Abstract
Brittle and “tiger-tail” hair is the diagnostic hallmark of trichothiodystrophy (TTD), a rare recessive disease associated with a wide spectrum of clinical features including ichthyosis, intellectual disability, decreased fertility, and short stature. As a result of premature abrogation of terminal differentiation, the hair is brittle and fragile and contains reduced cysteine content. Hypersensitivity to UV light is found in about half of individuals with TTD; all of these individuals harbor bi-allelic mutations in components of the basal transcription factor TFIIH, and these mutations lead to impaired nucleotide excision repair and basal transcription. Different genes have been found to be associated with non-photosensitive TTD (NPS-TTD); these include MPLKIP (also called TTDN1), GTF2E2 (also called TFIIEβ), and RNF113A. However, a relatively large group of these individuals with NPS-TTD have remained genetically uncharacterized. Here we present the identification of an NPS-TTD-associated gene, threonyl-tRNA synthetase (TARS), found by next-generation sequencing of a group of uncharacterized individuals with NPS-TTD. One individual has compound heterozygous TARS variants, c.826A>G (p.Lys276Glu) and c.1912C>T (p.Arg638∗), whereas a second individual is homozygous for the TARS variant: c.680T>C (p.Leu227Pro). We showed that these variants have a profound effect on TARS protein stability and enzymatic function. Our results expand the spectrum of genes involved in TTD to include genes implicated in amino acid charging of tRNA, which is required for the last step in gene expression, namely protein translation. We previously proposed that some of the TTD-specific features derive from subtle transcription defects as a consequence of unstable transcription factors. We now extend the definition of TTD from a transcription syndrome to a “gene-expression” syndrome.
Keywords: non-photosensitive trichothiodystrophy, aminoacyl tRNA synthetase, protein translation, transcription, tiger tail, TTD, brittle hair
Main Text
The multisystem neuroectodermal disease trichothiodystrophy (TTD) is a rare recessive genetic trait whose diagnostic hallmark is brittle hair.1 The sparse and fragile hair is characterized by its typical “tiger-tail” banding pattern, which is seen when the hair is examined under a polarized light microscope; this pattern is associated with low cysteine content. Individuals with TTD express a wide and heterogeneous range of additional clinical features affecting multiple organs and tissues.2 These features include ichthyosis, brittle nails, impaired intelligence, decreased fertility, short stature, anemia, and recurrent infections. Approximately 50% of individuals with TTD exhibit additional clinical and cellular photosensitivity. All of these photosensitive individuals with TTD (PS-TTD [MIM: 601675, 606390, 616395]) have bi-allelic mutations within the excision repair cross-complementation group 2 (ERCC2 also called XPD; MIM: 126340]) gene,3 the excision repair cross-complementation group 3 (ERCC3 also called XPB; MIM: 133510]) gene,4 or the general transcription factor II H, polypeptide 5 (GTF2H5 also called TTDA; MIM: 608780]) gene.5, 6, 7 These three genes encode for distinct subunits of the basal transcription factor IIH (TFIIH). In addition to its essential role in transcription initiation, TFIIH is also pivotal for nucleotide excision repair (NER), the sole DNA repair process in mammals that is able to remove DNA lesions induced by UV light (which is present in sunlight);8 this explains the photosensitive phenotype. The dual function of TFIIH led to the hypothesis that part of the additional TTD features derive from tissue-specific impairments of transcription.9, 10, 11, 12, 13 Indeed, reduced expression of tissue-specific genes has been observed in primary cells from individuals with TTD14, 15 as well as in neuronal cells or terminally differentiated keratinocytes derived from an Ercc2 mutant mouse model that carries a single amino acid missense TTD-specific point mutation.16, 17 Importantly, TTD-associated pathogenic mutations cause severe instability of the mutant protein itself as well as the entire TFIIH complex in which they reside, resulting in reduced steady-state protein amounts in subject-derived primary fibroblasts.18, 19, 20 In addition, we previously identified a thermo-labile single amino acid missense mutation in ERCC2 in an individual with mild TTD. We found a strong decline in the total TFIIH amount, repair capacity, and transcription levels upon culturing subject-derived fibroblasts at slightly elevated temperatures; this mimics the striking aggravation of TTD-specific features this individual has experienced after episodes of fever.11 These findings further corroborate the idea that some of the TTD-associated transcription defects are derived from transcription factor instability. The other half of the TTD-affected individuals who were studied are non-photosensitive and NER-proficient. The non-photosensitive TTD (NPS-TTD [MIM: 234050, 616943, 300953]) in these individuals is caused by mutations in MPLKIP (also called TTDN1; MIM: 609188);21, 22 in GTF2E2 (also called TFIIEβ; MIM: 189964);23, 24 in the only X-linked TTD-associated gene, RNF113A (OMIM: 300951); 25 or in an as-yet-unidentified gene or genes.
Strikingly, a strong reduction of the total amount of both subunits of the transcription initiation factor TFIIE was observed in primary fibroblasts of GTF2E2-mutated subjects, but this reduction nevertheless did not significantly affect the overall basal transcription capacity. However, these mutations caused reduced TFIIH-mediated phosphorylation of the TFIIEα subunit.23 Induced pluripotent stem cell (iPS) reprogramming of these fibroblasts and subsequent in vitro differentiation into a red blood cell lineage revealed a hematopoietic defect during late-stage differentiation; this defect is associated with hemoglobin subunit imbalance, recapitulating the subjects’ anemia.24, 26 In these differentiated cells, fragile TFIIE becomes limited in its ability to efficiently support transcription of globin genes. Hemoglobin subunit imbalance is commonly induced by impaired transcription, thus corroborating the idea that some of the typical TTD-features are not only due to altered transcription but may also be further aggravated by unstable transcription factors. Recently, the biological function of another NPS-TTD-associated protein, RNF113A, was identified and shown to be associated with the splicing machinery.27 This observation seems to broaden the hypothesis that, in addition to transcription initiation impairments, TTD features may also be caused by mRNA processing alterations. Currently, many individuals with NPS-TTD are still genetically uncharacterized, so there is potential for expanding the spectrum of possible TTD-associated genes. Discovering additional NPS-TTD-associated genes will further increase our understanding of how gene expression defects cause TTD-associated phenotypes.
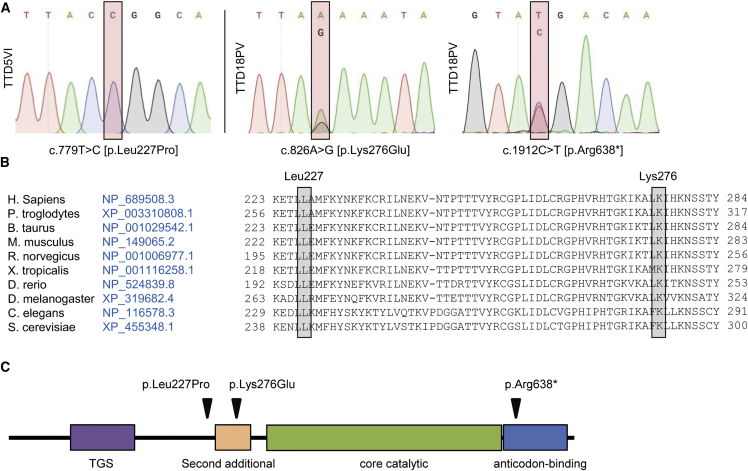
To identify disease-associated genes and the molecular basis of NPS-TTD, we performed whole-genome sequencing (WGS)28, 29 and whole-exome sequencing (WES) in a selected group of 24 unassigned individuals with NPS-TTD. Biological samples were obtained with appropriate informed consent and according to the protocols approved by institute-specific ethical review boards. Sequence data were aligned to the human reference genome and provided as lists of sequence variants (SNPs and short indels) relative to this reference genome. Analysis of the sequence variants revealed that two affected individuals were carriers of homozygous or compound-heterozygous variants within the threonyl-tRNA synthetase (TARS [MIM: 187790]) gene (GenBank: NM_152295.4 transcript variant 1 and GenBank: NP_689508.3). Subject TTD18PV was born in 1990 to non-consanguineous Italian parents and clinically diagnosed with TTD in 1992 on the basis of hair analysis, which revealed that hairs from this individual had lower cysteine content (8.5%) than did hairs from healthy donors (20.8%). Intriguingly, a significantly lower level of the amino acid threonine (Thr) was also found in this individual’s hair (3% compared to 6.9% in hair from healthy donors), which was not observed for any other amino acid (Table S1). The hair showed fractures (trichoschisis) and the typical, for TTD, appearance of an alternating light and dark banding pattern (“tiger tail”) when analyzed under polarization microscopy. There were no signs of sun-sensitive skin, but ichthyosis and follicular keratosis were reported. Additional associated features were delayed physical development, recurrent infections of the respiratory tract, and acromandibular dysplasia. No later records exist for this individual. Two allelic TARS variants, c.826A>G (p.Lys276Glu) and c.1912C>T (p.Arg638∗) (GenBank NM_152295.4) were identified in this individual (Figure 1). Subject TTD5VI was also clinically diagnosed with TTD based on hair analysis, which revealed an abnormal structure of the hair shaft both visually and under the microscope (“tiger-tail” banding pattern). She was born encased in a tight, shiny membrane (”collodion baby”) and had ichthyosis, both commonly associated with TTD.2 Also for this case, no follow-up clinical report is available. In subject TTD5VI, we identified a homozygous variant within the TARS gene: c.680T>C (p.Leu227Pro]) (Figure 1) (GenBank: NM_152295.4). We confirmed the absence of DNA repair deficiency by assessing the UV-irradiation sensitivity and the NER capacity; we did this through the use of clonogenic UV-survival and unscheduled DNA-repair synthesis assays, respectively (Figure S1). The presence of all three TARS variants was confirmed by Sanger sequencing (Figure 1A). The missense variant c.826A>G (p.Lys276Glu) is not present in public databases, and it obtained high pathogenicity scores in three prediction algorithms (PolyPhen-2 score 0.763 = possibly damaging; Mutation Taster score = disease causing with p = 0.999; CADD_phred score = 25.7). The nonsense variant c.1912C>T is reported in the ExAC database but only in the heterozygous condition (allele frequency = 0.00008661). The missense variant c.680T>C (p.Leu227Pro) in TTD5VI is also not present in public databases, and it obtained high pathogenicity scores in three prediction algorithms (PolyPhen-2 score 0.994 = damaging; Mutation Taster score = disease causing with p = 1; CADD_phred score = 32). All three variants are thus predicted to be pathogenic and to produce either a C-terminal truncated TARS protein (in case of p.Arg638∗) or an amino acid substitution in a highly conserved residue (in the cases of p.Leu227Pro and p.Lys276Glu) (Figure 1B). After identification of the TARS mutations in TTD18PV and TTD5VI, we performed Sanger sequencing of TARS cDNA in 24 additional individuals with unsolved NPS-TTD. No bi-allelic mutations were identified. This indicates that in our combined cohort of 47 affected individuals (17 cases in the Dutch cohort; 16 cases from 14 families in the Italian cohort; 16 cases in the UK cohort), defective TARS accounts for about 4% of individuals with NPS-TTD.
Figure 1.
Identification of TARS Variants
(A) Sanger sequencing profiles of TARS cDNA. Subject TTD5VI shows homozygosity for the c.680T>C missense variant (right panel), and subject TTD18PV shows compound heterozygosity for missense variant c.826A>G (left panel) and nonsense variant c.1912C>T (middle panel).
(B) Amino acid conservation across different species orthologs.
(C) Schematic representation (created with InterPro) of the domain structure of the TARS protein with the location of the subjects’ variants.
TARS is an essential component of the translation machinery, and it belongs to the aminoacyl-tRNA synthetase (ARS) protein family.30, 31 This family is divided into two subgroups, class I and class II ARS proteins, based on their structure and presence of shared functional domains.32, 33, 34 TARS is a class II ARS protein and catalyzes the covalent binding of threonine to a threonine-specific tRNA (tRNAThr) to form the aminoacyl Thr-tRNAThr complex.35, 36 The inactive monomeric form of TARS needs to dimerize first in order to establish a catalytically active configuration, which allows specific binding of each monomer to the corresponding uncharged tRNA and free amino acid Thr. Amino acid charging is highly specific and facilitated by several conserved domains within TARS (Figure 1C) that include the C-terminal anticodon-binding domain, the catalytic core domain, a putative N-terminal editing domain, a mode of the so-called second additional domain, and a structurally uncharacterized TGS (for ThrRS, GTPase, and SpoT) domain. The second additional domain consists of several antiparallel beta sheets that surround the dimeric and catalytically active core, whereas the TGS domain appears to facilitate nucleotide binding.36 The identified amino acid substitutions in subjects TTD18PV and TTD5VI are close to the core catalytic domain and might have an impact on the catalytic activity of the protein.
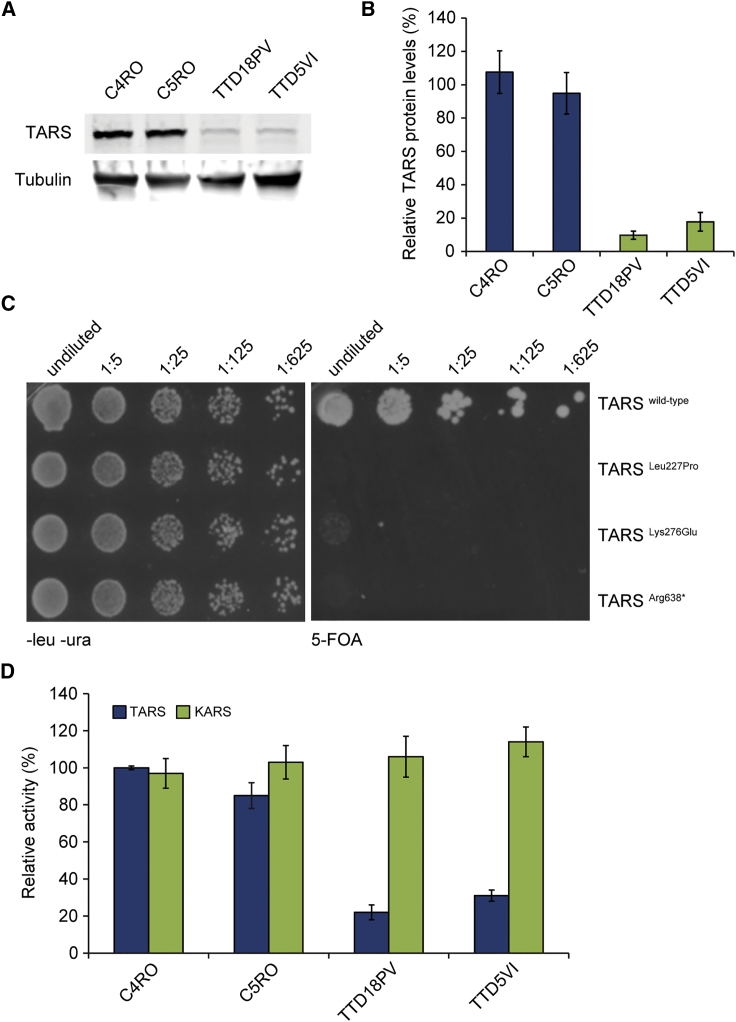
Immunoblot analysis of whole-cell extracts from TTD18PV and TTD5VI subject fibroblasts revealed a strong reduction in the total cellular amount of full-length TARS in both subjects’ lysates (to approximately 20% of the amount seen in controls) (Figure 2A and 2B). The TARS-specific protein bands were of the same molecular mass as that seen in control lysates, and this mass corresponded to the predicted protein mass containing the amino substitutions p.Lys276Glu or p.Leu227Pro (Figure 2A and 2B). The TARS variant c.1912C>T (p.Arg638∗) is predicted to lack the last 85 amino acid residues at the C-terminal end; this would almost completely delete the anticodon binding domain. However, this predicted lower-sized protein band was not visible in immunoblots, indicating that this mutant protein might be less abundantly synthetized and/or severely unstable as a result of the C-terminal truncation. Total TARS mRNA levels, as measured by real-time quantitative reverse-transcription PCR (qRT-PCR), were only slightly reduced in TTD18PV primary fibroblasts compared to control cells (Figure S2). Allele-specific qRT-PCR showed that only ∼10% of the total TARS mRNA molecules in TTD18PV primary fibroblasts originated from the c.1912C>T variant allele, suggesting that in TTD18PV, most TARS proteins contain the missense Lys276Glu TARS variant. Altogether, these data show that the single amino acid substitutions p.Leu227Pro and p.Lys276Glu in TARS cause severe protein instability.
Figure 2.
Functional Consequence of TARS Variants
(A) Immunoblot analysis used for determining TARS protein amounts in lysates of primary fibroblasts TTD5VI and TTD18PV and two wild-type controls; anti-tubulin was used as a loading control.
(B) Quantification of the immunoblot. The band intensities of TARS were normalized to tubulin and expressed as percentage of the intensities for control cells (C4RO). The error bars indicate the SEM of three independent experiments.
(C) Representative picture of a yeast complementation assay. Wild-type and mutant strains were grown on solid medium without leucine and uracil (−LU; left panels) or in the presence of 5-FOA (right panels). Cells were spotted with serial dilutions and pictures were taken after 96 h at 30°C.
(D) The cytosolic fraction from TTD5VI, TTD18PV, and two control fibroblasts was used for determining TARS (blue) and KARS (green; internal control) amino acid charging on tRNA. Threonine and lysine charging in control fibroblast C4RO was set at 100%, and error bars indicate SD of three independent experiments.
Next, we tested the functionality of the identified TARS variants by using a well-established complementation yeast assay.37 It has been recently demonstrated that the haploid yeast knockout Saccharomyces cerevisiae strain deprived of the TARS ortholog THS1 essential gene (Δths1) can be complemented by the human TARS cDNA.38 We could therefore use plasmid shuffling experiments to test the ability of c.680T>C (p.Leu227Pro), c.826A>G (p.Lys276Glu), and c.1912C>T (p.Arg638∗) variants to rescue the cell lethality associated with the deletion of the endogenous yeast THS1. In comparison with those expressing wild-type TARS, yeast cells expressing the variants showed dramatically reduced growth, strongly supporting the notion that these protein alterations severely impact TARS functionality (Figure 2C).
To further investigate the functional consequences of the TARS variants in the context of mammalian cells, we measured steady-state aminoacylation reactions on protein lysates from subjects’ fibroblasts (from TTD5VI and TTD18PV) and control fibroblasts (from C4RO and C5RO). KARS (for Lysine-tRNA synthetase) activity was simultaneously measured as an internal control. Intra-assay variation was <15%, and no decreases were observed for KARS (internal control) activity. In contrast, the results showed an approximate 70%–80% reduction of threonine tRNA charging activity in both subject cells as compared to control fibroblasts (Figure 2D). These data corroborate the results obtained from the yeast-based complementation assays and demonstrate the loss-of-function (LoF) effect of TARS variants on tRNA charging activity.
In summary, we identified bi-allelic TARS variants affecting highly conserved amino acids in two unrelated individuals with NPS-TTD (Figures 1A and 1B): compound-heterozygous variants in subject TTD18PV (c.826A>G [p.Lys276Glu] and c.1912C>T [p.Arg638∗]), and a homozygous variant in subject TTD5VI (c.680T>C [p.Leu227Pro]). Immunoblotting revealed an approximately 80% reduction of steady-state protein amounts of full-length TARS in fibroblast lysates from subjects TTD18PV and TTD5VI when these cells were compared to control fibroblasts. We are tempted to speculate that the identified amino acid substitutions compromise the structural integrity of the dimeric and catalytically active TARS complex and perhaps force the dimeric TARS protein complex into its monomeric and inactive form, which might be unstable and more susceptible to protein degradation. Protein instability seems to be a common cellular hallmark observed in TTD cells; it was also observed for TTD-subject cells carrying mutations in either TFIIH (ERCC3, ERCC2, or TFB5/TTD [also called TTDA]) or TFIIE (GTF2E2).7, 11, 19, 20, 23, 24, 39
The canonical function of TARS in tRNA charging strongly depends on its structural conformation and correct assembly of a catalytically active dimeric TARS complex. It is thus likely that along with haploinsufficiency caused by protein instability the catalytic activity of mutant TARS is also affected, and this contributes to the phenotype. The lack of complementation in yeast strains overexpressing the missense TARS variants (Figure 2C) supports this hypothesis. This reduced activity will decrease the number of charged tRNAThr molecules available for translation in subjects’ cells, impairing protein production in general. However, it is important to understand the degree to which translation is impaired and contributes to tissue-specific phenotypes. There is also evidence suggesting that proofreading deficiencies might increase the number of mischarged tRNA molecules, and such an increase might lead to accumulation of misfolded proteins in the brain and thus cause progressive neurological phenotypes.40
Bi-allelic mutations in other ARS-related genes typically cause severe, early-onset, recessive diseases that affect a wide range of tissues.41 The vast majority of these bi-allelic mutations also show LoF effects, reduction in ARS protein amounts,41, 42 and severe decreases in enzyme activity. Mutations in TARS have not yet been associated with human disorders. The notion that TARS variants are associated with clinical features such as brittle hair and ichthyosis is somewhat surprising because ARS-mediated LoF effects were thus far not linked to brittle hair or ichthyosis.43 However, very recently, inactivation mutations in the cysteinyl-tRNA synthetase (CARS [MIM: 123859]) have been identified in three unrelated families with wide clinical spectra and some TTD-phenotype-reminiscent features, including microcephaly, developmental delay, and brittle hair with weak or moderate “tiger-tail” banding.44 The few cases identified so far do not allow one to determine whether the clinical differences between TARS- and CARS-defective individuals are due to the type of mutations or to the impact that alterations in specific ARSs may have on cellular metabolism.
The notion that faulty tRNA charging, which would likely reduce protein translation, is associated with TTD-specific features suggests that gene expression defects (i.e., the net result of transcription, transcript maturation and stability, and protein synthesis and stability) comprise the underlying molecular mechanism that drives segmental TTD features. Our observation that TTD is also associated with affected protein translation, together with the notion that another TTD-causing mutation was found in an mRNA splicing factor,27 prompted us to extend the definition of TTD from being a transcription syndrome to a gene-expression syndrome.
This expanding spectrum of TTD-associated genes will contribute to a better understanding of the molecular mechanisms underlying the rare genetic disorder TTD. Besides known impairments in genome maintenance and transcription failure, anomalies in translation also appear to contribute to TTD-specific clinical features such as brittle hair.
Acknowledgments
This work was supported by grants from Associazione Italiana Ricerca sul Cancro (17710 and 21737) to D.O., from the Telethon Foundation (GEP13022) to E.B., from the European Research Council (ERC; grant number 233424) to J.H., and from the Dutch Science Organization (NWO) ZonMW division (grant number 912.12.132) and the European Research Council (ERC; grant number 340988) to W.V. We thank the Optical Imaging Centre (OIC) of the Erasmus University Medical Center for microscopy support and Xiao-Long Zhou (Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Shanghai, China) for providing the Δths1 yeast strain and TARS expressing vectors. The bioinformatics work was financially supported by an H2020 grant (BigMedilytics, big data for medical analysis). We would like to thank Joost Verlouw, Pascal Arp, and Mila Jhamai for help with WES and variant calling.
Published: August 1, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.06.017.
Contributor Information
Wim Vermeulen, Email: w.vermeulen@erasmusmc.nl.
Donata Orioli, Email: orioli@igm.cnr.it.
Web Resources
Alamut Visual, https://www.interactive-biosoftware.com/alamut-visual/
Complete Genomics, https://www.completegenomics.com/sequence-data/cgatools/
Human Genome Variation Society, http://www.hgvs.org/mutnomen/
Illumina, https://www.illumina.com/
Ingenuity Variant Analysis, https://www.ingenuity.com/products/variant-analysis
Interpro, https://www.ebi.ac.uk/interpro/
MutationTaster, http://www.mutationtaster.org/
NCBI HomoloGene, https://www.ncbi.nlm.nih.gov/homologene
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
OMIM, https://www.omim.org/
Sequence Variant Nomenclature, http://varnomen.hgvs.org/
Declaration of Interests
The authors declare no competing interests.
Supplemental Data
References
- 1.Price V.H., Odom R.B., Ward W.H., Jones F.T. Trichothiodystrophy: sulfur-deficient brittle hair as a marker for a neuroectodermal symptom complex. Arch. Dermatol. 1980;116:1375–1384. doi: 10.1001/archderm.116.12.1375. [DOI] [PubMed] [Google Scholar]
- 2.Faghri S., Tamura D., Kraemer K.H., Digiovanna J.J. Trichothiodystrophy: a systematic review of 112 published cases characterises a wide spectrum of clinical manifestations. J. Med. Genet. 2008;45:609–621. doi: 10.1136/jmg.2008.058743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stefanini M., Lagomarsini P., Arlett C.F., Marinoni S., Borrone C., Crovato F., Trevisan G., Cordone G., Nuzzo F. Xeroderma pigmentosum (complementation group D) mutation is present in patients affected by trichothiodystrophy with photosensitivity. Hum. Genet. 1986;74:107–112. doi: 10.1007/BF00282072. [DOI] [PubMed] [Google Scholar]
- 4.Weeda G., Rossignol M., Fraser R.A., Winkler G.S., Vermeulen W., van ’t Veer L.J., Ma L., Hoeijmakers J.H., Egly J.M. The XPB subunit of repair/transcription factor TFIIH directly interacts with SUG1, a subunit of the 26S proteasome and putative transcription factor. Nucleic Acids Res. 1997;25:2274–2283. doi: 10.1093/nar/25.12.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giglia-Mari G., Coin F., Ranish J.A., Hoogstraten D., Theil A., Wijgers N., Jaspers N.G., Raams A., Argentini M., van der Spek P.J. A new, tenth subunit of TFIIH is responsible for the DNA repair syndrome trichothiodystrophy group A. Nat. Genet. 2004;36:714–719. doi: 10.1038/ng1387. [DOI] [PubMed] [Google Scholar]
- 6.Stefanini M., Botta E., Lanzafame M., Orioli D. Trichothiodystrophy: From basic mechanisms to clinical implications. DNA Repair (Amst.) 2010;9:2–10. doi: 10.1016/j.dnarep.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 7.Theil A.F., Hoeijmakers J.H., Vermeulen W. TTDA: Big impact of a small protein. Exp. Cell Res. 2014;329:61–68. doi: 10.1016/j.yexcr.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 8.Marteijn J.A., Lans H., Vermeulen W., Hoeijmakers J.H. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat. Rev. Mol. Cell Biol. 2014;15:465–481. doi: 10.1038/nrm3822. [DOI] [PubMed] [Google Scholar]
- 9.Vermeulen W., van Vuuren A.J., Chipoulet M., Schaeffer L., Appeldoorn E., Weeda G., Jaspers N.G., Priestley A., Arlett C.F., Lehmann A.R. Three unusual repair deficiencies associated with transcription factor BTF2(TFIIH): Evidence for the existence of a transcription syndrome. Cold Spring Harb. Symp. Quant. Biol. 1994;59:317–329. doi: 10.1101/sqb.1994.059.01.036. [DOI] [PubMed] [Google Scholar]
- 10.Bergmann E., Egly J.M. Trichothiodystrophy, a transcription syndrome. Trends Genet. 2001;17:279–286. doi: 10.1016/s0168-9525(01)02280-6. [DOI] [PubMed] [Google Scholar]
- 11.Vermeulen W., Rademakers S., Jaspers N.G., Appeldoorn E., Raams A., Klein B., Kleijer W.J., Hansen L.K., Hoeijmakers J.H. A temperature-sensitive disorder in basal transcription and DNA repair in humans. Nat. Genet. 2001;27:299–303. doi: 10.1038/85864. [DOI] [PubMed] [Google Scholar]
- 12.Dubaele S., Proietti De Santis L., Bienstock R.J., Keriel A., Stefanini M., Van Houten B., Egly J.M. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol. Cell. 2003;11:1635–1646. doi: 10.1016/s1097-2765(03)00182-5. [DOI] [PubMed] [Google Scholar]
- 13.Keriel A., Stary A., Sarasin A., Rochette-Egly C., Egly J.M. XPD mutations prevent TFIIH-dependent transactivation by nuclear receptors and phosphorylation of RARalpha. Cell. 2002;109:125–135. doi: 10.1016/s0092-8674(02)00692-x. [DOI] [PubMed] [Google Scholar]
- 14.Orioli D., Compe E., Nardo T., Mura M., Giraudon C., Botta E., Arrigoni L., Peverali F.A., Egly J.M., Stefanini M. XPD mutations in trichothiodystrophy hamper collagen VI expression and reveal a role of TFIIH in transcription derepression. Hum. Mol. Genet. 2013;22:1061–1073. doi: 10.1093/hmg/dds508. [DOI] [PubMed] [Google Scholar]
- 15.Arseni L., Lanzafame M., Compe E., Fortugno P., Afonso-Barroso A., Peverali F.A., Lehmann A.R., Zambruno G., Egly J.M., Stefanini M., Orioli D. TFIIH-dependent MMP-1 overexpression in trichothiodystrophy leads to extracellular matrix alterations in patient skin. Proc. Natl. Acad. Sci. USA. 2015;112:1499–1504. doi: 10.1073/pnas.1416181112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Boer J., de Wit J., van Steeg H., Berg R.J., Morreau H., Visser P., Lehmann A.R., Duran M., Hoeijmakers J.H., Weeda G. A mouse model for the basal transcription/DNA repair syndrome trichothiodystrophy. Mol. Cell. 1998;1:981–990. doi: 10.1016/s1097-2765(00)80098-2. [DOI] [PubMed] [Google Scholar]
- 17.Compe E., Malerba M., Soler L., Marescaux J., Borrelli E., Egly J.M. Neurological defects in trichothiodystrophy reveal a coactivator function of TFIIH. Nat. Neurosci. 2007;10:1414–1422. doi: 10.1038/nn1990. [DOI] [PubMed] [Google Scholar]
- 18.Theil A.F., Nonnekens J., Steurer B., Mari P.O., de Wit J., Lemaitre C., Marteijn J.A., Raams A., Maas A., Vermeij M. Disruption of TTDA results in complete nucleotide excision repair deficiency and embryonic lethality. PLoS Genet. 2013;9:e1003431. doi: 10.1371/journal.pgen.1003431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Botta E., Nardo T., Lehmann A.R., Egly J.M., Pedrini A.M., Stefanini M. Reduced level of the repair/transcription factor TFIIH in trichothiodystrophy. Hum. Mol. Genet. 2002;11:2919–2928. doi: 10.1093/hmg/11.23.2919. [DOI] [PubMed] [Google Scholar]
- 20.Vermeulen W., Bergmann E., Auriol J., Rademakers S., Frit P., Appeldoorn E., Hoeijmakers J.H., Egly J.M. Sublimiting concentration of TFIIH transcription/DNA repair factor causes TTD-A trichothiodystrophy disorder. Nat. Genet. 2000;26:307–313. doi: 10.1038/81603. [DOI] [PubMed] [Google Scholar]
- 21.Botta E., Offman J., Nardo T., Ricotti R., Zambruno G., Sansone D., Balestri P., Raams A., Kleijer W.J., Jaspers N.G. Mutations in the C7orf11 (TTDN1) gene in six nonphotosensitive trichothiodystrophy patients: No obvious genotype-phenotype relationships. Hum. Mutat. 2007;28:92–96. doi: 10.1002/humu.20419. [DOI] [PubMed] [Google Scholar]
- 22.Heller E.R., Khan S.G., Kuschal C., Tamura D., DiGiovanna J.J., Kraemer K.H. Mutations in the TTDN1 gene are associated with a distinct trichothiodystrophy phenotype. J. Invest. Dermatol. 2015;135:734–741. doi: 10.1038/jid.2014.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuschal C., Botta E., Orioli D., Digiovanna J.J., Seneca S., Keymolen K., Tamura D., Heller E., Khan S.G., Caligiuri G. GTF2E2 mutations destabilize the general transcription factor complex TFIIE in individuals with DNA repair-proficient trichothiodystrophy. Am. J. Hum. Genet. 2016;98:627–642. doi: 10.1016/j.ajhg.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Theil A.F., Mandemaker I.K., van den Akker E., Swagemakers S.M.A., Raams A., Wüst T., Marteijn J.A., Giltay J.C., Colombijn R.M., Moog U. Trichothiodystrophy causative TFIIEβ mutation affects transcription in highly differentiated tissue. Hum. Mol. Genet. 2017;26:4689–4698. doi: 10.1093/hmg/ddx351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corbett M.A., Dudding-Byth T., Crock P.A., Botta E., Christie L.M., Nardo T., Caligiuri G., Hobson L., Boyle J., Mansour A. A novel X-linked trichothiodystrophy associated with a nonsense mutation in RNF113A. J. Med. Genet. 2015;52:269–274. doi: 10.1136/jmedgenet-2014-102418. [DOI] [PubMed] [Google Scholar]
- 26.Viprakasit V., Gibbons R.J., Broughton B.C., Tolmie J.L., Brown D., Lunt P., Winter R.M., Marinoni S., Stefanini M., Brueton L. Mutations in the general transcription factor TFIIH result in beta-thalassaemia in individuals with trichothiodystrophy. Hum. Mol. Genet. 2001;10:2797–2802. doi: 10.1093/hmg/10.24.2797. [DOI] [PubMed] [Google Scholar]
- 27.Wu N.Y., Chung C.S., Cheng S.C. Role of Cwc24 in the first catalytic step of splicing and fidelity of 5′ splice site selection. Mol. Cell. Biol. 2017;37 doi: 10.1128/MCB.00580-16. e00580–e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drmanac R., Sparks A.B., Callow M.J., Halpern A.L., Burns N.L., Kermani B.G., Carnevali P., Nazarenko I., Nilsen G.B., Yeung G. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 29.Swagemakers S.M., Jaspers N.G., Raams A., Heijsman D., Vermeulen W., Troelstra C., Kremer A., Lincoln S.E., Tearle R., Hoeijmakers J.H., van der Spek P.J. Pollitt syndrome patients carry mutation in TTDN1. Meta Gene. 2014;2:616–618. doi: 10.1016/j.mgene.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ibba M., Soll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 31.Rajendran V., Kalita P., Shukla H., Kumar A., Tripathi T. Aminoacyl-tRNA synthetases: Structure, function, and drug discovery. Int. J. Biol. Macromol. 2018;111:400–414. doi: 10.1016/j.ijbiomac.2017.12.157. [DOI] [PubMed] [Google Scholar]
- 32.Bryson D.I., Fan C., Guo L.T., Miller C., Söll D., Liu D.R. Continuous directed evolution of aminoacyl-tRNA synthetases. Nat. Chem. Biol. 2017;13:1253–1260. doi: 10.1038/nchembio.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chaliotis A., Vlastaridis P., Mossialos D., Ibba M., Becker H.D., Stathopoulos C., Amoutzias G.D. The complex evolutionary history of aminoacyl-tRNA synthetases. Nucleic Acids Res. 2017;45:1059–1068. doi: 10.1093/nar/gkw1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 35.Smith T.F., Hartman H. The evolution of class II aminoacyl-tRNA synthetases and the first code. FEBS Lett. 2015;589:3499–3507. doi: 10.1016/j.febslet.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 36.Sankaranarayanan R., Dock-Bregeon A.C., Romby P., Caillet J., Springer M., Rees B., Ehresmann C., Ehresmann B., Moras D. The structure of threonyl-tRNA synthetase-tRNA(Thr) complex enlightens its repressor activity and reveals an essential zinc ion in the active site. Cell. 1999;97:371–381. doi: 10.1016/s0092-8674(00)80746-1. [DOI] [PubMed] [Google Scholar]
- 37.Oprescu S.N., Griffin L.B., Beg A.A., Antonellis A. Predicting the pathogenicity of aminoacyl-tRNA synthetase mutations. Methods. 2017;113:139–151. doi: 10.1016/j.ymeth.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruan Z.R., Fang Z.P., Ye Q., Lei H.Y., Eriani G., Zhou X.L., Wang E.D. Identification of lethal mutations in yeast threonyl-tRNA synthetase revealing critical residues in its human homolog. J. Biol. Chem. 2015;290:1664–1678. doi: 10.1074/jbc.M114.599886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Botta E., Nardo T., Orioli D., Guglielmino R., Ricotti R., Bondanza S., Benedicenti F., Zambruno G., Stefanini M. Genotype-phenotype relationships in trichothiodystrophy patients with novel splicing mutations in the XPD gene. Hum. Mutat. 2009;30:438–445. doi: 10.1002/humu.20912. [DOI] [PubMed] [Google Scholar]
- 40.Lee J.W., Beebe K., Nangle L.A., Jang J., Longo-Guess C.M., Cook S.A., Davisson M.T., Sundberg J.P., Schimmel P., Ackerman S.L. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443:50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 41.Antonellis A., Oprescu S.N., Griffin L.B., Heider A., Amalfitano A., Innis J.W. Compound heterozygosity for loss-of-function FARSB variants in a patient with classic features of recessive aminoacyl-tRNA synthetase-related disease. Hum. Mutat. 2018;39:834–840. doi: 10.1002/humu.23424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakayama T., Wu J., Galvin-Parton P., Weiss J., Andriola M.R., Hill R.S., Vaughan D.J., El-Quessny M., Barry B.J., Partlow J.N. Deficient activity of alanyl-tRNA synthetase underlies an autosomal recessive syndrome of progressive microcephaly, hypomyelination, and epileptic encephalopathy. Hum. Mutat. 2017;38:1348–1354. doi: 10.1002/humu.23250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyer-Schuman R., Antonellis A. Emerging mechanisms of aminoacyl-tRNA synthetase mutations in recessive and dominant human disease. Hum. Mol. Genet. 2017;26(R2):R114–R127. doi: 10.1093/hmg/ddx231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuo M.E., Theil A.F., Kievit A., Malicdan M.C., Introne W.J., Christian T., Verheijen F.W., Smith D.E.C., Mendes M.I., Hussaarts-Odijk L. Cysteinyl-tRNA synthetase mutations cause a multi-system, recessive disease that includes microcephaly, developmental delay, and brittle hair and nails. Am. J. Hum. Genet. 2019;104:520–529. doi: 10.1016/j.ajhg.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.