Abstract
While genetic causes are known for many syndromes involving developmental anomalies, a large number of individuals with overlapping phenotypes remain undiagnosed. Using exome-sequencing analysis and review of matchmaker databases, we have discovered four de novo missense variants predicted to affect the N-terminal region of WDR37—p.Ser119Phe, p.Thr125Ile, p.Ser129Cys, and p.Thr130Ile—in unrelated individuals with a previously unrecognized syndrome. Features of WDR37 syndrome include the following: ocular anomalies such as corneal opacity/Peters anomaly, coloboma, and microcornea; dysmorphic facial features; significant neurological impairment with structural brain defects and seizures; poor feeding; poor post-natal growth; variable skeletal, cardiac, and genitourinary defects; and death in infancy in one individual. WDR37 encodes a protein of unknown function with seven predicted WD40 domains and no previously reported human pathogenic variants. Immunocytochemistry and western blot studies showed that wild-type WDR37 is localized predominantly in the cytoplasm and mutant proteins demonstrate similar protein levels and localization. CRISPR-Cas9-mediated genome editing generated zebrafish mutants with novel missense and frameshift alleles: p.Ser129Phe, p.Ser129Cys (which replicates one of the human variants), p.Ser129Tyr, p.Lys127Cysfs, and p.Gln95Argfs. Zebrafish carrying heterozygous missense variants demonstrated poor growth and larval lethality, while heterozygotes with frameshift alleles survived to adulthood, suggesting a potential dominant-negative mechanism for the missense variants. RNA-seq analysis of zebrafish embryos carrying a missense variant detected significant upregulation of cholesterol biosynthesis pathways. This study identifies variants in WDR37 associated with human disease and provides insight into its essential role in vertebrate development and possible molecular functions.
Keywords: WDR37, Peters, coloboma, syndrome, seizures, CRISPR-Cas9, zebrafish, Peters plus, Walker-Warburg, dominant
Main Text
Developmental ocular conditions such as Peters anomaly and coloboma are often accompanied by additional syndromic anomalies.1, 2 Some syndromes are well defined with known genetic causes, including Peters-plus syndrome (B3GLCT [MIM: 261540]),3 CHARGE syndrome (CHD7 [MIM: 214800]),4 Temtamy syndrome (C12orf57 [MIM: 218340]),5 and Walker-Warburg syndrome (POMT1 [MIM: 236670], POMT2 [MIM: 613150], and other genes).6, 7 Many similarly affected individuals, however, have features that partially overlap these known syndromes and remain causally unexplained.1, 8, 9
In this study we report human disease-causing variants in WDR37 (WD repeat domain 37; GenBank: NM_014023.3) in several individuals with previously uncharacterized syndromic malformations. This human study was approved by the Institutional Review Board of the Children’s Hospital of Wisconsin with written informed consent obtained for participation and publication for every participant. Detailed case descriptions are provided in the Supplemental Note. Exome sequencing was performed (Table S1) and analyzed as previously reported1 with in silico predictions of the effect of missense variants obtained from dbNSFP for SIFT, PolyPhen2, MutationTaster, MutationAssessor, and FATHMM MKL.
Trio analysis in individual 1 (Figure 1) and his parents identified a de novo missense variant in WDR37, c.356C>T (p.Ser119Phe) (Figure S1). The variant is not present in gnomAD and is predicted damaging by all five in silico programs. Review of exome data from other individuals with unexplained syndromic eye disorders1, 9 enrolled in our study identified an additional de novo missense variant in WDR37 in individual 2, c.389C>T (p.Thr130Ile) (Figures 1 and S1), also not present in gnomAD and predicted damaging by all five programs. Both affected individuals were referred to the study with possible clinical diagnoses of either Peters plus or Walker-Warburg syndromes. Both were non-verbal with limited or no ambulation and seizure onset shortly after birth and showed overlapping clinical features including ocular anomalies (corneal opacity/Peters anomaly, microcornea, coloboma, and lens defects), dysmorphic craniofacial features (microcephaly, thin upper lip, broad nasal bridge, abnormal ears), structural brain anomalies (Dandy-Walker spectrum malformation, abnormal myelination), congenital heart defects, skeletal anomalies (severe short stature, scoliosis, contractures), genitourinary anomalies (cryptorchidism, micropenis, hypoplastic uterus, and kidney defects), and feeding disorders (poor intake/feeding intolerance) (Table 1, Supplemental Note). Individual 1 is now 30 years old; individual 2 died at age 22 months from respiratory failure due to pneumonia and cardiac anomalies. Several other rare variants of uncertain significance were also identified within the exome data, but based on testing of family members, general population data, and in silico predictions, all are considered likely benign (Table S2).
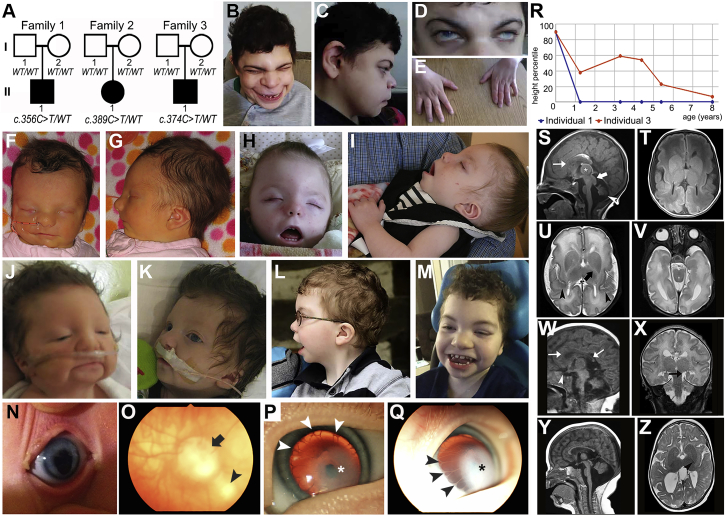
Figure 1.
Images of Affected Individuals
(A) Pedigrees from individuals 1–3 with WDR37 variants indicated.
(B–E) Photographs of individual 1 at age 30 years showing microcephaly, broad nasal bridge, thin upper lip, protruding jaw, and ear anomalies (B, C); eye anomalies including corneal opacity and microcornea (D); and small hands (E).
(F–I) Photographs of individual 2 at age 4 days (F, G) and 22 months (H, I) showing microcephaly, apparent hypertelorism, broad nasal bridge, thin upper lip, micrognathia, and ear anomalies.
(J–M) Facial photographs of individual 3 at ages 1 day (J), 3 weeks (K), 5 years (L), and 8 years (M) showing microcephaly, broad nasal bridge, thin upper lip, broad chin, and abnormal ears.
(N–Q) Ocular images of individual 3 taken at age 5 months showing right eye with iris coloboma (N) and mild inferior optic nerve dysplasia (arrow) and chorioretinal coloboma (arrowhead) (O) and left eye (pupil dilated) with Peters anomaly (central corneal leukoma [∗] and iris adhesions to the endothelium [arrowheads]), iris coloboma, and microcornea (P–Q).
(R) Height centiles for Individuals 1 and 3 by year of age, showing normal birth length but decreasing height centiles over time.
(S–V) Brain MRI images from individual 2 at age 5 days. Sagittal noncontrast T1 image (S) shows a prominent massa intermedia (asterisk), large midbrain tectum (large white arrow), small pons, hypoplastic corpus callosum (thin white arrow) with a small rostrum and splenium, bright pericallosal lipoma, and Dandy-Walker variant with vermian hypoplasia (white arrowhead) and dilation of the fourth ventricle. Axial noncontrast T1 (T) demonstrates an abnormal gyral pattern and abnormal configuration of the basal ganglia. Axial T2 images (U and V) show abnormal diffuse white matter hyperintensity (decreased myelination), particularly notable in the posterior limb of the internal capsule (large black arrow) and dysplastic, thickened cortex in the sylvian fissures (black arrowheads), as well as left microphthalmia with aphakia and small, incompletely inverted hippocampi.
(W–Z) Brain MRI images from individual 3. Sagittal noncontrast T1 image at age 11 days (W) shows thinned corpus callosum (white arrows), enlarged massa intermedia, mildly prominent midbrain tectum, and a Dandy-Walker variant with hypoplastic cerebellar vermis. The optic chiasm and nerves appear atrophic (white arrowhead) and the pituitary gland is small with absent posterior bright spot. Concurrent coronal T2 image (X) shows simplified gyral pattern, diffuse white matter hyperintensity, and small and incompletely inverted hippocampi (black arrow). Follow-up sagittal noncontrast T1 at age 11 months (Y) shows increased dilation of the posterior fossa and relatively small brainstem while the concurrent T2 image (Z) shows delayed myelination of the posterior limb of the internal capsule (black arrowhead) and extensive white matter volume loss with enlarged ventricles. Detailed clinical and MRI descriptions are included in the Supplemental Note.
Table 1.
Clinical Features of Affected Individuals with WDR37 Variants
| Individual 1 | Individual 2 | Individual 3a | Individual 4b | |
|---|---|---|---|---|
| De novo WDR37 variant | c.356C>T (p.Ser119Phe) | c.389C>T (p.Thr130Ile) | c.374C>T (p.Thr125Ile) | c.386C>G (p.Ser129Cys) |
| Age | 30 years | 22 months (dec) | 8 years | U |
| Ocular Features | ||||
| Microcornea∗ | + | + | + | U |
| Corneal opacity/Peters∗b | + | + | + | U |
| Coloboma∗ | ? | + | + | + |
| Other | glaucoma, juvenile cataract | aphakia, ONH, retinal dysplasia | cataract, ONH, retinal dysplasia | U |
| Craniofacial/Neck Features | ||||
| Microcephaly∗ | + | + | + | + |
| Protruding jaw/broad chin | + | − | + | U |
| Thin upper lip∗ | + | + | + | U |
| Broad nasal bridge∗ | + | + | + | + |
| Ear anomalies∗ | + | + | + | + |
| Excess nuchal skin/webbed neck | U | + | + | U |
| Other | arched eyebrows | hearing loss, hypertelorism | hearing loss | hypertelorism |
| Neurological Anomalies | ||||
| Seizures∗ | + | + | + | + |
| Dandy-Walker variant∗ | + | + | + | ? |
| Delayed myelination/dysmyelination∗ | + | + | + | U |
| Abnormal gyration pattern | − | + | + | U |
| Absent/hypoplastic corpus callosum | − | + | + | U |
| Non-verbal, severe global delay∗ | + | + | + | ? |
| Cardiovascular Anomalies | ||||
| Septal defect (ASD/VSD) | + | + | − | ? |
| Patent ductus arteriosus∗ | + | + | + | ? |
| Genitourinary Anomalies | ||||
| Cryptorchidism, micropenis | + | NA (female) | + | U |
| Hypoplastic uterus | NA (male) | + | NA (male) | U |
| Hydronephrosis/ureterocele | − | + | + | U |
| Other | incomp. puberty | renal dysplasia | − | U |
| Skeletal/Spinal Anomalies | ||||
| Short stature∗ | + | + | − | + |
| Spine anomalies | + | + | − | U |
| Syndactyly | − | + | + | U |
| Contractures∗ | + | + | + | U |
| Other | hip dislocation, pectus anomaly | − | hip dislocation, coxa valga | U |
| Gastrointestinal | ||||
| Feeding disorder∗ | + | + | + | U |
| Other | − | pancreatic lipomatosis | small stomach, malrotation, GERD | U |
Abbreviations: U, unknown; ?, features possibly consistent with this diagnosis but insufficient details available; dec, deceased; ONH, optic nerve hypoplasia; GERD, gastresophageal reflux disease; NA, not applicable; features with asterisk (∗) are observed in three or more affected individuals; GenBank: NM_014023.3 used as reference sequence.
Individual 3 also has a maternally inherited 17p13.2 deletion
Clinical features as noted in MyGene2
A search of the matchmaker database MyGene2 (see Web Resources) identified two additional individuals with de novo, novel variants in WDR37 within the same region; both variants were similarly predicted damaging by all five in silico programs. Individual 3, with a c.374C>T (p.Thr125Ile) variant, was subsequently enrolled in the study for confirmation of the variant and detailed review of medical records (Figures 1 and S1; Table 1; Supplemental Note). The fourth individual, with a c.386C>G (p.Ser129Cys) variant, is presented with data drawn from MyGene2 (Table 1, Supplemental Note). The referring provider confirmed the submission, but the individual could not be enrolled in our study for detailed phenotypic evaluation. Comparison of all four affected individuals showed overlapping phenotypes consisting of severe complex ocular anomalies, dysmorphic craniofacial features, structural brain anomalies with severe neurological impairment, congenital heart defects, skeletal/spinal anomalies, genitourinary anomalies, feeding disorders with post-natal growth deficiency, structural gastroenterological anomalies, excess nuchal skin/cystic hygroma, hearing loss with recurrent otitis media, hip dysplasia, syndactyly, and contractures (Table 1). Height centiles were plotted for individuals 1 and 3 and showed normal length at birth with decreasing height centiles over time (Figure 1R); while a specific birth length was not available for individual 2, she was also reported to have a normal length at birth but fell to the 1st centile by age 2 months, similar to individual 1.
WDR37 is predicted to encode a 494-amino acid protein of unknown function (Figure S2). Literature review identified only two reports about this gene: one connecting WDR37 with osteoarthritis in rats and humans11 and the other identifying it as one of several possible loci affecting renal function in humans.12 Additional data collected via publicly available databases suggest a broad pattern of expression for Wdr37 in mice (BioGPS; Figure S3) with enrichment in ocular and brain tissues. Review of phenotype data for mice homozygous for a Wdr37 tm1a (“knockout first”) allele (Wdr37tm1a(KOMP)Wtsi) from the International Mouse Phenotyping Consortium (IMPC) showed significant associations (p < .0001) with decreased body weight, decreased grip strength, and skeletal abnormalities of the spine, as well as a possible increase (p ≤ 0.05) in ocular (lens and corneal) anomalies, hearing defects, and changes in the size of the ulna, radius, and hindpaw. In addition, statistical analysis of the number of Wdr37 wild type, heterozygous, and homozygous pups from the IMPC indicates a significant decrease in the number of homozygous pups compared to expected Mendelian ratios (p < .001), suggesting decreased in utero survival. This line has not been characterized to determine how the tm1a allele affects the function of Wdr37.
Protein sequence analysis predicts seven WD domains in the WDR37 C-terminal region from amino acid 154 to amino acid 493 (Figures 2A and S2) (Uniprot).13 All the amino acids affected by the identified de novo variants are located upstream of the predicted WD domains and show high conservation among various species (Figure 2B). Analysis of wild-type WDR37 and its variants in silico using I-TASSER Suite and PyMOL predicted a seven-bladed propeller-like structure in the C-terminal region (corresponding to the seven WD-domains) with all four human variants located in the N-terminal region (Figure 2C); the missense variants are predicted to result in a conformational change affecting both the N- and C-terminal regions of the protein including the possible addition of an eighth propeller blade (Figure 2C). To further explore the functional role(s) of WDR37, we performed experiments in human cell culture and zebrafish (Supplemental Material and Methods).
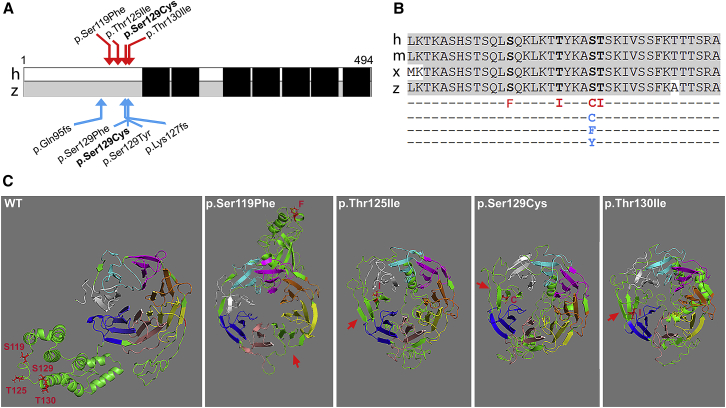
Figure 2.
Characterization of WDR37 Variants
(A) Schematic of human (h) and zebrafish (z) WDR37/wdr37. Human variants are indicated above with red arrows and zebrafish variants are indicated below with blue arrows, WD domains are indicated with black boxes.
(B) Protein sequence alignment of the affected region. Sequence alignment shown for human (h; GenBank: NP_054742.2), mouse (m; GenBank: NP_001034477.1), Xenopus (x; GenBank: NP_001096465.1) and zebrafish (z; GenBank: NP_001161736.1) proteins. Human variants are indicated in red in line 5; zebrafish missense variants are shown in blue in lines 6, 7, and 8.
(C) Predicted WDR37 structure. Protein structure shown for WDR37 wild-type (WT) and mutant sequence, based on I-TASSER top predicted models; the top identified structural analog for the WT protein was RCSB Protein Data Bank assembly 5o9zl, a pre-catalytic human spliceosome primed for activation. Amino acids affected by human variants are indicated in red in WT structure; the red arrow indicates a predicted additional blade in mutant proteins. WT, wild-type; S, Ser; F, Phe; T, Thr; I, Ile; C, Cys; Y, Tyr.
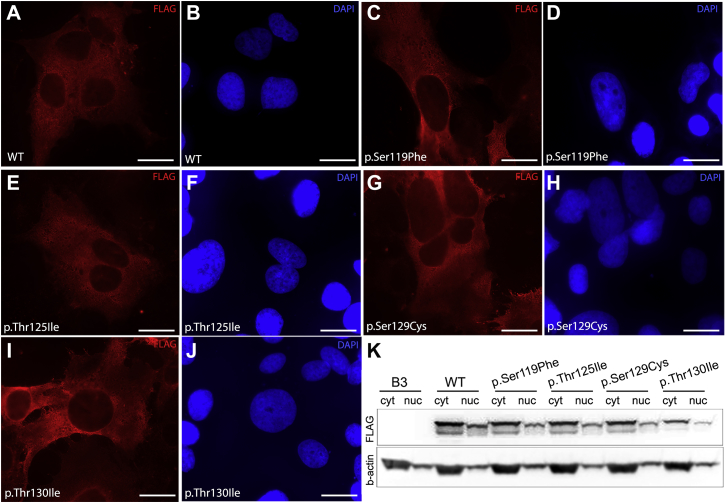
Wild-type (WT) and mutant WDR37 (WDR37_Ser119Phe, WDR37_Thr125Ile, WDR37_Ser129Cys, and WDR37_Thr130Ile) FLAG-tagged expression constructs were generated and transfected into human lens epithelial cells (B-3 (ATCC CRL-11421)) to examine protein levels and cellular localization (Figure 3). Immunocytochemistry with anti-FLAG revealed a predominantly cytoplasmic staining for WT and all mutant proteins (Figures 3A–3J). These results were further verified using anti-FLAG western blotting of cytoplasmic- and nuclei-enriched fractions prepared from lens cells transfected with the above constructs. All mutant proteins demonstrated comparable levels and cellular distribution to wild-type WDR37 (Figures 3K and S4).
Figure 3.
Protein Studies of Wild-Type and Mutant WDR37
(A–J) Immunocytochemistry demonstrating cellular localization of WT and mutant WDR37. Anti-FLAG staining (red) revealed predominantly cytoplasmic staining for wild type and all mutant proteins with missense variants. Specific variants are indicated in the lower left corner. DAPI staining in blue highlights the nucleus. Scale bar = 20 microns.
(K) Western blot analysis of cytoplasmic (cyt) and nuclear (nuc) fractions from human lens epithelial cells transfected with either WT or mutant WDR37 expression plasmids confirmed predominantly cytoplasmic presence with comparable patterns between mutant and wild type proteins. B3, untransfected B3 human lens epithelial cells; WT, wild-type.
Analysis of the zebrafish genome identified a single ortholog for human WDR37, wdr37, located on chromosome 24. Analogous to humans, zebrafish wdr37 contains 14 exons with similar exon/intron structure and is also predicted to encode a 494-aa protein; the human and zebrafish proteins show 86% identity (Figure S2) and conservation for all positions affected in the presented individuals (Figure 2B). We performed expression studies of wdr37 using in situ hybridization. This analysis revealed robust and broad expression for wdr37 in zebrafish embryos (Figure S3), indicating a possible role in development.
We first analyzed the effects of mutant WDR37 on embryo development via injections of the mRNAs encoding for wild-type or mutant proteins (for the p.Thr125Ile and p.Ser129Cys variants) into fertilized zebrafish eggs at the 1- to 4-cells stage followed by observation of the developing embryos until 5 dpf (days post fertilization). Expression of exogenous human WDR37 and encoded protein was confirmed via RT-PCR and western blot using 2-dpf embryos (Figure S4), but no visible effect on embryo development up to 5 dpf was observed.
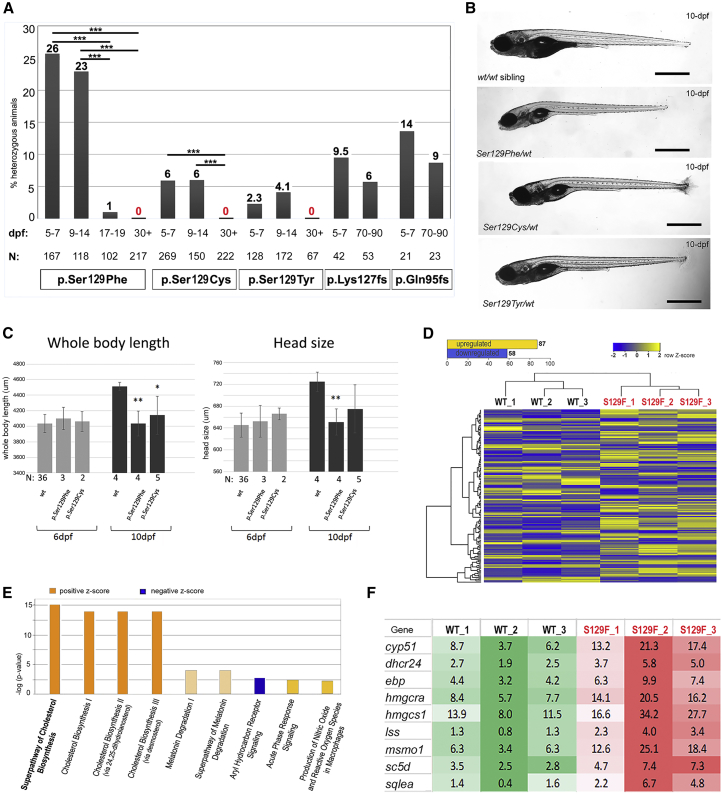
We next utilized CRISPR-Cas9-mediated genome editing to generate novel alleles in zebrafish wdr37, both frameshift and missense. Three missense variants were identified in the offspring (F1) of mosaic founders (F0): c.386C>T (p.Ser129Phe), c.386C>G (p.Ser129Cys), and c. 386C>A (p.Ser129Tyr) (Figures 2A, 2B, and S1). All changes modify the Ser129 affected in individual 4 (with p.Ser129Cys being the exact replica of the human substitution) and were recovered in ∼26%, 6%, and 2%, respectively, upon testing of 5- to 7-dpf progeny of various F0 mosaic parents. In addition, two frameshift alleles were isolated, c.377_396+4del p.Lys127Cysfs and c.282_291del p.Gln95Argfs, in ∼10% and 14% of embryos produced by mosaic founders. The mosaic parents were bred multiple times to generate additional progeny carrying the above variants, which were analyzed for their survival and phenotype. Similar to the results from injection of human mutant mRNA, no obvious phenotype was detected at 1–5 dpf. The most striking feature identified in F1 fish carrying the p.Ser129Phe, p.Ser129Cys, or p.Ser129Tyr heterozygous alleles was their larval lethality as no heterozygous animals were recovered after 1 month post fertilization (Figure 4A); in contrast, fish heterozygous for the c.377_396+4del and c.282_291del frameshift alleles were identified in 6% and 9% of their respective adult populations (at 2–3 months post fertilization). Statistical analysis confirmed significant differences between the proportion of embryos heterozygous for the p.Ser129Phe allele at either 5–7 or 9–14 dpf and at 17–19 or 30+ dpf (p < 0.0001), as well as the proportion of embryos heterozygous for the p.Ser129Cys alleles at either 5–7 or 9–14 dpf and at 30+ dpf (p < 0.0005) (Figure 4A). Similarly, no heterozygous larvae older than 30 dpf were detected for the p.Ser129Tyr allele (Figure 4A). Conversely, there was no significant difference between the proportion of heterozygous fish with frameshift wdr37 variants at different stages with many animals surviving to adulthood. Careful observation under a stereomicroscope of mutant larvae carrying missense alleles at 6 dpf (n = 31) and 10 dpf (n = 33) did not identify any obvious defects that explain their early death; no coloboma, corneal anomalies, or consistent craniofacial abnormalities were observed. One common feature of fish carrying missense alleles was their poor growth prior to death (Figure 4B); measurements were undertaken for larvae heterozygous for p.Ser129Phe or p.Ser129Cys variants and confirmed a statistically significant decrease in overall body length (p < 0.01 and p < 0.05, correspondingly) as well as head size (significant for p.Ser129Phe only, p < 0.01) at 10 dpf, while body length and head size appeared normal at 6-dpf for both mutants (Figure 4C). The ratio of head to body length was calculated for both the missense mutants and wild-type embryos; comparison of these ratios did not identify any significant difference, indicating that the observed reduction in head size was not specific to the anterior structures but was proportionate to the overall decrease in body size. While additional studies are needed to determine the cause of the early lethality, overall, these data support a critical role for wdr37 in normal vertebrate development and pathogenicity of the missense alleles affecting the N-terminal region of WDR37/wdr37. The presence of a severe larval phenotype (poor growth, lethality) in heterozygous animals carrying missense but not frameshift variants suggests a dominant-negative mechanism for the WDR37/wdr37 missense alleles.
Figure 4.
wdr37 Function in Zebrafish
(A) Effect of wdr37 variants on larval survival. Note that no larvae heterozygous for any missense allele were detected at 30+ dpf; conversely, similar percentages of animals carrying frameshift alleles were recovered at embryonic (5–7 dpf) and adult (70–90 dpf) stages. Black bars indicate the percentage of fish with heterozygous variants identified per stage, dpf corresponds to age (days post fertilization); N, total number of offspring genotyped; statistically significant differences indicated (∗∗∗p < 0.0005).
(B) 10-dpf WT larvae and fish heterozygous for wdr37 p.Ser129Cys, p.Ser129Phe, p.Ser129Tyr variants are shown; fish carrying missense variants in general looked smaller and less viable; same magnification images are shown with scale bar = 775 microns.
(C) Total body length and head size data (in μm) for wild-type and mutant larvae at 6- and 10-dpf; N, total number of offspring measured; statistically significant differences indicated (∗∗p < 0.01 and ∗p < 0.05).
(D) RNA-seq data analysis of 5-dpf wdr37 heterozygous fish carrying the p.Ser129Phe allele in comparison to their wild-type siblings. 87 targets were upregulated and 58 were downregulated with p < 0.05 (see Table S1). Heatmap of the one-way hierarchical clustering using Z-score for normalized value (log2 based) that includes all 145 significantly changed transcripts is shown.
(E) Graphical representation of the top modified pathways identified through Ingenuity Pathway Analysis of transcripts altered in zebrafish heterozygous for the p.Ser129Phe variant shows cholesterol biosynthesis to be the top upregulated pathways.
(F) Heatmap of specific upregulated transcripts within the Super Pathway of Cholesterol Biosynthesis (Ingenuity Pathway Analysis). Out of the 28 transcripts included in the Super Pathway of Cholesterol Synthesis, 9 were upregulated (activation z-score = 3 and p value 6.37E−16) in heterozygous wdr37 p.Ser129Phe mutants in comparison to their wild-type siblings. The averaged FPKM (fragments per kilobase of transcript per million mapped reads) values are shown. S129F, p.Ser129Phe; WT, wild-type.
To identify possible pathways involved in WDR37-associated disease, RNA-seq was performed on RNA extracted from embryos heterozygous for the p.Ser129Phe variant and their wild-type siblings at 5 dpf. The expression levels of 145 zebrafish transcripts were affected, and human orthologs were identified for 92 of these (Figure 4D; Table S3). Analysis of these 92 transcripts using the Core Analysis function of Ingenuity Pathway Analysis identified upregulation of genes involved in cholesterol biosynthesis as the most significantly modified pathway (Figure 4E). Out of the 28 transcripts included in the Super Pathway of Cholesterol Synthesis, 9 were upregulated (activation z-score = 3 and p value 6.37E−16) (Figure 4F). The most affected cellular functions were steroid metabolism, which was significantly increased (z-score = 2.207), and accumulation of lipids, which was decreased (z-score = 1.747).
Based on the data presented here, heterozygous missense variants in WDR37 in humans are causally associated with a multisystemic disorder characterized by poor growth and anomalies of the ocular, craniofacial, neurological, cardiovascular, genitourinary, skeletal, and gastrointestinal systems. Early lethality at less than 2 years of age occurred in one of four presented individuals. Zebrafish with CRISPR-Cas9-induced de novo missense variants similarly showed poor growth with shortened whole body length and proportionate small head evident by 10 dpf along with striking early lethality.
Two of the four individuals described here had clinical diagnoses of possible Peters-plus (PPS) or Walker-Warburg syndromes (WWS). The ocular phenotype in affected individuals is characterized by coloboma, lens anomalies, microcornea, and corneal opacity most consistent with Peters anomaly type 1, recently also described as kerato-irido-lenticular dysgenesis (KILD).14 Each of the affected persons displayed multiple additional eye anomalies suggesting generalized ocular dysgenesis. Classic PPS is characterized by Peters anomaly along with other developmental ocular anomalies in some cases, short stature, brachydactyly, dysmorphic features, variable cleft lip/palate, cardiac, and genitourinary anomalies8 and is caused by recessive pathogenic variants in glucosyltransferase B3GLCT.3 Numerous individuals with similar, overlapping features that do not meet classic criteria are largely unexplained genetically.1 WWS is a dystroglycanopathy characterized by both anterior and posterior segment ocular anomalies, muscular dystrophy, and severe structural brain anomalies with early death.6 Recessive variants in 18 different genes have been identified in individuals with dystroglycanopathies, the majority of which are glycosyltransferases, with additional cases still unexplained.7 Since both PPS and WWS are autosomal-recessive disorders while WDR37 syndrome is caused by de novo, dominantly expressed variants, accurate diagnosis is important for genetic counseling regarding recurrence risks.
The function of WDR37 is currently unknown. WD40 repeat (WD) domains represent a common protein interaction domain in humans, generally mediating interaction with other proteins, with more than 360 proteins identified that contain these regions.15, 16 Pathogenic variants in various WD domain-containing proteins have been linked to both dominant and recessive human ocular, neurological, skeletal, and genitourinary syndromes.17, 18 All of the variants reported here in WDR37 are upstream of the WD40 domains identified in Uniprot.13 Similar dominant missense variants upstream of the identified WD domains have been identified in other WD proteins including WDR26 and TBL1XR1.19 However, further structural modeling of one of these factors, WDR26, identified additional “imperfect” WD40 domains within the affected upstream region.18
RNA-seq data from mutant zebrafish suggest that WDR37 may play a role in the cholesterol biosynthesis pathway, with expression of dhcr24, sc5d, and ebp upregulated, among other transcripts. Comparison of the WDR37 phenotypes to known cholesterol biosynthesis disorders such as desmosterolosis (DHCR24 [MIM: 602398]), lathosterolosis (SC5D [MIM: 607330]), and MEND syndrome (EBP [MIM: 300960]) shows substantial overlap. Both groups are characterized by significant growth failure, structural brain defects, seizures, intellectual disability, microcephaly, ear and jaw anomalies, ophthalmological abnormalities (cataracts, microphthalmia, glaucoma, corneal clouding), defects of the cardiac, genitourinary, and skeletal systems, and early lethality.20 Further work is needed to determine how WDR37 interacts with the cholesterol pathway.
Based on the data presented in this paper, pathogenic missense WDR37 variants appear to produce stable proteins while causing conformational changes disruptive to normal WDR37 activity. It is possible that WDR37 must dimerize to perform its function and that mutant WDR37 forms steady, yet functionally impaired, dimers with wild-type protein, thus exerting a dominant-negative effect. Further analysis is needed to determine the exact functional impact of the variants as well as the roles that WDR37/wdr37 plays in vertebrate development.
Declaration of Interests
The authors declare no competing interests
Acknowledgments
The authors would like to thank the affected individuals and their families for their participation in this research study as well as the contributors to MyGene2 and the University of Washington Center for Mendelian Genomics for use of data. The authors also wish to acknowledge the expert work of Deborah Costakos, MD in the Department of Ophthalmology at the Medical College of Wisconsin for her review of ocular images and Leigh B. Thorne, MD, Thomas W. Bouldin, MD, and Vincent J. Moylan, Jr., MS, PA(ASCP)CM in the UNC Department of Pathology for their detailed post-mortem anatomic description of individual 2. This work was supported by the National Institutes of Health awards R01EY015518 and R01EY025718 and funds provided by the Children's Hospital of Wisconsin (E.V.S.).
Published: July 18, 2019
Footnotes
Supplemental Data can be found online at https://doi.org/10.1016/j.ajhg.2019.06.015.
Accession Numbers
The accession number for the RNA-seq data presented in this manuscript is ArrayExpress: E-MTAB-8029.
Web Resources
ArrayExpress, https://www.ebi.ac.uk/arrayexpress/
bioGPS, http://biogps.org/
gnomAD Browser, https://gnomad.broadinstitute.org/
MyGene2, https://mygene2.org/MyGene2/
OMIM, https://omim.org/
PyMOL, https://pymol.org/
UniProt, https://www.uniprot.org/
Supplemental Data
References
- 1.Weh E., Reis L.M., Happ H.C., Levin A.V., Wheeler P.G., David K.L., Carney E., Angle B., Hauser N., Semina E.V. Whole exome sequence analysis of Peters anomaly. Hum. Genet. 2014;133:1497–1511. doi: 10.1007/s00439-014-1481-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reis L.M., Semina E.V. Conserved genetic pathways associated with microphthalmia, anophthalmia, and coloboma. Birth Defects Res. C Embryo Today. 2015;105:96–113. doi: 10.1002/bdrc.21097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lesnik Oberstein S.A., Kriek M., White S.J., Kalf M.E., Szuhai K., den Dunnen J.T., Breuning M.H., Hennekam R.C. Peters Plus syndrome is caused by mutations in B3GALTL, a putative glycosyltransferase. Am. J. Hum. Genet. 2006;79:562–566. doi: 10.1086/507567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vissers L.E., van Ravenswaaij C.M., Admiraal R., Hurst J.A., de Vries B.B., Janssen I.M., van der Vliet W.A., Huys E.H., de Jong P.J., Hamel B.C. Mutations in a new member of the chromodomain gene family cause CHARGE syndrome. Nat. Genet. 2004;36:955–957. doi: 10.1038/ng1407. [DOI] [PubMed] [Google Scholar]
- 5.Alrakaf L., Al-Owain M.A., Busehail M., Alotaibi M.A., Monies D., Aldhalaan H.M., Alhashem A., Al-Hassnan Z.N., Rahbeeni Z.A., Murshedi F.A. Further delineation of Temtamy syndrome of corpus callosum and ocular abnormalities. Am. J. Med. Genet. A. 2018;176:715–721. doi: 10.1002/ajmg.a.38615. [DOI] [PubMed] [Google Scholar]
- 6.Vajsar J., Schachter H. Walker-Warburg syndrome. Orphanet J. Rare Dis. 2006;1:29. doi: 10.1186/1750-1172-1-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouchet-Séraphin C., Vuillaumier-Barrot S., Seta N. Dystroglycanopathies: About Numerous Genes Involved in Glycosylation of One Single Glycoprotein. J. Neuromuscul. Dis. 2015;2:27–38. [PubMed] [Google Scholar]
- 8.Weh E., Reis L.M., Tyler R.C., Bick D., Rhead W.J., Wallace S., McGregor T.L., Dills S.K., Chao M.C., Murray J.C., Semina E.V. Novel B3GALTL mutations in classic Peters plus syndrome and lack of mutations in a large cohort of patients with similar phenotypes. Clin. Genet. 2014;86:142–148. doi: 10.1111/cge.12241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deml B., Reis L.M., Lemyre E., Clark R.D., Kariminejad A., Semina E.V. Novel mutations in PAX6, OTX2 and NDP in anophthalmia, microphthalmia and coloboma. Eur. J. Hum. Genet. 2016;24:535–541. doi: 10.1038/ejhg.2015.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korostyński M., Małek N., Piechota M., Starowicz K. Blood Transcriptional Signatures for Disease Progression in a Rat Model of Osteoarthritis. Int. J. Genomics. 2017;2017:1746426. doi: 10.1155/2017/1746426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Köttgen A., Pattaro C., Böger C.A., Fuchsberger C., Olden M., Glazer N.L., Parsa A., Gao X., Yang Q., Smith A.V. New loci associated with kidney function and chronic kidney disease. Nat. Genet. 2010;42:376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017;45(D1):D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nischal K.K. Genetics of Congenital Corneal Opacification--Impact on Diagnosis and Treatment. Cornea. 2015;34(Suppl 10):S24–S34. doi: 10.1097/ICO.0000000000000552. [DOI] [PubMed] [Google Scholar]
- 15.Schapira M., Tyers M., Torrent M., Arrowsmith C.H. WD40 repeat domain proteins: a novel target class? Nat. Rev. Drug Discov. 2017;16:773–786. doi: 10.1038/nrd.2017.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu C., Min J. Structure and function of WD40 domain proteins. Protein Cell. 2011;2:202–214. doi: 10.1007/s13238-011-1018-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Rakan M.A., Abothnain M.D., Alrifai M.T., Alfadhel M. Extending the ophthalmological phenotype of Galloway-Mowat syndrome with distinct retinal dysfunction: a report and review of ocular findings. BMC Ophthalmol. 2018;18:147. doi: 10.1186/s12886-018-0820-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skraban C.M., Wells C.F., Markose P., Cho M.T., Nesbitt A.I., Au P.Y.B., Begtrup A., Bernat J.A., Bird L.M., Cao K., Deciphering Developmental Disorders Study WDR26 Haploinsufficiency Causes a Recognizable Syndrome of Intellectual Disability, Seizures, Abnormal Gait, and Distinctive Facial Features. Am. J. Hum. Genet. 2017;101:139–148. doi: 10.1016/j.ajhg.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A., Cooper D.N. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herman G.E., Kratz L. Disorders of sterol synthesis: beyond Smith-Lemli-Opitz syndrome. Am. J. Med. Genet. C. Semin. Med. Genet. 2012;160C:301–321. doi: 10.1002/ajmg.c.31340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.