Abstract
Many Gram‐negative bacteria pathogenic to plants and animals deploy the type III secretion system (T3SS) to inject virulence factors into their hosts. All bacteria that rely on the T3SS to cause infectious diseases in humans have developed antibiotic resistance. The T3SS is an attractive target for developing new antibiotics because it is essential in virulence, and part of its structural component is exposed on the bacterial surface. The structural component of the T3SS is the needle apparatus, which is assembled from over 20 different proteins and consists of a base, an extracellular needle, a tip, and a translocon. This review summarizes the current knowledge on the structure and assembly of the needle, tip, and translocon.
Keywords: bacteria, T3SS needle apparatus, tip complex, translocon, type III secretion system
Abbreviations
- cryo‐EM
cryo‐electron microscopy
- rmsd
root‐mean‐square deviation
- ssNMR
solid‐state NMR
- T3SS
type III secretion system
- TEM
transmission electron microscopy
1. INTRODUCTION
Pathogenic Gram‐negative bacteria use various types of protein secretion systems1, 2 to deliver virulence effector proteins into eukaryotic cells to establish infection. Human pathogens such as Salmonella spp., Shigella spp., Burkholderia spp., Yersinia pestis, Pseudomonas aeruginosa, Chlamydia spp.,3, 4 Vibrio cholerae,5 and enteropathogenic and enterohemorrhagic Escherichia coli 6 assemble and utilize the type III secretion system (T3SS) to infect their hosts, causing a variety of infectious diseases such as plague, typhoid fever, sexually transmitted diseases and dysentery.7, 8, 9, 10, 11 These bacteria that rely on the T3SS to cause infectious diseases have developed multidrug resistant strains.12, 13, 14 The T3SS is also used by plant pathogenic bacteria such as Pseudomonas syringae, Ralstonia solanacearum, Xanthomonas spp., and Erwinia spp. to establish diverse plant–bacteria interactions.15, 16, 17
The structural component of the T3SS is a syringe‐like needle apparatus (Figure 1) that is assembled from over 20 different proteins, forming a base, an extracellular needle, a tip complex, and a translocon.8, 10, 18, 19, 20, 21, 22 The base spans the bacterial inner and outer membranes whereas the translocon spans the host cell plasma membrane. Results from biophysical, biochemical and genetic approaches have provided insights into the architecture and formation of the needle apparatus.10, 23, 24, 25, 26 This structural framework is conserved across the T3SS of animal and plant bacterial pathogens. The proteins that form the T3SS needle apparatus (Figure 1) are described using the proposed unified nomenclature based on Sct (for Secretion and cellular translocation).7, 9, 20, 22 The basal body consists of ring‐like structures spanning the inner and outer membranes of bacteria (Figure 1). The inner membrane concentric rings are known as SctJ and SctD. The outer membrane ring is formed by the secretin SctC. The export apparatus is located just beneath the basal body and consists of five membrane proteins SctR, SctS, SctT, SctU, and SctV. The C‐ring lies below the export apparatus, and is made up of the protein SctQ and the ATPase complex. The ATPase complex consists of the ATPase SctN, the stator protein SctL, the stalk protein SctO and the cofactor SctK. Both the C‐ring and the ATPase complex form the sorting platform of the T3SS for substrate recruitment and secretion.
Figure 1.
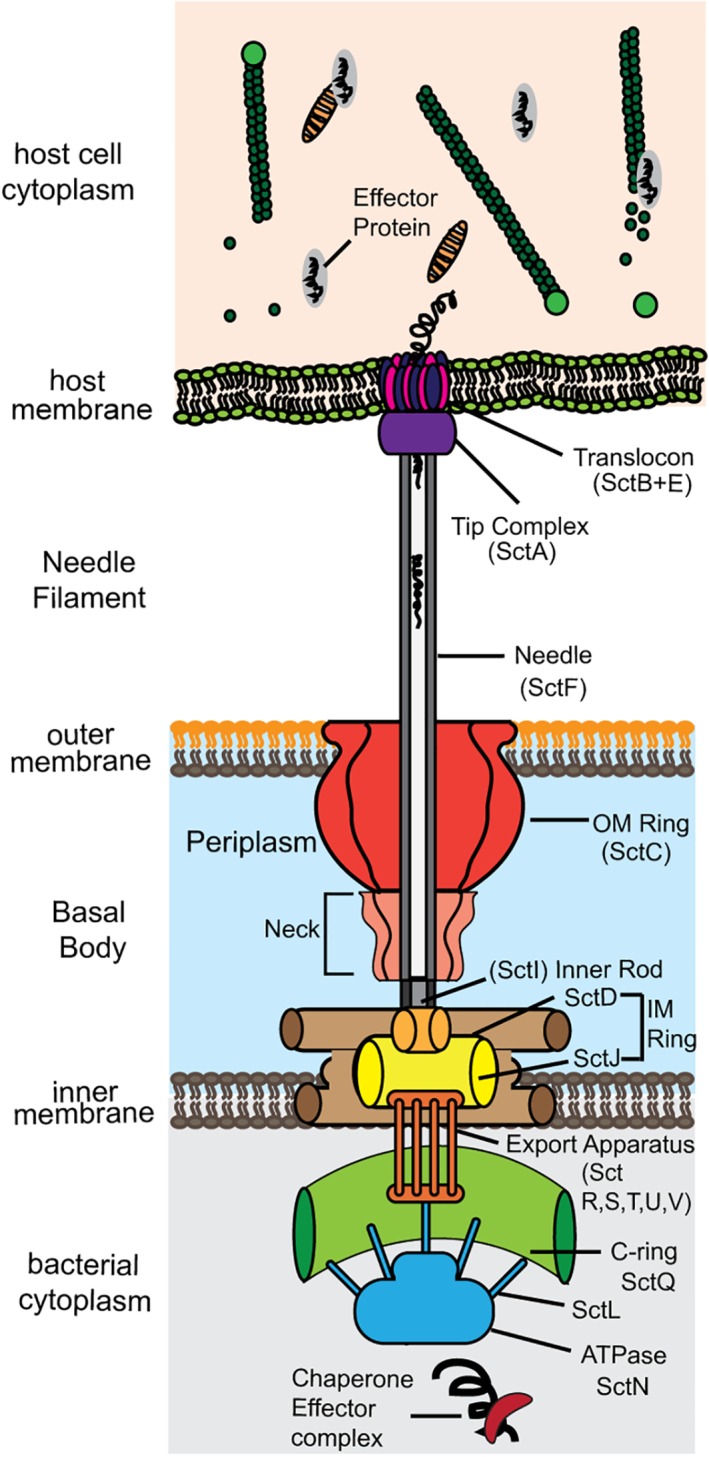
Names of proteins that form the components of the T3SS needle apparatus using the Sct nomenclature (for secretion and cellular translocation)
The needle, a cylindrical structure formed by oligomers of SctF, extends from the bacterial surface to the external environment (Figure 1). The inner rod protein SctI is connected to the inner membrane ring of the basal body, and possibly helps to anchor the needle to the basal body. The external end of the needle is capped with SctA, the tip protein, which is thought to form a five‐membered ring complex at the tip of the needle. The tip complex serves as a platform for the assembly of the translocon, which forms a translocon pore in the host cell membrane. The translocon is formed by SctB and SctE, and the precise stoichiometry and copy numbers of SctB and SctE in the translocon remains unclear. The needle apparatus forms a continuous conduit between the bacterial cytosol and the host cytoplasm, and this allows the injection of bacterial effector proteins into the host.
The T3SS among bacteria can be divided into the following subfamilies (Table 1): (a) the Inv‐Mxi‐Spa family, which includes the T3SS of Salmonella spp. (encoded by the Salmonella Pathogenicity Island‐1, or SPI‐1), Shigella and Burkholderia; (b) the Ysc‐family, which includes the Ysc system of Yersinia spp., the Psc system of Pseudomonas aeruginosa and the Asc system of Aeromonas spp.; (c) the Ssa‐Esc family, which includes the SPI‐2 from Salmonella enterica, enteropathogenic and enterohemorrhagic Escherichia coli (EPEC, EHEC); and (d) Hrp‐1 and Hrp‐2 from plant T3SS, and others like Desulfovibrionales, Chlamydiales, Myxococcales, and Rhizobiales. In this review, we highlight the structure and assembly of the needle, tip, and translocon of the T3SS from the Invi‐Mxi‐Spa and Ysc family (Table 1).
Table 1.
Names of the needle, tip, and translocon proteins of the Inv‐Mxi‐Spa and Ysc family of T3SS
| Proteins | Scta | Inv‐Mxi‐Spa family | Ysc family | ||||
|---|---|---|---|---|---|---|---|
| Salmonella (SPI‐1)b | Shigella | Burkholderia T3SS‐3c | Yersinia Ysc | P. aeruginosa Psc | Aeromonas Asc | ||
| Needle | SctF | PrgI | MxiH | BsaL | YscF | PscF | AscF |
| Tip | SctA | SipD | IpaD | BipD | LcrV | PcrV | AcrV |
| Translocon (major) | SctE | SipB | IpaB | BipB | YopB | PopB | AopB |
| Translocon (minor) | SctB | SipC | IpaC | BipC | YopD | PopD | AopD |
Sct, for secretion and cellular translocation.
SPI‐1, Salmonella Pathogenicity Island‐1 (Salmonella has SPI‐1 and SPI‐2).
Burkholderia pseudomallei has three T3SS.
2. STRUCTURE OF THE ASSEMBLED NEEDLES AND NEEDLE MONOMERS
Currently, the atomic structures of the assembled needles (Figure 2a) are only available for Salmonella and Shigella needles of the Inv‐Mxi‐Spa family of T3SS.27, 29, 33, 34, 35, 36 The atomic structure of the assembled needle for any of the Ysc family remains unknown. The needle has nanoscale dimensions of ~50 nm long, ~7 nm wide, and a lumen diameter from 1.5 nm36 to 2.5 nm27, 37 Electron microscopy of partially purified needles from P. aeruginosa showed structures that were 60–80 nm long and 6–8 nm wide.38 The needle is assembled from the polymerization of many copies of a small needle protein of about 80 residues, and arranged in a helical symmetry along the needle axis. The copy numbers of needle monomers that assemble into needles vary from ~120 copies for Salmonella 26 to about 200–300 copies for Yersinia 39 The T3SS needle was first visualized at low resolution by electron microscopy in 1998.40 The atomic‐resolution structures of the Salmonella and Shigella needles reconstituted from recombinant needle monomers, PrgI and MxiH, respectively27, 34, 35 were determined by a combination of solid‐state NMR (ssNMR), electron microscopy, and Rosetta modeling (Figure 2a).27, 34, 35
Figure 2.
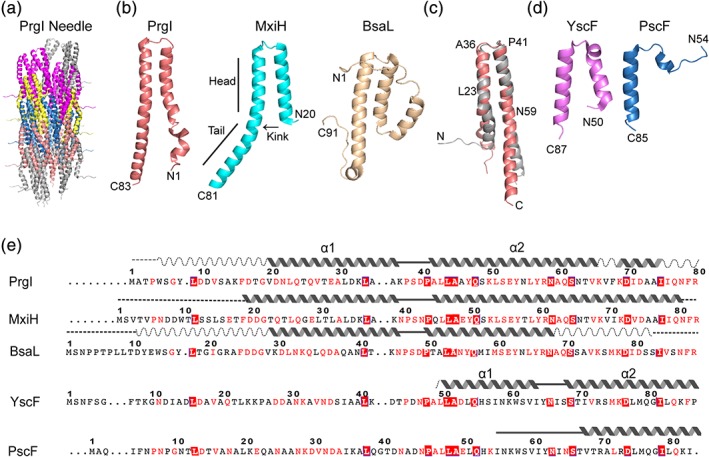
Structures of T3SS needle and needle proteins from various bacteria. (a) Atomic structure of the Salmonella needle derived by solid‐state NMR.27 Groups of PrgI monomer are colored differently (gray, purple, yellow, blue, and pink) for clarity. (b) Structure of Salmonella PrgI.28 Shigella MxiH29 and Burkholderia BsaL.30 (c) Superposition of a PrgI subunit from the ssNMR structure of the PrgI needle from (a) (gray) and the solution NMR structure of a monomeric PrgI (light orange) from (b). (d) Crystal structures of Yersinia YscF31 and P. aeruginosa PscF.32 (e) Sequence alignment of T3SS needle proteins that have been structurally characterized shown with well‐structured helices (solid wavy lines), regions with partial helices (dashed wavy lines), ordered loops (solid lines), and random coil regions (dashed lines). Amino acid residues are colored based on sequence identity (boxed solid red) or sequence conservation (red letters)
2.1. Atomic structures of the T3SS needles
In the assembled Salmonella needle (Figure 2a), each PrgI monomer forms an α‐helical hairpin comprising of an N‐terminal helix (residues 9–35), a turn at the PxxP motif, and a C‐terminal helix (residues 42–80).27 The α‐hairpin is flanked by flexible regions at the N‐ and C‐termini. To assemble the needle, 11 monomers are arranged in a right‐handed helical symmetry per turn around the lumen of the needle. The N‐terminal helix forms the outside wall of the needle, positioning the flexible N‐terminal eight residues on the outer surface of the needle. The C‐terminal helix is located inside the needle, forming the wall of the lumen.27
The ssNMR‐derived structure (Figure 2a) is the current atomic model of the needle27, 34, 35 and has since been confirmed,41 and further refined by cryo‐EM.35, 36 There are major differences between the current ssNMR structure of the needle from earlier models derived by crystallography and electron microscopy.29, 33 In earlier models,29, 33 the N‐terminal helix is positioned inside the needle, forming the wall of the lumen; with the C‐terminal helix positioned on the outside surface of the needle. Likewise, in the current model,27 there are no β‐strands42 or β‐hairpin33, 43 in needle monomers as had been proposed earlier. A cryo‐EM density on the surface of the Shigella needle was assigned to a β‐hairpin (Q51–Q64) at the C‐terminal region of MxiH.33 The current model reassigned this density to a short α helix at the N‐terminal residues P6‐D9 of MxiH.35
2.2. Structures of PrgI, MxiH, and BsaL needle monomers
The atomic structures of needle proteins have also been determined in their monomeric forms (Figure 2b) for the Inv‐Mxi‐Spa family. The needle proteins Burkholderia BsaL,30 Salmonella PrgI28 and Shigella MxiH29 can be rendered monomeric by deletion of their C‐terminal five residues, allowing structural determination by solution NMR30 or crystallography.29 The C‐terminal deletions, however, are nonfunctional43 as they are incapable of assembling needles in vivo. A full length PrgI double mutant V65A/V67A is functional and monomeric below 0.3 mM concentration, allowing the determination of the crystal structure of PrgI.42
The structure of monomeric PrgI28, 42 and MxiH29 (Figure 2b) consists of α‐helical hairpins similar to the α‐helical hairpins of the PrgI27 and MxiH needles derived by ssNMR.35 The structure of the central portion of the α‐helical hairpin comprising of shorter segments of helices flanking the PxxP motif is similar to the structure of the PrgI or MxiH monomer determined by solution NMR or by crystallography and the structure of a subunit in the assembled PrgI or MxiH needle. The solution NMR structure of monomeric PrgI28, 42 can be superimposed with a subunit of the ssNMR‐derived structure of the PrgI needle27 with a Cα rmsd of 1.8 å for residues 23–36 and 41–59 (Figure 2c). The crystal structure of monomeric PrgI superimposes with a Cα rmsd of 1.3 å for residues 19–34 and 41–58. Likewise, the crystal structure of monomeric MxiH29 can be superimposed with a subunit of the ssNMR‐derived structure of the MxiH needle35 with Cα rmsd of 1.0 å for residues 26–57.
The NMR structure of monomeric BsaL30 also shows an α‐helical hairpin for residues (Figure 2b), flanked by flexible regions. Based on the structures of the PrgI and MxiH needles27, 35 and monomers,28, 29, 42 the flexible N‐ and C‐terminal regions of BsaL are expected to assume a more ordered α‐helical structures upon assembly into the BsaL needle. Despite having similar structural folds, the needle proteins MxiH, PrgI and BsaL have different electrostatic surfaces.28 In PrgI, the negatively charged surface is mostly located on the “side” of the α‐helical hairpin and runs almost the entire length of the N‐terminal helix, whereas for BsaL and MxiH, the negative charged surface is mostly located at the interface between the two helices.
2.3. Structures of YscF and PscF
The atomic structures of the assembled needles or the monomeric forms of the Ysc family, like the Yersinia YscF and the P. aeruginosa PscF, have not been determined. However, crystal structures are available for YscF31 and PscF32 in complex with their chaperones—YscG and YscE for Yersinia; and PscG and PscE for P. aeruginosa—respectively (Figure 2d). (In contrast, there are no known chaperones for the needle proteins of the Inv‐Mxi‐Spa family). For YscF, only the C‐terminal half of YscF, from residues 50–87, is visible in the crystal structure of the YscF‐YscE‐YscG complex (Figure 2D).31 YscF (residues 50–87) forms an α‐helical hairpin with helix 1 (residues 50–63) and helix 2 (residues 67–87) connected by a five‐residue loop (residues 64–68). Results of mutations show that YscF residues 27–30 are important for docking of the tip protein LcrV, while residues 64, 80, and 75 are primarily important for interacting with other YscF monomers.44 For PscF, only about a third of PscF—comprising of residue 55–85 at its C‐terminus (Figure 2d)—is visible in the crystal structure of the PscF‐PscE‐PscG complex.32 Residues 55–67 form an extended coil and residues 68–85 form an amphipathic 25 å α‐helix (Figure 2d). Residues in the extended coil interact with the needle chaperone PscG, whereas residues in the α‐helix are required for polymerization of the needle.32
These are currently the only available structures of YscF and PscF (Figure 2d), which have only been determined when bound to their chaperones. These atomic structures of YscF and PscF are different from the α‐helical hairpin structures of PrgI and MxiH in monomeric forms and in the assembled PrgI and MxiH needles (Figure 2e). These results suggest that the structures of YscF and PscF are different when bound to their chaperones, and when bound to other needle monomers in their respective assembled needles. The dearth of biophysical and structural data for monomeric YscF and PscF suggest challenges and difficulties in purifying and stabilizing YscF and PscF when separated from their chaperones. Because of this difficulty, we do not expect the atomic structures of YscF or PscF to be forthcoming in the near future. The structures of the YscF and PscF needles and monomers would have to be deduced from analogy with the PrgI and MxiH needles and monomers.
3. THE TIP PROTEINS
The tip protein forms a complex that caps the distal end of the needle.45, 46, 47 The tip protein complex serves as a platform for the assembly of the translocon pore.48 Crystal structures are available for the tip proteins SipD,49, 50 IpaD51 and BipD51, 52 of the Inv‐Mxi‐Spa family; and for LcrV of the Ysc family (Figure 3). A common structural feature of all tip proteins is the long central coiled‐coil motif (Figure 3). A major difference among tip proteins is the presence of the tip protein chaperone proteins for the Ysc family, whereas the Inv‐Mxi‐Spa family of tip proteins does not have chaperones.
Figure 3.
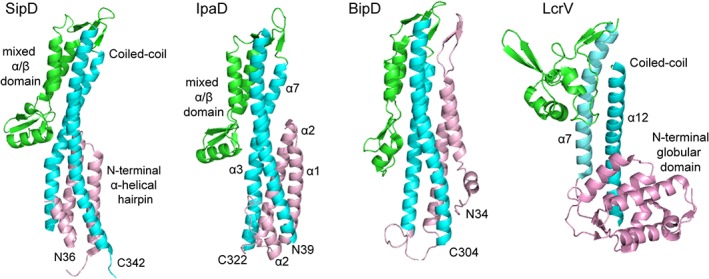
Crystal structures of the tip proteins SipD, IpaD, BipD of the Inv‐Mxi‐Spa family, and LcrV of the Ysc family of T3SS tip proteins, colored by their protein domains as follows: mixed α/β domain (green), coiled‐coil (cyan), and N‐terminal α‐helical hairpin or globular domain (pink)
In Salmonella and Shigella, the tip protein also functions as an environmental sensor by binding bile salts,49, 50, 53, 54, 55, 56 which controls the release of the translocon proteins. The tip protein complex is therefore hypothesized to exist in two states—first, a closed state, that prevents the egress of the translocon proteins from the needle; and second, an open state, which occurs upon contact of bacteria with the host eukaryotic cells where the translocon proteins exit the needle to form the translocon on the host cell membrane, and complete the assembly of the needle apparatus (Figure 4). Other small molecule scaffolds have since been identified that can bind to the tip proteins.57, 58
Figure 4.
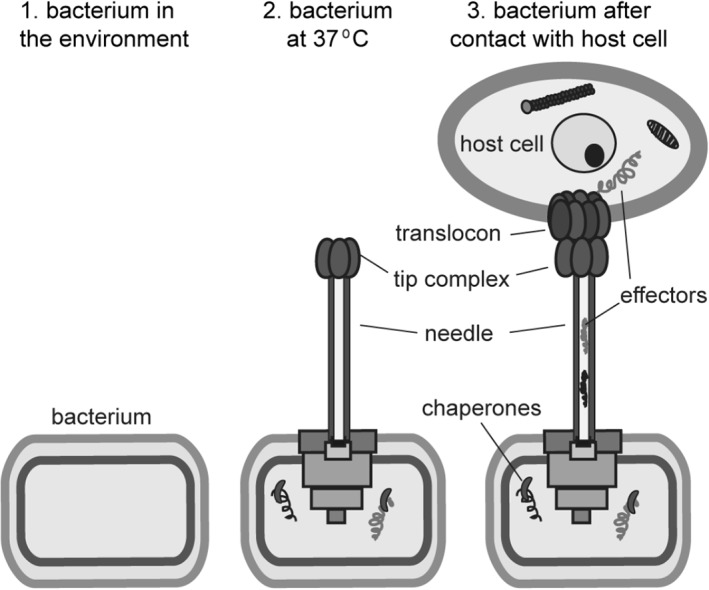
Cartoon depicting the stages in the assembly of the needle, tip and translocon. Increased temperature signals the assembly of the needle apparatus up to the tip complex. The translocon is assembled last, upon contact of bacteria with host cell
3.1. Structure of SipD, IpaD, and BipD
For the Inv‐Mxi‐Spa family of tip proteins, the crystal structures of Salmonella SipD,49, 50 Shigella IpaD,51 and Burkholderia BipD51, 52 show close structural homology (Figure 3). The Cα RMSD of SipD is 1.3 å with IpaD and 2.2 å with BipD, whereas IpaD and BipD show a Cα rmsd of 1.6 å. SipD, IpaD, and BipD share three common structural features – an N‐terminal α‐helical hairpin, a long central coiled‐coil, and a mixed α/β domain (Figure 3). The N‐terminal region forms an α‐helical hairpin that packs at one end of the long central coiled‐coil, forming a four‐helix bundle. The other end of the coil–coil leads to the mixed α/β domain, which in SipD, consists of three short α‐helices and five antiparallel β‐strands. Two α‐helices of the mixed α/β domain also pack against the central coiled‐coil region forming a four‐helix bundle at the other end of the coiled‐coil. There are structural differences among these tip proteins, notably, the N‐terminal hairpin of BipD is much longer compared to that of SipD and IpaD, and the orientation of the N‐terminal hairpins of SipD and IpaD with respect to the central coiled‐coil differs by 18°.
3.2. Structure of LcrV
For the Ysc family of tip proteins, the only available atomic structure is that of Yersinia LcrV (Figure 3).59, 60 Similar to the structures of the tip proteins above, LcrV contains a long central coiled‐coil consisting of two antiparallel α‐helices (formed by helix α7 and α12). A major difference among the tip proteins is that instead of an α‐helical hairpin seen in SipD, IpaD, or BipD (Figure 3), the N‐terminal region of LrcV (residues 25–147) folds into a globular domain. The LcrV N‐terminal globular domain consists of two antiparallel β‐strands and six α‐helices, and this domain packs at one end of the long central coiled‐coil. The other end of the coiled‐coil is connected to a second globular domain consisting of a mixture of four short α‐helices and four antiparallel β‐strands, similar to the mixed α/β domain of SipD, IpaD, and BipD. Thus, the structure of LcrV resembles a dumbbell with two globular domains on either side of a central coiled‐coil (Figure 2b). The atomic structure of the other member of the Ysc family, the Pseudomonas PcrV tip protein is currently unknown. PcrV shares 38% sequence identity (and 66% sequence similarity) with LcrV; it is expected to assume a similar structure.
Another major difference among the tip proteins is the presence of tip chaperone proteins for LcrV and PcrV whereas SipD, IpaD, and BipD are not known to have any chaperone proteins. LcrG61, 62 is the tip chaperone protein of LcrV, and PcrG63 is the tip chaperone protein of PcrV. LcrG64 and PcrG65 are small, partially folded proteins. They lack tertiary structures, however, they consist of partially formed alpha helices. The atomic structure of the LcrV‐LcrG or PcrV‐PcrG complex remains unknown.
4. THE TIP COMPLEX
At the distal end of the needle sits a tip complex, formed by several subunits of the tip protein. The Yersinia tip complex, formed by an estimated three to five copies of LcrV66 was first visualized by electron microscopy.45 Crosslinking data showed a direct association of LcrV with the needle protein YscF.67 The N‐terminal globular domain of LcrV is predicted to form the base of the tip complex, while the central globular domain forms the head.45, 66 Others reported that four copies of PcrV or LcrV can oligomerize in solution to form doughnut‐like structures with an internal diameter of 3–4 nm.68 In Shigella, a tip complex of IpaD forms at the tip of the MxiH needle.47, 69 The Shigella tip complex is estimated to be formed by five copies of IpaD at the needle tip.51, 70 Likewise, the Salmonella tip complex is estimated to be formed by pentameric SipD at the needle tip.49 The tip protein interacts with the needle protein, in order to assemble the tip complex. The IpaD N‐terminal α‐hairpin71 and the MxiH residues N43, P44, L47, Q51, and Y5029, 70, 71 are important for the IpaD–MxiH interaction. Likewise, in Salmonella, the SipD coiled‐coil region near its N‐terminal hairpin interacts with the needle protein PrgI.72, 73 Fitting the crystal structure of IpaD into electron density maps generated three‐dimensional models of the Shigella tip complex.24, 74
5. THE TRANSLOCON
The final step in the assembly of the T3SS needle apparatus is the formation of the translocon pore on the host cell membrane (Figure 5a), which then allows the injection of effector proteins directly into the host cytoplasm. The translocon pore is assembled by two hydrophobic membrane proteins—the minor translocon protein and the major translocon proteins, designated based on the relative sizes of these proteins to each other.77, 78, 79, 80 Each pair of major and minor translocon proteins share a common chaperone, for example, the Shigella IpaB and IpaC translocon proteins share the same chaperone, IpgC. The major translocon proteins are predicted to contain two transmembrane helices, whereas the minor translocon proteins are predicted to contain one transmembrane helix (Figure 5b). These proteins are predicted to contain coiled‐coils81 and intrinsically disordered regions.82, 83 Currently, the only known atomic structures of the translocon proteins are the crystal structures of the N‐terminal ectodomains of the major translocon proteins Shigella IpaB and Salmonella SipB,75 which form three helix bundles; and that of Aeromonas AopB76 in complex with its chaperone (Figure 5c). The structure of the translocon at atomic resolution is currently unknown. The highest‐resolution image of a T3SS translocon to date is the structure of the Salmonella translocon in a membrane environment (Figure 5a)48 obtained by cryo‐electron tomography at nanoscale resolution. The atomic structure for any minor translocon protein is currently unknown. The dearth of atomic‐level structural data on the translocon reflects the experimental challenges posed by these membrane proteins.
Figure 5.
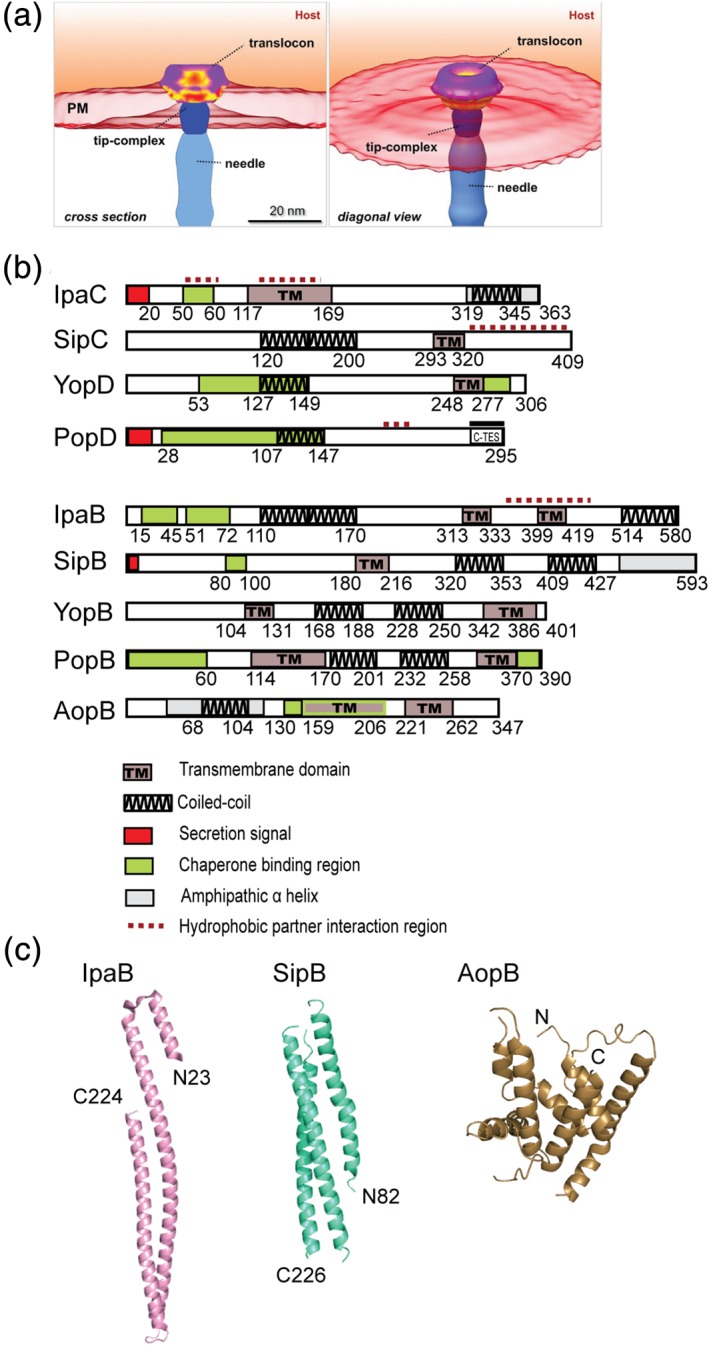
(a) Structure of the Salmonella translocon cryo‐electron tomography (images were from Park et al.48 and used with permission). (b) Domain maps of the minor and major translocon proteins. (c) Crystal structures of the major translocon proteins IpaB and SipB N‐terminal domains;75 and AopB76
5.1. The minor translocon proteins
Several functional domains have been identified or predicted for IpaC (Figure 5b), the Shigella minor translocon protein. The N‐terminal region (residues 1–116) contains the secretion signal and the binding sites for the IpaC chaperone, IpgC, and the Shigella major translocon protein, IpaB. IpaB competes with IpaC for binding IpgC. Residues 50–80 are important for the invasion function of IpaC.84 The transmembrane region was identified to reside in the middle of the protein, residues 117–169.85 This 53‐residue region was predicted to contain one transmembrane domain,85 and was reported to be involved in self‐association by yeast two‐hybrid.86 The C‐terminus (residues 319–345) is predicted to form into a coiled‐coil.87 The C‐terminal region is responsible for self‐oligomerization.85, 88 In addition to its structural role in the assembly of the translocon, IpaC also functions as an effector. The C‐terminal region of IpaC is important for its role as an effector, and is involved in cytoskeletal rearrangements and actin polymerization.87, 89, 90
For the Ysc family, YopD is the minor translocon protein in Yersinia, and it is predicted to contain a transmembrane domain.91 The chaperone‐binding regions of YopD are in the N‐terminal (residues 53–149) and the C‐terminal regions (residues 278–292).91 The C‐terminal region consists of a predicted coiled‐coil (residues 248–277)92 and an amphipathic α‐helix (residues 278–292), which is required for self‐oligomerization and interaction with the tip protein LcrV.93 The atomic structure of the amphipathic α‐helix was determined by NMR.94 The equivalent of YopD in P. aeruginosa, PopD, including the major translocon protein (PopB) has been reported to exist as molten globules.83, 95
5.2. The major translocon proteins
Results of secondary predictions, structural studies, deletions and point mutations have contributed toward our current understanding of the various structural and functional domains of the major translocon protein (Figure 5b).96, 97 For Shigella IpaB, its N‐terminal domain consists of the chaperone‐binding regions (residues 15–45 and 51–72);98, 99 and predicted coiled‐coil region (residues 110–170)100 and amphipathicα‐helical region (residues 240–280).96 Residues 227–236 and 297–306 of IpaB are required for pore formation.97 The crystal structure of the N‐terminal domain of IpaB (residues 74–224)75 shows three antiparallel α‐helices folded into a coiled‐coil structure (Figure 5c), and is structurally homologous (with Cα rmsd of 1.4 å) to the crystal structure of the IpaB homolog in Salmonella, SipB (residues 82–226).75 In the middle of IpaB are two predicted transmembrane domains (residues 313–333 and 399–419),100 one of which, overlaps with the reported IpaC‐binding region (residues 367–458) identified by yeast two‐hybrid and truncations.86 A long coiled‐coil region (residues 514–580) was predicted at the C‐terminus of IpaB100 and deletions of short three to nine residues in this region showed decreased association of IpaB with the needle.100
Currently, the only available atomic structure for a translocon protein that includes the transmembrane regions, is that of the crystal structure of Aeromonas hydrophilia major translocon protein AopB in complex with its chaperone AcrH (Figure 5c).76 Residues 40–264 of the 347‐residue AopB formed a complex with AcrH without requiring any membrane mimics. The protein–protein interactions of AcrH with the transmembrane helices of AopB allowed formation of the AopB‐AcrH complex in solution. The atomic structure for any of the major or minor translocon proteins in a membrane environment remains unknown.
5.3. Translocon–tip interaction
For Shigella, the tip protein IpaD is needed for the insertion of translocon IpaB and IpaC into the target cell membrane.77, 101, 102, 103 Antibodies targeted to the surface‐exposed N‐terminal region of IpaD reduce the ability of Shigella to insert IpaB and IpaC into erythrocyte membranes and form pores,69 blocking bacterial entry into epithelial cells. IpaB is localized at the needle tip via interaction with IpaD.70, 104 On binding to IpaD, the predicted coiled‐coil (residues 167–177) region of IpaB remains exposed, as they are easily accessible to IpaB antibodies.70 The stoichiometry of the IpaB–IpaD interaction is 1:4.51, 70 The role of bile salts in recruiting IpaB at the needle tip was proposed.104 Bile salts (deoxycholate, chenodeoxycholate, and taurodeoxycholate) are required for the localization of IpaB at the needle tip in presence of IpaD. Bile salts bind to IpaD where the N‐terminal domain meets the central coiled‐coil region, and induce conformational changes in other regions.53 The conformational changes in IpaD in presence of bile salts regulate its binding to IpaB.54, 56 The distal domain of IpaD moves further away from the C‐terminus in presence of deoxycholate105 to accommodate IpaB N‐terminal region (residues 11–226). IpaB residues 11–27 are important for binding to IpaD.105
Results from NMR106, 107 showed that the IpaD mixed α/β domain with the nearby coiled‐coil interacts with the N‐terminal domain of IpaB (residues 9–226). Similar results were obtained for the interaction of the Salmonella SipD tip protein and the SipB translocon protein.58, 106 A model of IpaB–IpaD interaction based on electron microscopy suggests a IpaB:IpaD stoichiometry of 1:4 at the needle tip.24 Initially, five copies IpaD form a tip complex at the tip of the needle tip, and upon signal to assemble the translocon, one of the subunits of IpaD is displaced by IpaB.24
The Yersinia minor (YopD) and major (YopB) translocon proteins interact with the tip protein LcrV,108 which forms a platform for the assembly of the translocon.66 YopD interacts with LcrV,93 and YopB interacts with the N‐terminal domain of LcrV and inserts into the host cell membrane.66 In P. aeruginosa, the C‐terminal region of PopD interacts with the N‐terminal globular domain of PcrV, and this interaction is required for controlling the secretion of effector proteins.109
5.4. Assembly of the translocon on membranes
Animal pathogens containing the T3SS form pores in erythrocyte membranes and liposomes. The Shigella translocon proteins IpaB and IpaC form pores on membranes estimated at 25 å diameter.77 Results from cryo‐electron tomography reveal that the Salmonella SipB/SipC translocon spanning the host membrane has a thickness of 8.1 nm and a pore diameter of 13.5 nm (Figure 5a).48 In Salmonella, SipB interacts with SipC through the SipC C‐terminal region (340–409) to form pores within host cells.110 SipB form trimers and hexamers on phospholipid vesicles through its N‐terminal coiled‐coil regions.111 The Yersinia translocon proteins YopB and YopD form pores in the host cell membranes estimated to be 1.6–2.3 nm diameter.78 The stoichiometry of YopD:YopB in the pore complex was estimated to be 2.4:1.112 Others have estimated the Y. enterocolitica pore complex (from native gel) to contain YopB‐YopD oligomers of 600 kDa, which is equivalent to 15–20 YopB‐YopD monomers.113 YopD is present as decamers and dimers in the pore complex.
PopB and PopD formed pores on erythrocyte membranes, estimated to be at 2.8–3.5 nm in diameter based on osmoprotection.114 Pores formed by recombinant PopB and PopD on liposomes were visualized by TEM,79 and showed ring‐like structures with inner diameter of 4 nm and outer diameter of 8 nm. PopB or PopD formed pores on liposomes individually, or after mixing in equimolar amounts, indicating that both proteins oligomerize into ring‐like structures on membranes.79 Others have observed PopB and PopD pores with 3.4–6.1 nm diameter,115 and PopD forming into hexameric structures while PopB assembles into 6–12 subunits on membranes.116 When both PopB and PopD were added together, eight subunits each of PopB and PopD associated and formed into a 16‐subunit complex on membranes.116 Acidic pH changes the protonation of PopD residues 63–81, making this segment more hydrophobic, and together with the predicted transmembrane segment (residues 119–137), assists PopD in the formation of the translocon pore.117 Recently, it has been observed that the PopB‐PopD translocon remains attached to the host cell membrane even after bacterium has left the site of contact.118
6. CONCLUSIONS
There has been much progress in understanding the atomic structures of the needle, tip, and translocon proteins since the visualization of the needle apparatus over two decades ago. The atomic structures of the needle and the tip proteins are known. However major challenges remain regarding the atomic structures of the translocon proteins, partly, because they are membrane proteins. Crystal structures are available for soluble parts of the major translocon proteins; however, the atomic structures for any of the minor translocon proteins remain unknown. A major motivation in understanding the T3SS is because of its essential role in virulence. The needle, tip, and translocon are exposed on the bacterial surface, making them attractive targets for developing new anti‐infectives. Structural results have identified binding sites and hotspots in the tip and translocon proteins for binding small molecules that could potentially disrupt the assembly of the needle apparatus. Major challenges remain in turning these small molecules into inhibitors of type III secretion to combat the growing threat of antibiotic resistance in many bacterial pathogens.
Dey S, Chakravarty A, Guha Biswas P, De Guzman RN. The type III secretion system needle, tip, and translocon. Protein Science. 2019;28:1582–1593. 10.1002/pro.3682
Funding information National Institute of Allergy and Infectious Diseases, Grant/Award Number: AI074856
REFERENCES
- 1. Green ER, Mecsas J. Bacterial secretion systems: An overview. Microbiol Spectr. 2016;4:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Costa TR, Felisberto‐Rodrigues C, Meir A, et al. Secretion systems in Gram‐negative bacteria: Structural and mechanistic insights. Nat Rev Microbiol. 2015;13:343–359. [DOI] [PubMed] [Google Scholar]
- 3. Dai W, Li Z. Conserved type III secretion system exerts important roles in Chlamydia trachomatis . Int J Clin Exp Pathol. 2014;7:5404–5414. [PMC free article] [PubMed] [Google Scholar]
- 4. Mueller KE, Plano GV, Fields KA. New frontiers in type III secretion biology: The Chlamydia perspective. Infect Immun. 2014;82:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Alam A, Miller KA, Chaand M, Butler JS, Dziejman M. Identification of Vibrio cholerae type III secretion system effector proteins. Infect Immun. 2011;79:1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gaytan MO, Martinez‐Santos VI, Soto E, Gonzalez‐Pedrajo B. Type three secretion system in attaching and effacing pathogens. Front Cell Infect Microbiol. 2016;6:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Deng W, Marshall NC, Rowland JL, et al. Assembly, structure, function and regulation of type III secretion systems. Nat Rev Microbiol. 2017;15:323–337. [DOI] [PubMed] [Google Scholar]
- 8. Burkinshaw BJ, Strynadka NC. Assembly and structure of the T3SS. Biochim Biophys Acta. 2014;1843:1649–1663. [DOI] [PubMed] [Google Scholar]
- 9. Galan JE, Lara‐Tejero M, Marlovits TC, Wagner S. Bacterial type III secretion systems: Specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol. 2014;68:415–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buttner D. Protein export according to schedule: Architecture, assembly, and regulation of type III secretion systems from plant‐ and animal‐pathogenic bacteria. Microbiol Mol Biol Rev. 2012;76:262–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Coburn B, Sekirov I, Finlay BB. Type III secretion systems and disease. Clin Microbiol Rev. 2007;20:535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wellington EM, Boxall AB, Cross P, et al. The role of the natural environment in the emergence of antibiotic resistance in Gram‐negative bacteria. Lancet Infect Dis. 2013;13:155–165. [DOI] [PubMed] [Google Scholar]
- 13. Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allen HK, Donato J, Wang HH, Cloud‐Hansen KA, Davies J, Handelsman J. Call of the wild: Antibiotic resistance genes in natural environments. Nat Rev Microbiol. 2010;8:251–259. [DOI] [PubMed] [Google Scholar]
- 15. Buttner D, He SY. Type III protein secretion in plant pathogenic bacteria. Plant Physiol. 2009;150:1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Buttner D, Bonas U. Who comes first? How plant pathogenic bacteria orchestrate type III secretion. Curr Opin Microbiol. 2006;9:193–200. [DOI] [PubMed] [Google Scholar]
- 17. Alfano JR, Collmer A. Type III secretion system effector proteins: Double agents in bacterial disease and plant defense. Annu Rev Phytopathol. 2004;42:385–414. [DOI] [PubMed] [Google Scholar]
- 18. Notti RQ, Stebbins CE. The structure and function of type III secretion systems. Microbiol Spectr. 2016;4:VMBF‐0004‐2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Portaliou AG, Tsolis KC, Loos MS, Zorzini V, Economou A. Type III secretion: Building and operating a remarkable nanomachine. Trends Biochem Sci. 2016;41:175–189. [DOI] [PubMed] [Google Scholar]
- 20. Diepold A, Wagner S. Assembly of the bacterial type III secretion machinery. FEMS Microbiol Rev. 2014;38:802–822. [DOI] [PubMed] [Google Scholar]
- 21. Schraidt O, Marlovits TC. Three‐dimensional model of Salmonella's needle complex at subnanometer resolution. Science. 2011;331:1192–1195. [DOI] [PubMed] [Google Scholar]
- 22. Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nans A, Kudryashev M, Saibil HR, Hayward RD. Structure of a bacterial type III secretion system in contact with a host membrane in situ. Nat Commun. 2015;6:10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheung M, Shen DK, Makino F, et al. Three‐dimensional electron microscopy reconstruction and cysteine‐mediated crosslinking provide a model of the type III secretion system needle tip complex. Mol Microbiol. 2015;95:31–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Radics J, Konigsmaier L, Marlovits TC. Structure of a pathogenic type 3 secretion system in action. Nat Struct Mol Biol. 2014;21:82–87. [DOI] [PubMed] [Google Scholar]
- 26. Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galán JE, Unger VM. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306:1040–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Loquet A, Sgourakis NG, Gupta R, et al. Atomic model of the type III secretion system needle. Nature. 2012;486:276–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang Y, Ouellette AN, Egan CW, Rathinavelan T, Im W, De Guzman RN. Differences in the electrostatic surfaces of the type III secretion needle proteins PrgI, BsaL, and MxiH. J Mol Biol. 2007;371:1304–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Deane JE, Roversi P, Cordes FS, et al. Molecular model of a type III secretion system needle: Implications for host‐cell sensing. Proc Natl Acad Sci U S A. 2006;103:12529–12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang L, Wang Y, Picking WL, Picking WD, De Guzman RN. Solution structure of monomeric BsaL, the type III secretion needle protein of Burkholderia pseudomallei . J Mol Biol. 2006;359:322–330. [DOI] [PubMed] [Google Scholar]
- 31. Sun P, Tropea JE, Austin BP, Cherry S, Waugh DS. Structural characterization of the Yersinia pestis type III secretion system needle protein YscF in complex with its heterodimeric chaperone YscE/YscG. J Mol Biol. 2008;377:819–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Quinaud M, Ple S, Job V, et al. Structure of the heterotrimeric complex that regulates type III secretion needle formation. Proc Natl Acad Sci U S A. 2007;104:7803–7808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fujii T, Cheung M, Blanco A, Kato T, Blocker AJ, Namba K. Structure of a type III secretion needle at 7‐å resolution provides insights into its assembly and signaling mechanisms. Proc Natl Acad Sci U S A. 2012;109:4461–4466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Demers JP, Sgourakis NG, Gupta R, et al. The common structural architecture of Shigella flexneri and Salmonella typhimurium type three secretion needles. PLoS Pathog. 2013;9:e1003245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Demers JP, Habenstein B, Loquet A, et al. High‐resolution structure of the Shigella type‐III secretion needle by solid‐state NMR and cryo‐electron microscopy. Nat Commun. 2014;5:4976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hu J, Worrall LJ, Hong C, et al. Cryo‐EM analysis of the T3S injectisome reveals the structure of the needle and open secretin. Nat Commun. 2018;9:3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Cordes FS, Komoriya K, Larquet E, et al. Helical structure of the needle of the type III secretion system of Shigella flexneri . J Biol Chem. 2003;278:17103–17107. [DOI] [PubMed] [Google Scholar]
- 38. Pastor A, Chabert J, Louwagie M, Garin J, Attree I. PscF is a major component of the Pseudomonas aeruginosa type III secretion needle. FEMS Microbiol Lett. 2005;253:95–101. [DOI] [PubMed] [Google Scholar]
- 39. Cornelis GR, Boland A, Boyd AP, et al. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kubori T, Matsushima Y, Nakamura D, et al. Supramolecular structure of the Salmonella typhimurium type III protein secretion system. Science. 1998;280:602–605. [DOI] [PubMed] [Google Scholar]
- 41. Verasdonck J, Shen DK, Treadgold A, et al. Reassessment of MxiH subunit orientation and fold within native Shigella T3SS needles using surface labelling and solid‐state NMR. J Struct Biol. 2015;192:441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Poyraz O, Schmidt H, Seidel K, et al. Protein refolding is required for assembly of the type three secretion needle. Nat Struct Mol Biol. 2010;17:788–792. [DOI] [PubMed] [Google Scholar]
- 43. Kenjale R, Wilson J, Zenk SF, et al. The needle component of the type III secreton of Shigella regulates the activity of the secretion apparatus. J Biol Chem. 2005;280:42929–42937. [DOI] [PubMed] [Google Scholar]
- 44. Davis AJ, Mecsas J. Mutations in the Yersinia pseudotuberculosis type III secretion system needle protein, YscF, that specifically abrogate effector translocation into host cells. J Bacteriol. 2007;189:83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mueller CA, Broz P, Müller SA, et al. The V‐antigen of Yersinia forms a distinct structure at the tip of injectisome needles. Science. 2005;310:674–676. [DOI] [PubMed] [Google Scholar]
- 46. Lara‐Tejero M, Galan JE. Salmonella enterica serovar typhimurium pathogenicity island 1‐encoded type III secretion system translocases mediate intimate attachment to nonphagocytic cells. Infect Immun. 2009;77:2635–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Espina M, Olive AJ, Kenjale R, et al. IpaD localizes to the tip of the type III secretion system needle of Shigella flexneri . Infect Immun. 2006;74:4391–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Park D, Lara‐Tejero M, Waxham MN, et al. Visualization of the type III secretion mediated Salmonella‐host cell interface using cryo‐electron tomography. Elife. 2018;7:e39514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lunelli M, Hurwitz R, Lambers J, Kolbe M. Crystal structure of PrgI‐SipD: insight into a secretion competent state of the type three secretion system needle tip and its interaction with host ligands. PLoS Pathog. 2011;7:e1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chatterjee S, Zhong D, Nordhues BA, Battaile KP, Lovell S, De Guzman RN. The crystal structures of the Salmonella type III secretion system tip protein SipD in complex with deoxycholate and chenodeoxycholate. Protein Sci. 2011;20:75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Johnson S, Roversi P, Espina M, et al. Self‐chaperoning of the type III secretion system needle tip proteins IpaD and BipD. J Biol Chem. 2007;282:4035–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Erskine PT, Knight MJ, Ruaux A, et al. High resolution structure of BipD: an invasion protein associated with the type III secretion system of Burkholderia pseudomallei . J Mol Biol. 2006;363:125–136. [DOI] [PubMed] [Google Scholar]
- 53. Barta ML, Guragain M, Adam P, et al. Identification of the bile salt binding site on IpaD from Shigella flexneri and the influence of ligand binding on IpaD structure. Proteins. 2012;80:935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dickenson NE, Zhang L, Epler CR, Adam PR, Picking WL, Picking WD. Conformational changes in IpaD from Shigella flexneri upon binding bile salts provide insight into the second step of type III secretion. Biochemistry. 2011;50:172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang Y, Nordhues BA, Zhong D, De Guzman RN. NMR characterization of the interaction of the Salmonella type III secretion system protein SipD and bile salts. Biochemistry. 2010;49:4220–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stensrud KF, Adam PR, La Mar CD, et al. Deoxycholate interacts with IpaD of Shigella flexneri in inducing the recruitment of IpaB to the type III secretion apparatus needle tip. J Biol Chem. 2008;283:18646–18654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Dey S, Anbanandam A, Mumford BE, De Guzman RN. Characterization of small‐molecule scaffolds that bind to the Shigella type III secretion system protein IpaD. ChemMedChem. 2017;12:1534–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. McShan AC, Anbanandam A, Patnaik S, De Guzman RN. Characterization of the binding of hydroxyindole, indoleacetic acid, and morpholinoaniline to the Salmonella type III secretion system proteins SipD and SipB. ChemMedChem. 2016;11:963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Derewenda U, Mateja A, Devedjiev Y, et al. The structure of Yersinia pestis V‐antigen, an essential virulence factor and mediator of immunity against plague. Structure. 2004;12:301–306. [DOI] [PubMed] [Google Scholar]
- 60. Chaudhury S, Battaile KP, Lovell S, Plano GV, De Guzman RN. Structure of the Yersinia pestis tip protein LcrV refined to 1.65 å resolution. Acta Crystallogr. 2013;F69:477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. DeBord KL, Lee VT, Schneewind O. Roles of LcrG and LcrV during type III targeting of effector Yops by Yersinia enterocolitica . J Bacteriol. 2001;183:4588–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Matson JS, Nilles ML. LcrG‐LcrV interaction is required for control of Yops secretion in Yersinia pestis . J Bacteriol. 2001;183:5082–5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee PC, Zmina SE, Stopford CM, Toska J, Rietsch A. Control of type III secretion activity and substrate specificity by the cytoplasmic regulator PcrG. Proc Natl Acad Sci U S A. 2014;111:E2027–E2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chaudhury S, de Azevedo SC, Plano GV, De Guzman RN. The LcrG tip chaperone protein of the Yersinia pestis type III secretion system is partially folded. J Mol Biol. 2015;427:3096–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chaudhury S, Nordhues BA, Kaur K, Zhang N, De Guzman RN. Nuclear magnetic resonance characterization of the type III secretion system tip chaperone protein PcrG of Pseudomonas aeruginosa . Biochemistry. 2015;54:6576–6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Broz P, Mueller CA, Muller SA, et al. Function and molecular architecture of the Yersinia injectisome tip complex. Mol Microbiol. 2007;65:1311–1320. [DOI] [PubMed] [Google Scholar]
- 67. Harmon DE, Murphy JL, Davis AJ, Mecsas J. A mutant with aberrant extracellular LcrV‐YscF interactions fails to form pores and translocate Yop effector proteins but retains the ability to trigger Yop secretion in response to host cell contact. J Bacteriol. 2013;195:2244–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gebus C, Faudry E, Bohn YS, Elsen S, Attree I. Oligomerization of PcrV and LcrV, protective antigens of Pseudomonas aeruginosa and Yersinia pestis . J Biol Chem. 2008;283:23940–23949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Sani M, Botteaux A, Parsot C, Sansonetti P, Boekema EJ, Allaoui A. IpaD is localized at the tip of the Shigella flexneri type III secretion apparatus. Biochim Biophys Acta. 2007;1770:307–311. [DOI] [PubMed] [Google Scholar]
- 70. Veenendaal AK, Hodgkinson JL, Schwarzer L, Stabat D, Zenk SF, Blocker AJ. The type III secretion system needle tip complex mediates host cell sensing and translocon insertion. Mol Microbiol. 2007;63:1719–1730. [DOI] [PubMed] [Google Scholar]
- 71. Zhang L, Wang Y, Olive AJ, et al. Identification of the MxiH needle protein residues responsible for anchoring invasion plasmid antigen D to the type III secretion needle tip. J Biol Chem. 2007;282:32144–32151. [DOI] [PubMed] [Google Scholar]
- 72. Rathinavelan T, Lara‐Tejero M, Lefebre M, et al. NMR model of PrgI‐SipD interaction and its implications in the needle‐tip assembly of the Salmonella type III secretion system. J Mol Biol. 2014;426:2958–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rathinavelan T, Tang C, De Guzman RN. Characterization of the interaction between the Salmonella type III secretion system tip protein SipD and the needle protein PrgI by paramagnetic relaxation enhancement. J Biol Chem. 2011;286:4922–4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Epler CR, Dickenson NE, Bullitt E, Picking WL. Ultrastructural analysis of IpaD at the tip of the nascent MxiH type III secretion apparatus of Shigella flexneri . J Mol Biol. 2012;420:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Barta ML, Dickenson NE, Patil M, et al. The structures of coiled‐coil domains from type III secretion system translocators reveal homology to pore‐forming toxins. J Mol Biol. 2012;417:395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Nguyen VS, Jobichen C, Tan KW, et al. Structure of AcrH‐AopB chaperone‐translocator complex reveals a role for membrane hairpins in type III secretion system translocon assembly. Structure. 2015;23:2022–2031. [DOI] [PubMed] [Google Scholar]
- 77. Blocker A, Gounon P, Larquet E, et al. The tripartite type III secreton of Shigella flexneri inserts Ipab and Ipac into host membranes. J Cell Biol. 1999;147:683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Neyt C, Cornelis GR. Insertion of a Yop translocation pore into the macrophage plasma membrane by Yersinia enterocolitica: requirement for translocators YopB and YopD, but not LcrG. Mol Microbiol. 1999;33:971–981. [DOI] [PubMed] [Google Scholar]
- 79. Schoehn G, Di Guilmi AM, Lemaire D, Attree I, Weissenhorn W, Dessen A. Oligomerization of type III secretion proteins PopB and PopD precedes pore formation in Pseudomonas . EMBO J. 2003;22:4957–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ide T, Laarmann S, Greune L, Schillers H, Oberleithner H, Schmidt MA. Characterization of translocation pores inserted into plasma membranes by type III‐secreted Esp proteins of enteropathogenic Escherichia coli . Cell Microbiol. 2001;3:669–679. [DOI] [PubMed] [Google Scholar]
- 81. Delahay RM, Frankel G. Coiled‐coil proteins associated with type III secretion systems: a versatile domain revisited. Mol Microbiol. 2002;45:905–916. [DOI] [PubMed] [Google Scholar]
- 82. Gazi AD, Charova SN, Panopoulos NJ, Kokkinidis M. Coiled‐coils in type III secretion systems: structural flexibility, disorder and biological implications. Cell Microbiol. 2009;11:719–729. [DOI] [PubMed] [Google Scholar]
- 83. Dey S, Basu A, Datta S. Characterization of molten globule PopB in absence and presence of its chaperone PcrH. Protein J. 2012;31:401–416. [DOI] [PubMed] [Google Scholar]
- 84. Harrington AT, Hearn PD, Picking WL, Barker JR, Wessel A, Picking WD. Structural characterization of the N terminus of IpaC from Shigella flexneri . Infect Immun. 2003;71:1255–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kuwae A, Yoshida S, Tamano K, Mimuro H, Suzuki T, Sasakawa C. Shigella invasion of macrophage requires the insertion of IpaC into the host plasma membrane. Functional analysis of IpaC. J Biol Chem. 2001;276:32230–32239. [DOI] [PubMed] [Google Scholar]
- 86. Page A‐L, Fromont‐Racine M, Sansonetti P, Legrain P, Parsot C. Characterization of the interaction partners of secreted proteins and chaperones of Shigella flexneri . Mol Microbiol. 2001;42:1133–1145. [DOI] [PubMed] [Google Scholar]
- 87. Terry CM, Picking WL, Birket SE, et al. The C‐terminus of IpaC is required for effector activities related to Shigella invasion of host cells. Microb Pathog. 2008;45:282–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Picking WL, Coye L, Osiecki JC, Barnoski Serfis A, Schaper E, Picking WD. Identification of functional regions within invasion plasmid antigen C (IpaC) of Shigella flexneri . Mol Microbiol. 2001;39:100–111. [DOI] [PubMed] [Google Scholar]
- 89. Mounier J, Popoff MR, Enninga J, Frame MC, Sansonetti PJ, Van Nhieu GT. The IpaC carboxyterminal effector domain mediates Src‐dependent actin polymerization during Shigella invasion of epithelial cells. PLoS Pathog. 2009;5:e1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kueltzo LA, Osiecki J, Barker J, et al. Structure‐function analysis of invasion plasmid antigen C (IpaC) from Shigella flexneri . J Biol Chem. 2003;278:2792–2798. [DOI] [PubMed] [Google Scholar]
- 91. Francis MS, Aili M, Wiklund M‐L, Wolf‐Watz H. A study of the YopD–LcrH interaction from Yersinia pseudotuberculosis reveals a role for hydrophobic residues within the amphipathic domain of YopD. Mol Microbiol. 2000;38:85–102. [DOI] [PubMed] [Google Scholar]
- 92. Pallen MJ, Dougan G, Frankel G. Coiled‐coil domains in proteins secreted by type III secretion systems. Mol Microbiol. 1997;25:423–425. [DOI] [PubMed] [Google Scholar]
- 93. Costa TR, Edqvist PJ, Broms JE, Ahlund MK, Forsberg A, Francis MS. YopD self‐assembly and binding to LcrV facilitate type III secretion activity by Yersinia pseudotuberculosis . J Biol Chem. 2010;285:25269–25284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Tengel T, Sethson I, Francis MS. Conformational analysis by CD and NMR spectroscopy of a peptide encompassing the amphipathic domain of YopD from Yersinia . Eur J Biochem. 2002;269:3659–3668. [DOI] [PubMed] [Google Scholar]
- 95. Faudry E, Job V, Dessen A, Attree I, Forge V. Type III secretion system translocator has a molten globule conformation both in its free and chaperone‐bound forms. FEBS J. 2007;274:3601–3610. [DOI] [PubMed] [Google Scholar]
- 96. Guichon A, Hersh D, Smith MR, Zychlinsky A. Structure‐function analysis of the Shigella virulence factor IpaB. J Bacteriol. 2001;183:1269–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Shen DK, Saurya S, Wagner C, Nishioka H, Blocker AJ. Domains of the Shigella flexneri type III secretion system IpaB protein involved in secretion regulation. Infect Immun. 2010;78:4999–5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lunelli M, Lokareddy RK, Zychlinsky A, Kolbe M. IpaB‐IpgC interaction defines binding motif for type III secretion translocator. Proc Natl Acad Sci U S A. 2009;106:9661–9666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lokareddy RK, Lunelli M, Eilers B, Wolter V, Kolbe M. Combination of two separate binding domains defines stoichiometry between type III secretion system chaperone IpgC and translocator protein IpaB. J Biol Chem. 2010;285:39965–39975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hume PJ, McGhie EJ, Hayward RD, Koronakis V. The purified Shigella IpaB and Salmonella SipB translocators share biochemical properties and membrane topology. Mol Microbiol. 2003;49:425–439. [DOI] [PubMed] [Google Scholar]
- 101. Tran Van Nhieu G, Bourdet‐Sicard R, Duménil G, Blocker A, Sansonetti PJ. Bacterial signals and cell responses during Shigella entry into epithelial cells. Cell Microbiol. 2000;2:187–193. [DOI] [PubMed] [Google Scholar]
- 102. Ménard R, Sansonetti P, Parsot C. The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 1994;13:5293–5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Picking WL, Nishioka H, Hearn PD, et al. IpaD of Shigella flexneri is independently required for regulation of Ipa protein secretion and efficient insertion of IpaB and IpaC into host membranes. Infect Immun. 2005;73:1432–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Olive AJ, Kenjale R, Espina M, Moore DS, Picking WL, Picking WD. Bile salts stimulate recruitment of IpaB to the Shigella flexneri surface, where it colocalizes with IpaD at the tip of the type III secretion needle. Infect Immun. 2007;75:2626–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Dickenson NE, Arizmendi O, Patil MK, et al. N‐terminus of IpaB provides a potential anchor to the Shigella type III secretion system tip complex protein IpaD. Biochemistry. 2013;52:8790–8799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Kaur K, Chatterjee S, De Guzman RN. Characterization of the Shigella and Salmonella type III secretion system tip–translocon protein–protein interaction by paramagnetic relaxation enhancement. Chembiochem. 2016;17:745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. McShan AC, Kaur K, Chatterjee S, Knight KM, De Guzman RN. NMR identification of the binding surfaces involved in the Salmonella and Shigella type III secretion tip–translocon protein–protein interactions. Proteins. 2016;84:1097–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Sarker MR, Neyt C, Stainier I, Cornelis GR. The Yersinia Yop virulon: LcrV is required for extrusion of the translocators YopB and YopD. J Bacteriol. 1998;180:1207–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Armentrout EI, Rietsch A. The type III secretion translocation pore senses host cell contact. PLoS Pathog. 2016;12:e1005530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Myeni SK, Wang L, Zhou D. SipB‐SipC complex is essential for translocon formation. PLoS One. 2013;8:e60499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Hayward RD, McGhie EJ, Koronakis V. Membrane fusion activity of purified SipB, a Salmonella surface protein essential for mammalian cell invasion. Mol Microbiol. 2000;37:727–739. [DOI] [PubMed] [Google Scholar]
- 112. Thorslund SE, Edgren T, Pettersson J, et al. The RACK1 signaling scaffold protein selectively interacts with Yersinia pseudotuberculosis virulence function. PLoS One. 2011;6:e16784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Montagner C, Arquint C, Cornelis GR. Translocators YopB and YopD from Yersinia enterocolitica form a multimeric integral membrane complex in eukaryotic cell membranes. J Bacteriol. 2011;193:6923–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Dacheux D, Goure J, Chabert J, Usson Y, Attree I. Pore‐forming activity of type III system‐secreted proteins leads to oncosis of Pseudomonas aeruginosa‐infected macrophages. Mol Microbiol. 2001;40:76–85. [DOI] [PubMed] [Google Scholar]
- 115. Faudry E, Vernier G, Neumann E, Forge V, Attree I. Synergistic pore formation by type III toxin translocators of Pseudomonas aeruginosa . Biochemistry. 2006;45:8117–8123. [DOI] [PubMed] [Google Scholar]
- 116. Romano FB, Tang Y, Rossi KC, Monopoli KR, Ross JL, Heuck AP. Type 3 secretion translocators spontaneously assemble a hexadecameric transmembrane complex. J Biol Chem. 2016;291:6304–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Tang Y, Romano FB, Brena M, Heuck AP. The Pseudomonas aeruginosa type III secretion translocator PopB assists the insertion of the PopD translocator into host cell membranes. J Biol Chem. 2018;293:8982–8993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Dortet L, Lombardi C, Cretin F, Dessen A, Filloux A. Pore‐forming activity of the Pseudomonas aeruginosa type III secretion system translocon alters the host epigenome. Nat Microbiol. 2018;3:378–386. [DOI] [PubMed] [Google Scholar]


