Abstract
Progression of cell cycle is regulated by sequential expression of cyclins, which associate with distinct cyclin kinases to drive the transition between different cell cycle phases. The complex of Cyclin A with cyclin‐dependent kinase 2 (CDK2) controls the DNA replication activity through phosphorylation of a set of chromatin factors, which critically influences the S phase transition. It has been shown that the direct interaction between the Cyclin A‐CDK2 complex and origin recognition complex subunit 1 (ORC1) mediates the localization of ORC1 to centrosomes, where ORC1 inhibits cyclin E‐mediated centrosome reduplication. However, the molecular basis underlying the specific recognition between ORC1 and cyclins remains elusive. Here we report the crystal structure of Cyclin A‐CDK2 complex bound to a peptide derived from ORC1 at 2.54 å resolution. The structure revealed that the ORC1 peptide interacts with a hydrophobic groove, termed cyclin binding groove (CBG), of Cyclin A via a KXL motif. Distinct from other identified CBG‐binding sequences, an arginine residue flanking the KXL motif of ORC1 inserts into a neighboring acidic pocket, contributing to the strong ORC1‐Cyclin A association. Furthermore, structural and sequence analysis of cyclins reveals divergence on the ORC1‐binding sites, which may underpin their differential ORC1‐binding activities. This study provides a structural basis of the specific ORC1‐cyclins recognition, with implication in development of novel inhibitors against the cyclin/CDK complexes.
Keywords: cyclin‐inhibitor complex, ORC1, Cyclin A, Cyclin E, cell cycle regulation
1. INTRODUCTION
In eukaryotes, the regulation of cell cycle progression is achieved by the sequential expression of cyclin proteins, which associate with cognate cyclin‐dependent kinases (CDK) to mediate the phosphorylation of phase‐specific cellular factors.1, 2, 3, 4 For instance, cyclin E associates with cyclin‐dependent kinase 2 (CDK2) to activate its phosphorylation of p220 (a.k.a. NPAT) protein,5, 6, 7 retinoblastoma protein (Rb),8 and p27,9, 10, 11 which regulates gene transcription and DNA replication initiation during the G1/S phase transition. Subsequently, newly formed Cyclin A‐CDK2 complex mediates the phosphorylation of CDC6, the component of DNA pre‐replication complex,12 leading to inhibition of the DNA replication reentry and transition of S/G2 phases.13 Given the important roles of these proteins in cell replication, dysregulations of the cyclin‐CDK activities have been linked to various human diseases.14, 15
Origin recognition complex subunit 1 (ORC1), one of the core subunits of the ORC, plays a critical role in regulating DNA replication.16 In addition, ORC1 has been implicated in genomic maintenance through interacting with a number of cyclins.17, 18, 19, 20 The direct interaction of ORC1 with Cyclin A mediates the localization of ORC1 to the centrosomes, where it inhibits Cyclin E‐dependent reduplication of centrioles and centrosomes.17, 21 Dysregulation of the ORC1 and cyclins activity led to serious human diseases such as Meier–Gorlin syndrome.18 However, the molecular basis for the ORC1‐cyclins interaction remains elusive.
To understand the structural basis of the ORC1‐cyclins interaction, we solved the crystal structure of Cyclin A‐CDK2 complex bound to an ORC1 fragment. The structure reveals that the ORC1 peptide presents two leucine residues for interaction with a hydrophobic groove of Cyclin A, with another flanking arginine inserting into a neighboring acidic cavity of Cyclin A. This binding mode is similar to but distinct from those of other Cyclin A‐inhibitor complexes, providing important implication to development of novel inhibitors against Cyclin A and other cyclin proteins.
2. RESULTS AND DISCUSSION
2.1. Crystal structure of the Cyclin A‐CDK2‐ORC1 complex
A previous study has indicated that the ORC1‐Cyclin A interaction is mediated by the N‐terminal fragment of ORC1, with residues K235 and L237 indispensable for the binding18 (Figure 1a). To further identify the Cyclin A‐binding region of ORC1, we have compared the Cyclin A‐binding affinities of an ORC1 N‐terminal fragment (residues 1–250, ORC11–250) and a peptide derived from the region C‐terminal to the bromo‐ajacent‐homology (BAH) domain, comprised of residues A233–G240 of ORC1 (ORC1233–240), by isothermal titration calorimetry (ITC). ORC11–250 and ORC1233–240 bind to Cyclin A with a dissociation constant (K d) of 0.49 and 0.63 μM, respectively (Figure 1b), suggesting the ORC1233–240 fragment as a major Cyclin A‐binding site (CBS) of ORC1. On the other hand, the fact that the titration with ORC11–250 gave higher enthalpy and entropy changes (Figure 1b) than that with the ORC1233–240 peptide implies distinct chemical environments between the two ORC1 fragments.
Figure 1.
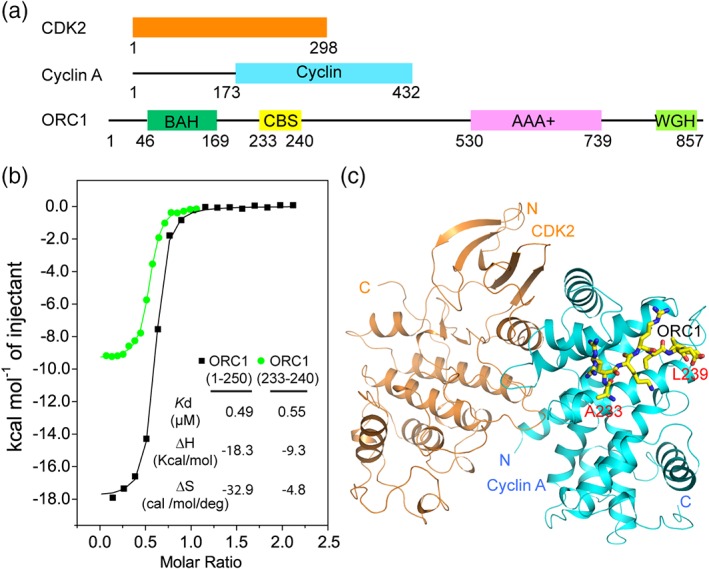
Crystal structure of the Cyclin A‐CDK2‐ORC233–240 complex. (a) Domain architecture of CDK2, Cyclin A, and ORC1. (b) ITC binding curves of ORC11–250 and ORC1233–240 titrated against Cyclin A. (c) Structural overview of the Cyclin A‐CDK2‐ORC1233–240 complex, with Cyclin A and CKD2 colored in cyan and orange, respectively. The ORC1 peptide is shown in stick representation. CDK2, cyclin‐dependent kinase 2; ORC1, origin recognition complex subunit 1
Next, we determined the crystal structure of the Cyclin A‐CDK2 complex [Figure S1(a,b)] bound to ORC1233–240 at 2.54 å resolution (Figure 1c, Table 1). The ORC1233–240 peptide is primarily docked into a hydrophobic groove of Cyclin A, located next to the Cyclin A‐CDK2 interface, but not directly involved in any interaction with CDK2 (Figures 1c and 2a–d). Structural alignment of the Cyclin A‐CDK2‐ORC1 complex with CDK2 alone (PDB ID: 4KD1) [Figure S1(c)] or the Cyclin A‐CDK2 binary complex (PDB ID: 1FIN) [Figure S1(d)] indicated that the ORC1 binding does not appreciably affect the conformation of CDK2, including the activation loop around its active site. Consistent with the structural analysis, our ITC binding assays indicated that monomeric Cyclin A and Cyclin A‐CDK2 show similar binding affinity for ORC11–250 [Figures 1b and S1(e)].
Table 1.
Data collection and refinement statistics (PDB 6P3W)
| Data collection | |
|---|---|
| Space group | P 6 2 2 2 |
| Cell dimensions | |
| a, b, c (å) | 186.396, 186.396, 214.707 |
| α, β, γ (°) | 90, 90, 120 |
| Wavelength | 0.9774 |
| Resolution (å) | 20.07–2.54 (2.63–2.54)a |
| R sym or R merge | 0.1438 (2.349) |
| I/σI | 9.84 (0.82) |
| Completeness (%) | 99.51 (98.77) |
| Redundancy | 8.1 (8.7) |
| CC1/2 | 0.997 (0.46) |
| Refinement | |
| Resolution (å) | 20.07–2.54 (2.63–2.54) |
| No. reflections | 72513 (7055) |
| R work/R free (%) | 20.5/23.9 (35.3/39.2) |
| No. atoms | |
| Macromolecules | 8857 |
| Solvent | 50 |
| Ligands | 60 |
| B factors (å2) | |
| Macromolecules | 86.03 |
| Solvent | 68.67 |
| Ligands | 197.46 |
| R.M.S. deviations | |
| Bond lengths (å) | 0.003 |
| Bond angles (°) | 0.78 |
Values in parentheses are for highest resolution shell.
Figure 2.

Structural and biochemical analysis of the ORC1‐Cyclin A interaction. (a) Close‐up view of the ORC1233–240–Cyclin A interactions. The hydrogen bonds are shown as dashed lines. (b) Electrostatic surface view of the ORC1233–240–Cyclin association. (c) Schematic view of the ORC1233–240 (magenta)–Cyclin A (black) interactions. Hydrogen bonding and electrostatic interactions are shown as dashed lines. Hydrophobic interactions are indicated as gears. (d) The Fo‐Fc omit map (blue mesh) of the ORC1233–240 peptide bound to Cyclin A‐CDK, generated after initial refinement using the molecular replacement solution, contoured at 1.7 σ level. (e, f) ITC binding analysis of the WT and mutant proteins of ORC11–250 (e) and Cyclin A (f). CDK, cyclin‐dependent kinase; ITC, isothermal titration calorimetry; N.D.B., no detectable binding, ORC1, origin recognition complex subunit 1
The association of the ORC1233–240 peptide with Cyclin A is dominated by nonpolar contacts, with the side chains of L237 and L239 inserting into the CBG to interact with the side chains of M210, I213, L214, W217, R250, and L253 of Cyclin A (Figure 2a,c). Outside the CBG, the side chain of ORC1 R234 packs against the aromatic ring of Cyclin A Y280 with two alternative conformations in similar occupancy, with one of which also forming salt bridges with D283 and D284 of Cyclin A; ORC1 K235 interacts with Cyclin A E220 via electrostatic attraction; ORC1 R236 extends toward an opposite direction, involving van der Waals contacts with the side chains of residues D283 and T285 of Cyclin A [Figures 2a,c and S1(f)]. At the N‐terminus of the peptide, ORC1 A233 is anchored to an acidic groove cavity, formed by W217, E220, E224, and I281 of Cyclin A. In addition, the backbone of ORC1 forms a number of hydrogen bonds with Cyclin A: the backbone amide groups of K235 and L237 are hydrogen bonded to the backbone and side‐chain carbonyl groups of I281 and Q254 of Cyclin A, respectively; while the backbone carbonyl group of K235 receives a hydrogen bond from the side‐chain amino group of Cyclin A Q254. ORC1 E238 is fully exposed to the solvent and does not engage any interaction with Cyclin A (Figure. 2a,c).
Guided by the structural observations, we have selected a number of key interacting residues, including R234, K235, and L237 of ORC1 and W217, E220, and E224 of Cyclin A, for mutagenesis and performed ITC experiments. Introduction of ORC1 L237A and Cyclin W217A mutations both led to completely abolished ORC1‐Cyclin A interaction (Figure 2e,f). In addition, introduction of the ORC1 R234A and K235A and Cyclin A E220A and E224A mutations all led to 3–30‐fold reduction in the ORC1‐Cyclin A binding, supporting that these residues play important roles in the ORC1‐Cyclin A association. Together, these data lend a strong support for the structural observation of the Cyclin A‐CDK2‐ORC1233–240 complex.
The ORC1‐interacting groove, termed cyclin binding groove (CBG), has previously been shown to mediate the interaction of Cyclin A with a wide spectrum of substrates and inhibitors, such as p53.22, 23, 24, 25, 26 However, these Cyclin A‐binding proteins do not share significant sequence homology, except for an Arg/Lys‐X‐Leu (“RXL” or “KXL”) sequence motif [Figure S2(a)]. Indeed, sequence and structural comparison of the ORC1233–240 fragment with other Cyclin A‐binding proteins reveals a highly conserved Leu at P0 site and another Arg or Lys at the P−2 site (following a previously developed nomenclature26) (Figure S2). The P0‐Leu is invariably anchored into the hydrophobic groove and the P−2‐Arg/Lys points the side chain toward E220 [Figure S2(b–e)].22, 23, 24, 25, 26, 27, 28 However, the P−3 site is not conserved and structurally divergent among these proteins [Figure S2(b–e)]. In this study, we showed that the P−3‐Arg (R234) of ORC1 plays an important role in the Cyclin A binding, which opens up a possibility of designing a more robust inhibitor against Cyclin A.
In addition to Cyclin A, the crystal structures of other cyclins, including Cyclin B,29 Cyclin D,30 and Cyclin E31 have been reported. Sequence and structural comparison of these cyclins indicates that the ORC1‐binding residues of Cyclin A are relatively conserved across the cyclin family (Figure 3a). Nevertheless, structural divergence of the corresponding regions was observed. Most notably, residue E224 of Cyclin A, which interacts with residue A233 of ORC1, is replaced by a valine in Cyclin E; residue T285 of Cyclin A, which makes contact with ORC1 R236, is replaced by an alanine in Cyclin E (Figure 3b). In addition, residue D284 of Cyclin A, which forms a salt bridge with residue R234 of ORC, is replaced by a glycine in Cyclin E (Figure 3b). These structural differences support a previous observation that ORC1 interacts with Cyclin E through its BAH domain, instead of the KXL motif.18 Indeed, unlike Cyclin A that interacts with ORC1233–240 strongly (Figure 1b), our ITC binding assays indicated that the ORC1233–240 does not interact with Cyclin E‐CDK2 appreciably (Figure 3c). In contrast, a strong binding was observed when Cyclin E‐CDK2 was titrated with ORC11–250, which gave a K d of 0.48 μM (Figure 3d). Likewise, although the CBG of Cyclin B contains the majority of the ORC1‐binding residues, the ORC1 A233‐interacting sites are not conserved: residues E220 and E224 of Cyclin A are replaced by a lysine (K51) and a glutamine (Q47) in Cyclin B, respectively (Figure 3e). On the other hand, the CBG of Cyclin D shares closer sequence and structural homology with that of Cyclin A (Figure 3f). The major difference between the CBGs of Cyclins A and D lies in the region that interacts with residue R234 of ORC1: the residues corresponding to Y280 and D284 of Cyclin A are I126 and N130 in Cyclin D, respectively (Figure 3f). Together, the structural and sequence variations in the CBG region of cyclins may underpin their differential binding activities for ORC1.
Figure 3.

Sequence and structural analysis the ORC1‐cyclin interaction. (a) Sequence alignment of cyclins using the ENDscript server,32 with the ORC1‐binding sites of Cyclin A marked by asterisks. (b) Structural overlay of Cyclin E (PDB 1W98) with ORC1233–240‐bound Cyclin A, with the side chains of the interacting residues shown in stick representation. (c, d) ITC binding curves of Cyclin E with ORC1233–240 (c) and ORC11–250 (d). (e) Structural overlay of Cyclin B (PDB 2B9R) with ORC1233–240‐bound Cyclin A. (f) Structural overlay of Cyclin D (PDB 2W96) with ORC1233–240‐bound Cyclin A. ORC1, origin recognition complex subunit 1
2.2. Summary
Cyclin A and other cyclin proteins are essential for cell cycle regulation, hence providing better targets than CDK2 for drug design in cancer therapy.33, 34 This study determined the structural basis for the interaction between ORC1 and Cyclin A, thereby providing a framework for functional understanding of how ORC1 regulates the cell cycle progress in the Cyclin‐CDK axis. More importantly, a comprehensive understanding of the molecular basis of cyclin‐inhibitor interaction might ultimately benefit the development of more potent cyclin‐targeting drugs.
3. EXPERIMENTAL PROCEDURES
3.1. Protein expression and production
The DNA fragments encoding human Cyclin A (residues 173–432), Cyclin E (residues 96–378), and ORC11–250 were inserted into a modified pRSFDuet‐1 vector (Novagen), preceded by an N‐terminal His6‐SUMO tag and a ULP1 (ubiquitin‐like protease) cleavage site, and the cDNA encoding CDK2 was inserted into a pGEX6P‐1 vector. The resulting plasmids were then transformed into BL21 (DE3) RIL cell strain (Agilent Technologies) for protein expression. The cells were initially grown at 37°C. After the cell density reached an optical density at 600 nm (A600) of 0.6, the temperature was shifted to 16°C, followed by addition of 0.4 mM isopropyl β‐d‐galactoside for protein expression induction. Cyclin A (173–432), Cyclin E (96–378), and ORC11–250 were purified subsequently through a nickel column, removal of His6‐SUMO tag via ULP1 cleavage, a Phenyl Sepharose column (GE Healthcare) (for Cyclin A) or a Q HP column (GE Healthcare) (for ORC1), and a Superdex 75 16/600 column (GE Healthcare). The GST‐CDK2 fusion protein was purified using a Glutathione Sepharose 4 Fast Flow column, followed by Heparin column (GE Healthcare) and a Superdex 75 16/600 column. To prepare the CyclinA‐CDK2 complex, purified Cyclin A was mixed with CDK2 in a 1:1 molar ratio and subjected into size‐exclusion chromatography. The protein identity and purity were confirmed by size‐exclusion chromatography and SDS‐PAGE analysis [Fig. S1(a,b)]. The protein complex was concentrated to 16.5 mg/mL in buffer containing 25 mM Tris–HCl (pH 7.5), 200 mM NaCl, 5 mM DTT, and 5% glycerol containing and stored at −80°C. The Cyclin A and ORC1 mutants were constructed through site‐directed mutagenesis and produced in the same way as wild‐type proteins.
3.2. Crystallization and structure determination
The Cyclin A/CDK2 complex was mixed with the ORC1233–240 peptide, containing a C‐terminal tyrosine, in a 1:4 molar ratio. The crystals were generated under the condition containing 0.49 M NaH2PO4 and 0.91 M K2HPO4 by hanging‐drop vapor diffusion method at 4°C. Crystals were soaked in the cryoprotectant solution containing mother liquor supplemented with 25% glycerol in a single step before flash frozen in liquid nitrogen. X‐ray diffraction data was collected on the 24‐ID‐E beamline at the Advanced Photon Source, Argonne National Laboratory. The data were indexed, integrated and scaled utilizing the HKL2000 program.35 The structure was solved by molecular replacement using the PHASER program,36 with the structure of the Cyclin A/CDK2 complex (PDB ID: 1FIN) as the search model. Iterative cycles of model rebuilding and refinement were carried out using COOT37 and PHENIX,38 respectively. The same R‐free test set was used throughout the refinement. Data collection and structure refinement statistics were summarized in Table 1.
3.3. ITC measurements
The protein samples were dialyzed against buffer containing 25 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1 mM 2‐Mercaptoethanol and 5% glycerol at 4°C overnight. For the sake of protein solubility, the salt concentration was increased to 200 mM for the binding between ORC11–250 and Cyclin A‐CDK2 [Fig. S1(e)], for comparison of WT and mutant ORC1/Cyclin A proteins (Figure 2d,e) and for titration of ORC1233–240 against Cyclin E (Figure 3c), and increased to 300 mM for titration of ORC11–250 against Cyclin E (Figure 3d). A MicroCal iTC200 system (GE Healthcare) was used to perform the ITC measurements, with 500 μM ORC1 proteins titrated against 50 μM Cyclin A. A total of 17 injections with a spacing of 180 s and a reference power of 5 μcal s−1 were carried out at 25°C. The ITC curves were processed with software ORIGEN (MicroCal) using a one‐site fitting model.
Supporting information
Appendix S1: Supplementary Material
ACKNOWLEDGMENTS
This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). The Eiger 16M detector on 24‐ID‐E beam line is funded by a NIH‐ORIP HEI grant (S10OD021527). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE‐AC02‐06CH11357. This work was supported by March of Dimes Foundation (1‐FY15‐345 to J.S.) and NIH (R35GM119721 to J.S).
Wang B, Song J. Structural basis for the ORC1‐Cyclin A association. Protein Science. 2019;28:1727–1733. 10.1002/pro.3689
Funding information National Institute of General Medical Sciences, Grant/Award Numbers: R35GM119721, P30 GM124165; March of Dimes Foundation, Grant/Award Number: 1‐FY15‐345; NIH‐ORIP, Grant/Award Number: S10OD021527; National Institutes of Health, Grant/Award Number: R35GM119721
REFERENCES
- 1. Ewen ME, Faha B, Harlow E, Livingston DM. Interaction of p107 with cyclin A independent of complex formation with viral oncoproteins. Science. 1992;255:85–87. [DOI] [PubMed] [Google Scholar]
- 2. Harper JW, Adams PD. Cyclin‐dependent kinases. Chem Rev. 2001;101:2511–2526. [DOI] [PubMed] [Google Scholar]
- 3. Morgan DO. Cyclin‐dependent kinases: engines, clocks, and microprocessors. Ann Rev Cell Devel Biol. 1997;13:261–291. [DOI] [PubMed] [Google Scholar]
- 4. Wood DJ, Endicott JA. Structural insights into the functional diversity of the CDK‐cyclin family. Open Biol. 2018;8:180112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao J, Dynlacht B, Imai T, Hori T, Harlow E. Expression of NPAT, a novel substrate of cyclin E‐CDK2, promotes S‐phase entry. Genes Dev. 1998;12:456–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao J, Kennedy BK, Lawrence BD, et al. NPAT links cyclin E‐Cdk2 to the regulation of replication‐dependent histone gene transcription. Genes Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]
- 7. Ma T, Van Tine BA, Wei Y, et al. Cell cycle‐regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes Dev. 2000;14:2298–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zarkowska T, Mittnacht S. Differential phosphorylation of the retinoblastoma protein by G1/S cyclin‐dependent kinases. J Biol Chem. 1997;272:12738–12746. [DOI] [PubMed] [Google Scholar]
- 9. Muller D, Bouchard C, Rudolph B, et al. Cdk2‐dependent phosphorylation of p27 facilitates its Myc‐induced release from cyclin E/cdk2 complexes. Oncogene. 1997;15:2561–2576. [DOI] [PubMed] [Google Scholar]
- 10. Vlach J, Hennecke S, Amati B. Phosphorylation‐dependent degradation of the cyclin‐dependent kinase inhibitor p27. EMBO J. 1997;16:5334–5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sheaff RJ, Groudine M, Gordon M, Roberts JM, Clurman BE. Cyclin E‐CDK2 is a regulator of p27Kip1. Genes Dev. 1997;11:1464–1478. [DOI] [PubMed] [Google Scholar]
- 12. Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. [DOI] [PubMed] [Google Scholar]
- 13. Petersen BO, Lukas J, Sorensen CS, Bartek J, Helin K. Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J. 1999;18:396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Casimiro MC, Crosariol M, Loro E, Li Z, Pestell RG. Cyclins and cell cycle control in cancer and disease. Genes Cancer. 2012;3:649–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vermeulen K, Van Bockstaele DR, Berneman ZN. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. 2003;36:131–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bell SP. The origin recognition complex: from simple origins to complex functions. Genes Dev. 2002;16:659–672. [DOI] [PubMed] [Google Scholar]
- 17. Hemerly AS, Prasanth SG, Siddiqui K, Stillman B. Orc1 controls centriole and centrosome copy number in human cells. Science. 2009;323:789–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hossain M, Stillman B. Meier‐Gorlin syndrome mutations disrupt an Orc1 CDK inhibitory domain and cause centrosome reduplication. Genes Dev. 2012;26:1797–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Laman H, Coverley D, Krude T, Laskey R, Jones N. Viral cyclin‐cyclin‐dependent kinase 6 complexes initiate nuclear DNA replication. Mol Cell Biol. 2001;21:624–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li CJ, Vassilev A, DePamphilis ML. Role for Cdk1 (Cdc2)/cyclin A in preventing the mammalian origin recognition complex's largest subunit (Orc1) from binding to chromatin during mitosis. Mol Cell Biol. 2004;24:5875–5886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ferguson RL, Pascreau G, Maller JL. The cyclin A centrosomal localization sequence recruits MCM5 and Orc1 to regulate centrosome reduplication. J Cell Sci. 2010;123:2743–2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen J, Saha P, Kornbluth S, Dynlacht BD, Dutta A. Cyclin‐binding motifs are essential for the function of p21CIP1. Mol Cell Biol. 1996;16:4673–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luciani MG, Hutchins JR, Zheleva D, Hupp TR. The C‐terminal regulatory domain of p53 contains a functional docking site for cyclin A. J Mol Biol. 2000;300:503–518. [DOI] [PubMed] [Google Scholar]
- 24. Russo AA, Jeffrey PD, Patten AK, Massague J, Pavletich NP. Crystal structure of the p27Kip1 cyclin‐dependent‐kinase inhibitor bound to the cyclin A‐Cdk2 complex. Nature. 1996;382:325–331. [DOI] [PubMed] [Google Scholar]
- 25. Brown NR, Noble ME, Endicott JA, Johnson LN. The structural basis for specificity of substrate and recruitment peptides for cyclin‐dependent kinases. Nat Cell Biol. 1999;1:438–443. [DOI] [PubMed] [Google Scholar]
- 26. Lowe ED, Tews I, Cheng KY, et al. Specificity determinants of recruitment peptides bound to phospho‐CDK2/cyclin A. Biochemistry. 2002;41:15625–15634. [DOI] [PubMed] [Google Scholar]
- 27. Adams PD, Li X, Sellers WR, et al. Retinoblastoma protein contains a C‐terminal motif that targets it for phosphorylation by cyclin‐cdk complexes. Mol Cell Biol. 1999;19:1068–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adams PD, Sellers WR, Sharma SK, Wu AD, Nalin CM, Kaelin WG Jr. Identification of a cyclin‐cdk2 recognition motif present in substrates and p21‐like cyclin‐dependent kinase inhibitors. Mol Cell Biol. 1996;16:6623–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Petri ET, Errico A, Escobedo L, Hunt T, Basavappa R. The crystal structure of human cyclin B. Cell Cycle. 2007;6:1342–1349. [DOI] [PubMed] [Google Scholar]
- 30. Day PJ, Cleasby A, Tickle IJ, et al. Crystal structure of human CDK4 in complex with a D‐type cyclin. Proc Natl Acad Sci USA. 2009;106:4166–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Honda R, Lowe ED, Dubinina E, et al. The structure of cyclin E1/CDK2: implications for CDK2 activation and CDK2‐independent roles. EMBO J. 2005;24:452–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murphy M, Stinnakre MG, Senamaud‐Beaufort C, et al. Delayed early embryonic lethality following disruption of the murine cyclin A2 gene. Nat Genet. 1997;15:83–86. [DOI] [PubMed] [Google Scholar]
- 34. Andrews MJ, Kontopidis G, McInnes C, et al. REPLACE: a strategy for iterative design of cyclin‐binding groove inhibitors. Chembiochem. 2006;7:1909–1915. [DOI] [PubMed] [Google Scholar]
- 35. Otwinowski Z, Minor W. Processing of X‐ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. [DOI] [PubMed] [Google Scholar]
- 36. McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emsley P, Cowtan K. Coot: model‐building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 38. Adams PD, Grosse‐Kunstleve RW, Hung LW, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Cryst. 2002;D58:1948–1954. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supplementary Material


