oncogenesis. Int. J. Mol. Sci. 2018;19:1217. doi: 10.3390/ijms19041217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murata T. Regulation of Epstein-Barr virus reactivation from latency. Microbiol. Immunol. 2014;58:307–317. doi: 10.1111/1348-0421.12155. [DOI] [PubMed] [Google Scholar]
- 31.Sinclair A.J. Epigenetic control of Epstein-Barr virus transcription—relevance to viral life cycle? Front. Genet. 2013;4:161. doi: 10.3389/fgene.2013.00161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrei G., Trompet E., Snoeck R. Novel Therapeutics for Epstein–Barr Virus. Molecules. 2019;24:e997. doi: 10.3390/molecules24050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dugan J.P., Coleman C.B., Haverkos B. Opportunities to Target the Life Cycle of Epstein-Barr Virus (EBV) in EBV-Associated Lymphoproliferative Disorders. Front. Oncol. 2019;9:127. doi: 10.3389/fonc.2019.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong M., Chen J.N., Huang J.T., Gong L.P., Shao C.K. The roles of EBV-encoded microRNAs in EBV-associated tumors. Crit. Rev. Oncol. Hematol. 2019;135:30–38. doi: 10.1016/j.critrevonc.2019.01.014. [DOI] [PubMed] [Google Scholar]
- 35.Rowe M., Rowe D.T., Gregory C.D., Young L.S., Farrell P.J., Rupani H., Rickinson A.B. Differences in B cell growth phenotype reflect novel patterns of Epstein-Barr virus latent gene expression in Burkitt’s lymphoma cells. EMBO J. 1987;6:2743–2751. doi: 10.1002/j.1460-2075.1987.tb02568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brooks L., Yao Q.Y., Rickinson A.B., Young L.S. Epstein-Barr virus latent gene transcription in nasopharyngeal carcinoma cells: coexpression of EBNA1, LMP1, and LMP2 transcripts. J. Virol. 1992;66:2689–2697. doi: 10.1128/jvi.66.5.2689-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorley-Lawson D.A., Gross A. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med. 2004;350:1328–1337. doi: 10.1056/NEJMra032015. [DOI] [PubMed] [Google Scholar]
- 38.He L., Hannon G.J. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 39.Shukla G.C., Singh J., Barik S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol. Cell. Pharmacol. 2011;3:83–92. [PMC free article] [PubMed] [Google Scholar]
- 40.Hammond S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015;87:3–14. doi: 10.1016/j.addr.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chong M.M., Zhang G., Cheloufi S., Neubert T.A., Hannon G.J., Littman D.R. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24:1951–1960. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daugaard I., Hansen T.B. Biogenesis and Function of Ago-Associated RNAs. Trends Genet. 2017;33:208–219. doi: 10.1016/j.tig.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 43.Lee Y., Kim M., Han J., Yeom K.H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borchert G.M., Lanier W., Davidson B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 45.Shivdasani R.A. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108:3646–3653. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berkhout B., Jeang K.T. RISCy business: MicroRNAs, pathogenesis, and viruses. J. Biol. Chem. 2007;282:26641–26645. doi: 10.1074/jbc.R700023200. [DOI] [PubMed] [Google Scholar]
- 47.Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li M., Marin-Muller C., Bharadwaj U., Chow K.H., Yao Q., Chen C. MicroRNAs: control and loss of control in human physiology and disease. World J. Surg. 2009;33:667–684. doi: 10.1007/s00268-008-9836-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng Y., Croce C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hayes J., Peruzzi P.P., Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol. Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 51.Adams B.D., Anastasiadou E., Esteller M., He L., Slack F.J. The Inescapable Influence of Noncoding RNAs in Cancer. Cancer Res. 2015;75:5206–5210. doi: 10.1158/0008-5472.CAN-15-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klinke O., Feederle R., Delecluse H.J. Genetics of Epstein-Barr virus microRNAs. Semin. Cancer Biol. 2014;26:52–59. doi: 10.1016/j.semcancer.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 53.Kang M.S., Kieff E. Epstein-Barr virus latent genes. Exp. Mol. Med. 2015;47:e131. doi: 10.1038/emm.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Navari M., Fuligni F., Laginestra M.A., Etebari M., Ambrosio M.R., Sapienza M.R., Rossi M., De Falco G., Gibellini D., Tripodo C. Molecular signature of Epstein Barr virus-positive Burkitt lymphoma and post-transplant lymphoproliferative disorder suggest different roles for Epstein Barr virus. Front. Microbiol. 2014;5:728. doi: 10.3389/fmicb.2014.00728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakamoto K., Sekizuka T., Uehara T., Hishima T., Mine S., Fukumoto H., Sato Y., Hasegawa H., Kuroda M., Katano H. Next-generation sequencing of miRNAs in clinical samples of Epstein-Barr virus-associated B-cell lymphomas. Cancer Med. 2017;6:605–618. doi: 10.1002/cam4.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amoroso R., Fitzsimmons L., Thomas W.A., Kelly G.L., Rowe M., Bell A.I. Quantitative studies of Epstein-Barr virus-encoded microRNAs provide novel insights into their regulation. J. Virol. 2011;85:996–1010. doi: 10.1128/JVI.01528-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cosmopoulos K., Pegtel M., Hawkins J., Moffett H., Novina C., Middeldorp J., Thorley-Lawson D.A. Comprehensive profiling of Epstein-Barr virus microRNAs in nasopharyngeal carcinoma. J. Virol. 2009;83:2357–2367. doi: 10.1128/JVI.02104-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu J.Y., Pfuhl T., Motsch N., Barth S., Nicholls J., Grässer F., Meister G. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J. Virol. 2009;83:3333–3341. doi: 10.1128/JVI.01689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen S.J., Chen G.H., Chen Y.H., Liu C.Y., Chang K.P., Chang Y.S., Chen H.C. Characterization of Epstein-Barr virus miRNAome in nasopharyngeal carcinoma by deep sequencing. PLoS ONE. 2010;5:e12745. doi: 10.1371/journal.pone.0012745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gourzones C., Jimenez A.S., Busson P. Profiling of Epstein-Barr virus-encoded microRNAs in nasopharyngeal carcinoma reveals potential biomarkers and oncomirs. Cancer. 2012;118:4634. doi: 10.1002/cncr.26514. author reply 4634–4635. [DOI] [PubMed] [Google Scholar]
- 61.Wan X.X., Yi H., Qu J.Q., He Q.Y., Xiao Z.Q. Integrated analysis of the differential cellular and EBV miRNA expression profiles in microdissected nasopharyngeal carcinoma and non-cancerous nasopharyngeal tissues. Oncol. Rep. 2015;34:2585–2601. doi: 10.3892/or.2015.4237. [DOI] [PubMed] [Google Scholar]
- 62.Marquitz A.R., Mathur A., Chugh P.E., Dittmer D.P., Raab-Traub N. Expression profile of microRNAs in Epstein-Barr virus-infected AGS gastric carcinoma cells. J. Virol. 2014;88:1389–1393. doi: 10.1128/JVI.02662-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shinozaki-Ushiku A., Kunita A., Isogai M., Hibiya T., Ushiku T., Takada K., Fukayama M. Profiling of Virus-Encoded MicroRNAs in Epstein-Barr Virus-Associated Gastric Carcinoma and Their Roles in Gastric Carcinogenesis. J. Virol. 2015;89:5581–5591. doi: 10.1128/JVI.03639-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsai C.Y., Liu Y.Y., Liu K.H., Hsu J.T., Chen T.C., Chiu C.T., Yeh T.S. Comprehensive profiling of virus microRNAs of Epstein-Barr virus-associated gastric carcinoma: highlighting the interactions of ebv-Bart9 and host tumor cells. J. Gastroenterol. Hepatol. 2017;32:82–91. doi: 10.1111/jgh.13432. [DOI] [PubMed] [Google Scholar]
- 65.Imig J., Motsch N., Zhu J.Y., Barth S., Okoniewski M., Reineke T., Tinguely M., Faggioni A., Trivedi P., Meister G. microRNA profiling in Epstein-Barr virus-associated B-cell lymphoma. Nucleic Acids Res. 2011;39:1880–1893. doi: 10.1093/nar/gkq1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Motsch N., Alles J., Imig J., Zhu J., Barth S., Reineke T., Tinguely M., Cogliatti S., Dueck A., Meister G. MicroRNA profiling of Epstein-Barr virus-associated NK/T-cell lymphomas by deep sequencing. PLoS ONE. 2012;7:e42193. doi: 10.1371/journal.pone.0042193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oduor C.I., Movassagh M., Kaymaz Y., Chelimo K., Otieno J., Ong’echa J.M., Moormann A.M., Bailey J.A. Human and Epstein-Barr Virus miRNA Profiling as Predictive Biomarkers for Endemic Burkitt Lymphoma. Front. Microbiol. 2017;8:501. doi: 10.3389/fmicb.2017.00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xia T., O’Hara A., Araujo I., Barreto J., Carvalho E., Sapucaia J.B., Ramos J.C., Luz E., Pedroso C., Manrique M. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3. Cancer Res. 2008;68:1436–1442. doi: 10.1158/0008-5472.CAN-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fink S.E., Gandhi M.K., Nourse J.P., Keane C., Jones K., Crooks P., Jöhrens K., Korfel A., Schmidt H., Neumann S. A comprehensive analysis of the cellular and EBV-specific microRNAome in primary CNS PTLD identifies different patterns among EBV-associated tumors. Am. J. Transplant. 2014;14:2577–2587. doi: 10.1111/ajt.12858. [DOI] [PubMed] [Google Scholar]
- 70.Tsai M.H., Lin X., Shumilov A., Bernhardt K., Feederle R., Poirey R., Kopp-Schneider A., Pereira B., Almeida R., Delecluse H.J. The biological properties of different Epstein-Barr virus strains explain their association with various types of cancers. Oncotarget. 2017;8:10238–10254. doi: 10.18632/oncotarget.14380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Correia S., Palser A., Elgueta Karstegl C., Middeldorp J.M., Ramayanti O., Cohen J.I., Hildesheim A., Fellner M.D., Wiels J., White R.E. Natural Variation of Epstein-Barr Virus Genes, Proteins, and Primary MicroRNA. J. Virol. 2017;91 doi: 10.1128/JVI.00375-17. e00375-e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Verhoeven R.J., Tong S., Zhang G., Zong J., Chen Y., Jin D.Y., Chen M.R., Pan J., Chen H. NF-κB Signaling Regulates Expression of Epstein-Barr Virus BART MicroRNAs and Long Noncoding RNAs in Nasopharyngeal Carcinoma. J. Virol. 2016;90:6475–6488. doi: 10.1128/JVI.00613-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thompson M.P., Kurzrock R. Epstein-Barr virus and cancer. Clin. Cancer Res. 2004;10:803–821. doi: 10.1158/1078-0432.ccr-0670-3. [DOI] [PubMed] [Google Scholar]
- 74.Khan G., Hashim M.J. Global burden of deaths from Epstein-Barr virus attributable malignancies 1990-2010. Infect. Agent. Cancer. 2014;9:38. doi: 10.1186/1750-9378-9-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Skalsky R.L., Cullen B.R. EBV Noncoding RNAs. Curr. Top. Microbiol. Immunol. 2015;391:181–217. doi: 10.1007/978-3-319-22834-1_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qiu J., Smith P., Leahy L., Thorley-Lawson D.A. The Epstein-Barr virus encoded BART miRNAs potentiate tumor growth in vivo. PLoS Pathog. 2015;11:e1004561. doi: 10.1371/journal.ppat.1004561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xie Y.J., Long Z.F., He X.S. Involvement of EBV-encoded BART-miRNAs and dysregulated cellular miRNAs in nasopharyngeal carcinoma genesis. Asian Pac. J. Cancer Prev. 2013;14:5637–5644. doi: 10.7314/apjcp.2013.14.10.5637. [DOI] [PubMed] [Google Scholar]
- 78.Chan J.Y., Gao W., Ho W.K., Wei W.I., Wong T.S. Overexpression of Epstein-Barr virus-encoded microRNA-BART7 in undifferentiated nasopharyngeal carcinoma. Anticancer Res. 2012;32:3201–3210. [PubMed] [Google Scholar]
- 79.Riley K.J., Rabinowitz G.S., Yario T.A., Luna J.M., Darnell R.B., Steitz J.A. EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency. EMBO J. 2012;31:2207–2221. doi: 10.1038/emboj.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Skalsky R.L., Corcoran D.L., Gottwein E., Frank C.L., Kang D., Hafner M., Nusbaum J.D., Feederle R., Delecluse H.J., Luftig M.A. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012;8:e1002484. doi: 10.1371/journal.ppat.1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zeng Z., Huang H., Huang L., Sun M., Yan Q., Song Y., Wei F., Bo H., Gong Z., Zeng Y. Regulation network and expression profiles of Epstein-Barr virus-encoded microRNAs and their potential target host genes in nasopharyngeal carcinomas. Sci. China Life Sci. 2014;57:315–326. doi: 10.1007/s11427-013-4577-y. [DOI] [PubMed] [Google Scholar]
- 82.Yan Q., Zeng Z., Gong Z., Zhang W., Li X., He B., Song Y., Li Q., Zeng Y., Liao Q. EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC. Oncotarget. 2015;6:41766–41782. doi: 10.18632/oncotarget.6155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cai L., Li J., Zhang X., Lu Y., Wang J., Lyu X., Chen Y., Liu J., Cai H., Wang Y., Li X. Gold nano-particles (AuNPs) carrying anti-EBV-miR-BART7-3p inhibit growth of EBV-positive nasopharyngeal carcinoma. Oncotarget. 2015;6:7838–7850. doi: 10.18632/oncotarget.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lei T., Yuen K.S., Xu R., Tsao S.W., Chen H., Li M., Kok K.H., Jin D.Y. Targeting of DICE1 tumor suppressor by Epstein-Barr virus-encoded miR-BART3* microRNA in nasopharyngeal carcinoma. Int. J. Cancer. 2013;133:79–87. doi: 10.1002/ijc.28007. [DOI] [PubMed] [Google Scholar]
- 85.Zhao Z., Liu W., Liu J., Wang J., Luo B. The effect of EBV on WIF1, NLK, and APC gene methylation and expression in gastric carcinoma and nasopharyngeal cancer. J. Med. Virol. 2017;89:1844–1851. doi: 10.1002/jmv.24863. [DOI] [PubMed] [Google Scholar]
- 86.Zhou L., Bu Y., Liang Y., Zhang F., Zhang H., Li S. Epstein-Barr Virus (EBV)-BamHI-A Rightward Transcript (BART)-6 and Cellular MicroRNA-142 Synergistically Compromise Immune Defense of Host Cells in EBV-Positive Burkitt Lymphoma. Med. Sci. Monit. 2016;22:4114–4120. doi: 10.12659/MSM.897306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ambrosio M.R., Navari M., Di Lisio L., Leon E.A., Onnis A., Gazaneo S., Mundo L., Ulivieri C., Gomez G., Lazzi S. The Epstein Barr-encoded BART-6-3p microRNA affects regulation of cell growth and immuno response in Burkitt lymphoma. Infect. Agent. Cancer. 2014;9:12. doi: 10.1186/1750-9378-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Boi M., Zucca E., Inghirami G., Bertoni F. PRDM1/BLIMP1: a tumor suppressor gene in B and T cell lymphomas. Leuk. Lymphoma. 2015;56:1223–1228. doi: 10.3109/10428194.2014.953155. [DOI] [PubMed] [Google Scholar]
- 89.Mandelbaum J., Bhagat G., Tang H., Mo T., Brahmachary M., Shen Q., Chadburn A., Rajewsky K., Tarakhovsky A., Pasqualucci L., Dalla-Favera R. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18:568–579. doi: 10.1016/j.ccr.2010.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ma J., Nie K., Redmond D., Liu Y., Elemento O., Knowles D.M., Tam W. EBV-miR-BHRF1-2 targets PRDM1/Blimp1: potential role in EBV lymphomagenesis. Leukemia. 2016;30:594–604. doi: 10.1038/leu.2015.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dölken L., Malterer G., Erhard F., Kothe S., Friedel C.C., Suffert G., Marcinowski L., Motsch N., Barth S., Beitzinger M. Systematic analysis of viral and cellular microRNA targets in cells latently infected with human gamma-herpesviruses by RISC immunoprecipitation assay. Cell Host Microbe. 2010;7:324–334. doi: 10.1016/j.chom.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 92.Bellot G., Cartron P.F., Er E., Oliver L., Juin P., Armstrong L.C., Bornstein P., Mihara K., Manon S., Vallette F.M. TOM22, a core component of the mitochondria outer membrane protein translocation pore, is a mitochondrial receptor for the proapoptotic protein Bax. Cell Death Differ. 2007;14:785–794. doi: 10.1038/sj.cdd.4402055. [DOI] [PubMed] [Google Scholar]
- 93.Kim H., Choi H., Lee S.K. Epstein-Barr virus miR-BART20-5p regulates cell proliferation and apoptosis by targeting BAD. Cancer Lett. 2015;356(2 Pt B):733–742. doi: 10.1016/j.canlet.2014.10.023. [DOI] [PubMed] [Google Scholar]
- 94.Kim H., Choi H., Lee S.K. Epstein-Barr Virus MicroRNA miR-BART20-5p Suppresses Lytic Induction by Inhibiting BAD-Mediated caspase-3-Dependent Apoptosis. J. Virol. 2015;90:1359–1368. doi: 10.1128/JVI.02794-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Marquitz A.R., Mathur A., Nam C.S., Raab-Traub N. The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim. Virology. 2011;412:392–400. doi: 10.1016/j.virol.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Choy E.Y., Siu K.L., Kok K.H., Lung R.W., Tsang C.M., To K.F., Kwong D.L., Tsao S.W., Jin D.Y. An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival. J. Exp. Med. 2008;205:2551–2560. doi: 10.1084/jem.20072581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harold C., Cox D., Riley K.J. Epstein-Barr viral microRNAs target caspase 3. Virol. J. 2016;13:145. doi: 10.1186/s12985-016-0602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Choi H., Lee H., Kim S.R., Gho Y.S., Lee S.K. Epstein-Barr virus-encoded microRNA BART15-3p promotes cell apoptosis partially by targeting BRUCE. J. Virol. 2013;87:8135–8144. doi: 10.1128/JVI.03159-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Choi H., Lee S.K. TAX1BP1 downregulation by EBV-miR-BART15-3p enhances chemosensitivity of gastric cancer cells to 5-FU. Arch. Virol. 2017;162:369–377. doi: 10.1007/s00705-016-3109-z. [DOI] [PubMed] [Google Scholar]
- 100.Richard A., Tulasne D. Caspase cleavage of viral proteins, another way for viruses to make the best of apoptosis. Cell Death Dis. 2012;3:e277. doi: 10.1038/cddis.2012.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ramakrishnan R., Donahue H., Garcia D., Tan J., Shimizu N., Rice A.P., Ling P.D. Epstein-Barr virus BART9 miRNA modulates LMP1 levels and affects growth rate of nasal NK T cell lymphomas. PLoS ONE. 2011;6:e27271. doi: 10.1371/journal.pone.0027271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Desbien A.L., Kappler J.W., Marrack P. The Epstein-Barr virus Bcl-2 homolog, BHRF1, blocks apoptosis by binding to a limited amount of Bim. Proc. Natl. Acad. Sci. USA. 2009;106:5663–5668. doi: 10.1073/pnas.0901036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fukayama M., Ushiku T. Epstein-Barr virus-associated gastric carcinoma. Pathol. Res. Pract. 2011;207:529–537. doi: 10.1016/j.prp.2011.07.004. [DOI] [PubMed] [Google Scholar]
- 104.Song Y., Li X., Zeng Z., Li Q., Gong Z., Liao Q., Li X., Chen P., Xiang B., Zhang W. Epstein-Barr virus encoded miR-BART11 promotes inflammation-induced carcinogenesis by targeting FOXP1. Oncotarget. 2016;7:36783–36799. doi: 10.18632/oncotarget.9170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kumar S., Weaver V.M. Mechanics, malignancy, and metastasis: the force journey of a tumor cell. Cancer Metastasis Rev. 2009;28:113–127. doi: 10.1007/s10555-008-9173-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Allday M.J., Sinclair A., Parker G., Crawford D.H., Farrell P.J. Epstein-Barr virus efficiently immortalizes human B cells without neutralizing the function of p53. EMBO J. 1995;14:1382–1391. doi: 10.1002/j.1460-2075.1995.tb07124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Seto E., Moosmann A., Grömminger S., Walz N., Grundhoff A., Hammerschmidt W. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010;6:e1001063. doi: 10.1371/journal.ppat.1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Feederle R., Linnstaedt S.D., Bannert H., Lips H., Bencun M., Cullen B.R., Delecluse H.J. A viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog. 2011;7:e1001294. doi: 10.1371/journal.ppat.1001294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Feederle R., Haar J., Bernhardt K., Linnstaedt S.D., Bannert H., Lips H., Cullen B.R., Delecluse H.J. The members of an Epstein-Barr virus microRNA cluster cooperate to transform B lymphocytes. J. Virol. 2011;85:9801–9810. doi: 10.1128/JVI.05100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bernhardt K., Haar J., Tsai M.H., Poirey R., Feederle R., Delecluse H.J. A Viral microRNA Cluster Regulates the Expression of PTEN, p27 and of a bcl-2 Homolog. PLoS Pathog. 2016;12:e1005405. doi: 10.1371/journal.ppat.1005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Szymula A., Palermo R.D., Bayoumy A., Groves I.J., Ba Abdullah M., Holder B., White R.E. Epstein-Barr virus nuclear antigen EBNA-LP is essential for transforming naïve B cells, and facilitates recruitment of transcription factors to the viral genome. PLoS Pathog. 2018;14:e1006890. doi: 10.1371/journal.ppat.1006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Poling B.C., Price A.M., Luftig M.A., Cullen B.R. The Epstein-Barr virus miR-BHRF1 microRNAs regulate viral gene expression in cis. Virology. 2017;512:113–123. doi: 10.1016/j.virol.2017.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Mo X., Wei F., Tong Y., Ding L., Zhu Q., Du S., Tan F., Zhu C., Wang Y., Yu Q. Lactic Acid Downregulates Viral MicroRNA To Promote Epstein-Barr Virus-Immortalized B Lymphoblastic Cell Adhesion and Growth. J. Virol. 2018;92 doi: 10.1128/JVI.00033-18. e00033-e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hugo H., Ackland M.L., Blick T., Lawrence M.G., Clements J.A., Williams E.D., Thompson E.W. Epithelial—mesenchymal and mesenchymal—epithelial transitions in carcinoma progression. J. Cell. Physiol. 2007;213:374–383. doi: 10.1002/jcp.21223. [DOI] [PubMed] [Google Scholar]
- 116.Valastyan S., Weinberg R.A. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shinozaki A., Sakatani T., Ushiku T., Hino R., Isogai M., Ishikawa S., Uozaki H., Takada K., Fukayama M. Downregulation of microRNA-200 in EBV-associated gastric carcinoma. Cancer Res. 2010;70:4719–4727. doi: 10.1158/0008-5472.CAN-09-4620. [DOI] [PubMed] [Google Scholar]
- 118.Hsu C.Y., Yi Y.H., Chang K.P., Chang Y.S., Chen S.J., Chen H.C. The Epstein-Barr virus-encoded microRNA MiR-BART9 promotes tumor metastasis by targeting E-cadherin in nasopharyngeal carcinoma. PLoS Pathog. 2014;10:e1003974. doi: 10.1371/journal.ppat.1003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cai L., Ye Y., Jiang Q., Chen Y., Lyu X., Li J., Wang S., Liu T., Cai H., Yao K. Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma. Nat. Commun. 2015;6:7353. doi: 10.1038/ncomms8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kanda T., Miyata M., Kano M., Kondo S., Yoshizaki T., Iizasa H. Clustered microRNAs of the Epstein-Barr virus cooperatively downregulate an epithelial cell-specific metastasis suppressor. J. Virol. 2015;89:2684–2697. doi: 10.1128/JVI.03189-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lin T.C., Liu T.Y., Hsu S.M., Lin C.W. Epstein-Barr virus-encoded miR-BART20-5p inhibits T-bet translation with secondary suppression of p53 in invasive nasal NK/T-cell lymphoma. Am. J. Pathol. 2013;182:1865–1875. doi: 10.1016/j.ajpath.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 122.He B., Li W., Wu Y., Wei F., Gong Z., Bo H., Wang Y., Li X., Xiang B., Guo C. Epstein-Barr virus-encoded miR-BART6-3p inhibits cancer cell metastasis and invasion by targeting long non-coding RNA LOC553103. Cell Death Dis. 2016;7:e2353. doi: 10.1038/cddis.2016.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Altiok A., Bejarano M.T., Klein G., Klein E. Effect of TGF-beta 1 on the EBV-induced transformation of human lymphocyte cultures. Int. J. Cancer. 1992;50:772–776. doi: 10.1002/ijc.2910500518. [DOI] [PubMed] [Google Scholar]
- 124.Fahmi H., Cochet C., Hmama Z., Opolon P., Joab I. Transforming growth factor beta 1 stimulates expression of the Epstein-Barr virus BZLF1 immediate-early gene product ZEBRA by an indirect mechanism which requires the MAPK kinase pathway. J. Virol. 2000;74:5810–5818. doi: 10.1128/jvi.74.13.5810-5818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Callegari S., Gastaldello S., Faridani O.R., Masucci M.G. Epstein-Barr virus encoded microRNAs target SUMO-regulated cellular functions. FEBS J. 2014;281:4935–4950. doi: 10.1111/febs.13040. [DOI] [PubMed] [Google Scholar]
- 126.Pandya D., Mariani M., He S., Andreoli M., Spennato M., Dowell-Martino C., Fiedler P., Ferlini C. Epstein-Barr Virus MicroRNA Expression Increases Aggressiveness of Solid Malignancies. PLoS ONE. 2015;10:e0136058. doi: 10.1371/journal.pone.0136058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hooykaas M.J.G., van Gent M., Soppe J.A., Kruse E., Boer I.G.J., van Leenen D., Groot Koerkamp M.J.A., Holstege F.C.P., Ressing M.E., Wiertz E.J.H.J., Lebbink R.J. EBV MicroRNA BART16 Suppresses Type I IFN Signaling. J. Immunol. 2017;198:4062–4073. doi: 10.4049/jimmunol.1501605. [DOI] [PubMed] [Google Scholar]
- 128.Garlanda C., Dinarello C.A., Mantovani A. The interleukin-1 family: back to the future. Immunity. 2013;39:1003–1018. doi: 10.1016/j.immuni.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mayer-Barber K.D., Yan B. Clash of the Cytokine Titans: counter-regulation of interleukin-1 and type I interferon-mediated inflammatory responses. Cell. Mol. Immunol. 2017;14:22–35. doi: 10.1038/cmi.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Skinner C.M., Ivanov N.S., Barr S.A., Chen Y., Skalsky R.L. An Epstein-Barr Virus MicroRNA Blocks Interleukin-1 (IL-1) Signaling by Targeting IL-1 Receptor 1. J. Virol. 2017;91 doi: 10.1128/JVI.00530-17. e00530-e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhang Y.M., Yu Y., Zhao H.P. EBV-BART-6-3p and cellular microRNA-197 compromise the immune defense of host cells in EBV-positive Burkitt lymphoma. Mol. Med. Rep. 2017;15:1877–1883. doi: 10.3892/mmr.2017.6173. [DOI] [PubMed] [Google Scholar]
- 132.Matsumiya T., Stafforini D.M. Function and regulation of retinoic acid-inducible gene-I. Crit. Rev. Immunol. 2010;30:489–513. doi: 10.1615/critrevimmunol.v30.i6.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lu Y., Qin Z., Wang J., Zheng X., Lu J., Zhang X., Wei L., Peng Q., Zheng Y., Ou C. Epstein-Barr Virus miR-BART6-3p Inhibits the RIG-I Pathway. J. Innate Immun. 2017;9:574–586. doi: 10.1159/000479749. [DOI] [PubMed] [Google Scholar]
- 134.Young H.A., Bream J.H. IFN-gamma: recent advances in understanding regulation of expression, biological functions, and clinical applications. Curr. Top. Microbiol. Immunol. 2007;316:97–117. doi: 10.1007/978-3-540-71329-6_6. [DOI] [PubMed] [Google Scholar]
- 135.Lanier L.L. Evolutionary struggles between NK cells and viruses. Nat. Rev. Immunol. 2008;8:259–268. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Reich N.C. STAT dynamics. Cytokine Growth Factor Rev. 2007;18:511–518. doi: 10.1016/j.cytogfr.2007.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huang W.T., Lin C.W. EBV-encoded miR-BART20-5p and miR-BART8 inhibit the IFN-γ-STAT1 pathway associated with disease progression in nasal NK-cell lymphoma. Am. J. Pathol. 2014;184:1185–1197. doi: 10.1016/j.ajpath.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 138.Boissel N., Rea D., Tieng V., Dulphy N., Brun M., Cayuela J.M., Rousselot P., Tamouza R., Le Bouteiller P., Mahon F.X. BCR/ABL oncogene directly controls MHC class I chain-related molecule A expression in chronic myelogenous leukemia. J. Immunol. 2006;176:5108–5116. doi: 10.4049/jimmunol.176.8.5108. [DOI] [PubMed] [Google Scholar]
- 139.Nachmani D., Stern-Ginossar N., Sarid R., Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5:376–385. doi: 10.1016/j.chom.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 140.Haneklaus M., Gerlic M., Kurowska-Stolarska M., Rainey A.A., Pich D., McInnes I.B., Hammerschmidt W., O’Neill L.A., Masters S.L. Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1β production. J. Immunol. 2012;189:3795–3799. doi: 10.4049/jimmunol.1200312. [DOI] [PubMed] [Google Scholar]
- 141.Tagawa T., Albanese M., Bouvet M., Moosmann A., Mautner J., Heissmeyer V., Zielinski C., Lutter D., Hoser J., Hastreiter M. Epstein-Barr viral miRNAs inhibit antiviral CD4+ T cell responses targeting IL-12 and peptide processing. J. Exp. Med. 2016;213:2065–2080. doi: 10.1084/jem.20160248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Blum J.S., Wearsch P.A., Cresswell P. Pathways of antigen processing. Annu. Rev. Immunol. 2013;31:443–473. doi: 10.1146/annurev-immunol-032712-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Albanese M., Tagawa T., Bouvet M., Maliqi L., Lutter D., Hoser J., Hastreiter M., Hayes M., Sugden B., Martin L. Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific CD8+ T cells. Proc. Natl. Acad. Sci. USA. 2016;113:E6467–E6475. doi: 10.1073/pnas.1605884113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hanna S., Etzioni A. MHC class I and II deficiencies. J. Allergy Clin. Immunol. 2014;134:269–275. doi: 10.1016/j.jaci.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 145.Su Z., Peluso M.V., Raffegerst S.H., Schendel D.J., Roskrow M.A. The generation of LMP2a-specific cytotoxic T lymphocytes for the treatment of patients with Epstein-Barr virus-positive Hodgkin disease. Eur. J. Immunol. 2001;31:947–958. doi: 10.1002/1521-4141(200103)31:3<947::aid-immu947>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 146.Lung R.W., Tong J.H., Sung Y.M., Leung P.S., Ng D.C., Chau S.L., Chan A.W., Ng E.K., Lo K.W., To K.F. Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22. Neoplasia. 2009;11:1174–1184. doi: 10.1593/neo.09888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Teow S.Y., Liew K., Khoo A.S., Peh S.C. Pathogenic Role of Exosomes in Epstein-Barr Virus (EBV)-Associated Cancers. Int. J. Biol. Sci. 2017;13:1276–1286. doi: 10.7150/ijbs.19531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Meckes D.G., Jr., Shair K.H., Marquitz A.R., Kung C.P., Edwards R.H., Raab-Traub N. Human tumor virus utilizes exosomes for intercellular communication. Proc. Natl. Acad. Sci. USA. 2010;107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pegtel D.M., Cosmopoulos K., Thorley-Lawson D.A., van Eijndhoven M.A., Hopmans E.S., Lindenberg J.L., de Gruijl T.D., Würdinger T., Middeldorp J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gourzones C., Gelin A., Bombik I., Klibi J., Vérillaud B., Guigay J., Lang P., Témam S., Schneider V., Amiel C. Extra-cellular release and blood diffusion of BART viral micro-RNAs produced by EBV-infected nasopharyngeal carcinoma cells. Virol. J. 2010;7:271. doi: 10.1186/1743-422X-7-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rechavi O., Erlich Y., Amram H., Flomenblit L., Karginov F.V., Goldstein I., Hannon G.J., Kloog Y. Cell contact-dependent acquisition of cellular and viral nonautonomously encoded small RNAs. Genes Dev. 2009;23:1971–1979. doi: 10.1101/gad.1789609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nanbo A., Kawanishi E., Yoshida R., Yoshiyama H. Exosomes derived from Epstein-Barr virus-infected cells are internalized via caveola-dependent endocytosis and promote phenotypic modulation in target cells. J. Virol. 2013;87:10334–10347. doi: 10.1128/JVI.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Komabayashi Y., Kishibe K., Nagato T., Ueda S., Takahara M., Harabuchi Y. Circulating Epstein-Barr virus-encoded micro-RNAs as potential biomarkers for nasal natural killer/T-cell lymphoma. Hematol. Oncol. 2017;35:655–663. doi: 10.1002/hon.2360. [DOI] [PubMed] [Google Scholar]
- 154.Gu A.D., Lu L.X., Xie Y.B., Chen L.Z., Feng Q.S., Kang T., Jia W.H., Zeng Y.X. Clinical values of multiple Epstein-Barr virus (EBV) serological biomarkers detected by xMAP technology. J. Transl. Med. 2009;7:73. doi: 10.1186/1479-5876-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wong A.M., Kong K.L., Tsang J.W., Kwong D.L., Guan X.Y. Profiling of Epstein-Barr virus-encoded microRNAs in nasopharyngeal carcinoma reveals potential biomarkers and oncomirs. Cancer. 2012;118:698–710. doi: 10.1002/cncr.26309. [DOI] [PubMed] [Google Scholar]
- 156.Zhang G., Zong J., Lin S., Verhoeven R.J., Tong S., Chen Y., Ji M., Cheng W., Tsao S.W., Lung M. Circulating Epstein-Barr virus microRNAs miR-BART7 and miR-BART13 as biomarkers for nasopharyngeal carcinoma diagnosis and treatment. Int. J. Cancer. 2015;136:E301–E312. doi: 10.1002/ijc.29206. [DOI] [PubMed] [Google Scholar]
- 157.Zheng X.H., Lu L.X., Cui C., Chen M.Y., Li X.Z., Jia W.H. Epstein-Barr virus mir-bart1-5p detection via nasopharyngeal brush sampling is effective for diagnosing nasopharyngeal carcinoma. Oncotarget. 2016;7:4972–4980. doi: 10.18632/oncotarget.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jiang C., Chen J., Xie S., Zhang L., Xiang Y., Lung M., Kam N.W., Kwong D.L., Cao S., Guan X.Y. Evaluation of circulating EBV microRNA BART2-5p in facilitating early detection and screening of nasopharyngeal carcinoma. Int. J. Cancer. 2018;143:3209–3217. doi: 10.1002/ijc.31642. [DOI] [PubMed] [Google Scholar]
- 159.Kang B.W., Choi Y., Kwon O.K., Lee S.S., Chung H.Y., Yu W., Bae H.I., Seo A.N., Kang H., Lee S.K. High level of viral microRNA-BART20-5p expression is associated with worse survival of patients with Epstein-Barr virus-associated gastric cancer. Oncotarget. 2017;8:14988–14994. doi: 10.18632/oncotarget.14744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Choy E.Y., Kok K.H., Tsao S.W., Jin D.Y. Utility of Epstein-Barr virus-encoded small RNA promoters for driving the expression of fusion transcripts harboring short hairpin RNAs. Gene Ther. 2008;15:191–202. doi: 10.1038/sj.gt.3303055. [DOI] [PubMed] [Google Scholar]